Abstract
As anti-HIV therapy becomes more widely available in developing nations, it is clear that drug resistance will continue to be a major problem. The related viruses HIV-1 and HIV-2 share many of the same resistance pathways to nucleoside reverse transcriptase inhibitors (NRTIs). However, clinical data suggest that while HIV-1 reverse transcriptase (RT) usually uses an ATP-dependent excision pathway to develop resistance to the nucleoside analog zidovudine (AZT), HIV-2 RT does not appear to use this pathway. We previously described data that suggested that wild-type (WT) HIV-2 RT has a much lower ability to excise AZT monophosphate (AZTMP) than does WT HIV-1 RT and suggested that this is the reason that HIV-2 RT more readily adopts an exclusion pathway against AZT triphosphate (AZTTP), while HIV-1 RT is better able to exploit the ATP-dependent pyrophosphorolysis mechanism. However, we have now done additional experiments, which show that while HIV-1 RT can adopt either an exclusion- or excision-based resistance mechanism against AZT, HIV-2 RT can use only the exclusion mechanism. All of our attempts to make HIV-2 RT excision competent did not produce an AZT-resistant RT but instead yielded RTs that were less able to polymerize than the WT. This suggests that the exclusion pathway is the only pathway available to HIV-2.
INTRODUCTION
HIV-1 infection has been the target of various multidrug therapies, but to date, the effectiveness of all anti-HIV drugs has been blunted by drug-resistant mutations that arise in the genome of the virus. Reverse transcriptase (RT) is an enzyme that contains two enzymatic activities, a DNA polymerase that can copy either an RNA or DNA template, and an RNase H, which degrades RNA if the RNA is part of an RNA/DNA duplex. RT uses these two enzymatic activities to convert the single-stranded RNA genome of the virus into a double-stranded DNA that can be integrated into the genome of the host cell. The synthesis of a DNA copy of the viral genome is a crucial step in the life cycle of the virus, and RT has, for that reason, been the target of a number of different anti-HIV drugs (for reviews, e.g., see references 2, 12, 17, 19, 21, 23, 31, and 32). The earliest anti-HIV therapies involved nucleoside RT inhibitors (NRTIs). These analogs enter the cell and are converted to the triphosphate form (nucleoside RT inhibitor triphosphates [NRTI-TPs]) by host cell kinases. Because the NRTI-TPs are analogs of the normal deoxynucleoside triphosphates (dNTPs), NRTI-TPs are incorporated into the primer strand by RT. However, because the NRTI-TPs do not have a 3′-OH group on the sugar or pseudosugar moiety, an NRTI monophosphate (NRTI-MP) that has been incorporated into viral DNA cannot support continued DNA synthesis and the primer chain is terminated. Decreased susceptibility to NRTIs means that the mutant RT has an enhanced ability to select normal dNTPs over the NRTI-TPs. Two primary mechanisms have been identified by which HIV-1 RT becomes less susceptible to the NRTI-TPs. One mechanism is exclusion, in which the mutant RT has a reduced ability to bind and incorporate the NRTI-TP. This exclusion can involve either active exclusion of the analog by steric hindrance or a reduction in the binding/incorporation of the analog. The mutations M184I and -V lead to a steric clash between the oxathiolane ring of lamivudine triphosphate (3TCTP) and the β-branch on the side chain of the isoleucine or valine (11, 31). In contrast, Q151M has been reported to alter the hydrogen bonding between RT and the 3′-OH group of the incoming dNTP (31). This altered hydrogen bond network helps the Q151M RT bind and incorporate a normal dNTP better than an NRTI-TP lacking the 3′-OH. In effect, the NRTI-TP is discriminated against because it cannot efficiently compete against the normal substrate. The second mechanism is ATP-mediated pyrophosphorolysis. In this mechanism, the NRTI-TP is still bound and incorporated efficiently by the mutant HIV-1 RT. However, the mutant RT is able to remove the chain-terminating nucleoside RT inhibitor monophosphate (NRTI-MP) from the end of the primer, using ATP as a pyrophosphate donor, in a process similar to polymerization run in reverse (2, 14, 17, 21, 23, 31, 32).
Mutations in HIV-1 RT can cause zidovudine (AZT) resistance by either of these two mechanisms. An early treatment protocol used AZT and dideoxyinosine (ddI) in combination; ddI is converted into ddATP by the host cell (1). This combination therapy selected drug-resistance mutations in HIV-1 RT, many of which included the primary mutation Q151M. Other mutations (A62V, V75I, F77L, and F116Y) were found in various combinations with Q151M. The fact that these mutations reduce the ability of RT to incorporate some NRTI-TPs, including AZT triphosphate (AZTTP), was first described by the laboratory of H. Mitsuya (20, 39). For simplicity, we have called the cluster of five mutations the Q151M complex. These mutations cause resistance by the exclusion mechanism. In the mutants, AZTTP/ddATP binding and incorporation are reduced compared to those in the normal substrates TTP and dATP. As was just discussed, it has been suggested that the Q151M mutation alters the hydrogen bond interactions between HIV-1 RT and the 3′-OH of the incoming dNTP, causing an increased discrimination against the NRTI-TPs (31). The purpose of the other mutations in the Q151M complex is less clear. They could be compensatory mutations that counteract any deleterious effects of the Q151M mutation, or they could further increase the ability of the mutant RT to discriminate against the NRTI-TPs. It has been suggested, based on structural analysis, that mutations at V75 may alter interactions of RT with the nucleic acid substrate (16, 22).
Most other therapies for HIV-1 that include AZT select for resistance via the excision pathway. The two primary excision resistance mutations, T215Y/F and K70R, enhance the ability of the mutant RT to bind the ATP that serves as the pyrophosphate donor in the excision reaction (38). Excision requires that the AZT monophosphate (AZT-MP) at the 3′ end of the primer is bound in the dNTP binding site (N-site) rather than the priming site (P-site). If the end of the primer is in the N-site, an appropriately bound ATP can be used to remove the AZT-MP by pyrophosphorolysis, leaving a free 3′ primer end and generating a free dinucleoside-tetraphosphate (AZT-ppppA) (2, 14, 17, 21, 23, 31, 32, 36). Recently, A62V was proposed to stabilize a configuration of the binary complex (RT and nucleic acid) in which the end of the primer is in the pretranslocated position (N-site), which would favor excision (33).
A related virus, HIV-2, is endemic to West Africa, and there are more localized foci of infections in other areas, such as India. Like HIV-1, HIV-2 can cause AIDS; however, compared to HIV-1, the disease progression is slower, the transmission rate and plasma viral load are also lower, and many patients are asymptomatic (13, 19, 24–26, 29, 30, 36, 37). As resources and drugs have become more widely available, HIV-2 infections have been treated with NRTIs. There are similarities in the drug resistance mutations that are selected when HIV-1 RT and HIV-2 RT are treated with the same drugs. However, it is apparent that HIV-2 RT favors the Q151M exclusion pathway for resistance to AZT, and there is little evidence for selection of mutations in HIV-2 that could cause resistance by the excision pathway (13, 19, 24–26, 29, 30, 36, 37). As described above, the Q151M exclusion pathway mutations are selected for HIV-1 RT only with a defined treatment protocol involving both AZT and ddI. Other regimens involving AZT select for excision pathway mutations. We previously compared wild-type (WT) HIV-1 RT (BH10) and HIV-2 RT (ROD) both biochemically and structurally. The sequences of the polymerase domains of these two isolates were also compared (5). We showed that the WT HIV-1 RT has a significantly greater capability to excise AZT-MP than WT HIV-2 RT and that HIV-2 RT is better able to exclude (avoid incorporating) AZTTP compared to HIV-1 RT. We suggested, based on a comparison of models of the structures of the two RTs, that the mutations that create the ATP binding site in HIV-1 RT are not likely to generate an equivalent ATP binding site in HIV-2 RT. We also suggested that the RTs selected the pathway that reinforced the native properties of the WT RTs (5). Smith et al. (36) have examined the effects of various mutations in HIV-1 and HIV-2 using both a cell culture infection assay and purified RTs and reached similar conclusions. In this report, we use purified RTs to examine in more detail the effects of the mutations that HIV-1 RT and HIV-2 RT do and do not choose to develop resistance to NRTIs.
MATERIALS AND METHODS
Preparation of HIV-1 RT.
The open reading frames encoding wild-type HIV-1 RT and each of the RT mutants were cloned into a plasmid containing the HIV-1 protease (PR) open reading frame as previously described (5, 40). The RT coding region was derived from HIV-1 strain BH10 (GenBank accession no. HIVBH102). The plasmid is based on the expression vector pT5m and was introduced into Escherichia coli strain BL21(DE3)(pLysE). A similar plasmid containing the coding regions of HIV-2 RT and HIV-2 protease was the kind gift of Amnon Hizi (Tel Aviv University). The HIV-2 RT coding region was derived from strain ROD (GenBank accession no. HIV2ROD). After induction with isopropyl-β-d-thiogalactopyranoside (IPTG), the plasmids express both RT and PR. Approximately half of the RT in the bacteria is converted into the small subunit by PR. The bacteria were collected by centrifugation, and the cells were frozen at −70°C. Both the HIV-1 and HIV-2 RTs were purified in the same manner.
Frozen cells (Approximately 2 g) were thawed and homogenized with 4 ml of a mixture of cold 50 mM NaPO4, 50 mM NaCl, 0.75 mg/ml lysozyme, and 1.5 mM phenylmethylsulfonyl fluoride (PMSF) (pH 8.0). After a 25-min incubation on ice, 0.43 ml of 4 M NaCl was mixed in with the extract. The suspension was sonicated three times for 30 s each at 50% output with a 70% pulse. The maximum output of sonicator was 350 W. Following sonication, the sample was centrifuged at 85,000 × g for 60 min, and the supernatant was separated from the pellet and diluted with an equal volume of 66 mM NaPO4–300 mM NaCl (pH 6.8). The sample was then loaded onto a 1.5-ml nickel-nitrilotriacetic acid (Ni-NTA) metal affinity column, which was equilibrated with 50 mM NaPO4–300 mM NaCl (pH 7.0). After loading the sample, the column was washed with 50 ml of equilibration buffer. Next, the column was washed with 90 ml of a mixture of 50 mM NaPO4, 300 mM NaCl, 10% glycerol, and 20 mM imidazole (pH 6.0). The RT was eluted with a 15-ml by 15-ml, 20 mM to 500 mM imidazole gradient with the other pH 6.0 buffer components present. Fractions of 1.5 ml were collected and analyzed by Coomassie-stained SDS-PAGE. Fractions containing significant amounts of RT were pooled.
The pool was dialyzed against Q-B buffer (25 mM Tris acid–25 mM Tris base [pH 8.3]) over an 8- to 12-h period. The dialysate was centrifuged at 14,000 × g for 30 min. The supernatant was loaded onto a 3-ml Q-Sepharose column equilibrated with Q-B buffer. After loading was complete, the column was washed with 25 ml of Q-B buffer. A pH gradient consisting of 15 ml of the Q-B buffer and 15 ml of 50 mM HEPES–25 mM MES (morpholineethanesulfonic acid) (pH 6.25) was run through the column. Samples were pooled after analysis with silver-stained SDS-PAGE. One-tenth volume of 2 M NaCl was added to samples, which were then concentrated by centrifugal ultrafiltration with a 30-kDa cutoff. The concentration of the sample was measured by the Bradford method, using bovine gamma globulin as the standard. Except where otherwise noted, all procedures in the purification were carried out at 4°C.
Processivity and polymerase assays.
The processivity and polymerase assays have been previously described (5, 4, 40). In brief, the −47 sequencing primer (5′-CGCCAGGGTTTTCCCAGTCACGAC-3′) (New England BioLabs) was 5′-end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to single-stranded M13mp18 DNA (1.0 μg of a DNA stock at a concentration of 0.25 μg/μl for each sample to be assayed) by heating and slow cooling. The RNA template was generated using a clone containing a T7 RNA polymerase promoter, the HIV-1 polypurine tract (PPT), U3, R, U5, and the HIV-1 primer binding site was linearized with NotI. The RNA transcript that contains the HIV-1 PPT, long terminal repeat (LTR), and primer binding site sequences (sense strand) was synthesized using the T7 MEGAScript kit (Ambion) and a plasmid containing sequences derived from pNL 4-3. A synthetic DNA oligonucleotide (Biosource) complementary to the primer binding site sequence (5′-GTCCCTGTTCGGGCGCCA-3′) was 5′-end labeled and then annealed to the RNA template.
The labeled RNA template/DNA primer or DNA template/DNA primer combination was resuspended in a mixture containing 25 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg of bovine serum albumin (BSA) per ml, 10.0 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 2.0 mM dithiothreitol (DTT). One microgram of wild-type RT or an RT variant was added to each tube and allowed to bind to the labeled template/primer for 2 min. Extension was initiated by the addition of dNTPs, to a final concentration of 10.0 μM each. For the processivity assays, 0.5 U of poly(rC) · oligo(dG) per ml was added to the reaction mixtures after the RT binding step, but before the dNTPs. The polymerase assays do not contain this “cold trap.” The poly(rC) · oligo(dG) prevents RT from rebinding to the labeled primer by binding the RT after it disassociates from the labeled template/primer. The reactions were allowed to proceed at 37°C for 10 min and were then halted by the addition of EDTA. The samples were precipitated by the addition of 2 volumes of ethanol. The samples were fractionated by electrophoresis on a 6.0% polyacrylamide gel, and the gel was autoradiographed.
Low-dNTP extension assay.
The low-dNTP extension assay has been described previously (5, 4, 40). Briefly, the DNA template/DNA primer and RNA template/DNA primer were generated as described above. For each sample, 1.0 μg of wild-type RT or an RT variant was added to the labeled template/primer in a mixture containing 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA per ml, and 10.0 mM CHAPS. The reaction mixture was supplemented with 0.1, 0.2, 0.5, or 1.0 μM (each) dATP, dCTP, dGTP, and dTTP. The reactions were allowed to proceed at 37°C for 15 min and were halted by the addition of EDTA. The samples were precipitated by the addition of 2 volumes of ethanol and fractionated by electrophoresis on a 6.0% polyacrylamide gel, and the gel was autoradiographed.
Pyrophosphorolysis.
ATP-dependent pyrophosphorolysis analysis was done as previously described (5, 4, 40). A synthetic DNA oligonucleotide (5′-CAGGTCACTGTTCGAGCACCA-3′; Biosource, Camarillo, CA) was 5′-end labeled and then annealed to the DNA template (5′-GCGCAGTGTAGACAATCCCTAGCTATGGTGCTCGAACAGTGACCTG-3′) or the RNA template (5′-GCGCAGUGUAGACAAUCCCTAGCUAUGGUGCUCGAACAGUGACCUG-3′). The 3′ end of the primer was then blocked by the addition of AZTMP, using a purified HIV-1 RT that contains mutations in the RNase H active site (D443A and D549A). The underlined letter indicates the position on the template that specifies the position at which the AZTMP was added. After purification of the blocked template/primer, the template/primer was incubated with HIV-1 RT or the RT variant in 50.0 μl of a mixture of 50.0 mM HEPES (pH 7.5), 75 mM KCl, 16.0 mM MgCl2, 100.0 μg of bovine serum albumin per ml, 10.0 mM CHAPS, and 10.0 μM (each) dATP, dTTP, dGTP and dCTP; and 3.0 mM ATP for the lengths of time indicated in the figure. The ATP used for the experiments shown in Fig. 5C was obtained from Sigma. ATP for the other reactions was obtained from ChemCyte. We have consistently seen more excision in assays that are performed with ChemCyte ATP; however, the rank order of the ability of the various mutant RTs to carry out excision is not affected. The reactions were halted by the addition of EDTA: the salts and nucleosides were removed by passage through a CentriSpin-10 column (Princeton Separations), and the template/primer was precipitated by the addition of ethanol. The products were fractionated on a 15% polyacrylamide sequencing gel. The total amount of template/primer (blocked and unextended plus deblocked and extended) and the amount of full-length product (deblocked and extended to the end of the template) were determined using a PhosphorImager.
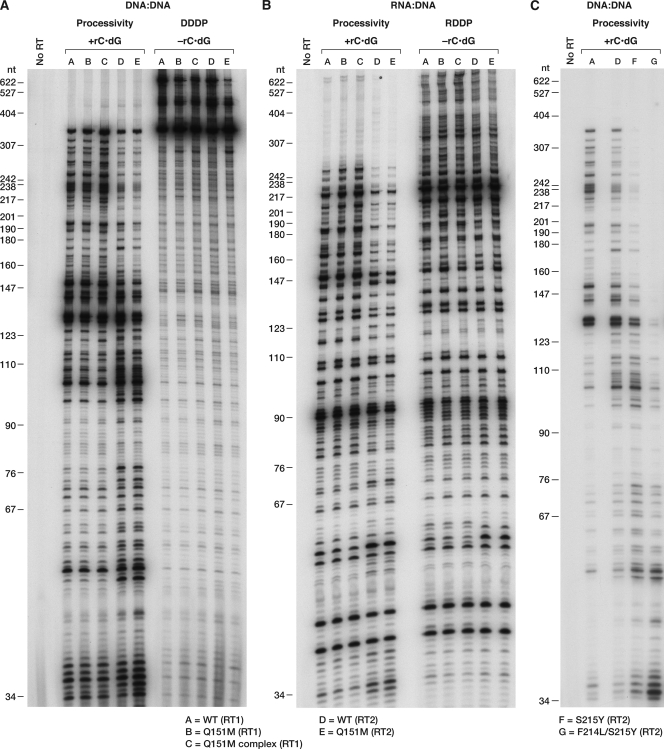
Fig 5.
Excision/extension assays. As described in Materials and Methods, a 5′-end-labeled primer was annealed to a template. The 3′ end of the primer was then blocked by the addition of AZTMP. The excision/extension assays were done in the presence of 3.0 mM ATP as the pyrophosphate donor and 10.0 μM each dNTP. All experiments were done at least twice at different times. When experiments were repeated with different batches of enzymes, the results were reproducible. Typical results are shown. The ATP used for the experiments shown in Fig. 5C was obtained from Sigma. ATP for other reactions was obtained from ChemCyte. We have consistently seen more excision in assays that are performed with ChemCyte ATP; however, the rank order of the ability of the various mutant RTs to carry out excision is not affected. (A) Excision and extension of an AZTMP-blocked primer by Q151M-containing RT variants on a DNA template/DNA primer. (B) Excision and extension of an AZTMP-blocked primer by Q151M-containing RT variants on an RNA template-DNA primer. (C) Excision and extension of an AZTMP-blocked primer by S215Y-containing HIV-2 RT variants on a DNA template/DNA primer. AZT-R is an HIV-1 variant that contains the mutations M41L, D67N, K70R, T215Y, and K219Y and is known to be ATP-dependent excision competent.
NRTI-TP inhibition assay.
A synthetic DNA oligonucleotide (5′-CAGGTCACTGTTCGAGCACCA-3′; Biosource, Camarillo, CA) was 5′-end labeled and then annealed to the DNA template (5′-GCGCAGTGTAGACAATCCCTAGCTATGGTGCTCGAACAGTGACCTG-3′) or the RNA template (5′-GCGCAGUGUAGACAAUCCCTAGCUAUGGUGCUCGAACAGUGACCUG-3′). The annealed template/primer was suspended in 50.0 μl of a mixture of 25 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg of bovine serum albumin per ml, 10.0 mM CHAPS, and 10.0 μM (each) dATP, dTTP, dGTP, and dCTP and then was incubated with 1.0 μg of RT in the presence of various concentrations (as described in the figure legends) of the triphosphate form of the analog for 10 min at 37°C. The products were purified and then fractionated on a 15% polyacrylamide sequencing gel. The total amount of template/primer (analog blocked products and full-length products) and the amount of full-length product (extended to the end of the template) were determined by using a PhosphorImager. The amount of full-length product in the absence of any NRTI-TP was considered 100% activity. The activity with the NRTI-TPs was normalized to that value. The percentage of full-length product was calculated by determining the amount of primer that was extended to the end of the template and dividing this amount by the amount of all extension products (full-length and NRTI-MP-blocked primers).
RNase H assay.
The RNase H assay has been previously described (6). The RNA oligonucleotide was obtained from Dharmacon Research, Inc., and has the sequence 5′-GGGCCACUUUUUAAAAGAAAAGGGGGGACUGGAAGGGCUAAUUCACUCAC-3′. This sequence matches the sequence of the HIV-1 RNA genome and extends from just 5′ of the poly(U) tract, through the PPT, and into the U3 region. The RNA oligonucleotide was 5′-end labeled and then annealed to a synthetic DNA oligonucleotide (5′-GAGTGAATTAGCCCTTCCAGTCCC-3′) by heating and slow cooling. A 0.2 μM concentration of template/primer was suspended in a mixture of 25 mM Tris (pH 8.0), 50 mM NaCl, 5.0 mM MgCl2, 100 μg of bovine serum albumin/ml, 10 mM CHAPS, and 1 U of Superasin (Ambion)/μl. The final reaction volume was 12 μl. The reactions were initiated by the addition of 35.0 nM RT and were incubated at 37°C. Aliquots of 2.5 μl were removed at the indicated time points, and the reactions were halted by the addition of 4.0 μl gel loading buffer (Ambion). The reaction products were fractionated on a 15% polyacrylamide sequencing gel. Products were visualized by exposure to X-ray film.
RESULTS
Polymerase assays.
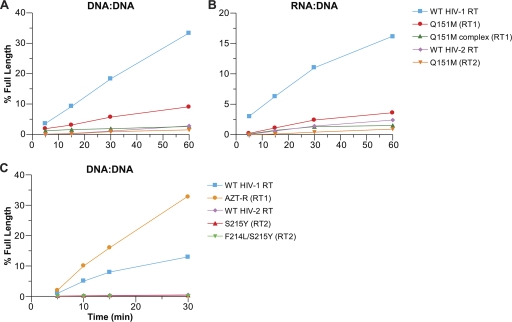
We initially tested the various RTs for their polymerase activities using both a standard polymerase assay and a processivity assay (Fig. 1). In the standard assay, RT is free to dissociate from the template primer and then rebind and continue synthesis. The processivity assay includes an unlabeled poly(rC) · oligo(dG) “cold trap,” which binds any RT that dissociates from the template/primer, limiting RT to one round of polymerization. If the RT disassociates from the labeled template/primer, it will bind to the “cold trap,” which is present in excess. The standard polymerase assay does not have the “cold trap,” and the RT can disassociate and reassociate with the labeled template/primer during the course of the reaction. To simplify the nomenclature, the wild-type forms of HIV-1 RT and HIV-2 RT are designated (RT1) and (RT2), respectively. HIV-1 RT and HIV-2 RT carrying the Q151M mutation are designated Q151M (RT1) and Q151M (RT2), respectively. The Q151M family of mutations (A62V, V75I, F77L, F116Y, and Q151M) in HIV-1 RT is designated the Q151M complex (RT1). It should be noted that in wild-type (WT) HIV-2 RT, the sequence normally contains an I75 residue. WT HIV-1 RT, Q151M (RT1), and Q151M complex (RT1) had similar processivities with a DNA template/DNA primer (single-stranded M13mp18 DNA plus a 5′-end-labeled primer) (Fig. 1A); WT HIV-2 RT had a lower processivity than did WT HIV-1 RT, which is in agreement with our previous results (5). Q151M (RT2) had a polymerase activity similar to that of WT HIV-2 RT. However, in the standard polymerase assay, it appeared that Q151M (RT2) was slightly less active than WT HIV-2 RT (Fig. 1A). The assays were also done with an RNA template/DNA primer; the results were similar (Fig. 1B).
Fig 1.
Processivity and polymerase activities of the wild-type and mutant HIV-1 and HIV-2 RTs. As described in Materials and Methods, a 5′-end-labeled primer was annealed to single-stranded M13mp18 DNA and then extended with wild-type RT or an RT variant in the presence of a 10.0 μM concentration of each dNTP. Processivity assays include unlabeled poly(rC) · oligo(dG), while the polymerase assays do not. The cold trap limits extension to one round of polymerization. The location of the size markers is shown on the left. (A) Processivity and polymerase activity of the Q151M-containing RT variants on a DNA template/DNA primer. (B) Processivity and polymerase activity of Q151M-containing RT variants on an RNA template/DNA primer. (C) Processivity activity of HIV-2 mutants containing S215Y and F214L/S215Y on a DNA template/DNA primer.
We also modified the region at amino acids (aa) 214 and 215 in HIV-2 RT. The serine at aa 215 was converted to a tyrosine [S215Y (RT2)]. This change is equivalent to one of the mutations known to confer AZT resistance in HIV-1 RT by increasing the ability of the mutant RT to excise AZTMP using ATP-mediated pyrophosphorolysis (described above). The HIV-2 RT we used in these experiments contains a phenylalanine at position 214; the HIV-1 RT we are using (derived from BH10) contains a leucine at this position. Therefore, we also altered the sequence in the HIV-2 RT coding region so that it would contain a leucine at position 214 [F214L S215Y (RT2)]. L214 is a common polymorphism in HIV-1 RT, but a leucine at this position appears to have a deleterious effect on AZT resistance when combined with T215Y (28). These mutant RTs were also tested for their polymerase activity. Mutations at the 214 and 215 positions in the HIV-2 RT had deleterious effects on the processivity of the polymerase: S215Y (RT2) had a slightly lower polymerase activity than WT (RT2), and F214L/S215Y (RT2) had a significantly lower activity (Fig. 1C).
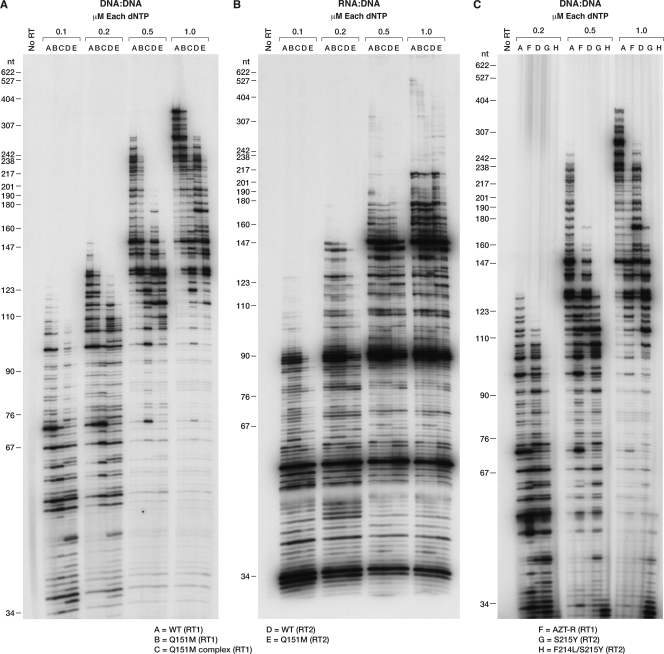
To test the effects of the mutations on the ability of RT to use suboptimal concentrations of dNTPs, a low-dNTP assay was performed. In these assays, the concentrations of each of the four dNTPs were 0.1, 0.2, 0.5, and 1.0 μM. With a DNA template/DNA primer, WT HIV-1 RT was better able to extend the primer than the other RTs we tested. This was true for all of the dNTP concentrations tested. The Q151M mutant (RT1) was slightly less able to extend the primer than WT HIV-1 RT; however, the polymerase activity of the Q151M complex (RT) was significantly impaired at the lowest dNTP concentrations (Fig. 2A). The WT HIV-2 RT was less able to extend the primer than WT HIV-1 RT, while Q151M (RT2) had a slight impairment of the polymerase activity compared to WT HIV-2 RT (Fig. 2A). The experiment was repeated using the RNA template/DNA primer combination (Fig. 2B). The differences between the various HIV-1 RTs were less apparent than when a DNA template/DNA primer was used. The polymerase activity of WT HIV-2 RT was lower than that of the HIV-1 RTs, while Q151M (RT2) was the most impaired RT (Fig. 2B).
Fig 2.
Extension of the primer in the presence of low dNTP concentrations. A 5′-end-labeled primer was annealed to single-stranded M13mp18 DNA or single-stranded RNA and extended with wild-type RT or an RT variant in the presence of 0.1, 0.2, 0.5, and 1.0 μM concentrations of each dNTP. The location of the size marker bands is shown on the left. (A) Low-dNTP polymerase activity of Q151M-containing RT variants on a DNA template/DNA primer. (B) Low dNTP polymerase activity of Q151M-containing RT variants on an RNA template/DNA primer. (C) Low dNTP polymerase activity of HIV-2 mutants containing S215Y and F214L/S215Y on a DNA template/DNA primer.
The S215Y (RT2) mutation in HIV-2 RT also had deleterious effects on the activity of the polymerase; this mutant was less active than WT (RT2) (Fig. 2C). The effects are particularly pronounced for the F214L/S215Y (RT2) double mutant, which is barely able to extend the primer.
Drug resistance assays. (i) NRTI-TP inhibition assays.
We previously described an NRTI inhibition assay that measured the incorporation of [α-32P]dCTP into the primer strand as the RT was polymerizing in either the presence or absence of an NRTI-TP (5). However, this assay may skew the apparent resistance of the RTs because the amount of radioactivity incorporated into the nucleic acid depends on how far the primer is extended. The RT that could, on average, extend the primer for the greatest distance would be the most vulnerable to inhibition by an NRTI-TP. This is because incorporation (on average) of 1 NRTI-TP every 50 nucleotides (nt) is sufficient to cause 50% decrease in incorporation if the product is 100 nt in length, but the incorporation of one NRTI-TP every 500 nt is sufficient to cause a 50% decrease if the product is 1,000 nt long. To circumvent this problem, synthetic oligonucleotide template/primers were designed. The primer is 5′-end labeled with [γ-32P]ATP, so that all of the template/primers have only one label per duplex. The amount of full-length product was measured in the absence of NRTI-TP, and this amount was set as 100%. The amount of full-length product in the presence of NRTI-TPs was normalized to this value. Both DNA template/DNA primer and RNA template/DNA primer were used in these assays, and both of the substrates have the same sequence.
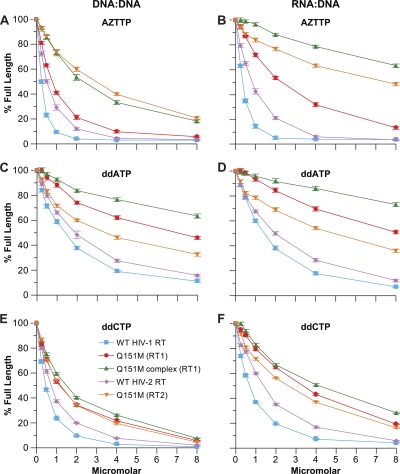
Because the Q151M mutation is known to selectively interfere with the incorporation of AZTTP (31), we first examined the effects of the mutations on the ability of the RTs to incorporate AZTTP. With a DNA template/DNA primer, Q151M (RT2) and Q151M complex (RT1) were the least susceptible RTs to inhibition by AZTTP, followed by Q151M (RT1), WT HIV-2 RT, and, lastly, WT HIV-1 RT, which was the most sensitive to AZTTP (Fig. 3A). A similar pattern is seen when RNA template/DNA primer was used, except in this case, the Q151M complex (RT1) was less susceptible to AZTTP than Q151M (RT2) (Fig. 3B). Two dideoxynucleotide NRTI-TPs, ddATP and ddCTP, were examined for their ability to inhibit the various RTs. For ddATP, the exclusion capability of the RTs was Q151M complex (RT1) > Q151M (RT1) > Q151M (RT2) > WT (RT2) > WT (RT1) for both a DNA template/DNA primer substrate and RNA template/DNA primer substrate (Fig. 3C and D). Similar results were obtained for ddCTP; however, the RT mutants we tested were less able, relative to WT, to avoid inhibition by ddCTP compared to ddATP (Fig. 3E and F).
Fig 3.
Inhibition of polymerization by the various NRTI-TPs. The assay was done as described in Materials and Methods. Normal dNTPs were present in the reaction mixture at 10.0 μM each. Increasing concentrations of the triphosphate form of the analog were added as shown. The amount of full-length product in the absence of analog was considered 100% activity; the other reactions were normalized to this value. The amount of full-length product in the absence of any NRTI-TP was considered 100% activity. The activity with the NRTI-TPs was normalized to that value. The percentage of full-length product was calculated by determining the amount of primer that was extended to the end of the template and dividing this amount by the amount of all extension products (full-length and NRTI-MP-blocked primers). Assays were done in triplicate. The error bars represent the standard deviation. (A) Inhibition by AZTTP on a DNA template/DNA primer. (B) Inhibition by AZTTP on an RNA template/DNA primer. (C) Inhibition by ddATP on a DNA template/DNA primer. (D) Inhibition by ddATP on an RNA template/DNA primer. (E) Inhibition by ddCTP on a DNA template/DNA primer. (F) Inhibition by ddCTP on an RNA template/DNA primer.
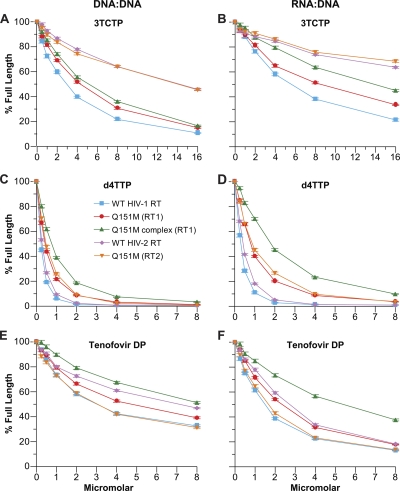
The Q151M-based mutations in HIV-1 RT were originally selected by treatments that included AZT and ddI. Since that time, other NRTIs have been approved for the treatment of HIV-1 infection. As described in the introduction, several of these NRTIs have also been used for the treatment of HIV-2 infection, and Smith et al. (36) have examined the effects of the compounds on HIV-2, primarily in virus replication assays in cell culture. We tested the triphosphate forms of some of these inhibitors in vitro to see how their presence would affect the ability of the RT mutants to polymerize. Like the dideoxynucleotides described above, 3TC does not contain a 3′ OH group and acts as a chain terminator (e.g., see references 11 and 31). The Q151M mutation does not appear to be selected by 3TC treatment; 3TC selects for M184I/V. In our polymerization assays, using a DNA template/DNA primer substrate, all of the HIV-1 RTs were sensitive to inhibition by 3TC triphosphate (3TCTP); however, Q151M (RT1) and Q151M complex (RT1) were slightly less sensitive than WT (RT1) (Fig. 4A). WT HIV-2 RT and Q151M (RT2) excluded 3TCTP more effectively than the corresponding HIV-1 RT variants. A similar pattern was seen using an RNA template/DNA primer substrate, except that Q151M (RT1) and the Q151M complex (RT1) were slightly more resistant to 3TCTP. Both WT HIV-2 RT and Q151M (RT2) were moderately resistant to inhibition by 3TCTP (Fig. 4B), which agrees with the data reported by Smith et al. (35). Stavudine triphosphate (d4TTP), which also lacks a 3′-OH group, and acts as a chain terminator, was able to inhibit DNA synthesis by all of the RTs when a DNA template/DNA primer substrate was used. d4TTP yields results similar to ddATP and ddCTP: Q151M complex (RT1) > Q151M (RT2) ≥ Q151M (RT1) > WT (RT2) > WT (RT1) (Fig. 4C). Similar results were obtained with an RNA template/DNA primer substrate, except that there was a larger difference between Q151M complex (RT1) and the WT RTs (Fig. 4D). The pattern with tenofovir diphosphate (tenofovir-DP) was somewhat different. While Q151M complex (RT1) was the most resistant to the analog, WT (RT2) was also able to exclude the analog to a greater extent than Q151M (RT1). Q151M (RT2) was relatively susceptible to inhibition by tenofovir-DP. The susceptibility of this RT2 mutant was similar to that of WT (RT1) (Fig. 4E and F).
Fig 4.
Inhibition of polymerization by the various NRTI-TPs. The assay was done as described in Materials and Methods. Normal dNTPs were present in the reaction mixture at 10.0 μM each. Increasing concentrations of the triphosphate form of the analog were added as shown. The amount of full-length product in the absence of analog was considered 100% activity; the other reactions were normalized to this value. The amount of full-length product in the absence of any NRTI-TP was considered 100% activity. The activity with the NRTI-TPs was normalized to that value. The percentage of full-length product was calculated by determining the amount of primer that was extended to the end of the template and dividing this amount by the amount of all extension products (full-length and NRTI-MP-blocked primers). Assays were done in triplicate. The error bars represent the standard deviation. (A) Inhibition by 3TCTP on a DNA template/DNA primer. (B) Inhibition by 3TCTP on an RNA template/DNA primer. (C) Inhibition by d4TTP on a DNA template/DNA primer. (D) Inhibition by d4TTP on an RNA template/DNA primer. (E) Inhibition by tenofovir-DP on a DNA template/DNA primer. (F) Inhibition by tenofovir-DP on an RNA template/DNA primer.
(ii) Excision assays.
To verify that the mode of action of these resistant mutants was exclusion of AZTTP, the Q151M RT variants were tested for their ability to excise AZTMP from the end of the primer, using ATP as the pyrophosphate donor. As shown in Fig. 5A and B, while WT (RT1) was able to excise AZTMP to some extent and extend the newly unblocked primer with either an RNA or a DNA template, both the RT2 variants and WT (RT2) had a very limited ability to excise AZTMP (Fig. 5A and B). Therefore, it is unlikely that the resistance seen with any of these mutants involves excision of the analog from the primer. In a reciprocal experiment, the WT (RT2) coding region was altered to contain the mutation S215Y. In HIV-1 RT, T215Y is one of the primary resistance mutations; T215Y, by itself, is sufficient to cause a measurable increase in AZTMP excision (4). AZT-R, an HIV-1 variant containing the mutations M41L, D67N, K70R, T215Y, and K219Q, is known to be excision competent and was included as a positive control. The S215Y mutation did not increase the ability of the HIV-2 RT to excise AZTMP from the end of a primer. Because the HIV-1 RT was derived from BH10, which contains L214, the HIV-2 ROD RT was modified so that it also contained the double mutation F214L/S215Y. This mutant was unable to excise AZTMP from the end of the primer (Fig. 5C), suggesting that HIV-2 RT cannot use these mutations to improve ATP-mediated excision.
It has been suggested that mutations that cause a reduction in RNase H activity give RT more time to carry out excision of a nucleoside analog during the synthesis of the first DNA strand, which enhances resistance via the excision mechanism (9). To make certain that the mutations we are testing do not significantly affect the RNase H activity, an RNA template/DNA primer combination was treated with the various RTs for various lengths of time. All of the RT1 and RT2 variants have RNase H activities similar to their respective WT RT (data not shown). The RNase H activity of the HIV-2 RTs is slightly less than that seen for the HIV-1 RTs, which matches published data (15).
DISCUSSION
Any drug-resistant mutant represents a compromise between a gain in the ability of the target protein to evade the inhibitor and a potential loss in the protein's ability to perform the function(s) necessary for the replication of the virus. As previously described, HIV-1 RT and HIV-2 RT are sufficiently different from each other that challenges with NRTIs can elicit mutations that are in different resistance pathways (5, 35). Treatment of HIV-1 infections with AZT alone tends to select for a family of mutations that usually involve the T215Y/F and/or K70R amino acid change. There is good evidence that the mechanism of resistance to AZT caused by these mutations involves the excision of the blocking AZTMP group from the end of the primer, in a reaction that uses ATP as the pyrophosphate donor. In contrast, the Q151M complex in HIV-1 was originally selected in humans by a drug treatment containing both AZT and ddI (20, 39). These mutations allow HIV-1 RT to preferentially select and incorporate normal dNTPs instead of the NRTI-TPs while retaining sufficient polymerase activity to complete the conversion of the RNA genome into DNA. Unlike HIV-1RT, HIV-2 RT preferentially uses the Q151M mutation for AZT resistance; examination of the Stanford Resistance Database reveals few examples of mutations associated with the ATP-dependent excision mechanism (K70R or T215Y/F) being selected in the RT of HIV-2.
With HIV-1 RT, our results indicate that the mutations that are selected involve a tradeoff between a loss of polymerase activity and acquisition of drug resistance. In terms of resistance, WT HIV-1 RT was the least able to selectively incorporate TTP relative to AZTTP (i.e., to exclude AZTTP) or to choose dATP over ddATP regardless of whether the template was RNA or DNA. The Q151M (RT1) mutation caused an increase in the ability of RT to exclude the analogs AZTTP and ddATP compared to WT (RT1), while the Q151M complex (RT1) was significantly better able to exclude the analogs while retaining the ability to incorporate the normal dNTPs. Broadly speaking, similar results were obtained when ddCTP was used, but the degree of discrimination between the ddCTP and dCTP was lower with a DNA template than an RNA template. The degree of selectivity conferred by the Q151M mutation appears to depend on the exact nature of the NRTI-TP. HIV-1 RT variants containing Q151M appeared to be susceptible to inhibition by d4TTP or 3TCTP when the polymerization template was DNA. However, the RT variants containing Q151M appeared to cause a modest reduction in susceptibility to d4TTP and 3TCTP when the template was RNA, suggesting that any effect of the mutations on the susceptibility of HIV-1 to these compounds may preferentially occur during first-strand DNA synthesis. It appeared that the HIV-1 RT Q151M variants were not susceptible to inhibition by tenofovir-DP during polymerization with either a DNA or RNA template.
In terms of the impact of the mutations on polymerase activity, there was little or no difference between WT (RT1), Q151M (RT1), and the Q151M complex (RT1) in the processivity or the standard polymerase assay (Fig. 1A and B), although there was a distinct decrease in the ability of the mutant RTs to polymerize at low dNTP concentrations (Fig. 2A and B). This difference was especially marked when the reaction mixtures contained a DNA template/DNA primer combination. We originally thought that the secondary mutations in the Q151M complex (RT1) were compensatory mutations, whose primary role was to improve polymerase activity of RTs that had acquired the Q151M mutation. However, it appears that the opposite is true. As stated above, Q151M forms part of the dNTP binding site of HIV-1 RT, and F116Y is located nearby. A62V, V75I, and F77L are located in the β3-β4 loop of the fingers subdomain. This portion of the p66 fingers subdomain closes down onto the incoming dNTP after it binds at the N site (30). The Q151M mutation has been proposed to alter the hydrogen bond interactions between HIV-1 RT and the incoming dNTP or incoming NRTI-TP in a way that emphasizes the importance of the 3′-OH, allowing the Q151M mutant to reduce the incorporation of the NRTI-TP while retaining the ability to incorporate the correct dNTP. However, these altered interactions also decrease the overall interaction with a normal dNTP, an effect that was most evident in the experiments done at low dNTP concentrations. In actively dividing cells, this effect may not be as important; however, HIV-1 RT is known to infect nondividing cells, where the concentration of the four dNTPs can be much lower (3, 7, 10, 33, 34). Thus, with the Q151M complex (RT1), high levels of drug resistance may come at the price of decreasing the ability of the virus to replicate in nondividing cells. Because the Q151M complex (RT1) mutations have deleterious effects on the polymerase activity of RT, the alternative resistance mechanism, the excision pathway, which has only a modest impact on polymerase activity, will be the pathway that is preferentially selected.
It is also clear that the mechanism of NRTI resistance caused by the Q151M mutation is exclusively due to exclusion. The Q151M mutants are actually worse at excision than WT RT1. This result makes sense in light of the fact that the excision reaction is closely related to the polymerase reaction run in reverse. None of the mutations in the Q151M complex are in positions where they would be expected to enhance ATP binding (37). Without enhanced ATP binding to HIV-1 RT, the excision reaction would depend primarily on the polymerase activity of the RT variants. Because the Q151M mutation is less active than WT HIV-1 RT and the Q151M complex (RT1) is even less active, it is not surprising that these mutant RTs are also less proficient at excision than WT RT.
WT HIV-2 RT has a decreased level of polymerase activity compared to WT HIV-1 RT. The processivity of WT HIV-2 RT is lower than that of WT RT1, and WT HIV-2 RT is less able to polymerize at low dNTP concentrations. The presence of the Q151M mutation in HIV-2 RT (WT RT2 normally contains I75) caused a further reduction in the polymerase activity at low dNTP concentrations. However, the Q151M mutation (perhaps acting together with I75) greatly increased the ability of HIV-2 RT to exclude AZTTP to levels comparable to those of Q151M complex (RT1). The RT2 Q151M mutant was also able to exclude ddATP to a significant extent. Because the mutation impairs the ability of HIV-2 RT to incorporate dNTPs at low concentrations, it is likely that the HIV-2 virus carrying the Q151M mutation would encounter many of the same problems in the host that an HIV-1 virus carrying the Q151M complex mutations would have. We originally proposed that the different RTs chose the two different pathways based on the properties of the respective WT enzymes. We showed that the polymerase activity of WT HIV-2 RT was somewhat less susceptible to inhibition by AZTTP than WT HIV-1 RT, while WT HIV-1 RT had a higher level of excision activity than WT HIV-2 RT. This proposal was based on the idea that these two different RTs chose their preferred resistance pathways based primarily on which of the two resistance mechanisms was a better match to the properties of the respective WT enzymes. The data presented here clarify the processes that affect the choice of a resistance mechanism by these two RTs and suggest that the driving force for HIV-2 RT to use the exclusion pathway based on the Q151M mutation is that the excision pathway is simply not available to HIV-2 RT. In HIV-2 RT, the presence of the S215Y mutation does not enhance ATP-dependent pyrophosphorolysis to any detectable extent, and this mutation has a negative impact on polymerase activity. As might be expected, very few HIV-2 isolates carrying this mutation were found in the Stanford database. The addition of F214L, which would make the sequence of HIV-2 RT in the region where ATP binds more similar to the sequence found in the BH10 derivative of HIV-1 RT, had no effect on ATP-dependent pyrophosphorylysis and had an even more profound negative effect on polymerase activity. Because the excision pathway is apparently unavailable to HIV-2 RT, the Q151M exclusion pathway is used, despite the fact that it has a negative impact on polymerase activity. In contrast, HIV-1 RT seems to use the Q151M exclusion pathway only in response to certain defined NRTI combinations, presumably because these combinations contain NRTIs that are not susceptible to the excision mechanism, and when it can be used, the excision pathway is the preferred resistance mechanism.
Better access to anti-HIV drugs in areas in which HIV-2 is endemic has led to therapies in which HIV-2 RT is challenged by various NRTIs. Interestingly, in an in vitro polymerization assay, WT HIV-2 RT appears to have only a modest susceptibility to 3TCTP with either an RNA or DNA template. However, the fact that 3TC treatment selects the M184V mutation in HIV-2 suggests that the modest susceptibility of HIV-2 RT to 3TC is not sufficient. It would appear that for 3TC, only the essentially complete loss of susceptibility conferred by the M184V/I mutation is sufficient. Presumably, the value of an essentially complete loss of susceptibility outweighs the known fitness costs of the M184V mutation (8, 18, 27).
These results highlight the fact that HIV-1 and HIV-2 RTs are different, and despite the fact that some resistance mutations are found in common between the two (such as K65R and M184V), treatments containing certain NRTIs (for example AZT) can not only select for different resistance mutations, but also select for mutations that involve distinct resistance pathways. This reinforces the need to monitor the genotype of the infecting virus and determine how the genotype correlates with the level of drug susceptibility, and points to the need to develop and use animal models for NRTI testing in which the virus used in the experiment replicates using HIV-1 RT.
ACKNOWLEDGMENTS
This study was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research, and in part with federal funds from the National Cancer Institute, NIH, under contract HHSN26120080001E.
We thank Alan Kane for help with the figures and Teresa Burdette with assistance with the preparation of the manuscript.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 21 March 2012
REFERENCES
- 1. Ahluwalia G, et al. 1987. Initial studies on the cellular pharmacology of 2′,3′-dideoxyinosine, an inhibitor of HIV infectivity. Biochem. Pharmacol. 36:3797–3800 [DOI] [PubMed] [Google Scholar]
- 2. Basavapathruni A, Anderson KS. 2007. Reverse transcription of the HIV-1 pandemic. FASEB J. 21:3795–3808 [DOI] [PubMed] [Google Scholar]
- 3. Bofill M, Fairbanks LD, Ruckemann K, Lipman M, Simmonds HA. 1995. T-lymphocytes from AIDS patients are unable to synthesize ribonucleotides de novo in response to mitogenic stimulation. Impaired pyrimidine responses are already evident at early stages of HIV-1 infection. J. Biol. Chem. 270:29690–29697 [PubMed] [Google Scholar]
- 4. Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. 2006. Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? PLoS Pathog. 2:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boyer PL, Stenbak CR, Clark PK, Linial ML, Hughes SH. 2004. Characterization of the polymerase and RNase H activities of human foamy virus reverse transcriptase. J. Virol. 78:6112–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cohen A, Barankiewicz J, Lederman HM, Gelfand EW. 1983. Purine and pyrimidine metabolism in human T lymphocytes. Regulation of deoxyribonucleotide metabolism. J. Biol. Chem. 258:12334–12340 [PubMed] [Google Scholar]
- 8. Deeks SG, et al. 2005. Interruption of treatment with individual therapeutic drug classes in adults with multidrug-resistant HIV-1 infection. J. Infect. Dis. 192:1537–1544 [DOI] [PubMed] [Google Scholar]
- 9. Delviks-Frankenberry KA, et al. 2008. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc. Natl. Acad. Sci. U. S. A. 105:10943–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fairbanks LD, Bofill M, Ruckemann K, Simmonds HA. 1995. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J. Biol. Chem. 270:29682–29689 [PubMed] [Google Scholar]
- 11. Gao HQ, Boyer PL, Sarafianos SG, Arnold E, Hughes SH. 2000. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J. Mol. Biol. 300:403–418 [DOI] [PubMed] [Google Scholar]
- 12. Gotte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SF. 2010. Reverse transcriptase in motion: conformational dynamics of enzyme-substrate interactions. Biochim. Biophys. Acta 1804:1202–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gottlieb GS, et al. 2009. Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resource-limited West Africa. Clin. Infect. Dis. 48:476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herschhorn A, Hizi A. 2010. Retroviral reverse transcriptases. Cell. Mol. Life Sci. 67:2717–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hizi A, Tal R, Shaharabany M, Loya S. 1991. Catalytic properties of the reverse transcriptases of human immunodeficiency viruses type 1 and type 2. J. Biol. Chem. 266:6230–6239 [PubMed] [Google Scholar]
- 16. Huang H, Chopra R, Verdine GL, Harrison SC. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669–1675 [DOI] [PubMed] [Google Scholar]
- 17. Ibe S, Sugiura W. 2011. Clinical significance of HIV reverse-transcriptase inhibitor-resistance mutations. Future Microbiol. 6:295–315 [DOI] [PubMed] [Google Scholar]
- 18. Jain V, et al. 2011. Differential persistence of transmitted HIV-1 drug resistance mutation classes. J. Infect. Dis. 203:1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jallow S, et al. 2009. Virological response to highly active antiretroviral therapy in patients infected with human immunodeficiency virus type 2 (HIV-2) and in patients dually infected with HIV-1 and HIV-2 in the Gambia and emergence of drug-resistant variants. J. Clin. Microbiol. 47:2200–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maeda Y, Venzon DJ, Mitsuya H. 1998. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J. Infect. Dis. 177:1207–1213 [DOI] [PubMed] [Google Scholar]
- 21. Martinez-Picado J, Martinez MA. 2008. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res. 134:104–123 [DOI] [PubMed] [Google Scholar]
- 22. Matamoros T, Nevot M, Martinez MA, Menendez-Arias L. 2009. Thymidine analogue resistance suppression by V75I of HIV-1 reverse transcriptase: effects of substituting valine 75 on stavudine excision and discrimination. J. Biol. Chem. 284:32792–32802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Menendez-Arias L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 134:124–146 [DOI] [PubMed] [Google Scholar]
- 24. Ndembi N, et al. 2008. Molecular characterization of human immunodeficiency virus type 1 (HIV-1) and HIV-2 in Yaounde, Cameroon: evidence of major drug resistance mutations in newly diagnosed patients infected with subtypes other than subtype B. J. Clin. Microbiol. 46:177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ntemgwa ML, d'Aquin Toni T, Brenner BG, Camacho RJ, Wainberg MA. 2009. Antiretroviral drug resistance in human immunodeficiency virus type 2. Antimicrob. Agents Chemother. 53:3611–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ntemgwa ML, et al. 2009. Nucleoside and nucleotide analogs select in culture for different patterns of drug resistance in human immunodeficiency virus types 1 and 2. Antimicrob. Agents Chemother. 53:708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Paredes R, et al. 2009. In vivo fitness cost of the M184V mutation in multidrug-resistant human immunodeficiency virus type 1 in the absence of lamivudine. J. Virol. 83:2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Puertas MC, et al. 2009. Effect of the human immunodeficiency virus type 1 reverse transcriptase polymorphism Leu-214 on replication capacity and drug susceptibility. J. Virol. 83:7434–7439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruelle J, et al. 2008. Transmitted drug resistance, selection of resistance mutations and moderate antiretroviral efficacy in HIV-2: analysis of the HIV-2 Belgium and Luxembourg database. BMC Infect. Dis. 8:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruelle J, et al. 2007. Genetic polymorphisms and resistance mutations of HIV type 2 in antiretroviral-naive patients in Burkina Faso. AIDS Res. Hum. Retroviruses 23:955–964 [DOI] [PubMed] [Google Scholar]
- 31. Sarafianos SG, Das K, Hughes SH, Arnold E. 2004. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr. Opin. Struct. Biol. 14:716–730 [DOI] [PubMed] [Google Scholar]
- 32. Sarafianos SG, et al. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scarth B, McCormick S, Gotte M. 2011. Effects of mutations F61A and A62V in the fingers subdomain of HIV-1 reverse transcriptase on the translocational equilibrium. J. Mol. Biol. 405:349–360 [DOI] [PubMed] [Google Scholar]
- 34. Smith AJ, Meyer PR, Asthana D, Ashman MR, Scott WA. 2005. Intracellular substrates for the primer-unblocking reaction by human immunodeficiency virus type 1 reverse transcriptase: detection and quantitation in extracts from quiescent- and activated-lymphocyte subpopulations. Antimicrob. Agents Chemother. 49:1761–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith AJ, Scott WA. 2006. The influence of natural substrates and inhibitors on the nucleotide-dependent excision activity of HIV-1 reverse transcriptase in the infected cell. Curr. Pharm. Des. 12:1827–1841 [DOI] [PubMed] [Google Scholar]
- 36. Smith RA, Anderson DJ, Pyrak CL, Preston BD, Gottlieb GS. 2009. Antiretroviral drug resistance in HIV-2: three amino acid changes are sufficient for classwide nucleoside analogue resistance. J. Infect. Dis. 199:1323–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trevino A, et al. 2011. Drug resistance mutations in patients infected with HIV-2 living in Spain. J. Antimicrob. Chemother. 66:1484–1488 [DOI] [PubMed] [Google Scholar]
- 38. Tu X, et al. 2010. Structural basis of HIV-1 resistance to AZT by excision. Nat. Struct. Mol. Biol. 17:1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ueno T, Shirasaka T, Mitsuya H. 1995. Enzymatic characterization of human immunodeficiency virus type 1 reverse transcriptase resistant to multiple 2′,3′-dideoxynucleoside 5′-triphosphates. J. Biol. Chem. 270:23605–23611 [DOI] [PubMed] [Google Scholar]
- 40. Vu BC, Boyer PL, Siddiqui MA, Marquez VE, Hughes SH. 2011. 4′-C-methyl-2′-deoxyadenosine and 4′-C-ethyl-2′-deoxyadenosine inhibit HIV-1 replication. Antimicrob. Agents Chemother. 55:2379–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]