Abstract
The Epstein-Barr virus (EBV) predominantly establishes latent infection in B cells, and the reactivation of the virus from latency is dependent on the expression of the viral BZLF1 protein. The BZLF1 promoter (Zp) normally exhibits only low basal activity but is activated in response to chemical or biological inducers, such as 12-O-tetradecanoylphorbol-13-acetate (TPA), calcium ionophores, or histone deacetylase (HDAC) inhibitors. In some cell lines latently infected with EBV, an HDAC inhibitor alone can induce BZLF1 transcription, while the treatment does not enhance expression in other cell lines, such as B95-8 or Raji cells, suggesting unknown suppressive mechanisms besides histone deacetylation in those cells. Here, we found the epigenetic modification of the BZLF1 promoter in latent Raji cells by histone H3 lysine 27 trimethylation (H3K27me3), H3K9me2/me3, and H4K20me3. Levels of active markers such as histone acetylation and H3K4me3 were low in latent cells but increased upon reactivation. Treatment with 3-deazaneplanocin A (DZNep), an inhibitor of H3K27me3 and H4K20me3, significantly enhanced the BZLF1 transcription in Raji cells when in combination with an HDAC inhibitor, trichostatin A (TSA). The knockdown of Ezh2 or Suv420h1, histone methyltransferases for H3K27me3 or H4K20me3, respectively, further proved the suppression of Zp by the methylations. Taken together, the results indicate that H3K27 methylation and H4K20 methylation are involved, at least partly, in the maintenance of latency, and histone acetylation and H3K4 methylation correlate with the reactivation of the virus in Raji cells.
INTRODUCTION
The Epstein-Barr virus (EBV) is a human gammaherpesvirus that establishes latent infection predominantly in B lymphocytes. Only a small percentage of infected cells switch from the latent stage into the lytic cycle and produce progeny viruses. Although the mechanism of EBV reactivation in vivo is not fully understood, it is known to be elicited in vitro by the treatment of latently infected B cells with chemical or biological reagents, such as 12-O-tetradecanoylphorbol-13-acetate (TPA), calcium ionophore, sodium butyrate, and anti-immunoglobulin (Ig). The stimulation of the EBV lytic cascade by these reagents leads to the expression of two presumed viral immediate-early genes, BZLF1 and BRLF1. The BZLF1 protein is a transcriptional activator that shares structural similarities to basic leucine zipper (b-Zip) family transcriptional factors, and BZLF1 expression alone can trigger the entire reactivation cascade (1, 50, 53).
The expression of the BZLF1 gene is tightly controlled at the transcriptional level. The BZLF1 promoter (Zp) normally exhibits low basal activity and is activated in response to TPA or the other reagents described above. The promoter is activated by transcriptional factors, including myocyte enhancer factor 2D (MEF2D) (35) and Sp1/3 (34). Cellular b-Zip-type transcription factors, such as the cyclic AMP response element binding protein (CREB), activating transcription factor (ATF), activator protein 1 (AP-1) (33, 42, 43, 48), or a spliced form of X-box binding protein 1 [XBP-1(s)] (2), also play crucial roles in promoter activation. We previously showed the importance of CREB and its calcineurin-dependent activation by transducer of regulated CREB 2 (TORC2) (43). Once produced, BZLF1 itself can bind to and activate its own promoter (16, 41). Most of the positive factors have been demonstrated or are presumed to upregulate the BZLF1 promoter by recruiting transcriptional coactivators, such as histone acetylases. On the other hand, the activity of Zp is restricted by repressive factors, including Jun dimerization protein 2 (JDP2) (42), Zinc finger E-box binding factor (ZEB) (58), Yin Yang 1 (YY1) (40), and sumoylation of BZLF1 (19, 41), since those factors facilitate the access of repressive transcriptional cofactors, such as histone deacetylase (HDAC), to the promoter and/or block the binding or functions of the transcriptional activators noted above.
The silencing of the BZLF1 promoter in latently infected cells is mediated, at least in some cell lines, such as Akata, by low levels of histone acetylation, since inhibitors of HDAC, like sodium butyrate or trichostatin A (TSA), can reverse the silencing (37, 38). However, treatment with butyrate or TSA alone does not efficiently induce BZLF1 transcription in cell lines like B95-8 or Raji, suggesting that the molecular mechanisms that govern the suppression of BZLF1 transcription in those cells must be more than just the low acetylation levels of the promoter (10, 11, 17). 5′-CG-3′ dinucleotide (CpG) DNA methylation was one possible cause of promoter repression, because inhibitors of DNA methylation, such as 5-aza-2′-deoxycytidine (5-Aza), elicit BZLF1 transcription from the promoter (57). Nevertheless, it is highly likely that CpG methylation is not involved in the process, because the methylation of the Zp has not been detected in any cells, including B95-8, Raji, and Akata (15). In addition, treatment with 5-Aza induces BZLF1 transcription within a very short period of time (15 min or less) (10), although it takes days to bring about the hypomethylation of the CpG DNA, because 5-Aza is an inhibitor of DNA methyltransferase and does not actively reverse or abolish methylation without de novo DNA amplification. Therefore, as Countryman and others discuss in their report (11), it is likely that 5-Aza activates EBV lytic gene expression by an unknown mechanism that does not require viral DNA replication or DNA demethylation.
Possible epigenetic modifications for silencing the promoter other than CpG methylation include histone modification, such as histone H3 lysine 27 trimethylation (H3K27me3), H4K20me3, or H3K9me2/me3 (28). H3K27me3 is a histone modification that is involved in the suppression of a wide variety of genes (29). The methylation is mediated by enhancer of Zeste 2 (Ezh2), a member of polycomb repressor complex 2 (PRC2) (5). H4K20me3 also is a repressive chromatin marker that is frequently associated with heterochromatin. The H4K20me3 methyltransferase is a member of the SET domain-containing proteins, suppressor of variegation 420 h (Suv420h) (49). H3K9me2, catalyzed by G9a, is a typical repressive marker of facultative heterochromatin, whereas H3K9me3 methylation, predominantly found in constitutive heterochromatin, is mediated by enzymes including Suv39h.
In the present study, we found that the Zp is modified by negative markers, such as H3K27me3 and H4K20me3, in latently infected cells and upon reactivation modification by active markers, such as histone acetylation, and H3K4 methylation is increased. The treatment of cells with TSA and 3-deazaneplanocin A (DZNep), an inhibitor of H3K27me3 and H4K20me3 (39, 51), augmented levels of BZLF1 in Raji cells. The knockdown of Ezh2 or Suv420h1 by RNA interference markedly increased BZLF1 induction when treated with TSA. These results indicate that the Zp promoter in Raji cells is silenced, at least to some extent, by H3K27me3 and H4K20me3 during latency.
MATERIALS AND METHODS
Cell culture and reagents.
293EBV-bacterial artificial chromosome (BAC) epithelial cells (43) were maintained in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum. Akata, Raji, and lymphoblastoid cell line (LCL) EBV-BAC cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum. SNK6 cells (44) were cultured in RPMI 1640 medium supplemented with 10% human serum and interleukin-2 (IL-2). Horseradish peroxidase-linked goat antibodies to mouse/rabbit IgG were from Amersham Biosciences. Anti-histone H3 (ab1791) and anti-Suv420H1 (ab49251), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (14C10 and 2118), and anti-H3K4me3 (17-614) were purchased from Abcam, Cell Signaling, and Millipore, respectively. Anti-H3K9Ac (39137), anti-H3K9me2 (39375), anti-H3K9me3 (39161), anti-H3K27me3 (39155 and 39535), anti-H4K20me3 (39180), and anti-Ezh2 (39875) antibodies were from Active Motif.
Immunoblotting and ChIP assay.
Immunoblotting was carried out as described previously (43). Chromatin IP (ChIP) assays were performed essentially as described previously (43) with formaldehyde cross-linked chromatin from 1 × 106 cells for each reaction. Cells were lysed, and chromatin was sonicated to obtain DNA fragments with an average length of 300 bp. Following centrifugation, the chromatin was diluted 10-fold with ChIP dilution buffer and precleared with protein A agarose beads containing salmon sperm DNA (Upstate). Immune complexes were collected by the addition of protein A agarose beads, and DNA was purified using a QIAquick PCR purification kit (Qiagen) after the uncoupling of the cross-linking and proteinase K digestion. The PCR products were then analyzed by real-time PCR for the quantification of DNA sequences using the following primers and SYBR Premix Ex Taq II (TaKaRa). The recovered DNA was amplified by PCR using the following specific primers: for Zp (Zp0), 5′-TAGCCTCGAGGCCATGCATATTTCAACTGG-3′ and 5′-GCCAAGCTTCAAGGTGCAATGTTTAGTGAG-3′; for Zp-3000, 5′-ACCTCACTACACAAACAGAC-3′ and 5′-TTCAACACAGCAGGCCTCTC-3′; for Zp-2000, 5′-CCACTTCGGGATAGTGTTTC-3′ and 5′-TTCCTTGTTGAGGACGTTGC-3′; for Zp-1200, 5′-GACAGAGGAGCTACGTGAG-3′ and 5′-ATGAAACTGTCCGGACTCCG-3′; for Zp-600, 5′-AGGTATGTTCCTGCCAAAGC-3′ and 5′-GTTCATGGACAGGTCCTGTG-3′; for Zp +500, 5′-GGAGAAGCACCTCAACCTG-3′ and 5′-CTCCTTACCGATTCTGGCTG-3′; for the BRLF1 promoter (Rp), 5′-TAAGATCTTGGGGACGATGG-3′ and 5′-ACCATTAAAATCTTTCCTCC-3′; for the origin of lytic DNA replication (oriLyt), 5′-CCGGCTCGCCTTCTTTTATCCTC-3′ and 5′-CCTGGTTCAACCCTATGGAGGGGAC-3′; for the BMRF1 promoter (Mp), 5′-TAAAGCAGTTTCTGGAGGCC-3′ and 5′-GCCCAGAAACCTGAGCAAGT-3′; for the Q promoter of EBNA (Qp), 5′-GGCTCACGAAGCGAGAC-3′ and 5′-GTCGTCACCCAATTTCTGTC-3′; for the dyad symmetry in the origin of latent replication (oriP DS), 5′-GTGACAGCTCATGGGGTGGG-3′ and 5′-GATAAGCGGACCCTCAAGAG-3′; for the C promoter of EBNA (Cp), 5′-AGTTGGTGTAAACACGCCGT-3′ and 5′-TCCACCTCTAAGGTCCCACG-3′; for the β-globin promoter (Globinp), 5′-AGGACAGGTACGGCTGTCATC-3′ and 5′-TTTATGCCCAGCCCTGGCTC-3′; and for the GAPDH promoter (GAPDHp), 5′-CGTGCCCAGTTGAACCAGG-3′ and 5′-AGGAGGAGCAGAGAGCGAAG-3′. Real-time PCR was performed in 10 μl of solution containing 0.2 μM primers, 0.2 μl ROX dye, and the sample DNA in 1× One Step SYBR reverse transcription-PCR (RT-PCR) buffer. The intensity of ROX dye was used to compensate for volume fluctuations among the tubes. PCR included 10 s at 95°C and 40 cycles at 95°C for 5 s, followed by 45 s at 60°C. Immediately after the PCR, we carried out dissociation curve analysis and confirmed the specificity of each PCR product. A standard curve was constructed using serial dilutions of DNA and was used to quantitate the amount of DNA.
siRNA.
Duplexes of 21-nucleotide small interfering RNA (siRNA) specific to human Ezh2 or Suv420h1 mRNA, including two nucleotides of deoxythymidine (dTdT) at the 3′ end, were synthesized and annealed (Gene Design, Inc.). The sense and antisense sequences of the duplex were the following: for Ezh2, 5′-CCAUGUUUACAACUAUCAAdTdT-3′ and 5′-UUGAUAGUUGUAAACAUGGdTdT-3′; for Suv420h1, 5′-CCAUGAUUGCAGACCUAAUdTdT-3′ and 5′-AUUAGGUCUGCAAUCAUGGdTdT-3′; and for control siRNA, 5′-GCAGAGCUGGUUUAGUGAAdTdT-3′ and 5′-UUCACUAAACCAGCUCUGCdTdT-3′. Raji cells (1 × 105) were transfected with 50 pmol of the duplex RNA per well of a 24-well plate using a microporator (Digital Bio). Two days after transfection, TPA was added for Raji cells, followed by incubation for another day.
RT-PCR.
Total cell RNA was purified using TriPure isolation reagent (Roche) and subjected to real-time RT-PCR using a One Step SYBR PrimeScript RT-PCR kit II (TaKaRa) and real-time PCR system 7300 according to the manufacturer's instructions. PCR was performed in 10 μl of solution containing 0.2 μM primers, 0.2 μl ROX dye, and the sample RNA in 1× One Step SYBR RT-PCR buffer. The intensity of the ROX dye was used to compensate for volume fluctuations among the tubes. PCR included 5 min at 42°C, 10 s at 95°C, and 40 cycles at 95°C for 5 s, followed by 40 s at 60°C. Immediately after RT-PCR, we carried out dissociation curve analysis and confirmed the specificity of each PCR product. An arbitrary RNA value was set to 1.0, and a standard curve was constructed using serial dilutions of RNA from the RNA set to 1.0. The amount of mRNA was quantitated based on the standard curve. Real-time PCR with GAPDH primers was also performed to serve as an internal control for input RNA. Primers used for RT-PCR were the following: for GAPDH mRNA, 5′-TGCACCACCAACTGCTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′; for BZLF1 mRNA, 5′-AACAGCCAGAATCGCTGGAG-3′ and 5′-GGCACATCTGCTTCAACAGG-3′; and for EBNA2 mRNA, 5′-TTAGAGAGTGGCTGCTACGCATT-3′ and 5′-TCACAAATCACCTGGCTAAG-3′.
RESULTS
Epigenetic histone modification of the BZLF1 promoter during latency and reactivation by chemical inducers.
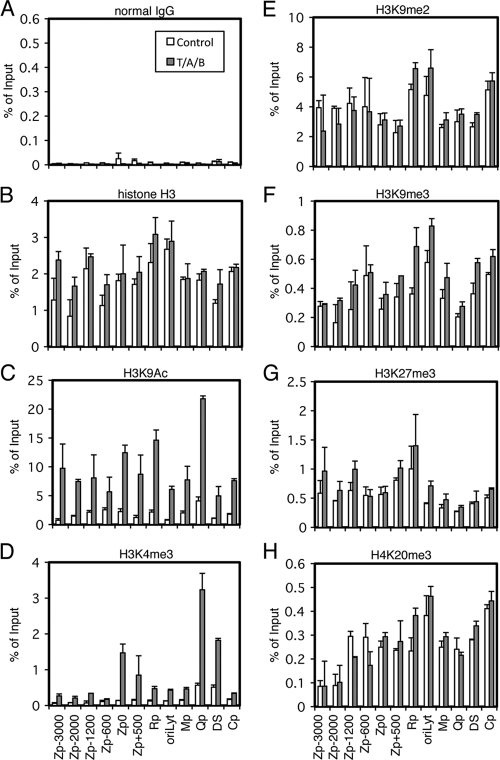
The reactivation of EBV from latency is tightly blocked at the level of the transcription of the BZLF1 gene. It has been reported that the silencing of the gene is dependent only on low levels of histone acetylation in some cell lines, such as Akata (37, 38). However, because TSA alone does not induce BZLF1 transcription in certain cells, like B95-8 or Raji, it has been estimated that there must be other molecular mechanisms that are responsible for the silencing of BZLF1 transcription (10, 11, 17). To analyze the mechanisms that govern BZLF1 transcription besides histone acetylation in such cell lines, we first examined levels of various epigenetic histone modifications at certain regions of EBV DNA, including −3000 to +500 of Zp, the BRLF1 promoter oriLyt, the BMRF1 promoter (Mp), the Q promoter of EBNA (Qp), the dyad symmetry in the origin of latent replication (oriP DS), and the C promoter of EBNA (Cp). We chose Raji cells for two reasons. First, the cells have a repressive mechanism besides low-level acetylation. Second, as EBV in Raji cells lacks the single-stranded DNA binding protein BALF2 and thus is replication incompetent (20), we can observe epigenetic alterations that affect transcriptions of BZLF1 or other genes without the complication associated with viral genome amplification. To achieve lytic induction, Raji cells were treated with TPA, A23187, and sodium butyrate (T/A/B) (Fig. 1, gray bars), which efficiently induces the lytic cycle in the cell line. As shown in Fig. 1A and B, background precipitation with normal IgG could be ignored, and histone H3 levels were fairly constant.
Fig 1.
Histone modification pattern of EBV Zp upon lytic reactivation. Raji cells were treated with TPA (20 ng/ml), A23187 (1 μM), and sodium butyrate (5 mM) (T/A/B; gray bar) or the vehicle (Control; white bar) for 20 h. Cells then were cross-linked, and ChIP experiments were performed as described in Materials and Methods using normal IgG (A), anti-histone H3 (B), anti-H3K9Ac (C), anti-H3K4me3 (D), anti-H3K9me2 (E), anti-H3K9me3 (F), anti-H3K27me3 (G), or anti-H4K20me3 (H) antibody, followed by DNA extraction and real-time PCR to detect DNA fragments using the primers as indicated. Zp, BZLF1 promoter; Rp, BRLF1 promoter; oriLyt, origin of lytic DNA replication; Mp, BMRF1 promoter; Qp, one of the EBNA promoters; DS, dyad symmetry, a part of oriP (origin of plasmid replication), containing multiple EBNA1 binding sites; Cp, one of the EBNA promoters. The number of Zp indicates sequence position relative to the transcription start site.
Active chromatin markers, such as H3K9Ac or H3K4me3, were present at lower levels in the viral genome (Fig. 1C and D, white bars). It is noteworthy that the levels of those active marks were intrinsically higher at the commonly active latent EBNA promoter Qp (Fig. 1C and D, white bars). Elevated levels of histone H3/H4 acetylation and H3K4 methylation at the Qp in latent Raji cells was previously demonstrated by Day et al. as well (12). Relatively high histone H3K4me3 levels in the latent DS, the EBNA1 binding sites in oriP (Fig. 1D, white bars) (12) may reflect EBNA1's function as a transcriptional activator (24).
Lytic induction by TPA, A23187, and sodium butyrate markedly elevated the active histone markers H3K9Ac and H3K4me3 in the viral genome (Fig. 1C and D). The enhancement of H3K4me3 upon the induction of the EBV lytic cycle has never been reported to our knowledge, although such modification is reported for herpes simplex virus (HSV) (21, 45) and Kaposi's sarcoma-associated herpesvirus (KSHV) (18, 52). The enhancement of H3K4me3 at the BZLF1 transcription start site Zp0 (−221 to +12) and Zp +500 upon the induction of the EBV lytic cycle was notably higher (1.5 and 0.85% of input, respectively) than that of the other part of the BZLF1 promoter (less than 0.35%) (Fig. 1D, gray bars). This enhancement of H3K4me3 levels at the proximal part of Zp will be discussed later.
On the other hand, repressive markers, including H3K9me2, H3K9me3, H3K27me3, and H4K20me3, were present overall in latency (Fig. 1E to H). The presence of histones H3K9me3 and H3K27me3 at the Zp and oriLyt in the latent EBV genome of Akata cells was reported quite recently (47). H3K9me2 and H4K20me3 modifications have not been reported for the BZLF1 promoter of EBV to our knowledge.
Unexpectedly, lytic induction did not significantly diminish levels of those repressive markers (Fig. 1E to H), if any (see Fig. 7E). The reason why repressive marks did not noticeably diminish upon lytic induction in Raji cells is not clear, because the lytic induction of KSHV by sodium butyrate (18) or K-Rta (52) caused the loss of H3K27me3 at the K-Rta (Orf50) promoter region, at least to some extent. We speculate that this was because EBV in Raji cells cannot replicate at all due to the lack of the BALF2 gene, while the elimination of negative marks might need lytic viral DNA replication. Although Toth and others paid attention to the possible effect of viral genome replication and performed assays within 12 h after K-Rta induction (52), this induction might still touch off undetectable levels of replication. We normalized levels of immunoprecipitated DNA fragments to input levels, which is a very common method of normalization. Because the normalization of the data to a certain internal control, such as the GAPDH promoter (7), might provide better resolution for comparison between the control and lytic induction, we measured the levels of H3K27me3 (Fig. 2A) and H4K20me3 (Fig. 2B) modifications at the β-globin promoter (Globinp) and GAPDH promoter (GAPDHp) in Raji cells. The levels of histone modifications at the cellular promoters, however, did not appreciably change by lytic induction, thus the normalization of the ChIP data to an internal control do not make a significant change in the result that levels of the repressive marks did not diminish (Fig. 1E to H). Recently, Ramasubramanyan and others proved the presence of histone H3K9me3 and H3K27me3 at the Zp and oryLyt in the latent EBV genome of Akata cells (47), where they clearly demonstrated that lytic induction caused an increase of phosphorylated histone H2AX; however, they did not show the decrease of H3K9me3 and H3K27me3.
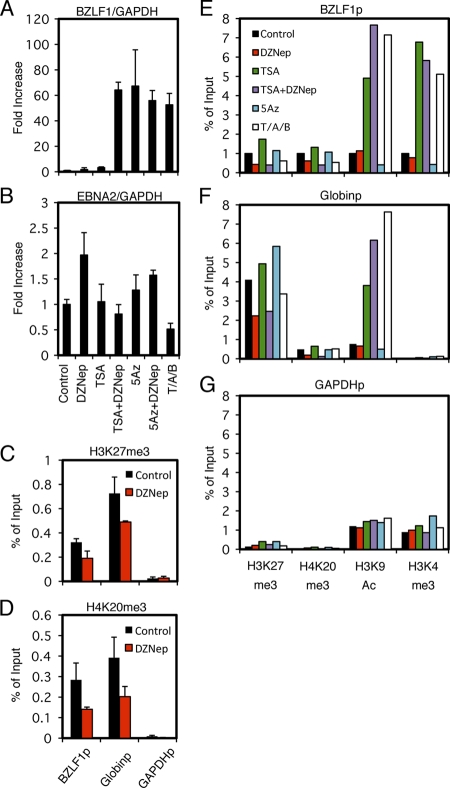
Fig 7.
Effects of pharmacological inhibitors on BZLF1 expression. (A and B) Effects of pharmacological inhibitors on BZLF1 expression. Raji cells were treated with either vehicle (Control), 10 μM DZNep (DZ), 300 nM trichostatin A (TSA), 1 μM 5-aza-2′-deoxycytidine (5Az), or in combinations as indicated. As a positive control (T/A/B), Raji cells were treated with 20 ng/ml TPA, 1 μM A23187, and 5 mM sodium butyrate. For DZNep or 5-Aza-2′-deoxycytidine (5Az) treatment, cells were exposed to the reagent daily for 3 days. Treatment with other chemicals was conducted for 24 h. Real-time RT-PCR was carried out as described in Materials and Methods. Levels of BZLF1 (A) and EBNA2 (B) mRNAs were normalized to GAPDH mRNA levels and are shown as fold increase. Each bar represents the means and SD from three independent treatments. (C to G) Effects of pharmacological inhibitors on epigenetic modifications. (C and D) Raji cells treated with DZNep were subjected to ChIP assays using anti-H3K27me3 (C) and anti-H4K20me3 (D) antibodies, followed by DNA extraction and real-time PCR to detect DNA fragments using the indicated primers. (E to G) Raji cells treated with inhibitors in the same way were subjected to ChIP assays using anti-H3K27me3, anti-H4K20me3, anti-H3K9Ac, or anti-H3K4me3 antibody, followed by DNA extraction and real-time PCR to detect DNA fragments using the indicated primers.
Fig 2.
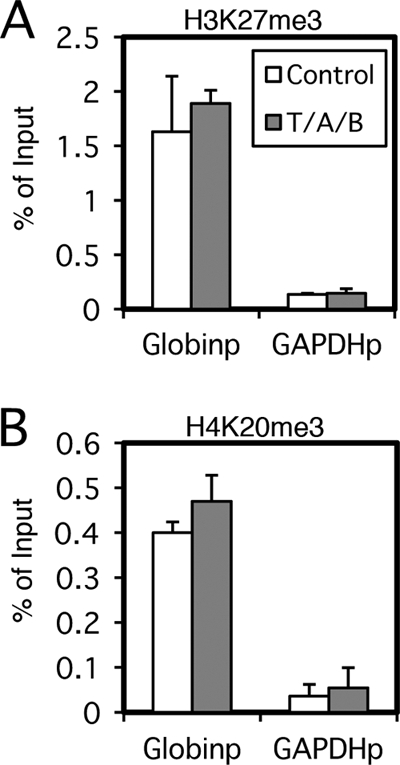
Effect of lytic induction on cellular promoters. Raji cells were treated with TPA (20 ng/ml), A23187 (1 μM), and sodium butyrate (5 mM) (T/A/B; gray bar) or with the vehicle (Control; white bar) for 20 h. Cells then were cross-linked, and ChIP experiments were performed using anti-histone H3K27me3 (A) or anti-H4K20me3 (B) antibody, followed by DNA extraction and real-time PCR to detect DNA fragments using the primers as indicated.
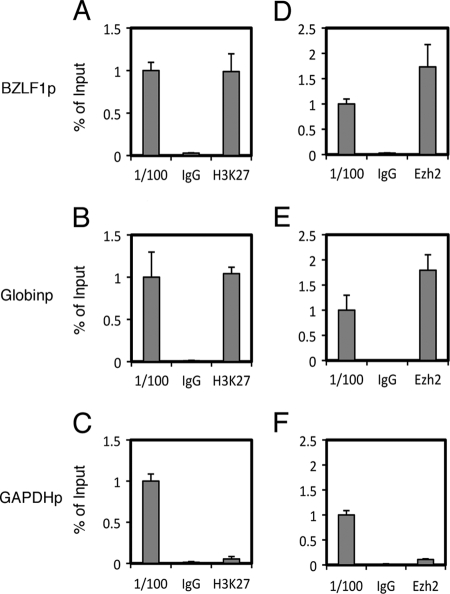
Presence of the repressive H3K27me3 marker and Ezh2 in the BZLF1 promoter.
Since epigenetic analyses of other herpesviruses, including HSV (7, 32) and KSHV (18, 52), have implicated the repressive histone H3K27me3 marker in the maintenance of viral latency, we sought to further examine this modification in the BZLF1 promoter of EBV by ChIP assays (Fig. 3). For detection, we used a set of specific primers that amplify minimal the BZLF1 promoter (−221 to +12; identical to Zp0 in Fig. 1), which is required for the transcriptional activation of the gene in response to TPA or anti-Ig. The histone H3K27me3 modification level of the Zp (0.99% of input) (Fig. 3A) was almost comparable to that of the β-globin promoter (1.0) (Fig. 3B) when the modification of the GAPDH promoter was markedly low (0.053%) (Fig. 3C) in Raji cells. Since it is known that the β-globin locus is marked by a high level of H3K27me3 in nonerythroid cells and the locus of GAPDH, a typical housekeeping gene, is highly demethylated at H3K27 (26), we conclude that the Zp is modified by H3K27me3.
Fig 3.
Presence of H3K27me3 and Ezh2 in Raji cells. ChIP experiments were carried out using lysates from Raji cells latently infected with EBV. Cross-linked DNA-protein complexes were precipitated using normal IgG, anti-H3K27me3 antibody (A to C), or anti-Ezh2 antibody (D to F), followed by DNA extraction and real-time PCR analysis for quantification using the indicated primers.
To further test this conclusion, we tested if Ezh2, the enzyme responsible for H3K27 trimethylation (5), associates with Zp. Ezh2 was detected at a very low level at the GAPDHp (0.11% of input) (Fig. 3F), but the detection levels were substantially higher at the Zp (1.7%) (Fig. 3D) and Globinp (1.8%) (Fig. 3E). Therefore, the association of Ezh2 was correlated with the levels of the H3K27me3 marker in Raji cells.
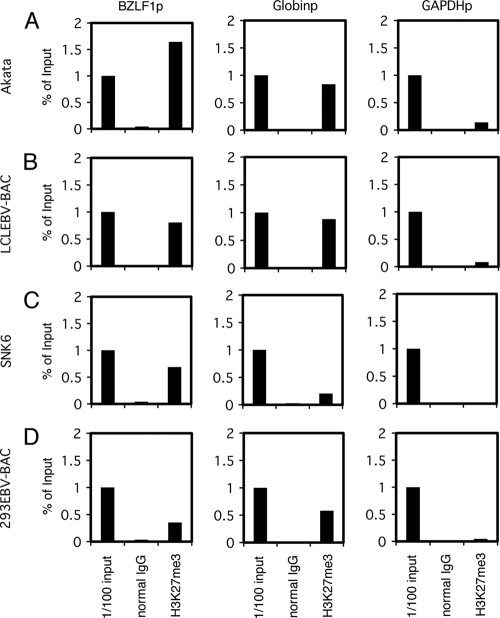
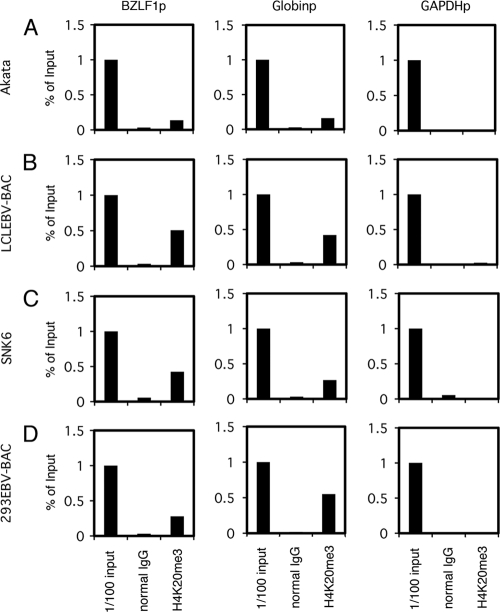
Panels of cell lines latently infected with EBV were tested for H3K27me3: B cells (Akata and LCL EBV-BAC), NK cells (SNK6), and epithelial cells (293EBV-BAC). Depending on the cell type, 0.35 to 1.64% of the promoter region in the input lysate was coprecipitated with H3K27me3 antibody, while normal IgG failed to pull down the promoter sequence (Fig. 4). Histone H3K27me3 levels at the Zp were comparable to those at the β-globin promoter and were notably higher than those at the GAPDH promoter (Fig. 4). Therefore, histone H3K27me3 modification is present in the Zp of EBV, as in other herpesviruses.
Fig 4.
H3K27me3 status in EBV-positive cells. ChIP experiments were carried out using lysates from latent Akata (A), LCLEBV-BAC (B), SNK6 (C), and 293EBV-BAC (D) cells. Cross-linked DNA-protein complexes were precipitated using normal IgG or anti-H3K27me3 antibody, followed by DNA extraction and real-time PCR analysis for quantification using the indicated primers.
Presence of the repressive H4K20me3 marker in the BZLF1 promoter.
We then proved that another repressive marker, histone H4K20me3, is present at the Zp by comparing to control promoters in Raji cells (Fig. 5). The presence of the mark at the Zp (1.1% of input) (Fig. 5A) as well as at the Globinp (0.81%) (Fig. 5B) was confirmed, whereas the GAPDH promoter was not efficiently coprecipitated with H4K20me3 antibody (0.065%) (Fig. 5C).
Fig 5.
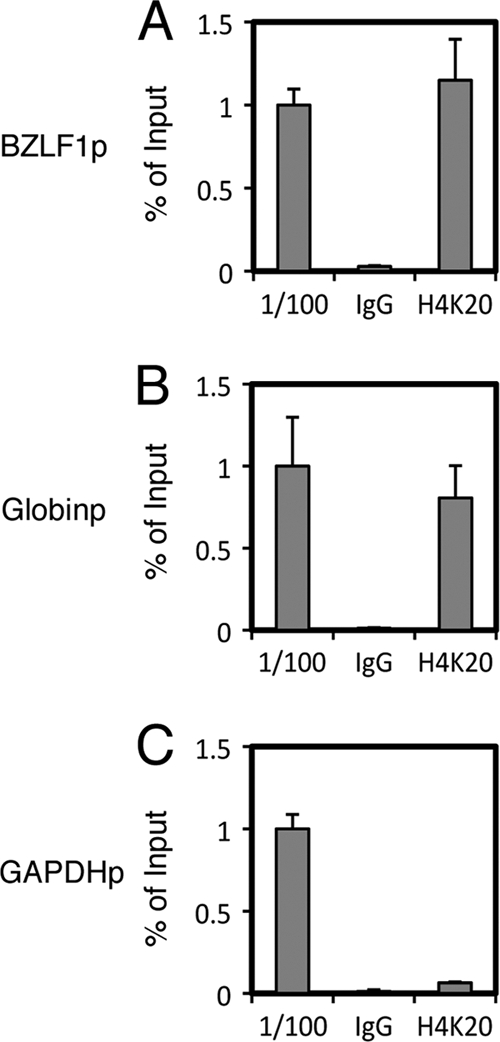
Presence of H4K20me3 in Raji cells. (A to C) ChIP experiments were carried out using lysates from Raji cells latently infected with EBV. Cross-linked DNA-protein complexes were precipitated using normal IgG or anti-H4K20me3 antibody, followed by DNA extraction and real-time PCR analysis for quantification using the indicated primers.
We then checked if the H4K20me3 modification was present in other EBV-positive cell lines (Fig. 6). To our knowledge, the presence of the H4K20me3 marker of promoters of key molecules that regulate reactivation from latency of any herpesviruses, including EBV, has never been clearly confirmed. ChIP analysis demonstrated that the H4K20me3 methylation in the Zp of EBV in Akata, LCL EBV-BAC, SNK6, and 293EBV-BAC cells was present and was comparable to that of the β-globin promoter, whereas the GAPDH promoter was hardly modified by the marker (Fig. 6).
Fig 6.
H4K20me3 status of EBV-positive cells. ChIP experiments were carried out using lysates from latent Akata (A), LCLEBV-BAC (B), SNK6 (C), and 293EBV-BAC (D) cells. Cross-linked DNA-protein complexes were precipitated using normal IgG or anti-H4K20me3 antibody, followed by DNA extraction and real-time PCR analysis for quantification using the indicated primers.
Pharmacological inhibition of H3K27me3 and H4K20me3 by DZNep augments BZLF1 expression with TSA in Raji cells.
After confirming the presence of histone H3K27me3 and H4K20me3 in the Zp of EBV, to examine if the repressive mark is actually functional we next used a small molecule, 3-deazaneplanocin A (DZNep), which has been reported to suppress H3K27me3 and H4K20me3 histone modification (39, 51). While the treatment of Raji cells with either DZNep or TSA alone had only minor effects on BZLF1 levels (1.8- and 3.3-fold increase, respectively), the use of the two inhibitors in combination (TSA plus DZNep) stimulated the expression 64.2-fold (Fig. 7A). This result suggests that in Raji cells, not only histone deacetylation but also histone H3K27me3 and H4K20me3 serve to inhibit BZLF1 transcription in collaboration. In addition, we confirmed that 5-Aza, an inhibitor of CpG DNA methylation, clearly upregulates the BZLF1 transcription levels (Fig. 7A) as previously demonstrated (57), yet it is very likely that CpG methylation was not involved in the process (10, 15). As a control, we also examined the levels of EBNA2 mRNA in the same samples (Fig. 7B). EBNA2 is a latent gene abundantly expressed in type III latency, including in Raji cells. As expected, none of the pharmacological inhibitions tested here further markedly induce EBNA2 expression (Fig. 7B), as the levels were intrinsically high. We then confirmed that DZNep treatment effectively and significantly (P < 0.05) reduced the levels of H3K27me3 (Fig. 7C) and H4K20me3 (Fig. 7D) in the BZLF1 and β-globin promoters. In addition, we checked H3K27me3, H4K20me3, H3K9Ac, and H3K4me3 markers in the BZLF1 (Fig. 7E), β-globin (Fig. 7F), and GAPDH (Fig. 7G) promoters in Raji cells treated with the inhibitors used for Fig. 7A. DZNep or TSA plus DZNep treatment reduced the H3K27me3 and H4K20me3 levels of the Zp and Globinp (Fig. 7E, F, H3K27me3 and H4K20me3, red and purple bars), while other treatments did not noticeably decrease the modification, confirming the specificity and efficacy of DZNep. The presence of TSA or sodium butyrate (T/A/B) strikingly elevated levels of the active marks H3K9Ac and H3K4me3 at the Zp (Fig. 7E, H3K9Ac and H3K4me3, green, purple, or white). Interestingly, those HDAC inhibitors caused a remarkable increase of H3K9Ac modification at the β-globin promoter, but the H3K4me3 modification of the promoter was not affected (Fig. 7F). We speculate that the behavior of active markers varies a great deal depending on the promoters, and notably the activation of the BZLF1 promoter may be characterized by H3K4me3.
To extend the experiment, we checked BZLF1 expression in other typical EBV-positive cell lines, Akata and B95-8 (Fig. 8). As reported previously (37, 38), the treatment of Akata cells with an HDAC inhibitor, TSA, alone was sufficient to induce BZLF1 transcription (52.6-fold), suggesting that low levels of histone acetylation play an important role in the silencing. On the other hand, TSA alone did not induce BZLF1 transcription in B95-8, suggesting that the molecular mechanisms that govern the suppression of BZLF1 transcription in those cells is more complicated (10, 11, 17). An inhibitor of CpG methylation, 5-Aza, enhanced BZLF1 expression in both cell lines as previously reported (57). Unlike Raji cells, both Akata and B95-8 cells did not respond to DZNep, even in combination with TSA or 5-Aza. Notably, EBNA2 is hardly expressed in Akata cells, because the virus takes type I latency and then is induced by 5-Aza as reported previously (15), suggesting that the inhibitor is working as intended.
Fig 8.
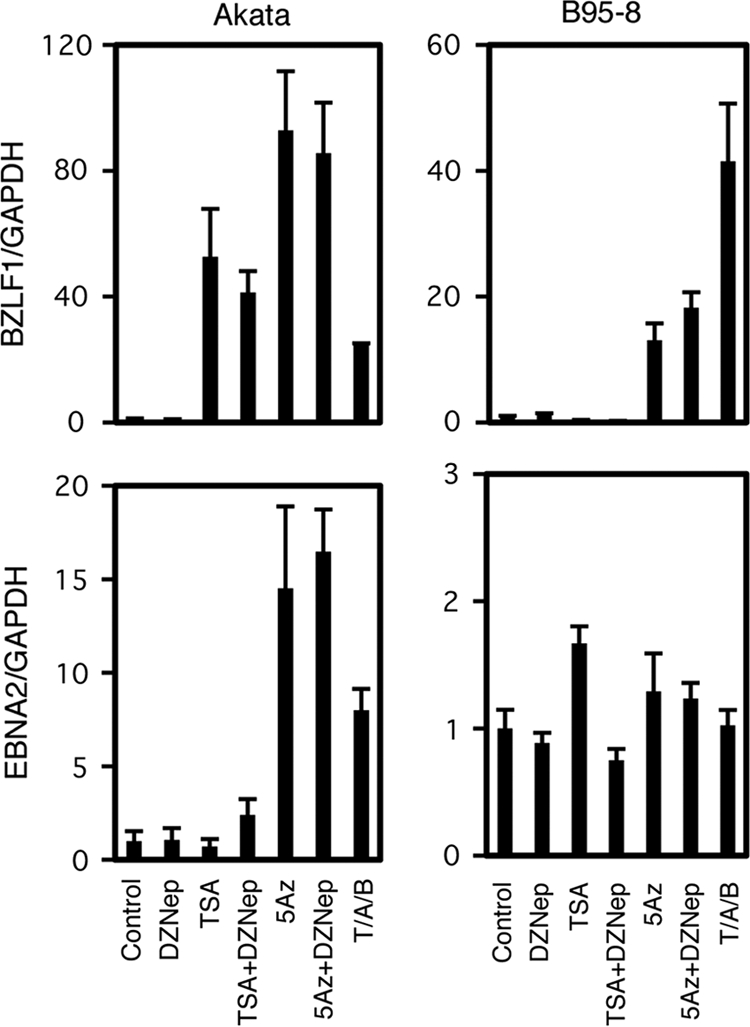
Effects of pharmacological inhibitors on BZLF1 expression in Akata and B95-8 cells. Akata or B95-8 cells were treated with either vehicle (Control), 10 μM DZNep (DZ), 300 nM Trichostatin A (TSA), 1 μM 5-aza-2′-deoxycytidine (5Az), or in combinations as indicated. As positive controls (PC), Akata cells were treated with anti-IgG (10 μg/ml), and B95-8 cells were treated with 20 ng/ml TPA, 1 μM A23187, and 5 mM sodium butyrate. For DZNep or 5Az treatment, cells were exposed to the reagent daily for 3 days. Treatments with other chemicals were conducted for 24 h. Real-time RT-PCR was carried out as described in Materials and Methods. Levels of BZLF1 (upper panels) and EBNA2 (lower panels) mRNAs were normalized to GAPDH mRNA levels and are shown as fold increase. Each bar represents the means and SD from three independent treatments.
Knockdown of H3K27me3 methyltransferase Ezh2.
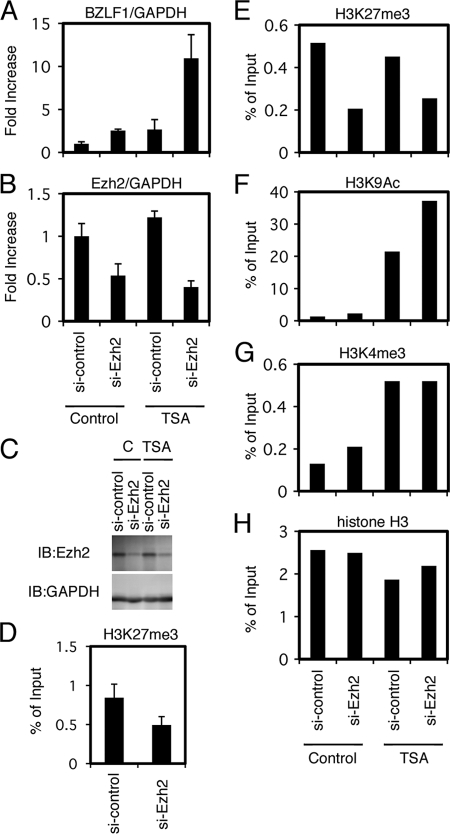
We tested small-molecule epigenetic inhibitors in the previous section, and the results strongly suggested the involvement of histone H3K27me3 and H4K20me3 methylations in the maintenance of latency in Raji cells. We next carried out the knockdown of methylation enzymes (Fig. 9 and 10). First, to further confirm the physiological significance of the epigenetic silencing of the BZLF1 gene by histone H3K27me3 in Raji cells, we knocked down Ezh2, the methyltransferase enzyme responsible for the modification (5). As shown in Fig. 9B, the transfection of siRNA against Ezh2 caused a decrease in Ezh2 mRNA levels (54 or 40% of control, without or with TSA, respectively). Accordingly, immunoblotting showed protein levels of Ezh2 to be markedly decreased by the siRNA treatment (Fig. 9C). The silencing of Ezh2 increased BZLF1 levels by 2.5-fold even without TSA, and the addition of TSA elevated this to 10.9-fold (Fig. 9A). The induction of BZLF1 mRNA by si-Ezh2 and TSA was correlated with the reduction of repressive H3K27me3 marker and the increment of active H3K9Ac and H3K4me3 markers (Fig. 9D to H). Taking the results of inhibitor experiments in Fig. 7 into consideration, this result pointed to the involvement of Ezh2 methyltransferase and the histone H3K27me3 mark in the silencing of BZLF1 gene expression during EBV latency. In addition, we note that histone acetylation is also needed for the efficient expression of BZLF1.
Fig 9.
Knockdown of H3K27me3 methyltransferase Ezh2. (A to C) Raji cells transfected with siRNA against Ezh2 (si-Ezh2) or a control siRNA (si-control) were cultured for 48 h and then treated with TSA (300 nM) or vehicle DMSO (Control) for an additional 24 h. Levels of BZLF1 (A) and Ezh2 (B) mRNA were checked by real-time RT-PCR. (C) Levels of Ezh2 protein were examined by immunoblotting. (D to H) Epigenetic status of the Zp after Ezh2 knockdown. (D) Raji cells treated with siRNA against Ezh2 were subjected to ChIP assays using anti-H3K27me3 antibody, followed by DNA extraction and real-time PCR to detect the DNA fragment of the Zp. (E to H) Raji cells treated with siRNA and TSA in the same manner were subjected to ChIP assays using anti-H3K27me3, anti-H3K9Ac, anti-H3K4me3, and anti-histone H3 antibodies, followed by DNA extraction and real-time PCR to detect the DNA fragment of the Zp.
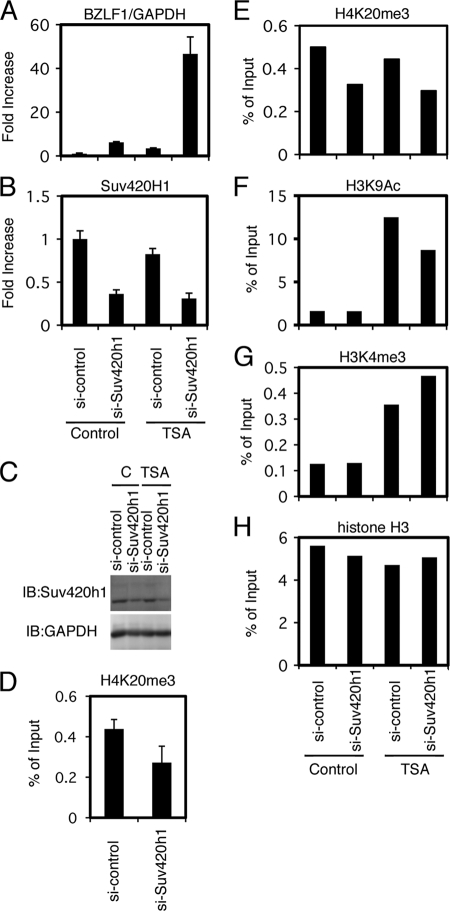
Fig 10.
Knockdown of H4K20me3 methyltransferase Suv420h1. (A to C) Raji cells transfected with siRNA against Suv420h1 (si-Suv420h1) or a control siRNA (si-control) were cultured for 48 h and then treated with TSA (300 nM) or vehicle DMSO (Control) for an additional 24 h. Levels of BZLF1 (A) and Suv420h1 (B) mRNA were checked by real-time RT-PCR. (C) Levels of Suv420h1 protein were examined by immunoblotting. (D to H) Epigenetic status of the Zp after Suv420h1 knockdown. (D) Raji cells treated with siRNA against Suv420h1 were subjected to ChIP assays using anti-H4K20me3 antibody, followed by DNA extraction and real-time PCR to detect the DNA fragment of the Zp. (E to H) Raji cells treated with siRNA and TSA in the same manner were subjected to ChIP assays using anti-H4K20me3, anti-H3K9Ac, anti-H3K4me3, and anti-histone H3 antibodies, followed by DNA extraction and real-time PCR to detect the DNA fragment of the Zp.
Knockdown of H4K20me3 methyltransferase Suv420h1.
Results in the previous sections strongly suggested the significance of the repressive H4K20me3 marker for the suppression of Zp. To specifically examine the effect of H4K20me3 methylation on the silencing of the BZLF1 gene, Suv420h1, the methyltransferase responsible for the modification, was knocked down by siRNA technology. When the transfection of si-Suv420h1 reduced the mRNA levels of the methyltransferase down to 36 or 31% of the control level (Fig. 10B), the 6.2- or 46.6-fold induction of BZLF1 mRNA was observed in the absence or presence of TSA, respectively (Fig. 10A). The remarkable induction of the BZLF1 gene by Suv420h1 knockdown and TSA (Fig. 10A) corresponded with the reduction of H4K20me3 levels and the elevation of active H3K9Ac and H3K4me3 markers (Fig. 10D to H). This indicates that the silencing of the BZLF1 promoter in Raji cells is brought about by histone H4K20me3 methylation as well. Therefore, histone acetylation, in addition to H4K20me3 demethylation, enhances promoter activity.
DISCUSSION
In this report, we document evidence that histone H3K27me3 and H4K20me3 play a crucial role in the suppression of the BZLF1 gene in Raji cells, mainly by using a small-molecule inhibitor of epigenetic modifications, DZNep. The pharmacological inhibitor reduced the levels of histone H3K27me3 and H4K20me3 markers, which coincides with BZLF1 induction when combined with an HDAC inhibitor, TSA.
Because DZNep exhibited potent inducing effects on BZLF1 gene transcription (Fig. 7), we also tested BIX01294, a specific inhibitor of G9a, the methyltransferase that is responsible for histone H3K9me2 methylation (30). However, the treatment of Raji cells or other EBV-positive cells with BIX01294 alone or in combination with TSA, DZNep, or 5-Aza did not increase BZLF1 expression at all or caused a very modest increase at most (Fig. 11). Although H3K9me2 methylation is ubiquitously detected in the EBV genome (Fig. 1E) (6, 12) and other herpesvirus genomes (18, 32, 52), the data imply that K3K9me2 does not play an important role in the suppression of BZLF1, at least in Raji cells. Further knockdown experiments are needed to provide conclusive evidence of the involvement of H3K9 methylation in BZLF1 silencing.
Fig 11.
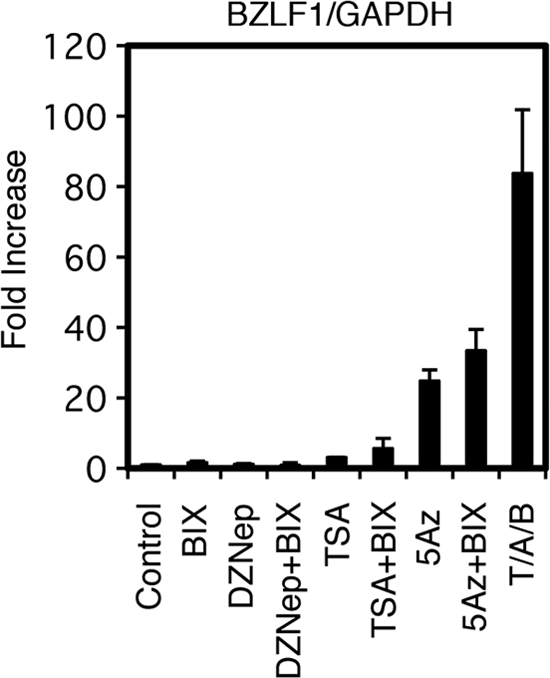
Effect of BIX01294, an inhibitor of G9a, a methyltransferase that is responsible for histone H3K9me2, on BZLF1 expression. Raji cells were treated with either vehicle (Control), 10 μM BIX01294 (BIX), 10 μM DZNep (DZ), 300 nM trichostatin A (TSA), 1 μM 5-aza-2′-deoxycytidine (5Az), or in combinations as indicated. As a positive control, Raji cells were treated with 20 ng/ml TPA, 1 μM A23187, and 5 mM sodium butyrate (T/A/B). For BIX01294, DZNep, or 5Az treatment, cells were exposed to the reagent daily for 3 days. Treatment with other chemicals were conducted for 24 h. Real-time RT-PCR was carried out as described in Materials and Methods. Levels of BZLF1 mRNAs were normalized to GAPDH mRNA levels and are shown as fold increases. Each bar represents the means and SD from three independent treatments.
Interestingly, whereas the treatment with TPA, A23187, and sodium butyrate did not alleviate repression markers, such as H3K9me2/me3, H3K27me3, or H4K20me3, at all (Fig. 1E to H), it did elicit the expression of BZLF1 (Fig. 7A). We suggest a hypothesis to explain this. It is known that multiple (50 to 100) copies of the episomal EBV genome are present per latent Raji cell (13, 56). If TPA-A23187-butyrate treatment induced reductions of such repressive histone methylations in only a few copies and allow the efficient expression of BZLF1 from the limited copies, the reduction of the repressive modifications must be hard to detect, because the histone methylations in most of the genome copies remain intact. Meanwhile, since the induction of active histone marks, like histone acetylation or H3K4me3 methylation, was clearly observed (Fig. 1C and D), such active marks may take place more broadly and efficiently among the viral copies.
A facultative repression marker, H3K27me3, is broadly found in EBV (Fig. 1G) (6, 12) and other herpesvirus (7, 18, 32, 52) genomes, and it functions to maintain latency (Fig. 7 and 9). We found here that H3K27me3, at least partly, functions in this regard by suppressing BZLF1 gene expression. On the other hand, another repression marker, H4K20me3, has not been implicated elsewhere in the suppression of herpesvirus gene expression.
The reduction of the H3K27me3 and H4K20me3 modifications by either siRNA or with an inhibitor caused the notable induction of the BZLF1 gene when treated with TSA (Fig. 7 and 10), indicating the importance of the methylations in silencing the BZLF1 promoter. However, we have to mention that the knockdown or use of inhibitors might cause unexpected influences on cellular genes, because histone H3K27me3- and H4K20me3-repressive marks are crucial regulators of many cellular genes. Thus, we cannot deny the possibility that the cellular gene(s) induced by the knockdown of the methylases or the inhibitor act on BZLF1 expression, and thus the enhanced BZLF1 expression we observe here might be a secondary effect of histone H3K27me3 or H4K20me3 downregulation.
EBV generally establishes latency in B cells in which BZLF1 expression is restricted by the epigenetic modification of histones but not CpG methylations (15). Similarly, lytic genes of HSV also are suppressed primarily by repressive histone marks during latency in sensory neurons (4, 27, 54). On the other hand, it is estimated from HSV data (9, 46) that the herpesvirus DNA genome inside the nucleocapsid is not appreciably associated with histones, although the state of the EBV genome in viral particles has not been dissected yet. Upon infection, the herpesvirus genome becomes rapidly associated with cellular histones and nucleosomes, albeit irregularly (8, 25, 36, 46). Interestingly, upon the primary infection of EBV in naive B cells, the virus transcribes the BZLF1 gene (23, 55), although it cannot fully trigger the execution of the viral lytic replication cycle. These facts suggest that although the EBV genomic DNA in the incoming virus particle does not bear histones, it rapidly becomes incorporated into nucleosomes with active histone markers upon infection, which allows the expression of the BZLF1 gene at least for the first several days. Later, however, the expression is silenced by acquired repressive epigenetic modifications, such as histone H3K27me3 and H4K20me3, implying that the acquisition of repressive histone markers takes longer than that of active markers. Without such epigenetic silencing of BZLF1, the lytic replication of EBV causes the arrest of cell cycle progression (31) and eventual cell death. Besides those repressive histone modifications, however, other possible mechanisms, such as the absence of required activators, signal transductions, or the presence of repressors, can play a role in the restriction of the gene. In any case, the restriction of BZLF1 transcription in infected cells is a very crucial point for the life cycle of the virus and for the infinite proliferation capacity of infected cells.
It must be emphasized that the response of BZLF1 promoter activity to a certain epigenetic inhibitor depends largely on the cell type. To take one example, levels of BZLF1 mRNA in Akata cells were induced 53-fold by TSA treatment alone. Combination treatment of Akata cells with TSA plus DZNep did not further induce gene expression. Therefore, it is inferred that the BZLF1 gene in Akata cells is suppressed mostly by low acetylation levels of histones, even though the promoter in Akata was still modified by H3K27me3 (47) (Fig. 4) and H4K20me3 (Fig. 6). At the same time, the BZLF1 gene was hardly induced (1.0- and 3.3-fold) by TSA alone in B95-8 (Fig. 8) and Raji cells (Fig. 7A), respectively. The simultaneous use of the two inhibitors (TSA plus DZNep) induced the BZLF1 gene by 64-fold in Raji cells (Fig. 7A), but combined application (TSA plus DZNep) failed to increase the BZLF1 level at all in B95-8 cells (Fig. 8), suggesting the presence of other suppression mechanisms besides low acetylation, H3K27me3, and H4K20me3 in B95-8 cells. We speculate that this variety of silencing mechanisms means that BZLF1 gene expression somehow is suppressed in order to proliferate and achieve cancerous growth, but the mechanisms of the suppression do not matter, so long as it is not a constitutive, deep repression, such as CpG methylation (15, 28), which is generally irreversible.
The transcriptional activation of EBV regulatory genes by histone acetylation has been extensively reported elsewhere (11, 22), thus we focus here on H3K4me3, which has not been reported for the EBV BZLF1 promoter to our knowledge. Lines of evidence indicate the crucial role of histone H3K4me3 modification for the execution of the lytic replication process of HSV (21, 36, 45) and KSHV (18, 52) genomes. In Fig. 1C, of note, while histone H3K9 acetylation took place overall in the EBV genome in response to HDAC inhibitors, H3K4me3 occurred in a different manner: histones at Zp0 (−221 to +12) and Zp +500 were efficiently trimethylated at H3K4 by TPA-A23187-butyrate (Fig. 1D, Zp0 and Zp +500), but this was not true for other distal parts of the Zp (Fig. 1D, Zp-3000, Zp-2000, Zp-1200, and Zp-600). Because the Zp0 (−221 to +12) region coincides with the known minimal promoter sequence required for the reactivation of EBV from latency (3), this specificity may bring about the specific transcriptional activation of the BZLF1 gene by a particular transcription factor. We speculate that Zp-specific transcription factors, including Sp1, MEF2D, CREB, ATF, and/or XBP-1(s), recruit the H3K4 methyltransferase Set1 or MLL. We plan to further analyze the significance of this modification by using siRNA, inhibitor, and mutant EBV-BAC cells with mutations at arbitrary sites of the BZLF1 promoter, such as ZI or ZII (42), in future work.
Collectively, our results indicate that two histone modifications, H3K27me3 and H4K20me3, which are associated with the latent EBV BZLF1 promoter region, are at least partly involved in the maintenance of virus latency in Raji cells. With these silencing mechanisms, EBV restricts the expression of viral lytic genes and avoids surveillance by the host immune system. The involvement of two active markers, histone acetylation and H3K4 trimethylation, at the BZLF1 promoter is implicated in reactivation from viral latency. In addition, since the execution of the viral lytic program arrests cell cycle progression in EBV-infected cells (31), the induction of EBV lytic replication in EBV-positive cancers by epigenetic inhibitors, such as HDAC inhibitors, 5-Aza, and/or DZNep, may offer clinical applications as a type of oncolytic therapy in the future, especially when combined with antiviral drugs, such as ganciclovir (14).
ACKNOWLEDGMENTS
We thank W. Hammerschmidt, H. J. Delecluse, and N. Shimizu for providing the EBV-BAC system, HEK293 cells, and SNK6 cells, respectively. We also thank C. Noda and T. Gamano for technical assistance.
This work was supported by grants in aid for Scientific Research from the Ministry of Education, Science, Sports, Culture and Technology (no. 20390137 and 21022055 to T.T. and no. 20790362 and 22790448 to T.M.), the Ministry of Health, Labour and Welfare (to T.T.), and partly by the Uehara Memorial Research Fund (to T.T.), the Japan Leukaemia Research Fund, and the Yasuda Medical Foundation (to T.M.).
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Amon W, Farrell PJ. 2005. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15:149–156 [DOI] [PubMed] [Google Scholar]
- 2. Bhende PM, Dickerson SJ, Sun X, Feng WH, Kenney SC. 2007. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J. Virol. 81:7363–7370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Binne UK, Amon W, Farrell PJ. 2002. Promoter sequences required for reactivation of Epstein-Barr virus from latency. J. Virol. 76:10282–10289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloom DC, Giordani NV, Kwiatkowski DL. 2010. Epigenetic regulation of latent HSV-1 gene expression. Biochim. Biophys. Acta 1799:246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao R, et al. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039–1043 [DOI] [PubMed] [Google Scholar]
- 6. Chau CM, Lieberman PM. 2004. Dynamic chromatin boundaries delineate a latency control region of Epstein-Barr virus. J. Virol. 78:12308–12319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cliffe AR, Garber DA, Knipe DM. 2009. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 83:8182–8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cliffe AR, Knipe DM. 2008. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82:12030–12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cohen GH, et al. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34:521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Countryman J, et al. 2009. Stimulus duration and response time independently influence the kinetics of lytic cycle reactivation of Epstein-Barr virus. J. Virol. 83:10694–10709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Countryman JK, Gradoville L, Miller G. 2008. Histone hyperacetylation occurs on promoters of lytic cycle regulatory genes in Epstein-Barr virus-infected cell lines which are refractory to disruption of latency by histone deacetylase inhibitors. J. Virol. 82:4706–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Day L, et al. 2007. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 81:6389–6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delecluse HJ, Bartnizke S, Hammerschmidt W, Bullerdiek J, Bornkamm GW. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J. Virol. 67:1292–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng WH, Hong G, Delecluse HJ, Kenney SC. 2004. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J. Virol. 78:1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fernandez AF, et al. 2009. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 19:438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flemington E, Speck SH. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gradoville L, Kwa D, El-Guindy A, Miller G. 2002. Protein kinase C-independent activation of the Epstein-Barr virus lytic cycle. J. Virol. 76:5612–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gunther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6:e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagemeier SR, et al. 2010. Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase. J. Virol. 84:4383–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hatfull G, Bankier AT, Barrell BG, Farrell PJ. 1988. Sequence analysis of Raji Epstein-Barr virus DNA. Virology 164:334–340 [DOI] [PubMed] [Google Scholar]
- 21. Huang J, et al. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 80:5740–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jenkins PJ, Binne UK, Farrell PJ. 2000. Histone acetylation and reactivation of Epstein-Barr virus from latency. J. Virol. 74:710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. 2010. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl. Acad. Sci. U. S. A. 107:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kennedy G, Sugden B. 2003. EBNA-1, a bifunctional transcriptional activator. Mol. Cell. Biol. 23:6901–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kent JR, et al. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 78:10178–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim YW, Kim A. 2011. Characterization of histone H3K27 modifications in the beta-globin locus. Biochem. Biophys. Res. Commun. 405:210–215 [DOI] [PubMed] [Google Scholar]
- 27. Knipe DM, Cliffe A. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6:211–221 [DOI] [PubMed] [Google Scholar]
- 28. Kondo Y. 2009. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med. J. 50:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kondo Y, et al. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 40:741–750 [DOI] [PubMed] [Google Scholar]
- 30. Kubicek S, et al. 2007. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25:473–481 [DOI] [PubMed] [Google Scholar]
- 31. Kudoh A, et al. 2003. Reactivation of lytic replication from B cells latently infected with Epstein-Barr virus occurs with high S-phase cyclin-dependent kinase activity while inhibiting cellular DNA replication. J. Virol. 77:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwiatkowski DL, Thompson HW, Bloom DC. 2009. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 83:8173–8181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu P, Liu S, Speck SH. 1998. Identification of a negative cis element within the ZII domain of the Epstein-Barr virus lytic switch BZLF1 gene promoter. J. Virol. 72:8230–8239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu S, Borras AM, Liu P, Suske G, Speck SH. 1997. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 gene promoter. Virology 228:11–18 [DOI] [PubMed] [Google Scholar]
- 35. Liu S, Liu P, Borras A, Chatila T, Speck SH. 1997. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 16:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu X, Triezenberg SJ. 2010. Chromatin assembly on herpes simplex virus genomes during lytic infection. Biochim. Biophys. Acta 1799:217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luka J, Kallin B, Klein G. 1979. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94:228–231 [DOI] [PubMed] [Google Scholar]
- 38. Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97:81–109 [DOI] [PubMed] [Google Scholar]
- 39. Miranda TB, et al. 2009. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8:1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montalvo EA, Cottam M, Hill S, Wang YJ. 1995. YY1 binds to and regulates cis-acting negative elements in the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:4158–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murata T, et al. 2010. Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase. J. Biol. Chem. 285:23925–23935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murata T, et al. 2011. Involvement of Jun dimerization protein 2 (JDP2) in the maintenance of Epstein-Barr virus latency. J. Biol. Chem. 286:22007–22016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murata T, et al. 2009. TORC2, a coactivator of cAMP-response element-binding protein, promotes Epstein-Barr virus reactivation from latency through interaction with viral BZLF1 protein. J. Biol. Chem. 284:8033–8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nagata H, et al. 2001. Characterization of novel natural killer (NK)-cell and gammadelta T-cell lines established from primary lesions of nasal T/NK-cell lymphomas associated with the Epstein-Barr virus. Blood 97:708–713 [DOI] [PubMed] [Google Scholar]
- 45. Narayanan A, Ruyechan WT, Kristie TM. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oh J, Fraser NW. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 82:3530–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramasubramanyan S, Osborn K, Flower K, Sinclair AJ. 2012. Dynamic chromatin environment of key lytic cycle regulatory regions of the Epstein-Barr virus genome. J. Virol. 86:1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruf IK, Rawlins DR. 1995. Identification and characterization of ZIIBC, a complex formed by cellular factors and the ZII site of the Epstein-Barr virus BZLF1 promoter. J. Virol. 69:7648–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schotta G, et al. 2004. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18:1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Speck SH, Chatila T, Flemington E. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399–405 [DOI] [PubMed] [Google Scholar]
- 51. Tan J, et al. 2007. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 21:1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Toth Z, et al. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6:e1001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tsurumi T, Fujita M, Kudoh A. 2005. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 15:3–15 [DOI] [PubMed] [Google Scholar]
- 54. Wang QY, et al. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wen W, et al. 2007. Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J. Virol. 81:1037–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yamamoto N, Bister K, zur Hausen H. 1979. Retinoic acid inhibition of Epstein-Barr virus induction. Nature 278:553–554 [DOI] [PubMed] [Google Scholar]
- 57. Ye J, Gradoville L, Daigle D, Miller G. 2007. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi's sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J. Virol. 81:9279–9291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu X, Wang Z, Mertz JE. 2007. ZEB1 regulates the latent-lytic switch in infection by Epstein-Barr virus. PLoS Pathog. 3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]