Abstract
Human enterovirus species A (HEV-A) consists of at least 16 members of different serotypes that are known to be the causative agents of hand, foot, and mouth disease (HFMD), herpangina, and other diseases, such as respiratory disease and polio-like flaccid paralysis. Enterovirus 71 (EV71) and coxsackievirus A16 (CVA16) are the major causative agents of HFMD. CVA5, CVA6, CVA10, and CVA12 mainly cause herpangina or are occasionally involved with sporadic cases of HFMD. We have previously shown that human scavenger receptor class B, member 2 (SCARB2) is a cellular receptor for EV71 and CVA16. Using a large number of clinical isolates of HEV-A, we explored whether all clinical isolates of EV71 and other serotypes of HEV-A infected cells via SCARB2. We tested this possibility by infecting L-SCARB2 cells, which are L929 cells expressing human SCARB2, by infecting human RD cells that had been treated with small interfering RNAs for SCARB2 and by directly binding the viruses to a soluble SCARB2 protein. We showed that all 162 clinical isolates of EV71 propagated in L-SCARB2 cells, suggesting that SCARB2 is the critical receptor common to all EV71 strains. In addition, CVA7, CVA14, and CVA16, which are most closely related to each other, also utilized SCARB2 for infection. EV71, CVA14, and CVA16 are highly associated with HFMD, and EV71 and CVA7 are occasionally associated with neurological diseases, suggesting that SCARB2 plays important roles in the development of these diseases. In contrast, another group of viruses, such as CVA2, CVA3, CVA4, CVA5, CVA6, CVA8, CVA10, and CVA12, which are relatively distant from the EV71 group, is associated mainly with herpangina. None of these clinical isolates infected via the SCARB2-dependent pathway. HEV-A viruses can be divided into at least two groups depending on the use of SCARB2, and the receptor usage plays an important role in developing the specific diseases for each group.
INTRODUCTION
Human enteroviruses (HEVs) are one of the large families of human pathogens belonging to the Picornaviridae, and they can cause a variety of diseases, such as poliomyelitis, meningitis, acute flaccid paralysis, gastroenteritis, diarrhea, respiratory diseases, myocarditis, pancreatitis, hand, foot, and mouth disease (HFMD), and herpangina (40). HEVs are classified into species A (HEV-A) to species D (HEV-D) according to the similarity of the amino acid sequence of the capsid protein. HEV-A is composed of at least 16 members of different serotypes: coxsackievirus A2 (CVA2), CVA3, CVA4, CVA5, CVA6, CVA7, CVA8, CVA10, CVA12, CVA14, and CVA16 and enterovirus 71 (EV71), EV76, EV89, EV90, and EV91 (40). The prototype strain of EV71, strain BrCr, was isolated from a patient with neurological diseases between 1969 and 1972 (45). CVA7 prototype strain Parker was isolated in the United State in 1949 (10). CVA14 prototype strain G-14 and CVA16 prototype strain G-10 were isolated in the Republic of South Africa in 1950 and 1951, respectively (47). EV76 was isolated in France in 1991, and EV89, EV90, and EV91 were isolated in Bangladesh in 1999-2000 (35). Other prototype strains of HEV-A were isolated in the United States between 1947 and 1950 (10). These prototype strains were repeatedly subcultured using cultured cells or suckling mice. The members of HEV-A are increasing because of the isolation and characterization of new viruses (34, 36).
Members of HEV-A are known as causative agents of HFMD, herpangina, respiratory disease, meningitis, and polio-like flaccid paralysis (46). Phylogenetic analysis of HEV-A based on the capsid protein sequences revealed that these viruses were clustered into three major groups (35). Interestingly, there is an association between viral serotype and diseases. One of the groups consists of CVA7, CVA14, CVA16, and EV71 (called “the EV71 group” in this report). CVA16 and EV71 are the major causative agents of HFMD, which is characterized by fever and vesicular exanthema, mostly in the hands, feet, and oral mucosa (29). CVA7 and CVA14 infrequently cause sporadic cases of HFMD (1, 2, 41). Other HEV-As, such as CVA2, CVA4, CVA5, CVA6, and CVA10, which belong to another group (“the CVA2 group”), are mainly associated with herpangina in infants, which is also caused by coxsackie B viruses or echoviruses (9, 23, 28, 32, 50). It has been reported that CVA6 and CVA10 occasionally cause epidemic outbreaks of HFMD (6, 39). HFMD is usually mild, but neurological complications and even fatalities occur during HFMD outbreaks when the causative agent is EV71 (29). From 2008 to 2011, epidemic outbreaks of neurovirulent EV71 in China resulted in a total of approximately 3 million HFMD cases, including approximately 1,500 fatal cases (http://www.moh.gov.cn/publicfiles/business/htmlfiles/mohjbyfkzj/s2907/index.htm) (53). CVA7 is also neurotropic and can cause paralytic poliomyelitis (16, 17, 42). EV76, EV89, EV90, and EV91 were isolated from patients with gastroenteritis or polio-like acute flaccid paralysis and belong to the third group (“the EV76 group”) (35). However, these viruses are not well characterized because there have been few reports and limited numbers of clinical cases.
Virus receptors play important roles in developing disease characteristics for each virus because they are the primary determinant of cell, tissue, and species tropism. Because the ability of the virus to bind to its receptor and the subsequent processes that lead to infection are determined by interactions between the viral proteins and the receptor, we thought that receptor usage in HEV-As might be closely related to the clinical symptoms caused by the viruses. So far, two molecules have been proposed as the receptors for EV71 and CVA16 but not for other viruses belonging to HEV-A. We previously reported that scavenger receptor class B, member 2 (SCARB2, also known as lysosomal integral membrane protein II or CD36b like-2) permits efficient infection of mouse L929 cells by EV71 (52). SCARB2 belongs to the CD36 family and has two transmembrane domains (13). Physiologically, it plays a role in the reorganization of the endosomal/lysosomal compartment (21) and works as the receptor for the mannose-6-phosphate-independent transport of β-glucocerebrosidase (β-GC) to the lysosome (5, 43). The expression of SCARB2 has been observed in monolayer culture cells of primate origin, which are susceptible to EV71 infection (52), and in almost all organs in humans (13). The SCARB2 region important for EV71 binding was mapped to amino acids 142 to 204 using a series of chimeric receptors of human and mouse SCARB2 (51). We established a cell line, L-SCARB2, that expresses human SCARB2 constitutively (52). It is possible to determine the SCARB2 dependency for infection by testing the infectivity of the viruses on L-SCARB2 cells and parental L929 cells. We tested the infectivities of 8 laboratory strains of EV71 in the L-SCARB2 cells. Infection of all of these strains of EV71 in L-SCARB2 cells but not in L929 cells was as efficient as that in the RD cells (52). P-selectin glycoprotein ligand 1 (PSGL-1, also known as selectin P ligand [SELPLG]), was identified as an EV71 receptor from human T cell leukemia Jurkat cells using the panning assay, which detected molecules having a strong binding affinity for EV71 particles (33). PSGL-1 is expressed primarily on leukocytes and is involved in leukocyte interactions with vascular endothelium (22). L929 cells expressing PSGL-1 were susceptible only to some EV71 strains (PSGL-1 binding strains) (33). Viral propagation and cytopathic effect (CPE) occurred much more slowly than in RD cells (33). However, it is not known whether SCARB2 can serve as a receptor for all EV71 strains. We therefore examined whether all EV71 strains infect via a SCARB2-dependent pathway. In addition, we found that the prototype strain of CVA16 also utilized SCARB2 (52). This result led us to the idea that SCARB2 is widely used as the receptor for the members of HEV-A that cause the same clinical symptoms. It was not possible, however, to determine whether prototype strains of CVA7, CVA10, and CVA14 infect cells via a SCARB2-dependent pathway using the above strategy because these three viruses propagated well in the parental L929 cells (52).
In this work, we hypothesized that SCARB2 is used as a common receptor for “the EV71 group” of HEV-As that mainly cause HFMD, and we conducted experiments to show the following: (i) that all clinical isolates of EV71 use SCARB2 as a receptor and (ii) that EV71, CVA7, CVA14, and CVA16, which are most closely related to each other and are highly associated with HFMD, also use SCARB2. Because the prototype strains of HEV-As have undergone a number of passages in cultured cells or suckling mice, they might have accumulated mutations that affect virus-receptor interaction. To overcome this potential problem, we collected a number of clinical isolates of HEV-As. We used L-SCARB2 cells to investigate SCARB2 dependency for HEV-A infection. In addition, we adopted the additional methods of small interfering RNA (siRNA) techniques to downregulate SCARB2 expression in RD cells to examine the SCARB2 dependency of infection and coimmunoprecipitation to examine direct virus-receptor binding.
MATERIALS AND METHODS
Ethics statement.
All specimens were collected after the parents of the enrolled children had given oral or written informed consent. Demographic and clinical information was extracted from the patient record by the attending physician for each specimen. The anonymous samples and information were sent to Prefectural Institutes of Public Health and used for viral isolation. The use of isolated viral strains at Tokyo Metropolitan Institute of Medical Science was approved by the institutional committee for experiments of recombinant DNA and pathogens.
Cells.
Human RD cells were cultured in Dulbecco's modified Eagle medium (Sigma) supplemented with 5% fetal bovine serum (FBS) and a penicillin-streptomycin solution (Invitrogen) (5% FBS-DMEM). L-Empty cells (52) and L-SCARB2 cells (52) were cultured in 5% FBS-DMEM supplemented with puromycin (4 μg ml−1; Calbiochem).
Viruses.
EV71 and CVs, which belong to HEV-A, are listed in Tables S1 and S2 in the supplemental material, with year, place of isolation, clinical diagnosis or symptoms, subgenogroup, and reference, if known. These strains were isolated between 1985 and 2010 at the Shimane Prefectural Institute of Public Health and Environmental Science, Aichi Prefectural Institute of Public Health, Yamagata Prefectural Institute of Public Health (30, 31), or Virus Research Center, Sendai Medical Center in Japan. Viruses were isolated from original clinical specimens using a variety of cell lines. The isolates were typed by a neutralization assay with virus-specific antisera or by sequences of the capsid region. Prototype viruses of CVA7 (strain Parker) and CVA14 (strain G14) were propagated in RD cells. EV71-GFP, which expresses green fluorescent protein (GFP) upon viral replication, was recovered from an infectious cDNA clone, pSVA-EV71-GFP (51, 52).
Virus inoculation.
RD cells, L-Empty cells, and L-SCARB2 cells were infected with each virus listed in Tables S1 and S2 in the supplemental material. These cells were incubated at 37°C for a week to check for the appearance of a cytopathic effect (CPE).
Virus titration.
Viral titers were determined by the microtitration method using RD cells or L-SCARB2 cells and are expressed as the 50% tissue culture infectious dose (TCID50) according to the Reed-Muench method (44).
Sequence of EV71 VP1 for genotyping.
Viral RNA was extracted from the infected cell culture supernatant using the QIAamp viral RNA minikit (Qiagen). Reverse transcription-PCR (RT-PCR) was performed using the SuperScript III One-Step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen) as follows: after 30 min of cDNA synthesis at 55°C and 2 min of denaturation at 94°C, samples were subjected to 40 cycles of amplification, consisting of 30 s at 94°C, 30 s at 42°C, and 1 min at 72°C, with a final additional extension step at 72°C for 5 min. The PCR products were purified with a QIAquick PCR purification kit (Qiagen) or a QIAquick gel extraction kit (Qiagen) and then sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) on an ABI 3730xl sequencer (Applied Biosystems). For PCR and sequencing analysis, primers 159 (sense, 5′-ACYATGAAAYTGTGCAAGG-3′, nucleotides 2387 to 2405 of EV71/SK-EV006/Malaysia/97; accession no. AB469182) (7), 161 (sense, 5′-CTGGGACATAGAYATAACWGG-3′, nucleotides 2764 to 2784) (7), 162 (antisense, 5′-CCRGTAGGKGTRCACGCRAC-3′, nucleotides 2871 to 2852) (7), 16R-Y (antisense, 5′-GARAAACTGACTGGRTAGTG-3′, nucleotides 3561 to 3542), 159-190 (sense, 5′-AGCAACACTCACTACAGAGC-3′, nucleotides 2231 to 2250), and 162-2 (antisense, 5′-CCGGTGGGCGTRCATGCAAC-3′, nucleotides 2871 to 2852) were used. The subgenogroup was determined by a phylogenetic tree constructed by the neighbor-joining method using the GENETYX software program, version 9.0.1 (Genetyx).
Sequence of the P1 region.
Viral RNA was extracted from the infected cell culture supernatant using a QIAamp viral RNA minikit. The viral RNA was then transcribed into cDNA with ReverTra Ace (Toyobo) and a random primer. By using cDNA as a template, the P1 region was amplified by PCR using PfuUltra High Fidelity DNA polymerase (Agilent Technologies) or PfuUltra II Fusion HS DNA polymerase (Agilent Technologies). The PCR products were purified with a QIAquick PCR purification kit or a QIAquick gel extraction kit and then sequenced as described above. The sequence data were analyzed by GENETYX software. The primers used for PCR and sequencing analysis are listed in Table 1.
Table 1.
Oligonucleotide primers used for RT-PCR and sequencing
| Primer | Sequence (5′–3′)a | Application(s) |
|---|---|---|
| EV146-600(+) | TTACCATATAGCTATTGGATTGG | PCR/SEQb |
| EV146-2100(+) | ATGCTCATCGCCTACACCCCACC | PCR/SEQ |
| EV146-3400(-) | GCTGTCCTCCCATACAAGATTTGCCC | PCR/SEQ |
| EV146-4444(-) | GGTGTTTGCTCTTGAACTGC | PCR/SEQ |
| EV71-1050(+) | GGTTATGGTGAGTGGCCYTC | PCR/SEQ |
| EV71-1150(-) | CCCTTAGATGATTTYTCCCACA | PCR |
| EV71-1700(+) | TCTGAGTTTGCAGGTCTCAGRCAAGC | PCR/SEQ |
| EV71-1800(-) | GGGTGGRAGTTTGGTAGAATGGG | PCR |
| EV71-2100(+) | ATGCTCATAGCTTATACACCTCC | PCR |
| EV71-2200(-) | GTAATACCATGGATCAGCAACACTCACTAC | PCR |
| EV71-2700(+) | GGAGAGATAGAYCTCCCTCTTGARGG | PCR |
| EV71-2800(-) | CGCATGTAGGTGAACAGCTCCAC | PCR/SEQ |
Y, C or T; R, A or G.
SEQ, sequencing.
Phylogenetic tree based on P1 region of HEV-A.
The phylogenetic tree of prototype strains of HEV-A was constructed using the neighbor-joining method with the GENETYX software program. The amino acid sequences of the P1 region of CVA2 (strain Freetwood; accession no. AY421760), CVA3 (Olson; AY421761), CVA4 (High Point; AY421762), CVA5 (Swartz; AY421763), CVA6 (Gdula; AY421764), CVA7 (Parker; AY421765), CVA8 (Donovan; AY421766), CVA10 (Kowalik; AY421767), CVA12 (Texas12; AY421768), CVA14 (G-14; AY421769), CVA16 (G-10; U05876), EV71 (BrCr; U22521), EV76 (10226; AY697458), EV89 (10359; AY697459), EV90 (10399; AY697460), EV91 (10406; AY697461), PV (Mahony; V01149) and EV70 (J670/71; EV70CG) were obtained from GenBank.
Coimmunoprecipitation assay.
A coimmunoprecipitation assay was performed as described previously (51). Briefly, the indicated viruses were incubated with control Fc (Fc portion of human IgG) (3 μg; R&D systems) or human SCARB2-Fc (3 μg; R&D systems) and anti-human IgG (Fc specific)-agarose (Sigma) in 1 ml of 5% FBS-DMEM for 2 h at 4°C. The beads were then washed twice with 5% FBS-DMEM, suspended in SDS sample buffer, and incubated for 10 min at 95°C. After the beads were removed, the samples were loaded on 12% Mini-PROTEAN TGX precast gels (Bio-Rad), followed by Western blotting with the monoclonal antibody against VP2 (clone 422-8D-4C-4D; Millipore), a rabbit anti-CVA6 serum, a rabbit anti-CVA10 serum (27), or the anti-human IgG Fcγ fragment-specific antibody (Jackson ImmunoResearch).
RNAi.
RNA interference (RNAi) was performed using the following Accell siRNA sequences targeting the human SCARB2 coding region, GCAAUAUGAUUAAUGGAAC (#13), GUAUCGAGAAGAAAAUUGU (#14), CCCUUAUCCAUGUUUUCAG (#15), or UGGGUGTGUUCUUUGGUUU (#16) (Dharmacon), or using a control nontargeting siRNA (Dharmacon) according to the manufacturer's recommendations. Briefly, RD cells were treated with 1 μM siRNA in Accell siRNA delivery medium (Dharmacon) for 48 h. In some cases, RD cells were transfected with an empty vector or pCA-M(H4)-F (51) before siRNA treatment. These cells were harvested for Western blot analysis with a goat anti-SCARB2 antibody (R&D systems), a rabbit anti-FLAG antibody (Sigma), or a mouse anti-β-actin (ACTB) antibody (clone AC-74; Sigma) as an internal control or infected with the indicated viruses and incubated for another 24 h at 37°C. The cells infected with EV71-GFP were imaged with an IX70 microscope with a DP70 camera (Olympus) and analyzed using DP controller software (OLYMPUS). The cells infected with other viruses were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and strained with the monoclonal antibody against VP2, the rabbit anti-CVA6 serum, or the rabbit anti-CVA10 serum, followed by incubation with an Alexa Fluor 488 donkey anti-mouse IgG (Invitrogen) or an Alexa Fluor 488 donkey anti-rabbit IgG (Invitrogen). Images were acquired using the IX70 microscope. In some cases, viral titers in the cell culture were measured with RD cells.
RESULTS
All clinical isolates of EV71 induced CPE in L-SCARB2 cells.
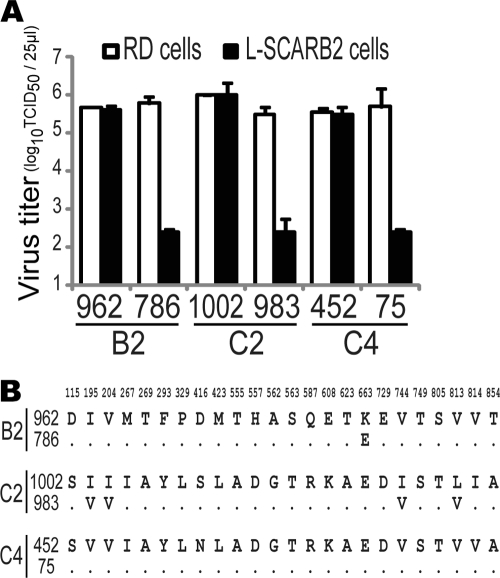
Eight laboratory strains of EV71, including the prototype strain BrCr, infected cells via a SCARB2-dependent pathway as described previously (52). To determine whether all EV71 isolates use SCARB2 for infection, we collected 162 clinical isolates of EV71 isolated between 1990 and 2010 in the Yamagata (30, 31), Miyagi, Aichi, and Shimane prefectures of Japan and identified their subgenogroups by their VP1 sequences (see Table S1 in the supplemental material). A total of 2, 18, 19, 12, 37 and 73 isolates were identified as subgenogroups B2, B4, B5, C1, C2, and C4, respectively. To confirm whether these isolates utilize SCARB2 for infection, each isolate was inoculated into L-SCARB2 cells, RD cells as a positive control, and L-Empty cells as a negative control. As a result of inoculation, all 162 isolates, irrespective of the subgenogroup, induced CPE in RD cells and L-SCARB2 cells but not in L-Empty cells (Table 2). However, a few isolates (approximately 10%) showed a low propagation efficiency in L-SCARB2 cells. To exclude the possibility that these viruses failed to use SCARB2 properly, we selected three strains as representative isolates from the isolates with a low growth phenotype (786-Yamagata [subgenogroup B4], 983-Yamagata [C2], and 75-Yamagata [C4]) and from the isolates with a high-growth phenotype (962-Yamagata [B4], 1002-Yamagata [C2], and 452-Yamagata [C4]). We compared the viral propagation in L-SCARB2 cells with that in RD cells by determining the viral titers of these 6 isolates using the microtitration method (Fig. 1A). The viral titers of the strains 962-Yamagata, 1002-Yamagata, and 452-Yamagata in L-SCARB2 cells were similar to those in RD cells, whereas the viral titers of the strains 786-Yamagata, 983-Yamagata, and 75-Yamagata in L-SCARB2 cells were approximately 1,000-fold lower than those in RD cells. We found a very small number of amino acid substitutions in the capsid region of these three viruses with the low-growth phenotype compared with those of the high-growth phenotype (Fig. 1B). Notably, there were no differences in the capsid region between strains 452-Yamagata and 75-Yamagata. We speculated that isolates of both growth phenotypes utilized human SCARB2 with similar efficiencies and that poor replication efficiency in the L-SCARB2 cells was due to the incompatibility with some host factors in mouse cells.
Table 2.
Induction of CPE by EV71
| Subgenogroup | No. of isolates | No. of isolates associated with CPE |
||
|---|---|---|---|---|
| RD cells | L-Empty cells | L-SCARB2 cells | ||
| B2 | 2 | 2 | 0 | 2 |
| B4 | 18 | 18 | 0 | 18 |
| B5 | 19 | 19 | 0 | 19 |
| C1 | 12 | 12 | 0 | 12 |
| C2 | 38 | 38 | 0 | 38 |
| C4 | 73 | 73 | 0 | 73 |
| Total | 162 | 162 | 0 | 162 |
Fig 1.
Comparisons of EV71 clinical isolates. (A) Viral titers of EV71 clinical isolates in RD cells and L-SCARB2 cells. Viral titers of clinical isolates of the EV71 strains 962-Yamagata (962), 786-Yamagata (786), 1002-Yamagata (1002), 983-Yamagata (983), 452-Yamagata (452), and 75-Yamagata (75) were determined with RD cells and L-SCARB2 cells. The data are shown as mean viral titers with SD (n = 3). (B) Comparison of amino acid sequences of the P1 region. Amino acid sequences of EV71 strains 962-Yamagata (subgenogroup B4), 1002-Yamagata (C2), and 452-Yamagata (C4) (upper lines) were compared with those of strains 786-Yamagata (B4), 983-Yamagata (C2), and 75-Yamagata (C4) (lower lines). Only amino acids divergent among the 6 isolates are shown.
Evaluation of SCARB2-dependent infection of EV71 in RD cells.
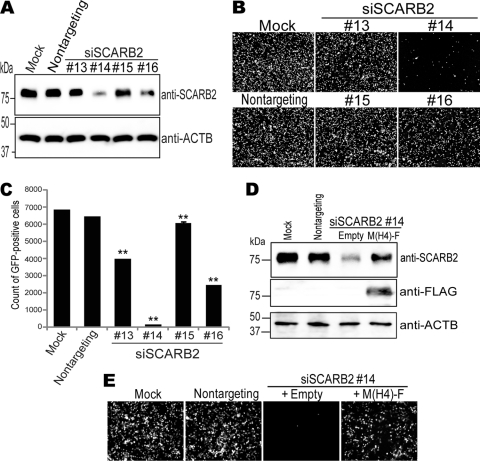
To confirm that these isolates were able to infect to human RD cells using SCARB2, we employed RNA interference (RNAi) technology to downregulate SCARB2 expression. First, using a recombinant EV71 strain, EV71-GFP, which was already characterized as a SCARB2-dependent virus (51, 52), we conducted an experiment to determine which of 4 kinds of small interfering RNAs (siRNAs) were able to downregulate SCARB2 expression and whether the downregulation of SCARB2 really inhibited EV71 infection. RD cells were treated with nontargeting siRNA or siRNAs against human SCARB2 (siSCARB2 #13, #14, #15, or #16) for 48 h, and the SCARB2 expression of these cells was evaluated by Western blotting (Fig. 2A). SCARB2 expression in RD cells treated with siSCARB2 #14 or #16 was appreciably or intermediately downregulated compared with that in mock-treated cells, evaluated by Western blotting. The RD cells were then infected with EV71-GFP, which expressed GFP upon infection, and imaged at 24 h postinfection (Fig. 2B). Compared with the mock treatment, the numbers of GFP-positive cells in the cell culture that had been treated with siSCARB2 #14 or #16 were greatly or slightly reduced, respectively, whereas those in the cell culture treated with nontargeting siRNA or siSCARB2 #13 or #15 were not affected. To quantify the microscopic observations, these cells (a total of 10,000 cells) were analyzed by fluorescence-activated cell sorting (FACS) to count the number of GFP-positive cells (Fig. 2C). There was no remarkable difference in the number of GFP-positive cells between the mock-treated cells (6,841 ± 17) and the cells treated with nontargeting siRNA (6,449 ± 26). The numbers of GFP-positive cells in siSCARB2 #13-, #14-, #15-, and #16-treated cells were 3,970 ± 15, 171 ± 7.0, 6,090 ± 68, and 2,453 ± 7.0, respectively (Fig. 2C). These results showed that siSCARB2 #14 and #16 were effective both for the downregulation of SCARB2 expression and for the inhibition of EV71-GFP infection. The results also indicated that the efficiency of EV71-GFP infection correlated with the expression level of SCARB2. We confirmed that these siSCARB2 siRNAs did not affect poliovirus infection of RD cells (data not shown).
Fig 2.
Downregulation of SCARB2 expression inhibited EV71-GFP infection. (A) Cell lysates were prepared from RD cells mock treated or treated with nontargeting siRNA or siSCARB2 #13, #14, #15, or #16 and were analyzed by Western blotting using an anti-SCARB2 antibody or an anti-ACTB (β-actin) antibody. (B and C) The siRNA-treated RD cells were infected with EV71-GFP and imaged via fluorescence microscopy at 24 h postinfection (B) and then analyzed by FACS to quantify the number of GFP-positive cells (C). A total of 10,000 cells were analyzed by FACS, and data are shown as mean counts with SD (n = 3). (D) SCARB2 expression was restored by exogenous M(H4)-F expression. Cell lysates were prepared from RD cells mock treated or treated with nontargeting siRNA or siSCARB2 #14, which were transfected with either empty vector or pCA-M(H4)-F and analyzed by Western blotting using an anti-SCARB2 antibody, an anti-FLAG antibody, or an anti-ACTB antibody. (E) M(H4)-F expression rescued the EV71-GFP infection that was inhibited by treatment with siSCARB2 #14. The siRNA-treated RD cells with/without plasmid transfection were infected with EV71-GFP. After 24 h, the cells were imaged via fluorescence microscopy.
To confirm the specificity of the inhibition of EV71 infection that was putatively achieved by the repression of SCARB2 expression, RD cells were transfected with either the empty plasmid or a plasmid encoding a human-mouse chimeric SCARB2, M(H4)-F (51), before treatment with siSCARB2 #14. M(H4)-F is a chimeric mouse Scarb2 wherein amino acids 142 to 204, the EV71 binding site, have been replaced with the corresponding sequence of human SCARB2. The chimera functions as an EV71 receptor to virtually the same extent as human SCARB2 and was able to escape downregulation by siSCARB2 #14 because siSCARB2 #14 did not match the mouse Scarb2 sequence perfectly. Endogenous SCARB2 expression repressed by siSCARB2 #14 treatment was restored by the exogenous expression of M(H4)-F but not by transfection with the empty plasmid (Fig. 2D). Similarly, the number of GFP-positive cells, reduced by treatment with siSCARB2 #14, was markedly restored by M(H4)-F expression (Fig. 2E). These results indicate that it is possible to evaluate SCARB2-dependent infection of RD cells using RNAi with siSCARB2 #14 and #16.
SCARB2-dependent infection of EV71 clinical isolates in RD cells.
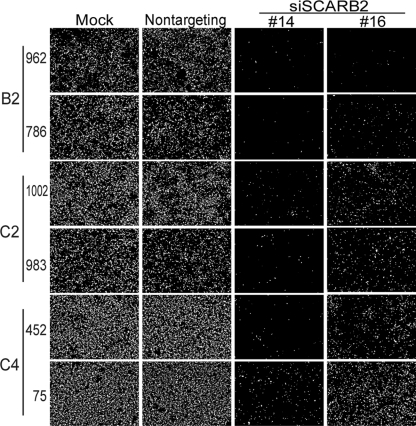
We tested the SCARB2-dependent infection of representative clinical isolates of EV71 with the siRNAs. RD cells treated with siSCARB2 #14 or #16 were infected with the 3 clinical isolates of EV71 shown in Fig. 1 that had a high- or low-growth phenotype in L-SCARB2 cells, fixed at 24 h postinfection, and then stained with an anti-VP2 monoclonal antibody to detect the EV71-infected cells (Fig. 3). Upon infection with 3 isolates of the high-growth phenotype in L-SCARB2 cells, VP2-positive cells in RD cell culture treated with siSCARB2 #14 or #16 were markedly reduced compared with those in mock-treated or nontargeting siRNA-treated cell culture. Similarly, when 3 isolates with the low-growth phenotype in L-SCARB2 cells were used, the number of VP2-positive cells in the RD cell culture treated with siSCARB2 #14 or #16 was reduced. The reduced infection of each clinical isolate with the high- or low-growth phenotype was restored by exogenous M(H4)-F expression (data not shown). These results show that at least 3 EV71 isolates that exhibited the low-growth phenotype in L-SCARB2 cells infect RD cells via the SCARB2-dependent pathway. The results suggest that all clinical isolates of EV71 are able to infect RD cells via a SCARB2-dependent pathway.
Fig 3.
SCARB2-dependent infection of RD cells by EV71. siRNA-treated RD cells were infected with EV71 strains 962-Yamagata, 786-Yamagata, 1002-Yamagata, 983-Yamagata, 452-Yamagata, and 75-Yamagata. After 24 h, the cells were fixed and stained with monoclonal antibody against VP2.
Binding of clinical isolates of EV71 to SCARB2.
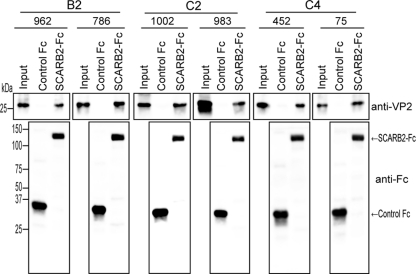
We then directly examined the binding of the 3 clinical isolates of EV71 with the high- or low-growth phenotype in L-SCARB2 cells to a soluble SCARB2 protein by using a coimmunoprecipitation assay. Briefly, each clinical isolate of EV71 used for Fig. 1 was mixed with 3 μg of control Fc or 3 μg of SCARB2-Fc together with anti-Fc-Agarose beads, and precipitated proteins were analyzed by Western blotting using the anti-VP2 or an anti-Fc antibody (Fig. 4). Similar amounts of control Fc and SCARB2-Fc were precipitated by anti-Fc-agarose beads (Fig. 4, lower panels). As expected, similar amounts of VP2 from each clinical isolate were detected in the SCARB2-Fc lane, and VP2 was not detected in the control Fc lane (Fig. 4, upper panels). These results indicate that all 6 clinical isolates of both growth phenotypes in L-SCARB2 cells bind to SCARB2 with similar efficiencies. Together, the data suggested that all clinical isolates of EV71 tested in this study are able to use SCARB2 as a receptor.
Fig 4.
Binding of clinical isolates of EV71 to SCARB2-Fc. The EV71 strains 962-Yamagata, 786-Yamagata, 1002-Yamagata, 983-Yamagata, 452-Yamagata, and 75-Yamagata were incubated with control Fc (3 μg) or SCARB2-Fc (3 μg) bound to the anti-human Fc-agarose. Precipitated proteins were analyzed by Western blotting with monoclonal antibody against VP2 and anti-Fc antibody.
Correlation between viral serotypes and clinical symptoms.
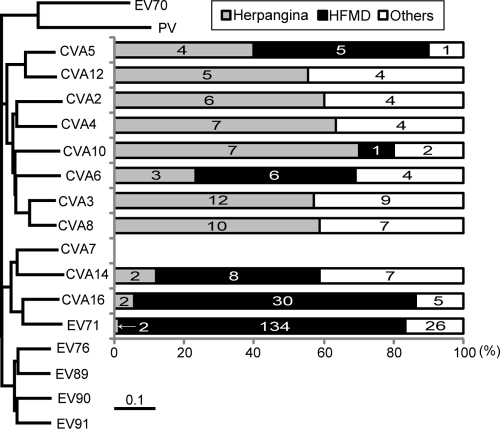
Next, to determine whether other members of HEV-As use SCARB2 as a receptor, we collected 9 to 22 clinical isolates of CVA2, CVA3, CVA4, CVA5, CVA6, CVA8, CVA10, CVA12, CVA14, and 37 clinical isolates of CVA16 isolated between 1985 and 2010 in the Yamagata, Aichi, and Shimane prefectures of Japan (see Table S2 in the supplemental material). These viruses were isolated from patients with diseases, including HFMD, herpangina, acute upper respiratory infection, pharyngitis, and tonsillitis. We were unable to obtain any clinical isolates of CVA7, EV76, EV89, EV90, and EV91. The diseases caused by the clinical isolates belonging to each serotype are listed in Table S2 in the supplemental material and summarized in Fig. 5 together with the phylogenetic tree of prototype strains of HEV-A based on the amino acid sequences of capsid proteins (37). The viruses were classified into three clusters in the phylogenetic tree: the EV71 group included CVA7, CVA14, CVA16, and EV71; the CVA2 group included CVA5, CVA12, CVA2, CVA4, CVA10, CVA6, CVA3, and CVA8; and the EV76 group included EV76, EV89, EV90, and EV91. In general, most enterovirus infections are asymptomatic, although the same receptors are presumably used in asymptomatic and symptomatic cases. In addition, many enteroviruses use different receptors but cause somewhat similar symptoms. However, in this case, we found an association between viral serotypes and clinical symptoms. The viruses in the EV71 group were highly associated with HFMD. They were isolated from patients with HFMD (79.3%) or herpangina (2.8%). In contrast, there is a tendency for the viruses in the CVA2 group to be associated with herpangina. More than 50% of CVA12, CVA2, CVA4, CVA3, and CVA8 isolates were obtained from patients with herpangina, and none were obtained from HFMD patients. CVA5, CVA6, and CVA10 were isolated mainly from herpangina patients, but some were from HFMD patients; CVA5 was isolated from patients with herpangina (4/10), HFMD (5/10), and other diseases (1/10); CVA6 was isolated from patients with herpangina (3/13), HFMD (6/13), and other diseases (4/13); and CVA10 was isolated from patients with herpangina (7/10), HFMD (1/10), and other diseases (2/10). The viruses in the CVA2 group were isolated from patients with herpangina (53.5%) and HFMD (11.9%). The relationships between the serotypes and the clinical symptoms or disease observed in our collected samples were similar to what has been previously reported elsewhere. Because we could not obtain any viruses belonging to the EV76 group, we focused on the EV71 group and the CVA2 group.
Fig 5.
Association of viral serotypes and clinical symptoms. The phylogenetic tree was drawn based on the amino acid sequences of the capsid regions of the HEV-A prototype strains. PV and EV70 are defined as outgroups. The numbers of clinical isolates from patients with indicated diseases are based on data in Table S2 in the supplemental material.
All clinical isolates of CVA14 and CVA16 propagate in L-SCARB2 cells.
To confirm whether each isolate of the 156 HEV-A viruses listed in Table S2 in the supplemental material utilizes SCARB2 for infection, isolates were inoculated into L-SCARB2 cells, RD cells, and L-Empty cells. As a result of the inoculations, all 17 and 37 isolates of CVA14 and CVA16, respectively, induced CPE in both RD cells and L-SCARB2 cells but not in L-Empty cells (Table 3). All clinical isolates of CVA2, CVA3, CVA4, CVA5, CVA6, CVA8, CVA10, and CVA12 produced CPE in RD cells but not in L-SCARB2 cells and L-Empty cells (Table 3). These results clearly indicated that all tested clinical isolates of CVA14 and CVA16 showed SCARB2-dependent infection, whereas the clinical isolates of CVA2, CVA3, CVA4, CVA5, CVA6, CVA8, CVA10, and CVA12 did not. It is likely that HEV-A can be clustered at least into two groups depending on the use of SCARB2.
Table 3.
Induction of CPE by coxsackieviruses
| Virus | No. of isolates | No. of isolates associated with CPE |
||
|---|---|---|---|---|
| RD cells | L-Empty cells | L-SCARB2 cells | ||
| CVA2 | 10 | 10 | 0 | 0 |
| CVA3 | 22 | 22 | 0 | 0 |
| CVA4 | 11 | 11 | 0 | 0 |
| CVA5 | 10 | 10 | 0 | 0 |
| CVA6 | 13 | 13 | 0 | 0 |
| CVA8 | 17 | 17 | 0 | 0 |
| CVA10 | 10 | 10 | 0 | 0 |
| CVA12 | 9 | 9 | 0 | 0 |
| CVA14 | 17 | 17 | 0 | 17 |
| CVA16 | 37 | 37 | 0 | 37 |
SCARB2-dependent infection of CVA7, CVA14, and CVA16 in RD cells.
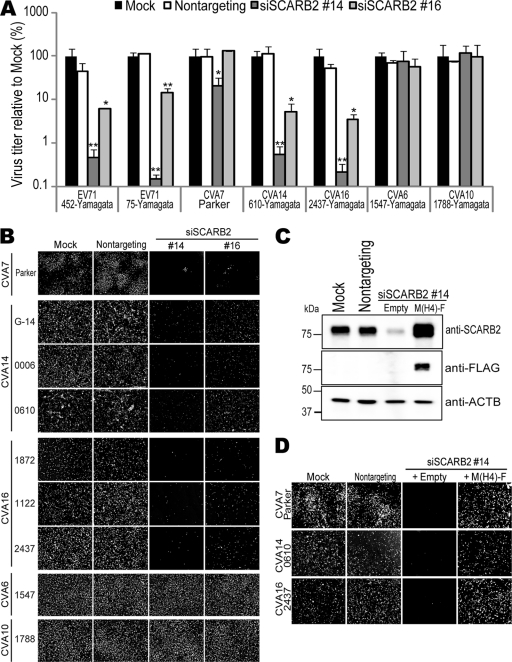
All clinical isolates of CVA14, CVA16, and EV71 showed SCARB2-dependent infection (Table 3). The prototype strains of CVA7, CVA14, CVA16, and EV71 are monophyletic based on the analysis of the amino acid sequences of capsid proteins (Fig. 5). We therefore hypothesized that CVA7 also utilizes SCARB2 as a receptor for infection and added the CVA7 prototype strain Parker to further analyses. We then evaluated the dependency on SCARB2 for infection of RD cells using siRNA techniques. RD cells treated with siSCARB2 #14 or #16 were infected with EV71 strains 452-Yamagata and 75-Yamagata as positive controls and CVA16 strain 2437-Yamagata, CVA14 strain 0006-Yamagata, CVA7 strain Parker, CVA6 strain 1547-Yamagata, or CVA10 strain 1788-Yamagata as a negative control at a multiplicity of infection (MOI) of 0.1 (Fig. 6A). Consistent with the results in Table 3, the viral titers of EV71s, CVA16, and CVA14 in siSCARB2 #14- or #16-treated cells were approximately 10- or 100-fold lower than those in mock or nontargeting siRNA-treated cells. The viral titer of CVA7 in RD cells treated with siSCARB2 #14 was significantly decreased, but the decrease was not evident in siSCARB2 #16-treated cells. siSCARB2 #14 and #16 did not affect the propagation of CVA6 and CVA10 in RD cells. These results showed that CVA14 and CVA16 utilize SCARB2 for infection, whereas CVA6 and CVA10 do not. The SCARB2 dependency of CVA7 was not apparent under this condition.
Fig 6.
SCARB2-dependent infection of CVA7, CVA14, and CVA16. (A) Infection of CVA16 and CVA14 was inhibited by repression of SCARB2 expression. siRNA-treated RD cells were infected with the indicated viruses, and the viral titers were determined at 24 h postinfection. The data are shown as mean relative viral titers with SD (n = 2). Statistical significance was determined by Student's t test. (*, P < 0.05; **, P < 0.01) (B) Infection by CVA7, CVA14, and CVA16 was inhibited by downregulation of SCARB2. The siRNA-treated RD cells were infected with the CVA7 strain Parker, the CVA14 strains G-14, 0006-Yamagata, and 0610-Yamagata, the CVA16 strains 1872-Yamagata, 1122-Yamagata, and 2437-Yamagata, the CVA6 strain 1547-Yamagata, or the CVA10 strain 1788-Yamagata. After 24 h, the cells were fixed and stained with monoclonal antibody against VP2, an anti-CVA6 serum, or an anti-CVA10 serum. (C) Rescue of SCARB2 expression by exogenous M(H4)-F expression. Cell lysates were prepared from RD cells mock treated or treated with nontargeting siRNA or siSCARB2 #14, which were transfected with either empty vector or pCA-M(H4)-F and analyzed by Western blotting using anti-SCARB2 antibody, anti-FLAG antibody, or the -ACTB antibody. (D) M(H4)-F expression rescued the inhibition of CVA7, CVA14, and CVA16 infection. siRNA-treated RD cells with/without plasmid transfection were infected with CVA7 strain Parker, CVA14 strain 0610-Yamagata, and CVA16 strain 2437-Yamagata. After 24 h, the cells were fixed and stained with monoclonal antibody against VP2.
For further analysis of SCARB2 dependency for infection, we infected RD cells pretreated with siSCARB2s with the CVA7 strain Parker, the CVA14 strains G-14, 0006-Yamagata, and 0610-Yamagata, the CVA16 strains 1872-Yamagata, 1122-Yamagata, and 2437-Yamagata, the CVA6 strain 1547-Yamagata, and the CVA10 strain 1788-Yamagata and identified the infected cells with anti-VP2 antibody, anti-CVA6 antibody, or anti-CVA10 antibody (Fig. 6B). In CVA7-infected cells, the number of VP2-positive cells in RD cells treated with siSCARB2 #14 or #16 was obviously reduced compared with those of mock- or nontargeting siRNA-treated cells. The SCARB2 dependency of CVA7 infection was evident under this condition. The inhibition of viral infection by siSCARB2 treatment was similarly observed for each of the 3 isolates of CVA14 and CVA16, whereas infection by CVA6 and CVA10 was not affected by siSCARB2 treatment.
To evaluate the specificity of siSCARB2, we transfected RD cells with pCA-M(H4)-F or the empty plasmid before treatment with siSCARB2 #14. The repression of endogenous SCARB2 expression by siSCARB2 #14 treatment was restored by exogenous expression of M(H4)-F but not by transfection of the empty plasmid (Fig. 6C). After infection with CVA7 strain Parker, CVA14 strain 0610-Yamagata, or CVA16 strain 2437-Yamagata, a number of VP2-positive cells were observed (Fig. 6D). We found that the number of VP2-positive cells in the cells transfected with pCA-M(H4)-F was increased compared with that of the cells transfected with empty plasmid. These data showed that the reduction of the number of VP2-positive cells by siSCARB2 treatment is specifically caused by downregulation of SCARB2 expression, indicating that CVA7, CVA14, and CVA16 utilize SCARB2 for infection of RD cells.
Binding of CVA7, CVA14, and CVA16 to SCARB2.
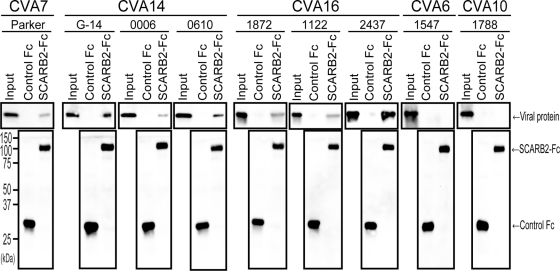
To confirm that CVA7, CVA14, and CVA16 bound directly to SCARB2, we conducted the coimmunoprecipitation assays shown in Fig. 4 using the isolates of CVA7, CVA14, CVA16, CVA6, and CVA10 used in Fig. 6B (Fig. 7). Similar amounts of control Fc and SCARB2-Fc were precipitated by anti-Fc-agarose beads (Fig. 7, lower panels). VP2 from CVA7, CVA14, or CVA16 was detected in each SCARB2-Fc lane (Fig. 7, upper panels), although the amounts of precipitated VP2 varied among the viruses. Viral protein of CVA6 or CVA10 was not detected in either the control Fc or SCARB2-Fc lanes. These results indicate that CVA7, CVA14, and CVA16 bind to SCARB2 specifically but that the binding efficiencies may not be equal among the virus strains.
Fig 7.
Binding of CVA7, CVA14, and CVA16 to SCARB2-Fc. CVA7 strain Parker, CVA14 strains G-14, 0006-Yamagata, and 0610-Yamagata, CVA16 strains 1872-Yamagata, 1122-Yamagata, and 2437-Yamagata, CVA6 strain 1547-Yamagata, and CVA10 strain 1788-Yamagata were incubated with control Fc (3 μg) or SCARB2-Fc (3 μg) bound to anti-human Fc-agarose. Bound viruses were analyzed by Western blotting with monoclonal antibody against VP2, anti-CVA6 serum, or anti-CVA10 serum.
DISCUSSION
In this study, we examined the relationship between the use of SCARB2 as the receptor and differences in diseases caused by the viruses. First, we investigated whether all EV71 clinical isolates used SCARB2 as a receptor. EV71 was genetically classified into 3 genogroups based on the VP1 nucleotide sequence: genogroups A, B, and C (7). Genogroup A contains a single member, prototype stain BrCr, and genogroups B and C are further subdivided into clusters: subgenogroups B1, B2, B3, B4, B5, C1, C2, C3, C4, and C5 (7). We collected a total of 162 clinical isolates of EV71 classified into subgenogroups B2, B4, B5, C1, C2, and C4. Viruses belonging to B1, B3, C3, and C5 were not included in this collection. We have shown that all 162 isolates of EV71 infected cells via a SCARB2-dependent pathway by testing the infectivity in L-SCARB2 cells. A few isolates (approximately 10%) propagated in L-SCARB2 cells less efficiently than other isolates. However, additional experiments of SCARB2 depletion in RD cells and a coimmunoprecipitation assay clearly showed that these isolates infected via a SCARB2-dependent pathway. These isolates with a low-growth phenotype in L-SCARB2 cells may have some incompatibility in the interaction of viral proteins/genome with the host factors (24–26, 48, 49) in mouse cells but not in the interaction with SCARB2. We have previously reported that EV71 strains BrCr/USA/70 (genogroup A), Nagoya/Japan/73 (subgenogroup B1), 258/Bulgaria/75 (subgenogroup B1), Hungary/78 (subgenogroup B1), and SK-EV006/Malaysia/97 (subgenogroup B3) infect cells via a SCARB2-dependent pathway (52). Although we have not tested the viruses belonging to subgenogroups C3 and C5, the data strongly suggested that EV71 universally utilizes SCARB2 as the receptor. On the contrary, PSGL-1 mediates infection of a subset of EV71 strains and is expressed primarily on leukocytes (22, 33). It is therefore unlikely that PSGL-1 is the receptor that plays a critical role in causing HFMD, but it may contribute to modulating EV71 pathogenicity for PSGL-1 binding strains under some circumstances. SCARB2 is widely expressed in vivo (13) and could be directly involved in systemic infection. Taken together, these results suggested that SCARB2 is the most probable candidate for the primary receptor and that it plays a critical role in EV71 infections.
Second, we also investigated whether other members of HEV-A used SCARB2 as a receptor. The results indicated that HEV-A viruses are divided into at least two groups: viruses whose infection is dependent on SCARB2 and viruses not dependent on SCARB2. The phylogenetic tree based on the amino acid sequences of capsid proteins revealed that HEV-A viruses diverged into three clusters, as described previously (37). We have explored the receptor usage of two of three groups. We showed that viruses in the EV71 group infect cells via a SCARB2-dependent pathway and viruses in the CVA2 group infect cells via a SCARB2-independent pathway. Each cluster roughly correlated with clinical outcomes; viruses in the EV71 group were mainly isolated from patients with HFMD, whereas viruses in the CVA2 group were generally isolated from patients with herpangina. Thus, the SCARB2-dependent viruses tend to cause HFMD, and the SCARB2-independent viruses tend to cause herpangina. There are some exceptions, however: CVA5, CVA6, and CVA10 have occasionally been isolated from patients with HFMD (3, 11, 14, 20, 38, 39). An outbreak of HFMD caused by CVA6 occurred in Japan in 2011. During this outbreak, clinicians reported that HFMD caused by CVA6 was different from typical HFMD; the exanthema caused by CVA6 was larger than the typical one and appeared in the thigh and abdomen as well as in hands and feet. It was also reported that onychomadesis was a characteristic feature in patients during the HFMD outbreak caused by CVA6 in Finland in 2008; parents and clinicians reported that their children shed fingernails and/or toenails within 1 to 2 months after HFMD (39). Although the clinical symptoms of HFMD caused by CVA5, CVA6, or CVA10 look similar to those caused by CVA16 and EV71, the molecular basis of the disease might be different from that for those caused by SCARB2-dependent viruses.
Among HEV-As, the viruses in the EV71 group infect cells via a SCARB2-dependent pathway, but the viruses in the CVA2 group do not. Among HEV-Bs, CVB1-6 uses coxsackie-adenovirus receptor (CAR), while other members of HEV-B do not (4). Similarly, only polioviruses 1 to 3 use CD155 as the receptor among the HEV-Cs (4). In contrast, major group human rhinoviruses (HRVs) that use intercellular cell adhesion molecule 1 (ICAM-1) as the receptor are present in both HRV-A and HRV-B, and minor group HRVs are all in HRV-A (4, 15). The monophyletic use of SCARB2 by CVA7, CVA14, CVA16, and EV71 is similar to the use of CAR by CVB1-6 and the use of CD155 by polioviruses 1 to 3.
CVA7, CVA14, CVA16, and EV71 share the same receptor and are frequently associated with HFMD. In addition, neurological disease caused by CVA7 and EV71 has been reported (16–18, 29, 42, 53). These data suggest that this group of viruses, which infect via a SCARB2-dependent pathway, is capable of invading the central nervous system (CNS) using this receptor. Supporting this idea, our preliminary experiments showed that SCARB2 is expressed in neurons in the CNS in humans, monkeys, and transgenic mice expressing human SCARB2 and that the adult transgenic mice showed encephalitis after infection with EV71. Although these viruses are able to utilize SCARB2 as a receptor, the occurrence of severe neurological disease caused by EV71 was quite low before recent outbreaks in the eastern Asian countries, and severe neurological diseases associated with CVA16 have not been reported. It is possible that the EV71 strains currently circulating in countries that suffer from severe EV71 outbreaks obtained an unidentified neurovirulence determinant(s) in the viral genome and became more neurovirulent than those that circulated previously. It is also possible that CVA16 has the potential to cause neurological disease, because it has been reported, in the case of poliovirus, that an increase in neurovirulence levels can be caused by point mutations or by genetic recombination between avirulent poliovirus vaccine strains and nonpolio enteroviruses (49, 50, 51, 52, 53, 54). Because CVA7, CVA14, CVA16, and EV71 utilize the same receptor and because SCARB2-dependent viruses sometimes cocirculate during an epidemic of HFMD (1, 12), these viruses might have a high potential to undergo an intertypic recombination by coinfection of a SCARB2-expressing cell in vivo. Indeed, it has been reported that intertypic recombination occurred between EV71 and CVA16 (8, 19, 54–56). Fortunately, neither severe neurological diseases caused by the intertypic recombinants between EV71 and CVA16 nor recombinant viruses of EV71 and CVA7 or CVA14 have been reported. These viruses might appear as an emerging infectious pathogen, however, and may have unexpectedly high virulence. Careful and continuous surveillance of this group of viruses is important for public health.
In summary, SCARB2 is used as a receptor for certain members of the HEV-A viruses and may play important roles in the pathogenesis of HFMD and neurological diseases. We are now investigating the precise interaction between EV71 and SCARB2 and generating a mouse model expressing human SCARB2. These further studies will help to elucidate the molecular basis of HFMD and the neurological disease caused by these viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Shimizu and Y. Nishimura (National Institute of Infectious Diseases of Japan) for providing antiserum and helpful discussions and M. Agoh (Nagasaki Prefectural Institute for Environmental Research and Public Health) for helpful discussions.
This work was supported in part by a Grant-in-aid for Scientific Research (B) (23390116) and a Grant-in-aid for Scientific Research (C) (23590557) from the Japan Society for the Promotion of Science and in part by a Grant-in-aid for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor and Welfare, Japan.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 21 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Ang LW, et al. 2009. Epidemiology and control of hand, foot and mouth disease in Singapore, 2001–2007. Ann. Acad. Med. Singapore. 38:106–112 [PubMed] [Google Scholar]
- 2. Baker DA, Phillips CA. 1979. Fatal hand-foot-and-mouth disease in an adult caused by coxsackievirus A7. JAMA 242:1065. [PubMed] [Google Scholar]
- 3. Barlean L, Avram G, Pavlov E, Cotor F. 1994. Investigation of five cases of vesicular enteroviral stomatitis with exanthema induced by coxsackie A5 virus. Rev. Roum. Virol. 45:3–9 [PubMed] [Google Scholar]
- 4. Bergelson JM. 2010. Receptors, p 73–86 In Ehrenfeld E, Domingo E, Roos RP. (ed), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 5. Blanz J, et al. 2010. Disease-causing mutations within the lysosomal integral membrane protein type 2 (LIMP-2) reveal the nature of binding to its ligand beta-glucocerebrosidase. Hum. Mol. Genet. 19:563–572 [DOI] [PubMed] [Google Scholar]
- 6. Blomqvist S, et al. 2010. Co-circulation of coxsackieviruses A6 and A10 in hand, foot and mouth disease outbreak in Finland. J. Clin. Virol. 48:49–54 [DOI] [PubMed] [Google Scholar]
- 7. Brown BA, Oberste MS, Alexander JP, Jr, Kennett ML, Pallansch MA. 1999. Molecular epidemiology and evolution of enterovirus 71 strains isolated from 1970 to 1998. J. Virol. 73:9969–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan YF, AbuBaker S. 2004. Recombinant human enterovirus 71 in hand, foot and mouth disease patients. Emerg. Infect. Dis. 10:1468–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen SP, et al. 2010. Comparison of clinical features between coxsackievirus A2 and enterovirus 71 during the enterovirus outbreak in Taiwan, 2008: a children's hospital experience. J. Microbiol. Immunol. Infect. 43:99–104 [DOI] [PubMed] [Google Scholar]
- 10. Dalldorf G. 1953. The coxsackie virus group. Ann. N. Y. Acad. Sci. 56:583–586 [DOI] [PubMed] [Google Scholar]
- 11. Davia JL, et al. 2011. Onychomadesis outbreak in Valencia, Spain associated with hand, foot, and mouth disease caused by enteroviruses. Pediatr. Dermatol. 28:1–5 [DOI] [PubMed] [Google Scholar]
- 12. De W, et al. 2011. A large outbreak of hand, foot, and mouth disease caused by EV71 and CAV16 in Guangdong, China, 2009. Arch. Virol. 156:945–953 [DOI] [PubMed] [Google Scholar]
- 13. Eskelinen EL, Tanaka Y, Saftig P. 2003. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 13:137–145 [DOI] [PubMed] [Google Scholar]
- 14. Flewett TH, Warin RP, Clarke SK. 1963. ‘Hand, foot, and mouth disease’ associated with Coxsackie A5 virus. J. Clin. Pathol. 16:53–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuchs R, Blaas D. 2010. Uncoating of human rhinoviruses. Rev. Med. Virol. 20:281–297 [DOI] [PubMed] [Google Scholar]
- 16. Gear JH. 1984. Nonpolio causes of polio-like paralytic syndromes. Rev. Infect. Dis. 6(Suppl 2):S379–S384 [DOI] [PubMed] [Google Scholar]
- 17. Grist NR, Bell EJ. 1984. Paralytic poliomyelitis and nonpolio enteroviruses: studies in Scotland. Rev. Infect. Dis. 6(Suppl 2):S385–S386 [DOI] [PubMed] [Google Scholar]
- 18. Helin I, Widell A, Borulf S, Walder M, Ulmsten U. 1987. Outbreak of coxsackievirus A-14 meningitis among newborns in a maternity hospital ward. Acta Paediatr. Scand. 76:234–238 [DOI] [PubMed] [Google Scholar]
- 19. Huang SC, et al. 2008. Appearance of intratypic recombination of enterovirus 71 in Taiwan from 2002 to 2005. Virus Res. 131:250–259 [DOI] [PubMed] [Google Scholar]
- 20. Itagaki A, et al. 1983. A clustering outbreak of hand, foot, and mouth disease caused by coxsackie virus A10. Microbiol. Immunol. 27:929–935 [DOI] [PubMed] [Google Scholar]
- 21. Kuronita T, et al. 2002. A role for the lysosomal membrane protein LGP85 in the biogenesis and maintenance of endosomal and lysosomal morphology. J. Cell Sci. 115:4117–4131 [DOI] [PubMed] [Google Scholar]
- 22. Laszik Z, et al. 1996. P-selectin glycoprotein ligand-1 is broadly expressed in cells of myeloid, lymphoid, and dendritic lineage and in some nonhematopoietic cells. Blood 88:3010–3021 [PubMed] [Google Scholar]
- 23. Lee HY, et al. 2010. Clinical features of echovirus 6 and 9 infections in children. J. Clin. Virol. 49:175–179 [DOI] [PubMed] [Google Scholar]
- 24. Lin JY, et al. 2008. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 89:2540–2549 [DOI] [PubMed] [Google Scholar]
- 25. Lin JY, Li ML, Shih SR. 2009. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res. 37:47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin JY, et al. 2009. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 83:6106–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin TL, et al. 2008. Rapid and highly sensitive coxsackievirus A indirect immunofluorescence assay typing kit for enterovirus serotyping. J. Clin. Microbiol. 46:785–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lo SH, et al. 2011. Clinical and epidemiologic features of coxsackievirus A6 infection in children in northern Taiwan between 2004 and 2009. J. Microbiol. Immunol. Infect. 44:252–257 [DOI] [PubMed] [Google Scholar]
- 29. McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107 [DOI] [PubMed] [Google Scholar]
- 30. Mizuta K, et al. 2005. Frequent importation of enterovirus 71 from surrounding countries into the local community of Yamagata, Japan, between 1998 and 2003. J. Clin. Microbiol. 43:6171–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mizuta K, et al. 2009. Cross-antigenicity among EV71 strains from different genogroups isolated in Yamagata, Japan, between 1990 and 2007. Vaccine 27:3153–3158 [DOI] [PubMed] [Google Scholar]
- 32. Nakayama T, et al. 1989. Outbreak of herpangina associated with coxsackievirus B3 infection. Pediatr. Infect. Dis. J. 8:495–498 [DOI] [PubMed] [Google Scholar]
- 33. Nishimura Y, et al. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 15:794–797 [DOI] [PubMed] [Google Scholar]
- 34. Oberste MS, Jiang X, Maher K, Nix WA, Jiang B. 2008. The complete genome sequences for three simian enteroviruses isolated from captive primates. Arch. Virol. 153:2117–2122 [DOI] [PubMed] [Google Scholar]
- 35. Oberste MS, et al. 2005. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species human enterovirus A. J. Gen. Virol. 86:445–451 [DOI] [PubMed] [Google Scholar]
- 36. Oberste MS, Maher K, Pallansch MA. 2007. Complete genome sequences for nine simian enteroviruses. J. Gen. Virol. 88:3360–3372 [DOI] [PubMed] [Google Scholar]
- 37. Oberste MS, Penaranda S, Maher K, Pallansch MA. 2004. Complete genome sequences of all members of the species human enterovirus A. J. Gen. Virol. 85:1597–1607 [DOI] [PubMed] [Google Scholar]
- 38. Ooi MH, et al. 2007. Evaluation of different clinical sample types in diagnosis of human enterovirus 71-associated hand-foot-and-mouth disease. J. Clin. Microbiol. 45:1858–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Osterback R, et al. 2009. Coxsackievirus A6 and hand, foot, and mouth disease, Finland. Emerg. Infect. Dis. 15:1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pallansch M, Roos R. 2007. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p 839–893 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 41. Podin Y, et al. 2006. Sentinel surveillance for human enterovirus 71 in Sarawak, Malaysia: lessons from the first 7 years. BMC Public Health 6:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ranzenhofer ER, Dizon FC, Lipton MM, Steigman AJ. 1958. Clinical paralytic poliomyelitis due to coxsackie virus group A, type 7. New Engl. J. Med. 259:182. [DOI] [PubMed] [Google Scholar]
- 43. Reczek D, et al. 2007. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of beta-glucocerebrosidase. Cell 131:770–783 [DOI] [PubMed] [Google Scholar]
- 44. Reed LJ, Muench H. 1938. A simple method of estimating 50 percent endpoints. Am. J. Hyg. (Lond.) 27:493–499 [Google Scholar]
- 45. Schmidt NJ, Lennette EH, Ho HH. 1974. An apparently new enterovirus isolated from patients with disease of the central nervous system. J. Infect. Dis. 129:304–309 [DOI] [PubMed] [Google Scholar]
- 46. Semler B, Wimmer E. 2002. Molecular biology of picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 47. Sickles GM, Mutterer M, Feorino P, Plager H. 1955. Recently classified types of coxsackie virus, group A; behavior in tissue culture. Proc. Soc. Exp. Biol. Med. 90:529–531 [DOI] [PubMed] [Google Scholar]
- 48. Tang WF, et al. 2007. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J. Biol. Chem. 282:5888–5898 [DOI] [PubMed] [Google Scholar]
- 49. Weng KF, Li ML, Hung CT, Shih SR. 2009. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 5:e1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yamashita T, Ito M, Taniguchi A, Sakae K. 2005. Prevalence of coxsackievirus A5, A6, and A10 in patients with herpangina in Aichi Prefecture, 2005. Jpn. J. Infect. Dis. 58:390–391 [PubMed] [Google Scholar]
- 51. Yamayoshi S, Koike S. 2011. Identification of a human SCARB2 region that is important for enterovirus 71 binding and infection. J. Virol. 85:4937–4946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamayoshi S, et al. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15:798–801 [DOI] [PubMed] [Google Scholar]
- 53. Yang F, et al. 2009. Enterovirus 71 outbreak in the People's Republic of China in 2008. J. Clin. Microbiol. 47:2351–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yip CC, et al. 2010. Emergence of enterovirus 71 “double-recombinant” strains belonging to a novel genotype D originating from southern China: first evidence for combination of intratypic and intertypic recombination events in EV71. Arch. Virol. 155:1413–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yoke-Fun C, AbuBakar S. 2006. Phylogenetic evidence for inter-typic recombination in the emergence of human enterovirus 71 subgenotypes. BMC Microbiol. 6:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang Y, et al. 2010. An emerging recombinant human enterovirus 71 responsible for the 2008 outbreak of hand foot and mouth disease in Fuyang city of China. Virol. J. 7:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.