Abstract
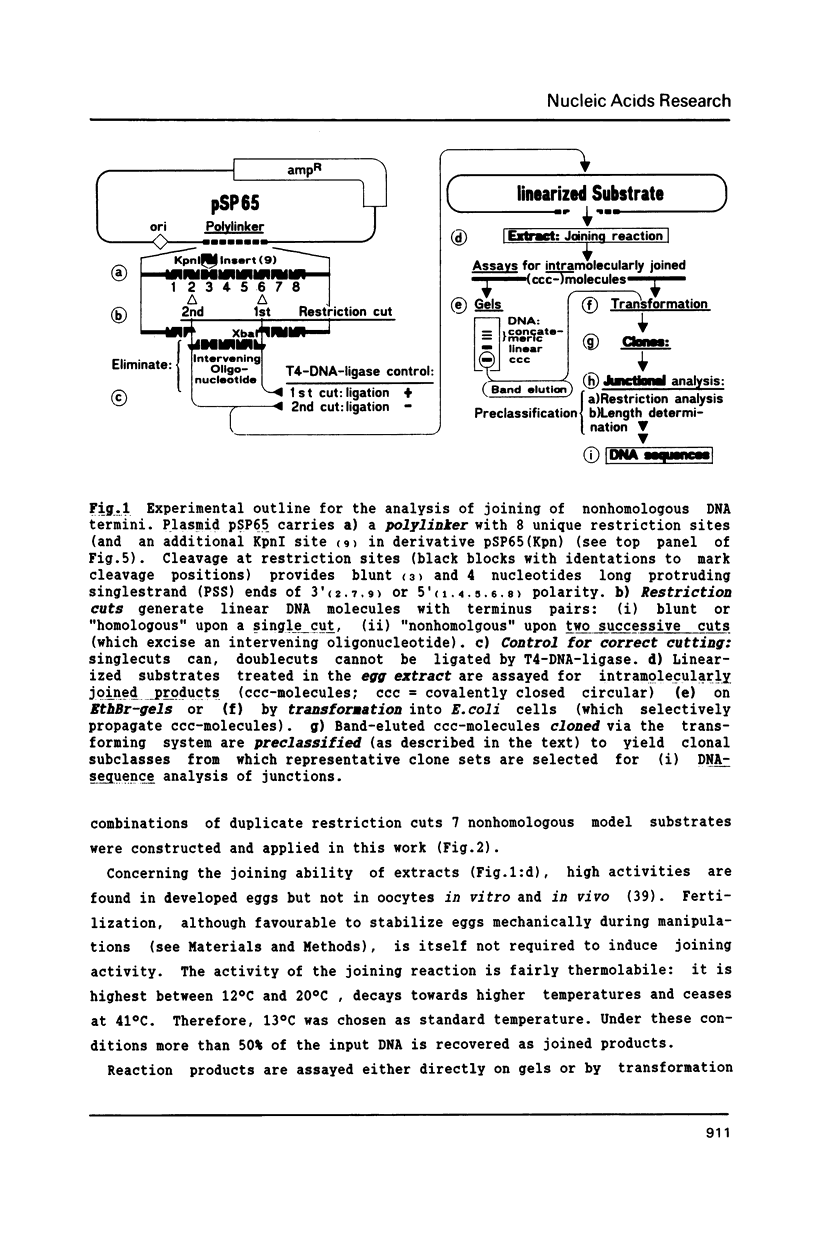
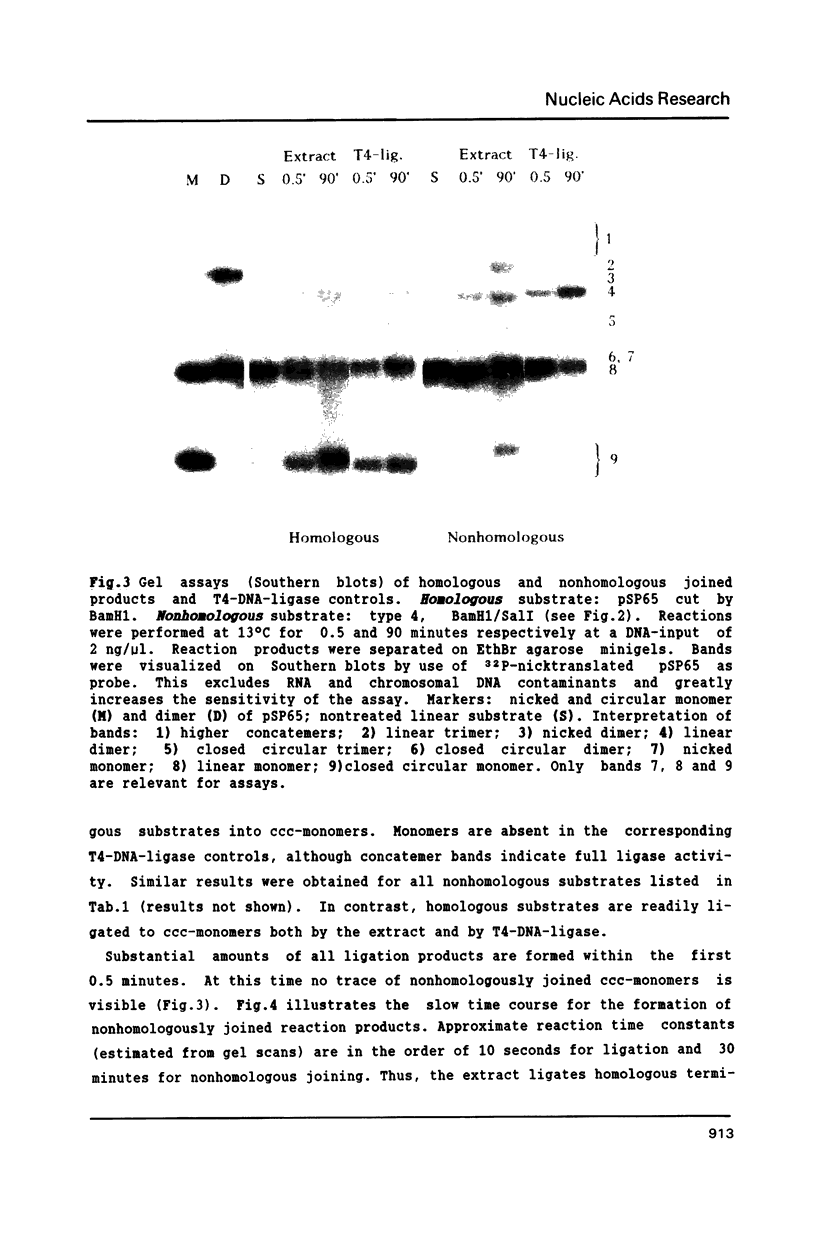
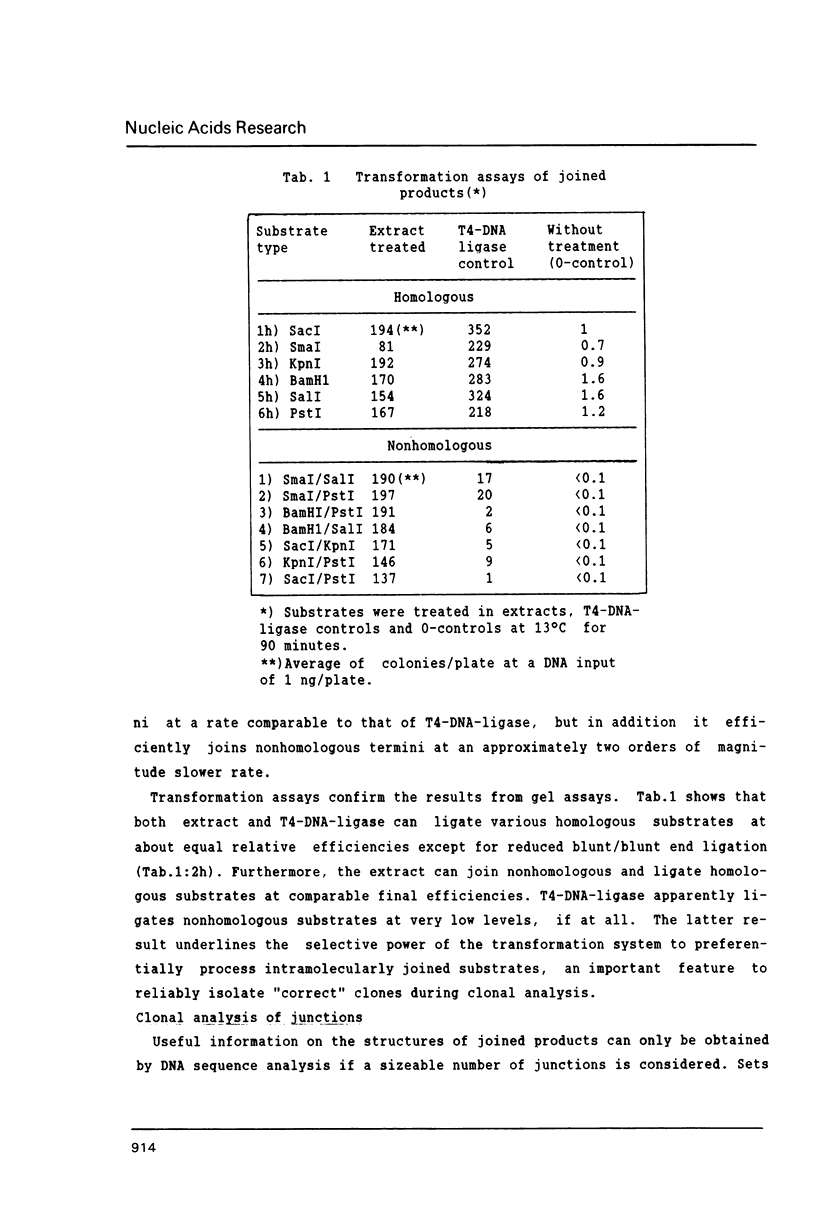
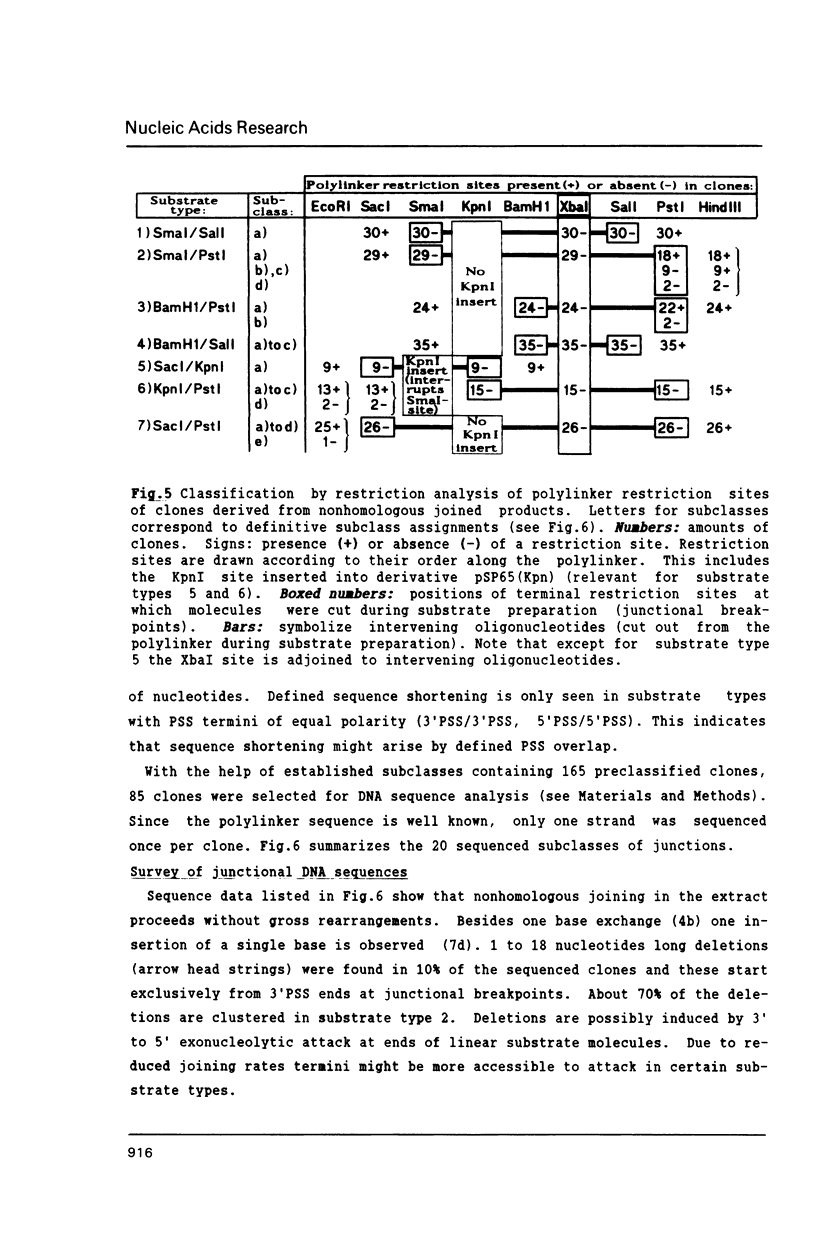
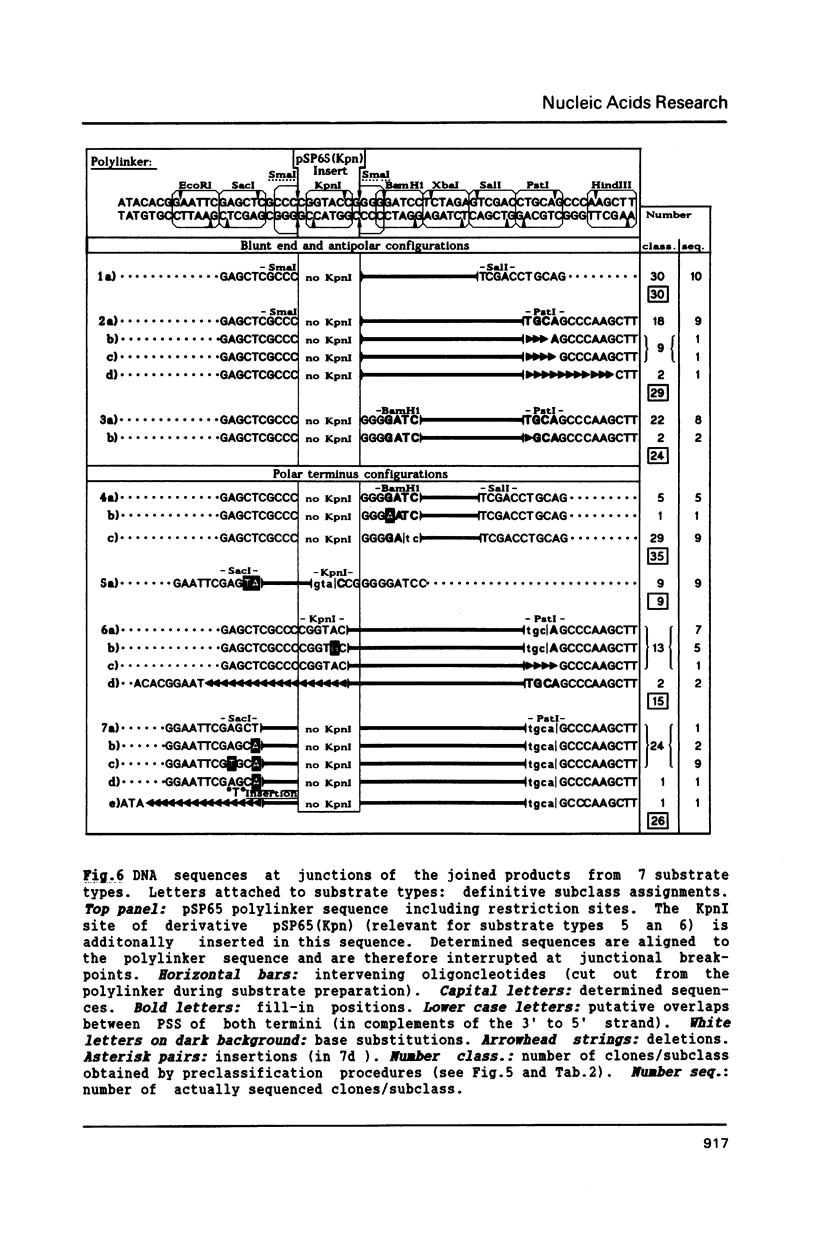
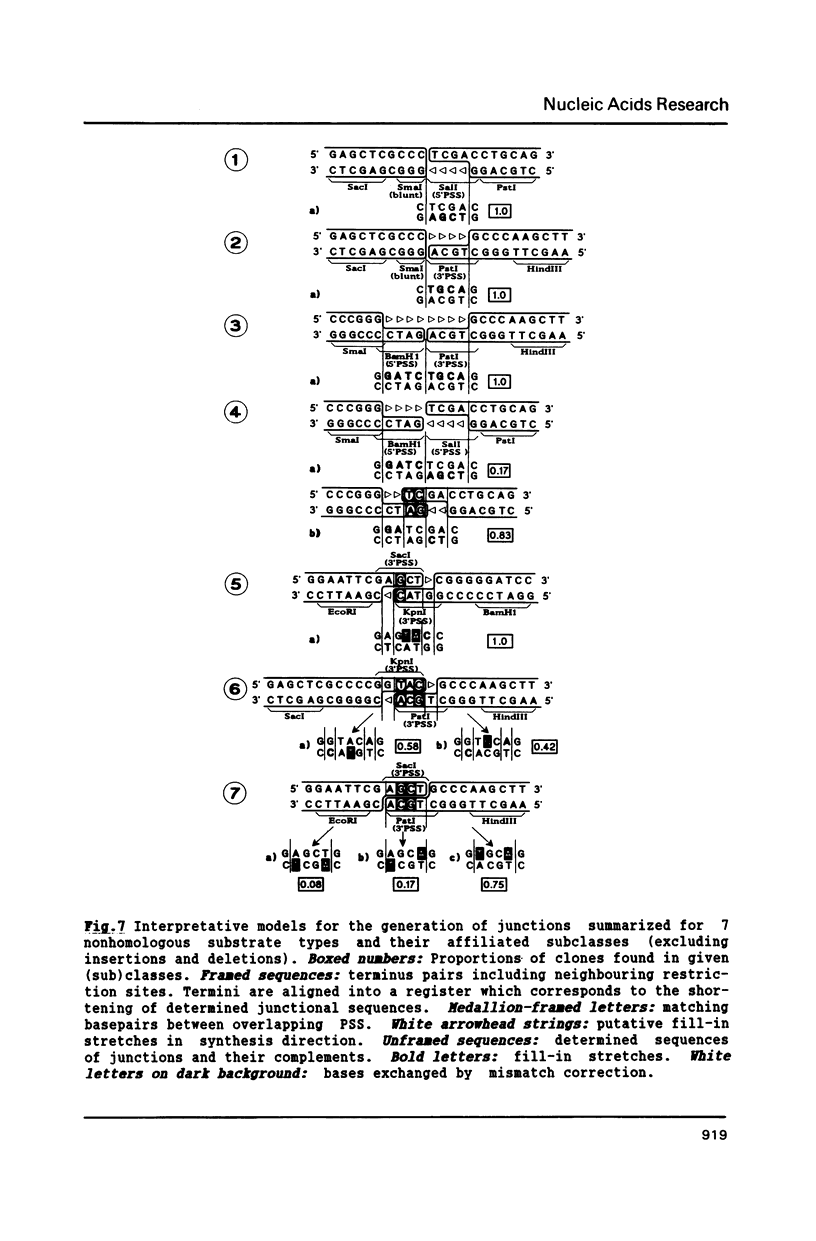
Extracts of Xenopus laevis eggs can efficiently join ends of duplex DNA that differ in structure and sequence. This was analysed by recircularisation of linear plasmid DNA molecules with dissimilar termini, generated by successive cuts with two different restriction enzymes within the pSP65 polylinker. Use of various enzymes provided blunt ended or 4 nucleotides long 3' and 5' protruding single strand (PSS) termini which were successfully joined in vitro in any tested combination. Sequence analysis of numerous junctions from cloned reaction products of 7 terminus combinations reveal: apart from very rare base exchanges and single nucleotide insertions less than 10% deletions (1 to 18 nucleotides long) were detected. Blunt/PSS or 3'PSS/5'PSS terminus pairs undergo simple "blunt end" joining which preserves PSS ends by fill-in. In contrast, equally polar 3'PSS/3'PSS or 5'PSS/5'PSS terminus pairs are joined by a complex mode: PSS ends overlap by a defined number of nucleotides, set by matching basepairs. Even one basematch suffices to define the setting. This then determines the final mismatch repair and fill-in pattern. We propose that yet unknown terminal DNA-binding proteins stabilize the energetically highly unfavorable configuration of single matching basepairs and help to support defined overlap structures.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arrand J. E., Willis A. E., Goldsmith I., Lindahl T. Different substrate specificities of the two DNA ligases of mammalian cells. J Biol Chem. 1986 Jul 15;261(20):9079–9082. [PubMed] [Google Scholar]
- Ashman C. R., Davidson R. L. High spontaneous mutation frequency in shuttle vector sequences recovered from mammalian cellular DNA. Mol Cell Biol. 1984 Nov;4(11):2266–2272. doi: 10.1128/mcb.4.11.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne M. L., Alexander R. F., Benbow R. M. DNA binding protein from ovaries of the frog, Xenopus laevis which promotes concatenation of linear DNA. J Mol Biol. 1984 Jan 5;172(1):87–108. doi: 10.1016/0022-2836(84)90416-9. [DOI] [PubMed] [Google Scholar]
- Benbow R. M., Krauss M. R. Recombinant DNA formation in a cell-free system from Xenopus laevis eggs. Cell. 1977 Sep;12(1):191–204. doi: 10.1016/0092-8674(77)90197-0. [DOI] [PubMed] [Google Scholar]
- Bendig M. M. Persistence and expression of histone genes injected into Xenopus eggs in early development. Nature. 1981 Jul 2;292(5818):65–67. doi: 10.1038/292065a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Carroll D., Wright S. H., Wolff R. K., Grzesiuk E., Maryon E. B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986 Jun;6(6):2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouse G. F., Frischauf A., Lehrach H. An integrated and simplified approach to cloning into plasmids and single-stranded phages. Methods Enzymol. 1983;101:78–89. doi: 10.1016/0076-6879(83)01006-x. [DOI] [PubMed] [Google Scholar]
- Dugaiczyk A., Boyer H. W., Goodman H. M. Ligation of EcoRI endonuclease-generated DNA fragments into linear and circular structures. J Mol Biol. 1975 Jul 25;96(1):171–184. doi: 10.1016/0022-2836(75)90189-8. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):59–69. doi: 10.1128/mcb.5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Mortimer R. K. A genetic study of x-ray sensitive mutants in yeast. Mutat Res. 1974 Sep;24(3):281–292. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- Gerondakis S., Cory S., Adams J. M. Translocation of the myc cellular oncogene to the immunoglobulin heavy chain locus in murine plasmacytomas is an imprecise reciprocal exchange. Cell. 1984 Apr;36(4):973–982. doi: 10.1016/0092-8674(84)90047-3. [DOI] [PubMed] [Google Scholar]
- Glikin G. C., Ruberti I., Worcel A. Chromatin assembly in Xenopus oocytes: in vitro studies. Cell. 1984 May;37(1):33–41. doi: 10.1016/0092-8674(84)90298-8. [DOI] [PubMed] [Google Scholar]
- Grzesiuk E., Carroll D. Recombination of DNAs in Xenopus oocytes based on short homologous overlaps. Nucleic Acids Res. 1987 Feb 11;15(3):971–985. doi: 10.1093/nar/15.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Barkan A., Mertz J. E. Nucleotide sequence analysis of the recombinant joints in 16 naturally arising deletion mutants of simian virus 40. Virology. 1982 Dec;123(2):464–469. doi: 10.1016/0042-6822(82)90281-1. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasin F., Hutchinson F. Repair of DNA double-strand breaks in Escherichia coli, which requires recA function and the presence of a duplicate genome. J Mol Biol. 1977 Oct 15;116(1):81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- Leach D., Lindsey J. In vivo loss of supercoiled DNA carrying a palindromic sequence. Mol Gen Genet. 1986 Aug;204(2):322–327. doi: 10.1007/BF00425517. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Mosbaugh D. W., Linn S. Gap-filling DNA synthesis by HeLa DNA polymerase alpha in an in vitro base excision DNA repair scheme. J Biol Chem. 1984 Aug 25;259(16):10247–10251. [PubMed] [Google Scholar]
- Newport J., Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982 Oct;30(3):675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Porter T. N., Wilson J. H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985 Oct;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi S., Schaffner W. Transformation of frog embryos with a rabbit beta-globin gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5051–5055. doi: 10.1073/pnas.78.8.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf K., Krämmer G., Dürst M., Suhai S., Röwekamp W. G. Human papillomavirus type 16 DNA sequence. Virology. 1985 Aug;145(1):181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Kobayashi I., Stahl M. M. In phage lambda, cos is a recombinator in the red pathway. J Mol Biol. 1985 Jan 20;181(2):199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- Subramani S., Berg P. Homologous and nonhomologous recombination in monkey cells. Mol Cell Biol. 1983 Jun;3(6):1040–1052. doi: 10.1128/mcb.3.6.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Plasmid recombination intermediates generated in a Saccharomyces cerevisiae cell-free recombination system. Mol Cell Biol. 1985 Sep;5(9):2361–2368. doi: 10.1128/mcb.5.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg R. A. Integrated genomes of animal viruses. Annu Rev Biochem. 1980;49:197–226. doi: 10.1146/annurev.bi.49.070180.001213. [DOI] [PubMed] [Google Scholar]
- Wiaderkiewicz R., Ruiz-Carrillo A. Mismatch and blunt to protruding-end joining by DNA ligases. Nucleic Acids Res. 1987 Oct 12;15(19):7831–7848. doi: 10.1093/nar/15.19.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]