Abstract
TNPO3 is a nuclear importer required for HIV-1 infection. Here, we show that depletion of TNPO3 leads to an HIV-1 block after nuclear import but prior to integration. To investigate the mechanistic requirement of TNPO3 in HIV-1 infection, we tested the binding of TNPO3 to the HIV-1 core and found that TNPO3 binds to the HIV-1 core. Overall, this work suggests that TNPO3 interacts with the incoming HIV-1 core in the cytoplasm to assist a process that is important for HIV-1 infection after nuclear import.
TEXT
The host factor TNPO3 is required for the infection of several lentiviruses (1, 3, 9, 10, 13, 19, 21). To assess the role of TNPO3 in retroviral infection, we stably knocked down TNPO3 expression in HeLa cells by transduction of the pLKO.1-shRNA vector that specifically targets the TNPO3 transcript. HeLa cell lines stably expressing pLKO.1-shRNA-TNPO3 presented a remarkable reduction of TNPO3 expression (see Fig. S1 in the supplemental material). Next, we tested the ability of different retroviruses to infect cells that are deficient in TNPO3. For this purpose, we used a set of retroviruses pseudotyped with vesicular stomatitis virus G protein (VSV-G) containing a green fluorescent protein (GFP) as a reporter for infectivity (5). HeLa TNPO3 knockdown (K.D.) cells reduced simian immunodeficiency virus SIVmac, human immunodeficiency virus type 2 (HIV-2), HIV-1, bovine immunodeficiency virus (BIV), and equine infectious anemia virus (EIAV) infection 17-fold, 15-fold, 12-fold, 4-fold, and 3-fold, respectively (see Table S1 in the supplemental material). In contrast, TNPO3-deficient cells did not reduce the infection by Moloney murine leukemia virus (MMLV) or feline immunodeficiency virus (FIV) (see Table S1). These results were in agreement with previous observations (10, 14, 19).
Previous work suggested that depletion of TNPO3 prevents the nuclear import of HIV-1 (3); therefore, we analyzed the requirement of TNPO3 at a cellular stage that resembles a nondividing cell by the use of aphidicolin, which blocks the cell cycle at early S phase (20). We hypothesized that the HIV-1 dependency on TNPO3 is greater in nondividing cells. Contrary to our expectations, aphidicolin treatment of cells did not increase retrovirus dependency on TNPO3 for infection by HIV-1, HIV-2, and SIV (see Table S1 in the supplemental material). As expected, infection of MMLV was completely inhibited by aphidicolin. These findings raised the possibility that the role of TNPO3 is after the nuclear import of the HIV-1 preintegration complex (PIC), as recently suggested (22).
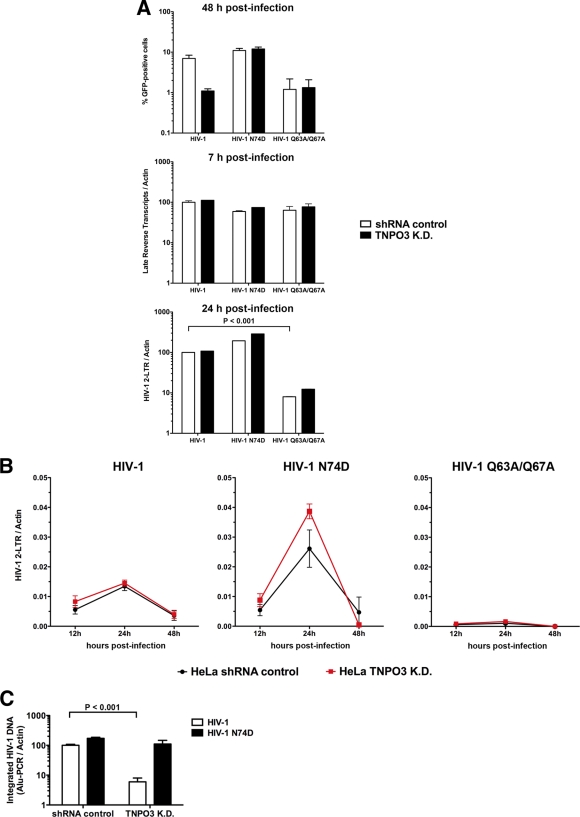
To test the hypothesis that TNPO3 assists HIV-1 replication after nuclear import of the PIC, we measured the levels of HIV-1 two-long terminal repeat (2-LTR) circles in TNPO3-depleted cells. Formation of 2-LTR circles demonstrates nuclear transport of the PIC and subsequent circularization of the viral genome by the host's DNA repair machinery (2). We infected small hairpin RNA (shRNA) control and TNPO3 K.D. cells with the indicated GFP reporter viruses and monitored infection by measuring GFP-positive cells 48 h postinfection (Fig. 1A, upper panel). HIV-1, HIV-1 with an N74D mutation (HIV-1 N74D), and HIV-1 Q63A/Q67A were normalized by quantifying the particle-associated reverse transcriptase activity of viral supernatants, as described previously (18). In parallel, similar infections were used to monitor late reverse transcripts 7 h postinfection and HIV-1 2-LTR circles 24 h postinfection (Fig. 1A). In contrast to results for HIV-1 N74D, infection by wild-type (wt) HIV-1 was decreased ∼10-fold in TNPO3 K.D. cells (Fig. 1A, upper panel). In agreement with previous results, we did not observe infection of HIV-1 Q63A/Q67A, which is a mutant impaired in nuclear import (Fig. 1A, upper panel) (7). As expected, the levels of late reverse transcripts 7 h postinfection in HeLa TNPO3 K.D. cells were similar to those in shRNA control cells (Fig. 1A, middle panel) (1, 3). Intriguingly, HIV-1 and HIV-1 N74D infections generated the same levels of 2-LTR circles in TNPO3 K.D. cells as that in shRNA control cells at 24 h postinfection, suggesting that TNPO3-depleted cells block HIV-1 infection after nuclear import (Fig. 1A, lower panel). As expected, infection of cells by HIV-1 Q63A/Q67A exhibited reduced levels of 2-LTR circles due to its defect on nuclear import (Fig. 1A, lower panel) (7). Together, these results suggest that depletion of TNPO3 is required for HIV-1 infection after PIC nuclear import.
Fig 1.
TNPO3-deficient cells block HIV-1 infection after nuclear import but prior to integration. (A) HeLa TNPO3 K.D. cells (TNPO3 K.D.) and HeLa cells transduced with the empty vector pLKO.1 (shRNA control) were challenged with DNase-pretreated HIV-1-GFP, HIV-1-N74D-GFP, and HIV-1-Q63A/Q67A-GFP as indicated. Infection was determined by measuring the percentage of GFP-positive cells by flow cytometry 48 h postinfection (upper panel). In parallel, cells from similar infections were lysed at 7 and 24 h postinfection and total DNA was extracted. The DNA samples collected at 7 h postinfection were used to determine the levels of late reverse transcripts by real-time PCR (middle panel). HIV-1 2-LTR circles were quantified by real-time PCR in DNA samples collected at 24 h postinfection (lower panel). Late reverse transcripts and 2-LTR circle levels were normalized on a per-cell basis relative to HeLa shRNA control cells infected with HIV-1. (B) Similarly, TNPO3 K.D. and shRNA control cells were infected by HIV-1-GFP, HIV-1-N74D-GFP, and HIV-1-Q63A/Q67A-GFP normalized by quantifying particle-associated reverse transcriptase activity on viral supernatants. At 12, 24, and 48 h postinfection, total DNA was extracted and used to determine the levels of HIV-1 2-LTR circles. (C) DNA extracted at 24 h was also used to determine integration into the genome by measuring integrated proviruses using real-time Alu-PCR products relative to HeLa shRNA control cells infected with HIV-1; Alu-PCR products are achieved by using primers complementary to the HIV LTR U3 region and chromosomal Alu repeats. Similar results were obtained in three independent experiments, and standard deviations are shown. Statistical differences were given as P values of <0.001 by two-way analysis of variance (ANOVA) followed by the Bonferroni post test.
To better understand this process, we monitored 2-LTR circle formation over time (Fig. 1B). Consistently, HIV-1 infection of TNPO3 K.D. cells generated levels of 2-LTR circles similar to those generated by wt HIV-1 infection of shRNA control cells over time (Fig. 1B, left panel). As expected for HIV-1 infection, the levels of HIV-1 2-LTR circles reached maximum levels at 24 h and decreased by 48 h postinfection (Fig. 1B, left panel) (2). Infection of TNPO3 K.D. and shRNA control cells by HIV-1 N74D generated similar levels of HIV-1 2-LTR circles (Fig. 1B, middle panel). As expected, infection by HIV-1 Q63A/Q67A exhibited reduced levels of HIV-1 2-LTR circles throughout (Fig. 1B, right panel). Altogether, these results suggested that the PIC is transported into the nucleus of TNPO3 K.D. cells with similar kinetics and that TNPO3 is required in a subsequent step of the retroviral life cycle.
Next, we measured the ability of HIV-1 to be integrated in TNPO3 K.D. cells. To test for integration, we performed real-time PCR using primers complementary to the HIV-1 LTR and chromosomal Alu sequence repeats in the human genome (2). Remarkably, wt HIV-1 infection of TNPO3 K.D. cells led to a 20-fold decrease in integration compared to the Alu-PCR products from HIV-1-infected shRNA control cells (Fig. 1C). As expected, the integration of HIV-1 N74D was not affected (Fig. 1C). We therefore conclude that the loss of TNPO3 impacts HIV-1 replication during a step after nuclear import but prior to integration.
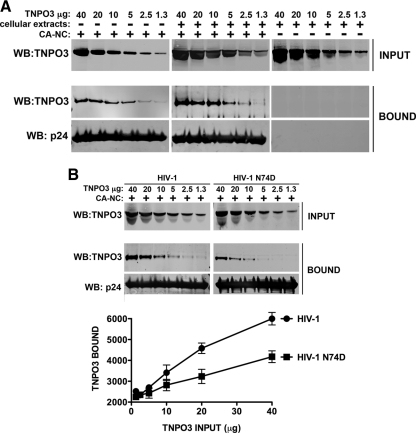
Previous reports have pointed to the HIV-1 capsid as a possible genetic determinant for the requirement of TNPO3 in HIV-1 infection (10). To test this hypothesis, we measured the ability of TNPO3 to directly interact with in vitro-assembled HIV-1 capsid-nucleocapsid (CA-NC) complexes (6). Intriguingly, we found that recombinant TNPO3 binds to in vitro-assembled HIV-1 CA-NC complexes in the presence or absence of cellular extracts (Fig. 2A), demonstrating an interaction between TNPO3 and assembled HIV-1 CA-NC complexes. Moreover, as shown in Fig. 2A, we observed that TNPO3 bound to the capsid in a concentration-dependent manner. As a control, we incubated purified TNPO3 in cellular extracts without CA-NC complexes and showed that it was unable to traverse the 70% sucrose cushion (Fig. 2A). Previous observations have established that the HIV-1 viral capsid N74D mutant is insensitive to TNPO3 loss during infection (12, 19). One possible explanation for these observations is that the N74D mutant capsid binds to TNPO3 with a changed affinity. To test this hypothesis, we measured the ability of purified TNPO3 to interact with in vitro-assembled HIV-1 CA-NC complexes bearing the N74D mutation (Fig. 2B). In agreement with our hypothesis, TNPO3 bound with lower affinity to in vitro-assembled HIV-1 CA-NC complexes bearing the N74D mutation than to the wt. Therefore, we conclude that TNPO3 binds to in vitro-assembled HIV-1 CA-NC complexes. However, TNPO3 bound to in vitro-assembled HIV-1-N74D CA-NC complexes with lower affinity, suggesting that the binding is specific.
Fig 2.
Binding of TNPO3 to HIV-1 capsid-nucleocapsid complexes. (A) The indicated amounts of recombinant TNPO3 protein were incubated with HIV-1 CA-NC complexes at room temperature for 1 h in the presence of cellular extracts or capsid-binding buffer. The mixtures were applied onto a 70% sucrose cushion and centrifuged. INPUT represents the mixtures analyzed by Western blotting before being applied to the 70% cushion. The INPUT mixtures were Western blotted using anti-TNPO3 antibodies. The pellet from the 70% cushion (BOUND) was analyzed by Western blotting using anti-TNPO3 and anti-capsid antibodies. To demonstrate that TNPO3 is unable to traverse the cushion by itself, we applied the different amounts of TNPO3 incubated in cellular extracts without HIV-1 CA-NC complexes onto a 70% sucrose cushion. The results of three independent experiments were similar; the result of a single experiment is shown. (B) Similar amounts of recombinant TNPO3 were incubated with HIV-1 wt or mutant CA-NC complexes for 1 h in the presence of cellular extracts. As described above, input and bound fractions were analyzed by fluorescent Western blotting using anti-TNPO3 and anti-capsid antibodies. Quantifications of TNPO3 bound versus TNPO3 input fractions of three independent experiments were plotted with their respective standard deviations on the lower panel. Statistical differences were given as P values of <0.001 for an input fraction of 40 μg by two-way ANOVA followed by the Bonferroni post test.
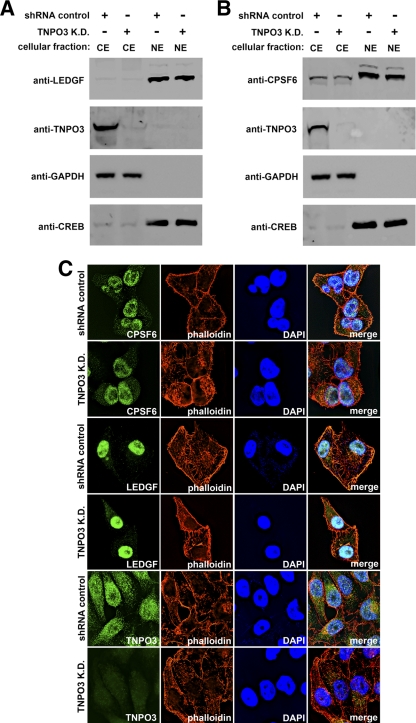
Our collective observations suggest that TNPO3 binds to the HIV-1 core in the cytoplasm and that this interaction is ultimately important for the virus's integration into the host genome. Interestingly, depletion of TNPO3 has recently been shown to alter the pattern of HIV-1 site integration in the host genome (16). Therefore, one possibility is that TNPO3 is needed for the nuclear import of host factors that are required for viral integration. To test this hypothesis, we depleted TNPO3 and assessed the subcellular localization of the lens epithelium-derived growth factor (LEDGF/p75), a chromatin-associated host factor that is critical for HIV-1 integration (4). As shown in Fig. 3A, the subcellular localization of LEDGF/p75 was not altered in TNPO3 K.D. cells compared to that in control cells (Fig. 3A). In agreement, immunofluorescence (IF) experiments also showed that the localization of LEDGF/p75 was not altered in TNPO3 K.D. cells compared to control cells (Fig. 3C). These experiments show that TNPO3 depletion does not affect the localization of the HIV-1 integration cofactor LEDGF/p75.
Fig 3.
Subcellular localization of LEDGF and CPSF6 in TNPO3-depleted cells. (A and B) To determine the cellular localization of LEDGF and CPSF6, HeLa TNPO3 knockdown cells (TNPO3 K.D.) or HeLa cells transduced with the empty vector pLKO.1 (shRNA control) were biochemically fractionated to separate nuclear from cytosolic proteins. Cytosolic extracts (CE) and nuclear extracts (NE) were blotted using antibodies against LEDGF, CPSF6, and TNPO3. To assay the bona fide origin of the extracts, the fractions were Western blotted using anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) and anti-CREB as cellular markers for the cytosol and nucleus, respectively. (C) Subcellular localization of CPSF6 and LEDGF by immunofluorescence. HeLa TNPO3 knockdown cells (TNPO3 K.D.) or HeLa cells transduced with the empty vector pLKO.1 (shRNA control) were fixed and stained using antibodies against CPSF6, LEDGF, and TNPO3. The nuclear compartment and F-actin was labeled using 4′,6-diamidino-2-phenylindole (DAPI) and phalloidin, respectively. Similar results were obtained in three independent experiments, and a representative experiment is shown.
TNPO3 also mediates the nuclear import of the serine/arginine-rich family of splicing factors (SR proteins), with loss of TNPO3 producing a cytosolic accumulation of SR proteins (11). Interestingly, a truncated SR protein, CPSF6, which is defective in nuclear localization, was also found to potently block wt HIV-1 nuclear import (12). Therefore, we investigated the cellular localization of endogenous full-length CPSF6 in TNPO3 K.D. cells by biochemical fractionation and IF. Analysis of TNPO3 K.D. cells revealed that the distribution of CPSF6 was not altered compared to that in control cells (Fig. 3B and C). As shown in Fig. 3B, CPSF6 is predominantly in the nuclear fraction. Thus, levels of TNPO3 depletion that decreased HIV-1 integration 20-fold in HeLa cells did not change the subcellular localization of CPSF6, suggesting that such an event does not explain HIV-1's TNPO3 dependency.
Here, we report several novel findings regarding the role of the host cell nuclear importer TNPO3 in lentiviral replication. We found that TNPO3 physically binds to in vitro-assembled HIV-1 CA-NC complexes, which recapitulate the surface of the viral core and are composed of capsid, nucleocapsid, and RNA (8). Our work revealed that the interaction between TNPO3 and the assembled HIV-1 CA-NC complexes bearing the capsid mutation N74D is diminished compared to that of the wt, suggesting that binding of TNPO3 to the HIV-1 core is specific. Depletion of TNPO3 inhibits HIV-1 viral replication after nuclear import but prior to proviral integration, which is consistent with a recent observation (22). TNPO3 thus represents a previously unappreciated class of HIV-dependency factor, which physically associates with the HIV-1 CA-NC complexes and is required for successful viral integration and genome site selection. Our results and those of others are consistent with a model wherein TNPO3 binding to the HIV-1 core in the cytoplasm is needed for the successful completion of a process that is subsequently required for integration. One possibility is that TNPO3 binding to the HIV-1 core permits the proper maturation of the PIC in the cytosol. For example, the 3′ processing activity of viral integrase on the HIV LTRs is required for efficient integration and has been suggested to occur in the cytosol (15, 17). Therefore, the cytosolic binding of TNPO3 to the HIV-1 core could play a role in ensuring the proper 3′ processing of the viral LTRs, setting the stage for effective viral integration subsequent to nuclear import. Future experiments will test whether depletion of TNPO3 is able to affect the 3′ processing of the viral LTRs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andre Rosowsky for critical readings of the manuscript. We are grateful to Chris Aiken, Vaibhav Shah, Eric Poeschla, Peter Cherepanov, and Goedele Maertens for providing reagents.
Funding to A.L.B. for this effort was provided by the Harvard Center for AIDS Research (CFAR), the Phillip T. and Susan M. Ragon Institute Foundation, and the Bill and Melinda Gates Foundation's Global Health Program. This work was funded by NIH grant R01 AI087390 to F.D.-G. and a K99/R00 Pathway to Independence Award to F.D.-G. from the National Institutes of Health, grant 4R00MH086162-02. F.D.N. and N.A. were supported by the Fondation pour la Recherche Médicale (FRM) and the Centre National de la Recherche Scientifique (CNRS).
Footnotes
Published ahead of print 7 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Brass AL, et al. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319:921–926 [DOI] [PubMed] [Google Scholar]
- 2. Butler SL, Hansen MS, Bushman FD. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631–634 [DOI] [PubMed] [Google Scholar]
- 3. Christ F, et al. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192–1202 [DOI] [PubMed] [Google Scholar]
- 4. Ciuffi A, et al. 2005. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287–1289 [DOI] [PubMed] [Google Scholar]
- 5. Diaz-Griffero F, et al. 2008. A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology 378:233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diaz-Griffero F, et al. 2009. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83:10737–10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dismuke DJ, Aiken C. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83 [DOI] [PubMed] [Google Scholar]
- 9. Konig R, et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135:49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krishnan L, et al. 2010. The requirement for cellular transportin 3 (TNPO3 or TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J. Virol. 84:397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. 2000. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275:7950–7957 [DOI] [PubMed] [Google Scholar]
- 12. Lee K, et al. 2010. Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levin A, Hayouka Z, Friedler A, Loyter A. 2010. Transportin 3 and importin alpha are required for effective nuclear import of HIV-1 integrase in virus-infected cells. Nucleus 1:422–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Logue EC, Taylor KT, Goff PH, Landau NR. 2011. The cargo-binding domain of transportin 3 is required for lentivirus nuclear import. J. Virol. 85:12950–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miller MD, Farnet CM, Bushman FD. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ocwieja KE, et al. 2011. HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 7:e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pauza CD. 1990. Two bases are deleted from the termini of HIV-1 linear DNA during integrative recombination. Virology 179:886–889 [DOI] [PubMed] [Google Scholar]
- 18. Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360 [DOI] [PubMed] [Google Scholar]
- 19. Thys W, et al. 2011. Interplay between HIV entry and transportin-SR2 dependency. Retrovirology 8:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamashita M, Emerman M. 2005. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 1:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou H, et al. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504 [DOI] [PubMed] [Google Scholar]
- 22. Zhou L, et al. 2011. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 7:e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.