Abstract
There are no known RNA viruses that infect Archaea. Filling this gap in our knowledge of viruses will enhance our understanding of the relationships between RNA viruses from the three domains of cellular life and, in particular, could shed light on the origin of the enormous diversity of RNA viruses infecting eukaryotes. We describe here the identification of novel RNA viral genome segments from high-temperature acidic hot springs in Yellowstone National Park in the United States. These hot springs harbor low-complexity cellular communities dominated by several species of hyperthermophilic Archaea. A viral metagenomics approach was taken to assemble segments of these RNA virus genomes from viral populations isolated directly from hot spring samples. Analysis of these RNA metagenomes demonstrated unique gene content that is not generally related to known RNA viruses of Bacteria and Eukarya. However, genes for RNA-dependent RNA polymerase (RdRp), a hallmark of positive-strand RNA viruses, were identified in two contigs. One of these contigs is approximately 5,600 nucleotides in length and encodes a polyprotein that also contains a region homologous to the capsid protein of nodaviruses, tetraviruses, and birnaviruses. Phylogenetic analyses of the RdRps encoded in these contigs indicate that the putative archaeal viruses form a unique group that is distinct from the RdRps of RNA viruses of Eukarya and Bacteria. Collectively, our findings suggest the existence of novel positive-strand RNA viruses that probably replicate in hyperthermophilic archaeal hosts and are highly divergent from RNA viruses that infect eukaryotes and even more distant from known bacterial RNA viruses. These positive-strand RNA viruses might be direct ancestors of RNA viruses of eukaryotes.
INTRODUCTION
In contrast to viruses that infect Bacteria and Eukarya, little is known about the viruses that infect Archaea. Fewer than 50 archaeal viruses have been discovered, compared to more than 5,500 viruses known to infect hosts from the other two domains of life (2). Of the few archaeal viruses that have been characterized, mainly those infecting members of the phylum Crenarchaeota, many have revealed both unusual gene content (56) and unique virion morphologies (42, 54, 55, 65). To date, all archaeal viruses possess double-stranded DNA (dsDNA) genomes, with the exception of one single-stranded DNA (ssDNA) virus infecting archaea of the genus Halorubrum (52). No RNA viruses infecting archaea have been described. Eukaryotic RNA viruses are a dominant viral form: they are numerous, diverse, and widespread, found to infect animals, plants, and many unicellular forms (17, 28, 38, 40, 70). Only two narrow groups of bacterial RNA viruses are known, and these do not show a close relationship with the viruses infecting eukaryotes (38, 40). Thus, the origin of the RNA viruses of eukaryotes remains an enigma. In this context, the search for RNA viruses infecting archaea appears to be of special interest because the discovery of such viruses would have the potential to shed new light on the origin of viruses of eukaryotes.
One factor limiting the discovery of archaeal viruses has been the reliance on culture-dependent approaches for virus isolation. Many Archaea inhabit extreme environments, such as those with high-temperature and high-salinity conditions, making culture-dependent approaches for virus discovery difficult. Over the past decade, direct sequencing of viral communities from environmental samples (viral metagenomics) has been used to more fully understand viral diversity in natural environments, including marine (13), sedimentary (10), human and animal fecal (11, 14, 33), gut (23), and freshwater (60) environments. Viral metagenomics overcomes many of the limitations of traditional culture-based approaches for the detection of new viruses. In general, viral metagenomics studies have revealed an enormous diversity and abundance of viruses. For example, more than 5,000 different viral genotypes were identified in 200 liters of seawater (12). Although most viral metagenomics studies have focused primarily on dsDNA viruses (5, 72), recent advances in methodology have enabled the implementation of RNA viral metagenomics as an approach for investigating RNA viral communities. Studies in marine environments have revealed a diverse community of RNA viruses broadly distributed throughout multiple viral taxa (16, 17). In these metagenomic studies, RNA viruses infecting bacteria or archaea have not been identified, supporting the view that the major hosts for RNA viruses in the marine environment are unicellular eukaryotes (73). Metagenomic analyses of RNA viruses in fresh water have shown that the majority of sequences did not show significant similarity to known viruses in public databases, following the trend of marine environments. Those sequences that did match were broadly distributed to nearly 30 families of viruses infecting a variety of eukaryotes but did not include the few groups of identified RNA viruses infecting bacteria or putative RNA viruses of archaea (19). The absence of evidence of RNA viruses infecting archaea in both marine and freshwater environments (5, 13, 17, 19) appears surprising in light of the indications that archaea comprise up to 40% of the planet's microbial biomass and up to one-third of the ocean's prokaryotes (34).
The recently described clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immunity system can be used to link viral genome sequences to cellular hosts. The CRISPR system appears to be an active defense mechanism against invading nucleic acids, including viruses, through a genetic interference pathway (7, 29, 44, 45, 68). The CRISPR system is present in ∼40% and ∼90% of sequenced bacterial and archaeal genomes, respectively (41). A CRISPR locus is composed of a leader sequence that directs the transcription of a noncoding RNA that spans multiple copies of an ∼25-nucleotide (nt) direct repeat (DR) region that are separated by ∼35-nt spacer sequences that can form long arrays. Each spacer sequence within a CRISPR array is unique. Immunity requires a sequence match between the invading nucleic acid and the spacer sequences that lie between DRs. The sequence content of the DRs can often be used to assign a CRISPR type at the cellular family level (22). It has been shown that the CRISPR loci provide a record of a cell's interaction with viruses (45, 64). Accordingly, the CRISPR DR and spacer content present in an environmental sample can be used to connect the viruses present in that environment with bacterial or archaeal hosts.
The acidic hot springs in Yellowstone National Park (YNP) in the United States offer an opportunity to search for archaeal RNA viruses in an environment where Archaea predominate. Based on previous rRNA gene sequence analysis, these environments are inhabited by microbial communities of low complexity that are dominated by fewer than 10 archaeal species (30). In these low-pH (pH < 4), high-temperature (>80°C) springs, bacteria and eukaryotes are scarce or, in many cases, absent (9, 30, 58). Using traditional culture-dependent approaches, many dsDNA archaeal viruses have been isolated from these high-temperature, acidic hot springs in YNP (59) and other thermal hot springs worldwide (6, 48, 53). We used a viral metagenomic approach to search directly for archaeal RNA viruses in several acidic hot springs found in YNP. We report here the detection of putative archaeal RNA virus genomes.
MATERIALS AND METHODS
YNP hot spring sample sites.
Twenty-eight YNP high-temperature, acidic hot springs were initially screened for the presence of RNA in the enriched viral fraction (see Table S1 in the supplemental material) between October and November 2008. In-depth sampling for metagenomic analysis was performed on 3 of these sites: NL10, NL17, and NL18. A total of 7 paired DNA viral and RNA viral samples were collected. Site NL10 in the Nymph Lake hot springs was sampled in October 2009, February 2010, and June 2010. Nymph Lake hot spring sites NL17 and NL18 were sampled in October 2009 and June 2010. Nymph Lake hot spring site NL10 was also sampled in August 2008 and 2009 for total cellular DNA.
Initial screening of enriched viral samples for RNA genomes by 32P-labeling experiments (reverse transcriptase [RT]-dependent signal from RNA viral fraction).
An enriched viral fraction was created from each sample by filtration of 26 ml of hot spring water through two successive 0.8-/0.2-μm Acrodisc 25-mm PF filters (Millipore) to remove cells, collection of the virus particles in the flowthrough, centrifugation at 30,000 × g for 2 h, and resuspension of the virus pellet into 200 μl sterile water. Total viral nucleic acids were extracted from the virus-enriched fraction using the mirVana miRNA (microRNA) isolation kit (Ambion). Following nucleic acid extraction, DNA was removed by DNase I (Ambion) treatment. Aliquots of RNA-enriched extracted viral nucleic acids were treated (controls) or not treated with RNase One (Promega) prior to cDNA synthesis. First-strand cDNA synthesis reactions were carried out using 1 mM random hexamers in the presence of 10 μCi of [α-32P]dCTP (MP BioMedicals). Reaction mixtures were incubated at 25°C for 10 min and at 50°C for 90 min and then terminated at 85°C for 5 min, followed by RNaseH treatment. Trichloroacetic acid (TCA)-precipitated nucleic acids were collected on glass fiber filters (Whatman G/F), and scintillation counts of precipitated and nonprecipitated material were collected.
Isolation of cellular fraction for community sequence analysis.
For community sequence analysis (see Fig. S1 in the supplemental material), cells were isolated from ∼500 ml of hot spring water by filtration onto 0.45-μm Pall filters (Millipore) and stored at −20 C. Total DNA was isolated from cells using phenol-chloroform-isoamyl alcohol 25:24:1 extraction of the filters followed by ethanol precipitation. DNA was amplified using GenomePlex whole-genome amplification (Sigma). Amplification products were purified using a QIAquick PCR purification kit (Qiagen). Approximately 10 μg of cellular DNA was provided to the University of Illinois Champaign-Urbana Sequencing Center (Urbana, IL) and subjected to 454 Titanium pyrosequencing (Roche).
Isolation of enriched viral nucleic acids.
Approximately 1.2 liters of hot spring water was filtered through two successive Pall 0.8-/0.2-μm filters (Millipore). The filtrate flowthrough was concentrated with Ultra centrifugal filters (Millipore) using a 100,000 molecular weight cutoff (100K MWCO) to a volume of 500 μl. Total viral nucleic acids were extracted from 200 μl of each sample with the Purelink viral RNA/DNA minikit (Invitrogen) and eluted to a final volume of 60 μl.
Isolation, preparation and sequencing of enriched RNA viral fraction.
An improved method to isolate nucleic acids from the enriched viral fraction was developed after initial RT-PCR screening. To obtain the enriched RNA viral fractions, 30 μl of total extracted nucleic acid was treated with 3 U RNase-free DNase I (Ambion) at 37°C for 20 min, followed by the addition of ethyl alcohol (EtOH) to 37%. This mixture was applied to the RNA/DNA minikit columns (Purelink), eluted, and re-extracted following the manufacturer's protocols. The eluted nucleic acids were amplified with the Transplex whole-transcriptome amplification kit (Sigma). The reaction mixtures were purified using QIAquick PCR purification kits (Qiagen) followed by passage through Amicon Ultra-0.5 filter columns (100K MWCO). Approximately 100 ng of purified, amplified viral cDNA products were provided to the Broad Institute (Cambridge, MA) for sequencing using the Titanium chemistry on the 454 FLX pyrosequencer (Roche).
Isolation, preparation, and amplification of enriched DNA viral fraction.
To obtain the enriched DNA viral fraction, 10 μl of enriched viral total nucleic acids was amplified with the GenomePlex whole-genome amplification kit (Sigma). The reaction mixtures were purified as described above for the enriched RNA viral fractions. Approximately 5 μg of purified, amplified viral DNA was provided to the Broad Institute (Cambridge, MA) for sequencing as with the enriched RNA viral fractions.
Sequence assembly and analysis.
DNA sequences were trimmed of both primer and tag sequences. To aid in assembly, highly represented duplicated sequences were identified and removed using CD-HIT-454 (50) with a 40-bp overlap and 98% sequence identity. All 7 viral RNA data sets were then cross-assembled using gsAssembler version 2.5 (Roche) at 98% sequence identity with a 50-bp overlap. Cellular and DNA viral metagenomic data sets were assembled using the same procedures. The assembled RNA viral contigs (greater than 100 bp) were compared against the Nymph Lake viral DNA metagenomes, as well as the GreenGenes rRNA data set (18). Sequences matching either DNA viral reads or ribosomal sequences were excluded from future analysis. These filtered RNA viral contigs were subsequently analyzed against the NCBI-nr and -nt databases using BLASTN, BLASTX, TBLASTX, BLASTP, and PSI-BLAST for iterative search of protein sequence databases (3). The RPSTBLASTN program from the NCBI BLAST package was used to compare contigs against the conserved domain database. The resulting metagenomes were also analyzed using the MG-RAST metagenomics analysis server (49) to assign taxonomies. Contigs showing significant matches (E-value of 1 × 10−3) to RNA-dependent RNA polymerases (RdRps) and structural proteins were subjected to a more rigorous examination using HHpred (67) against the pdb70 database (November 2010 release). Sequence assembly and analysis are outlined in Fig. S2 in the supplemental material.
Protein structure analysis.
Secondary structure predictions for RdRp and CP sequences were performed using the HHpred software. Sequence-structure threading was performed using the Phyre server (35) and HHpred, the three-dimensional (3-D) model was built with MODELLER (21), and visualization was rendered using PyMOL (PyMOL Molecular Graphics System, version 1.3r1). Multiple alignment of 3-D structures of capsid proteins was extracted from the DALI database (27).
Phylogenetic analysis of RdRps.
Seed alignments of viral RdRp (cd01699) and group II intron reverse transcriptases (cd01651) were downloaded from the NCBI CDD (Conserved Domain Database) (47). These alignments were used to generate position-specific scoring matrices (PSSM) that served as queries for PSI-BLAST iterative search (4) against positive-strand ssRNA virus sequences in the NCBI RefSeq database, producing 988 matches. The detected RdRp sequences were clustered into 29 groups using the NCBI BLASTClust program (http://www.ncbi.nlm.nih.gov/Web/Newsltr/Spring04/blastlab.html), roughly corresponding to various taxa of ssRNA viruses. Additionally, a cluster of 31 nonredundant sequences of group II intron reverse transcriptases and a cluster with two RdRp sequences from contig00002 and contig00228 were added to the data set. Multiple sequence alignments within clusters were produced using the MUSCLE program (20). Cluster alignments were progressively aligned using the HHalign program (66); at each step, the highest-scoring pair of alignments replaced the two “parent” alignments; alignment positions containing less than 33% nongap characters were removed. For the purpose of phylogenetic analysis, alignment positions containing less than 50% of nongap characters were further eliminated from the final alignment. The phylogenetic tree of the full data set (1,021 sequences) was reconstructed using the FastTree program with default parameters (57). The optimal sequence evolution model for the data set was selected using the ProtTest program (1). A representative data set of 104 sequences was selected using the full tree for guidance; a phylogenetic tree was reconstructed using the RAxML program (69) with the PROTGAMMALGF evolutionary model, as indicated by the ProtTest analysis. Bootstrap analysis with 100 replications and the Shimodaira-Hasegawa log-likelihood test for alternative tree topologies derived from constraint trees were performed using RAxML.
Validation of selected assembled contigs from enriched RNA viral metagenomes by RT-PCR and DNA sequencing.
Up to 2 years after the original samples were collected for the production of the viral and cellular metagenomes, additional NL10, NL17, and NL18 cellular and viral fraction samples were collected in June 2011 and September 2011 as described above to determine whether selected RNA viral genomes were still present in these hot springs. Viral nucleic acids were extracted using the ZR viral RNA kit (Zymo Research) in combination with on-column DNase digestion, as recommended by the manufacturer. Forward and reverse PCR primers designed from selected metagenomic contigs (see Table S2 in the supplemental material) were used with SuperScript III one-step RT-PCR with Platinum Taq (Invitrogen). To test for RT dependency, the RT was excluded from the procedure. To test for strand specificity, only one primer was added during the cDNA synthesis stage and the second primer was added after the 94°C heat kill of the reverse transcriptase and activation of Platinum Taq. PCR products were cloned into the pCR2.1 vector using the TopoTA cloning kit (Invitrogen). Sequencing of the products was performed using Sanger sequencing with BigDye Terminator version 3.1 on an ABI 3100.
Analysis of CRISPR in cellular and viral metagenomes.
Reads from the cellular metagenomes were analyzed for CRISPR-related sequences with the CRISPR Recognition Tool (CRT) (8), using the program's default parameters. Reads identified as containing CRISPRs using CRT were then analyzed using CRISPRFinder (25). All reads identified as CRISPRs with CRT were also identified as CRISPRs with CRISPRFinder. CRISPR-containing reads were parsed for direct repeats (DRs) and spacers generated from CRT. The resulting CRISPR spacers were compared against the viral RNA and DNA metagenomes using BLASTN with an ungapped alignment and 100% nucleotide identity across the entire length of the spacer. Reads with spacers matching RNA contigs were further analyzed by extraction of their associated DRs and identification of their closest reference microbial genome.
Examining the viability of a eukaryotic virus in YNP hot springs.
To test for possible external contamination of eukaryotic viruses in the hot springs examined, 10 ng (4,800 genomes) of purified cowpea chlorotic mottle virus (CCMV), a highly stable ssRNA plant virus, was mixed with 50 ml of hot spring water in a Falcon tube and placed in the hot springs. Aliquots of 50 μl were taken from the sample at various time intervals and analyzed with quantitative RT-PCR (qRT-PCR) using SSoFast EvaGreen supermix (Life Sciences) on a RotorGene-Q system (Qiagen).
Comparing RNA metagenomes to other environments.
To assess whether similar RNA viral genomes were present in YNP high-temperature, near-neutral hot spring environments that are dominated by bacteria, viral RNA metagenomes were compared against transcriptome libraries of Mushroom and Octopus Springs in YNP (36, 43) using BLASTN.
Nucleotide sequence accession numbers.
The nucleotide sequences of contig0002 and contig0028 have been submitted to GenBank under accession numbers JQ756122 and JQ756123, respectively.
RESULTS
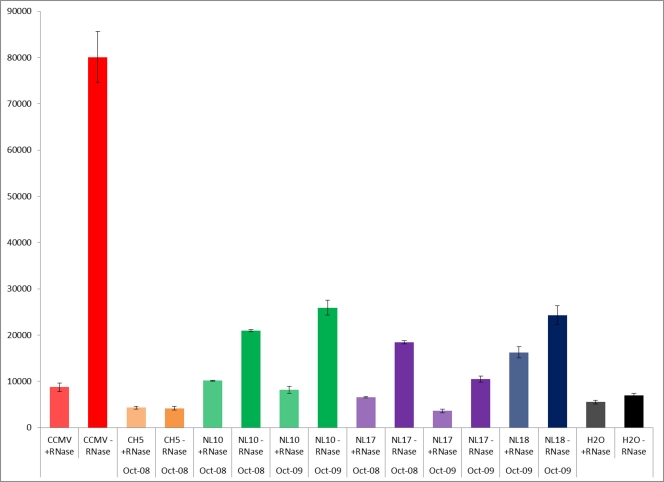
An initial survey of 28 YNP hot springs was performed to determine which hot springs contained a detectable RNA signal in the isolated RNA viral fraction (see Table S1 in the supplemental material). The surveyed sites were selected based on being high-temperature (>80°C) and low-pH (<4.0) springs likely to be dominated by Archaea. This initial screen of viral fractions was based on an RNA-dependent reverse transcriptase (RT) assay where [32P]dCTP incorporation into first-strand cDNA was determined in RNase-treated controls and untreated enriched viral samples, both of which had previously been DNase treated. While 24 sites showed little difference between the amounts of [32P]dCTP incorporated by RNase-treated and untreated samples, 3 hot springs showed reproducible and statistically significant differences (Fig. 1). Resampling of these hot springs 12 months later resulted in nearly identical detection of the RNA signal in the RNA viral fraction, suggesting that RNA viral communities were being maintained in these hot springs. The three sites (NL10, NL17, and NL18) with the highest RT-dependent signals were selected for generating metagenomic libraries.
Fig 1.
Selected examples of the results of screening hot springs for the presence of RNA templates in the RNA virus-enriched fractions. [32P]dCTP incorporation into RT-dependent first-strand cDNA synthesis is shown. For each hot spring sample, results are shown for prior RNase treatment of the sample (+RNase), which was performed as a control. Resampling of selected hot springs 12 months later demonstrated the maintenance of RNA signal in the viral fraction. The positive control was a known positive-strand ssRNA virus, cowpea chlorotic mottle virus (CCMV), and the negative control was water (H2O). Error bars show standard deviations.
A stringent approach was taken to assemble the RNA viral and DNA viral metagenomes. Removal of highly duplicated sequence reads at 98% similarity with CD-HIT-454 resulted in a 41.6% reduction in reads across the viral metagenomes (Table 1). These duplicated sequences are probably indicators of both the amplification bias and the high sequencing depth of the relatively low-complexity viral communities found in these hot springs. Removal of these highly represented sequencing reads significantly improved the overall assembly, resulting in much higher confidence contigs.
Table 1.
Cellular, DNA viral, and RNA viral assembly statistics
| Parameter | DNA metagenomesa | RNA metagenomesa |
Cellular metagenomesb | |
|---|---|---|---|---|
| Pre-DNA filtration | Post-DNA filtration | |||
| Total no. of reads | 1,075,407 | 3,568,86 | 90,227 | 632,479 |
| Total no. of bases (Mb) | 34.0 | 4.2 | 1.1 | 229 |
| % of reads assembled | 72.90 | 34.1 | 34.1 | 64.6 |
| Largest contig (bp) | 21,541 | 5,846 | 5,846 | 23,333 |
| No. of large contigs (>1,000 bp) | 3,411 | 519 | 79 | 4,820 |
| Total no. of contigs >100 bp | 27,746 | 9,696 | 2,636 | 22,649 |
| Average contig length (bp) | 573.9 | 434.3 | 409.5 | 716.8 |
| Avg coverage (fold) | 37.8 | 36.8 | 34.2 | 22.7 |
Sampled from hot spring sites NL10, NL17, and NL18 in October 2009 and February and June 2010.
Sampled from hot spring site NL10 in August of 2008 and 2009.
The initial analysis of the viral RNA metagenomes indicated that they contained a high proportion of sequences found in the paired viral DNA metagenomes. This finding suggests that the initial treatment of the isolated viral nucleic acid with DNase was insufficient to remove 100% of the viral DNA sequences, probably due to its vast excess compared to the amount of RNA in the viral fraction. To address this bias toward DNA viral sequences, RNA contigs were compared against the DNA viral metagenomes using BLASTN. Matches of RNA contigs (E-value of 1 × 10−30) against reads from the DNA metagenomes were excluded from further analysis. More than 72% of the contigs in the initial RNA metagenomic data sets were removed as a result of matching sequences in the viral DNA data sets (7,060 of the initial 9,696 contigs). This screening procedure, while removing the majority of contigs, provided confidence that the remaining contigs assembled from the RNA viral metagenomes were derived from RNA viruses present in that hot spring and not derived from viral DNA sequences.
The assembly of sequences from the Nymph Lake hot spring sites resulted in a large number of contigs under stringent assembly conditions (98% identity within at least 50 bp of overlap) (Table 1). The 14 viral samples (7 enriched RNA viral samples and 7 enriched DNA viral samples) generated ∼2.8 million reads and 807 Mb of sequence. After deduplication, there were 877,000 reads and 590 Mb of unique sequence. The assembled viral DNA metagenomes generated 27,746 total contigs, with an average large-contig size (>1,000 bp) of 1,898 bp. The assembled viral RNA metagenomes generated 9,696 total contigs, with an average large-contig length of 1,361 bp. Nearly 73% of the sequence reads in the DNA viral metagenomes were either fully or partially assembled into contigs, whereas 36.8% of sequence reads in the RNA viral metagenomes assembled into contigs. The assembled viral DNA metagenomic contigs had an average length of 574 bp with an average read depth of 38-fold. The final assembled viral RNA metagenomes had an average contig length of 410 bp with an average read depth of 34-fold base coverage.
Cellular DNA metagenomes, comprising 632,000 sequencing reads which represented 229 Mb of sequence, were assembled under the same high-stringency parameters as the viral data sets (Table 1). The NL0808 and NL0908 cellular assemblies generated 22,649 contigs with an average contig length of 716 bp and an average read depth of 36-fold base coverage. Sixty-five percent of the reads assembled into contigs.
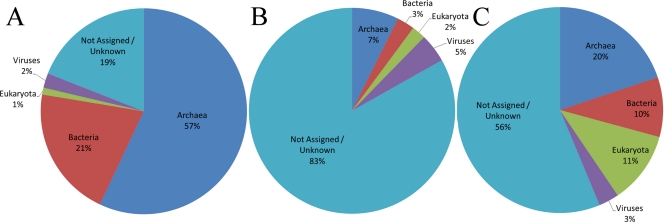
The analysis of the assembled cellular metagenomes revealed that 81.0% of the contigs had a significant match against the server's protein databases (Fig. 2A). The majority of the sequences (57%) matched to Archaea, primarily to the Crenarchaeota (86.9%) and to a lesser degree to the Euryarchaeota (11.6%). Matches to Nanoarchaeota, Korarchaeota, and Thaumarchaeota were also detected. A smaller portion of the cellular contigs matched to Bacteria (20.6%), mostly to Delta- and Gammaproteobacteria and Firmicutes (clostridia and bacilli). However, overall these matches to the Bacteria were weak compared to the matches to the Archaea. This could reflect shared genes between Archaea and Bacteria and/or a statistical bias toward bacterial sequences due to their overrepresentation compared to the representation of archaeal sequences in the public databases. In support of this possibility, further analysis of the cellular metagenomes using the ribosomal databases available on the MG-RAST server (Ribosomal Database Project, SILVA rRNA Database Project, and Greengenes) revealed 4 matches against the Crenarchaeota (Thermoprotei), one match to the Nanoarchaea, and a single distal match to Sulfurihydrogenibium yellowstonense, a bacterium of the phylum Aquificae originally isolated from a YNP hot spring. Overall, these results confirm that the YNP hot springs analyzed here are dominated by Archaea.
Fig 2.
Hierarchical classification of contigs with MG-RAST. Classification of cellular (A), viral DNA (B), and viral RNA (C) sequences based on the M5NR database in MG-RAST. Sequences with insufficient significance or those that did not match were classified as “Not Assigned/Unknown.”
Examination of the putative viral RNA contigs showed that the majority of sequences (56%) did not align to known sequences (Fig. 2C). A small fraction of the sequences matched known viruses (3%). The remaining sequences matched sequences from Archaea (20%), Bacteria (10%), and Eukaryota (11%).
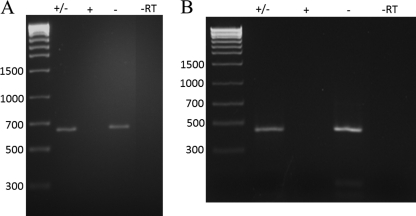
Analysis of the RNA viral contigs using NCBI's protein and nucleotide nonredundant databases and the CDD, as well as Uniprot/Swissprot (31), detected several sequences with similarity to RNA-dependent RNA polymerases (RdRps), a positive-strand single-stranded RNA virus “hallmark gene” (39). The largest contig in the RNA viral data set (5.6 kb) and smaller contigs with similarity to RdRps were confirmed to be present and persistent in the hot springs over time. Resampling of the NL10 and NL18 RNA viral fractions and extraction of the total RNA from the cellular fractions 18 months after the last sampling for the metagenomic analysis showed the continued presence of the RNA genome segments. An RT-PCR assay with primers designed from these RNA contigs was performed on samples taken from the same hot springs over an 18-month period (Fig. 3). These assays demonstrated that sequences were single stranded because amplifiable product was generated only when cDNA synthesis used a specific, directional primer. The longest RT-dependent contig, contig00002, is 5,662 nt in length, composed of 258 reads, and contains a single large open reading frame (ORF) that encodes a putative viral polyprotein encompassing an RdRp and a putative capsid protein as detailed below. Omission of the RT or template or pretreatment of samples with RNase prior to the RT-PCR assay eliminated the signal, indicating that the source of the signal was an RNA template in the viral fraction (Fig. 3). This analysis supported the original contig assemblies and demonstrated that these putative RNA viruses are stable members of the viral community within the hot springs explored.
Fig 3.
Detection and single-stranded nature of RNA viral sequences within hot springs months after sampling for metagenomic analysis. Total nucleic acids of samples obtained 18 months after the original sampling were extracted from either the viral fraction or total cellular fraction of NL10 (B) and NL18 (A), DNase treated, and subjected to RT-PCR for metagenomic analysis. The detection and strand-specific nature of contig00009 (A) and contig00002 (B) were determined using contig-specific primer sets (see Table S2 in the supplemental material). Either forward (+), reverse (−), or both (+/−) primers were added to the initial first-strand cDNA synthesis mix and the mixture incubated as described in Materials and Methods. After first-strand synthesis, the complete primer pair was added (if necessary) for the PCR stage. The control, where reverse transcriptase was excluded from the procedure (−RT), and the molecular-size marker (bp) are indicated.
Further search for RdRps, as well as viral structural (capsid) proteins, was performed using a combination of BLAST, MG-RAST, and HHpred. Two contigs were identified as containing significant similarity to known RdRps. A similar search examining the DNA-enriched viral metagenome data set revealed no similarity to RdRps but did show many hits to capsid proteins, mainly those of archaeal DNA viruses, as expected.
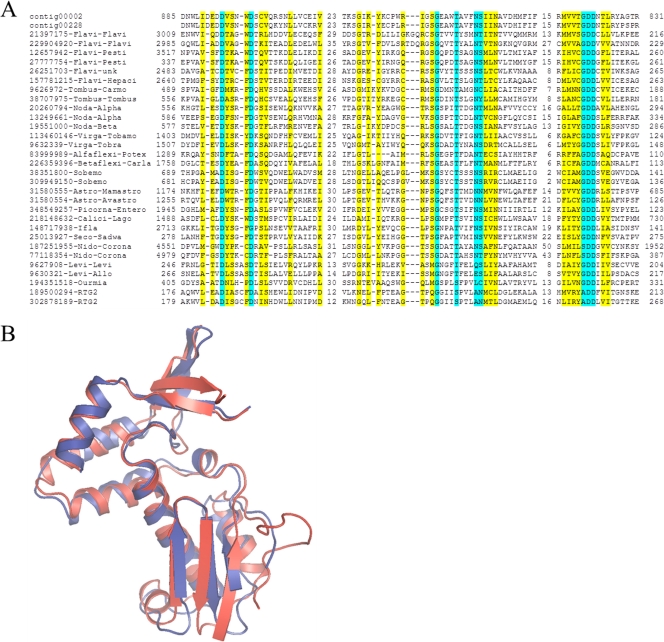
Contig00002 contains a single long ORF potentially coding for a 198-kDa (1,809 amino acids) polyprotein that spans almost the entire length of the contig, suggesting that a complete or nearly complete RNA viral genome was assembled. In the middle part of this polyprotein (approximately between amino acids 800 and 1020), a putative RdRp domain was identified using several search methods. A search of the Conserved Domain Database at the NCBI using the RPS-BLAST program detected a highly significant similarity (E-value of approximately 6 × 10−11) to the RdRp domain profile (no significant similarity to any other domain was detected for this or other parts of the polyprotein). A BLASTP search of the nonredundant protein database at the NCBI yielded statistically significant similarity (E-value of 2 × 10−4, 23% identity in an alignment of 341 amino acid residues) to the RdRp of Pariacato virus, a nodavirus, and limited, not-significant (E-value of approximately 0.1) similarity to the RdRps of several other RNA viruses, including tombusviruses and pestiviruses. The second iteration of the PSI-BLAST search resulted in highly significant similarity to the RdRps of a variety of positive-strand RNA viruses of eukaryotes. This putative RdRp sequence contains all three highly conserved motifs, A (D-X[4,5]-D) and B ([S/T]G-X[3]-T-X[4]-N[S/T)], which are involved in nucleotide selection as well as metal binding (24), and C, which contains the highly conserved GDD motif and is an essential part of the Mg2+-binding site (Fig. 4A) (32, 37). Modeling of the inferred protein RdRp onto the X-ray structure of the positive-strand single-stranded Norwalk virus RdRp structure showed that the putative archaeal virus RdRp contained the principal structural elements of the Palm domain of the RdRps of eukaryotic positive-strand RNA viruses (Fig. 4B). Matches to RdRps were also detected for contig00228 (comprising 10 reads), which encompassed three overlapping short ORFs, each of which showed approximately 70% amino acid sequence identity to the predicted RdRp of contig00002 (Fig. 4A). Thus, this contig appears to encode an RdRp of a putative archaeal virus that is related to but clearly distinct from the putative virus encoded by contig00002.
Fig 4.
The RdRp of the putative archaeal viruses. (A) Multiple sequence alignment of the putative archaeal virus RdRps with homologs from positive-strand RNA viruses of eukaryotes and bacteria and reverse transcriptases from bacterial group 2 introns. The (nearly) universally conserved amino acid residues in the three signature motifs of the RdRps are highlighted in cyan, and partially conserved residues are highlighted in yellow. The top two sequences are from the putative archaeal viruses identified in this work. The rest of the sequences include representatives of the 29 clusters of viral RdRps identified as described in Materials and Methods. The two sequences at the bottom (RTG2) are reverse transcriptases. Each sequence is denoted by the GenBank identifier (GI number) and the name of the virus group (typically family) and subgroup (typically genus). The numbers denote the lengths (number of amino acids) in less-well-conserved regions between the conserved motifs and the distances between the ends of the respective proteins and the aligned segments. (B) Structural model of the central core region of the putative archaeal virus RdRp from contig00002 (blue) using the calicivirus RdRp as a template (red; PDB ID 3bso) (74).
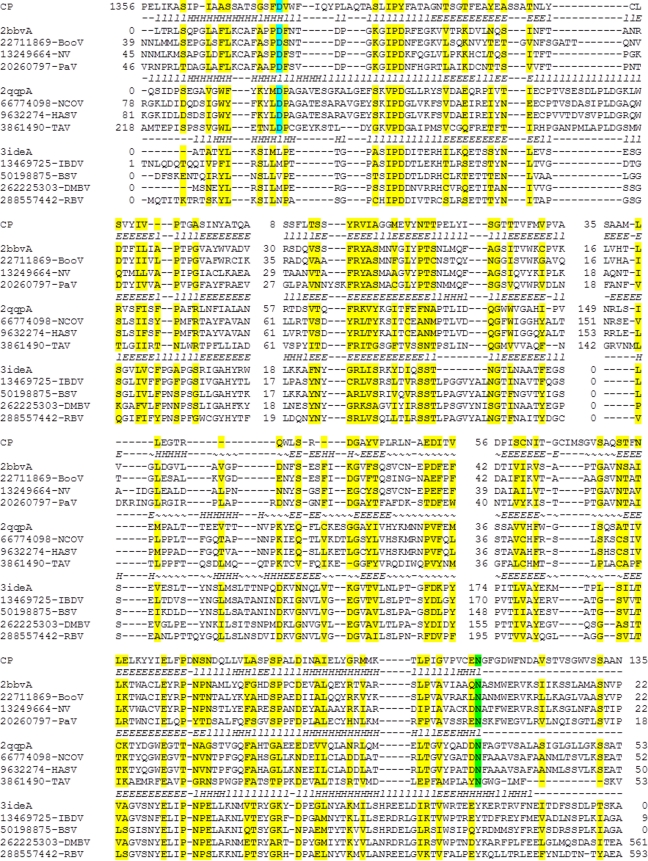
A comparison of the contig00002 polyprotein sequence to databases of protein family profiles using the HHPred program supported the finding of highly significant similarity to viral RdRps and, additionally, led to the detection of sequences that were similar to capsid proteins of positive-strand eukaryotic RNA viruses within the nodavirus family. The sequence with the highest similarity to capsid proteins is approximately 90 amino acids in length and is located C terminal to the RdRp, spanning amino acid residues 1562 to 1652 of the polyprotein. The alignment between this portion of the polyprotein and the capsid proteins of eukaryotic viruses demonstrated a level of similarity that was not statistically significant, although the alignments included approximately 30% identical amino acid residues. Nevertheless, a more detailed, direct comparison of the contig00002 polyprotein sequence to the nodavirus capsid protein sequences, as well as the related capsid protein sequences of tetraviruses and nodaviruses, allowed us to extend the alignment and identify the main structural elements of the capsid proteins (Fig. 5). The nodavirus capsid protein adopts an 8-strand jelly roll fold that is distantly related to the structures of the capsid proteins of other small icosahedral positive-strand RNA viruses. In addition, unlike other well-characterized viral capsid proteins, nodaviruses possess an autoproteolytic activity that is involved in the processing of the capsid protein precursor (the α protein) into the mature large (β) and small (γ) capsid proteins (61, 63). The nodavirus capsid protein is the founding member of the A6 peptidase superfamily that employs an unusual catalytic mechanism involving an interaction between the active aspartate of the capsid protein precursor and a conserved asparagine that is proximal to the scissile peptide bond in the C-terminal part of the protein, at the boundary between β and γ capsid proteins (75). These residues, which are essential for autocatalytic cleavage of the nodavirus capsid protein precursor, are also conserved in the sequences of capsid proteins of tetraviruses. Counterparts to both the catalytic aspartate and the cleavage site asparagine were detected in the contig00002 polyprotein, and regions surrounding these two conserved amino acids were found to have the highest levels of sequence similarity (Fig. 5). Exhaustive analysis of the parts of the contig00002 polyprotein outside the RdRp and capsid protein regions failed to detect similarity to any other known proteins or domains. Collectively, the observation of a large polyprotein (a common feature of eukaryotic positive-strand RNA viruses) containing putative RdRp and capsid proteins in addition to a possible autoproteolytic activity suggest that this contig corresponds to a near full-length genome of a previously unknown positive-strand RNA virus.
Fig 5.
The predicted capsid protein of the putative archaeal virus and its potential autoproteolytic activity. Multiple alignment of the putative archaeal virus capsid protein with the capsid protein sequences of nodaviruses, tetraviruses, and birnaviruses. Sequences are identified either by PDB ID or by GenBank GI numbers. Secondary structure is shown for the three available crystal structures denoted by the corresponding PDB codes (l, loop; H, helix; E, beta-strand). Alignment columns with homogeneity of 0.4 or greater (see Materials and Methods) are highlighted in yellow. The catalytic Asp is highlighted in cyan, and the cleavage site Asn is highlighted in green.
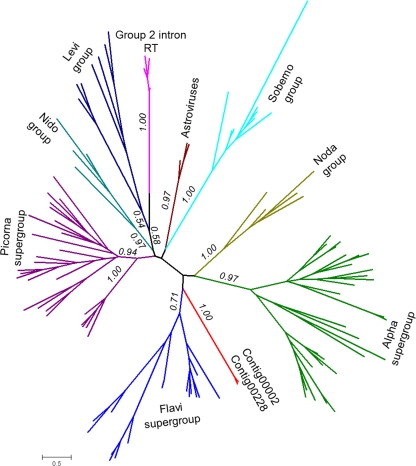
Phylogenetic analysis of the identified RdRp sequences showed that they formed a lineage distinct from known RdRps of eukaryotic and bacterial RNA viruses (Fig. 6). The putative RdRps encoded by contig00002 and contig00228 did not show specific affinity with any of the three superfamilies of eukaryote positive-strand RNA viruses (picornavirus-like, alphavirus-like, and flavivirus-like) or the only known bacterial lineage (leviviruses), and statistical tests showed that association with any of these major lineages or additional distinct lineages, such as nodaviruses, could not be convincingly ruled out (see the supplemental material). These results are compatible with a central position of the new RdRp in the tree and, hence, with its possible ancestral status.
Fig 6.
Phylogenetic tree of the RdRps. The unrooted phylogenetic tree was generated as described in Materials and Methods. Bootstrap support values greater than 0.5 are indicated for deep internal branches. Large groups of positive-strand RNA viruses from eukaryotes and bacteria (Levi group) and the bacterial retrotranscribing elements (RT) are indicated and shown by unique colors.
To investigate the possibility that the RNA virus genomes detected were contaminants of the hot springs introduced from external sources, we tested the ability of an RNA plant virus known for its high stability to be maintained within the hot spring environment. Samples of CCMV were mixed with hot spring water in 50-ml Falcon tubes and placed back into the hot springs. Aliquots were taken at regular intervals for up to 30 min and quantified using a qRT-PCR assay which can detect as few as 10 viral genome copies. Within 1 min of sampling, there was no detectable amount of the CCMV RNA (data not shown). This experiment indicates that it is unlikely that an externally introduced RNA virus can survive long enough to be detected, especially in the form of long RNA segments, under the high-temperature acidic conditions of the hot springs from which the putative archaeal virus sequences were obtained.
To further probe the possibility that the detected RNA viral genomes replicate in archaeal hosts, these sequences were compared to the sequences produced by environmental transcriptomic analysis of two high-temperature YNP hot springs at neutral or alkaline pHs (Mushroom and Octopus Springs) that are known to be dominated by thermophilic bacteria. These hot springs harbor a thermophilic bacterial community of relatively high complexity that consists of Chloroflexi, Cyanobacteria, Acidobacteria, and Chlorobi (36). A BLASTN analysis found that the great majority of the contigs (97% of the total RNA viral contigs) did not have statistically significant matches to the metatranscriptome of these neutral or alkaline YNP hot springs. Three percent of the RNA viral contigs did show a significant match. However, the average length of the matching contigs was 342 bp, much shorter than the overall average contig length. This analysis indicates that there is little overlap between the RNA viral community present in the archaea-dominated acidic hot springs and the bacteria-dominated neutral or alkaline hot springs and provides further evidence that the putative viral RNA genomes detected are associated with archaeal hosts.
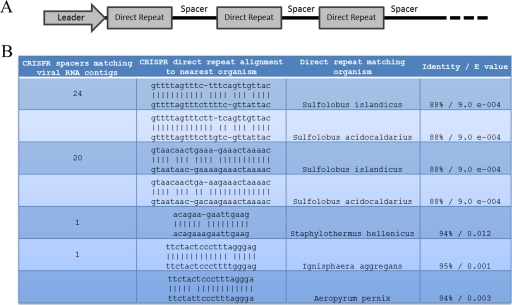
Finally, we analyzed the CRISPR direct repeat (DR) and spacer content present in our cellular metagenomic data sets in an attempt to link the putative RNA viral genomes directly to a specific host type and to investigate potential host immunity to RNA viruses. CRISPR DRs and spacers were extracted from the cellular metagenomic data sets. The spacer sequences (10,349) were compared with the assembled viral RNA and DNA metagenomes. Forty-six spacers (0.44%), associated with 4 types of DRs, were identical to RNA sequences within the viral metagenomic data set (Fig. 7). A similar comparison of the spacers to the DNA viral metagenome revealed 628 (6.07%) identical spacers (data not shown). The majority of matching spacer sequences of the RNA metagenome (44/46) were related to DRs of the archaeal species Sulfolobus islandicus and Sulfolobus acidocaldarius. These were the only significant matches found in the CRISPRfinder direct repeat database. These findings suggest that the RNA viral genomes replicate in an archaeal host belonging to the Sulfolobales, a cell type commonly found in NL10 and acidic hot springs worldwide, and elicit a CRISPR-mediated immune response. However, we cannot categorically rule out the unlikely possibility that the CRISPR spacer matches to the RNA viral metagenome are in fact to DNA viral sequences that are still present in our viral RNA metagenomic data set.
Fig 7.
Analysis of cellular CRISPR direct repeat units and numbers of unique spacers matching archaeal species and RNA viral metagenome contigs. (A) Schematic representation of the CRISPR loci indicating the direct repeat structure interspaced with spacer regions. (B) The number of CRISPR spacer sequences with 100% match to viral RNA contigs and the alignment of the direct repeat structures (top sequence) with the closest sequenced organism (bottom sequence). Percentages of identity and E values are indicated.
DISCUSSION
To our knowledge, this work is the first attempt at viral RNA community analysis in extreme environments. The results demonstrate the presence of putative RNA viral genomes in high-temperature acidic hot springs dominated by Archaea. Several lines of experimental and comparative genomic evidence support the conclusion that the assembled genome segments belong to RNA viral genomes. In particular, two contigs encoded protein sequences with a specific, significant similarity to the RdRp, a hallmark protein of positive-strand RNA viruses. One of these, the 5.6-kb contig00002, might comprise a (nearly) complete viral genome with signature features of positive-strand RNA viruses of eukaryotes. Specifically, this contig encodes a polyprotein that is similar in size to the polyproteins of diverse eukaryote positive-strand RNA viruses that contain both the RdRp and capsid protein domains. Polyproteins of positive-strand RNA viruses of eukaryotes, in particular viruses of the picornavirus-like superfamily, are processed by viral proteases to yield individual functional proteins. A distinct protease domain was not detected in the virus polyprotein encoded in contig00002. However, the catalytic site and the cleavage site of the nodavirus capsid autoprotease were conserved in the region of the putative archaeal virus polyprotein that is similar to the viral capsid proteins (Fig. 5). In nodaviruses, the capsid protein precursor is encoded in a distinct segment of genomic RNA such that the protease is involved only in the autocatalytic maturation of the capsid proteins (15, 62, 75). In the novel virus described here, this protease might perform more versatile roles by catalyzing additional cleavage events and releasing the RdRp, the capsid protein, and possibly, other mature viral proteins that remain to be identified.
The results of this study do not unequivocally demonstrate the existence of archaeal RNA viruses. It cannot be ruled out that the detected RNA genome segments originate from viruses replicating in bacterial hosts present in these hot springs. However, several lines of independent experimental and comparative genomic evidence suggest that the RNA genome segments identified probably originate from viruses replicating in archaeal hosts. First, analysis of both the overall composition of the cellular metagenomes by MG-RAST and the 16S rRNA gene content shows that the microbiota of the sampled hot springs consists primarily of Archaea. Second, the putative RNA viral sequences were recovered from multiple hot springs at multiple time points. Third, experimental exposure of a plant RNA virus known for high stability to the acidic hot springs environment shows that the virus is rapidly degraded under these conditions. Fourth, when we compared the RNA viral sequences detected in the YNP acidic hot springs with the transcriptomes of YNP neutral or alkaline hot springs that are known to be dominated by bacteria rather than archaea, there was no overlap. Taken together, this experimental evidence suggests that any RNA virus sequence that is consistently recovered from the hot springs replicates in archaeal cells.
This evidence is complemented by the sequence analysis results which clearly indicate that the novel virus detected in this study (i) is a positive-strand RNA virus related to positive-strand RNA viruses of eukaryotes and (ii) does not belong to any known virus family. This conclusion is supported both by the topology of the phylogenetic tree of the RdRps (Fig. 6) and by the unique arrangement of protein domains in the polyprotein. It should be further noted that the only known family of bacterial positive-strand RNA viruses, the Leviviridae, is a group of limited diversity that does not show evolutionary affinity to any particular groups of viruses infecting eukaryotes and might not even share an origin with eukaryotic viruses (40). Thus, should the novel virus described here derive from bacteria, its genome organization would be completely unprecedented among genetic elements so far isolated from bacterial hosts.
Finally, when we compared the RNA viral sequences detected in the YNP acidic hot springs with the transcriptomes of YNP neutral or alkaline hot springs that are known to be dominated by bacterial and not archaeal communities, there was no overlap. This supports the notion that the RNA viral communities present in Archaea-dominated hot springs are distinct from the viral communities present in Bacteria-dominated hot spring environments.
Interestingly, we observed multiple matches between archaeal CRISPR spacers and the RNA metagenomes isolated from the hot springs. The interpretation of this observation is complicated by the fact that none of these matches were to the contigs containing sequences homologous to RNA viruses of eukaryotes. Nevertheless, the identification of these spacers might indicate that archaeal RNA viruses elicit CRISPR-mediated immunity, a possibility that is of particular interest given that at least some archaeal CRISPR systems have been shown to target RNA, in contrast to bacterial CRISPRs that appear to only target DNA (26, 71).
The definitive demonstration of the existence of archaeal RNA viruses awaits the isolation of viral particles capable of infecting archaeal hosts and producing infectious progeny. However, assuming that the preliminary indications presented here are born out, the implications for the origin and evolution of positive-strand RNA viruses of eukaryotes will be fundamental. At present, the prokaryotic ancestry of eukaryotic RNA viruses remains unclear. The leviviruses, the only known group of positive-strand RNA viruses of prokaryotes (bacteria), do not seem to be direct ancestors of the viruses of eukaryotes, and the closest homolog of the latter in prokaryotes appears to be the RT of bacterial retrotranscribing elements (40). In contrast, the putative RNA virus of Archaea identified here could be related to the direct ancestors of eukaryotic viruses, as indicated by the homology of the RdRps and the capsid proteins. Moreover, it is notable that the strongest similarity detected for both of these proteins was with the respective proteins of nodaviruses, a family of viruses of eukaryotes that is only distantly related to other positive-strand RNA viruses and is among the simplest known groups of viruses with respect to genome architecture and the protein repertoire (40). The nodavirus family seems to be a good candidate for the ancestral group of eukaryotic RNA viruses that might have evolved directly from archaeal progenitors. Notably, the putative direct ancestral relationship between RNA viruses of archaea and eukaryotes mimics similar relationships that have recently been described for some of the key functional systems of the eukaryotic cells, such as the ubiquitin network (51) or the membrane remodeling and cell division apparatus (46). From the methodological standpoint, this study demonstrates that viral metagenomic approaches have the potential to yield information that is essential to ultimately isolate and characterize novel RNA viruses and their cellular hosts.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grant numbers DEB-0936178 and EF-080220 and National Aeronautics and Space Administration grant number NNA-08CN85A. Y.I.W. and E.V.K. are supported by the Department of Health and Human Services intramural program (NIH, National Library of Medicine).
We thank Mary Bateson, Jamie Snyder, Martin Lawrence, Nikki Dellas, and Brian Bother for critical reading of the manuscript.
Footnotes
Published ahead of print 29 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 [DOI] [PubMed] [Google Scholar]
- 2. Ackermann HW. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227–243 [DOI] [PubMed] [Google Scholar]
- 3. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Angly FE, et al. 2006. The marine viromes of four oceanic regions. Plos Biol. 4:2121–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arnold HP, Ziese U, Zillig W. 2000. SNDV, a novel virus of the extremely thermophilic and acidophilic archaeon Sulfolobus. Virology 272:409–416 [DOI] [PubMed] [Google Scholar]
- 7. Barrangou R, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 [DOI] [PubMed] [Google Scholar]
- 8. Bland C, et al. 2007. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blank CE, Cady SL, Pace NR. 2002. Microbial composition of near-boiling silica-depositing thermal springs throughout Yellowstone National Park. Appl. Environ. Microbiol. 68:5123–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Breitbart M, et al. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 271:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Breitbart M, et al. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Breitbart M, Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278–284 [DOI] [PubMed] [Google Scholar]
- 13. Breitbart M, et al. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cann AJ, Fandrich SE, Heaphy S. 2005. Analysis of the virus population present in equine faeces indicates the presence of hundreds of uncharacterized virus genomes. Virus Genes 30:151–156 [DOI] [PubMed] [Google Scholar]
- 15. Cheng RH, et al. 1994. Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure 2:271–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Culley AI, Lang AS, Suttle CA. 2003. High diversity of unknown picorna-like viruses in the sea. Nature 424:1054–1057 [DOI] [PubMed] [Google Scholar]
- 17. Culley AI, Lang AS, Suttle CA. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795–1798 [DOI] [PubMed] [Google Scholar]
- 18. DeSantis TZ, et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. 2009. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One 4:e7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eswar N, et al. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 5:5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garrett RA, et al. 2011. CRISPR-based immune systems of the Sulfolobales: complexity and diversity. Biochem. Soc. Trans. 39:51–57 [DOI] [PubMed] [Google Scholar]
- 23. Gill SR, et al. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gohara DW, et al. 2000. Poliovirus RNA-dependent RNA polymerase (3D(pol))—structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 275:25523–25532 [DOI] [PubMed] [Google Scholar]
- 25. Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hale CR, et al. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Holmes EC. 2009. RNA virus genomics: a world of possibilities. J. Clin. Invest. 119:2488–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horvath P, et al. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Inskeep WP, et al. 2010. Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5:e9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jain E, et al. 2009. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics 10:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kamer G, Argos P. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial-viruses. Nucleic Acids Res. 12:7269–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kapoor A, et al. 2008. A highly divergent picornavirus in a marine mammal. J. Virol. 82:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karner MB, DeLong EF, Karl DM. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510 [DOI] [PubMed] [Google Scholar]
- 35. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 36. Klatt CG, et al. 2011. Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J. 5:1262–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koonin EV, Boyko VP, Dolja VV. 1991. Small cysteine-rich proteins of different groups of plant RNA viruses are related to different families of nucleic acid-binding proteins. Virology 181:395–398 [DOI] [PubMed] [Google Scholar]
- 38. Koonin EV, Dolja VV. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375–430 [DOI] [PubMed] [Google Scholar]
- 39. Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient virus world and evolution of cells. Biol. Direct 1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koonin EV, Wolf YI, Nagasaki K, Dolja VV. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925–939 [DOI] [PubMed] [Google Scholar]
- 41. Kunin V, Sorek R, Hugenholtz P. 2007. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 8:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lawrence CM, et al. 2009. Structural and functional studies of archaeal viruses. J. Biol. Chem. 284:12599–12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Z, et al. 2011. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J. 5:1279–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Makarova KS, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Makarova KS, Yutin N, Bell SD, Koonin EV. 2010. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat. Rev. Microbiol. 8:731–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marchler-Bauer A, et al. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martin A, et al. 1984. Sav-1, a temperate UV-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 3:2165–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Meyer F, et al. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niu B, Fu L, Sun S, Li W. 2010. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nunoura T, et al. 2011. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 39:3204–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pietila MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72:307–319 [DOI] [PubMed] [Google Scholar]
- 53. Prangishvili D, et al. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the Sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prangishvili D, Garrett RA. 2004. Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses. Biochem. Soc. Trans. 32:204–208 [DOI] [PubMed] [Google Scholar]
- 55. Prangishvili D, Garrett RA. 2005. Viruses of hyperthermophilic Crenarchaea. Trends Microbiol. 13:535–542 [DOI] [PubMed] [Google Scholar]
- 56. Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52–67 [DOI] [PubMed] [Google Scholar]
- 57. Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Reysenbach AL, Wickham GS, Pace NR. 1994. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 60:2113–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rice G, et al. 2001. Viruses from extreme thermal environments. Proc. Natl. Acad. Sci. U. S. A. 98:13341–13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodriguez-Brito B, et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4:739–751 [DOI] [PubMed] [Google Scholar]
- 61. Schneemann A, Gallagher TM, Rueckert RR. 1994. Reconstitution of Flock House provirions: a model system for studying structure and assembly. J. Virol. 68:4547–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schneemann A, Marshall D. 1998. Specific encapsidation of nodavirus RNAs is mediated through the C terminus of capsid precursor protein alpha. J. Virol. 72:8738–8746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schneemann A, Zhong W, Gallagher TM, Rueckert RR. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Snyder JC, Bateson MM, Lavin M, Young MJ. 2010. Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl. Environ. Microbiol. 76:7251–7258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Snyder JC, et al. 2003. Viruses of hyperthermophilic Archaea. Res. Microbiol. 154:474–482 [DOI] [PubMed] [Google Scholar]
- 66. Soding J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21:951–960 [DOI] [PubMed] [Google Scholar]
- 67. Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR–a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186 [DOI] [PubMed] [Google Scholar]
- 69. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 70. Suttle CA. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 71. Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14:321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Venter JC, et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 73. Weinbauer MG. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127–181 [DOI] [PubMed] [Google Scholar]
- 74. Zamyatkin DF, et al. 2008. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283:7705–7712 [DOI] [PubMed] [Google Scholar]
- 75. Zlotnick A, et al. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J. Biol. Chem. 269:13680–13684 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.