Abstract
A unique feature of retroviruses is the packaging of two copies of their genome, noncovalently linked at their 5′ ends. In vitro, dimerization of human immunodeficiency virus type 2 (HIV-2) RNA occurs by interaction of a self-complementary sequence exposed in the loop of stem-loop 1 (SL-1), also termed the dimer initiation site (DIS). However, in virions, HIV-2 genome dimerization does not depend on the DIS. Instead, a palindrome located within the packaging signal (Psi) is the essential motif for genome dimerization. We reported previously that a mutation within Psi decreasing genome dimerization and packaging also resulted in a reduced proportion of mature particles (A. L'Hernault, J. S. Greatorex, R. A. Crowther, and A. M. Lever, Retrovirology 4:90, 2007). In this study, we investigated further the relationship between HIV-2 genome dimerization, particle maturation, and infectivity by using a series of targeted mutations in SL-1. Our results show that disruption of a purine-rich (392-GGAG-395) motif within Psi causes a severe reduction in genome dimerization and a replication defect. Maintaining the extended SL-1 structure in combination with the 392-GGAG-395 motif enhanced packaging. Unlike that of HIV-1, which can replicate despite mutation of the DIS, HIV-2 replication depends critically on genome dimerization rather than just packaging efficiency. Gag processing was altered in the HIV-2 dimerization mutants, resulting in the accumulation of the MA-CA-p2 processing intermediate and suggesting a link between genome dimerization and particle assembly. Analysis of revertant SL-1 mutant viruses revealed that a compensatory mutation in matrix (70TI) could rescue viral replication and partially restore genome dimerization and Gag processing. Our results are consistent with interdependence between HIV-2 RNA dimerization and the correct proteolytic cleavage of the Gag polyprotein.
INTRODUCTION
All known exogenous retroviruses except for spumaretroviruses encapsidate two copies of their RNA genome, linked at their 5′ ends through the dimer linkage site (DLS) (8). Genome dimerization plays an important role during several steps of the virus life cycle, including reverse transcription and packaging (reviewed in references 25 and 51), and allows for greater genetic diversity by facilitating recombination during reverse transcription (30). In human immunodeficiency virus type 1 (HIV-1), the cis-acting elements required for genome dimerization and encapsidation overlap and have been mapped to four stem-loops (SL-1 to SL-4) located in the 5′ leader of the genome, suggesting that the two processes may be linked (6, 7, 12, 13, 28, 39, 50, 54). The commonly accepted model for the initiation of HIV-1 genome dimerization is through a loop-loop interaction at the dimer initiation site (DIS)—a palindromic sequence located in the apical loop of SL-1 (14, 38, 47, 52, 57)—though there is some suggestion that the DIS may not be required for HIV-1 dimer formation (7, 29). Although in vitro studies suggested that a similarly sited palindrome in the loop of SL-1 might initiate HIV-2 RNA dimerization (20, 35), previous work in our laboratory demonstrated that the DIS is dispensable for dimerization of the HIV-2 genome in vivo (41). Instead, a second palindrome, termed pal (392-GGAGUGCUCC-401), located within the encapsidation signal Psi and part of SL-1, can promote RNA dimerization both in vitro and in vivo (34, 41).
In both HIV-1 and HIV-2, mutations within the packaging signal have been shown to impair genomic RNA dimerization and packaging and to adversely affect viral replication (26, 34, 36, 41). Long-term culture and phenotypic reversions of pal-mutated HIV-2 have led to the suggestion that an extended SL-1, formed through the base-pairing of stem B, is essential for efficient HIV-2 replication and RNA packaging (36). The 3′ end of pal (GCUCC-3′) is required for the formation of stem B and the extended SL-1. However, infectivity and viral replication are significantly impaired by the sole mutation of the 5′ end of pal (5′-GGAGU), which is not part of stem B (5, 41). Therefore, the precise sequence and structural elements of SL-1 required for HIV-2 genome dimerization, packaging, and viral replication remain incompletely defined.
Rous sarcoma virus and HIV-1 Gag have been shown to assemble in vitro in the absence of nucleic acid (10); however, it has been suggested that the retroviral RNA genome plays a role in particle assembly (9, 24), and there is increasing evidence that the Gag protein needs to bind both the RNA genome and the plasma membrane for the correct folding and assembly of the polyprotein to occur (17, 48). RNA dimerization, in particular the maturation step, depends on the viral protease and the proteolytic processing of Gag (22). Specifically, the primary cleavage of Gag between p2 (also known as SP1) and the nucleocapsid (NC) is required for the maturation of the HIV-1 RNA dimer (55). HIV-1 dimerization mutants have been shown to have a delay in the cleavage of capsid (CA)-p2 (42), and simian immunodeficiency virus macaque (SIVmac239) SL-1 mutants with reduced genome dimerization and encapsidation showed increases in levels of the intermediate Gag cleavage products matrix (MA)-CA and CA-NC, as well as aberrant morphology (59). It was recently proposed that HIV-1 p2 undergoes a coil-to-helix structural refolding required for particle assembly (18), although the elements or trigger necessary for this structural change remain unknown. However, the suggestion that interactions of Gag with itself, nucleic acid, and the plasma membrane are all required for full extension of the protein during assembly (17), together with evidence that the introduction of a leucine zipper could supplant the NC-RNA interaction and lead to particle assembly (15, 31, 60), has raised the possibility that the dimeric genome serves as a scaffold during assembly, helping to bring the Gag and Gag-Pol molecules into close proximity and the correct orientation to allow the required conformational change to occur.
In this study, we set out to investigate further the exact sequences and/or structures involved in HIV-2 genome dimerization and viral replication and to analyze the role of the dimeric genome during HIV-2 particle assembly and maturation. Using a series of substitution mutations in SL-1, we analyzed the conformation of the genome and the packaging efficiencies of mutant viruses and assessed their replication in T cells. Our results show that a purine-rich motif within the Psi region (392-GGAG-395) is required for HIV-2 replication, as suggested previously by an in vivo selection approach (5), and that this motif is essential for genome dimerization and encapsidation. Independently of dimerization, stem B enhances encapsidation. However, we demonstrate that it is genome dimerization rather than packaging that is critical for HIV-2 replication. A moderate packaging defect with a dimeric genome is compatible with efficient replication.
Analysis of Gag processing in the SL-1 mutant viruses revealed alteration of the rate of Gag processing in the dimerization mutants, resulting in the accumulation of the intermediate product MA-CA-p2. Revertant viruses harboring compensatory mutations in Gag were identified, including a matrix threonine-to-isoleucine mutation at position 70 (MA 70TI), which had been reported previously in SIVmac239 in the context of an SL-1 dimerization mutant (58) and which in HIV-1 rescues a membrane-binding and assembly defect (49). Introduction of the MA 70TI mutation into the HIV-2 dimerization mutants rescued viral replication, significantly restored the ability of the virus to form a dimeric genome, and corrected the processing defect of Gag. Taken together, the data presented here highlight the dependence of HIV-2 replication on the process of RNA dimerization and support a model in which the dimeric conformation of the packaged genome is required for the correct assembly of HIV-2 virions, possibly by acting as a scaffold to allow the correct folding and processing of the Gag polyprotein.
MATERIALS AND METHODS
Plasmid constructs.
pSVR is an infectious proviral clone of HIV-2 ROD containing a simian virus 40 origin of replication (44). pSVRΔNB contains a 550-nucleotide (nt) deletion in the env open reading frame (ORF) between positions 6369 and 6919 (26). Restriction sites and nucleotide numbering, where given, are relative to the first nucleotide of the viral RNA. Mutations in the 5′ leader were introduced into a subclone of HIV-2, pGRAXS (26), by site-directed mutagenesis using the QuikChange II kit (Stratagene). The sequences of the mutagenic primers are available on request. Sequences from the resulting subclones were introduced into the provirus by exchanging an AatII-XhoI fragment, generating the corresponding proviral constructs. All proviral constructs were verified by sequencing. pSVRNCm contains alanine substitutions for all cysteine and histidine residues in the two zinc fingers of the NC domain of Gag and has been described previously (32). Plasmid KS2ES, used to generate the antisense riboprobe to detect HIV-2 genomic RNA (positions 4915 to 5284) by Northern blotting, has been described previously (33).
Cell culture.
293T (European Collection of Cell Cultures [ECACC]) and C-33A (American Type Culture Collection [ATCC]) cells were maintained in Dulbecco's modified Eagle medium (DMEM) (PAA Laboratories) supplemented with 10% fetal calf serum (FCS; PAA), penicillin, and streptomycin (PAA) (complete DMEM). PM1 T cells (Centre for AIDS Reagents [CFAR], National Institute for Biological Standards and Control [NIBSC], United Kingdom) were maintained in RPMI 1640 (PAA) supplemented with 10% FCS, penicillin, and streptomycin (complete RPMI).
Virus preparation.
293T or C-33A cells were transfected with proviral constructs by using TransIT-LT1 (Mirus Bio) according to the manufacturer's instructions. Cells and supernatants were harvested 48 h later, and virus production was assessed with a reverse transcriptase (RT) assay (53). Virus-containing supernatants were purified through an 8.4% OptiPrep (Sigma-Aldrich) cushion by ultracentrifugation at 98,000 × g for 1 h at 4°C. Virus pellets were resuspended either in 500 μl of complete RPMI for infection or in 200 μl of proteinase K buffer for RNA extraction.
Virus infection and evolution study.
Wild-type (WT) and mutant viruses were prepared as described above by transfection of 293T cells with the appropriate proviral plasmid. An equivalent of 500 RT units of virus was used to infect 1 × 106 PM1 cells in a total volume of 5 ml. Viral replication was followed by measurement of the RT activity in the culture supernatant every 2 to 4 days, and cells were passaged 1/5 every 4 days. During the evolution study, upon replication of previously replication incompetent virus, 1 ml of cells was harvested, and 0.5 ml of the cell-free supernatant was used to infect 1 × 106 fresh PM1 cells as described above, while the cell pellet and remaining supernatant were stored at −20°C until reversion was confirmed by an improved replication kinetic. The cell pellet was then resuspended in 0.5 ml phosphate-buffered saline (PBS) and was lysed by the addition of 100 μl of lysis buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.5% Tween 20) and 6 μl of 20-mg/ml proteinase K. The mixture was mixed gently and was incubated at 56°C for 1 h. After inactivation of the proteinase K for 10 min at 95°C, 1 μl of the genomic DNA preparation was used in PCR with different sets of HIV-2-specific primers (sequences available on request) and high-fidelity Accuzyme DNA polymerase (Bioline). PCR products were purified by using the PCR purification kit (Qiagen) and were sent for sequencing analysis.
RNA preparation.
293T cells were transfected with a WT or mutant proviral plasmid as described above, and cytoplasmic RNA was harvested 48 h later by using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Viral RNA was extracted from purified virions by lysis of virus particles in 200 μl of proteinase K buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 100 μg/ml of proteinase K, 100 μg/ml of tRNA) for 30 min at 37°C. After extraction with acid-buffered phenol-chloroform, followed by one extraction with chloroform-isoamyl alcohol (IAA), the RNA was precipitated with ethanol at −80°C. Cytoplasmic and virion RNAs used for quantitative RT- PCR (qRT-PCR) were subsequently treated with 30 U of Turbo DNase (Life Technologies) for 2 h at 37°C, extracted once with phenol-chloroform and chloroform-IAA, and resuspended in 50 μl RNase-free H2O. Virion RNAs used in Northern blotting were resuspended in 20 μl of a buffer containing 140 mM NaCl, 10 mM Tris-HCl [pH 7.5], and 5 mM MgCl2.
Northern blotting.
Northern blotting was performed using the NorthernMax-Gly kit (Life Technologies) as recommended by the manufacturer. Virion-extracted RNAs (20 μl) were separated on a 0.8% agarose gel for 4 h at 4°C in 1× Gel Prep/Running Buffer, alongside the RiboRuler high-range RNA ladder (Fermentas). RNAs were transferred to a BrightStar-Plus positively charged nylon membrane (Life Technologies), cross-linked by baking for 20 min at 80°C, and detected using the BrightStar Biodetect kit according to the manufacturer's instructions. The biotinylated riboprobe used for the detection of HIV-2 genomic RNA was transcribed in vitro from the linearized (BamHI) KS2ES plasmid using T3 RNA polymerase (Promega) and a biotin RNA labeling mix (Roche). Following DNase treatment (with 10 U of Turbo DNase for 30 min at 37°C), the riboprobe was purified through a G-50 Sephadex column, aliquoted, and stored at −20°C.
Quantification of genomic RNA by qRT-PCR.
Cytoplasmic and virion RNAs were normalized to total-RNA concentration and RT activity, respectively, prior to quantification by one-step qRT-PCR using the SuperScript III One-Step RT-PCR system (Life Technologies) in a Rotor-Gene Q instrument (Qiagen). HIV-2 genomic RNA was detected using a gag-specific fluorescent probe, 6-carboxyfluorescein (FAM)-CCCAAGTCCCGCAG-minor groove binder (MGB) (Life Technologies), and primers 5′-GCAGGGCTGCTGGAAGTGTGGTAA-3′ and 5′-ACTGGGGGTGCTGTTGGTGTCA-3′ (Sigma-Aldrich). Human PGK1 (Phosphoglycerate Kinase 1) Endogenous Control (VIC/MGB probe, primer limited; Life Technologies) was used as an endogenous control in the cytoplasmic RNA samples. Cycling conditions were as follows: an initial reverse transcription step of 30 min at 50°C, followed by 2 min at 95°C and 45 cycles of 95°C for 15 s and 60°C for 1 min. The specific amplification of a 225-nt fragment (positions 1772 to 1997 of the HIV-2 genome) was validated by gel electrophoresis.
Western blotting.
Forty-eight hours after the transfection of 293T cells, virions were purified as described above, resuspended in 50 μl PBS, and lysed by the addition of 2× Laemmli buffer (Sigma-Aldrich). Samples were boiled and proteins resolved on 12% SDS-polyacrylamide gel electrophoresis (PAGE) gels. Gag was detected using a mouse anti-SIVp57/p27 antibody (ARP3005; CFAR, NIBSC, United Kingdom) followed by a horseradish peroxidase (HRP)-conjugated anti-mouse antibody (Santa Cruz Biotechnology). A mouse anti-β-actin antibody (Abcam) was used to verify equal loading of the cytoplasmic fractions.
Pulse-chase metabolic labeling of Gag.
C-33A cells were transfected with env-deleted proviral constructs in 6-well plates as described above. After 24 h, cells were washed twice in PBS and were starved for 60 min in 0.5 ml of DMEM lacking methionine and cysteine (Met-Cys-free DMEM; Sigma-Aldrich) supplemented with 4 mM glutamine (Gln; PAA), 10% FCS, penicillin, and streptomycin. Cells were then pulse-labeled for 30 min with 0.5 ml of Met-Cys-free DMEM supplemented with 110 μCi of EasyTag Express 35S protein labeling mix (11 mCi/ml; Perkin-Elmer) and 4 mM Gln, 10% FCS, penicillin, and streptomycin. The radioactive medium was removed (time point zero), and cells were chased in 0.5 ml of complete DMEM supplemented with 2 mM Met and 2 mM Cys for 30, 60, 90, and 120 min. At each time point, the virus-containing supernatants were harvested and their concentrations adjusted to 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS. The capsid (CA) and Gag proteins were immunoprecipitated using an anti-SIVp57/p27 antibody (ARP3061; CFAR, NIBSC United Kingdom) diluted at 1/150 overnight at 4°C. One hundred microliters of a mixture of protein A- and protein G-Sepharose beads (Sigma-Aldrich) prepared in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, protease inhibitor cocktail [Roche]) was added, and samples were rotated for a further 1 h 30 min at 4°C. Samples were spun at 6,000 × g for 2 min, washed three times in RIPA buffer, and resuspended in 50 μl 2× Laemmli buffer. Samples were boiled for 5 min, and 25 μl was loaded onto a 13% acrylamide gel alongside a Precision Plus protein marker (Bio-Rad). Gels were fixed in 40% methanol–10% acetic acid for 30 min, incubated in Amplify (GE Healthcare) for 30 min, and dried prior to autoradiography. Quantification (densitometry analysis) was performed using ImageJ software.
RESULTS
Mutation of the HIV-2 leader RNA.
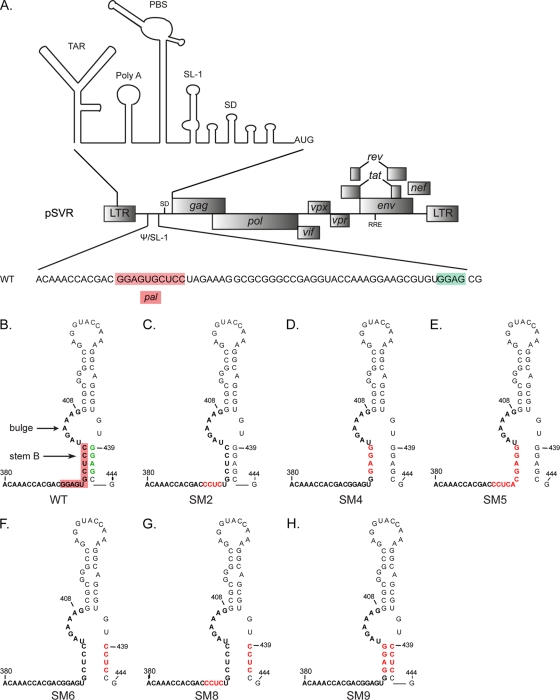
To investigate the exact requirement for the palindrome pal and the SL-1 structure and in particular to establish the precise role of stem B in HIV-2 viral replication, we introduced a series of mutations into the SL-1 region of the leader RNA of an infectious HIV-2 molecular clone (pSVR) (Fig. 1A), disrupting either the pal sequence or the structure of SL-1, or both (Fig. 1B to H). A previously characterized replication- and dimerization-deficient mutant, SM2 (392-CCUC-395) (41), was included for comparison (Fig. 1C). The SM4 and SM5 mutations both lead to disruption of stem B, with SM4 also disrupting pal, while SM5 retains a palindromic sequence (Fig. 1D and E). In SM6, a GGAG motif at the 3′ end of SL-1 was mutated to disrupt stem B while leaving pal unaffected (Fig. 1F). To assess the relative importance of the stem B structure compared to its constituent sequences, the mutations of SM4 and SM6 were combined (SM9) (Fig. 1H). Finally, since several GGAG motifs are present in SL-1 and similar purine tetrads have been implicated in RNA packaging and Gag binding in both HIV-1 (1, 3, 11, 16, 19, 40, 56) and HIV-2 (16), the mutations of SM2 and SM6 were combined (SM8) (Fig. 1G).
Fig 1.
Structure of stem-loop 1 in wild-type and mutant HIV-2. (A) Schematic of the molecular clone pSVR of the HIV-2 ROD genome, with the structure of the 5′ leader RNA (20) shown above and the sequence of the packaging signal (Psi)/stem-loop 1 (SL-1) region detailed below. The palindrome pal (residues 392 to 401) and the GGAG motif at position 439 are highlighted in red and green, respectively. TAR, trans-activator response; PBS, primer binding site; SD, major splice donor. (B to H) Structures of SL-1 in the WT (B) and substitution mutants (C to H) based on the previously described structure of SL-1 (4). The palindrome pal is highlighted in red on the WT structure. Stem B and the distal bulge (residues 402 to 409) are indicated. SM2 (C) (residues 392 to 395) has been described and characterized previously (41). Red letters represent mutated residues.
Defects in HIV-2 genome dimerization, but not RNA encapsidation, correlate with impaired viral replication.
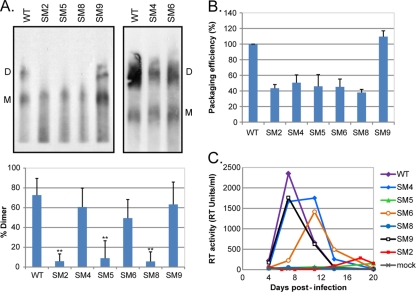
We wanted to clarify and reconcile previous in vitro and in vivo data on HIV-2 dimerization (37, 41) and the association with viral infectivity (41). Thus, we analyzed the ability of the mutant viruses to form genomic RNA dimers in vivo. WT and mutant virions were purified from transfected cells, and the viral genomic RNA was analyzed by native Northern blotting (Fig. 2A). Disruption of stem B (mutants SM4 and SM6) had no significant effect on the proportion of dimeric genomes present in the virions, indicating that the structure of SL-1 does not play a role in HIV-2 RNA dimerization. Replacement of the original pal sequence by an alternative palindrome (SM5) did affect RNA dimerization, suggesting that the native sequence is important. However, the self-complementarity of pal is disrupted in SM9, which dimerized as efficiently as the WT. To clarify this discrepancy, we compared the dimerization of two pal mutants, the previously described SM2 mutant (41), in which the 5′ end of pal is mutated, and SM4, in which the 3′ end of pal is mutated. The dimerization efficiencies of these mutants indicated that only the 5′ end of pal (392-GGAG-395) was required for HIV-2 genome dimerization. The level of dimers detected for the SM2 mutant differed slightly between the current study and our previously published study (41). This is most likely due to differences in the RNA extraction protocol. In this study, a more stringent extraction method was used, which would potentially increase the dissociation of loose dimers but would have little or no effect on the integrity of tight dimers. The levels of wild-type dimer are similar in the two studies, consistent with this interpretation. Mutations of both GGAG motifs present in Psi/SL-1 (SM8 virus) drastically reduced genome dimerization. Comparison of the SM2 and SM6 viruses (Fig. 2A) (41) strongly suggests that 392-GGAG-395 is the critical element for HIV-2 genome dimerization (Table 1). While mutants lacking the 392-GGAG-395 motif have an unequivocal dimerization defect, it should be noted that the method used does not allow us to determine whether the RNA extracted from virions is truly monomeric or represents unstable dimer that is disrupted by the extraction procedure. Increasing evidence that the HIV-1 genome is packaged as a dimer (45, 46), together with the disparities in the levels of dimer obtained for the SM2 mutant by different extraction methods, would suggest that loose dimers are initially formed but are not stable enough to withstand the more stringent extraction procedure.
Fig 2.
HIV-2 replication is dependent on efficient genome dimerization and the presence of the 392-GGAG-395 motif in the RNA leader. (A) (Top) Native Northern blot analysis of HIV-2 genomic RNA; (bottom) quantification (mean + standard deviation) (n = 3) of the percentage of dimeric RNA in the virions. 293T cells were transfected with WT and mutant proviral constructs, and viral RNA was extracted at 48 h posttransfection and was analyzed by native Northern blotting using an antisense riboprobe specific for pol. The positions of the dimeric (D) and monomeric (M) RNA species are indicated. Asterisks indicate a significant difference from the WT (**, P < 0.002) by an unpaired Student t test. (B) Packaging efficiencies of the mutant viruses relative to that of the WT, measured by qRT-PCR using the standard-curve method of quantification. The packaging efficiency of the WT was set to 100%. (C) WT and mutant HIV-2 virions were produced by transfection of 293T cells with the corresponding proviral plasmid, purified through 8.4% OptiPrep, and used to infect 1 × 106 PM1 T cells. Virus input was normalized on the reverse transcriptase (RT) activity of the virus preparation, and an equivalent of 500 RT units was used. Viral replication was followed by measuring the RT activity in the culture supernatant every 4 days. Mock, mock-infected cells.
Table 1.
Correlation between viral replication, RNA dimerization, and packaging and the sequences and structural elements located within HIV-2 SL-1
| Virus | Viral replicationa | Genome dimerization (%)b | Packaging efficiencyc | Presence or absence of: |
|||
|---|---|---|---|---|---|---|---|
| pal | Stem B | 392-GGAG-395 | 439-GGAG-442 | ||||
| WT | ++ | 72.7 | 1.00 | + | + | + | + |
| SM4 | ++ | 60.6 | 0.51 | − | − | + | + |
| SM6 | + | 49.4 | 0.45 | + | − | + | − |
| SM9 | ++ | 63.2 | 1.10 | − | + | + | − |
| SM2 | − | 5.8 | 0.44 | − | + | − | + |
| SM5 | − | 8.9 | 0.46 | +d | − | − | + |
| SM8 | − | 5.6 | 0.38 | − | − | − | − |
Replication in PM1 T cells was followed by measuring the RT activity in the culture supernatant. Symbols indicate the presence (++) or absence (−) of detectable RT activity at 4 to 15 days postinfection (n = 3).
Percentage of dimeric genome in the virion as measured by native Northern blotting (n = 3).
Relative to that of the WT (set to 1) as measured by qRT-PCR (n = 3).
Alternative palindromic sequence in place of WT pal.
It was proposed previously that stem B is required for efficient packaging of the HIV-2 genome and viral replication (36). Since stem B does not appear to play a role in genome dimerization, we decided to measure the packaging efficiencies of our Psi/SL-1 mutants relative to that of the WT (Fig. 2B). Our data confirmed the requirement for stem B in HIV-2 RNA encapsidation, but only in combination with the 392-GGAG-395 motif (Table 1). Indeed, the formation of stem B alone is not sufficient to promote efficient RNA packaging, as illustrated by the poor packaging efficiency of the SM2 mutant (5, 41; this study).
We next assessed the abilities of the SL-1 mutant viruses to replicate in T cells (Fig. 2C). We observed a striking direct correlation between the efficiency of genome dimerization and viral replication, although a packaging efficiency of 100% is not absolutely necessary for HIV-2 propagation. A 2-fold reduction in RNA packaging had no significant effect on the viral replication of the SM4 and SM6 mutants, whereas a major reduction in the level of dimeric genome abrogated replication of the SM2, SM5, and SM8 viruses. These results are in agreement with previous data showing that a GGRG motif at position 392 is required for HIV-2 replication, but they do not support an essential role for stem B in viral replication (5). The data presented here do strongly suggest that the conformation of the genomic RNA encapsidated in the virion, rather than the packaging efficiency, is critical for HIV-2 replication.
A compensatory mutation in matrix can rescue the replication of dimerization mutants.
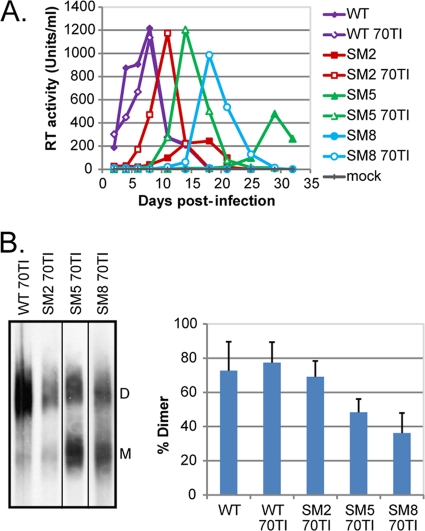
To seek reversion mutations with restored replication kinetics, T cells infected with the dimerization mutants were cultured for more than 3 weeks; eventually, replicating virus was detected in all three cultures (data not shown). Cell-free supernatant was used to infect fresh cells, and a faster replication kinetic than that with the original infection was observed, suggesting that a reversion event had occurred. The infected cells were harvested at the peak of infection; total DNA was isolated; and the 5′ untranslated region (5′ UTR) and gag gene of the integrated proviruses were sequenced. Every virus sequenced had retained the original mutation in the Psi/SL-1 region. However, several second-site mutations were identified in the gag gene, most notably a C-to-T mutation at position 754, resulting in a Thr-to-Ile mutation at position 70 of matrix (MA 70TI), which was identified in both SM5 and SM8 and had been observed previously in SIVmac239 in the context of an SL-1 dimerization-deficient mutant also harboring an 184MV mutation in RT (58).
The MA 70TI mutation was therefore introduced into the SM2, SM5, and SM8 proviral constructs, and the revertant viruses were phenotypically characterized (Fig. 3). The replication of WT HIV-2, the three dimerization mutants, and their corresponding revertant viruses was followed over a period of 4 weeks (Fig. 3A). The three MA 70TI revertant viruses were able to rescue the viral replication of SM2, SM5, and SM8, bringing it close to the WT level. Genome dimerization was then assessed by native Northern blotting (Fig. 3B). The MA mutation was able to rescue RNA genome dimerization by the SL-1 mutants (P, <0.05 by an unpaired Student t test). Introduction of the MA 70TI mutation into WT proviral DNA had no significant effect on viral replication or genome dimerization (Fig. 3A and B).
Fig 3.
A compensatory mutation in MA (70TI) can rescue the replication of dimerization mutants and partially restore genome dimerization. (A) PM1 T cells were infected with an equivalent of 500 RT units of WT, dimerization mutant, and MA 70TI mutant viruses. Viral replication was followed by measuring the RT activity in the culture supernatant every 2 to 4 days. 70TI, mutation in matrix at position 70; mock, mock-infected cells. (B) (Left) Native Northern blot analysis of MA 70TI mutant viruses; (right) quantification (means + standard deviations) (n = 2) of the percentages of dimeric RNA in the virions. The WT data are reported from Fig. 2A. D, dimer; M, monomer.
Overall, these data show that a second-site mutation at position 70 in matrix can rescue the replication of dimerization- and packaging-deficient mutant viruses and can partially restore genome dimerization.
During the course of our evolution study, other second-site mutations were identified, including MA 68VE, CA 26VI, CA 98TA, p6 35EK, p6 42EK, and p6 53EK. The p6 EK mutations alone are not sufficient to rescue infectivity—introduction of the 42EK mutation into the SM5 background did not rescue viral replication—indicating that other second-site mutations may have arisen elsewhere in the genome at the same time and may have been necessary for the improved replication fitness observed (data not shown). This corroborates a previous study with SIVmac239 in which a p6 49EK compensatory mutation was able to rescue the replication of an SL-1 dimerization mutant only in combination with two second-site mutations, nt 423AG in the DIS loop and CA 197KR (27). However, none of our evolution studies yielded reversion of the RNA 5′ UTR or any similar mutation in CA.
Dimerization-deficient mutants show a processing defect that is partially rescued by the MA 70TI reversion mutation.
Proteolytic processing of Gag is known to be associated with the RNA dimerization process, promoting the maturation of the dimer, as shown by analysis of protease-deficient HIV-1 and murine leukemia virus (MLV) virions (22, 23). Conversely, mutations reducing HIV-1 genome dimerization have been shown to impair Gag processing by delaying the CA-p2 cleavage (42). With this in mind, we investigated the effects of our SL-1 mutations on Gag synthesis and processing.
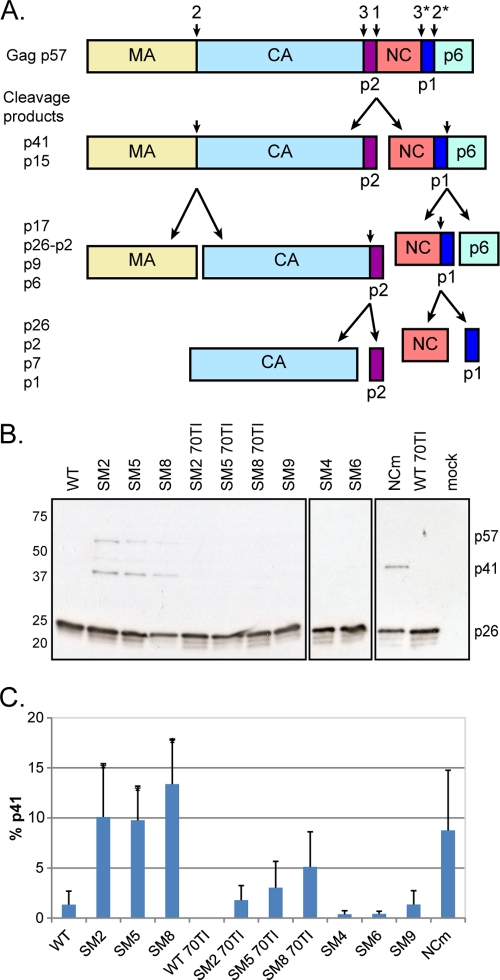
We first analyzed the levels of Gag and its cleavage products (Fig. 4A) in the particles released at 48 h posttransfection (Fig. 4B). While all the Gag protein has been cleaved into CA in the WT virus, the dimerization mutants showed a delay in Gag processing, with a small yet significant accumulation of the p41 (MA-CA-p2) intermediate (Fig. 4C). The replication-competent SL-1 mutants and the MA 70TI revertant viruses all showed normal processing of Gag (Fig. 4B and C). To examine further the contribution of dimer formation and Gag-RNA interaction to Gag processing, we analyzed an HIV-2 Gag mutant harboring cysteine-to-alanine and histidine-to-alanine substitutions in the two zinc finger domains of NC (NCm), which abrogate Gag-RNA binding (32). This mutant displayed the same phenotype as the SL-1 dimerization mutants (Fig. 4B and C), suggesting that binding of Gag to the genomic RNA is important for the correct processing of the protein.
Fig 4.
Mutations affecting RNA dimerization and Gag-RNA interaction result in suboptimal processing and the accumulation of the p41 intermediate. (A) Schematic of the Gag polyprotein (p57) and its proteolytic processing. The sites and order of cleavage are indicated above the p57 precursor and each intermediate product. p17, matrix (MA); p26, capsid (CA); p2, spacer peptide p2, or SP1; p7, nucleocapsid (NC); p1, spacer peptide p1, or SP2; p41, MA-CA-p2; p15, NC-p1-p6; p9, NC-p1. (B) Western blot analysis of virion proteins, extracted at 48 h posttransfection, by use of an anti-SIVp57/27 antibody. The positions of Gag (p57), MA-CA-p2 (p41), and CA (p26) are indicated on the right. WT, wild type; 70TI, mutation in matrix at position 70; NCm, mutation of all Cys and His residues to Ala in the two NC zinc fingers. (C) Quantification of the percentage of the p41 cleavage intermediate in the virion (means + standard deviations) (n = 3). Asterisks indicate significant differences from the wild type (*, P < 0.05) by the unpaired Student t test.
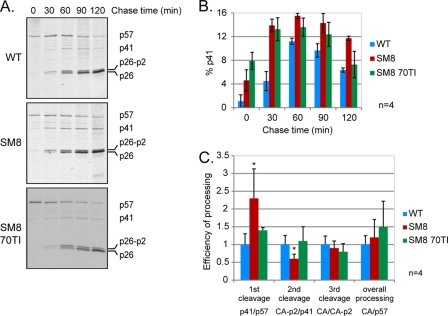
The precise step at which Gag processing is disrupted is difficult to ascertain with the “snapshot” approach described above, so we investigated this question further by use of pulse-chase metabolic labeling (Fig. 5). Transfected cells were pulse-labeled for 30 min and were chased for as long as 120 min. Gag and its CA-containing cleavage products were immunoprecipitated and analyzed by autoradiography (Fig. 5A). Confirming the results obtained by Western blotting (Fig. 4B), the dimerization-deficient mutant SM8 showed a significant accumulation of the p41 intermediate compared to that in the WT virus (Fig. 5A and B). The MA revertant of SM8, SM8 70TI, partially rescued this phenotype after 2 h. The efficiency of each individual cleavage, taken as the ratio of cleaved to uncleaved product, was calculated for each virus at 2 h postlabeling and is shown in Fig. 5C. The dimerization mutant SM8 was twice as efficient as the WT virus at cleaving Gag into p41 but was significantly slower at the second cleavage site, which explains the observed accumulation of the p41 intermediate. In accordance with the results shown in Fig. 4, the MA 70TI mutation was able to restore the correct rate of Gag processing in the SM8 mutant. The rates of the third cleavage of CA-p2 into CA and p2 were similar for all viruses, as were the overall processing efficiencies (CA/Gag ratios), suggesting that the improper rate of processing and the subsequent accumulation of p41—rather than decreases in the proportions of cleaved MA and CA—are responsible for the reduced infectivity.
Fig 5.
The rate of Gag processing is altered in a replication-incompetent SL-1 mutant with impaired RNA dimerization. Pulse-chase metabolic labeling and immunoprecipitation of Gag were performed 24 h after transfection of C33-A cells with the WT, SM8, and SM8 MA 70TI proviral plasmids. Cells were starved of methionine (Met) and cysteine (Cys) for 60 min, pulse-labeled with [35S]Met-Cys for 30 min, and chased in cold medium for 30 to 120 min. Capsid-containing Gag proteins were immunoprecipitated from the culture supernatant at each time point with anti-SIVp57/p27, resolved by SDS-PAGE, and visualized by autoradiography, prior to quantification by densitometry. (A) Representative results of pulse-chase metabolic labeling, immunoprecipitation, and gel electrophoresis are shown for each virus. The chase times are given above the gel, and the positions of Gag (p57) and its cleavage products MA-CA-p2 (p41), CA-p2 (p26-p2), and CA (p26) are indicated on the right. (B) Quantification of the percentage of the p41 cleavage intermediate for each virus (means ± standard deviations) (n = 4). (C) Efficiency of processing at 120 min postlabeling for each cleavage of Gag, as diagramed in Fig. 4A, corresponding to the ratio of the cleaved to the uncleaved product. Means + standard deviations are shown (n = 4). Asterisks indicate significant differences from the WT (*, P < 0.05) by an unpaired Student t test.
DISCUSSION
In this study, we evaluated the requirement for sequence and structural elements within the SL-1 region of the HIV-2 RNA leader for viral replication, genome dimerization, and encapsidation. Using substitution mutants that interfered with the palindrome pal or stem B, we have shown that neither of these elements is indispensable for HIV-2 replication (Table 1). However, the 392-GGAG-395 motif is critical for viral replication (as previously proposed [5]), and we show that this correlates precisely with genome dimerization. In addition, we have observed an absolute dependence on RNA dimerization, but not packaging, for HIV-2 replication. These findings posit a model in which in vivo dimerization occurs prior to RNA capture and encapsidation, as suggested for HIV-1 (46) and MLV (21). Surprisingly, the presence of stem B does not appear to influence viral replication, in contrast with the conclusions of a previous report in which the formation of an extended SL-1 was proposed to be important for viral replication and encapsidation (36). One explanation for this discrepancy lies in the fact that the role of stem B was derived from evolution studies of a randomized sequence, whereas our results were obtained from a mutagenesis approach whereby specific residues were replaced in order to alter the structure of SL-1. Our results do, however, confirm that stem B and the extended structure of SL-1 can enhance HIV-2 genome encapsidation, albeit exclusively in combination with the 392-GGAG-395 motif. Maintenance of the stem-loop structure immediately downstream of the nt 392-to-395 purine-rich motif may serve to ensure that the motif is optimally presented for Gag binding. The recent report that the HIV-1 leader RNA can adopt multiple conformations which are involved in the regulation of key processes in the virus life cycle (43) provides another explanation for the apparent differences between this and a previous study (36), as well as for those between the WT and SL-1 mutant viruses. Indeed, one could envisage a similar situation in HIV-2 where a structural switch in the genomic RNA regulates the transition between translation and genome dimerization and packaging.
Analysis of the HIV-2 dimerization mutants by Western blotting revealed an accumulation of the Gag intermediate cleavage product p41 (MA-CA-p2), a result confirmed by metabolic labeling and Gag immunoprecipitation. The rate of Gag processing was altered in the dimerization mutant from that in the WT, explaining the increase in the p41 level. Indeed, initial cleavage of the Gag polyprotein was faster, while cleavage of p41 was slower, in the mutant than in the WT. The overall processing efficiency remained the same, as did the level of CA, suggesting that the incorrect timing of processing leading to the accumulation of p41 is responsible for the poor infectivity of the mutant virus.
Evolution study of the dimerization mutants resulted in the emergence of several revertant viruses harboring compensatory mutations in the gag gene. Interestingly, a threonine-to-isoleucine mutation at position 70 of matrix, analogous to those previously described for HIV-1 (49) and SIVmac239 (58), was obtained in two out of three dimerization mutants. In HIV-1, a MA 69TI mutation was able to compensate for the viral release defect of the MA 6VR mutant, possibly by affecting the global conformation of the protein and correcting an alteration in the structure introduced by the original mutation. More relevant to the present study is the rescued replication of the SIVmac239 SL-1 mutant by the MA 70TI reversion. SIVmac239 is closely related to HIV-2, and it is interesting that, as with our findings for HIV-2, the SIV SL-1 mutant showed a lower level of dimeric genome and lower infectivity than the WT virus. However, the authors did not report on the effect of the compensatory mutation on genome dimerization.
When the MA 70TI compensatory mutation was introduced into the genomes of WT and mutant viruses, we observed rescue of the viral replication of the dimerization mutants but no significant effect on replication in the WT virus. This rescue was associated with a largely restored ability to form dimeric genomes and restoration of the correct rate of Gag processing, with levels of p41 similar to that of the WT at 2 h postlabeling. While residue 70 of matrix does not appear to be particularly involved in the structure of the protein, in the HIV-1 structure it is located near the MA-CA interface, and this spatial positioning may be responsible for the effect on Gag processing that was observed. In addition, it was shown recently that upon binding of HIV-1 matrix to the genomic RNA, certain residues (including residues 68 and 70) become more reactive (2), indicating a potential change of the conformation of this region upon MA-RNA interaction. The effects of the MA mutation on the replication, RNA dimerization, and Gag processing of HIV-2 mutants—together with the recently proposed model whereby HIV-1 NC and MA both bind to the genomic RNA and, following binding to the plasma membrane, the Gag protein extends and viral particles assemble (17)—provide additional insight into the role of matrix in HIV particle assembly.
Taking our findings together, this study provides further evidence of the role of the genomic RNA in retrovirus assembly and highlights the importance of genome dimerization for HIV-2 replication. Targeting this process through the cis-acting RNA elements located in the highly conserved (and thus less prone to escape mutations) 5′ UTR may open the way to new therapeutic approaches.
ACKNOWLEDGMENTS
We thank John Sinclair for critical reading of the manuscript.
The anti-SIVp57/p27 antibody (ARP3004) and the PM1 cells (ARP057) were obtained from the Programme EVA Centre for AIDS Reagents, NIBSC, United Kingdom, supported by the EC FP6/7 Europrise Network of Excellence, the AVIP and NGIN consortia, and the Bill and Melinda Gates GHRC-CAVD Project, and were donated by S. Osmanov and M. Reitz, respectively. This work was supported by the United Kingdom Medical Research Council (award G0800142) and the Biomedical Research Centre (grant RG52162).
Footnotes
Published ahead of print 14 March 2012
REFERENCES
- 1. Aldovini A, Young RA. 1990. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 64:1920–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alfadhli A, et al. 2011. HIV-1 matrix protein binding to RNA. J. Mol. Biol. 410:653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amarasinghe GK, et al. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491–511 [DOI] [PubMed] [Google Scholar]
- 4. Baig TT, Lanchy JM, Lodmell JS. 2007. HIV-2 RNA dimerization is regulated by intramolecular interactions in vitro. RNA 13:1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baig TT, Lanchy JM, Lodmell JS. 2009. Randomization and in vivo selection reveal a GGRG motif essential for packaging human immunodeficiency virus type 2 RNA. J. Virol. 83:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baudin F, et al. 1993. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J. Mol. Biol. 229:382–397 [DOI] [PubMed] [Google Scholar]
- 7. Berkhout B, van Wamel JL. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177–218 [DOI] [PubMed] [Google Scholar]
- 9. Campbell S, Rein A. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell S, Vogt VM. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clavel F, Orenstein JM. 1990. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 64:5230–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clever J, Sassetti C, Parslow TG. 1995. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J. Virol. 69:2101–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clever JL, Parslow TG. 1997. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 71:3407–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clever JL, Wong ML, Parslow TG. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70:5902–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crist RM, et al. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Damgaard CK, Dyhr-Mikkelsen H, Kjems J. 1998. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 26:3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Datta SA, et al. 2011. HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. J. Mol. Biol. 406:205–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Datta SA, et al. 2011. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? J. Virol. 85:4111–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Guzman RN, et al. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384–388 [DOI] [PubMed] [Google Scholar]
- 20. Dirac AM, Huthoff H, Kjems J, Berkhout B. 2001. The dimer initiation site hairpin mediates dimerization of the human immunodeficiency virus, type 2 RNA genome. J. Biol. Chem. 276:32345–32352 [DOI] [PubMed] [Google Scholar]
- 21. D'Souza V, Summers MF. 2004. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586–590 [DOI] [PubMed] [Google Scholar]
- 22. Fu W, Gorelick RJ, Rein A. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu W, Rein A. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83 [DOI] [PubMed] [Google Scholar]
- 25. Greatorex J. 2004. The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology 1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griffin SD, Allen JF, Lever AM. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058–12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guan Y, et al. 2001. Partial restoration of replication of simian immunodeficiency virus by point mutations in either the dimerization initiation site (DIS) or Gag region after deletion mutagenesis within the DIS. J. Virol. 75:11920–11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harrison GP, Lever AM. 1992. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 66:4144–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hill MK, et al. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 77:8329–8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu WS, Temin HM. 1990. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 87:1556–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson MC, Scobie HM, Ma YM, Vogt VM. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177–11185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaye JF, Lever AM. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023–3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kaye JF, Lever AM. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72:5877–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lanchy JM, Ivanovitch JD, Lodmell JS. 2003. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA 9:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lanchy JM, Lodmell JS. 2002. Alternate usage of two dimerization initiation sites in HIV-2 viral RNA in vitro. J. Mol. Biol. 319:637–648 [DOI] [PubMed] [Google Scholar]
- 36. Lanchy JM, Lodmell JS. 2007. An extended stem-loop 1 is necessary for human immunodeficiency virus type 2 replication and affects genomic RNA encapsidation. J. Virol. 81:3285–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lanchy JM, Rentz CA, Ivanovitch JD, Lodmell JS. 2003. Elements located upstream and downstream of the major splice donor site influence the ability of HIV-2 leader RNA to dimerize in vitro. Biochemistry 42:2634–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Laughrea M, Jette L. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464–13474 [DOI] [PubMed] [Google Scholar]
- 39. Laughrea M, et al. 1997. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 71:3397–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lever A, Gottlinger H, Haseltine W, Sodroski J. 1989. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 63:4085–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. L'Hernault A, Greatorex JS, Crowther RA, Lever AM. 2007. Dimerisation of HIV-2 genomic RNA is linked to efficient RNA packaging, normal particle maturation and viral infectivity. Retrovirology 4:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liang C, et al. 1999. Deletion mutagenesis within the dimerization initiation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J. Virol. 73:6147–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lu K, et al. 2011. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McCann EM, Lever AM. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71:4133–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moore MD, et al. 2007. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 81:4002–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moore MD, et al. 2009. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 5:e1000627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muriaux D, Girard PM, Bonnet-Mathoniere B, Paoletti J. 1995. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem. 270:8209–8216 [DOI] [PubMed] [Google Scholar]
- 48. Muriaux D, Mirro J, Harvin D, Rein A. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. U. S. A. 98:5246–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ono A, Huang M, Freed EO. 1997. Characterization of human immunodeficiency virus type 1 matrix revertants: effects on virus assembly, Gag processing, and Env incorporation into virions. J. Virol. 71:4409–4418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Paillart JC, et al. 1996. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 70:8348–8354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. 2004. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2:461–472 [DOI] [PubMed] [Google Scholar]
- 52. Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. U. S. A. 93:5572–5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Potts B. 1990. Mini RT assay, p 103–106 In Aldovini A, Walker B. (ed), Techniques in HIV research. Stockton Press, New York, NY [Google Scholar]
- 54. Sakuragi J, Ueda S, Iwamoto A, Shioda T. 2003. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J. Virol. 77:4060–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shehu-Xhilaga M, et al. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156–9164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shubsda MF, Paoletti AC, Hudson BS, Borer PN. 2002. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry 41:5276–5282 [DOI] [PubMed] [Google Scholar]
- 57. Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. U. S. A. 91:4945–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Whitney JB, Oliveira M, Detorio M, Guan Y, Wainberg MA. 2002. The M184V mutation in reverse transcriptase can delay reversion of attenuated variants of simian immunodeficiency virus. J. Virol. 76:8958–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Whitney JB, Wainberg MA. 2006. Impaired RNA incorporation and dimerization in live attenuated leader-variants of SIVmac239. Retrovirology 3:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]