Abstract
An in vitro culture system of primary cells from white-backed planthopper, an insect vector of Southern rice black-streaked dwarf virus (SRBSDV), a fijivirus, was established to study replication of the virus. Viroplasms, putative sites of viral replication, contained the nonstructural viral protein P9-1, viral RNA, outer-capsid proteins, and viral particles in virus-infected cultured insect vector cells, as revealed by transmission electron and confocal microscopy. Formation of viroplasm-like structures in non-host insect cells upon expression of P9-1 suggested that the matrix of viroplasms observed in virus-infected cells was composed basically of P9-1. In cultured insect vector cells, knockdown of P9-1 expression due to RNA interference (RNAi) induced by synthesized double-stranded RNA (dsRNA) from the P9-1 gene strongly inhibited viroplasm formation and viral infection. RNAi induced by ingestion of dsRNA strongly abolished viroplasm formation, preventing efficient viral spread in the body of intact vector insects. All these results demonstrated that P9-1 was essential for viroplasm formation and viral replication. This system, combining insect vector cell culture and RNA interference, can further advance our understanding of the biological activities of fijivirus replication proteins.
INTRODUCTION
Plant reoviruses, comprising the genera Phytoreovirus, Fijivirus, and Oryzavirus, cause diseases of numerous important crops and are transmitted by insect vectors in a persistently propagative manner (21, 23, 28). For control of such viruses, understanding their mode of transmission and replication is critical. For a persistently propagative plant virus to be transmitted by its vector, the virus ingested by insect feeding on infected plants must enter the epithelial cells of the alimentary canal and then replicate and assemble progeny virions to move into the salivary glands, from which the virus can be transmitted to more plants during feeding (10). Viral replication and assembly of progeny virions, critical for the propagation of plant reoviruses in their insect vectors, are thought to occur in viroplasms (21, 23, 28), which contain viral proteins, viral particles, and viral RNAs.
Southern rice black-streaked dwarf virus (SRBSDV), a tentatively identified species in the genus Fijivirus, which is transmitted by the white-backed planthopper (WBPH; Sogatella furcifera Horváth), has spread rapidly throughout southern China and northern Vietnam and can severely damage rice (16, 29, 33, 34). The icosahedral, double-layered particles of SRBSDV are ca. 70 nm in diameter and contain 10 segments of double-stranded RNA (dsRNA) (29, 34). Phylogenetic analyses showed that SRBSDV, the first WBPH-borne reovirus to be identified, is most closely related to but distinct from Rice black-streaked dwarf virus (RBSDV), also a fijivirus (29, 34). Comparing the different genomic segments of SRBSDV to their counterparts in RBSDV suggests that SRBSDV encodes at least six putative structural proteins (P1, P2, P3, P4, P8, and P10) and five putative nonstructural proteins (P6, P7-1, P7-2, P9-1, and P9-2) (29).
Among the putative structural proteins encoded by SRBSDV, P1, P2, and P4 are a putative RNA-dependent RNA polymerase, a core protein, and an outer-shell B-spike protein, respectively (29, 32); P3 is a putative capping enzyme (29, 32); and P8 and P10 are putative core and major outer capsid proteins, respectively (12, 29). Among the putative nonstructural proteins encoded by SRBSDV, P6 is a viral RNA-silencing suppressor (18); P7-1 is the major constitute of the tubules and has the intrinsic ability to self-interact to form tubules in non-host insect cells (16); and P9-1 of SRBSDV has about 77% amino acid identity with its counterpart, P9-1 of RBSDV (29). In RBSDV, P9-1 forms an octameric, cylindrical structure in vitro and accumulates in the matrix of viroplasms in virus-infected cells (1, 12). As a major constitute of the viroplasm, P9-1 is thus likely to play an important role in the formation of viroplasm (1). Therefore, P9-1 of SRBSDV may also be essential for viroplasm formation during viral infection in the host plant and insect vector. However, the precise function(s) of the proteins in viroplasm formation and viral replication of plant reoviruses is poorly understood due in part to the lack of a reverse-genetics system and useful culture systems for their respective insect vectors.
Insect vector cells in monolayer (VCM) is an in vitro experimental system with notable advantages over the use of whole intact insects for investigating plant viruses (5, 24). This is due to its capability of obtaining a uniform viral infection, which enables us to follow synchronous viral multiplication (24). The VCM also provides a very sensitive bioassay system for tracing the fate of viral infectivity under different conditions (24). We have already used VCMs derived from the leafhopper that transmits Rice dwarf virus (RDV), another phytoreovirus, to clarify that the Pns12 nonstructural protein of RDV plays a key role in the formation of viroplasms and in recruiting viral assembly complexes to the viroplasms in VCMs (31). We thus adapted the VCM system for WBPH, the vector of SRBSDV, to trace the infection and multiplication process of virus.
To further investigate the functional roles of viral proteins in the infection cycles of plant reoviruses in insect vectors, here we used RNA interference (RNAi), a conserved sequence-specific gene silencing mechanism that is induced by dsRNAs (6). By exploitation of its ability to efficiently silence gene expression, RNAi has been used in mammalian, insect, and plant cell studies to characterize the function of numerous genes (3, 4, 11, 20). It has also been used to interfere with the replication of animal reoviruses (7, 15, 17), which are closely related to plant reoviruses. We thus introduced dsRNA from the P9-1 gene of SRBSDV into VCMs or the intact insect to knock down the expression of the P9-1 gene and examine the subsequent effect on viroplasm formation and viral replication.
In this study, by growing a primary cell culture of WBPH in a monolayer (VCM) and using the RNAi strategy, we could elucidate that the P9-1 nonstructural protein of SRBSDV functions in the assembly of viroplasm and viral replication. The P9-1 nonstructural protein appeared to be the major constituent of the matrix of viroplasms where viral RNA, major outer-capsid protein P10, and viral particles accumulated in virus-infected VCMs. RNAi induced by dsRNA from the P9-1 gene in VCMs or the intact insect strongly inhibited such viroplasm formation, preventing efficient viral replication in vitro and in vivo. Development of RNAi and VCMs can thus overcome the lack of a reverse-genetics system for persistently propagative plant viruses and advance our understanding of the biological activities of viral proteins in the replication cycle in insect vectors.
MATERIALS AND METHODS
Virus and antibodies.
Rabbit polyclonal antisera against the P9-1 nonstructural protein and the P10 major outer capsid protein of SRBSDV (29, 33, 34) were prepared. The P9-1 and P10 genes from an SRBSDV isolate from Hunan Province, China, were amplified by reverse transcription-PCR (RT-PCR), and the products were purified and engineered into Gateway vector pDEST17 (Invitrogen). The resulting pDEST17-P9-1 and pDEST17-P10 plasmids were then used to transform Escherichia coli strain Rosetta and expressed by adding isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma) (1 mmol/liter). Cells were harvested and sonicated. The final suspension containing P9-1 or P10 protein was purified with nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen), and rabbits were immunized with the purified proteins, as described previously (27). IgG was isolated using specific polyclonal antiserum and a protein A-Sepharose affinity column. Eluted IgG was dialyzed exhaustively against phosphate-buffered saline (PBS). The IgG was conjugated directly to fluorescein-5-isothiocyanate (FITC) or rhodamine according to the manufacturer's instructions (Invitrogen).
To detect the specificity of P9-1 and P10 antibodies of SRBSDV, we extracted total plant proteins from 1 g of either SRBSDV-infected or healthy rice plants. SRBSDV was crudely purified from infected rice plants as previously described by Miyazaki et al. (22). The proteins and solution of purified viruses were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto a polyvinylidene difluoride (PVDF) membrane, and detected on immunoblots using prepared antibodies as described previously (27).
Establishment of primary cell cultures derived from WBPH.
Primary cell cultures derived from WBPH were established by adapting the protocol described by Kimura and Omura (14). In a preliminary test, WBPH embryos at the blastokinetic stage were found to be a suitable source for primary cell cultures. This stage can be recognized by the appearance of red eye spots on the eggs on day 8 after oviposition. Embryos at this stage were sterilized with 70% ethanol, washed with Tyrode's solution (13, 14), and then crushed with a sterilized pestle into tissue fragments. The tissue fragments were treated with 0.25% trypsin in Tyrode's solution and then incubated with Kimura's insect medium at 25°C (13, 14).
Examination of viral infection in primary cell cultures derived from WBPH by immunofluorescence microscopy.
SRBSDV inocula for infecting VCMs derived from WBPHs were prepared from infected plants, essentially as described previously (13, 14). VCMs growing on a coverslip and infected with SRBSDV were fixed in 4% paraformaldehyde 48 h postinoculation (p.i.), immunostained with P10-specific IgG conjugated to FITC (P10-FITC) and P9-1-specific IgG conjugated to rhodamine (P9-1–rhodamine), and then examined with a Leica TCS SP5 inverted confocal microscope, as described previously (30).
Immunofluorescence detection of newly made viral RNA in primary cell cultures derived from WBPH.
For determining whether the viroplasms are the sites of viral RNA synthesis, VCMs on a coverslip were either mock infected or infected with SRBSDV and then treated with actinomycin D (Sigma) for 1 h at 45 h p.i. and incubated with bromouridine 5′-triphosphate (BrUTP) for 2 h via the use of Cellfectin (Invitrogen). The samples were fixed 48 h p.i., stained with monoclonal anti-bromodeoxyuridine (anti-BrdU; Sigma), and then treated with anti-mouse IgG conjugated to FITC (Invitrogen) and P9-1–rhodamine for imaging with confocal microscopy, as described previously (30).
Baculovirus expression of nonstructural proteins of SRBSDV.
DNA fragments encoding full-length P9-1 and a carboxy-terminal (amino acids 346 though 368) deletion mutant (P9-1ΔC) were amplified by PCR. For the control, a DNA fragment of another nonstructural protein of SRBSDV, P7-1 (16), fused in frame with the sequence of Strep-tag II (P7-1–Strep-tag II), also was amplified by PCR. The products were purified and engineered into Gateway vector pDEST8 (Invitrogen) to generate plasmids pDEST8-P9-1, pDEST8-P9-1ΔC, and pDEST8-P7-1–Strep-tag II. The recombinant baculovirus vectors were introduced into E. coli DH10Bac (Invitrogen) for transposition into the bacmid. The recombinant bacmids were used to transfect Spodoptera frugiperda (Sf9) cells in the presence of Cellfectin. Sf9 cells, growing on a coverslip and infected with recombinant bacmids, were incubated for 72 h, fixed, and stained with the monoclonal anti-Strep-tag II (IBA) followed by Rhodamine anti-mouse IgG (Sigma) and P9-1-FITC for imaging with confocal microscopy as described previously (31).
Effect of synthesized dsRNAs on viral infection in primary cell cultures derived from WBPH.
We designed primers for PCR amplification of a 968-bp segment of the P9-1 gene and a 717-bp segment of a green fluorescence protein (GFP)-encoding gene as a control. The PCR products were used for dsRNA synthesis according to the protocol of a T7 RiboMAX Express RNAi System kit (Promega). VCMs were transfected with dsRNAs via Cellfectin, as reported by Rosa et al. (25). Briefly, VCMs were transfected with dsRNA (0.5 μg/μl) from the P9-1 gene (dsP9-1) or from the GFP gene (dsGFP) via the use of Cellfectin (Invitrogen) for 12 h and grown further in growth medium. The basic procedure of small interfering RNA (siRNA) detection was carried out essentially as described previously (26). Briefly, after a 72-h treatment with dsRNAs, total RNA was extracted from harvested VCMs. Digoxigenin (DIG)-labeled riboprobes corresponding to the negative-sense RNA of the P9-1 or GFP gene were generated with T7 RNA polymerase (Roche). Northern blotting was performed with a DIG Northern starter kit protocol (Roche).
For examining the effects of the dsRNAs on viral infection, after a 12-h treatment with dsRNAs, the VCMs were inoculated with SRBSDV and were fixed 3 days later with 4% paraformaldehyde and stained with P10-FITC and P9-1–rhodamine for visualization with confocal microscopy (30).
Effect of synthesized dsRNAs on viral infection in insect vectors.
A membrane-feeding approach for delivering dsRNAs to WBPHs was used. Second-instar nymphs of WBPH were fed with dsRNAs (0.5 μg/μl) diluted in an artificial diet, D-97, suitable for rearing planthoppers (8). This mixed diet was held between two layers of stretched parafilm covering one open end of the chamber employed. The nymphs were maintained on the diets for 1 day, allowed a 2-day period of acquisition on SRBSDV-infected rice plants, and then placed on healthy rice seedlings. Total RNAs from WBPHs receiving the dsRNA diet or the artificial diet alone were extracted by the use of TRIzol reagent (Invitrogen). The effects of dsRNAs on the transcript levels of virus P9-1 and P10 genes were determined by RT-PCR. To determine whether the treatment of dsP9-1 inhibited viral replication in intact insects, at 2, 6, and 12 days post-first access to diseased plants (padp), we dissected internal organs from 50 WBPHs that received dsRNAs. The organs were then fixed, stained with P10-FITC, P9-1–rhodamine, and the actin dye phalloidin-Alexa Fluor 647 carboxylic acid (Invitrogen), and processed for immunofluorescence as described previously (2).
Immunoelectron microscopy.
VCMs or Sf9 cells on coverslips were fixed, dehydrated, embedded, and cut as described previously (30). Cell sections were then incubated with antibodies against P9-1 or P10 and subjected to immunogold labeling with goat antibodies against rabbit IgG that had been conjugated with 15-nm-diameter gold particles (Sigma) (30).
RESULTS
Establishment of primary cell cultures derived from WBPH.
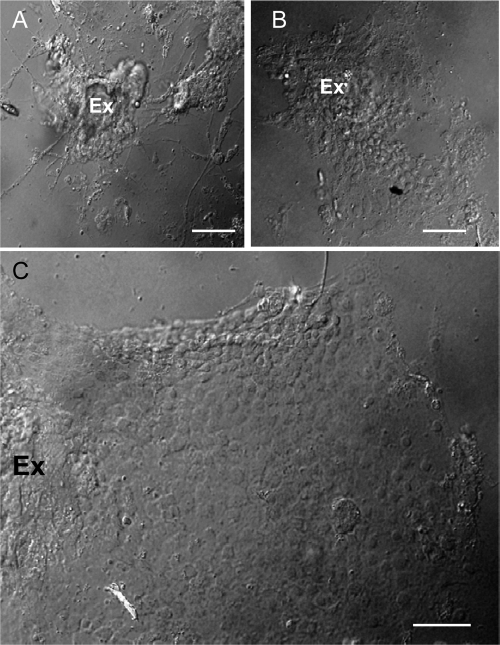
To establish primary cell cultures derived from WBPH, embryos at the blastokinetic stage were sterilized, washed with Tyrode's solution, and crushed into tissue fragments, which were then treated with trypsin and incubated with growth medium. Migration of fibroblast-like cells from the tissue fragments of embryos at the blastokinetic stage were observed as early as 36 h after setting up the cultures. Our observations suggested that tissue fragments of embryos at this stage could be used as explants for the migration of cells. Within 7 days of explanting, fibroblast-like cells had become the dominant cell type (Fig. 1A). Monolayers of epithelium-like cells started to form as cells migrated from the original explants at 9 to 12 days (Fig. 1B). They continued growing to form large epithelium-like cell sheets after 40 days (Fig. 1C). Our results indicated that the epithelium-like cells of WBPHs were able to self-propagate in controlled growth environments and formed monolayers on the surface of culture flasks. Such monolayers of primary cell cultures of WBPH, VCMs, were used to study the functional role of P9-1 in the replication of SRBSDV.
Fig 1.
Light micrograph of the primary cell cultures derived from WBPH in vitro. (A) Migration of fibroblast-like cells from explanted embryo fragments (Ex) of WBPH 7 days after preparation. (B) Migration of the monolayers of epithelium-like cells from the explanted embryo fragments (Ex) of WBPH 12 days after preparation. (C) A large epithelium-like cell sheet formed around explanted embryonic fragments (Ex) of WBPH 40 days after preparation. Bars, 100 μm (phase contrast).
Viral RNA and particles of SRBSDV accumulated in the viroplasm in virus-infected insect cell cultures.
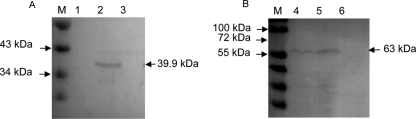
To study the localization of SRBSDV P9-1 and P10 proteins in virus-infected cells, we prepared the antibodies against these two proteins. In a Western blot analysis, the SRBSDV P9-1 and P10 antibodies recognized a 39.9-kDa and a 63-kDa protein, respectively (in accordance with the expected SRBSDV P9-1 and P10 proteins) present in protein extracts from infected rice plants (Fig. 2). Furthermore, P10 antibodies, rather than P9-1 antibodies, reacted with proteins from purified viral particles, confirming that P10 is the major outer capsid protein and P9-1 is the nonstructural protein of SRBSDV (Fig. 2). No reaction was observed with proteins from healthy plants (Fig. 2), confirming that the SRBSDV P9-1 and P10 antibodies were able to specifically recognize the proteins produced after SRBSDV infection.
Fig 2.
Western blot analyses of P9-1 and P10 proteins. Samples were separated by SDS-PAGE and detected with P9-1-specific (A) or P10-specific (B) antibodies. Lanes M, protein marker; lanes 1 and 4, purified SRBSDV particles; lanes 2 and 5, protein extracts from rice plants infected with SRBSDV; lanes 3 and 6, protein extracts from healthy rice plants.
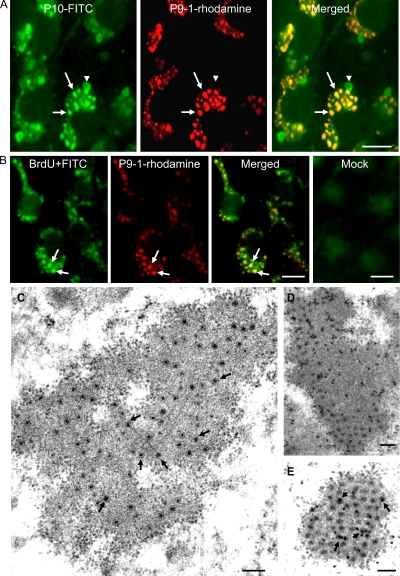
To study the replication of SRBSDV, we used immunofluorescence microscopy to observe the localization of P9-1 in virus-infected VCMs derived from WBPHs. VCMs on coverslip infected with SRBSDV were fixed 48 h p.i., stained with P10-FITC and P9-1–rhodamine, and then examined with confocal microscopy. In infected cells, P9-1 localized to punctate inclusions that colocalized with punctate inclusions of P10 in the cytoplasm (Fig. 3A). P10 also formed additional punctate inclusions (Fig. 3A). Our results suggested that the viral inclusions of P9-1 accumulated outer capsid proteins of SRBSDV during viral replication.
Fig 3.
Subcellular localization of P9-1 and P10 of SRBSDV in virus-infected VCMs 48 h p.i. (A) Immunostaining with P10-FITC (green) and P9-1–rhodamine (red). In merged images, colocalization of P9-1 and P10 is indicated in yellow (arrows). Arrowheads indicate indicate additional sites with P10. Bar, 10 μm. (B) Intracellular sites of RNA synthesis in mock- or SRBSDV-infected VCMs. BrUTP-labeled viral RNA was stained with anti-BrdU from mouse, followed by anti-mouse IgG conjugated to FITC (green). Viroplasm is stained with P9-1–rhodamine (red). Colocalization of P9-1–rhodamine and BrUTP-labeled SRBSDV RNA is indicated in yellow (arrows). Bars, 10 μm. (C) Electron micrograph of viroplasm in virus-infected VCMs. Arrows show viral particles in the matrix of viroplasm. Bars, 250 nm. (D) Immunogold labeling of P9-1 in the matrix of viroplasm. Bar, 250 nm. (E) Immunogold labeling of P10 with the viral particles (arrows) within the matrix of viroplasm. Bar, 250 nm. Cells were immunostained for P9-1 and P10 with P9-1- and P10-specific antibodies in panels D and E, respectively, as primary antibodies, followed by treatment with goat antibodies against rabbit IgG that had been conjugated with 15-nm-diameter gold particles as secondary antibodies.
The detection of newly made RNA in virus-infected VCMs by immunofluorescence showed that the BrUTP-labeled RNA was distributed in punctate inclusions that colocalized with viral inclusions of P9-1 (Fig. 3B). No reaction with special structures was observed in noninfected cells (Fig. 3B). All these results suggested that viral inclusions of P9-1 were the sites of viral RNA synthesis.
To confirm our observations, we fixed infected VCMs at 48 h p.i. and examined them by immunoelectron microscopy using P9-1 and P10 antibodies. P9-1 antibodies specifically reacted with the matrix of the viroplasm (Fig. 3C and D), corresponding to the viral inclusions of P9-1 seen with immunofluorescence (Fig. 3A). This result suggested that SRBSDV P9-1 was a constituent of the matrix of viroplasm in virus-infected cells. Furthermore, viral particles that were distributed within the matrix of the viroplasm were specifically labeled by P10 antibodies (Fig. 3C and E), suggesting that SRBSDV particles were assembled within the matrix during viral replication.
SRBSDV P9-1 was sufficient to induce formation of viroplasm-like structures in non-host insect cells.
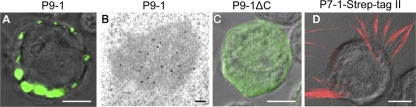
Immunofluorescence staining of P9-1 in Sf9 cells revealed punctate inclusions within the cells at 72 h p.i. (Fig. 4A). Furthermore, P9-1 specifically reacted with the granular inclusions, whose morphology was similar to that of the viroplasm matrix, as revealed by immunogold electron microscopy (Fig. 4B). It has been shown that the carboxy-terminal portion of P9-1 of two other fijiviruses, i.e., RBSDV and Mal de Río Cuarto virus (MRCV), contained important domains required for the formation of viroplasm-like structures (1, 19). To confirm a similar functional role for the carboxy-terminal portion of SRBSDV in the formation of viroplasm-like structures, we generated a P9-1 mutant in which the C-terminal 20 residues were deleted (P9-1ΔC) and found that this mutant was diffusely distributed in the cytoplasm (Fig. 4C), suggesting that the C terminus was required for the formation of viroplasm-like inclusions. Furthermore, another nonstructural protein, P7-1–Strep-tag II, formed tubule-like structures in Sf9 cells (Fig. 4D), as described previously (18), suggesting that the punctate inclusions formed by P9-1 were specific for this protein. No reaction with cellular structures after incubation with monoclonal anti-Strep-tag II in noninfected cells was observed (data not shown). All these results indicated that expression of P9-1 alone, in the absence of the viral multiplication process, was sufficient to induce the formation of the viroplasm-like structures in Sf9 cells, confirming an essential role of P9-1 in the biogenesis of the viroplasm in SRBSDV-infected cells.
Fig 4.
Subcellular localization of P9-1 and P7-1 of SRBSDV in recombinant baculovirus-infected Sf9 cells at 72 h p.i. (A) Immunofluorescence staining of P9-1-FITC revealed punctate inclusions of P9-1 of SRBSDV in Sf9 cells. Bar, 5 μm. (B) Immunogold labeling of P9-1 associated with electron-dense inclusion. Cells were immunostained with P9-1-specific polyclonal antibodies and goat antibodies against rabbit IgG that had been conjugated with 15-nm-diameter gold particles as secondary antibodies. Bar, 100 nm. (C) Immunofluorescence staining with P9-1-FITC revealed that a P9-1 mutant in which the C-terminal 20 residues had been deleted (P9-1ΔC) was diffusely distributed in the cytoplasm in Sf9 cells. Bar, 5 μm. (D) Immunofluorescence staining of monoclonal anti-Strep-tag II and rhodamine anti-mouse IgG as a secondary antibody revealed that P7-1–Strep-tag II formed the tubule-like structures in Sf9 cells. Bar, 5 μm.
RNAi induced by dsRNAs from the P9-1 gene inhibited the assembly of viroplasms and viral infection in insect vector cell cultures.
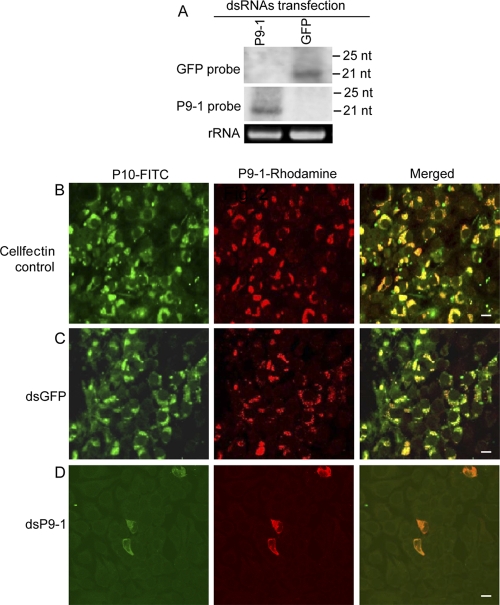
We then used an RNAi strategy to study the functional role of P9-1 in the viral replication cycle in VCMs. RNAi is induced by dsRNAs and manifested by the appearance of small interfering RNAs (siRNAs) that correspond to the mRNA target sequence (25). VCMs were transfected with dsP9-1 or dsGFP via Cellfectin-based transfection. Small RNAs approximately 21 nt in length were detected from RNAs extracted from dsP9-1- or dsGFP-transfected cells (Fig. 5A) at 72 h after transfection with the synthesized dsRNAs. These results showed that RNAi was induced by dsRNAs in VCMs.
Fig 5.
RNAi induced by dsP9-1 inhibited the assembly of viroplasm and the infection of SRBSDV in VCMs. (A) Detection of siRNAs in VCMs transfected with synthesized dsP9-1 or dsGFP at 72 h after transfection. Total RNA (ca. 5 μg) was probed with DIG-labeled riboprobes corresponding to the negative-sense RNA of the P9-1 or GFP gene. (Lower panel) 5.8S rRNA was used as a control to confirm loading of equal amounts of RNA in each lane. (B to D) Infection of SRBSDV was inhibited by RNAi induced by dsP9-1 in VCMs. At 24 h after transfection with Cellfectin transfection reagent (B), dsGFP (C), or dsP9-1 (D), the VCMs were inoculated with SRBSDV. At 48 h p.i., cells were stained with P10-FITC (green) and P9-1–rhodamine (red) and examined with confocal microscopy. Images are representative of the results of multiple experiments with multiple preparations. Bars, 10 μm.
To determine whether treatment with dsP9-1 would inhibit viroplasm formation, 24 h after transfection with dsRNAs, we inoculated VCMs with SRBSDV and processed them for immunofluorescence. Immunofluorescence showed that the dsGFP treatment did not prevent formation of viroplasm staining by P9-1–rhodamine and accumulation of viral particles staining by P10-FITC (Fig. 5B and C). In contrast, the dsP9-1 treatment led to a nearly complete inhibition of viroplasm formation and viral particle accumulation (Fig. 5B to D). These results suggested that the knockdown of P9-1 expression due to RNAi induced by dsP9-1 significantly inhibited viroplasm formation and viral infection in the VCMs.
Ingestion of dsRNAs from the P9-1 gene via membrane feeding knocked down P9-1 expression and strongly inhibited viral infection in intact WBPHs.
Our results obtained using the in vitro VCM-based system implied that RNAi induced by dsP9-1 can be used to examine the functional role of P9-1 in intact insects in vivo. The preliminary experiments showed that ingestion of dsRNAs via membrane feeding resulted in no phenotypic abnormalities in WBPHs (data not shown). The virus proliferates in the WBPH vector, and the insect becomes SRBSDV infective after a latent period of from 6 to 14 days (34). The effects of dsRNAs on the transcript levels of P9-1 and P10 viral genes were determined by RT-PCR. At 12 days post-first access to diseased plants (padp), RT-PCR analyses showed that ca. 87% (n = 100; 3 experiment repetitions) of WBPHs receiving dsGFP or diet alone contained the transcripts for P9-1 and P10 genes (Table 1). In contrast, ca. 11% (n = 100; 3 experiment repetitions) of WBPHs that received dsP9-1 contained the transcripts for these two genes (Table 1). These results confirmed that the P9-1 gene is susceptible to silencing by RNAi induced by dsP9-1 in vivo using a membrane-feeding approach.
Table 1.
RNAi induced by dsP9-1, ingested via membrane feeding, strongly inhibited infection by SRBSDV in the body of its insect vectors as detected by RT-PCR
| Insect dieta | No. of insects giving positive results (n = 100) |
||
|---|---|---|---|
| Expt I | Expt II | Expt III | |
| dsP9-1 | 11 | 10 | 12 |
| dsGFP | 86 | 86 | 89 |
| Control | 88 | 92 | 90 |
Second-instar nymphs of WBPHs were fed with dsP9-1 (0.5 μg/μl), dsGFP (0.5 μg/μl), or diet alone for 1 day, allowed a 2-day acquisition access period on virus-infected rice plants, and then placed on healthy rice seedlings for 10 days.
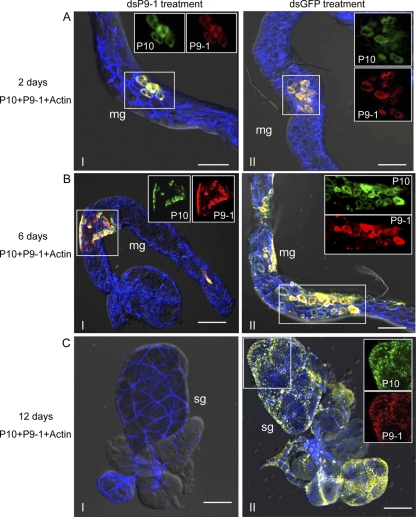
To determine whether the treatment of dsP9-1 would inhibit viral replication in intact insects, at 2, 6, and 12 days padp, we stained the internal organs from 50 WBPHs that had received dsRNAs with P10-FITC, P9-1–rhodamine, and actin dye phalloidin-Alexa Fluor 647 carboxylic acid. At 2 days padp, P9-1 and P10 were restricted to a few epithelial cells of the midgut in 84% of WBPHs receiving dsGFP but in only 12% of those receiving dsP9-1 (Fig. 6A and Table 2). The results suggested that the epithelial cells of the midgut were the initial entry site of SRBSDV. Furthermore, RNAi induced by dsP9-1 could specifically knock down P9-1 expression and significantly inhibit early SRBSDV infection in the epithelial cells of the midgut of the insect vectors. At 6 days padp, P9-1 and P10 were seen in an extensive area of the midgut in 83% of WBPHs receiving dsGFP but in only 6% of those receiving dsP9-1 (Fig. 6B and Table 2). However, in 12% of WBPHs that received dsP9-1, P9-1 and P10 were still restricted to a limited area of the midgut (Fig. 6B and Table 2). Therefore, the RNAi induced by dsP9-1 could inhibit efficient spread of virus from the initial infection site in the epithelial cells of the midgut. At 12 days padp, in 88% of the WBPHs receiving dsGFP, P9-1 and P10 were detected in an extensive area of the midgut and in the salivary glands of 78% of the insects (Fig. 6C and Table 2). However, much lower percentages of WBPHs that received dsP9-1 had P9-1 and P10 in the midgut, and none had the proteins in the salivary glands (Fig. 6C and Table 2). No significant difference in the numbers of positive samples was found between WBPHs that received dsGFP and those that received diet alone (data not shown). These results indicated that viral infection was restricted to the alimentary canal and that spread of virus to the salivary glands had been completely blocked in insect populations treated with dsP9-1.
Fig 6.
Ingestion of dsP9-1 via membrane feeding suppressed viral spread in vivo. Second-instar nymphs of WBPHs were fed with dsP9-1 (frames I) or dsGFP (frames II) via membrane feeding. At 2 days padp (A), 6 days padp (B), and 12 days padp (C), internal organs of WBPHs receiving dsP9-1 or dsGFP were stained for SRBSDV virions with P10-FITC (green), stained for viroplasm with P9-1–rhodamine (red), and stained for actin with actin dye phalloidin-Alexa Fluor 647 carboxylic acid (blue). The images with green fluorescence (P10 antigens), red fluorescence (P9-1 antigens), and blue fluorescence (actin dye) were merged under a background of transmitted light. Insets show green fluorescence (P10 antigens) and red fluorescence (P9-1 antigens) of the merged images in the boxed areas in each panel. Images are representative of the results of multiple experiments with multiple preparations. mg, midgut; sg, salivary gland. Bars, 70 μm.
Table 2.
Percentages of P9-1 and P10 antigens in tissues dissected from intact WBPHs after ingestion of dsP9-1 or dsGFP and acquisition feeding on virus-infected plants
| Tissuea | % of indicated antigen |
|||||
|---|---|---|---|---|---|---|
| 2 days padp |
6 days padp |
12 days padp |
||||
| dsP9-1 | dsGFP | dsP9-1 | dsGFP | dsP9-1 | dsGFP | |
| Limited area of midgut | 12 | 84 | 12 | 4 | 8 | 0 |
| Extensive area of midgut | 0 | 0 | 6 | 83 | 13 | 88 |
| Salivary gland | 0 | 0 | 0 | 20 | 0 | 78 |
Second-instar nymphs of WBPHs were fed with dsP9-1 (0.5 μg/μl) or dsGFP (0.5 μg/μl) via membrane feeding and then maintained on a mixed diet for 1 day, allowed a 2-day acquisition access period on SRBSDV-infected rice plants, and then placed on healthy rice seedlings. Fifty individuals were tested in each group.
DISCUSSION
VCMs enable us to study the molecular entities responsible for biological events during viral replication (30, 31). In the present study, using VCMs derived from WBPHs, our cytopathological analysis revealed that P9-1 of SRBSDV was a component of the viroplasm matrix and, for the first time, provided direct evidence to show that newly synthesized viral RNA, outer-capsid structural proteins, and viral particles accumulated together within the matrix of viroplasms that are induced by fijivirus infection (Fig. 3), confirming the hypothesis that the viroplasm is the site for the replication and assembly of fijiviruses. Our results extend the limited information from previous electron microscopic studies that showed that P9-1 of two other fijiviruses (RBSDV and MRCV) exclusively localized in the matrix of viroplasms in virus-infected cells (9, 12, 19). With the evidence indicating that P9-1 alone was necessary for the formation of viroplasm-like structures in nonhost cells (Fig. 4), our results suggest that P9-1 is the minimal viral factor required for viroplasm formation during SRBSDV infection.
To gain direct evidence that P9-1 plays a crucial role in viral replication, we used the RNAi strategy to knock down the expression of P9-1 in virus-infected VCMs and then analyzed its effect on viroplasm formation and viral infection. Because RNAi induced by dsRNA is a conserved sequence-specific gene silencing mechanism (6) and the sequences of P9-1 and other viral genes of SRBSDV had little homology (data not shown), we determined that the knockdown of P9-1 expression in VCMs was specifically caused by RNAi induced by dsP9-1. Our results indicated that the knockdown of P9-1 expression directly led to a significant loss in the function of P9-1 in the formation of viroplasms, with a significant simultaneous inhibition of the assembly of progeny virions within viroplasms in VCMs (Fig. 5). These results indicate that P9-1 was essential for the formation of the viroplasm and viral replication.
As a persistently propagative plant virus, SRBSDV, after moving into the epithelial cells of the alimentary canal, must replicate and assemble progeny virions, which eventually spread to the salivary glands in its insect vector. Our results showed that the knockdown of P9-1 expression due to RNAi induced by ingestion of dsP9-1 strongly inhibited the early formation of viroplasms and accumulation of viral particles in the epithelial cells of the midgut, which resulted in limited spread of the virus from the initial infection site to additional tissues, including the salivary glands in the insect (Table 2 and Fig. 6). Due to the essential role of P9-1 in the biogenesis of the viroplasm as shown above, we determined that the slow spread of SRBSDV in the body of the dsP9-1-treated insects was directly caused by a significant loss in the functioning of P9-1 in the assembly of viroplasm, leading to strong inhibition of viral replication in its insect vector. Our efforts also demonstrated for the first time that RNAi activity was inducible in WBPH.
Previously, due to the lack of insect vector cell cultures and a reverse-genetics system in fijiviruses, no clear function of virus-encoded proteins in the viral replicative cycle had been determined. In this study, the deployment of new biological tools, including VCMs derived from WBPH and RNAi induced by synthesized dsRNA in vitro and in vivo, helped overcome the lack of a reverse-genetics system in fijiviruses and advanced our understanding of SRBSDV-WBPH interactions. By combining these new approaches, the data demonstrated that P9-1 of SRBSDV is an essential protein for viral replication and suggested that this nonstructural protein may be a good target for disease control. In the viruses of family Reoviridae, RNAi of genes for viroplasm matrix proteins, including μNS of mammalian reovirus (15) and NSP5 of rotavirus (17), which are all orthologous to P9-1 of SRBSDV, had pleiotropic effects on the assembly of viroplasm and viral replication. Our results support the hypothesis that viroplasms play a pivotal role in replication of viruses in the family Reoviridae.
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (2010CB126203), the Specialized Research Fund for the Ministry of Agriculture (201003031), the National Natural Science Foundation of China (31130044 and 31070130), the Natural Science Foundation of Fujian Province (2011J06008), and the Doctoral Fund of the Ministry of Education of China (20113515130002).
Footnotes
Published ahead of print 7 March 2012
REFERENCES
- 1. Akita F, et al. 2012. Crystallographic analysis reveals octamerization of viroplasm matrix protein P9-1 of Rice black streaked dwarf virus. J. Virol. 86:746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen H, Chen Q, Omura T, Uehara-Ichiki T, Wei T. 2011. Sequential infection of Rice dwarf virus in the internal organs of its insect vector after ingestion of virus. Virus Res. 160:389–394 [DOI] [PubMed] [Google Scholar]
- 3. Cheng G, et al. 2010. A C-type lectin collaborates with a CD45 phosphatase homolog to facilitate West Nile virus infection of mosquitoes. Cell 142:714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherry S. 2011. RNAi screening for host factors involved in viral infection using Drosophila cells. Methods Mol. Biol. 721:375–382 [DOI] [PubMed] [Google Scholar]
- 5. Creamer R. 1993. Invertebrate tissue culture as a tool to study insect transmission of plant viruses. In Vitro Cell. Dev. Biol. 29:284–288 [DOI] [PubMed] [Google Scholar]
- 6. Fire A, et al. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811 [DOI] [PubMed] [Google Scholar]
- 7. Forzan M, Marsh M, Roy P. 2007. Bluetongue virus entry into cells. J. Virol. 81:4819–4827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Q, Zhang Z, Hu C, Lai F, Sun Z. 2001. A chemically defined diet enables continuous rearing of the brown planthopper, Nilaparvata lugens (Stål) (Homoptera: Delphacidae). Appl. Entomol. Zool. 36:111–116 [Google Scholar]
- 9. Guzmán FA, et al. 2010. Immunodetection and subcellular localization of Mal de Río Cuarto virus P9-1 protein in infected plant and insect host cells. Virus Genes 41:111–117 [DOI] [PubMed] [Google Scholar]
- 10. Hogenhout SA, Ammar ED, Whitfield AE, Redinbaugh MG. 2008. Insect vector interactions with persistently transmitted viruses. Annu. Rev. Phytopathol. 46:327–359 [DOI] [PubMed] [Google Scholar]
- 11. Huvenne H, Smagghe G. 2010. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J. Insect Physiol. 56:227–235 [DOI] [PubMed] [Google Scholar]
- 12. Isogai M, Uyeda I, Lee BC. 1998. Detection and assignment of proteins encoded by rice black streaked dwarf fijivirus S7, S8, S9 and S10. J. Gen. Virol. 79:1487–1494 [DOI] [PubMed] [Google Scholar]
- 13. Kimura I. 1986. A study of rice dwarf virus in vector cell monolayers by fluorescent antibody focus counting. J. Gen. Virol. 67:2119–2124 [Google Scholar]
- 14. Kimura I, Omura T. 1988. Leafhopper cell cultures as a means for phytoreovirus research. Adv. Dis. Vector Res. 5:111–135 [Google Scholar]
- 15. Kobayashi T, Chappell JD, Danthi P, Dermody TS. 2006. Gene-specific inhibition of reovirus replication by RNA interference. J. Virol. 80:9053–9063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y, et al. 2011. The P7-1 protein of southern rice black-streaked dwarf virus, a fijivirus, induces the formation of tubular structures in insect cells. Arch. Virol. 156:1729–1736 [DOI] [PubMed] [Google Scholar]
- 17. López T, Rojas M, Ayala-Bretón C, López S, Arias CF. 2005. Reduced expression of the rotavirus NSP5 gene has a pleiotropic effect on virus replication. J. Gen. Virol. 86:1609–1617 [DOI] [PubMed] [Google Scholar]
- 18. Lu YH, Zhang JF, Xiong RY, Xu QF, Zhou YJ. 2011. Identification of an RNA silencing suppressor encoded by southern rice black-streaked dwarf virus S6. Sci. Agric. Sinica 14:2909–2917 (In Chinese.) [Google Scholar]
- 19. Maroniche GA, et al. 2010. Functional and biochemical properties of Mal de Río Cuarto virus (Fijivirus, Reoviridae) P9-1 viroplasm protein show further similarities to animal reovirus counterparts. Virus Res. 152:96–103 [DOI] [PubMed] [Google Scholar]
- 20. McGinnis KM. 2010. RNAi for functional genomics in plants. Brief. Funct. Genomics 9:111–117 [DOI] [PubMed] [Google Scholar]
- 21. Milne RG, del Vas M, Harding RM, Marzachì R, Mertens PPC. 2005. Fijivirus, p 534–542 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the international committee on taxonomy of viruses Academic Press, Amsterdam, Holland [Google Scholar]
- 22. Miyazaki N, et al. 2008. Structural evolution of reoviridae revealed by oryzavirus in acquiring the second capsid shell. J. Virol. 82:11344–11353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Omura T, Mertens PPC. 2005. Phytoreovirus, p 543–549 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the international committee on taxonomy of viruses Academic Press, Amsterdam, Holland [Google Scholar]
- 24. Omura T, Kimura I. 1994. Leafhopper cell culture for virus research, p 91–107 In Maramorosch K, McIntosh AH. (ed), Arthropod cell culture systems. CRC Press, Philadelphia, PA [Google Scholar]
- 25. Rosa C, et al. 2010. RNAi effects on actin mRNAs in Homalodisca vitripennis cells. J. RNAi Gene Silencing 6:361–366 [PMC free article] [PubMed] [Google Scholar]
- 26. Shimizu T, Yoshii M, Wei T, Hirochika H, Omura T. 2009. Silencing by RNAi of the gene for Pns12, a viroplasm matrix protein of Rice dwarf virus, results in strong resistance of transgenic rice plants to the virus. Plant Biotechnol. J. 7:24–32 [DOI] [PubMed] [Google Scholar]
- 27. Spinelli S, et al. 2006. Modular structure of the receptor binding proteins of Lactococcus lactis phages. The RBP structure of the temperate phage TP901-1. J. Biol. Chem. 281:14256–14262 [DOI] [PubMed] [Google Scholar]
- 28. Upadhyaya NM, Mertens PPC. 2005. Oryzavirus, p 550–555 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the international committee on taxonomy of viruses Academic Press, Amsterdam, Holland [Google Scholar]
- 29. Wang Q, et al. 2010. The complete genome sequence of two isolates of Southern rice black-streaked dwarf virus, a new member of the Genus Fijivirus. J. Phytopathol. 158:733–737 [Google Scholar]
- 30. Wei T, et al. 2006. The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J. Virol. 80:8593–8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei T, et al. 2006. Pns12 protein of Rice dwarf virus is essential for formation of viroplasms and nucleation of viral-assembly complexes. J. Gen. Virol. 87:429–438 [DOI] [PubMed] [Google Scholar]
- 32. Zhang H, Chen J, Adams M. 2001. Molecular characterisation of segments 1 to 6 of rice black-streaked dwarf virus from China provides the complete genome. Arch. Virol. 146:2331–2339 [DOI] [PubMed] [Google Scholar]
- 33. Zhang HM, Yang J, Chen JP, Adams MJ. 2008. A black-streaked dwarf disease on rice in China is caused by a novel fijivirus. Arch. Virol. 153:1893–1898 [DOI] [PubMed] [Google Scholar]
- 34. Zhou G, et al. 2008. Southern rice black-streaked dwarf virus: a new proposed Fijivirus species in the family Reoviridae. Chin. Sci. Bull. 53:3677–3685 [Google Scholar]