Abstract
Viral pathogens usurp cell surface receptors to access clathrin endocytic structures, yet the mechanisms of virus incorporation into these structures remain incompletely understood. Here we used fluorescence microscopy to directly visualize the association of single canine parvovirus (CPV) capsids with cellular transferrin receptors (TfR) on the surfaces of live feline cells and to monitor how these CPV-TfR complexes access endocytic structures. We found that most capsids associated with fewer than five TfRs and that ∼25% of TfR-bound capsids laterally diffused into assembling clathrin-coated pits less than 30 s after attachment. Capsids that did not encounter a coated pit dissociated from the cell surface with a half-life of ∼30 s. Together, our results show how CPV exploits the natural mechanism of TfR endocytosis to engage the clathrin endocytic pathway and reveal that the low affinity of capsids for feline TfRs limits the residence time of capsids on the cell surface and thus the efficiency of virus internalization.
INTRODUCTION
Animal viruses exploit cellular endocytic pathways to invade target cells. Virus particles engage these pathways by binding cell surface receptor molecules that facilitate virus endocytosis. These viral receptors include a wide variety of transmembrane proteins that normally function in the endocytosis of physiological ligands, as well as other proteins and glycolipids without known endocytic functions. Although the primary receptors and pathways utilized by some viruses are now well defined (30), the mechanisms by which receptors direct virus uptake by a given endocytic structure or pathway remain poorly understood.
Canine parvovirus (CPV) is a nonenveloped virus that utilizes the cellular transferrin receptor type I (TfR) to bind, enter, and infect target cells (33). In nature, CPV is a pathogen of dogs, cats, and related hosts (36). The viral host range is strain specific and is dictated primarily by the affinity of capsids for TfRs expressed on host cells (18, 19). CPV particles measure 26 nm in diameter and consist of an icosahedral capsid that packages the ∼5-kb single-stranded DNA (ssDNA) genome (47). CPV binds to the TfR via raised regions that project from the 3-fold axes of capsid symmetry (14), and cells internalize the receptor-bound capsids by clathrin-dependent endocytosis (34). Capsids then penetrate endosomal compartments and deliver the viral genome to the nucleus to initiate replication (15).
The TfR is a type II, homodimeric transmembrane glycoprotein that delivers iron into cells by binding and internalizing iron-loaded transferrin (Tf) (46). The butterfly-shaped receptor ectodomain spans ∼11 nm and consists of three subdomains (23). Tf binds to residues in the membrane-proximal protease-like domain and the central helical domain (4), while CPV capsids contact residues in the membrane distal apical domain (12, 32). Structural modeling of CPV-TfR interactions suggested that a single capsid can bind up to 24 TfRs, but biochemical and cryo-electron microscopic analysis indicated that CPV capsids bind fewer than 7 TfR ectodomains in solution (14). It is currently unknown whether capsids cluster TfRs on the surfaces of target cells.
Cells internalize TfRs by clathrin-dependent endocytosis (3). This endocytic mechanism forms membrane vesicles that measure 40 to 120 nm in diameter and function to transport cargos from the cell surface to early endosomes (6, 21). Clathrin endocytic structures initiate upon clathrin recruitment to the plasma membrane by AP-2 adaptor complexes. Continued clathrin assembly invaginates the associated membrane to form a coated pit. Adaptor proteins within the assembling coated pits sequester cargos at the endocytic site by engaging the cytosolic domains of transmembrane receptor proteins, including those of the TfR (5, 45). The cargo-loaded pits then pinch off from the plasma membrane as coated vesicles, and the coat containing clathrin and the adaptors is rapidly disassembled to allow vesicle fusion with an endosome. Studies of clathrin-dependent endocytosis in live cells have shown that coated pits constitutively initiate on the cell surface and typically mature into coated vesicles within 30 to 90 s (10, 26, 41).
Numerous viruses utilize the clathrin endocytic pathway to enter host cells, yet the molecular events that govern virus incorporation into clathrin endocytic structures have not been characterized in detail. In this study, we imaged CPV entry with high temporal and spatial precision to dissect how CPV-TfR interactions influence the rate, efficiency, and mechanism of CPV incorporation into clathrin endocytic structures. Our analysis showed that CPV capsids bound to a low number of TfRs on the surfaces of live cells and rapidly engaged forming clathrin structures by a diffusion-based mechanism. Moreover, we found that capsids had a relatively short residence time on the cell surface, which in turn limited the efficiency of capsid internalization.
MATERIALS AND METHODS
Cells and viruses.
Feline kidney CRFK cells (7) were maintained at 37°C and 5% CO2 in a 1:1 mixture of McCoy's 5A and Liebovitz L15 media (Mediatech, Inc.; Manassas, VA) supplemented with 5% fetal bovine serum (FBS) (Tissue Culture Biologicals; Tulare, CA). Chinese hamster ovary cells lacking the hamster TfR (TRVb cells) (29) were maintained under similar conditions in Ham's F-12 nutrient mixture (Mediatech) containing 5% FBS (Tissue Culture Biologicals). CRFK and TRVb cells were transfected with plasmid DNA encoding the rat σ subunit of the AP-2 complex C-terminally tagged with enhanced green fluorescent protein (eGFP) (σ2-eGFP) (10). Plasmid-containing cells were selected with 0.5 (CRFK) or 0.75 (TRVb) mg/ml Geneticin (G418) (Invitrogen Corporation; Carlsbad, CA), and clones that expressed low levels of σ2-eGFP were isolated. TRVb cells were transfected with plasmids encoding wild-type (wt) feline TfR (GenBank accession no. AF276984.1) tagged with eGFP (fTfR-eGFP; see below for construction details) and selected in medium containing 0.75 mg/ml G418. Cell transfections were performed with FuGENE HD transfection reagent according to the manufacturer's (Roche Diagnostics; Indianapolis, IN) instructions, and selected populations of transfected cells were maintained in medium containing 0.4 mg/ml G418. For transient transfections of TRVb cells stably expressing σ2-eGFP with plasmids encoding fTfR-mCherry, cells were plated on 25-mm glass coverslips and transfected as above ∼20 h after plating. Cells expressing low levels of fTfR-mCherry were imaged ∼18 h after transfection.
Viruses were recovered from the infectious plasmid clone of CPV-2 (CPV-d) (35) in NLFK feline kidney cells, a derivative of CRFK cells. Virus capsids were concentrated by polyethylene glycol precipitation followed by sucrose gradient centrifugation, which separated the full (DNA-containing) and empty capsids. Purified capsids were dialyzed against either phosphate-buffered saline (PBS) or 20 mM Tris-HCl (pH 7.5) and stored at 4°C (1).
Fluorescently labeled virus.
Purified capsids were labeled with Alexa Fluor 647 succinimidyl ester (Invitrogen) (16), Atto 647N N-hydroxysuccinimide ester (NHS), or Atto 568N NHS (Sigma-Aldrich Inc.; St. Louis, MO) in PBS for 30 min at room temperature using ∼20% of the dye concentration recommended by the manufacturer. Labeled capsids were separated from free dye by passage through a Sephadex G25 column (GE Healthcare, Piscataway, NJ) in PBS, and eluted capsids were stored at 4°C. The number of dye molecules per capsid ranged from 6 to 10 for Alexa 647 and 3 to 6 for Atto 647, as measured spectroscopically (Alexa) or by capsid photobleaching analysis (Atto).
Plasmid construction.
The expression plasmids encoding fTfR-eGFP and fTfR-mCherry were generated as follows: (i) the wt feline TfR gene was PCR amplified from an existing pcDNA3.1(−) expression vector (33) using primers that contained restriction enzyme recognition sites for EcoRI (forward primer, 5′-GTCAGAATTCATGATGGATCAAGCCAGATC-3′) or AgeI (reverse primer, 5′-GACTACCGGTGGATCCCCAAACTCATTGTCAATATCCCAAATGTC-3′); (ii) the PCR product and peGFP-N1 (Clonetech Laboratories, Inc., Mountain View, CA) or an otherwise identical plasmid encoding mCherry in place of eGFP was digested with EcoRI and AgeI, and the cleaved DNA fragments were ligated to generate pfTfR-eGFP. This cloning strategy introduced a 7-amino-acid linker (GDPPVAT) between the TfR and the fluorescent protein open reading frames. The pAcGP67A baculovirus transfer vector encoding nonglycosylated (N413D, N611D) human transferrin with an N-terminal secretion signal followed by a hexahistidine tag (6His-Tf) was a kind gift of Pamela Bjorkman (California Institute of Technology, Pasadena, CA). Site-directed mutagenesis was used to introduce a single cysteine residue between the secretion signal and histidine tag, and the presence of the introduced cysteine was confirmed by sequence analysis. The plasmid encoding eGFP with a C-terminal hexahistidine tag (eGFP-6His) was a kind gift of Massimo Merighi (laboratory of Stephen Lory, Harvard Medical School).
Production of recombinant fluorescent proteins.
Baculoviruses expressing 6His-Tf were generated in Spodoptera frugiperda Sf9 cells using the BD BaculoGold transfection kit (BD Biosciences, San Diego, CA), and the recombinant protein was produced in Trichoplusia ni Hi5 cells. Secreted 6His-Tf was isolated from the cell supernatant using Talon metal affinity resin (Clontech). Isolated protein was eluted using 200 mM imidazole in PBS to a final concentration of 0.54 μM. Purified protein was labeled under nonreducing conditions with Atto 647N maleimide (Sigma-Aldrich) using a 4-fold molar excess of dye to protein for 1 h at room temperature. Labeled protein was separated from free dye using a G25 MicroSpin gel filtration column (GE Healthcare) and stored in PBS supplemented with 20% glycerol at −80°C. Spectroscopic analysis showed that ∼80% of 6His-Tf molecules were labeled with dye.
eGFP-6His was expressed in E. coli strain BL21 grown in LB medium containing 100 mg−1 ampicillin at 37°C to an optical density of 0.6 at 600 nm. Expression of eGFP-6His was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and the cells were grown for 3 h at 37°C. Cells were harvested by centrifugation (Sorvall RC3B rotor, 5,000 rpm, 10 min, 4°C) and resuspended in ice-cold lysis buffer (20 mM imidazole, 0.25 M NaCl, and 1 mM EDTA in PBS) in the presence of protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The resuspended cells were sonicated for 1 min on ice (Ultrasonic processor XL; Heat Systems, Farmingdale, NY). The cell debris was pelleted by centrifugation at 100,000 × g (Beckman 45Ti rotor, 40,000 rpm) for 45 min at 4°C, and the protein was purified by a passage over Talon metal affinity resin (Clontech). Isolated protein was eluted using 200 mM imidazole in PBS. After overnight dialysis (Slide-A-Lyzer, molecular weight cutoff [MWCO] of 104; Thermo Scientific, Barrington, IL) against filtered PBS; the final concentration of eGFP-6His was 2 mM. Purified eGFP was stored at −80°C in PBS containing 20% glycerol.
Microscopy. (i) Live cell imaging.
The total internal reflection fluorescence microscopy (TIRFM) and spinning disk confocal microscope systems used in this study have recently been described in detail (2, 9). The TIRFM system was modified prior to this study to include a Laser TIRF 3 motorized slider (Carl Zeiss, Inc.; Thornwood, NY), and the spherical aberration correction unit was removed from the emission path. The penetration depth of the evanescent field used in these studies was measured using a previously established method (11) and estimated to be ∼150 nm when samples were illuminated with a 488 laser. Slidebook 5.0.0.20 (TIRFM) or 4.2.13 (confocal) imaging software [Intelligent Imaging Innovations, Inc.; Denver, CO] was used to manipulate the hardware devices and visualize data.
Cells plated on 25-mm coverslips (no. 1.5; Electron Microscopy Sciences, Hatfield, PA) 16 to 20 h prior to imaging were placed in a preheated perfusion chamber on the microscope stage and overlaid with alpha minimal essential medium (α-MEM) (no phenol red; Invitrogen) containing 10 mM HEPES (pH 7.4) and 5% FBS. The microscope stage and objective lenses were maintained at 37°C within an environmental chamber, and the air above the cells was supplied with 5% CO2.
To image CPV- and Tf-TfR interactions on live cells, the virus or protein solution was cleared of aggregates by centrifugation for 1 min at 14,000 × g in a microcentrifuge, and cells were inoculated with a dose of material that ensured attachment of <200 objects to the visible cell surface during an imaging period. Tf imaging experiments were performed in the absence of FBS. Image acquisition was initiated immediately after CPV or Tf addition to cells, and images were captured at 1- to 3-s intervals following sequential illumination of the samples with the appropriate lasers for exposure times of 20 to 50 ms (confocal) or 10 to 20 ms (TIRFM). Time-lapse acquisitions typically spanned 3 to 4 min per cell in TIRFM experiments and 6 to 8 min in confocal experiments.
(ii) Fluorescent objects on glass.
Fluorescent CPV capsids were nonspecifically adsorbed to glass coverslips to yield ∼500 attached particles per field of view. Unbound virus was removed by washing, and single snapshots were acquired using exposure times equivalent to those in live cell imaging experiments. Recombinant eGFP molecules were diluted to ∼300 nM in PBS and briefly incubated with a glass coverslip that was modified with a copolymer of poly-l-lysine (PLL) and biotinylated polyethylene glycol (PEG) (SuSoS AG; Switzerland) and streptavidin (Sigma-Aldrich), as previously described (2). Nonspecifically adsorbed molecules were removed by washing, and bound molecules were photobleached by repeated illumination for 20 ms until a majority of the molecules no longer fluoresced when excited. The average time to photobleaching of single eGFP molecules was 40 ± 38 s (n = 576) under these imaging conditions.
Data analysis.
Slidebook 4.2.13 or 5.0.0.18 (III) was used to view and export images, and exported TIFF files were compiled into AVI movie files using the ImageJ software program (NIH). SigmaPlot 11.0 (SYSTAT; Point Richmond, CA) was used to plot data and perform statistical analyses.
(i) Fluorescence intensity measurements.
Fluorescence intensity measurements of adaptor proteins and CPV capsids imaged by confocal microscopy were performed as before (9) using a custom written software application (IMAB) (28) developed within MATLAB (Mathworks; Natick, MA). In all cases, the local background signal was subtracted from the pit or capsid signal. IMAB was used to record the lifetime and peak adaptor signal of the first ∼100 coated pits that initiated on the cell surface and did not capture a CPV capsid. Complete virus uptake events that occurred during the same time span were analyzed in an identical manner. Coated pits in which the adaptor assembly aborted within 15 s after initiation were excluded from further analysis, as were events in which the adaptor signal could not be accurately tracked for any portion of the pit lifetime.
The fluorescence intensity of eGFP molecules or CPV capsids bound to a glass coverslip and imaged by TIRFM was measured essentially as described previously (2) using software developed in MATLAB. Briefly, the objects were detected as local maxima in an averaged fluorescence image (average of first 3 frames), and the position of each object was determined by calculating the mean adjusted center of intensity at each local maximum. A 7 × 7 pixel mask was then centered on the position of each object, and objects with overlapping masks were eliminated from further analysis. The fluorescence intensity of the remaining objects was calculated in each image frame by summing the intensities of the pixels in the 7 × 7 object mask. The object fluorescence intensity was corrected for local background fluorescence using a routine that does the following: (i) generates a mask to exclude pixels that belong to all objects present in each image plane, (ii) computes the mean and standard deviation (SD) of the remaining pixels and excludes the hot pixels (intensity, 2.5 SD above the background mean), (iii) sums the first 100 pixels nearest to the center of intensity of the object, and (iv) subtracts the average intensity of those 100 pixels from each pixel within the object mask. A step-fitting function (2, 20) was employed to identify the time at which each single eGFP molecule photobleached and to measure the fluorescence intensity of each molecule before and after photobleaching. Photobleaching traces of eGFP molecules were manually sorted to exclude objects that showed more than one photobleaching step or steps in which the amplitude that was greater than that of a single molecule.
(ii) Analysis of CPV receptor binding, diffusion, dissociation, and entry.
The efficiency of clathrin-dependent CPV internalization was measured as follows: (i) each capsid that arrived in the field but was absent from the previous image frame was manually marked and tracked using Slidebook 5.0.0.18; (ii) the fate of each particle was tracked until the capsid fluorescence was no longer detectable above the local background level; (iii) particles that were only visible in ≤2 image frames (i.e., ≤6 s) or underwent directed motion (i.e., endosomal particles) were eliminated from further analysis; (iv) the number of particles that disappeared from view between 2 sequential frames (i.e., within 3 s) or entered by clathrin-dependent endocytosis was recorded.
Events of receptor engagement by CPV capsids were detected and analyzed in the following manner: (i) all CPV particles that appeared in the field after frame 1 but before frame 90 (2 to 180 s) of a given time-lapse acquisition were manually identified; (ii) the particle position in each frame was determined using Slidebook 5.0.0.18 as described below for analysis of virus diffusion; (iii) the MATLAB software used for analysis of objects on glass was modified to center a 7 × 7 pixel mask on the virus in each image using the x,y coordinates of the virus from Slidebook; (iv) the fluorescence intensity of the virus capsid and associated fTfR-eGFP molecules were measured in each image frame as for fluorescent objects on glass (see above); and (v) proper tracking of each virus particle was manually verified, and the fluorescence values for the virus and fTfR-eGFP were recorded from the frame in which the virus signal was most intense (i.e., the virus was closest to the glass). Frames in which the virus-receptor complex overlapped with irrelevant spots of receptor fluorescence were not included in the final data set, and in these cases, the receptor signal associated with the next-most-intense virus signal was recorded.
Slidebook 5.0.0.18 was used to track the motion of receptor-bound CPV or Tf on the cell surface. Only objects that arrived in the imaging field after the start of imaging and remained cell associated for at least 5 frames were considered for analysis. To map the object position in each frame, a roughly circular mask comprising 112 pixels was centered on the position of peak particle fluorescence, and the x,y coordinates of the mask centroid were recorded. The measured pixel size (0.106 × 0.106 μm2 [TIRFM]) was used to calculate the object displacement between frames and the mean squared displacement (MSD) (37) with respect to time. To calculate the diffusion coefficient (D) of each object, linear regression analysis was performed using the first 4 time points of a capsid MSD plot to identify the best-fit trend line. The formula <x2> = 4Dt (37), where <x2> is the slope of the best-fit line, was used to calculate D for each object.
CPV binding, uptake, and infection.
To determine the functionality of eGFP-tagged feline TfR, virus binding and uptake assays were performed with CRFK or TRVb fTfR-eGFP cells as previously described (12). Briefly, cells were detached from the plate, washed in PBS containing 1% ovalbumin (Sigma-Aldrich), and incubated in solution with 10 μg/ml of Alexa 647 CPV capsids for the indicated time intervals at 37°C. The level of cell-associated capsids was quantified in three independent experiments for at least 10,000 cells using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA).
The susceptibility of NLFK or TRVb fTfR-eGFP cells to infection with virus was quantified using previously established methods (31). Cells were inoculated with CPV-2 (multiplicity of infection [MOI] of 5) and incubated at 37°C for 48 h. Cells were fixed with 4% paraformaldehyde in PBS and then permeabilized with PBS containing 0.1% Triton X-100 and 1% bovine serum albumin (BSA). Permeabilized cells were immunostained with an Alexa 594-conjugated monoclonal antibody (CE-10) against nonstructural protein 1 (51).
RESULTS
Visualizing clathrin endocytic structure formation in feline cells.
We generated feline kidney CRFK cells that constitutively express an eGFP-tagged σ2 subunit of the AP-2 adaptor complex, a structural constituent of all clathrin endocytic structures on the plasma membrane (10). As in other mammalian cell types (41), AP-2 localized to the cytosol and to punctate structures on the plasma membrane of CRFK cells (Fig. 1A). Time-lapse images acquired using a spinning-disk confocal microscope revealed that the structures were dynamic and consisted mostly of diffraction-limited spots that assembled and disappeared from view with kinetics typical of clathrin-coated pits (Fig. 1A; see also Movie S1 in the supplemental material) (26, 41). In addition to the conventional coated pits, CRFK cells formed larger (i.e., not diffraction-limited) structures that likely correspond to clusters of coated pits and clathrin plaques observed previously (41, 44), since these structures often accumulated more adaptor proteins and had longer lifetimes. Thus, we isolated feline cells that express a fluorescent constituent of clathrin endocytic coats, and real-time imaging demonstrated that these cells form the full array of clathrin endocytic structures observed in other mammalian cell types.
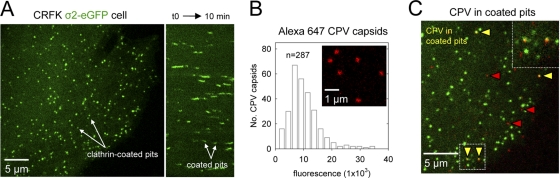
Fig 1.
CPV association with clathrin endocytic structures in feline cells. (A) Clathrin-coated vesicle formation in CRFK cells. Left, image of a CRFK σ2-eGFP cell extracted from frame 1 of a 10-min time-lapse acquisition. Images were captured at 3-s intervals using a spinning disk confocal microscope. Right, kymograph view of clathrin-coated vesicle formation by the cell shown at left. (B) Fluorescence intensity of Alexa 647 CPV capsids. Confocal images of labeled capsids on glass were acquired, and the fluorescence intensity of each capsid spot was quantified. Inset, enlarged view of capsids (red) on glass. (C) Colocalization of CPV capsids with clathrin-coated pits (see Movie S1 in the supplemental material). CRFK σ2-eGFP cells were inoculated with Alexa 647 capsids and imaged as for panel A. A merge of the AP-2 (green) and capsid (red) channels is shown, along with an enlarged view of 2 capsids in coated pits from the boxed region at left. Arrowheads highlight capsids in (yellow) or outside (red) coated pits.
Real-time imaging of clathrin-dependent CPV internalization.
We prepared fluorescent CPV capsids by conjugating Alexa Fluor 647 dye molecules to primary amines exposed on the surface of empty capsids (lacking DNA) using a previously established procedure that does not interfere with capsid-TfR interactions (16, 19). Structural and biochemical comparisons of empty and full (DNA-containing) capsids have shown that they are indistinguishable with regard to the structural elements that control TfR binding (47, 50) and their dependence on TfR for cell entry (19). Confocal images of the labeled capsids on glass revealed diffraction-limited, fluorescent spots that displayed a single peak distribution of intensity values, consistent with a population comprised mostly of single capsids (Fig. 1B).
To visualize CPV internalization by live CRFK σ2-eGFP cells, we incubated the cells with fluorescent capsids and acquired images at 3-s intervals using a spinning disk confocal microscope. Capsids attached to the cell surface and subsequently colocalized with clathrin endocytic structures containing σ2-eGFP (Fig. 1C; see also Movie S1 in the supplemental material). A representative example of clathrin-dependent CPV internalization is shown in Fig. 2 (see also Movie S2 in the supplemental material). A capsid attaches to the cell surface and moves laterally for ∼48 s before colliding with an assembling coated pit. The capsid and coated pit signals remain stably associated after the collision, and the adaptor fluorescence increases to a maximal value as the coated pit forms around the capsid (Fig. 2A and B). Shortly thereafter, the pit and capsid signals simultaneously disappear, signifying pit internalization, coat disassembly, and intracellular transport of the capsid-containing vesicle (Fig. 2A and B). This sequence of events is typical of most CPV internalization events and directly confirms that CPV capsids enter feline cells by the clathrin endocytic pathway.
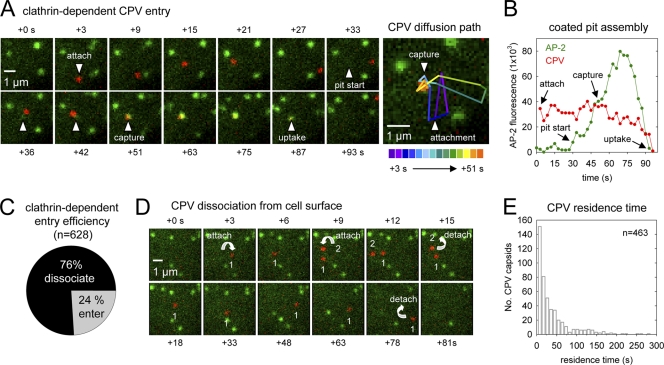
Fig 2.
Real-time imaging of clathrin-dependent CPV internalization. (A) Example of clathrin-dependent CPV endocytosis (see Movie S2 in the supplemental material). Left panels, CRFK σ2-eGFP cells (green, AP-2) were inoculated with fluorescent capsids (red) and imaged as before. The image frame prior to capsid attachment was set to t = 0 s. The capsid attaches in frame +3 and diffuses laterally on the cell surface. The coated pit that is the future site of capsid uptake initiates in frame +33, while the capsid is still undergoing diffusion. The capsid collides with the assembling pit in frame +51, and the capsid and pit signals remain colocalized, signifying capsid incorporation into the pit. Frame +87 depicts the capsid-containing pit during the process of internalization (note the decrease in the capsid and AP-2 fluorescence). Right panel, diffusion path of the capsid shown at left. A color-coded line trace of the capsid diffusion path is overlaid onto the t = 51-s image. (B) Plot of the background-corrected AP-2 (green) and capsid (red) fluorescence intensities with respect to time for the event in panel A. For frames prior to pit initiation, the AP-2 fluorescence intensity was quantified at the eventual site of pit initiation. (C) Efficiency of clathrin-dependent CPV entry. Efficiency is expressed as a percentage of the total particles that bound to cells during time-lapse imaging of 4 cells. The percentage of particles that entered by clathrin-dependent endocytosis was 8% (9 entering/108 total), 30% (38/125), 24% (52/220), and 27% (48/175). (D) Examples of CPV dissociation from the cell surface (see Movie S3 in the supplemental material). Time-lapse images showing the attachment (downward-facing arrows) of two capsids (red; no. 1, no. 2) and subsequent capsid dissociation (upward-facing arrows). (E) Residence time of CPV capsids that dissociated from CRFK cells. The elapsed time between capsid attachment and disappearance was measured for capsids that remained bound for ≥6 s. Events are from the same 4 cells that were analyzed for panel C.
We measured the efficiency of clathrin-dependent CPV internalization by detecting each capsid binding event and tracking the fate of capsids that remained bound to the cell for at least two consecutive frames (≥6 s). We found that 24% of attached capsids (n = 628) entered CRFK cells by clathrin-dependent endocytosis (Fig. 2C), while the majority (76%) of capsids abruptly disappeared from view before colliding with a coated pit (Fig. 2C and D; see also Movie S3 in the supplemental material). The latter capsid disappearance events occurred between consecutive image frames (i.e., in <3 s) and typically less than 30 s after attachment (Fig. 2D and E). Under the imaging conditions used here, photobleaching of the capsid-associated dyes was a gradual process that required at least 250 s (mean, 306 ± 36 s; n = 8). Moreover, the field of view was sufficiently deep (∼0.5 μm) to visualize the movement of cargo-containing vesicles toward the cell interior before the vesicles underwent intracellular transport. Thus, capsid disappearance prior to coated pit association was most probably due to capsid dissociation from TfR on the cell surface and not photobleaching or endocytic uptake of capsids via a clathrin-independent endocytic mechanism. These data show that clathrin-dependent endocytosis is the primary pathway of CPV internalization but that most capsids dissociate from receptors before the CPV-TfR complex encounters an endocytic structure.
CPV diffuses into newly formed clathrin-coated pits.
We next analyzed the entry process of capsids captured by forming coated pits in greater detail. Most capsids (80%; n = 347) bound to receptors on the free cell surface and then diffused laterally into an assembling coated pit (Fig. 3A and B). The remainder of capsids either attached to receptors in or near a forming coated pit (11%) or stopped diffusing at the future site of pit appearance (9%) (Fig. 3A and B). In these infrequent events, we could not resolve whether the capsids associated with coated pits by diffusion, since the capsid and coat signals were not optically separable (i.e., they were within ∼250 nm of one another) at the time of their initial colocalization.
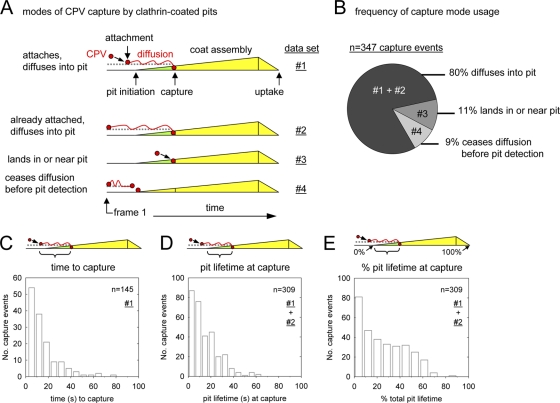
Fig 3.
CPV capsids diffuse into newly formed clathrin-coated pits. (A) Schematics depicting how CPV capsids incorporate into coated pits. For each clathrin-dependent CPV entry event, the process of capsid-pit association was analyzed and recorded. The events were assigned to one of the four observed categories (numbered 1 to 4). (B) Chart showing the frequency of each capture mode. Data are from 18 cells. The data set(s) that comprises each slice is indicated according to the nomenclature in panel A. (C) The time interval between particle attachment and capture by a coated pit. The mean time to capture was 15 ± 14 s. (D and E) Timing of CPV capture expressed relative to the time of coated pit initiation (D) or as a percentage of the total pit lifetime (E). In panel E, 0% = pit initiation; 100% = loss of adaptor signal.
The elapsed time between CPV attachment and capture by a coated pit (time to capture) was typically <20 s (Fig. 3C). Capsids preferentially associated with coated pits early in the coat assembly process, since the likelihood of particle incorporation was highest within the first 20% of the coated pit lifetime and lowest after a pit reached 70% of its total lifetime (Fig. 3D and E). Moreover, >90% of stable CPV-coated pit associations (i.e., lasting ≥6 s) led to capsid internalization. These results show that CPV-TfR complexes primarily diffuse into newly formed coated pits and that subsequent capsid uptake is highly efficient.
CPV incorporation does not alter the assembly kinetics or clathrin content of coated pits.
To assess whether CPV incorporation alters the adaptor composition or assembly kinetics of clathrin-coated pits in CRFK σ2-eGFP cells, we measured the maximum AP-2 content (an indicator of coat size) (10) and the total lifetime of pits that incorporated or lacked a CPV capsid. We excluded from our analysis the short-lived population of abortive clathrin coats (lifetimes < 15 s) that do not mature into complete coated vesicles, as well as objects that were not diffraction limited or could not accurately be tracked over their entire lifetime. Our results show that CPV incorporation does not alter the lifetime or AP-2 content of clathrin endocytic carriers, since coated vesicles formed in ∼54 ± 20 s (Fig. 4A) and had a similar range of peak AP-2 fluorescence values (Fig. 4B) regardless of whether they contained a CPV particle.
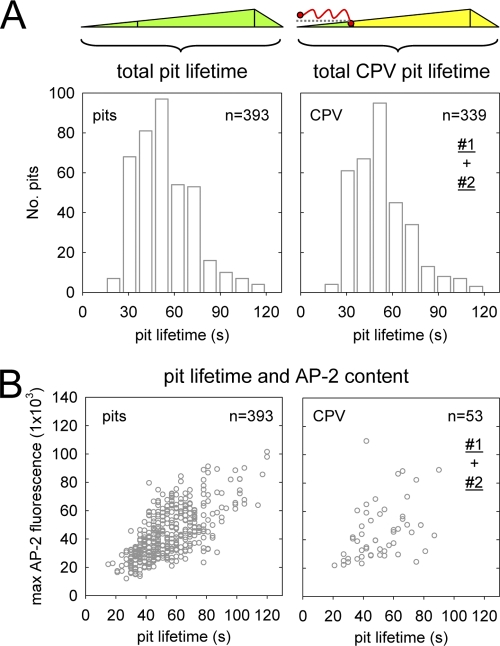
Fig 4.
CPV does not alter the assembly kinetics or adaptor content of coated pits. Confocal images of CRFK σ2-eGFP cells were acquired at 3-s intervals. (A) Total lifetime of conventional coated pits (left) or pits that internalized a CPV capsid (right). Data for standard coated pits are from 4 individual cells in the presence of CPV, and data for CPV-containing pits are from 17 cells and include events from data sets no. 1 and 2 (Fig. 3A). The average lifetimes of standard coated pits (54 ± 19 s) and CPV-containing pits (53 ± 19 s) are not significantly different (two-tailed Student's t test, P = 0.33). (B) Lifetime versus maximum AP-2 fluorescence of standard (left) or CPV-containing (right) coated pits. Data for standard coated pits are from 4 cells. CPV data are from the same 4 cells and include events in data sets no. 1 and 2. Each open circle represents a single coated pit. The average peaks of fluorescence of AP-2 in standard coated pits (43 × 103 ± 1.8 × 103) and CPV-containing pits (45 × 103 ± 2.0 × 103) are not significantly different (two-tailed Student's t test, P = 0.44).
The entry processes of empty and full CPV capsids are indistinguishable.
Next, we directly compared the entry processes of empty and full (DNA-containing) CPV capsids (summarized in Table 1). We labeled purified full capsids with Atto 647 and imaged their entry into CRFK σ2-eGFP cells alone (Table 1, experiment no. 2) or together with Atto 568-labeled empty capsids (Table 1, experiment no. 3). Similar proportions of full and empty capsids entered cells by clathrin-dependent endocytosis (∼25%) or dissociated from receptors on the cell surface before being captured by a forming coated pit (∼75%) (Table 1). As with empty capsids, a majority of the full capsids (>80%) diffused laterally into forming coated pits, and full capsid capture by coated pits preferentially occurred during the early stages of pit formation (Table 1). We observed modest (2- to 4-fold) variations in the time to capture of full or empty capsids compared to the results obtained for empty capsids in earlier experiments (Table 1, experiment no. 1). However, we observed these variations for both capsid forms, indicating that the variations did not correlate with the DNA content of the capsids. The residence time of both capsid forms was also longer in experiments no. 2 and 3 compared to that of the empty capsids analyzed in experiment no. 1, but this difference was also relatively minor (<2-fold). From these data, we conclude that full and empty CPV capsids access clathrin endocytic structures in the same way, a result that is consistent with the high degree of structural similarity between the two capsid forms (47, 50).
Table 1.
Properties of empty or full CPV capsid entry into CRFK or TRVb cellsa
| Parameter | Value for expt (nb) |
||||
|---|---|---|---|---|---|
| CRFK cells |
Expt 4, TRVbc cells (empty capsids) | ||||
| Expt 1 (empty capsids) | Expt 2 (full capsids) | Expt 3 |
|||
| Empty capsids | Full capsids | ||||
| % entry (no. of total CPV attachments; no. of cells analyzed) | 24 (628; no. of cells analyzed 4 cells) | 25 (121; no. of cells analyzed 6 cells) | 35 (54; no. of cells analyzed 3 cells) | 25 (68; no. of cells analyzed 3 cells) | 27 (118; no. of cells analyzed 4 cells) |
| Capture mode as % of entry events (no. of total entry events) | 80% diffuse into pit; 9% land in/near pit; 11% stop diffusing before pit detection (347) | 80% diffuse into pit; 0% land in/near pit; 20% stop diffusing before pit detection (30) | 88% diffuse into pit; 0% land in/near pit; 12% stop diffusing before pit detection (19) | 100% diffuse into pit; 0% land in/near pit; 0% stop diffusing before pit detection (17) | 95% diffuse into pit; 2% land in/near pit; 3% stop diffusing before pit detection (255) |
| Avg diffusion coefficient | 0.014 μm2 s−1 (101) | NT | NT | NT | NT |
| Time (avg no. of s ± SD) between CPV attachment and capture | 15 ± 14 (145) | 44 ± 47 (30)* | 67 ± 58 (22)* | 26 ± 26 (17)*# | 23 ± 19 (51)* |
| Time (avg no. of s ± SD) between pit detection and CPV capture | 13 ± 13 (309) | 13 ± 14 (30) | 13 ± 11 (20) | 15 ± 16 (17) | 10 ± 10 (231)* |
| Time between pit detection and CPV capture as % total pit lifetime | 25 ± 20 (309) | 19 ± 20 (30) | 20 ± 19 (20) | 21 ± 19 (17) | 25 ± 21 (231) |
| Total lifetime (avg no. of s ± SD) of CPV pits | 53 ± 19 (339) | 65 ± 19 (30)* | 60 ± 18 (20)* | 63 ± 27 (17)* | 40 ± 13 (231)* |
| Mean residence time (avg no. of s ± SD) of capsids that did not enter | 39 ± 44 (463) | 62 ± 55 (91)* | 71 ± 55 (35)* | 60 ± 46 (51)* | 46 ± 27 (71) |
*, statistical significance (P < 0.05, Student's t test) relative to corresponding data from experiment 1; #, statistical significance (P < 0.05, Student's t test) relative to corresponding data from empty capsids in experiments 1 and 3; NT, not tested.
n, number of events analyzed except where otherwise noted under “Parameters.”
TRVb cells expressing fTfR-eGFP.
Receptor-bound CPV capsids diffuse slower than single TfRs.
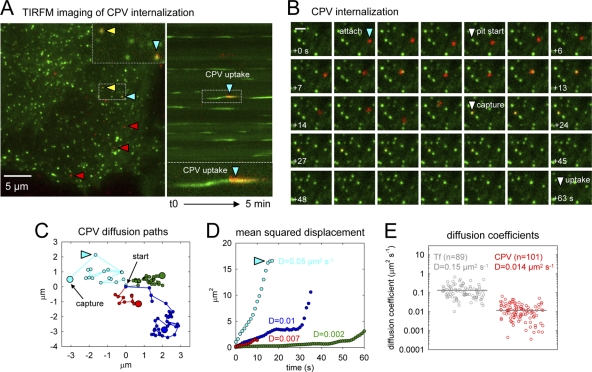
To address whether CPV binding alters the rate of TfR diffusion, we used total internal reflection fluorescence microscopy (TIRFM) to image CPV diffusion prior to coated pit association and compared the rate of capsid diffusion with that of individual TfRs. In these experiments, we inoculated cells with low doses of capsids or Atto 647-labeled Tf molecules (to label single TfR dimers) to allow unambiguous particle tracking. As observed by confocal microscopy, capsids explored large areas of the plasma membrane before encountering and being internalized by coated pits (Fig. 5A to C; see Movie S4 in the supplemental material). In some cases, capsids became transiently confined to submicron-sized areas of the plasma membrane (Fig. 5C), as previously seen for individual TfRs (43). Plots of the mean squared displacement (MSD) of the capsids were nearly always linear with respect to time during the early stages of virus diffusion, consistent with a random-walk mechanism of two-dimensional diffusion (Fig. 5D). Using the MSD plots, we calculated the macroscopic diffusion coefficient (D) for 104 individual CPV capsids and obtained a mean value of 0.014 μm2 s−1 (Fig. 5E). In contrast, Atto 647-labeled Tf molecules bound to single TfRs had a mean D value of 0.15 μm2 s−1(Fig. 5E; see Movie S5 in the supplemental material), consistent with previous reports (43). Thus, CPV-TfR complexes diffused slower than Tf-TfR complexes.
Fig 5.
Receptor-bound CPV capsids diffuse slower than single TfRs. (A) CPV diffusion and uptake imaged by TIRFM. CRFK σ2-eGFP cells were inoculated with fluorescent CPV capsids, and images were captured at 1-s intervals. Left, frame 194 of a 5-min time-lapse acquisition showing capsids (red) present on the free cell surface (red arrowheads) or colocalizing with AP-2 (green) in coated pits (yellow or blue arrowheads). The inset shows a zoomed view of two capsids in coated pits from the boxed region below. Right, kymograph view of coated pit formation over time in the cell shown at left. The blue arrowhead highlights internalization of the same capsid marked with a blue arrowhead at left. A zoomed view of the event is provided below. (B) Example CPV internalization event observed by TIRFM (see Movie S4 in the supplemental material). Frames prior to virus capture are spaced at 1-s intervals, and frames after virus capture are spaced at 3-s intervals. Scale bar, 1 μm. (C) Example traces of capsid diffusion. Traces of four capsids are displayed in separate colors. The start of each trace is centered at (x = 0, y = 0). Small points indicate the particle position in sequential frames spaced at 1-s intervals. Large dots indicate the location of particle capture by a coated pit. The light-blue trace corresponds to the diffusion path of the capsid highlighted by a blue arrowhead in panels A and B. (D) Mean square displacement plots for the objects shown in panel C. The diffusion coefficient (D) for each capsid is provided. Plot colors refer to traces of the same color in panel C. (E) Diffusion coefficients of receptor-bound Tf (gray; see Movie S5 in the supplemental material) or CPV (red). Gray bars designate the mean for each population. Tf and CPV data are from 6 and 8 cells, respectively.
CPV capsids cluster TfRs on the cell surface.
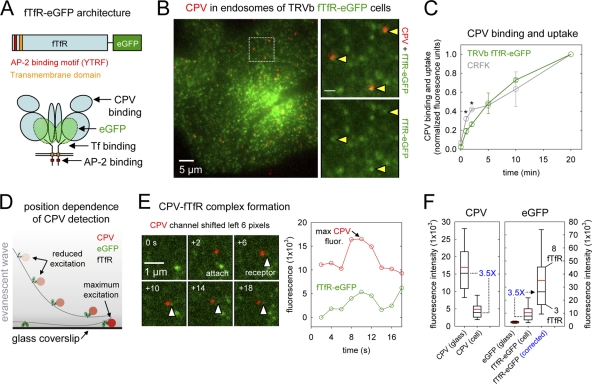
To directly visualize CPV-TfR interactions on the surfaces of live cells, we appended eGFP or mCherry to the C-terminal ectodomain of wt feline TfR (fTfR) (Fig. 6A). We then stably expressed the tagged receptors in Chinese hamster ovary TRVb cells, which lack endogenous TfRs and therefore do not support CPV attachment, entry, or infection (29, 33). CPV bound and entered TRVb cells expressing the tagged receptors with similar kinetics to those observed in CRFK cells, and capsids accumulated together with receptors in endosomal compartments (Fig. 6B and C). In TRVb cells coexpressing σ2-eGFP and fTfR-mCherry, we observed the binding, diffusion, and clathrin-dependent uptake of capsids (see Fig. S1A to D and Movie S6 in the supplemental material). The kinetics of capsid diffusion and capture, as well as the efficiency of capture and internalization, were indistinguishable from those observed in CRFK cells (Table 1, experiment no. 4). Moreover, fTfR-mCherry or fTfR-eGFP supported productive cell infection by CPV (data not shown), indicating that the fluorescent molecules did not prevent a downstream TfR-dependent step of virus entry. The tagged receptors also bound and internalized Tf, albeit less efficiently than wt TfRs (see Fig. S1E in the supplemental material). These data show that the fTfR fusion proteins support efficient CPV entry and infection in TRVb cells.
Fig 6.
CPV capsids cross-link TfRs on the surfaces of live cells. (A) Schematics of fTfR tagged with eGFP. The eGFP molecule was appended to the C-terminal ectodomain of the fTfR such that each receptor dimer contains two eGFPs. (B) Images of CPV capsids colocalizing with fTfR-eGFP in endosomal compartments. TRVb cells stably expressing fTfR-eGFP (green) were inoculated with Atto 647 CPV capsids (red) and incubated at 37°C for 10 min. Live cells were imaged by TIRFM, and a representative image from a time-lapse acquisition is shown. Right-hand panels show an expanded view of the boxed region at left. The fTfR-eGFP channel is shown as a merge with the Atto 647 CPV channel (top) or alone (bottom). Arrowheads highlight the receptor signal in endosomes that contain CPV. Scale bar in right panels, 1 μm. (C) Comparison of CPV binding and uptake in CRFK or TRVb fTfR-eGFP cells. Cells were inoculated with Alexa 647 CPV capsids at t = 0 and 37°C, and the cell-associated capsid fluorescence was quantified by flow cytometry at the indicated times for the receptor-expressing cells. At all times, 2-5-fold more capsids associated with TRVb fTfR-eGFP cells than with CRFK cells. To compare the rates of capsid association with both cell types, we expressed the raw fluorescence values as a fraction of the values obtained at 20 min postinoculation (p.i.) in each cell type. Data points are the means ± SD for 3 experiments. Asterisks indicate time points when the difference in the mean values is statistically significant (two-tailed Student's t test, P < 0.05). (D) Schematic illustrating the position dependence of CPV-fTfR-eGFP complex detection using TIRFM. The intensity of the evanescent wave (gray) generated from the totally internally reflected laser source decreases exponentially as the distance from the coverslip increases. Thus, CPV capsids (red) and the associated fTfR-eGFP molecules (green) are maximally excited when directly contacting the glass coverslip, but those on the upper cell surface are excited to a lesser extent and therefore emit a lower fluorescent signal. (E) Example of receptor engagement by a CPV capsid (see Movie S7 in the supplemental material). TRVb fTfR-eGFP cells were inoculated with Atto 647 CPV capsids, and images were acquired at 2-s intervals using TIRFM. Left, images showing attachment of a CPV capsid (red) and the spot of fTfR-eGFP (green, white arrowheads) that colocalizes with the particle. In each panel, the CPV channel was shifted to the left by 6 pixels to reveal the underlying receptor signal. Right, plot of the CPV and TfR fluorescence intensity for the images at left. (F) Box plots showing the fluorescence of Atto 647 CPV capsids or single eGFP molecules on glass compared to their signals in CPV-fTfR-eGFP complexes on the cell surface. Glass data for eGFP molecules and CPV are from Fig. S2 in the supplemental material, panels C and D. Cell data are from 76 events observed on 19 cells. Upper and lower bounds of boxes correspond to the 75th and 25th percentiles, while error bar whiskers show the 90th and 10th percentiles of the data. Red and black horizontal lines indicate the mean and median values of the data sets. The raw fluorescence intensity of fTfR-eGFP associated with each capsid on the cell surface (middle bar, right panel) was multiplied by a correction factor of 3.5 (right panel), derived from the difference in the median fluorescence intensities of capsids on glass compared to those on cells (left panel) due to the positional loss along the z axis in the strength of the evanescence field.
The intensity of the evanescent wave generated under TIR conditions decreases exponentially with increasing distance from the glass coverslip (Fig. 6D). Thus, prior to imaging CPV-TfR interactions on the cell surface, we measured the fluorescent signals of single eGFP molecules and CPV capsids bound to a coverslip to establish the maximum possible fluorescence intensity of the objects. Single eGFP molecules showed a normal distribution of fluorescence intensities centered at 3,082 ± 661 arbitrary fluorescence units (a.u.) (see Fig. S2A to C in the supplemental material), while the intensities of Atto 647-labeled capsids distributed across a broader range (16,911 ± 7,906 a.u.) that was equivalent to the fluorescence of one to eight Atto 647 dye molecules (see Fig. S2D to F in the supplemental material).
Next, we examined whether CPV capsids cluster receptors on the cell surface. We inoculated the cells with Atto 647 capsids as before and acquired images at 2-s intervals by TIRFM. We then detected events in which a capsid appeared in the field of view <3 min after the onset of imaging, a time at which most particles are still on the cell surface. An example of such an event is shown in Fig. 6E (see Movie S7 in the supplemental material). A capsid appears in the field of view and colocalizes with a spot of receptor that remains visible as the particle diffuses laterally on the cell surface. Importantly, the fluorescence intensities of the capsid and receptors varied together as a function of time, reflecting changes in the Z position of the capsid, as expected if the capsid was bound to a defined set of fTfR-eGFP molecules (Fig. 6E; see Figure S3 in the supplemental material for additional examples).
For 76 such events, we recorded the virus and receptor fluorescence values from the image frame in which the virus signal was most intense (i.e., when the CPV-TfR complex was closest to the glass) (Fig. 6E). The median signal of the cell-associated capsids imaged by TIRFM was 3.5-fold lower than that of capsids directly attached to the glass coverslip (Fig. 6F). This result indicates that the CPV-TfR complexes were close to (<400 nm) but not in direct contact with the coverslip and were therefore illuminated by a less-intense portion of the evanescent field (Fig. 6D). The eGFP fluorescence signal of most complexes (55/76) was ≤12,000 arbitrary fluorescence units (Fig. 6F), which corresponded to ≤4 eGFP molecules, or ≤2 receptor dimers. After correction for the positional offset of the complexes from the coverslip by a factor of 3.5, we determined that most capsids bound fewer than 5 receptors, with an upper limit of 8 receptors per capsid (Fig. 6F). We also observed a second class of objects in which the capsids colocalized with receptor spots of 2- to 3-fold higher intensity. In these cases, the capsid motion was often restricted or directed (see Fig. S3C in the supplemental material), suggesting that the particles were in a coated pit or an endocytic vesicle. We conclude that most CPV capsids cluster fewer than 5 fTfR dimers on the surfaces of live feline cells.
DISCUSSION
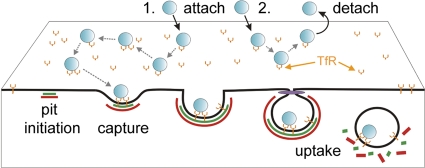
We investigated how receptor binding by a small nonenveloped virus, canine parvovirus (CPV), governs virus capsid uptake by clathrin endocytic structures. By visualizing the initial entry stages of individual capsids bound to TfR molecules, we resolved previously unappreciated aspects of capsid entry that support the following conclusions: (i) capsids clustered up to eight TfRs on the cell surface; (ii) ∼25% of attached capsids entered feline cells by clathrin-dependent endocytosis, while the remainder dissociated from receptors before engaging a clathrin-coated pit; and (iii) capsids engaged the clathrin system by diffusing into assembling coated pits. Based on these conclusions, we propose a model whereby TfRs tether CPV to target cells, and limited TfR clustering increases the probability of capsid association with a coated pit by allowing long-range diffusion of the CPV-TfR complex and by increasing the residence time of capsids on the cell surface (Fig. 7).
Fig 7.
Outcomes of CPV-TfR interactions identified in this study. CPV capsids (blue) attach to TfRs (orange) on the cell surface. The capsid-receptor complex diffuses laterally on the cell surface, and at times, capsids engage more than one receptor. Receptor-bound capsids can diffuse into an assembling clathrin-coated pit (green, AP-2; red, clathrin), leading to particle endocytosis upon pit scission by dynamin (purple) (outcome 1). Alternatively, capsids can detach from TfRs prior to association with a coated pit (outcome 2).
CPV capsids bound to TfRs entered feline cells by clathrin-dependent endocytosis. We did not observe evidence for clathrin-independent uptake of CPV. Instead, capsids that did not associate with a coated pit suddenly dissociated from receptors on the cell surface. Our results are consistent with previous electron microscopic images of CPV in coated pits but not other endocytic structures (34) and with the known clathrin-dependent uptake mechanism of TfRs (3). Thus, we have directly confirmed that CPV enters feline cells by clathrin-dependent endocytosis and further show that in this cell culture system, where capsids are not confined to the cell surface by adjacent cells, most capsids dissociate from feline TfRs prior to encountering a clathrin endocytic structure.
The half-life of CPV-TfR complexes calculated from the capsid residence times on CRFK cells was ∼30 s, which is ∼15-fold shorter than that of holotransferrin-TfR complexes (∼8 min) on other mammalian cells (17, 49). The short half-life of CPV-TfR complexes indicates that CPV has a much lower affinity for TfR than holotransferrin (Kd, ∼5 nM) (24, 49). We speculate that this low receptor affinity, combined with steric limitations imposed by the capsid architecture, restricts the number of receptors that simultaneously engage a capsid. In turn, this limited receptor clustering allows the CPV-TfR complex to laterally diffuse over large areas of the cell surface, thereby increasing the probability of capsid collision with an assembling coated pit. Whether capsid dissociation from TfRs is also required for intracellular steps of virus entry (e.g., endosomal membrane penetration) remains to be determined. Since CPV and the related parvovirus, feline panleukopenia virus (FPV), differ in their affinities for the feline and canine TfRs (18), it is of interest to explore how these differences in receptor affinity influence the virus entry process.
CPV capsids diffused into coated pits that had already initiated on the cell surface. This mode of cargo capture is similar to that of low-density lipoprotein (LDL)-receptor complexes (10) and to that of dengue virus serotype 2 (DENV) (48) but apparently distinct from that of vesicular stomatitis virus (VSV) and influenza A virus (8, 40). These latter viruses do not diffuse over large areas of the plasma membrane or engage existing coated pits. Instead, they incorporate into pits that initiate near or beneath the site of virus attachment (8, 40). These differences in virus mobility and capture mode correlate with the receptor binding capacity of the virus particles, since VSV and influenza A virus are larger and contain more receptor binding proteins than DENV or CPV. The fact that nearly all CPV capsids diffused into coated pits that had already initiated on the cell surface also shows that the low level of TfR clustering by CPV does not induce coated pit formation beneath attached capsids. Our results contrast with recent evidence suggesting that clustering of biotinylated TfRs (bTfRs) by tetravalent streptavidin complexes (SA) promotes coated pit initiation (25). Unlike our findings with CPV-TfR complexes, bTfR-SA complexes are presumably stable due to the high affinity of SA for biotin (Kd, ∼10−14 M) (13). Perhaps these or other properties of the ligand-receptor complexes influence whether they can serve as scaffolds for coated pit initiation.
CPV engaged coated pits faster than all other viruses studied to date. Most capsids associated with coated pits less than 20 s after receptor binding, while the average time to capture for DENV, influenza A virus, and VSV ranged from 20 s to several minutes (8, 40, 48). That coated pit assembly around these viruses typically requires at least 30 s indicates that the coat assembly process is the rate-limiting step for CPV uptake, while the time required for virus capture is often rate limiting for the other viruses. Although CPV capture was rapid, only 25% of attached capsids encountered a coated pit before dissociating from the cell surface. In contrast, approximately 65% of influenza A viruses and 90% of VSV particles associated with coated pits after attachment (8, 40). In the case of VSV, attached virions did not dissociate from receptors on the cell surface (8), suggesting that the stability of VSV-receptor complexes translates into a higher probability of capture by a coated pit than with CPV.
The potential importance of receptor clustering for virus entry has been inferred from the multivalent character of the particles themselves, but only the clustering of DC-SIGN by Uukuniemi virus has been directly visualized on the cell surface (27). Here we show that most CPV capsids cluster fewer than 5 TfRs on the plasma membranes of live feline cells. This finding is consistent with the presence of multiple receptor binding sites on one hemisphere of the capsid surface and with in vitro biochemical measurements showing that capsids in solution bind up to 7 TfR ectodomains (14). By comparing the diffusion rates of CPV and Tf bound to untagged TfRs, we found that CPV binding reduced the rate of TfR diffusion in the plasma membrane. We propose that the slower diffusion of the CPV-TfR complexes results from transient stages of receptor clustering by capsids that reduce either the free diffusion rate of the complexes or the rate at which the complexes migrate between submicron-sized compartments of membrane delimited by the underlying cell cytoskeleton (42). Indeed, previous studies showed that increasing the density of GM1 ganglioside receptors in an artificial membrane reduced the diffusion rate of simian virus 40 particles by inducing transient stages of virus confinement (22).
In summary, our data directly show that CPV-TfR interactions transport viral capsids into assembling clathrin endocytic structures by a relatively rapid diffusion-based mechanism. Given that CPV engages a low number of TfRs, this mechanism of cargo incorporation likely also extends to free and Tf-bound TfRs, as well as other viruses that might exploit the natural endocytic function of nonsignaling receptors to engage endocytic structures. Notably, TfRs also function as receptors for other viral pathogens, including mouse mammary tumor virus and certain New World arenaviruses (38, 39). These virus particles are significantly larger and have a greater receptor binding capacity than CPV. Thus, we anticipate that comparative analyses of these virus-receptor systems will reveal differences in the properties of virus-receptor complexes that modulate cargo capture by clathrin endocytic structures.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Wendy Weichert for expert technical assistance and Ramiro Massol for the development of the IMAB image analysis software. We also thank Eric Marino for maintaining the confocal microscope and TIRFM systems used in this study.
This work was funded by NIH grants AI092571 and AI028385 to C.R.P. and U54 AI057159 (New England Regional Center of Excellence in Biodefence and Emerging Infectious Diseases [NERCE BEID]) to T.K.
Footnotes
Published ahead of print 22 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. 1993. Structure determination of feline panleukopenia virus empty particles. Proteins 16:155–171 [DOI] [PubMed] [Google Scholar]
- 2. Bocking T, Aguet F, Harrison SC, Kirchhausen T. 2011. Single-molecule analysis of a molecular disassemblase reveals the mechanism of Hsc70-driven clathrin uncoating. Nat. Struct. Mol. Biol. 18:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Booth AG, Wilson MJ. 1981. Human placental coated vesicles contain receptor-bound transferrin. Biochem. J. 196:355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. 2004. Structure of the human transferrin receptor-transferrin complex. Cell 116:565–576 [DOI] [PubMed] [Google Scholar]
- 5. Collawn JF, et al. 1993. YTRF is the conserved internalization signal of the transferrin receptor, and a second YTRF signal at position 31–34 enhances endocytosis. J. Biol. Chem. 268:21686–21692 [PubMed] [Google Scholar]
- 6. Conner SD, Schmid SL. 2003. Regulated portals of entry into the cell. Nature 422:37–44 [DOI] [PubMed] [Google Scholar]
- 7. Crandell RA, Fabricant CG, Nelson-Rees WA. 1973. Development, characterization, and viral susceptibility of a feline (Felis catus) renal cell line (CRFK). In Vitro 9:176–185 [DOI] [PubMed] [Google Scholar]
- 8. Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5:e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cureton DK, Massol RH, Whelan SP, Kirchhausen T. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 6:e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ehrlich M, et al. 2004. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118:591–605 [DOI] [PubMed] [Google Scholar]
- 11. Gell C, Berndt M, Enderlein J, Diez S. 2009. TIRF microscopy evanescent field calibration using tilted fluorescent microtubules. J. Microsc. 234:38–46 [DOI] [PubMed] [Google Scholar]
- 12. Goodman LB, et al. 2010. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J. Virol. 84:4969–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green NM. 1975. Avidin. Adv. Protein Chem. 29:85–133 [DOI] [PubMed] [Google Scholar]
- 14. Hafenstein S, et al. 2007. Asymmetric binding of transferrin receptor to parvovirus capsids. Proc. Natl. Acad. Sci. U. S. A. 104:6585–6589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harbison CE, Chiorini JA, Parrish CR. 2008. The parvovirus capsid odyssey: from the cell surface to the nucleus. Trends Microbiol. 16:208–214 [DOI] [PubMed] [Google Scholar]
- 16. Harbison CE, Lyi SM, Weichert WS, Parrish CR. 2009. Early steps in cell infection by parvoviruses: host-specific differences in cell receptor binding but similar endosomal trafficking. J. Virol. 83:10504–10514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harding C, Heuser J, Stahl P. 1983. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 97:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hueffer K, Govindasamy L, Agbandje-McKenna M, Parrish CR. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 77:10099–10105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hueffer K, et al. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 77:1718–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kerssemakers JW, et al. 2006. Assembly dynamics of microtubules at molecular resolution. Nature 442:709–712 [DOI] [PubMed] [Google Scholar]
- 21. Kirchhausen T. 2009. Imaging endocytic clathrin structures in living cells. Trends Cell Biol. 19:596–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kukura P, et al. 2009. High-speed nanoscopic tracking of the position and orientation of a single virus. Nat. Methods 6:923–927 [DOI] [PubMed] [Google Scholar]
- 23. Lawrence CM, et al. 1999. Crystal structure of the ectodomain of human transferrin receptor. Science 286:779–782 [DOI] [PubMed] [Google Scholar]
- 24. Lebron JA, West AP, Jr, Bjorkman PJ. 1999. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J. Mol. Biol. 294:239–245 [DOI] [PubMed] [Google Scholar]
- 25. Liu AP, Aguet F, Danuser G, Schmid SL. 2010. Local clustering of transferrin receptors promotes clathrin-coated pit initiation. J. Cell Biol. 191:1381–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loerke D, et al. 2009. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lozach PY, et al. 2011. DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10:75–88 [DOI] [PubMed] [Google Scholar]
- 28. Massol RH, Boll W, Griffin AM, Kirchhausen T. 2006. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl. Acad. Sci. U. S. A. 103:10265–10270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McGraw TE, Greenfield L, Maxfield FR. 1987. Functional expression of the human transferrin receptor cDNA in Chinese hamster ovary cells deficient in endogenous transferrin receptor. J. Cell Biol. 105:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mercer J, Schelhaas M, Helenius A. 2010. Virus entry by endocytosis. Annu. Rev. Biochem. 79:803–833 [DOI] [PubMed] [Google Scholar]
- 31. Palermo LM, Hafenstein SL, Parrish CR. 2006. Purified feline and canine transferrin receptors reveal complex interactions with the capsids of canine and feline parvoviruses that correspond to their host ranges. J. Virol. 80:8482–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palermo LM, Hueffer K, Parrish CR. 2003. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J. Virol. 77:8915–8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parker JS, Murphy WJ, Wang D, O'Brien SJ, Parrish CR. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 75:3896–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parker JS, Parrish CR. 2000. Cellular uptake and infection by canine parvovirus involves rapid dynamin-regulated clathrin-mediated endocytosis, followed by slower intracellular trafficking. J. Virol. 74:1919–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parrish CR. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195–205 [DOI] [PubMed] [Google Scholar]
- 36. Parrish CR, Kawaoka Y. 2005. The origins of new pandemic viruses: the acquisition of new host ranges by canine parvovirus and influenza A viruses. Annu. Rev. Microbiol. 59:553–586 [DOI] [PubMed] [Google Scholar]
- 37. Qian H, Sheetz MP, Elson EL. 1991. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60:910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Radoshitzky SR, et al. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ross SR, Schofield JJ, Farr CJ, Bucan M. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. U. S. A. 99:12386–12390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rust MJ, Lakadamyali M, Zhang F, Zhuang X. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saffarian S, Cocucci E, Kirchhausen T. 2009. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 7:e1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sako Y, Kusumi A. 1995. Barriers for lateral diffusion of transferrin receptor in the plasma membrane as characterized by receptor dragging by laser tweezers: fence versus tether. J. Cell Biol. 129:1559–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sako Y, Kusumi A. 1994. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125:1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taylor MJ, Perrais D, Merrifield CJ. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Traub LM. 2009. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10:583–596 [DOI] [PubMed] [Google Scholar]
- 46. Trowbridge IS, Omary MB. 1981. Human cell surface glycoprotein related to cell proliferation is the receptor for transferrin. Proc. Natl. Acad. Sci. U. S. A. 78:3039–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tsao J, et al. 1991. The three-dimensional structure of canine parvovirus and its functional implications. Science 251:1456–1464 [DOI] [PubMed] [Google Scholar]
- 48. van der Schaar HM, et al. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4:e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ward JH, Kushner JP, Kaplan J. 1982. Transferrin receptors of human fibroblasts. Analysis of receptor properties and regulation. Biochem. J. 208:19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu H, Rossmann MG. 1993. The canine parvovirus empty capsid structure. J. Mol. Biol. 233:231–244 [DOI] [PubMed] [Google Scholar]
- 51. Yeung DE, et al. 1991. Monoclonal antibodies to the major nonstructural nuclear protein of minute virus of mice. Virology 181:35–45 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.