Abstract
Autophagy is a cellular process that sequesters cargo in double-membraned vesicles termed autophagosomes and delivers this cargo to lysosomes to be degraded. It is enhanced during nutrient starvation to increase the rate of amino acid turnover. Diverse roles for autophagy have been reported for viral infections, including the assembly of viral replication complexes on autophagic membranes and protection of host cells from cell death. Here, we show that autophagosomes accumulate in Semliki Forest virus (SFV)-infected cells. Despite this, disruption of autophagy had no effect on the viral replication rate or formation of viral replication complexes. Also, viral proteins rarely colocalized with autophagosome markers, suggesting that SFV did not utilize autophagic membranes for its replication. Further, we found that SFV infection, unlike nutrient starvation, did not inactivate the constitutive negative regulator of autophagosome formation, mammalian target of rapamycin, suggesting that SFV-dependent accumulation of autophagosomes was not a result of enhanced autophagosome formation. In starved cells, addition of NH4Cl, an inhibitor of lysosomal acidification, caused a dramatic accumulation of starvation-induced autophagosomes, while in SFV-infected cells, NH4Cl did not further increase levels of autophagosomes. These results suggest that accumulation of autophagosomes in SFV-infected cells is due to an inhibition of autophagosome degradation rather than enhanced rates of autophagosome formation. Finally, we show that the accumulation of autophagosomes in SFV-infected cells is dependent on the expression of the viral glycoprotein spike complex.
INTRODUCTION
Autophagy is a constitutive, dynamic, bulk degradation process that is necessary for a number of processes in living cells (reviewed in references 31 and 48). During autophagy, long-lived proteins and organelles are engulfed by specialized double membrane vesicles, termed autophagosomes, and are delivered to the lysosomes for subsequent degradation. The constant flow of autophagosomes to lysosomes is tightly regulated by a number of autophagy-related genes (atg), initially characterized in Saccharomyces cerevisiae. The mammalian homologue of the atg8 gene product, LC3 is found in two forms in the cell. LC3-I is the soluble cytoplasmic form and is not associated with the autophagic pathway. During assembly of autophagosomes, LC3-I becomes conjugated to a membrane lipid, phosphatidylethanolamine, by the ATG5-ATG12 complex. This conjugated form, LC3-II, is associated with autophagosomal membranes and is carried with the flow of autophagy to lysosomes and degraded therein. The cellular protein p62/SQSTM1 is also associated with autophagosomes and degraded in lysosomes. It acts as a link between LC3 and ubiquitinated protein aggregates and so may confer some level of specificity to the autophagic cargo (2). p62/SQSTM1 is also recruited to the intracellular bacterium Salmonella enterica serovar Typhimurium, targeting the bacterial cells for autophagic degradation and restricting intracellular bacterial replication (50). p62/SQSTM1 has also been reported to interact with the Sindbis virus (SINV) capsid protein, targeting it to autophagosomes, promoting survival of infected cells (35). p62/SQSTM1-mediated, pathogen-specific autophagy thus represents an innate immune defense mechanism.
Autophagy is a constitutive process, but it can be enhanced above basal levels by various stimuli, such as amino acid starvation. The regulation of autophagy by the protein kinase target of rapamycin (TOR) has been well studied in yeast (18). The mammalian TOR (mTOR), similar to yeast TOR, is a master sensor of nutrient status and regulates cell growth. Under nutrient-rich conditions, mTOR phosphorylates ULK1, the mammalian homologue of yeast Atg1, and negatively regulates the initial events in autophagosome formation (11). Amino acid starvation and rapamycin treatment both induce autophagy via inhibition of mTOR activity. The upregulation of autophagy during starvation allows a starved cell to increase its rate of amino acid recycling. Infection with certain pathogens also enhances autophagy (16, 17, 33), although whether pathogen-induced autophagy depends on mTOR inactivation has not been well studied. Two well-studied targets of mTOR are the S6 kinase and eukaryotic initiation factor (eIF) 4E-binding protein (4E-BP1), both of which are involved in translational control (14). While mTOR is the best-studied regulator of autophagy, the phosphorylation of eIF2α has also been reported to regulate autophagy (21, 43).
Autophagosomes accumulate in a wide range of RNA virus infections, and the effect of autophagy on viral replication varies between viruses. Autophagy was shown to enhance the replication of the picornaviruses poliovirus (16), coxsackievirus B3 (47), and foot-and-mouth disease virus (34), the flaviviruses dengue virus (24) and hepatitis C virus (HCV) (9, 39), and the togavirus Chikungunya virus (22). On the other hand, autophagy has no effect on the replication of other viruses. In human rhinovirus, coronavirus, and SINV infections, viral replication is independent of ATG5 (3, 35, 49). Influenza A virus (IAV), a negative-sense RNA virus that does not replicate in the cytoplasm, also induces the accumulation of autophagosomes, and its replication is also independent of ATG5 (13). Interestingly, despite a lack of a role for autophagy in controlling virus replication, in both SINV and IAV infections, autophagy promotes the survival of infected cells (13, 35).
As autophagosomes are continually formed and degraded, the accumulation of autophagosomes in infected cells may be due to an induction of autophagy, a blockade of autophagosome maturation, or both. Poliovirus simultaneously induces autophagy and blocks degradation of autophagosomes, as poliovirus 2BC expressed alone increases the lipidation of LC3 (45), while poliovirus 3A inhibits the movement of autophagosomes along microtubules (44). Coxsackievirus B3 infection did not increase the degradation rate of the autophagosome cargo p62/SQSTM1, suggesting that while autophagosome numbers increased, autophagosome maturation was impaired (47). In HCV-infected cells, accumulation of autophagosomes was shown to be due to a combination of autophagy induction via activation of the unfolded protein response (UPR) triggered by viral replication and the inhibition of autophagosome maturation by a factor(s) present in the infected cells (39). Interestingly, while both HCV and varicella-zoster virus have been reported to activate autophagy through endoplasmic reticulum (ER) stress (5, 39), thapsigargin, a pharmacologic inducer of ER stress, is reported to cause accumulation of autophagosomes not by inducing autophagy but by blocking autophagosome maturation (12). The IAV M2 protein also blocks autophagosome maturation, by preventing the fusion of autophagosomes with lysosomes (13). Finally, coronavirus-induced autophagy is not due to mTOR inactivation or ER stress (6), suggesting that the coronavirus-induced appearance of autophagosomes is due to unknown mechanisms of autophagy induction or due to a block in autophagosome maturation.
Semliki Forest virus (SFV) is an RNA virus of the Alphavirus genus, family Togaviridae. The viral particles are enveloped and contain a single-stranded positive-sense 42S RNA genome. The 5′ two-thirds of the capped and polyadenylated genome codes for the four viral nonstructural proteins (nsP1 to nsP4), whereas the structural protein-coding regions are carried on a subgenomic 26S mRNA, transcribed from the 3′ one-third of the 42S RNA. SFV infection induces many rapid and profound changes in the biology of its host cells (1, 28, 46). In this work, we investigated if the cellular autophagic apparatus was altered by SFV infection. We found that although SFV-infected cells accumulated LC3-II-positive autophagosomes in late times after infection, an active autophagic process was not necessary for efficient viral gene expression or replication. Viral proteins did not colocalize to any great extent with autophagosomal markers LC3 or p62/SQSTM1, and accumulation of autophagosomes during SFV infection did not depend on mTOR inactivation or eIF2α phosphorylation. Instead, we found that the accumulation of autophagosomes in SFV-infected cells was due to inhibition of autophagosome maturation and was dependent on the expression of the viral glycoproteins.
MATERIALS AND METHODS
Cell culture and virus propagation.
BHK-21 cells (ATCC) were cultured in Glasgow minimal essential medium supplemented with 10% fetal calf serum (FCS; Sigma), 20 mM HEPES, 10% tryptose phosphate broth, 2 mM l-glutamine, and penicillin-streptomycin (Invitrogen). Human osteosarcoma cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) with 10% FCS, 2 mM l-glutamine, and penicillin-streptomycin (Invitrogen). Amino acid and serum starvation were performed by incubation of cells in Earle's balanced salt solution (EBSS; Sigma). HOS-EGFP-LC3 and HOS-EGFP-LC3-G120A cells were generated by transfection of HOS cells with pEGFP-LC3 and pEGFP-LC3-G120A, respectively (as described previously [10]). atg5+/+ and atg5−/− mouse embryonic fibroblasts (MEFs; obtained from RIKEN Bioresource Center, Japan) and eIF2a-SS and -AA MEFs were maintained in DMEM with 10% FCS, 2 mM l-glutamine, and penicillin-streptomycin (Invitrogen). For inhibition of lysosomal degradation, cells were incubated in 15 mM ammonium chloride (NH4Cl) or 50 μM chloroquine (CQ). Wild-type (wt) SFV4 (referred to as SFV) was derived from the infectious clone pSP6-SFV4 as described previously (26) and titrated by quantification of plaque numbers. SFV-βGal, SFV-Δ6K, and SFV-ΔSpike were derived from the plasmids pSP6-SFV-lacZ (25), pSP6-SFV-Δ6K (26), and pSP6-SFV-C[am]-CWF (40), respectively, packaged as described previously (41), and titrated by immunofluorescence. Influenza A virus strain X31 was propagated in chicken eggs and titrated on Madin-Darby canine kidney cells. Virus stocks were used for infection as follows: cell monolayers were washed with phosphate-buffered saline (PBS), and virus was added in minimal essential medium (Invitrogen) supplemented with 0.2% bovine serum albumin, 2 mM l-glutamine, and 20 mM HEPES with periodic shaking for 1 h at 37°C. Virus solutions were then removed, and cells were washed with PBS before adding prewarmed complete medium.
Microscopy.
Cells grown on coverslips were fixed by incubation in 4% paraformaldehyde in PBS for 10 min at room temperature followed by incubation in ice-cold methanol for 10 min. Antibodies used were mouse anti-SFV nsP1, mouse anti-SFV nsP2 (23), rabbit anti-SFV nsP3 (a kind gift from Andres Merits, University of Tartu, Estonia), rabbit anti-total SFV, mouse anti-SFV capsid, mouse anti-LC3 (MBL International), and mouse and rabbit anti-p62/SQSTM1 (both from Santa Cruz Biotechnology). Coverslips were then incubated in PBS containing secondary antibodies and 1 μg/ml Hoechst 33258 (Invitrogen) for 1 h for identification of cell nuclei. Washed coverslips were then mounted in vinol mounting medium, and images were captured using a Leitz DM RB fluorescence microscope with a Hamamatsu cooled charge-coupled-device C4880 camera. For confocal microscopy, images were captured using a Perkin-Elmer Ultra-View spinning disc confocal microscope. Images were processed and compiled using Adobe Photoshop. Pearson's coefficients were calculated using the Manders coefficients plug-in in the ImageJ software.
Immunoblotting.
For identification of proteins by immunoblotting, cells were lysed on ice in lysis buffer (20 mM HEPES [pH 7.4], 110 mM KO-acetate [KOAc], 2 mM MgCl2, 0.1% Tween 20, 1% Triton X-100, 0.5% sodium deoxycholate, 0.5 M NaCl) and clarified by centrifugation at 6,000 × g for 5 min in a microcentrifuge at 4°C. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (GE Healthcare). Antibodies used were mouse monoclonal anti-SFV nsP2 (23), rabbit anti-nsP3, rabbit anti-S6 ribosomal protein, rabbit anti-phospho-S6 ribosomal protein, rabbit monoclonal anti-phospho-4E-BP1, and rabbit anti-4E-BP1 (Cell Signaling), rabbit anti-eIF2α and goat anti-actin (Santa Cruz), rabbit anti-phospho-eIF2α (Biosource), and rabbit anti-LC3 (Abgent). Chemiluminescence was detected using the enhanced chemiluminescence reagents (GE Healthcare).
Flow cytometry.
The fluorescence-activated cell sorting (FACS)-based assay for autophagy is described in detail in reference 10. HOS-EGFP-LC3 cells were harvested with trypsin, washed with PBS, and washed with PBS containing 0.05% saponin. Cells were then incubated with antibodies for intracellular staining for 20 min. These antibodies were mouse anti-SFV nsP1 and rabbit anti-IAV. Cells were then rinsed with PBS, incubated with goat anti-mouse secondary antibody conjugated to R-phycoerythrin (Southern Biotech) or goat anti-rabbit conjugated to Alexa Fluor 647 (Invitrogen) for 20 min, and rinsed twice with PBS. FACS data were collected using a FACSCalibur flow cytometer (Becton Dickinson) with CellQuest Pro software. More than 30,000 events were captured for every analysis. Data analysis was carried out with FlowJo.
RESULTS
Accumulation of autophagosomes in SFV-infected cells.
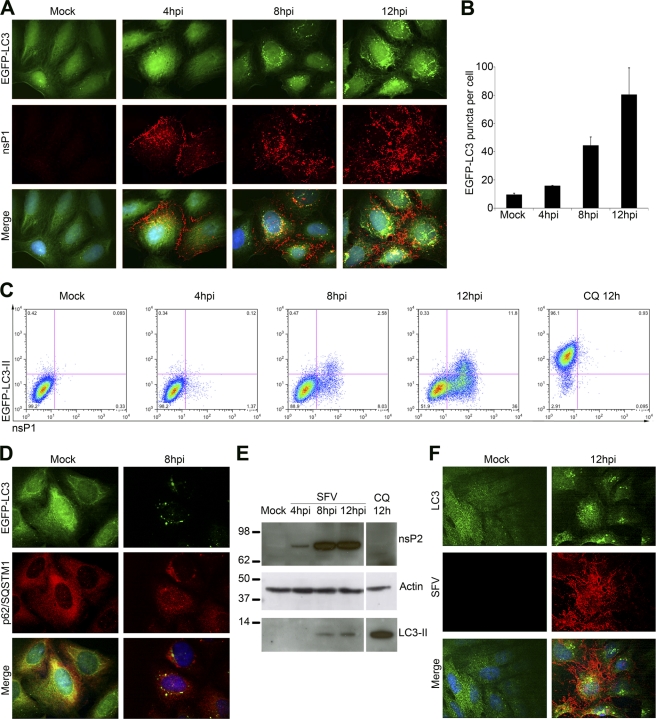
In order to quantify the number of autophagosomes in SFV-infected cells relative to noninfected cells, SFV-infected, or mock-infected HOS-EGFP-LC3 cells were fixed and stained for SFV nsP1, a component of the SFV RNA replication complex. We clearly observed EGFP-LC3 localization in SFV-infected cells that was more punctate than in noninfected cells, indicating that the reporter protein was assembled into autophagosomes (Fig. 1A). This punctate localization of EGFP-LC3 was evident in every cell that stained positive for nsP1. The number of EGFP-LC3-positive puncta per cell was quantified by manual counting and was shown to increase steadily during the time course of infection (Fig. 1B). In order to confirm that the EGFP-LC3 puncta in SFV-infected cells represent autophagosomal membrane-bound EGFP–LC3-II, we took advantage of our recently described quantitative FACS-based assay for autophagy (10). Because saponin extraction is specific for the soluble non-autophagosome-associated EGFP–LC3-I form of the protein, flow cytometry can be used to measure total fluorescence of saponin-extracted HOS-EGFP-LC3 cells as a measure of the level of autophagosome-associated EGFP–LC3-II. HOS-EGFP-LC3 cells were infected at a multiplicity of infection (MOI) of 1 with wt SFV, mock infected, or CQ treated, and processed for FACS analysis (Fig. 1C). During the course of the infection, a saponin-resistant EGFP–LC3-II–positive population emerged that was also SFV nsP1 positive, indicating that the EGFP-LC3 reporter accumulated in the EGFP–LC3-II form, preferentially in SFV-infected cells compared to the noninfected cells in the same culture or in mock-infected cells. CQ-treated HOS-EGFP-LC3 cells became strongly EGFP–LC3-II positive, as observed previously (10), indicating accumulation of reporter protein due to the block in acidification of lysosomes. With further characterization of SFV infection-induced EGFP–LC3-II puncta, we found that they colocalized moderately well with the autophagosomal cargo protein p62/SQSTM1 (Pearson's coefficient, 0.369) (Fig. 1D). Furthermore, infection of HOS-EGFP-LC3-G120A cells stably expressing a mutant form of EGFP-LC3 that cannot be lipidated and incorporated into autophagosomes did not result in accumulation of saponin-resistant EGFP-LC3 (data not shown).
Fig 1.
Accumulation of LC3-II-positive autophagosomes in SFV-infected cells. (A) HOS-EGFP-LC3 cells were infected with SFV at an MOI of 1. At 4, 8, or 12 hpi, cells were stained for SFV nsP1 (red) and analyzed for EGFP-LC3-positive puncta by confocal microscopy. Hoechst stain was used to identify cell nuclei (blue). Representative images are shown for each time point. (B) The number of EGFP-LC3-positive puncta in HOS-EGFP-LC3 cells infected and stained as described for panel A were manually counted in each of 50 cells. Infected cells were identified by their positive nsP1 staining. Data are means of three independent experiments. (C) HOS-EGFP-LC3 cells were infected with SFV at an MOI of 1 or treated with 50 μM CQ. At 4, 8, or 12 hpi or 12 h after CQ treatment, cells were washed briefly in 0.05% saponin in PBS, stained for nsP1, and analyzed by flow cytometry. Representative dot plots are shown. (D) HOS-EGFP-LC3 cells were infected with SFV at an MOI of 20. At 8 hpi, cells were stained for p62/SQSTM1 (red) and nuclei (blue) and analyzed by confocal microscopy. Representative images are shown. (E) HOS-EGFP-LC3 cells were infected with SFV. Lysates were obtained 4, 8, or 12 hpi and analyzed for SFV nsP2, actin, or LC3 by Western blotting. (F) HOS cells were infected with SFV at an MOI of 1. At 12 hpi, cells were stained for SFV antigen (red), endogenous LC3 (green), and nuclei (blue) and analyzed by confocal microscopy. Representative images are shown.
Since in other viral infection systems, the EGFP-LC3 reporter construct does not always behave as the endogenous protein (36), we wanted to confirm that endogenous LC3 also accumulates in the LC3-II form during SFV infection. To do this, lysates were prepared from SFV-infected HOS-EGFP-LC3 cells harvested at 4, 8, and 12 h postinfection (hpi) or CQ treated for 12 h and analyzed by immunoblotting for viral protein nsP2 and the cellular proteins actin and LC3 (Fig. 1E). We observed that the endogenous LC3-II form accumulated in SFV-infected cells and, in agreement with the microscopy and flow cytometric analyses with the EGFP-LC3 reporter protein, this form increased during the course of the infection. To determine the localization of endogenous LC3, parental HOS cells were infected with SFV or mock infected, fixed, and stained for endogenous LC3 and for total SFV antigen at 12 hpi. Again, LC3 was observed to relocalize in infected cells into puncta in the cytoplasm, which became larger and more numerous as the infection progressed (Fig. 1F). There is therefore good concordance between the results obtained with endogenous LC3 stainings and those obtained for EGFP-LC3. We also observed endogenous LC3-II accumulation in BHK cells and in MEFs infected with SFV for more than 8 h (data not shown). Taken together, the data in Fig. 1 show that LC3-II-containing autophagosomes accumulated in the cytoplasm of SFV-infected cells.
SFV replication in atg5−/− MEFs.
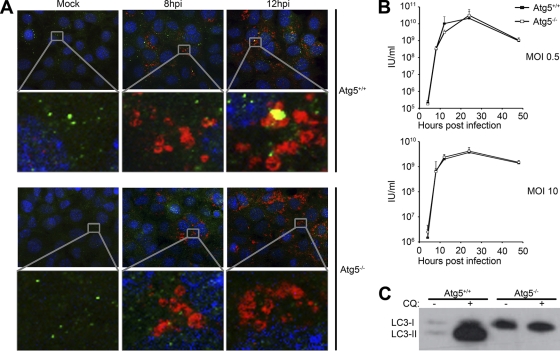
Accumulation of LC3-II-containing autophagosomes has been reported in a number of diverse virus infections (8). In many of these systems, autophagy has been shown to have various effects on virus replication, and we therefore wished to determine if the accumulation of autophagosomes in SFV-infected cells affected genome replication and growth kinetics. A number of RNA viruses have been shown to utilize autophagosomes as sources of membranes upon which they assemble RNA production complexes (16, 24, 36). To determine if the absence of ATG5 function affected the morphology of the SFV RNA replication complexes, we infected atg5−/− and atg5+/+ MEFs with SFV at an MOI of 10 and fixed and stained the cells at various times postinfection for SFV nsP3, another component of the SFV RNA replication complex. Figure 2A shows that the intensity of nsP3 staining was not significantly affected by the ablation of the gene for ATG5. Further, it was observed that the gross morphologies of nsP3-positive structures were similar in the presence or absence of ATG5, suggesting that autophagosomes are not necessary sources of membranes for SFV replication complex assembly.
Fig 2.
Genetic knockout of atg5 has no major effect on SFV gene expression or viral replication kinetics. (A) atg5+/+ or atg5−/− MEFs were infected with SFV at an MOI of 10. At 8 or 12 hpi, cells were stained for SFV nsP3 (red), LC3 (green), and nuclei (blue) and analyzed by fluorescence microscopy. (B) atg5+/+ or atg5−/− MEFs were infected with SFV at an MOI of 0.5 (top) or 10 (bottom). At 4, 8, 12, 24, or 48 hpi, supernatants were collected and SFV infectious units were quantified by plaque assay on BHK cells. Data are means of three independent experiments, and error bars indicate standard deviations. (C) atg5+/+ or atg5−/− MEFs were treated with 50 μM CQ for 8 h, and lysates were analyzed for LC3 by Western blotting. LC3-I and LC3-II can be distinguished by their apparent molecular masses of 18 kDa and 16 kDa, respectively.
To determine if the accumulation of autophagosomes in SFV-infected cells affected the production of infectious virus, we analyzed the kinetics of SFV replication in cells deficient for autophagy in growth curve experiments. We infected atg5−/− and atg5+/+ MEFs at an MOI of 0.5 or 10 with SFV and measured the production of virus in cell culture supernatants by plaque assay. No significant (Student's t test) difference was detected in the production of infectious virus between the two cell types after infection at a low or high MOI (Fig. 2B). To confirm that the cells used in these experiments were defective for autophagy, they were treated for 8 h with CQ, lysed, and analyzed by immunoblotting for endogenous LC3. As expected, LC3 remained in the LC3-I form in atg5−/− MEFs upon CQ treatment, while in atg5+/+ MEFs it accumulated in the LC3-II form (Fig. 2C). We conclude from the experiments shown in Fig. 2 that neither the accumulation of autophagosomes nor any other function of ATG5 plays an important role in the assembly of viral RNA replication complexes or production of infectious SFV in MEFs.
SFV nonstructural proteins colocalize with autophagosomes to a low degree.
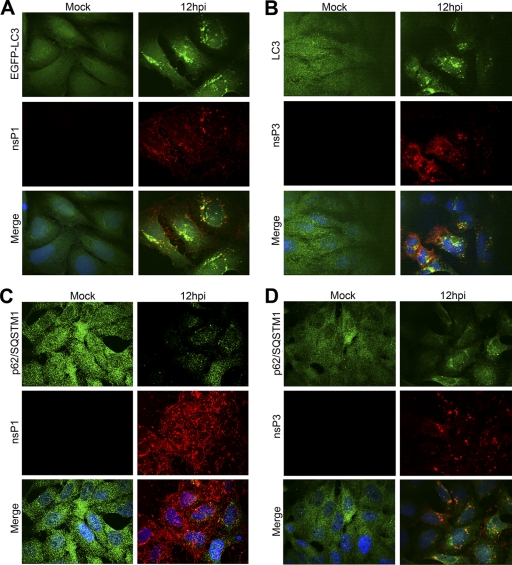
To further investigate the possibility that SFV uses autophagosomal membranes for replication complex assembly, we infected HOS-EGFP-LC3 cells with SFV, fixed cells at 4, 8, and 12 hpi, and stained them for SFV nsP1. In many instances, foci of nsP1-positive staining did not colocalize with EGFP-LC3 (Fig. 3A). Although it was evident that in some cases there was a moderate level of costaining of nsP1 foci with EGFP–LC3-II–positive autophagosomes (Pearson's coefficient, 0.36), the morphologies of the observed foci were different, suggesting juxtaposition rather than true colocalization. Further, this was a rare occurrence in cells that were heavily stained for both proteins. Shown in Fig. 3A are cells stained at 12 hpi, the time of peak nsP1 expression and of EGFP–LC3-II punctation. Since it was possible that the overexpression of the EGFP-LC3 reporter construct in the transfected cells affected the potential colocalization with replication complexes, we sought to examine the localization of endogenous LC3 relative to that of the viral replication complexes. HOS cells were therefore infected with SFV, fixed at 4, 8, and 12 hpi, and stained for SFV nsP3. As noted above (Fig. 1), the distribution of endogenous LC3 was similar to that of EGFP-LC3 in SFV-infected cells. Low to moderate levels of colocalization of nsP3 with LC3 (Pearson's coefficient, 0.253) were observed in infected cells (Fig. 3B), with the majority of nsP3 puncta being negative for LC3 and vice versa. Since the cellular protein p62/SQSTM1 was shown to target viral proteins for degradation via autophagy, another method to determine the involvement of autophagy with SFV replication complex formation was to determine the level of colocalization of this protein with SFV replication complexes. Accordingly, SFV-infected HOS cells were fixed at 4, 8, and 12 hpi and stained for SFV nsP1 or nsP3 and p62/SQSTM1. SFV infection also caused p62/SQSTM1 to accumulate in cytoplasmic puncta, although the extent of this was less than for LC3-II. Low and moderate levels of colocalization of p62/SQSTM1 with SFV nsP1 (Fig. 3C) and nsP3 (Fig. 3D) were observed (Pearson's coefficients, −0.017 and 0.311, respectively). SFV nsP2 staining at these times postinfection was strongest in the cell nuclei but was also found in cytoplasmic foci, which did not colocalize with p62/SQSTM1 (data not shown). Although we cannot exclude the possibility that some subset of autophagosomes can be utilized by SFV for replication complex assembly, we conclude from the experiments shown in Fig. 3 that autophagosomes are unlikely to represent a major source of membranes for SFV replication complex assembly.
Fig 3.
SFV replicase proteins rarely colocalize with autophagosomes in infected cells. (A) HOS-EGFP-LC3 cells were infected with SFV at an MOI of 1. At 12 hpi, cells were stained for nsP1 (red), and colocalization of nsP1 and EGFP-LC3 positive puncta was analyzed by confocal microscopy. (B) HOS cells were infected with SFV at an MOI of 1. At 12 hpi, cells were stained for nsP3 (red) and LC3 (green), and colocalization was analyzed by confocal microscopy. (C and D) HOS cells were infected with SFV. At 12 hpi, cells were stained for nsP1 (C) or nsP3 (D) (red) and p62/SQSTM1 (green). Colocalization was analyzed by confocal microscopy.
SFV structural proteins do not colocalize with autophagosomes.
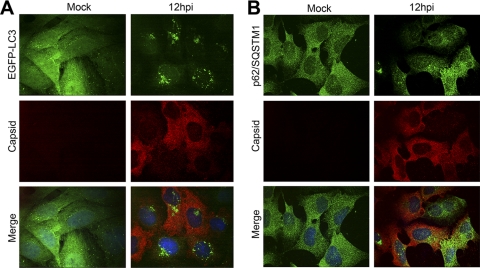
Orvedahl and coworkers recently reported that SINV capsid protein colocalized with p62/SQSTM1 in infected cells (35). To determine whether the capsid protein of SFV colocalized with autophagosomes in infected cells, we infected HOS-EGFP-LC3 cells with SFV and analyzed the extent of colocalization of the viral capsid protein with the EGFP-LC3 reporter protein at different times postinfection. At the time points when the cytoplasmic autophagosome accumulation was most pronounced, we were unable to detect any significant levels of colocalization of SFV capsid protein with the infection-induced EGFP-LC3 puncta (Pearson's coefficient, −0.02) (Fig. 4A). Capsid protein staining was observed in a diffuse pattern in all infected cells and was not enriched in the infection-induced EGFP-LC3 puncta. To determine whether p62/SQSTM1 targets this viral protein for autophagic degradation, we also determined the localization of p62/SQSTM1 relative to the capsid in infected HOS cells. Again, SFV capsid protein was found in a diffuse staining pattern and did not specifically colocalize with p62/SQSTM1 (Pearson's coefficient, 0.21) (Fig. 4B). Similarly, we did not detect any colocalization of SFV E1 or E2 spike proteins with EGFP-LC3, endogenous LC3, or p62/SQSTM1 in infected cells (data not shown). Since autophagy is a mechanism of degradation of cytoplasmic proteins and organelles, we reasoned that if viral proteins were degraded by this mechanism, we would need to inhibit the degradation in order to detect colocalization of viral proteins with autophagosomal markers. To this end, we infected HOS-EGFP-LC3 cells with SFV for 1 h before addition of NH4Cl or CQ for 7 h before fixing and staining the cells for viral capsid protein. Even with lysosomal degradation inhibited, we did not detect any colocalization of SFV capsid with EGFP-LC3 (see Fig. S1 in the supplemental material) nor any accumulation of capsid staining upon treatment with either NH4Cl or CQ. From the experiments presented in Fig. 4 and Fig. S1, we concluded that in SFV-infected cells, viral structural proteins are not targeted for degradation in the autophagic pathway.
Fig 4.
SFV capsid protein is not targeted to autophagosomes. HOS-EGFP-LC3 (A) or HOS (B) cells were infected with SFV at an MOI of 1. At 12 hpi, cells were stained for SFV capsid (red) (A) or SFV capsid (red) and p62/SQSTM1 (green) (B) and analyzed by confocal microscopy.
SFV-induced autophagosome accumulation is not dependent on inactivation of the mTOR complex.
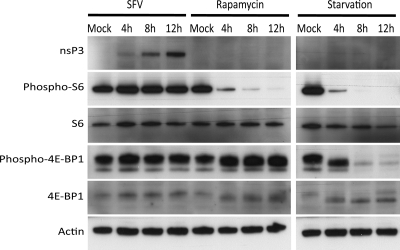
The mTOR kinase complex is a negative regulator of autophagy (11). When mTOR is inactivated, autophagy is induced above constitutive levels. In order to determine if the accumulation of autophagosomes in SFV-infected cells was a result of the induction of autophagy via inactivation of the mTOR complex, we analyzed the phosphorylation states of S6 ribosomal protein, which is phosphorylated by the mTOR substrate S6 kinase, and 4E-BP1, a direct substrate of mTOR. Under normal conditions, S6 is phosphorylated at serines 240 and 244 and 4E-BP1 is phosphorylated at threonines 37 and 46, but when mTOR is inactivated, both proteins become dephosphorylated. HOS cells were infected with SFV at an MOI of 20 and treated with rapamycin or starved of amino acids and serum. Lysates were taken at 4, 8, and 12 hpi or postinitiation of treatment, and the phosphorylation states of S6 and 4E-BP1 were analyzed by Western blotting (Fig. 5). When autophagy was induced by amino acid and serum starvation, both S6 and 4E-BP1 were rapidly dephosphorylated at the sites analyzed. Also, a reduction in the total levels of S6 was observed after 12 h of starvation, possibly resulting from specific autophagic degradation of ribosomes under these conditions. While rapamycin treatment also led to rapid dephosphorylation of S6 at serines 240 and 244, the signal for phosphothreonines 37 and 46 of 4E-BP1 was maintained during the course of the treatment. However, during the treatment the 4E-BP1 protein migrated slightly faster in the gel, suggesting that a different posttranslational modification is altered by rapamycin treatment. In contrast, in cells infected with SFV, neither of these proteins was modified from their status in mock-infected cells. S6 remained phosphorylated at serines 240 and 244 and 4E-BP1 remained as a slower-migrating band, phosphorylated at threonines 37 and 46. This result suggested that the accumulation of autophagosomes in SFV-infected cells is not due to induction of autophagy via inactivation of the mTOR complex.
Fig 5.
Autophagy is not induced via inactivation of mTOR during SFV infection. HOS-EGFP-LC3 cells were mock infected, infected with SFV at an MOI of 20, starved of amino acids and serum, or treated with 0.4 μM rapamycin. Lysates were obtained 4, 8, and 12 h later and analyzed for the indicated proteins by Western blotting.
SFV-induced autophagosome accumulation is not dependent on phosphorylation of eIF2α.
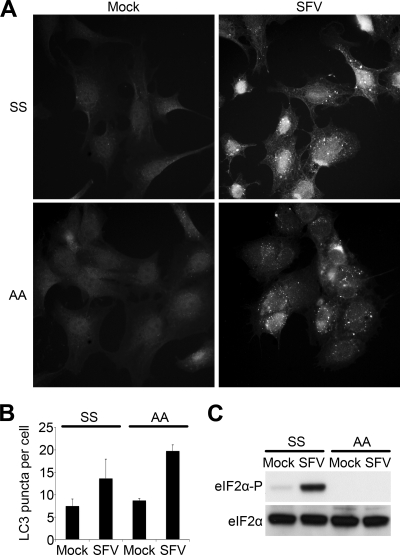
It has been reported that autophagy induction in the context of viral infection can occur via the phosphorylation of the α-subunit of the translation initiation factor eIF2 (43). We previously showed that this phosphorylation occurs rapidly upon infection of cells with SFV (28). To determine if this phosphorylation is necessary for SFV-induced accumulation of autophagosomes, we analyzed the localization of LC3 in SFV-infected MEFs from mice transgenic for an unphosphorylatable form of eIF2α (eIF2α-AA) and their wt counterparts (eIF2α-SS) (38). We found that LC3 localized into cytoplasmic puncta in SFV-infected eIF2α-AA cells in a similar manner to that observed in infected eIF2α-SS cells (Fig. 6A), indicating that eIF2α phosphorylation was not required for autophagosome accumulation in SFV-infected cells. The number of EGFP-LC3-positive puncta per cell was quantified by manual counting and was found to increase similarly in both cell lines during the infection (Fig. 6B). To confirm that the eIF2α-AA indeed expressed a nonphosphorylatable form of the protein, lysates from mock- or SFV-infected eIF2α-SS and eIF2α-AA cells were harvested 10 hpi and the phosphorylation state of the protein was analyzed by immunoblotting (Fig. 6C). As expected, eIF2α was phosphorylated after SFV infection of the eIF2α-SS cells but not the eIF2α-AA cells.
Fig 6.
Accumulation of autophagosomes is independent of eIF2α phosphorylation. (A) MEFs expressing wt eIF2α (eIF2α-SS) or a nonphosphorylatable mutant (eIF2α-AA) were infected with SFV at an MOI of 10. At 10 hpi, cells were fixed and stained for LC3 (shown here) or nsP3 (data not shown) and analyzed by fluorescence microscopy. (B) The number of LC3-positive puncta in cells infected and stained as described for panel A was manually counted in each of 50 cells. Infection was confirmed by positive nsP3 staining. Data are means of three independent experiments. (C) eIF2α-SS or eIF2α-AA MEFs were infected at an MOI of 10 with SFV for 10 h and analyzed for phosphorylated or total eIF2α by Western blotting.
SFV infection inhibits degradation of autophagosomes.
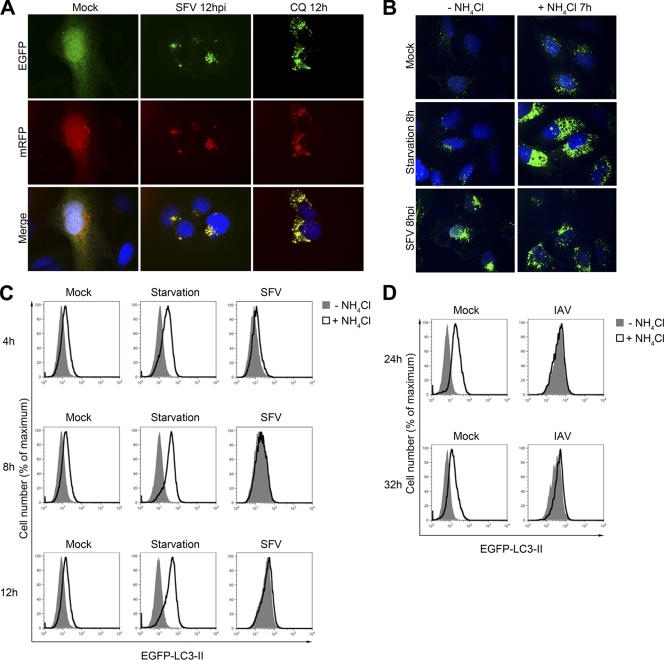
Autophagy is a system characterized by flux of autophagosomes from their initial formation and incorporation of cargo through trafficking to lysosomes and subsequent degradation of the associated cargo (32, 37). Since we have observed significantly increased levels of autophagosomes in infected cells, we sought to determine whether these autophagosomes were increased in number due to enhanced induction of their formation or to inhibition of their degradation. Accordingly, we analyzed the effect of SFV infection on the fluorescence of the tandem marker monomeric red fluorescent protein (mRFP)–EGFP-LC3. Due to the sensitivity of EGFP fluorescence to low pH, this signal is quenched soon after delivery of the marker to the lysosome, while the mRFP signal remains until the protein is degraded (19). By using this marker, autophagosomes that have fused with lysosomes can be distinguished from those that have not by virtue of their red or their red and green fluorescence, respectively. We transfected HOS cells with this reporter and subsequently infected them with SFV at an MOI of 20 or mock infected or treated them with CQ. In mock-infected cells, puncta were observed that were both mRFP and EGFP positive, as well as puncta that were only mRFP positive, indicating that autophagosomes are in constant flux from assembly to degradation (Fig. 7A). As expected, CQ treatment led to accumulation of large mRFP- and EGFP-positive puncta due to inhibition of lysosomal acidification. Infection with SFV also led to accumulation of large mRFP- and EGFP-positive puncta that resembled those in CQ-treated cells. In SFV-infected cells, we could also detect a small number of mRFP-positive puncta, although these appeared smaller in size and there were fewer than in mock-infected cells.
Fig 7.
Degradation of autophagosomes is blocked during SFV infection. (A) HOS cells were transfected with the tandem reporter construct mRFP-EGFP-LC3, grown in selective medium for 1 week, then mock infected, infected with SFV, or treated with 50 μM CQ. After 12 h, cells were fixed and stained for nuclei (blue) and processed for microscopy. (B) HOS-EGFP-LC3 cells were mock infected, starved of amino acids and serum, or infected with SFV. One hour later, cells were either left untreated or treated with NH4Cl. After a further 7 h, cells were fixed and stained for nuclei (blue) and processed for microscopy. (C) HOS-EGFP-LC3 cells were mock infected, starved or SFV infected for 4, 8, or 12 h, and treated at 1 h postinfection with NH4Cl for 3, 7, or 11 h, respectively, or left untreated. Cells were then washed briefly in 0.05% saponin in PBS, stained for nsP1, and analyzed by flow cytometry. Shown are representative histograms from four independent experiments. Histograms in the right-most column were gated on the nsP1-positive population and thus represent SFV-infected cells. (D) HOS-EGFP-LC3 cells were mock infected or infected with IAV X31 for 24 or 32 h and then treated at 1 hpi with NH4Cl for 23 or 31 h, respectively, or left untreated. Cells were then washed briefly in 0.05% saponin in PBS, stained with anti-IAV antibody, and analyzed by flow cytometry. Shown are representative histograms from two independent experiments. Histograms in the right-most column were gated on the IAV-positive population.
To further study autophagosomal flux during SFV infection, we analyzed the effect of the lysosomal acidification inhibitor NH4Cl on the accumulation of LC3-positive autophagosomes in HOS-EGFP-LC3 cells. In cells starved of amino acids and serum for 8 h, the number of autophagosomes was only slightly increased relative to mock-treated cells (Fig. 7B). This is due to the lysosomal degradation rate, which increases in tandem with the rate of autophagosome formation in starved HOS cells, as we described previously (10). While the number of autophagosomes in mock-treated cells was increased by the addition of NH4Cl, it was dramatically increased in starved cells by addition of NH4Cl. This increase represents the induction of autophagy under conditions of amino acid starvation. In contrast, autophagosome accumulation in SFV-infected cells was not enhanced by NH4Cl treatment (Fig. 7B).
To quantify the flux of autophagosomes in SFV-infected cells, we again employed our recently published flow cytometry-based assay for autophagic flux (10). Specifically, using flow cytometry we analyzed the kinetics of saponin-resistant (autophagosome-bound) EGFP–LC3-II accumulation in amino acid-starved cells or in SFV-infected cells in the presence or absence of NH4Cl. Again, amino acid starvation for 4, 8, or 12 h did not substantially increase the EGFP–LC3-II signal unless it was accompanied by NH4Cl treatment (Fig. 7C). The SFV-induced accumulation of EGFP–LC3-II fluorescence, however, was evident in the absence of NH4Cl and was not appreciably increased by its presence. Since an increase in LC3-II signal upon the addition of lysosomal acidification inhibitors is a hallmark of induction of autophagy (20), the data shown in Fig. 7B and C indicate that SFV-induced accumulation of autophagosomes was largely due to a deficiency in autophagosome degradation rather than induction of autophagy.
Since it was previously shown that IAV blocks the fusion of autophagosomes with lysosomes through the action of the M2 protein (13), we analyzed the effect of infection with this virus on autophagic flux in HOS-EGFP-LC3 cells. In a similar experimental setup to that shown in Fig. 7C, we infected HOS-EGFP-LC3 cells with IAV X31 and determined the levels of autophagosome-associated EGFP–LC3-II accumulation at different times postinfection in the presence and absence of NH4Cl (Fig. 7D). Similarly to SFV infection, IAV-infected cells maintained a strong saponin-resistant EGFP-LC3 signal, which was not increased by the addition of NH4Cl. Taken together, the data presented in Fig. 7 led us to conclude that the increase in levels of autophagosomes observed in SFV-infected cells is a result of their inefficient lysosomal degradation, as has been reported to be the case for autophagosome accumulation in some other viral systems (13, 39).
Accumulation of autophagosomes in SFV-infected cells is due to expression of the viral glycoprotein complex.
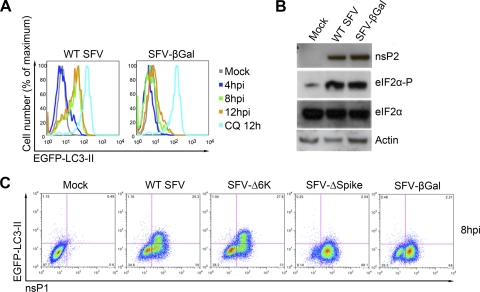
In order to identify the viral determinants of the inhibition of autophagosome/lysosome fusion, we took advantage of a recombinant SFV (rSFV) expression system, in which viral subgenomic replicons can be constructed which lack the structural protein-coding region and can be trans-packaged by using helper constructs (25). We infected HOS-EGFP-LC3 cells with wt SFV, with rSFV expressing Escherichia coli β-galactosidase (SFV-βGal), or mock infected them, and we analyzed the cells for LC3 accumulation by using FACS. As observed previously, wt SFV infection induced strong accumulation of EGFP–LC3-II, which was evident from 8 hpi onwards (Fig. 8A). Infection of cells with SFV-βGal, on the other hand, did not induce LC3 accumulation except in a very small number of cells. These data suggest that the expression of the viral structural proteins themselves, or a late stage in virus assembly, is responsible for the inhibition of autophagosome fusion with lysosomes. To confirm that the cells used in this experiment had been infected at equal levels, we took lysates from mock-, wt SFV-, and rSFV-βGal-infected HOS-EGFP-LC3 cells and analyzed them by immunoblotting for the viral protein nsP2 and cellular proteins eIF2α and actin (Fig. 8B). Similar levels of nsP2 and phosphorylated eIF2α were observed in cells infected with either virus, indicating equivalent levels of infection and confirming the absence of involvement of eIF2α phosphorylation in the accumulation of autophagosomes in SFV infected cells.
Fig 8.
Accumulation of autophagosomes depends on SFV-spike expression. (A) HOS-EGFP-LC3 cells were mock infected or infected with wt SFV or SFV-βGal at an MOI of 10 for 4, 8, or 12 h or treated with 50 μM CQ for 12 h. Cells were then washed briefly in 0.05% saponin in PBS, stained for nsP1 to confirm infection (data not shown), and analyzed by flow cytometry. Shown are representative histograms. (B) HOS-EGFP-LC3 cells were mock infected or infected with wt SFV or SFV-βGal at an MOI of 10 for 12 h. Lysates were analyzed for SFV nsP2, phosphorylated or total eIF2α, or actin by Western blotting. (C) HOS-EGFP-LC3 cells were mock infected, infected with wt SFV, or infected with rSFV variants as labeled, for 8 h. Cells were then washed briefly in 0.05% saponin in PBS, stained for nsP1, and analyzed by flow cytometry. Shown are representative dot plots from three independent experiments.
To further specify the viral determinants of the accumulation of autophagosomes, we determined the ability of various viral mutants to induce this effect. The wt SFV, SFV-Δ6K (lacking the small 6,000-kDa peptide [26]), SFV-ΔSpike (lacking the viral glycoprotein spike complex [40]) and SFV-βGal (lacking all structural proteins) were used to infect HOS-EGFP-LC3 cells, and the accumulation of EGFP–LC3-II was determined by flow cytometry. Identification of infected cell populations was achieved by costaining for nsP1. All viral mutants expressed similar levels of nsP1 in infected cells, but only wt SFV and SFV-Δ6K induced accumulation of a saponin-resistant EGFP–LC3-II –expressing population (Fig. 8C). Despite high levels of infection, neither SFV-ΔSpike-infected nor SFV-βGal-infected cells accumulated detectable levels of EGFP–LC3-II during the infection.
Since the SFV-ΔSpike vector directs the expression of an SFV capsid protein monomer that is competent for assembly into intracellular nucleocapsid particles (40), this result excludes that particle as the viral determinant of increased autophagosome formation. Microscopic analysis of HOS-EGFP-LC3 cells infected with either wt SFV or with SFV-ΔSpike revealed similar diffuse cytoplasmic SFV capsid staining but markedly different EGFP-LC3 staining (see Fig. S2 in the supplemental material). We conclude from the data presented in Fig. 8 that expression of the SFV glycoprotein complex is necessary for the accumulation of autophagosomes in infected cells.
DISCUSSION
In this work, we investigated if the autophagic pathway was modulated in SFV-infected cells. We found that although LC3-positive autophagosomes accumulate in the cytoplasm of SFV-infected cells, blocking this pathway did not affect viral replication. We detected low and no colocalization of nonstructural and structural viral proteins, respectively, with either of the autophagosomal markers LC3-II or p62/SQSTM1, suggesting that much of the viral proteins are not targeted to these organelles. The activity of an important negative regulator of autophagy, the mTOR complex, was maintained in infected cells, suggesting that there is no enhanced formation of autophagosomes via the lifting of constitutive mTOR repression during SFV infection. Furthermore, by using both the tandem mRFP-EGFP-LC3 reporter construct and a quantitative assay for autophagic flux, we showed that the autophagosomes that accumulated in SFV-infected cells were not degraded. Collectively, these results suggest that the autophagosomes formed during SFV infection accumulate as a result of their inefficient degradation rather than by an enhanced induction of their formation. Interestingly, although the viral glycoproteins did not colocalize with LC3-II, their expression was necessary for the block in autophagosome maturation in SFV-infected cells.
This work illustrates a fundamental concept in autophagy, discussed in many recent reviews (20, 32, 37), which is that, since the process is degradative, the accumulation of autophagosomes in the cytoplasm of a cell under certain conditions can result from inhibition of their fusion with lysosomes or through enhanced induction of autophagy. In fact, since lysosomal degradation rates increase concomitantly when autophagy is induced, as we observed under amino acid and serum starvation of HOS-EGFP-LC3 cells (reference 10 and this work), inhibition of autophagosome maturation leads to significantly higher levels of cytoplasmic autophagosomes than induction. Autophagosomes accumulate in the cytoplasm in a number of viral infections, and while for some of these, the mechanism of autophagosome accumulation has been well described, for others it has not. In the latter case, the conclusions that autophagy is induced by the infections are not convincing in the absence of assays demonstrating an increase in autophagic flux. Aside from LC3, another frequently used marker for autophagic degradation is p62/SQSTM1. Many studies have reported increased degradation of this reporter to support claims of autophagy induction. However, in many lytic viral infections, including SFV, the use of this reporter is complicated by the profound inhibition of host cell translation such that no new synthesis of the p62/SQSTM1 protein occurs during the infection. In SFV-infected cells, we have observed that the levels of this protein do not decrease significantly faster than cells treated with cycloheximide to simulate host cell translation inhibition (data not shown). Quantitative assays for autophagic flux remain the most reliable method to determine the origin of autophagosomes in virus infections.
The efficiency of SFV replication complex formation and the viral propagation rates in atg5−/− MEFs suggest that autophagy has little, if any, effect on intracellular SFV replication, either positively, through the provision of excess intracellular membranes, for example, or negatively, through targeting of viral protein complexes for degradation. We did detect some regions of colocalization between LC3 and viral nsPs (Fig. 1 to 3). This phenomenon was restricted to the nsPs, since we did not detect instances of colocalization of capsid or E1/E2 proteins with LC3 or with p62/SQSTM1 (Fig. 4 and data not shown; see also Fig. S1 in the supplemental material). The colocalization of viral nsPs with LC3 suggests that autophagosomes are in some cases used as structures for SFV RNA replication complex assembly, or that the replicase complexes occasionally are targeted for autophagic degradation. Recent work from Spuul and colleagues identified the plasma membrane as the principal source of membranes for SFV replication complex formation (42). Further, since the gross morphology of replication complexes appeared normal in MEFs lacking ATG5, we conclude that autophagosomes are unlikely to be utilized for replication complex formation by SFV to any large degree. We therefore consider the latter explanation for the colocalizations to be more likely. The observation that viral replication rates were not affected in atg5−/− MEFs showed that any possible inclusion of viral replicase complexes into autophagosomes does not measurably affect viral replication. In several other RNA virus systems, viral nsPs have been implicated in accumulation of autophagosomes. The poliovirus 2BC and 3A proteins, when expressed in the absence of the rest of the viral genome, localize to intracellular membranes as part of the assembly of the viral replication complexes and can induce the accumulation of autophagosomes (16). Similarly, coronavirus nsP6 is a membrane-associated protein which when expressed alone, localizes to ER membranes and can induce the accumulation of autophagsomes (6). However, the reduction in LC3-II accumulation in SFV-βGal-infected cells compared to wt SFV-infected cells suggests that the SFV nsPs, expressed at equal levels by wt SFV and SFV-βGal, are not involved in this phenomenon.
It is notable that the times when the greatest levels of LC3-II accumulation occurred were very late in infection, 8 h and later, when viral growth curves are starting to reach plateau levels. In other studies, we have observed cellular reactions to SFV infection at much earlier times. For example, the activation and nuclear translocation of the type I interferon transcription factors NF-κB p65 and interferon regulatory factor 3 are detectable at 3 hpi (4, 15), and the phosphorylation of eIF2α and assembly of stress granules are detectable at similarly early times (28). The latter is an example of a cellular stress pathway that is activated by virus infection but is then subverted by the virus for its own purposes (the efficient translation of viral mRNAs containing the translational enhancer element) (28). We consider therefore that the relatively late times in infection when autophagosome accumulation is observed are more reminiscent of a slow accumulation due to the inhibition of fusion with lysosomes, rather than an infection-induced cellular response.
Our experiments clearly implicate the structural region, specifically the viral glycoprotiens, in the inhibition of autophagosome maturation. The 6,000-kDa transmembrane protein has been reported to have ion channel activity and is important for the correct folding of the E1/E2 glycoproteins and for efficient budding of the viral particles (27, 29, 30). At the time points used in our experiments, SFV-Δ6K budding was between 1 and 10% as efficient as wt SFV (unpublished observations), and since wt SFV and SFV-Δ6K both induced similar levels of EGFP-LC3-II, we think it unlikely that viral budding, but rather a prebudding function of the glycoproteins, is responsible for the effect. The very high expression levels of the SFV spike proteins in infected cells have recently been shown to lead to ER stress and activation of the UPR. Although there have been some reports of involvement of the UPR in autophagy in other viral systems (5, 36, 39), we did not detect any contribution of SFV spike-dependent UPR induction to the accumulation of autophagosomes. The wt SFV-infected cells showed very modest alternative splicing of X-box-binding protien 1 (Xbp-1) mRNA at times of greatest accumulation of LC3-II, while thapsigargin treatment led to strong signal for spliced Xbp-1 but very weak induction of autophagosome assembly in HOS-EGFP-LC3 cells (data not shown). Another consequence of spike complex expression is the higher rate of cell death in cells infected with wt SFV than in recombinant viruses lacking this complex (such as SFV-βGal and SFV-ΔSpike) (reference 1 and unpublished observations). IAV infection blocks the maturation of autophagosomes in a manner that compromises the survival of infected cells (13). We have shown here that the accumulation of autophagosomes occurs to similar extents in IAV-infected and in SFV-infected HOS-EGFP-LC3 cells. Interestingly, the IAV M2 protein is alone responsible for the inhibition of autophagosome fusion with lysosomes, although not through its ion channel activity (13). Since the inhibition of autophagy in IAV-infected cells enhances cell death after infection, it is possible that the SFV spike-induced block in autophagosome maturation is a contributing factor to the enhanced cytopathogenicity of SFV variants that express this complex over those that do not (1). It may be that autophagy, induced as a cell survival mechanism, is blocked late in infection by certain viruses to allow the infected cells to go into apoptosis in order to limit cytokine production, for example, or otherwise avoid immune surveillance. Viruses modulate the autophagic pathway in many different ways. Since autophagy has several important roles in pathogen infection and immunity (7), studies of the regulation of autophagy during viral infections will lead to a more complete understanding of this aspect of the host-pathogen relationship.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Swedish Research Council to G.M.M. and to G.K.H. K.E. is a recipient of a scholarship from the Agency for Science, Technology and Research, Singapore (A*STAR).
We thank Peter Liljeström (Karolinska Institutet, Sweden), Andres Merits (University of Tartu, Estonia), Gerhard Hunsmann (Göttingen University, Germany), Anna Smed Sörensen (Karolinska Institutet, Sweden), and Angelo de Milito (Karolinska Institutet, Sweden) for provision of reagents.
Footnotes
Published ahead of print 21 March 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Barry G, et al. 2010. Semliki forest virus-induced endoplasmic reticulum stress accelerates apoptotic death of mammalian cells. J. Virol. 84:7369–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjorkoy G, et al. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brabec-Zaruba M, Berka U, Blaas D, Fuchs R. 2007. Induction of autophagy does not affect human rhinovirus type 2 production. J. Virol. 81:10815–10817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breakwell L, et al. 2007. Semliki Forest virus nonstructural protein 2 is involved in suppression of the type I interferon response. J. Virol. 81:8677–8684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carpenter JE, Jackson W, Benetti L, Grose C. 2011. Autophagosome formation during varicella-zoster virus infection following endoplasmic reticulum stress and the unfolded protein response. J. Virol. 85:9414–9424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cottam EM, et al. 2011. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 7:1335–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deretic V. 2011. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol. Rev. 240:92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dreux M, Chisari FV. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307 [DOI] [PubMed] [Google Scholar]
- 9. Dreux M, Gastaminza P, Wieland SF, Chisari FV. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046–14051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eng KE, Panas MD, Karlsson Hedestam GB, McInerney GM. 2010. A novel quantitative flow cytometry-based assay for autophagy. Autophagy 6:634–641 [DOI] [PubMed] [Google Scholar]
- 11. Ganley IG, et al. 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284:12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ganley IG, Wong PM, Gammoh N, Jiang X. 2011. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell 42:731–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gannage M, et al. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hay N, Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 [DOI] [PubMed] [Google Scholar]
- 15. Hidmark AS, et al. 2005. Early alpha/beta interferon production by myeloid dendritic cells in response to UV-inactivated virus requires viral entry and interferon regulatory factor 3 but not MyD88. J. Virol. 79:10376–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jackson WT, et al. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Joubert PE, et al. 2009. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe 6:354–366 [DOI] [PubMed] [Google Scholar]
- 18. Kamada Y, et al. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150:1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura S, Noda T, Yoshimori T. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3:452–460 [DOI] [PubMed] [Google Scholar]
- 20. Klionsky DJ, et al. 2008. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kouroku Y, et al. 2007. ER stress (PERK/eIF2α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14:230–239 [DOI] [PubMed] [Google Scholar]
- 22. Krejbich-Trotot P, et al. 2011. Chikungunya triggers an autophagic process which promotes viral replication. Virol. J. 8:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kujala P, et al. 1997. Monoclonal antibodies specific for Semliki Forest virus replicase protein nsP2. J. Gen. Virol. 78:343–351 [DOI] [PubMed] [Google Scholar]
- 24. Lee YR, et al. 2008. Autophagic machinery activated by dengue virus enhances virus replication. Virology 374:240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liljeström P, Garoff H. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (New York) 9:1356–1361 [DOI] [PubMed] [Google Scholar]
- 26. Liljeström P, Lusa S, Huylebroeck D, Garoff H. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loewy A, Smyth J, von Bonsdorff CH, Liljeström P, Schlesinger MJ. 1995. The 6-kilodalton membrane protein of Semliki Forest virus is involved in the budding process. J. Virol. 69:469–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McInerney GM, Smit JM, Liljeström P, Wilschut J. 2004. Semliki Forest virus produced in the absence of the 6K protein has an altered spike structure as revealed by decreased membrane fusion capacity. Virology 325:200–206 [DOI] [PubMed] [Google Scholar]
- 30. Melton JV, et al. 2002. Alphavirus 6K proteins form ion channels. J. Biol. Chem. 277:46923–46931 [DOI] [PubMed] [Google Scholar]
- 31. Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21:2861–2873 [DOI] [PubMed] [Google Scholar]
- 32. Mizushima N, Yoshimori T. 2007. How to interpret LC3 immunoblotting. Autophagy 3:542–545 [DOI] [PubMed] [Google Scholar]
- 33. Nakagawa I, et al. 2004. Autophagy defends cells against invading group A Streptococcus. Science 306:1037–1040 [DOI] [PubMed] [Google Scholar]
- 34. O'Donnell V, et al. 2011. Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology 410:142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Orvedahl A, et al. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reggiori F, et al. 2010. Coronaviruses hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe 7:500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rubinsztein DC, et al. 2009. In search of an “autophagomometer.” Autophagy 5:585–589 [DOI] [PubMed] [Google Scholar]
- 38. Scheuner D, et al. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7:1165–1176 [DOI] [PubMed] [Google Scholar]
- 39. Sir D, et al. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skoging U, Liljeström P. 1998. Role of the C-terminal tryptophan residue for the structure-function of the alphavirus capsid protein. J. Mol. Biol. 279:865–872 [DOI] [PubMed] [Google Scholar]
- 41. Smerdou C, Liljeström P. 1999. Two-helper RNA system for production of recombinant Semliki Forest virus particles. J. Virol. 73:1092–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Spuul P, Balistreri G, Kaariainen L, Ahola T. 2010. Phosphatidylinositol 3-kinase-, actin-, and microtubule-dependent transport of Semliki Forest virus replication complexes from the plasma membrane to modified lysosomes. J. Virol. 84:7543–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Talloczy Z, et al. 2002. Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 99:190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor MP, Burgon TB, Kirkegaard K, Jackson WT. 2009. Role of microtubules in extracellular release of poliovirus. J. Virol. 83:6599–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor MP, Kirkegaard K. 2007. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81:12543–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Urban C, et al. 2008. Apoptosis induced by Semliki Forest virus is RNA replication dependent and mediated via Bak. Cell Death Differ. 15:1396–1407 [DOI] [PubMed] [Google Scholar]
- 47. Wong J, et al. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang Z, Klionsky DJ. 2009. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 335:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao Z, et al. 2007. Coronavirus replication does not require the autophagy gene ATG5. Autophagy 3:581–585 [DOI] [PubMed] [Google Scholar]
- 50. Zheng YT, et al. 2009. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 183:5909–5916 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.