Abstract
Coronaviruses induce in infected cells the formation of replicative structures, consisting of double-membrane vesicles (DMVs) and convoluted membranes, where viral RNA synthesis supposedly takes place and to which the nonstructural proteins (nsp's) localize. Double-stranded RNA (dsRNA), the presumed intermediate in RNA synthesis, is localized to the DMV interior. However, as pores connecting the DMV interior with the cytoplasm have not been detected, it is unclear whether RNA synthesis occurs at these same sites. Here, we studied coronavirus RNA synthesis by feeding cells with a uridine analogue, after which nascent RNAs were detected using click chemistry. Early in infection, nascent viral RNA and nsp's colocalized with or occurred adjacent to dsRNA foci. Late in infection, the correlation between dsRNA dots, then found dispersed throughout the cytoplasm, and nsp's and nascent RNAs was less obvious. However, foci of nascent RNAs were always found to colocalize with the nsp12-encoded RNA-dependent RNA polymerase. These results demonstrate the feasibility of detecting viral RNA synthesis by using click chemistry and indicate that dsRNA dots do not necessarily correspond with sites of active viral RNA synthesis. Rather, late in infection many DMVs may harbor dsRNA molecules that are no longer functioning as intermediates in RNA synthesis.
INTRODUCTION
Plus-strand RNA viruses induce dramatic membrane rearrangements in infected cells, thereby generating a subcellular microenvironment that facilitates RNA replication. A general feature of the membrane rearrangements is the induction of invaginations, giving rise to the formation of vesicles, which are tethered to a cellular membrane. These spherules probably shield the viral double-stranded RNA (dsRNA) replication intermediates from immune surveillance, while at the same time providing access of cytoplasmic constituents to the replication machinery and means of exit for the newly synthesized RNA to enter the cytoplasm (7, 8).
Mouse hepatitis virus (MHV) belongs to the Coronaviridae, which is a family of enveloped, plus-strand RNA viruses that infect a wide variety of animals. MHV strain A59 is a prototype coronavirus (CoV) and serves as a model for the well-known severe acute respiratory syndrome (SARS)-CoV. CoVs have a very large genome (∼27 to 32 kb), the first two-thirds of which encode the replicase nonstructural proteins (nsp's) that collectively form the replication-transcription complexes (RTCs). The nsp's contain enzymatic activities required for RNA synthesis, including the RNA-dependent RNA polymerase (RdRp; residing in nsp12 [5]), but also transmembrane domains (residing in nsp3, 4, and 6 [1, 14, 21, 22]) that anchor the RTCs to the modified cellular membranes. The remaining part of the genome encodes the structural and accessory proteins, including the nucleocapsid protein (N), which is also found in association with the RTCs (4, 9, 38, 40, 41).
The CoV RTCs synthesize both full-length genomic RNA (gRNA; replication) and subgenomic RNAs (sgRNAs; transcription) (reviewed in reference 31). While a negative-sense full-length RNA strand functions as a template for new (positive-sense) gRNA, the 3′ coterminal nested set of sgRNAs are synthesized via a discontinuous transcription mechanism during subgenome-length minus strand synthesis (30); as a consequence, all sgRNAs contain a short 5′-leader sequence corresponding to the 5′ end of the genome. The plus-strand RNAs are synthesized more abundantly than their negative-sense counterparts, approximately in a 100-fold excess (28). During CoV replication and transcription, various complete and partially double-stranded RNA (dsRNA) intermediates, which are either active in transcription (termed transcriptive intermediates or forms) or active in replication (termed replicative intermediates or forms), are produced (28, 31). These dsRNA intermediates are generally considered to be a good marker for the location of the sites of active viral RNA synthesis in infected cells (31).
The replicative structures induced by CoVs in infected cells consist of a network of double-membrane vesicles (DMVs) and convoluted membranes (CMs), to which the nsp's localize (11, 12, 17, 22, 26, 35, 36, 38). dsRNA, which is considered a hallmark for plus-strand RNA virus genome replication, is localized to the DMV interior in SARS-CoV-infected cells (17). Strikingly, while tomography studies for flock house virus, dengue virus, and Kunjin virus reveal the presence of a neck between the virus-induced vesicles and host membranes (10, 18, 44), the inner vesicles of the coronavirus-induced DMVs appear to be closed structures (17).
Studies with other viruses show that viral genome replication occurs at the same sites where dsRNA is accumulated (reviewed in references 7 and 8). However, as no pores have been observed that connect the interior of the inner vesicles of the coronaviral DMVs with the cytoplasm, the role of the dsRNA-containing DMVs in coronavirus RNA synthesis remains obscure. Therefore, we explored the feasibility of studying coronavirus RNA synthesis using a recently developed chemical method. This method is based on the biosynthetic incorporation of the alkyne-modified uridine analog 5-ethynyl uridine (EU) into newly transcribed RNA. EU-labeled RNA is subsequently detected by using a copper(I)-catalyzed cycloaddition reaction (referred to as click chemistry) with azide-derivatized fluorophores, followed by microscopic imaging (13). By performing a time course analysis, we were able to visualize newly synthesized viral RNAs early in infection to localize at or adjacent to concentrated patches of dsRNA foci and nsp's, including the CoV RdRp. At later time points postinfection (p.i.), the dsRNA dots have become dispersed throughout the cytoplasm and colocalization between foci of newly synthesized RNA and dsRNA is less apparent. Many dsRNA dots are then no longer associated with newly synthesized RNA, indicating that they are not transcriptionally active. However, at all times, the foci of newly synthesized RNA had the nsp12-encoding RdRp associated with them, suggesting that these foci correspond with the active RTCs.
MATERIALS AND METHODS
Cells and viruses.
Murine LR7 fibroblast cells (19) were maintained as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM−/−; Cambrex BioScience) containing 10% fetal calf serum (FCS; Bodinco BV), 100 IU/ml of penicillin, and 100 μg/ml of streptomycin (both from Life Technologies; this medium is referred to as DMEM+/+). MHV strain A59, recombinant MHV-EFLM (6), and MHV-nsp2GFP (12), which express the firefly luciferase (FL) gene and the nsp2 coding sequence fused to that of green fluorescent protein (GFP) from an additional expression cassette, were propagated in LR7 cells.
Antibodies and plasmids.
The antibody directed against double-stranded RNA (dsRNA; K1 [33]) was purchased from English and Scientific Consulting Bt. The polyclonal antibodies VU145 (α-pol [5)]) and anti-D3 (34), which are directed against MHV nsp12 and nsp2/3, respectively, were kindly provided by Mark Denison and Susan Baker, respectively. The monoclonal antibody MN, recognizing the N-terminal domain of the MHV membrane (M) protein (37), was kindly provided by John Flemming. The anti-5-bromodeoxyuridine (BrdU) antibody was purchased from Sigma-Aldrich. The vector encoding the stress granule marker RasGAP-associated endoribonuclease fused to GFP (G3BP-GFP) was kindly provided by Paul Anderson and has been described previously (16).
Labeling cells with uridine analogs.
LR7 cells grown on glass coverslips were fed with different concentrations of 5-ethynyl uridine (EU; Invitrogen) or 5′-bromouridine (BrU; Sigma-Aldrich) in DMEM+/+ at the times and for the time periods indicated in the text and figure legends, after which they were fixed using a 4% paraformaldehyde (PFA) solution in phosphate-buffered saline (PBS). Where indicated in the text, cells were transfected with the G3BP-GFP expression plasmid as previously described (12). The fixed cells were washed with PBS and permeabilized using 0.1% Triton X-100 for 10 min at room temperature. BrU labeling was visualized by antibody staining as described below. EU labeling of cells was visualized according to the manufacturer's instructions (Invitrogen; Click-iT RNA imaging kits). Briefly, the samples were incubated with a 1× working solution of Click-iT reaction cocktail, containing among others the Alexa Fluor 594 azide and CuSO4, for 30 min at room temperature. After removal of the reaction cocktail, cells were washed once with Click-iT reaction rinse buffer. After this step, samples were washed with PBS and mounted on glass slides in FluorSave (Calbiochem), or samples were processed further for antibody staining as described below.
Antibody staining and fluorescence confocal microscopy.
Permeabilized cells were washed with PBS and incubated for 15 min in blocking buffer (PBS 10% normal goat serum), followed by 60 min of incubation with antibodies directed against nsp2/3, dsRNA, or BrdU. RNasin (0,125 U/ml) was added to all incubation and washing steps. After washing the cells three times with PBS containing 0.05% Tween 20, they were incubated for 60 min with Cy3-conjugated donkey anti-rabbit immunoglobulin G antibodies (Jackson Laboratories), fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G antibodies (ICN), or Cy3-conjugated donkey anti-mouse immunoglobulin G antibodies (Jackson Laboratories). After four washes with PBS, the samples were mounted on glass slides in FluorSave (Calbiochem). The samples were examined using a confocal fluorescence microscope (Leica TCS SP). Pearson's correlation coefficients of the different fluorescent signals were determined using the JACoP plugin (2) for the ImageJ processing and analyses software, version 1.41 (25).
Analysis of luciferase expression levels.
Virus replication was quantified by determining the virus-driven firefly luciferase expression levels. To this end, LR7 cells were infected with MHV-EFLM. At the indicated time points, the luciferase expression levels in the cells were determined using the firefly luciferase assay system (Promega) according to manufacturer's instructions and using a Berthold Centro LB 960 plate luminometer.
RESULTS
EU labeling of nascent viral RNA in CoV-infected cells.
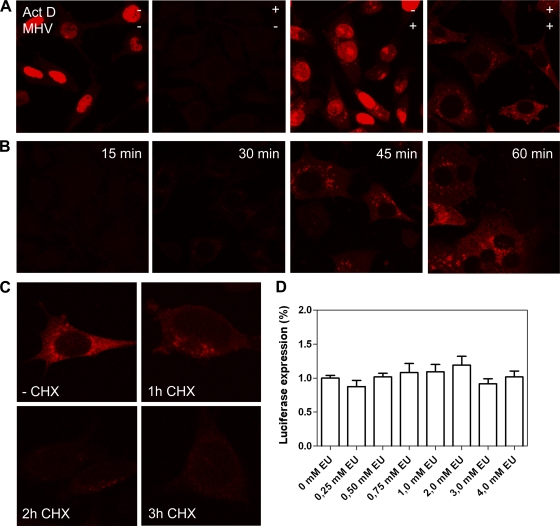
We started our experiments by analyzing the feasibility of labeling and detecting newly synthesized viral RNAs by metabolic labeling of cells with the uridine analog EU. To this end, cells were either infected with MHV or mock infected, after which they were (mock) treated with 20 μM actinomycin D, an inhibitor of (cellular) DNA-dependent RNA transcription. Replication of MHV is not inhibited by this drug at this concentration (23). At 5 h p.i., cells were fed with 1 mM EU for 1 h, after which they were fixed and prepared for detection of the incorporated alkyne-modified uridine analog by using click chemistry with azide-derivatized Alexa 594 fluorophores. As shown in Fig. 1A, in mock-infected cells, EU was incorporated into RNA in the nuclei, which was inhibited by the presence of actinomycin D. In infected cells, EU was incorporated into perinuclear cytoplasmic foci, besides the nuclei, while in the presence of actinomycin D, only the cytoplasmic foci were labeled. Apparently, this cytoplasmic staining corresponds to newly synthesized viral RNA. Next, we determined the minimal time of EU labeling needed to visualize these newly synthesized RNAs. As shown in Fig. 1B, hardly any labeling of viral RNAs could be detected when cells were fed the uridine analog for 15 or 30 min. However, viral RNAs could readily be detected after 45 min of labeling, which resulted in a staining pattern that was still very similar to the one obtained after a 60-min labeling.
Fig 1.
EU labeling of nascent viral RNA. MHV- or mock-infected LR7 cells were fed with 1 mM EU for 1 h (or for the indicated time period). Fixation of the cells was followed by detection of incorporated alkyne-modified EU with azide-derivatized Alexa 594 fluorophores by using click chemistry. (A) When indicated in the figure, 20 μM actinomycin D (Act D) was added to the infected cells at t = 1 h p.i. to inhibit cellular DNA-dependent RNA transcription. Cells were fed with EU at 5 h p.i. (B) Representative images of a time course analysis of EU labeling in the presence of actinomycin D in infected cells are shown, performed as described above, with the EU labeling times indicated. (C) Thirty-five micromolar cycloheximide (CHX) was added to the infected cells at 5 h p.i. to inhibit protein synthesis. Cells were fed with 1 mM EU at 5 h (1 h CHX), 6 h (2 h CHX), or 7 h (3 h CHX) p.i. Cells were also fed with EU in the absence of cycloheximide at 5 h p.i. (−CHX). At the end of the EU labeling period, cells were fixed. (D) LR7 cells were infected with MHV-EFLM at a multiplicity of infection (MOI) of 10, followed by treatment with different concentrations of EU ranging from 0 to 4 mM EU for 45 min starting at 5.15 h p.i. After lysis, the luciferase activity was determined and plotted as a percentage normalized to the control. The error bars indicate the standard error of the mean.
Subsequently, we studied whether labeling of newly synthesized viral RNAs could be inhibited by cycloheximide (CHX), an inhibitor of protein synthesis, shown previously to affect MHV RNA synthesis (29). In agreement with those results, the addition of CHX inhibited the labeling of viral RNAs (Fig. 1C) in a time-dependent manner. Next, we analyzed whether the addition of EU to infected cells would affect virus replication. Therefore, cells infected with a recombinant MHV expressing the luciferase reporter gene (MHV-EFLM) were treated with various concentrations of EU (0 to 4 mM) from 5.15 until 6 h p.i. The results (Fig. 1D) show that replication of MHV, as determined by the luciferase expression levels, was not inhibited by the addition of EU, at least for the time period tested. Taken together, our results indicate that in the presence of an inhibitor of cellular transcription, labeling of cells with EU can be used to specifically detect viral RNA synthesis.
Colocalization of nascent viral RNA with dsRNA and nsp2/3.
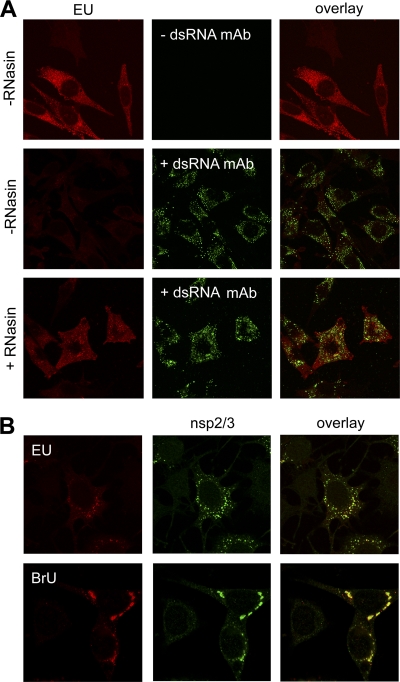
As a next step, we evaluated the possibility of combining the EU-mediated detection of nascent viral RNAs with immunocytochemistry using antibodies directed to viral components. Strikingly, the EU labeling was readily detected without but not with additional immunofluorescence staining of dsRNA in MHV-infected cells (Fig. 2A). However, when we added an inhibitor of RNase-A like enzymes (RNasin), the EU signal was preserved after immunofluorescence analysis of viral proteins and/or dsRNA. We hypothesize that the EU-containing viral RNAs are sensitive to RNases present in one of the components used in the immunocytochemical assay, presumably the FCS. Interestingly, we never observed a similar sensitivity when detecting dsRNA by immunofluorescence analysis. Therefore, we hypothesize that EU is mainly incorporated into RNase-sensitive, single-stranded RNA and not to detectable levels into dsRNA. This result in agreement with previous observations that coronavirus plus-strand RNAs are synthesized in ∼100-fold excess over their minus-strand templates (28), as a result of which the very large majority of the newly synthesized RNAs are single-stranded.
Fig 2.
Sensitivity of EU-labeled viral RNAs to RNases and comparison to BrU labeling of viral RNA. (A) LR7 cells were infected with MHV-wild type (WT) at an MOI of 10, followed by EU labeling for 45 min at 6 h p.i., fixed and processed for immunofluorescence analysis using antibodies directed against dsRNA (+dsRNA MAb) or mock treated (−dsRNA MAb), either in the presence (+) or in the absence (−) of RNase inhibitors (RNasin). (B) LR7 cells were infected with MHV-WT at an MOI of 10 for 5 h, followed by either EU or BrU labeling for 45 min and processed for immunofluorescence analysis as described in Materials and Methods. Antibodies directed against nsp2/3 were used to mark the replicative structures.
Another method to visualize newly synthesized viral RNAs is by metabolic labeling of infected cells using 5′ bromouridine (BrU; another uridine analog), followed by detection of incorporated BrU using BrdU antibodies. We performed a side-by-side comparison between the EU and BrU labeling of virus-infected cells. To this end, LR7 cells were infected with MHV-A59 in the presence of actinomycin D for 6 h. At 5.15 h p.i., the cells were fed with either 1 mM EU or BrU, and fixed, and nascent viral RNA were detected by click chemistry or immunocytochemistry, respectively. In addition, antibodies against nsp2/3, which were previously shown to exclusively label the replicative structures (38), were used. As can be seen in Fig. 2B, EU- and BrU-labeled RNAs were detected in the perinuclear region of the cell. These newly synthesized RNAs appeared to colocalize to the same extent with nsp2/3. Detection of BrU-labeled RNA was also critically dependent on the presence of an inhibitor of RNase-A like enzymes (data not shown).
Newly synthesized viral RNA localizes to RTCs but not to stress granules.
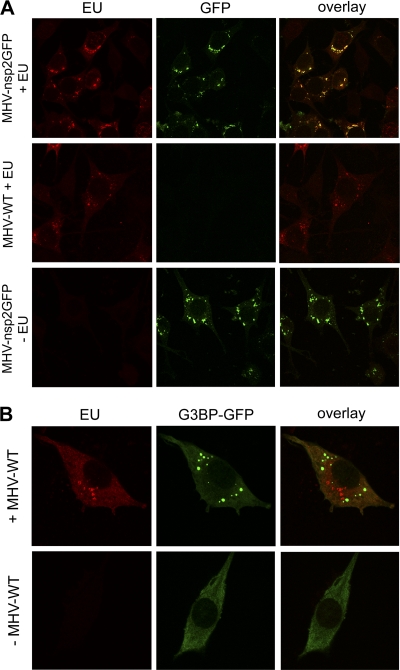
Copper(I)-catalyzed click chemistry has been reported to affect the detection of GFP fluorescence (Invitrogen). To study whether detection of EU labeling and of GFP fluorescence was compatible in our experiments, we analyzed the expression of newly synthesized RNAs by EU labeling in cells infected with a recombinant MHV, which expresses an additional expression cassette encoding GFP-tagged nsp2 (MHV-nsp2GFP). Nsp2-GFP was previously shown by electron microscopy to localize to the replicative structures (DMVs and CMs) induced by MHV in infected cells (12). As can be seen in Fig. 3A, perinuclear nsp2-GFP fluorescence was evident in combination with newly synthesized viral RNA using copper(I)-dependent click chemistry, although the fluorescent signal was somewhat lower than in nontreated cells (compare the top row with the bottom row of Fig. 3A). The newly synthesized RNA colocalized with the nsp2-GFP fluorescent foci, confirming that this fusion protein when expressed in trans localized to the structures involved in viral RNA synthesis (12).
Fig 3.
Nascent viral RNA localizes to nsp2GFP-positive foci but is excluded from stress granules. (A) LR7 cells were infected with either MHV-nsp2GFP or MHV-WT at an MOI of 10. At 7 h p.i., cells were labeled for 45 min with EU, fixed, and subsequently processed for immunofluorescence analysis. (B) Prior to infection with MHV-WT, cells were transfected with a plasmid expressing the stress granule marker G3BP-GFP, labeled for 45 min at 7 h p.i., fixed, and subsequently processed for immunofluorescence analysis.
The compatibility of fluorescence detection of GFP and viral RNAs allowed us to study to what extent the viral RNAs were localizing to stress granules in infected cells. Stress granules are cytoplasmic foci containing mRNAs stalled in translation (16). MHV is known to induce these cytoplasmic structures in infected cells (23). As is shown in Fig. 3B, the stress granule marker G3BP-GFP was distributed throughout the cytoplasm in noninfected cells, indicating the absence of stress granules. In infected cells, the protein concentrated in cytoplasmic foci corresponding to stress granules. Clearly, the newly synthesized RNA was not localizing to these foci, indicating that EU-positive foci do not correspond to stress granules.
Colocalization of nascent viral RNA with dsRNA and nsp2/3.
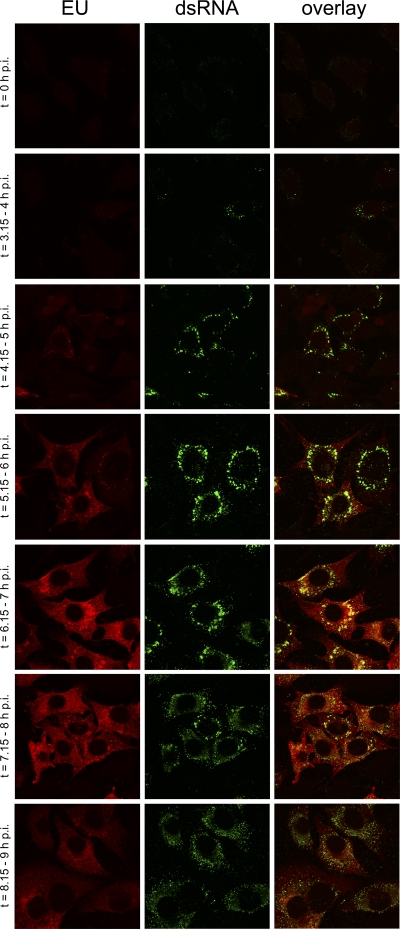
When performing the analyses described above, we noticed that the dsRNA foci were distributed in the infected cells in one of two distinct patterns. When cells were fixed at 6 h p.i., dsRNA foci were either spread throughout the cytoplasm or appeared to be more concentrated in perinuclear foci. Interestingly, the newly synthesized RNAs appeared to colocalize to a larger extent with the dsRNA foci when present in the concentrated patches (data not shown). To extend these observations, we performed a time course experiment in which cells were EU labeled for 45 min at different times p.i., after which the cells were fixed and processed for EU visualization and for immunofluorescence detection of dsRNA (see Fig. 4). At early time points p.i. (3.15 to 4 h p.i.), concentrated patches of dsRNA were observed, while at later time points (8.15 to 9 h p.i.), mainly the dispersed pattern of dsRNA foci was detected. Colocalization of newly synthesized RNA with dsRNA was more apparent at early than at later infection times. The total amount of EU-associated fluorescence seemed to decline at the later time points, in agreement with the kinetics of coronaviral RNA synthesis using similar experimental conditions (23).
Fig 4.
Time course analysis of dsRNA and nascent viral RNA in MHV-infected cells. LR7 cells were infected with MHV-WT at an MOI of 10, EU labeled at the indicated times p.i. for 45 min, fixed, and processed for immunofluorescence detection of EU and dsRNA.
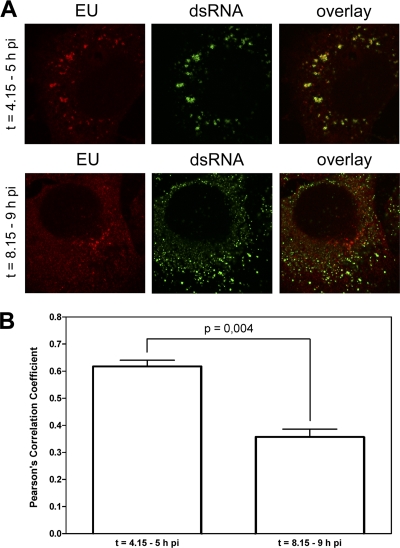
Close inspection of cells at higher magnification and with higher laser intensities (Fig. 5A) demonstrated that the EU labeling and dsRNA foci were present at closely adjacent locations rather than at identical sites. At the later times, the EU labeling did not appear to correlate to the same extent with the dsRNA foci now spread throughout the cell. Many dsRNA foci were not located adjacent to newly synthesized RNA, indicating that they were transcriptionally silent. In addition, the EU labeling was more spread throughout the cell, while some foci containing concentrated EU labeling were located at areas in the cell that were devoid of bright dsRNA dots. This observation was confirmed by determining the Pearson's correlation coefficients of the two fluorescent signals at early and late time points of infection (Fig. 5B). The Pearson's correlation coefficient is one of the standard parameters used in pattern recognition to describe the degree of overlap between two patterns. Using the JACoP plugin (2) in ImageJ (25), we calculated the Pearson's correlation coefficient to obtain an unbiased evaluation of the extent of colocalization between the two signals. The coefficients were significantly higher (P = 0.004) at the earlier (∼0,60) than at the later (∼0.33) time point p.i.
Fig 5.
Distinct dsRNA patterns at early and late times of infection. (A) Magnification of individual infected cells that were fed with EU either at an early (4.15 h p.i.) or a late (8.15 h p.i.) time point. EU labeling and dsRNA staining is shown. (B) Quantification of the colocalization by determining the Pearson's correlation coefficients of EU and dsRNA at both the early and the late time points (n = 4). Error bars indicate the standard error of the means.
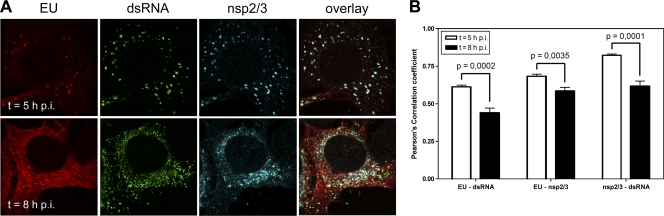
To further extend these observations, we performed a triple fluorescent labeling experiment in which, in addition to the EU labeling and dsRNA detection, the nsp2/3 proteins were also visualized (Fig. 6A). Antibodies to these nsp's label the coronavirus replicative structures (DMVs and CMs), as has been demonstrated by immunoelectron microscopy (38). While at the early time point, the signals for EU, dsRNA, and nsp2/3 labeling colocalized or were closely juxtaposed, this picture was much less apparent at the later times. The observation was again confirmed by determining the Pearson's correlation coefficients between the different staining patterns, which were all significantly lower at the later time points (see Fig. 6B). Similar results were obtained when antibodies against nsp8 (20) were used (data not shown).
Fig 6.
Temporal analysis of colocalization of EU with dsRNA and nsp2/3. LR7 cells were infected with MHV-WT at an MOI of 10 and labeled with EU for 45 min starting at 5 or 8 h p.i., fixed, and processed for EU visualization and immunofluorescence using antibodies against dsRNA or nsp2/3 (A). Quantification of the colocalization of EU-dsRNA, EU-nsp2/3, and nsp2/3-dsRNA was performed by calculation of the Pearson's correlation coefficients for both the early and the late times p.i. (n = 6). Error bars indicate the standard error of the mean.
Dispersal of dsRNA does not depend on a functional microtubular or actin network.
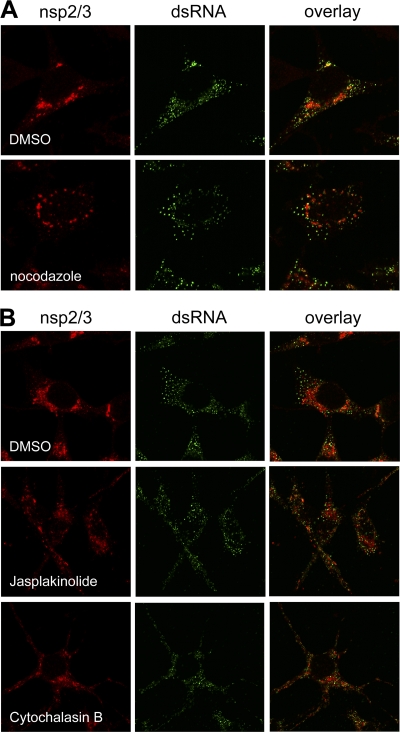
Previously, we demonstrated that nsp2-GFP-positive foci are mobile and move through MHV-infected cells using microtubular tracks (12). Hence, we hypothesized that the dispersal of the dsRNA foci might be dependent on an intact microtubular network as well. Therefore, the (co)localization of the EU labeling and the dsRNA foci was analyzed when cells were (mock) treated with 1 μM nocodazole, a microtubule-disturbing agent. Disruption of the microtubular network by the drug was confirmed by immunofluorescence analysis of α-tubulin (data not shown). As a control, we also stained for the nsp2/3 proteins and found, consistent with our previous results (12), the nsp2/3-positive foci to be concentrated in the perinuclear region in the absence but much less so in the presence of nocodazole, as illustrated in Fig. 7A. However, the dispersal of the dsRNA foci was not affected by nocodazole. From these results, we conclude that dispersal of the dsRNA foci is not dependent on an intact microtubular network. Subsequently, we investigated whether intact actin filaments were required for the dispersal of the dsRNA foci. To this end, we (mock) treated cells with 20 μM cytochalasin B or with 1 μM jasplakinolide to disrupt the actin network in MHV-infected cells. Complete disruption of the actin filaments was observed in these cells (data not shown). In mock-treated cells, the localization of nsp2/3 and dsRNA was similar to the previous observations in the earlier experiments (Fig. 7B). Treatment of infected cells with jasplakinolide or cytochalasin B did not inhibit the dispersal of the dsRNA foci at the late infection times (Fig. 7B). Furthermore, disruption of the actin filaments also resulted in the dispersal of the nsp2/3-positive fluorescent foci. In conclusion, dispersal of the dsRNA foci is not dependent on a functional microtubular or actin network, whereas both networks appear to be required for the efficient perinuclear accumulation of the nsp2/3-positive foci.
Fig 7.
Microtubule- and actin-independent movement of dsRNA foci. LR7 cells were infected with MHV-WT at an MOI of 10 and labeled for 45 min, starting at 7 h p.i., in the absence (dimethyl sulfoxide [DMSO] alone) or presence of either 1 μM nocodazole (A) or 20 μM cytochalasin B or 1 μM jasplakinolide (B) to disrupt the microtubular or actin network, respectively, followed by fixation and processing for EU visualization and immunofluorescence using antibodies against nsp2/3 or dsRNA. The cytoskeleton-disrupting drugs were added to the culture media from 2 h p.i. onwards.
Identifying the active sites of viral RNA replication.
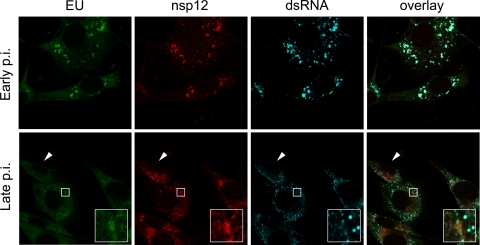
Our results indicated that not all replicative structures, despite them being the structures that harbor the DMV-containing dsRNAs, are actively involved in RNA synthesis. On the other hand, while the EU-positive foci may very well correspond with active RTCs, the EU-labeled nascent RNAs might as well be transported away from their original sites of synthesis. To confirm that EU-positive foci are indeed found at sites actively involved in RNA synthesis, we investigated whether the CoV RdRp (residing in nsp12) was also present at these sites. As shown in Fig. 8, early in infection nascent RNAs (EU labeling) and dsRNA dots colocalized with each other and with nsp12. At later time points, dsRNA dots were dispersed throughout the cell and RNA synthesis was decreased, although foci of EU labeling were readily detectable in many cells, which is in agreement with our earlier results. At this later time point, the nsp12 staining appeared less intense and somewhat more dispersed. However, EU-positive foci were still clearly colocalizing with nsp12 at this stage. Taken together, our results indicate that foci of EU-labeled viral RNAs correspond with active RTCs as they colocalize with the RdRp throughout the infection.
Fig 8.
Identifying the active sites of CoV RNA synthesis. LR7 cells were infected with MHV-WT at an MOI of 10, EU labeled at early and late times p.i., and fixed and processed for immunofluorescence detection of EU, dsRNA, and nsp12. Arrowheads indicate an nsp12-positive cell lacking most EU labeling, and the white boxes indicate examples of colocalization of EU and nsp12-positive foci at late times of infection.
DISCUSSION
To visualize viral RNA (synthesis) at the (sub)cellular level in infected cells, various methods have been used, like in situ hybridization, using fluorescently labeled probes, or dsRNA detection, using antibodies specific for dsRNA, and have been of great value but provide information only on the steady-state localization of specific RNA species. For the plus-strand RNA viruses, dsRNAs are thought to function as intermediates in RNA synthesis (22, 31) and have been shown to localize to the membrane-wrapped compartments dedicated to this process (17). Analysis of transcription rates of nascent RNAs, however, requires the labeling of newly synthesized RNAs with tritiated nucleotides or with uridine analogs, such as 5-bromouridine (BrU). In the present study, we have employed another, novel method suitable for the detection of nascent RNAs, based on the biosynthetic incorporation of an alkyne-modified EU, followed by click chemistry to attach fluorescent azide derivatives to the synthesized RNA. We show that this method is suitable for the specific visualization of newly synthesized, presumably full-length as well as subgenomic, coronaviral RNAs in the presence of actinomycin D. Furthermore, we show that this method can be combined with the detection of viral and host proteins by using GFP tags or immunohistochemistry. Our results indicate that foci of EU labeling correspond with active sites of coronaviral RNA synthesis throughout the infection. In contrast, at later times of infection, many dsRNA-positive foci, the presumed intermediates generated during plus-strand RNA virus replication, no longer correspond with coronaviral replicative structures actively involved in RNA synthesis.
Upon infection, CoVs induce a network of DMVs, which harbor dsRNA, and CMs. It is not clear, however, which of these structures are actually associated with the biosynthesis of the CoV RNA, even though BrUTP incorporation was detected in the vicinity of the presumed CoV-induced DMVs (11). Although dsRNA molecules function as intermediates of replication or transcription, their presence at certain sites per se does not imply (all) these structures to be actively involved in RNA synthesis. Likewise, the location of viral enzymes that are required for RNA synthesis does not need to correlate with active RTCs. Furthermore, we cannot exclude the possibility that nascent RNAs are not necessarily accumulated at their site of synthesis, as they may diffuse or be transported away to other subcellular locations. Even so, our results indicate that foci of nascent RNAs detected in MHV-infected cells correspond with sites of active coronaviral RNA synthesis. Although the colocalization of EU labeling with nsp2/3 and dsRNA, which was obvious early in infection (Fig. 5 and 8), appeared to decrease in time, the foci of nascent RNAs colocalized with the RdRp-containing nsp12 throughout the infection (Fig. 8). Furthermore, the EU-positive foci did not colocalize with a marker for stress granules (Fig. 3B), i.e., cytoplasmic structures containing mRNAs halted in their translation (16), which are induced upon infection with MHV (23). As an obvious next step, it will be interesting to further characterize the site(s) of coronaviral RNA synthesis at the ultrastructural level.
Simultaneous visualization of dsRNA, which is localized to the DMV interior (17), and EU incorporation revealed that while early in infection nascent RNAs were found colocalizing at or adjacent to patches of dsRNA dots, this correlation was much less apparent at later times when the dsRNA dots had spread throughout the cell. Many dsRNA dots were apparently not transcriptionally active, as no EU labeling was associated with them. The results may be explained by assuming that many DMVs are not actively involved in RNA synthesis but harbor dsRNAs that are no longer functioning as intermediates in RNA synthesis. In other words, dsRNA-containing DMVs may represent nonfunctional end-stage structures. According to this model, the DMVs might serve the important function of shielding dsRNA molecules from the innate immune system of the cell (15). In agreement herewith is the apparent lack of obvious pores that connect their inside with the cytoplasm. Also consistent are observations that CoVs prevent early induction of interferon synthesis (24, 43) presumably by keeping viral RNA away from host cell sensors (42), although this does not hold true for primary cell types such as plasmacytoid dendritic cells and macrophages (27). If DMVs would not be the active sites of RNA synthesis, the only plausible alternative would be the CMs. These latter structures harbor most nsp's studied to date (17, 38), although nsp12 (RdRp) has not yet been localized at the ultrastructural level, and may provide the RTCs with the membrane-protective environment (39). Of note, these different models do not necessarily exclude each other, as DMVs may be the initial site of active RNA synthesis, particularly early in infection, while at a later state the membranes become sealed and connections are lost and RNA synthesis shifts to the CM assemblies.
Whatever model is correct, our results suggest that the CoV replicative structures evolve over time as exemplified by the changing relations between different components (dsRNA, nsp's, and nascent RNA). Thus, while early in infection nascent viral RNA and nsp's colocalized with or occurred adjacent to dsRNA foci, at later time points the correlation between dsRNA dots, nsp2/3, and nascent RNAs was less obvious. However, foci of nascent RNAs were always found to colocalize with nsp12. Accordingly, others have reported the segregation of MHV replicase proteins and viral RNA into distinct populations of intracellular membranes (35), which became physically separated during the course of infection (3). Maturation of the CoV RTCs, which may involve processing of the replicase polyproteins, has also been suggested by others to be important in regulating the plus- and minus-strand RNA synthesizing capacity of the RTCs (29, 32). We expect that more extensive electron tomography studies as well as correlative light electron microscopy, ideally combined with labeling of nascent RNAs, will be required to fully understand the biogenesis, maturation, and function of these remarkable structures.
ACKNOWLEDGMENTS
We thank Richard Wubbolts and Esther van ‘t Veld from the Center for Cell Imaging of the Utrecht Faculty of Veterinary Medicine for technical support and Oliver Wicht, Christine Burkhard, Qiushi Wang, and Fulvio Reggiori for stimulating discussions. We thank Mark Denison and Susan Baker for kindly providing antibodies.
This work was supported by grants from the Netherlands Organization for Scientific Research (NWO-ALW) and Utrecht University (High Potential) to C.A.M.D.H. The funding sources had no role in the study design, in data collection, analysis, or interpretation, or in the writing of this article.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Baliji S, Cammer SA, Sobral B, Baker SC. 2009. Detection of nonstructural protein 6 in murine coronavirus-infected cells and analysis of the transmembrane topology by using bioinformatics and molecular approaches. J. Virol. 83:6957–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 [DOI] [PubMed] [Google Scholar]
- 3. Bost AG, Prentice E, Denison MR. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bost AG, Carnahan RH, Lu XT, Denison MR. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brockway SM, Clay CT, Lu XT, Denison MR. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515–10527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Haan CA, van Genne L, Stoop JN, Volders H, Rottier PJ. 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77:11312–11323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256 [DOI] [PubMed] [Google Scholar]
- 8. den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denison MR, et al. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862–6871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84:10438–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hagemeijer MC, et al. 2010. Dynamics of coronavirus replication-transcription complexes. J. Virol. 84:2134–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jao CY, Salic A. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. U. S. A. 105:15779–15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kanjanahaluethai A, Chen Z, Jukneliene D, Baker SC. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361:391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawai T, Akira S. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1–20 [DOI] [PubMed] [Google Scholar]
- 16. Kedersha N, et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knoops K, et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:1957–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopek BG, Perkins G, Miller DJ, Ellisman MH, Ahlquist P. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220 doi:10.1371/journal.pbio.0050220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuo L, Godeke GJ, Raamsman MJ, Masters PS, Rottier PJ. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu XT, Sims AC, Denison MR. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oostra M, et al. 2008. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 82:12392–12405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Oostra M, et al. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 81:12323–12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2007. Mouse hepatitis coronavirus replication induces host translational shutoff and mRNA decay, with concomitant formation of stress granules and processing bodies. Cell. Microbiol. 9:2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raaben M, Groot Koerkamp MJ, Rottier PJ, de Haan CA. 2009. Type I interferon receptor-independent and -dependent host transcriptional responses to mouse hepatitis coronavirus infection in vivo. BMC Genomics 10:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rasband W. 1997–2008. ImageJ. U.S. National Institutes of Health, Bethesda, MD: http;//rsbweb.nih.gov/ij [Google Scholar]
- 26. Reggiori F, et al. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rose KM, Weiss SR. 2009. Murine coronavirus cell type dependent interaction with the type i interferon response. Viruses 1:689–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sawicki D, Wang T, Sawicki S. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385–396 [DOI] [PubMed] [Google Scholar]
- 29. Sawicki SG, Sawicki DL. 1986. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J. Virol. 57:328–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sawicki SG, Sawicki DL. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499–506 [DOI] [PubMed] [Google Scholar]
- 31. Sawicki SG, Sawicki DL, Siddell SG. 2007. A contemporary view of coronavirus transcription. J. Virol. 81:20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sawicki SG, et al. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39 doi:10.1371/journal.ppat.0010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schönborn J, et al. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19:2993–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shi ST, et al. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. J. Virol. 73:5957–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sims AC, Ostermann J, Denison MR. 2000. Mouse hepatitis virus replicase proteins associate with two distinct populations of intracellular membranes. J. Virol. 74:5647–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Snijder EJ, et al. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taguchi F, Fleming JO. 1989. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology 169:233–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ulasli M, Verheije MH, de Haan CA, Reggiori F. 2010. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell. Microbiol. 12:844–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Hemert MJ, et al. 2008. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4:e1000054 doi:10.1371/journal.ppat.1000054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verheije MH, et al. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 4:e1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Verheije MH, et al. 2010. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J. Virol. 84:11575–11579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Versteeg GA, Slobodskaya O, Spaan WJ. 2006. Transcriptional profiling of acute cytopathic murine hepatitis virus infection in fibroblast-like cells. J. Gen. Virol. 87:1961–1975 [DOI] [PubMed] [Google Scholar]
- 44. Welsch S, et al. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375 [DOI] [PMC free article] [PubMed] [Google Scholar]