Abstract
Genetic drift of influenza virus genomic sequences occurs through the combined effects of sequence alterations introduced by a low-fidelity polymerase and the varying selective pressures experienced as the virus migrates through different host environments. While traditional phylogenetic analysis is useful in tracking the evolutionary heritage of these viruses, the specific genetic determinants that dictate important phenotypic characteristics are often difficult to discern within the complex genetic background arising through evolution. Here we describe a novel influenza virus sequence feature variant type (Flu-SFVT) approach, made available through the public Influenza Research Database resource (www.fludb.org), in which variant types (VTs) identified in defined influenza virus protein sequence features (SFs) are used for genotype-phenotype association studies. Since SFs have been defined for all influenza virus proteins based on known structural, functional, and immune epitope recognition properties, the Flu-SFVT approach allows the rapid identification of the molecular genetic determinants of important influenza virus characteristics and their connection to underlying biological functions. We demonstrate the use of the SFVT approach to obtain statistical evidence for effects of NS1 protein sequence variations in dictating influenza virus host range restriction.
INTRODUCTION
Influenza A virus belongs to the Orthomyxoviridae family (7, 22) and has an enveloped virion that contains a genome made of eight single-stranded negative-sense RNA segments that code for either 10 or 11 known proteins, including the surface glycoproteins hemagglutinin (HA) and neuraminidase (NA), matrix and ion-channel proteins (M1 and M2), RNA polymerase subunits (PB1, PB2, and PA), nucleoprotein (NP), and nonstructural proteins (NS1 and NS2/NEP), with some strains encoding an additional proapoptotic protein, PB1-F2. Reassortment among the various influenza virus genome segments occurs frequently (9) and is particularly observable among the surface glycoproteins. This phenomenon gives rise to different combinations of serologically distinct subtypes of HA and NA that circulate in host populations. Waterfowl are thought to be the natural reservoirs of influenza virus; however, the virus is also known to infect several other hosts, including human, swine, horse, dog, etc., in addition to a wide variety of avian species (14).
A thorough understanding of any pathogen, including influenza virus, requires an understanding of how variations in the sequence of the pathogen genome (genotype) are expressed as differences in the functional characteristics of the pathogen (phenotype). It is well known that influenza virus sequences constantly evolve by accumulating mutations through a process termed “genetic drift,” in which sequence variations introduced by the virus's low-fidelity polymerase are selected to preserve important structural and functional protein characteristics while attempting to evade host immune system recognition.
Comparative genomics studies have largely been restricted to phylogenetic analysis of whole-genome segments or statistical association of sequence variations at single residue positions and their effects on specific phenotypic characteristics. However, these traditional approaches to comparative genomics have certain limitations. Single-residue analysis does not take into account the impact of other genomic residues on the phenotype of interest. Whole-genome segment analysis does not highlight the specific regions responsible for the phenotypic effect. In addition, while the ancestry of genetic variants resulting from the cumulative effects of evolution can be revealed in the phylogenetic tree topology, phenotypic changes arising from convergent evolution are not revealed through phylogenetic tree reconstruction. Sequence variations can also influence virus traits that may not be subject to strong natural selective pressures in the reservoir host, including host range specificity (1, 15, 16), interspecies transmissibility (1, 4), altered replication (13, 21), virulence and pathogenicity in human (1, 15), and temperature sensitivity (19). Consequently, traditional whole-segment phylogenetic analysis may not reveal the most clinically and epidemiologically relevant sequence alterations, since the relationships between certain specific phenotypic changes and their underlying genotypic variations may be masked by the complex global effects of evolutionary selection on the entire viral genome.
To address these limitations, we have developed a novel method for studying the effects of sequence variation on organism phenotypes called the sequence feature variant type (SFVT) approach, wherein combinations of amino acid positions are defined as discrete sequence features (SFs) based on structural and functional characteristics. The extent of sequence variation can be determined for each SF independently as a set of variant types (VTs) for the SF, which can then be used for statistical analysis of genotype-phenotype associations. The SFVT approach was first described for HLA-disease association in the setting of human autoimmune disease (6). Here we describe the development and application of the SFVT approach for the study of influenza virus.
MATERIALS AND METHODS
Definition of influenza virus protein sequence features.
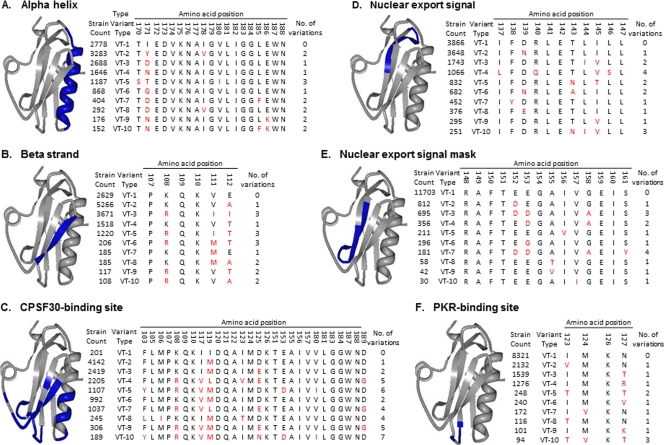
A sequence feature (SF) describes a specific region of a protein (or RNA or DNA molecule) possessing some characteristic of interest, for example a known structural property (e.g., a particular alpha helix or beta strand) (Fig. 1A and B), functional property (e.g., an enzyme active site, a nuclear localization signal, or a region mediating protein-protein interactions) (Fig. 1C to F), sequence alteration effect (e.g., a position in which a sequence alteration causes drug resistance), or immune epitope location. For proteins, the formal definition of an SF is the specific string of amino acid positions that makes up the defined region in a particular reference strain. A given SF can be a contiguous stretch of amino acids in the linear sequence (e.g., Fig. 1A) or can be discontinuous (e.g., Fig. 1C), as is often the case for enzyme active sites or antibody epitopes. There is no limitation on the size of an SF, as it can range from a single amino acid position with an interesting characteristic to a complete protein sequence. Individual SFs can also overlap each other such that a given amino acid position can be part of several different SFs (e.g., the CPSF30-binding site SF shown in Fig. 1C shares part of the beta-strand SF shown in Fig. 1B).
Fig 1.
Examples of selected NS1 SFs and corresponding VTs. Examples of two different structural and four different functional SFs are shown highlighted in blue on the 3-dimensional protein structure of the influenza A virus NS1 protein (PDB identifier 2GX9) on the left side of each panel: Influenza virus A_NS1_alpha-helix_170(19) (A), Influenza virus A_NS1_beta-strand_107(6) (B), Influenza virus A_NS1_CPSF30-binding-site_103(28) (C), Influenza virus A_NS1_nuclear-export-signal_137(11) (D), Influenza virus A_NS1_NES-mask_148(14) (E), and Influenza virus A_NS1_PKR-binding-site_123(4) (F). The right side of each panel shows tables of the first 10 VTs for each of these SFs with the amino acid residues that differ from the A/Udorn/1972 reference strain highlighted in red. The number of influenza virus strains that carry the given VT (strain count) is shown on the left side of the table. The number of amino acid positions that vary in comparison to the Udorn reference (no. of variations) is shown on the right side; the number of strains that bear the same amino acid sequence VT as the Udorn reference range from 201 for the CPSF30-binding site SF to 11,703 for the nuclear export signal mask SF.
SFs for influenza virus proteins were defined using information from the public domain databases UniProt (www.uniprot.org) and the Immune Epitope Database (IEDB, www.iedb.org), and from the published scientific literature. The initial list of SFs was defined by mining structural and functional definitions from UniProt records that include manually curated and computationally derived protein features that describe functional motifs, subcellular localization signals, domain structures, secondary structural elements, sites of posttranslational modifications, and sites of virus-virus and virus-host interactions. Experimentally determined immune epitopes (major histocompatibility complex [MHC] class I and II-dependent T-cell and B-cell/antibody epitopes) curated through the efforts of the IEDB were also included. Publications containing additional information about protein regions that impact antigenicity, virulence, pathogenicity, temperature sensitivity, transmission efficacy, host receptor binding, enzyme catalysis, and antiviral drug resistance were identified using PubMed searches (www.ncbi.nlm.nih.gov/pubmed). The resulting publications were then manually curated to identify the amino acid position(s) defining the described SF. This initial list of SFs was independently validated by scientists having expertise in each protein, who reviewed the SF names and positions for accuracy and added missing SFs to the list.
For each influenza virus protein, a standard reference strain was selected to define the specific amino acid positions that make up each SF. Reference strains were chosen based on the availability of experimentally determined crystal structures for the protein and their prominence in experimental studies (Table 1).
Table 1.
SF reference strains for each influenza protein
| Segment | Protein | Serotype | Reference strain | GenBank accession no. |
|---|---|---|---|---|
| 1 | PB2 | A/Vietnam/1203/2004 | EF467805 | |
| 2 | PB1 | A/Hong Kong/156/1997 | AF036362 | |
| 2 | PB1-F2 | A/WSN/1933 | CY034138 | |
| 3 | PA | A/WSN/1933 | CY034137 | |
| 4 | HA | H1 | A/California/04/2009 | FJ966082 |
| H2 | A/Japan/305/1957 | J02127 | ||
| H3 | A/Aichi/2/1968 | AB284320 | ||
| H5 | A/Vietnam/1203/2004 | AY818135 | ||
| H7 | A/Turkey/Italy/220158/2002 | AY586409 | ||
| 5 | NP | A/WSN/1933 | CY034135 | |
| 6 | NA | N1 | A/California/07/2009 | FJ984386 |
| N2 | A/Tokyo/3/1967 | U38242 | ||
| 7 | M | A/Udorn/1972 | J02167 | |
| 8 | NS | A/Udorn/1972 | V01102 |
Each SF was annotated with additional relevant information about its characteristics, including a descriptive name, the influenza virus segment and protein in which it occurs, the amino acid start and stop positions, the total length, SF category (i.e., structural, functional, sequence alteration, or immune epitope location), the name of the influenza virus strain in which it was characterized (which often differed from the reference strain), any known associated phenotype, evidence codes indicating how the SF was inferred (e.g., whether it is curated from literature or inferred from electronic annotations), and the publication or resource wherein it was initially described.
The SF descriptive name was assigned using the following syntax wherein every portion of the name is delimited by an underscore: Influenza virus type_protein symbol_sequence feature type_start position of the SF (total length of the SF). For example, the Influenza virus A_H1_cytoplasmic-domain_550(16) SF delineates the cytoplasmic domain of the hemagglutinin (HA) protein for subtype H1 of type A influenza virus, which starts at residue 550 and has a total length of 16 amino acids. In this case, the SF is made of a continuous string of amino acids between residues 550 and 565. Influenza virus A_H1_sialic-acid-binding-site_98(17) is an example of a discontinuous SF of the HA protein (defined as the set of amino acid positions 98, 134 to 138, 154, 156, 184, 191, 195, 196, and 225 to 229) that is involved in binding to sialic acid molecules on host cells. Sequential unique database identifiers were then assigned to each SF, starting with Influenza virus A_protein symbol_SF1 for each protein (e.g., Influenza virus A_PB1_SF1).
Determination of influenza virus sequence feature VTs.
The extent of sequence variation for each SF is determined as a collection of variant types (VTs) computed after multiple sequence alignments of all relevant influenza virus genomes available in the Influenza Research Database (IRD). Figure 1 shows the first 10 VTs for each of the SFs displayed. The first VT (VT-1) corresponds to the specific sequence string found in the reference strain for that protein. All influenza virus strains in IRD with amino acid sequences identical to those of the reference strain in that region are categorized as belonging to VT-1 for that SF. The rest of the VTs are ordered based on decreasing frequency of representation in the database as of November 2011. For example, in the NS1 nuclear export signal mask SF, the vast majority of virus strains (11,703) share the same amino acid sequences with the reference strain (A/Udorn/1972) and thus bear the VT-1 for this SF (Fig. 1E). In contrast, few strains (201) share the VT-1 reference amino acid sequence for the NS1 CPSF30-binding site SF (Fig. 1C). VT-unknown indicates that the sequence is either not completely defined or else artificially truncated; hence the SF cannot be defined for those strains.
The end result is that each set of unique amino acid residue combinations existing within any characterized region of the protein is defined as an individual sequence feature variant type (SFVT). The SFVT system is made freely available online to the influenza virus research community through the NIAID-funded, public Influenza Research Database (IRD, www.fludb.org) resource (18).
Statistical analyses of the SFVTs of Influenza virus A_NS1_SF18.
Statistical tests were performed using the R package (5) and the Microsoft Excel program. For convenience, and to maintain optimal statistical power, we grouped the virus hosts into seven broad categories: human, avian (excluding chicken), chicken, swine, equine, environmental samples, and other host species. We performed two sets of Pearson's chi-square tests of independence for the following hypotheses in order to identify any relationship existing between host groups and the first 16 VTs of Influenza virus A_NS1_SF18: (i) VT distribution across/between host groups (each host group has an equal probability of containing a particular VT [16 tests were performed]) and (ii) VT distribution within a host group (each host group has an equal probability of including all 16 VTs [6 tests were performed]). P values were then calculated for the above hypotheses.
Correction for geotemporal data bias.
To perform the corrections for temporal bias, we assumed that the total number of viruses for all host groups should be uniform across all years and therefore that the total number of sequence records derived from data collection should also be uniform. Each sequence record was assigned an initial temporal weight as follows: (i) calculate the annual proportion of records (number of records for each year/total number of records); (ii) calculate the cumulative proportion of records for all years; (iii) remove data for years in which cumulative proportion was <1% to eliminate years that were sparsely represented, as they would have been assigned an unreasonably high weight; (iv) randomly choose 90% of the records; (v) calculate the average for the remaining years; (vi) calculate the temporal weight for the records in a specific year (temporal weight = average/number of occurrences (occurrences) in that year); (vii) repeat steps iv to vi 1,000 times; and (viii) calculate the average temporal weight for each record.
Correction for geographic bias was done in a manner similar to that for temporal correction by assuming that major global regions of the world would have roughly the same influenza virus prevalence. Countries were grouped into 10 broad geographic regions, namely, North America, South America, Oceania, Europe, North Africa, sub-Saharan Africa, Northeast Asia, Southeast Asia, Southwest Asia, and unknown (which included records termed not applicable [N/A] or unknown for country). To compute the geographic weight, we calculated (i) the average number of records for each region (excluding unknown) and (ii) the geographic weight for the records in a specific region: geographic weight = average/number of occurrences (occurrences) in that region. The resultant new geographic weights for each record were multiplied by the temporal weights and normalized to get the final geotemporal weights. Normalization was necessary to guarantee that the sum of all weights was the same as the total number of records. A secondary analysis in which the regional bias was corrected based on population density using data for total population per region obtained from the Population Reference Bureau resource (www.prb.org) was also performed.
RESULTS
As of November 2011, a total of 3,875 SFs have been defined for the 11 known proteins of type A influenza virus (Table 2). For HA, which has 16 known subtypes, SFs are currently defined separately for the commonly studied subtypes H1, H2, H3, H5, and H7. For NA, which has 9 known subtypes, SFs are currently defined for N1 and N2. SF definitions will be expanded to the remaining subtypes in subsequent versions.
Table 2.
Number of SFs defined for each influenza A virus protein
| Protein | Subtypea | Functional | Structural | Immune epitopes | Sequence alterations | Total count |
|---|---|---|---|---|---|---|
| PB2 | 7 | 10 | 514 | 4 | 535 | |
| PB1-F2 | 2 | 2 | 2 | 6 | ||
| PB1 | 6 | 5 | 646 | 657 | ||
| PA | 1 | 29 | 459 | 1 | 490 | |
| HA | H1 | 4 | 37 | 323 | 364 | |
| HA | H2 | 7 | 7 | 19 | 33 | |
| HA | H3 | 2 | 59 | 362 | 30 | 453 |
| HA | H5 | 3 | 14 | 40 | 8 | 65 |
| HA | H7 | 1 | 2 | 3 | ||
| NP | 10 | 25 | 421 | 461 | ||
| NA | N1 | 10 | 26 | 101 | 4 | 141 |
| NA | N2 | 9 | 59 | 105 | 6 | 179 |
| M1 | 12 | 14 | 236 | 262 | ||
| M2 | 7 | 12 | 74 | 1 | 94 | |
| NS1 | 21 | 15 | 93 | 129 | ||
| NS2 | 2 | 3 | 62 | 67 | ||
| Total | 105 | 321 | 3,393 | 56 | 3,875 | |
SFs for HA protein are currently defined only for some of the commonly studied subtypes: H1, H2, H3, H5, and H7. For NA protein, SFs are defined for N1 and N2 subtypes.
The majority of SFs defined thus far belong to the immune epitope category (Table 2), reflecting the fact that immune epitopes are easier to define experimentally than other types of protein functional regions. The number of immune epitope SFs is roughly proportional to the size of the influenza virus protein, reflecting the fact that the majority are T-cell epitopes, which, unlike B-cell/antibody epitopes, are not focused on the surface of exposed proteins and are reasonably well dispersed along the entire length of proteins. Interestingly, the influenza virus protein with the largest number of functional SFs is the relatively small NS1, reflecting the intensity of molecular experimental study focused on this multifunctional protein (3).
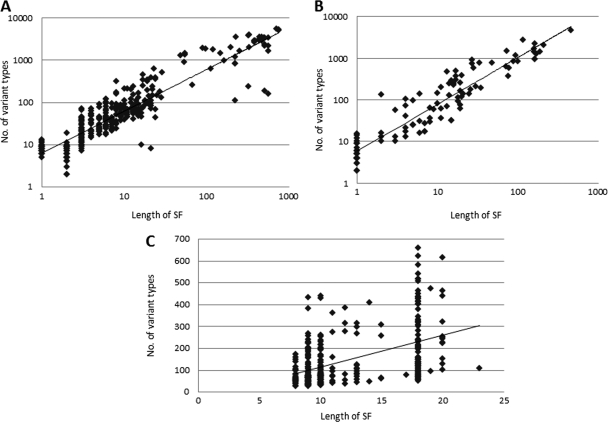
VTs of SFs are identified based on sequence variations that are observed between influenza virus strains within each SF region in the entire sequence record available in IRD. The number of VTs observed is roughly proportional to the length of the SF (Fig. 2). However, in some cases the number of observed VTs is smaller than would be expected based on SF size (points below the diagonal). These might correspond to regions of the protein that are under strong evolutionary constraint due to structural or functional requirements. In other cases, the number of VTs is larger than would be expected (points above the diagonal). These might correspond to regions under strong positive selective pressure to change rapidly (e.g., certain immune epitopes).
Fig 2.
Correlation between SF length and number of observed VTs. Scatter plots are used to display the correlations between the length of each SF and the total number of VTs it contains for all structural SFs (A), all functional SFs (B), and immune epitopes (C) of the H1 subtype of HA protein. A best-fit line is drawn for each graph. Graphs A and B are generated on a log-log scale to better represent the broad range of data.
Once unique VTs have been identified in the sequence record, influenza virus strains can be annotated and grouped based on VT membership. Since the VT definition is different for each SF, the grouping of influenza virus strains based on VT annotations will be different for each SF. Since influenza virus strain records in IRD have detailed metadata associated with them, such as country of isolation, host species, year of isolation, and subtype of the virus, the different SFVT groupings can be used to investigate genotype (VT grouping)-phenotype (metadata characteristic) associations.
Host range distribution of NS1 SFVTs.
To demonstrate the utility of the SFVT approach for genotype-phenotype association analysis, we selected an SF from the influenza virus NS1 protein Influenza virus A_NS1_SF18 to determine if certain VTs show a restricted host range. While the influence of surface protein subtypes (especially HA and NA) on host range constraints has been well studied, we chose to examine the nonstructural NS1 protein to determine the extent to which it may contribute to host range restriction. Influenza virus A_NS1_SF18 is an 11-amino-acid region involving residues 137 to 147 that is required for nuclear export of NS1 (8). VT-1 corresponds to the amino acid sequence found in A/Udorn/1972, which serves as the reference strain for all NS1 SFs. We examined the isolation host distribution for the first 16 VTs of Influenza virus A_NS1_SF18 since they contained at least 50 strains within each VT group.
For a given host, the proportion of strains carrying a given VT was plotted for the first 16 VTs of Influenza virus A_NS1_SF18 (Fig. 3). Four patterns emerged from this analysis. In some cases, sequences carrying a certain VT were found in restricted host groups. For example, virtually all equine strains (99%) were found to carry VT-8 for Influenza virus A_NS1_SF18, and the vast majority of VT-8-carrying strains (82%) were isolated from horse (Fig. 3A). Strains carrying VT-4, VT-7, VT-11, VT-12, VT-13, and VT-14 were predominantly found in avian/chicken hosts and virtually all, except VT-14, were excluded from human (Fig. 3B). In contrast, strains carrying VT-3, VT-5, VT-9, VT-10, and VT-16 were found predominantly in human and virtually excluded from avian/chicken (Fig. 3C). Interestingly, swine isolates appeared to be able to carry either avian-specific (VT-4) or human-specific (VT-3) VTs. Finally, some VTs (e.g., VT-1, VT-2, VT-6, VT-15) appeared in isolates from a wide range of hosts (Fig. 3D). The skewed distribution of many of the Influenza virus A_NS1_SF18 VTs across virus hosts suggested that sequence variations in NS1 might influence host range.
Fig 3.
Distribution of host proportions across VTs of Influenza virus A_NS1_SF18. VTs were divided into four groups based on their host representation patterns. (A) VT-8-carrying viruses were isolated predominantly from equine (and dog; included in the “Others” category). (B) VT-4-, 7-, 11-, 12-, 13-, and 14-carrying viruses were isolated predominantly from chicken and other avian species and environmental samples. (The majority of environmental samples appear to be derived from avian feces.) (C) VT-3-, 5-, 9-, 10-, and 16-carrying viruses were isolated predominantly from human hosts. (D) VT-1-, 2-, 6-, and 15-carrying viruses were isolated from a broad range of different hosts.
VT evolution.
To understand how the Influenza virus A_NS1_SF18 VTs evolved over time, we inferred their probable ancestry. We first generated a maximum likelihood-based phylogenetic tree using the RAxML (Randomized Axelerated Maximum Likelihood) program for the region containing the Influenza virus A_NS1_SF18 and then manually constructed the likely evolutionary history by tracking the accumulation of amino acid substitutions occurring in the SF over time (Fig. 4).
Fig 4.
SFVT lineage tree. The heritage of the first 16 VTs of NS1 Influenza virus A_NS1_SF18 was inferred from the phylogenetic analysis of the amino acid sequences and the apparent accumulation of amino acid substitutions at the time of first appearance of a particular VT in the sequence record (in parentheses) from 1902 to 2010. Each VT group is color coded based on the host in which it has circulated. Also shown are the amino acid substitutions (position and amino acid change) that caused each VT group to be assigned to subsequent “progeny” VTs.
From the inferred ancestor (VT-0), two major lineages (branches) emerged, VT-1 and VT-4, and these could be further separated into sublineages.
The VT-1 lineage began circulating as early as 1902 in diverse host groups as seen from the sequence records in the IRD resource. While viruses carrying VT-1 continue to circulate, VT-1 also gave rise to seven additional sublineages. Three sublineages were generated due to substitution at NS1 residue 139, including VT-15 (in 1943, D139G substitution), VT-8 (in 1963, D139E), and VT-2 (in 1963 with D139N). While the VT-8 sublineage is predominant in horse and dog, VT-1 and the other 139 variant sublineages show broad host range distributions. Two sublineages were generated due to substitution at residue 145—the VT-16 (in 1965, I145T) and VT-9 (in 1979, I145V) sublineages. These substitutions appear to exclude avian/chicken hosts. In contrast, sublineages with substitutions at residues 137 (VT-14) and 138 (VT-7) appear to prefer avian/chicken hosts, though VT-14 is also seen in human hosts.
VT-4 (I137L, R140Q, I145V, and L146S) appeared as early as 1949 and represents the second major lineage, with VT-11 (2001, with L141M added to the VT-4 substitutions) and VT-12 (1978, with D139N added to the VT-4 substitutions) as sublineages. All VT-4 lineage variants are predominantly restricted to avian/chicken isolates.
The major VT-1 and VT-4 lineages are consistent with previous studies defining the A and B alleles of NS1 protein based on nucleotide sequence homology, with allele B reported to be found exclusively in avian viruses and allele A found more broadly, including human, swine, and avian isolates (10, 20).
This ancestry analysis provides further evidence that sequence variations in Influenza virus A_NS1_SF18 show skewed distribution in certain host species, suggesting a possible involvement of NS1 in restricting the host range phenotype of influenza virus.
Statistical test of skewed host range distribution.
Two different sets of chi-square tests were performed to determine if the apparent skewed distributions are statistically significant: (i) tests of the distribution of each of the 16 VTs of Influenza virus A_NS1_SF18 across six host groups (Table 3) and (ii) tests of whether a particular host group has the same probability of containing all VT groups (Table 4). Using a significance cutoff level of 0.05, the first set of tests showed that there are significant and dramatic differences in the probabilities of host groups carrying a particular VT. Similarly, the second set of tests showed that there are also major significant differences in the probabilities of different VTs occurring in a specific host group.
Table 3.
Chi-square results and P values for unprocessed and processed datasets
| Variant type | dfa | Unprocessed data |
Processed data |
||
|---|---|---|---|---|---|
| Chi-square value | P value | Chi-square value | P value | ||
| VT-1 | 5 | 1,978.01 | <1.00E−320 | 2,307.53 | <1.00E−320 |
| VT-2 | 5 | 1,602.69 | <1.00E−320 | 370.55 | 6.57E−78 |
| VT-3 | 5 | 1,631.39 | <1.00E−320 | 604.83 | 1.83E−128 |
| VT-4 | 5 | 1,043.57 | 2.22E−223 | 1,269.96 | 2.06E−272 |
| VT-5 | 5 | 774.93 | 3.07E−165 | 354.07 | 2.33E−74 |
| VT-6 | 5 | 796.81 | 5.66E−170 | 294.23 | 1.74E−61 |
| VT-7 | 5 | 693.02 | 1.59E−147 | 3,364.64 | <1.00E−320 |
| VT-8 | 5 | 8,262.95 | <1.00E−320 | 9,573.24 | <1.00E−320 |
| VT-9 | 5 | 290.96 | 8.77E−61 | 404.03 | 4.01E−85 |
| VT-10 | 5 | 259.70 | 4.56E−54 | 104.45 | 6.09E−21 |
| VT-11 | 5 | 1,490.13 | 4.05E−320 | 584.51 | 4.49E−124 |
| VT-12 | 5 | 75.91 | 6.02E−15 | 104.86 | 4.99E−21 |
| VT-13 | 5 | 98.70 | 9.95E−20 | 15.52 | 0.008356874 |
| VT-14 | 5 | 101.94 | 2.06E−20 | 64.53 | 1.40E−12 |
| VT-15 | 5 | 70.61 | 7.66E−14 | 145.27 | 1.36E−29 |
| VT-16 | 5 | 48.80 | 2.44E−09 | 40.15 | 1.39E−07 |
df, degrees of freedom.
Table 4.
Chi-square results and P values for unprocessed and processed datasets
| Host type | dfa | Unprocessed data |
Processed data |
||
|---|---|---|---|---|---|
| Chi-square value | P value | Chi-square value | P value | ||
| Avian | 15 | 3,876.49 | <1.00E−320 | 2,263.90 | <1.00E−320 |
| Chicken | 15 | 1,869.13 | <1.00E−320 | 3,753.14 | <1.00E−320 |
| Equine | 15 | 8,254.37 | <1.00E−320 | 9,389.92 | <1.00E−320 |
| Human | 15 | 6,838.78 | <1.00E−320 | 3,994.25 | <1.00E−320 |
| Othersb | 15 | 1,457.60 | 6.68E−302 | 587.68 | 1.47E−115 |
| Swine | 15 | 300.45 | 4.51E−55 | 385.66 | 7.10E−73 |
df, degrees of freedom.
Includes environmental samples and other host species.
A known limitation of the chi-square test is that the absolute values of the statistic and resultant P values are influenced by the size of the data set, thereby artificially inflating their maximum/minimum values. Cramer's V test is an approach to normalize the chi-square statistic to control for data set size. Once again, Cramer's V measurement provided further support for the nominal association between VT and host (see Table S1 in the supplemental material). These results suggest that the chi-square test is reasonable, that the testing results are significant, and that they provide support for effects of NS1 genomic features on this key phenotypic characteristic.
Causes of apparent NS1 VT-associated host range specificity.
Skewed host distributions of the sort described above could be attributed to several factors. Phenotypic differences caused by sequence variations in NS1 could prevent influenza virus from infecting and/or replicating in particular host species. However, the skewed distribution could also arise due to founder effects resulting in restricted spatial-temporal distribution or due to ascertainment bias from focused oversampling phenomena. As sequence records in IRD are obtained from GenBank and from direct submissions, their numbers are largely influenced by the focus of sequencing efforts. For example, it is evident that more sequence data have been obtained during epidemic and pandemic outbreaks. The number of sequence records has also increased exponentially during the most recent decades, particularly in developed countries with more in-depth data collection processes and better surveillance infrastructure. Thus, the recent increase is likely due to increased data reporting rather than to an absolute increase in the population of influenza viruses circulating worldwide.
To assess the spatial-temporal distributions of the data used, the number and proportion of strains carrying each VT were further subdivided by time and geographic location of isolation (Tables 5 and 6, respectively). VT-13 and VT-14 are predominantly represented by viruses isolated from Southeast Asia (Vietnam) during the years 2003 to 2007, suggesting a probable founder effect. In other cases, skewing could be at least partly attributed to sampling bias, including the overreporting of VT-2 sequences in 2009 due to oversampling of the pandemic H1N1 viruses and the overreporting of VT-1 sequences from Southeast Asia due to oversampling of avian H5N1 strains. Similar arguments could also be made for the overrepresentation of VT-11 in avian hosts since most come from strains obtained through directed sampling of chickens and environmental samples in New York in 2005 and 2006.
Table 5.
Temporal distribution of sequence records in each VT group
| Variant type | Sequence record count for indicated year of isolationa |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1902-1960 | 1961-1970 | 1971-1980 | 1981-1990 | 1991-1995 | 1996-2000 | 2001-2005 | 2006-2010 | |
| VT-1 | 92 (0.0316) | 136 (0.0467) | 278 (0.0954) | 209 (0.0717) | 71 (0.0244) | 275 (0.0943) | 1,222 (0.4192) | 632 (0.2168) |
| VT-2 | 0 | 4 (0.002) | 7 (0.0034) | 52 (0.0256) | 12 (0.0059) | 60 (0.0295) | 132 (0.0649) | 1,768 (0.8688) |
| VT-3 | 0 | 0 | 0 | 0 | 0 | 334 (0.2387) | 705 (0.5039) | 360 (0.2573) |
| VT-4 | 3 (0.0039) | 2 (0.0026) | 52 (0.0676) | 51 (0.0663) | 58 (0.0754) | 231 (0.3004) | 202 (0.2627) | 170 (0.2211) |
| VT-5 | 0 | 0 | 0 | 0 | 0 | 67 (0.0999) | 147 (0.2191) | 457 (0.6811) |
| VT-6 | 0 | 0 | 0 | 0 | 3 (0.0048) | 169 (0.2713) | 416 (0.6677) | 35 (0.0562) |
| VT-7 | 0 | 0 | 0 | 0 | 0 | 0 | 76 (0.1886) | 327 (0.8114) |
| VT-8 | 0 | 4 (0.0245) | 25 (0.1534) | 44 (0.2699) | 26 (0.1595) | 3 (0.0184) | 16 (0.0982) | 45 (0.2761) |
| VT-9 | 0 | 0 | 8 (0.0279) | 42 (0.1463) | 174 (0.6063) | 57 (0.1986) | 3 (0.0105) | 3 (0.0105) |
| VT-10 | 0 | 0 | 0 | 0 | 0 | 0 | 234 (1) | 0 |
| VT-11 | 0 | 0 | 0 | 0 | 0 | 0 | 143 (0.8882) | 18 (0.1118) |
| VT-12 | 0 | 0 | 1 (0.0192) | 0 | 7 (0.1346) | 36 (0.6923) | 6 (0.1154) | 2 (0.0385) |
| VT-13 | 0 | 0 | 0 | 0 | 0 | 0 | 18 (0.2571) | 52 (0.7429) |
| VT-14 | 0 | 1 (0.0141) | 0 | 0 | 1 (0.0141) | 1 (0.0141) | 62 (0.8732) | 6 (0.0845) |
| VT-15 | 3 (0.0375) | 0 | 3 (0.0375) | 26 (0.325) | 2 (0.025) | 8 (0.1) | 24 (0.3) | 14 (0.175) |
| VT-16 | 0 | 1 (0.0154) | 2 (0.0308) | 1 (0.0154) | 10 (0.1538) | 33 (0.5077) | 18 (0.2769) | 0 |
The total record count is followed by the proportion of sequences in parentheses.
Table 6.
Geographic distributions of sequence records in each VT group
| Variant type | Sequence record count for indicated region of isolationa |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EUR | NAF | NAM | NEA | OCE | SAM | SEA | SSA | SWA | UNK | |
| VT-1 | 265 (0.0909) | 4 (0.0014) | 1,177 (0.4038) | 132 (0.0453) | 59 (0.0202) | 13 (0.0045) | 1,222 (0.4192) | 12 (0.0041) | 26 (0.0089) | 5 (0.0017) |
| VT-2 | 108 (0.0531) | 1 (0.0005) | 1,404 (0.6899) | 171 (0.084) | 68 (0.0334) | 49 (0.0241) | 222 (0.1091) | 0 | 12 (0.0059) | 0 |
| VT-3 | 63 (0.045) | 1 (0.0007) | 655 (0.4682) | 39 (0.0279) | 485 (0.3467) | 35 (0.025) | 104 (0.0743) | 0 | 17 (0.0122) | 0 |
| VT-4 | 86 (0.1118) | 0 | 520 (0.6762) | 47 (0.0611) | 8 (0.0104) | 21 (0.0273) | 75 (0.0975) | 6 (0.0078) | 6 (0.0078) | 0 |
| VT-5 | 20 (0.0298) | 0 | 386 (0.5753) | 46 (0.0686) | 157 (0.234) | 30 (0.0447) | 31 (0.0462) | 0 | 1 (0.0015) | 0 |
| VT-6 | 0 | 0 | 3 (0.0048) | 6 (0.0096) | 0 | 0 | 614 (0.9856) | 0 | 0 | 0 |
| VT-7 | 79 (0.196) | 20 (0.0496) | 0 | 64 (0.1588) | 0 | 12 (0.0298) | 43 (0.1067) | 122 (0.3027) | 63 (0.1563) | 0 |
| VT-8 | 6 (0.0209) | 0 | 236 (0.8223) | 10 (0.0348) | 0 | 0 | 33 (0.115) | 0 | 2 (0.007) | 0 |
| VT-9 | 48 (0.2945) | 1 (0.0061) | 74 (0.454) | 7 (0.0429) | 2 (0.0123) | 9 (0.0552) | 18 (0.1104) | 2 (0.0123) | 1 (0.0061) | 1 (0.0061) |
| VT-10 | 32 (0.136) | 0 | 82 (0.3504) | 0 | 117 (0.5) | 0 | 3 (0.0128) | 0 | 0 | 0 |
| VT-11 | 0 | 0 | 161 (1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VT-12 | 33 (0.6346) | 0 | 16 (0.3077) | 0 | 0 | 3 (0.0577) | 0 | 0 | 0 | 0 |
| VT-13 | 1 (0.0141) | 0 | 3 (0.0423) | 0 | 0 | 0 | 67 (0.9437) | 0 | 0 | 0 |
| VT-14 | 0 | 0 | 0 | 0 | 0 | 0 | 70 (1) | 0 | 0 | 0 |
| VT-15 | 8 (0.1) | 1 (0.0125) | 45 (0.5625) | 3 (0.0375) | 0 | 0 | 22 (0.275) | 1 (0.0125) | 0 | 0 |
| VT-16 | 4 (0.0615) | 0 | 26 (0.4) | 1 (0.0154) | 17 (0.2615) | 0 | 17 (0.2615) | 0 | 0 | 0 |
Abbreviations: EUR, Europe; NAF, North Africa; NAM, North America; NEA, Northeast Asia; OCE, Oceania; SAM, South America; SEA, Southeast Asia; SSA, sub-Saharan Africa; SWA, Southwest Asia; UNK, unknown. The total count is followed by the proportion of sequence records in parentheses.
However, these explanations of sampling bias and founder effects did not hold well for many VT groups that showed restricted host sources since they also demonstrated broad geographic and temporal representation. For example, equine VT-8 isolates were widespread geotemporally but still largely restricted to equine and canine hosts. Viruses carrying VT-8 had ample opportunities to infect other hosts as they moved across geographically diverse countries from 1963 through 2008-2009 and yet did not cross other host species barriers. Similarly, the avian-predominant VT-4 lineage and the human-predominant VT-9 and VT-16 groups were also broadly geotemporally diverse, suggesting that these VTs may indeed be examples of NS1-mediated host range restrictions.
Effect of debiasing on VT-host statistical associations.
In order to better estimate statistical significance by correcting for these geotemporal data collection biases, we developed a weighting method based on the underlying assumption that if the sampling of equal proportions of the influenza virus population had occurred in each year and each location, the total number of records would have been roughly the same in each year and each geographic region. Rather than discarding some data, we chose to give each record within the data set a new normalized weight to control for overreporting, as described in Materials and Methods.
The number of SFVT strain records was plotted against their reported year of isolation for both unprocessed and processed (weighted) data (see Fig. S1A in the supplemental material). The rise in the number of unprocessed records around 1997 is most likely due to the fact that sequencing efforts were extended to all influenza virus segments, rather than just HA and NA, during that time. The peaks at 2005-2006 and 2009-2010 correspond to the avian influenza outbreak and recent swine influenza pandemic, respectively. The curve for the processed data (dotted line) reflects how the oversampling bias associated with these episodic events has been reduced by this temporal weighting strategy. Similarly, the correction for geographic bias normalized the geographic distribution of the sequence records (see Fig. S1B).
The chi-square tests described earlier were recalculated using these processed (weighted) data sets (Table 3). Although the skewing was still substantial, data debiasing had a considerable impact on many of the P values calculated for these data sets. The P values for some tests increased dramatically, suggesting that, for these tests, data collection bias had contributed to the skewed distributions observed. Correction for temporal bias likely explains the change in the VT-2 P value, since this is the VT carried by the pandemic H1N1 2009 viruses, which have been the subject of massive sequencing scrutiny. Correction for geographic bias likely explains the change in P value for VT-6, VT-13, and VT-14, since these viruses were largely restricted to recent Southeast Asian isolates, indicating possible founder effects. VT-3 included records predominantly from New Zealand and the United States in 2005, so the calculated chi-square value dropped due to correction for Oceania. The change in the VT-11 P value was due to correction for both geographic and temporal bias, as it included samples mostly from the United States isolated during 2005 and 2006. Nevertheless all P values were found to be extremely small, both for the unprocessed data and for the processed data, suggesting that this region of NS1, alone or in conjunction with other SFs, appears to have a dramatic impact on host range specificity.
DISCUSSION
We have described an approach wherein the effects of sequence alterations in influenza virus proteins on virus phenotypic characteristics can be analyzed at a fine level of granularity, by defining combinations of specific amino acid residues that function as structural or functional units called sequence features (SFs). Recurrent sequence variations—variant types (VTs)—occurring within the defined SF region are computed by aligning each SF from a chosen reference strain to all other related sequences in the IRD resource (www.fludb.org). The SFVT module is freely available online to the influenza virus research community through IRD, which provides access to the complete list of SFs, SFVT alignments, and metadata associated with the VT sequences, including host, country and year of isolation, and virus subtype.
By compiling the set of all currently characterized SFs for influenza virus proteins, a valuable resource that can be used as a reference for all characterized influenza virus protein regions has been created. Each defined SF is associated with links to relevant publications, protein annotations, and protein structure records through PubMed, UniProt, PDB, and IEDB. The SFVT system is designed to support the addition of new SFs as they become available without altering the existing list. In an effort to make the SFVT component as comprehensive and up-to-date as possible, we are now creating a community-based annotation web interface to allow external researchers and IRD curators to submit new SFs to the system. The user interface for data capture will contain some required fields (e.g., virus name, SF positions, category and definition, submitter's name and affiliation, etc.) that the submitter will need to provide, while other fields will be populated automatically by the IRD system (e.g., SF identity [ID] and length) based on the primary data entered into the required fields. SFs submitted using this method will be internally validated for completeness, accuracy, and nonredundancy and subsequently reviewed by influenza virus experts before public release. In addition, IRD will automatically add new SFs from the UniProt and IEDB resources using custom parsing scripts.
In an effort to demonstrate the utility of this approach in studying genetic determinants of virus phenotypes, we performed computational and statistical analysis on the VTs of the Influenza virus A_NS1_SF18 of the NS1 protein for their potential correlation with host range restriction. Even after controlling for data collection biases, highly significant P values and dramatically skewed distributions of VTs were observed across different host groups, suggesting that sequence variations in Influenza virus A_NS1_SF18 appear to impact host range restriction with high statistical confidence.
For a virus to spread, it should have both the opportunity and capability to infect a given host. A virus infection within an isolated community, for example, might result in all individuals carrying a particular substitution carried by the founder virus; however, those living outside of that specific community may lack the substitution because of a lack of opportunity for the founder virus or its progeny to infect them. We checked to see if this kind of founder effect could explain the associations observed in our data set but found just the opposite to be true in many cases. For example, viruses carrying VT-8, which were found to infect predominantly horses and dogs, had ample opportunity to spread based on their worldwide occurrence over more than 45 years, and yet they continued to remain within their preferred host species. In fact, the only evidence of cross-species virus spreading from horses to another host group was at a racetrack in Miami, Florida, where dogs and horses raced at the same facility (2). Although most viruses in the VT-8 group also belong to the H3N8 subtype, it is clear from the sequence records that H3N8 viruses circulate effectively in birds and other hosts. Thus, the restriction of certain influenza viruses to equine and canine species appears to be dictated, at least in part, by sequence variations in the NS1 protein. Similar arguments can be made for the associations between the VT-4 lineage and avian host restriction and the VT-9 and VT-16 lineages and human host restriction.
It should be noted that for statistical inference, the data set used here cannot be considered as a random sample from the entire population of influenza virus-infected hosts, since the data records are from diverse sources and are based on free response data collection schemes and therefore may not be independent or random. One consequence of this observation is that the P values from chi-square analysis may be biased. Despite the fact that we cannot prove the independence and randomness of the data, we know that the records are at least from different regions of the world and were collected at different time points throughout several decades. Indeed, while our approach to control for geographic and temporal biases resulted in changes in the chi-square statistical values, the extreme skewing in VT-host distributions remained significant.
The geographic bias in the data could be attributed at least partially to difference in population density of host species. To control for this contribution to geographic bias, we repeated the chi-square analysis by also adjusting for virus prevalence per capita in human populations across different regions of the world (see Table S1 in the supplemental material). Once again, while the absolute values of the chi-square statistics and P value changed, the extreme skewing in VT-host distributions remained significant. However, it should be noted that this adjusts only for the effects of population density of the human host; population density information for the other influenza virus host species is not readily available in order to perform a similar adjustment of their effects.
As an alternative to the chi-square analysis, we also applied an association rule data-mining method to investigate the relationship between VTs and host groups. One advantage of this method is that it does not require independent data or random sampling to infer results. For this data-mining process, we separately considered two rules, namely, VT-to-host-type and host-type-to-VT relationships, and therefore for each direction we had 96 (16 · 6 = 96) possible rules. These rules were then assessed using two common evaluation criteria—support and confidence. Using 0.5 as a confidence cutoff, we ended up with two significant rules from host type to variant type (avian to VT-1 and equine to VT-8) and 11 significant rules from variant type to host type (VT-1, VT-4, VT-12, VT-13, and VT-14 to avian; VT-8 to equine; and VT-2, VT-3, VT-5, VT-10, and VT-16 to human), providing further support for the role of NS1 sequence variations in host range restriction (see Table S2 in the supplemental material).
To determine if sequence variations in Influenza virus A_NS1_SF18 are independent predictors of virus host range, we performed additional analysis of variance (ANOVA) tests to examine the main effects of host type, VT, HA subtype, NA subtype, and the two-way interactions of host type by VT, host type by HA, host type by NA, VT by HA, and VT by NA using the number of records as the response variable (see Table S3 in the supplemental material). In the ANOVA output, we see that the overall model is significant with P value F statistics of <0.0001, indicating that the model is valid. Since the interaction of host type and NA is significant (P value of 0.0037), we know that at least one pair of response variables for different combinations of host type and NA is different from each other. Hence, there is no need to look at their main effects because the possible significance of their main effects could be driven by their interactions. This finding agrees with the prior knowledge that certain host types are associated with certain virus NA subtypes. We also find that the P value for the interaction of VT and host is less than 0.0001, thus verifying our conclusion that VT is highly associated with host type. However, neither the interaction of VT and HA nor that of VT and NA is significant, implying that there is insufficient evidence of an association between VT and either NA or HA subtype. Therefore, virus subtype is not a confounding variable in our NS1 VT analysis, even though the NA subtype is also associated with host type. Thus, the association between host type and VT is verified and therefore NS1 sequence variation can be considered as an independent factor dictating host range restriction.
Several previous studies have provided evidence of a role for NS1 sequence variation in differential replication and pathogenicity in different host species, but most of the studies were focused on a whole-segment analysis strategy. Reassortant viruses carrying aberrant NS segments from the cold-adapted CR43 clone 3 virus were defective for replication in Madin-Darby canine kidney cells and ferrets (12). Reassortant A/Udorn/72 viruses carrying the B allele of NS1 from avian viruses were found to be attenuated for replication in the respiratory tract of squirrel monkeys in comparison with the same virus carrying the A allele of NS1 (20). The NS1 protein from the A/Goose/Guangdong/1/96 (H5N1) strain increased the replication efficiency of the A/FPV/Rostock/34 (H7N1) strain in human and mouse cell lines (11). These studies and others provide clear evidence of a role for NS1 in host range restriction at the segment level. More recently, the C-terminal ESEV/RSKV motif, conserved in avian and human viruses, respectively, was shown to affect replication in a species-specific manner in cell culture and animals (17). While all the statistical tests performed in this study provide strong evidence for the role of Influenza virus A_NS1_SF18 in contributing to host specificity, we cannot definitively conclude that the cellular nuclear export machinery is solely responsible for host restriction, as we have not yet completed an in-depth analysis of all other NS1 SFs for correlation with this phenotype. However, the SFVT strategy should allow us to rapidly identify the most likely candidates. Ultimately, experimental validations will be required to elucidate the connections between VT-mediated host range specificity and the relevant cellular/biochemical processes.
Supplementary Material
ACKNOWLEDGMENTS
This project is supported by NIAID under contract N01AI400041 as part of the Influenza Research Database (www.fludb.org) development. Work in Adolfo García-Sastre's laboratory is partly funded by CRIP (Center for Research in Influenza Pathogenesis) and the NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS), contract HHSN266200700010C. The authors have declared that no competing interests exist.
We thank Elizabeth McClellan and Al-Rawashdeh Ayman Ibrahim for their valuable suggestions on the preliminary statistical analysis of the SFVTs of Influenza virus A_NS1_SF18. We also thank Robert Lamb for reviewing the list of SFs defined for the matrix protein, M2.
Footnotes
Published ahead of print 7 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Baigent SJ, McCauley JW. 2003. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. BioEssays 25:657–671 [DOI] [PubMed] [Google Scholar]
- 2. Crawford PC, et al. 2005. Transmission of equine influenza virus to dogs. Science 310:482–485 [DOI] [PubMed] [Google Scholar]
- 3. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 4. Herlocher ML, et al. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res. 54:99–111 [DOI] [PubMed] [Google Scholar]
- 5. Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J. Comp. Graph. Stat. 5:299–314 [Google Scholar]
- 6. Karp DR, et al. 2010. Novel sequence feature variant type analysis of the HLA genetic association in systemic sclerosis. Hum. Mol. Genet. 19:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamb RA, Krug RM. 1996. Orthomyxoviridae: the viruses and their replication, p 1487–1531 In Knipe DM, Howley PM. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 8. Li Y, Yamakita Y, Krug R. 1998. Regulation of a nuclear export signal by an adjacent inhibitory sequence: the effector domain of the influenza virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 95:4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lindstrom SE, et al. 1998. Phylogenetic analysis of the entire genome of influenza A (H3N2) viruses from Japan: evidence for genetic reassortment of the six internal genes. J. Virol. 72:8021–8031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ludwig S, Schultz U, Mandler J, Fitch WM, Scholtissek C. 1991. Phylogenetic relationship of the nonstructural (NS) genes of influenza A viruses. Virology 183:566–577 [DOI] [PubMed] [Google Scholar]
- 11. Ma W, et al. 2010. The NS segment of an H5N1 highly pathogenic avian influenza virus (HPAIV) is sufficient to alter replication efficiency, cell tropism, and host range of an H7N1 HPAIV. J. Virol. 84:2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maassab HF, DeBorde DC. 1983. Characterization of an influenza A host range mutant. Virology 130:342–350 [DOI] [PubMed] [Google Scholar]
- 13. Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munster VJ, et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 3:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neumann G, Kawaoka Y. 2006. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg. Infect. Dis. 12:881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scholtissek C, Burger H, Kistner O, Shortridge KF. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287–294 [DOI] [PubMed] [Google Scholar]
- 17. Soubies SM, et al. 2010. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J. Virol. 84:6733–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Squires RB, et al. 2012. Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance. Influenza Other Respi. Viruses. doi:10.1111/j.1750-2659.2011.00331.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Treanor J, Perkins M, Battaglia R, Murphy BR. 1994. Evaluation of the genetic stability of the temperature-sensitive PB2 gene mutation of the influenza A/Ann Arbor/6/60 cold-adapted vaccine virus. J. Virol. 68:7684–7688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Treanor JJ, Snyder MH, London WT, Murphy BR. 1989. The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology 171:1–9 [DOI] [PubMed] [Google Scholar]
- 21. Wasilenko JL, et al. 2008. NP, PB1, and PB2 viral genes contribute to altered replication of H5N1 avian influenza viruses in chickens. J. Virol. 82:4544–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wright PF, Webster RG. 2001. Orthomyxoviruses, p 1533–1579 In Knipe DM, Howley PM. (ed), Fields virology, 4th ed, vol 1 Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.