Abstract
The Kaposi's sarcoma-associated herpesvirus (KSHV) LANA protein functions in latently infected cells as an essential participant in KSHV genome replication and as a driver of dysregulated cell growth. To identify novel LANA protein-cell protein interactions that could contribute to these activities, we performed a proteomic screen in which purified, adenovirus-expressed Flag-LANA protein was incubated with an array displaying 4,192 nonredundant human proteins. Sixty-one interacting cell proteins were consistently detected. LANA interactions with high-mobility group AT-hook 1 (HMGA1), HMGB1, telomeric repeat binding factor 1 (TRF1), xeroderma pigmentosum complementation group A (XPA), pygopus homolog 2 (PYGO2), protein phosphatase 2A (PP2A)B subunit, Tat-interactive protein 60 (TIP60), replication protein A1 (RPA1), and RPA2 proteins were confirmed in coimmunoprecipitation assays. LANA-associated TIP60 retained acetyltransferase activity and, unlike human papillomavirus E6 and HIV-1 TAT proteins, LANA did not reduce TIP60 stability. The LANA-bound PP2A B subunit was associated with the PP2A A subunit but not the catalytic C subunit, suggesting a disruption of PP2A phosphatase activity. This is reminiscent of the role of simian virus 40 (SV40) small t antigen. Chromatin immunoprecipitation (ChIP) assays showed binding of RPA1 and RPA2 to the KSHV terminal repeats. Interestingly, LANA expression ablated RPA1 and RPA2 binding to the cell telomeric repeats. In U2OS cells that rely on the alternative mechanism for telomere maintenance, LANA expression had minimal effect on telomere length. However, LANA expression in telomerase immortalized endothelial cells resulted in telomere shortening. In KSHV-infected cells, telomere shortening may be one more mechanism by which LANA contributes to the development of malignancy.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) was originally described in association with the endothelial lesion Kaposi's sarcoma and subsequently recognized to be associated with primary effusion lymphoma and multicentric Castleman disease (35). KSHV-associated malignancies occur with increased frequency in immunocompromised individuals, such as those undergoing organ transplantation and those with AIDS, and Kaposi's sarcoma is the most common AIDS-associated malignancy worldwide (11). In KSHV-associated malignancies, the majority of the tumor cells are latently infected and express the latency proteins LANA, vCyclin, viral FLICE inhibitory protein (vFLIP), and in some cases, viral interferon regulatory factor 3 (vIRF3) (115). A small proportion of the tumor cells express KSHV lytic proteins, such as viral G-protein coupled receptor (vGPCR) and viral interleukin 6 (vIL-6), that are also believed to contribute to disease pathogenesis (36).
LANA is essential for maintaining the latent form of KSHV infection and has multiple functions that contribute to the dysregulated growth and survival properties of KSHV-infected cells. In a transgenic mouse model, LANA induces B cell hyperproliferation (27). LANA serves as an origin binding protein to recruit cellular replication machinery to the KSHV latency origin of replication located in the KSHV terminal repeats (37, 39, 43, 45, 110). LANA also interacts with cellular chromatin to tether KSHV episomal genomes during cell division and ensure partitioning of the KSHV genomes to daughter cells during cell division (24, 49). LANA regulates both viral and cell gene transcription. LANA contributes to maintenance of latent infection by repressing expression of the KSHV RTA lytic regulator and replication at the lytic origin (56, 58, 62, 85). LANA modifies cell gene expression through interactions with transcription factors (1, 9, 54, 55, 60, 61, 65, 69, 80, 86, 101, 104, 107), by mediating epigenetic silencing (8, 91), and by modulating levels of cellular microRNAs (miRNAs) (112). LANA promotes cell cycle progression by binding to pRb (80), by stabilizing and activating c-Myc (8, 61), and by increasing β-catenin-regulated gene expression (32, 33). The cell survival-promoting properties of LANA are mediated in part through interaction with p53 and blocking of p53-mediated apoptosis (16, 30, 88).
Information on the mechanisms by which LANA promotes KSHV latency, cell survival, and cell proliferation can be obtained by identifying LANA-interacting partners. Previously, yeast two-hybrid screens (34, 52, 53, 79, 107), glutathione S-transferase (GST) affinity, and immunoprecipitation assays and chromatography coupled with mass spectroscopy (6, 15, 48, 78) have been applied to the identification of LANA binding proteins. Each approach has strengths and weaknesses, and different proteins tend to be identified by the different screening approaches. We took advantage of new protein array technology to discover novel LANA-interacting proteins. In this screen, Flag-LANA purified from cells infected with a recombinant adenovirus was incubated with a microarray that displays 4,191 nonredundant human proteins, including known and predicted transcription factors and representative proteins from other functional classes (46). The screen revealed multiple previously unrecognized interacting proteins, nine of which were selected for validation in coimmunoprecipitation assays. The eight validated interactions included Tat-interactive protein 60 (TIP60), protein phosphatase 2A (PP2A), and replication protein A (RPA) proteins. Interaction with TIP60 adds LANA to the list of viral-encoded proteins interacting with this modulator of chromatin structure and key player in the DNA damage response. LANA interaction with the phosphatase PP2A disrupted the PP2A holoenzyme complex, suggesting an additional mechanism for LANA-mediated alterations in the phosphorylation status of nuclear proteins in KSHV-infected cells, and the interactions with telomere binding proteins led to the observation that LANA can impact on cellular telomere length.
MATERIALS AND METHODS
Plasmids.
pTRE-LANA was cloned by digestion of DY52 (53) with BglII and ligation into the pTRE2pur BamHI site. The TRF1, xeroderma pigmentosum complementation group A (XPA), RPA1, and pygopus homolog 2 (PYGO2) open reading frames (ORFs; Open Biosystems) were subcloned into pDEST-Flag and hemagglutinin (HA) plasmids using the Gateway system (Invitrogen). pMF24 (31), HA–high-mobility group AT-hook 1 (HMGA1) (42), HA-HMGB1 (111), V5-TIP60, and Flag-TIP60 (59) were described previously.
Cell culture.
HEK 293, HEK 293T, and U2OS Tet-On cells were grown in Dulbecco's modified Eagles medium. BC3 and BCBL1 cells were grown in RPMI 1640 medium. Medium was supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. In order to generate U2OS-TRE and U2OS-LANA, U2OS Tet-On cells (Clontech) were transfected with pTRE2pur and pTRE-LANA, respectively. Stable cells were selected with 200 μg/ml G418 and 2 μg/ml puromycin.
Adenovirus expressed 3× Flag-LANA.
A 3× Flag-LANA-expressing adenovirus vector was generated using the AdEasy XL system (Stratagene). Briefly, 3× Flag-tagged full-length LANA DNA was cloned into pShuttle-internal ribosome entry site (IRES)-humanized recombinant green fluorescent protein 1 (hrGFP-1). The pShuttle-LANA-IRES-hrGFP-1 clone was linearized by PmeI digestion and then transformed into BJ5183-AD-1 competent cells. Transformants were selected using kanamycin resistance, and recombinant clones were selected and confirmed with restriction digestion. The confirmed recombinant pAd-1-LANA DNA was amplified in XL-10-GOLD competent cells. Purified pAd-1-LANA plasmid DNA was digested with PacI and transfected into AD-293 cells that express complementing proteins required for virus assembly. The cells were cultured for 7 to 10 days and viral stocks harvested.
To prepare 3× Flag-LANA, 293 cells were infected with Ad-1-LANA virus stock in a minimal volume for 2 h. Cells were supplemented with complete medium for 42 h. Upon confirmation of GFP expression, cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, 150 mM NaCl, 0.5% sodium deoxycholate, 0.5% NP-40, 1 mM dithiothreitol [DTT], and protease inhibitors). The cell lysate was precleared by centrifugation and loaded onto a column of anti-Flag M2 affinity gel (Sigma) at 4°C. After binding took place, the column was washed 3 times with prechilled Tris-buffered saline (TBS; 25 mM Tris-HCl [pH 7.4], 3 mM KCl, and 150 mM NaCl). 3× Flag-LANA was eluted with 100 μg/ml 3× Flag peptide (Sigma). Filtration with a 10,000 nominal molecular weight limit (NMWL) Centricon filtration unit (Millipore) was used to remove free 3× Flag peptide and concentrate 3× Flag-LANA.
Microarray probing with 3× Flag-LANA.
The protocol for protein interaction on the 4,191-human-protein array was modified from that of Hu et al. (46). Briefly, the protein arrays were blocked for 1 h at 4°C in Superblock (Pierce) supplemented with 0.5% bovine serum albumin and 5% normal goat serum. 3× Flag-LANA (1 μg) was diluted in blocking buffer (75 μl) and incubated on the coverslip-covered slides at 4°C for 4 h. Slides were subjected to three 5-min washes with TBST buffer (25 mM Tris-HCl [pH 7.4], 3 mM KCl, 150 mM NaCl, 0.05% Tween 20) and then incubated with mouse LANA antibody (Vector) in blocking buffer for 2 h at 4°C. Slides were washed as described before and then incubated with Cy3-labeled goat anti-mouse antibody in Superblock for 2 h at 4°C. After three 5-min washes, slides were rinsed briefly with doubly distilled water and spun dry. Control slides were incubated with primary and secondary antibodies only. The slides were scanned at 1,800 pixels/inch using a GenePix 4000 scanner (MDS Analytical Technologies) and analyzed using Genepix 3.0 (Molecular Devices). Each protein on the array is duplicated. Positive signals were defined as those signals with Z scores for both spots at or above the 4-standard-deviation (SD) cutoff in at least two of three independent assays.
Histone acetyltransferase activity.
TIP60 histone acetyltransferase (HAT) activity was assayed using a modification of the HAT assay kit (Millipore) protocol (98).
Immunoprecipitation assays.
For immunoprecipitation assays, HEK 293T cells in 10-cm dishes were transfected with 10 μg total DNA by using calcium phosphate precipitation. A total of 48 h after transfection, cells were lysed in 1 ml lysis buffer (50 mM Tris [pH 7.9], 100 mM NaCl, 0.5 mM EDTA, 2% glycerol, and 0.2% NP-40 plus protease inhibitors [0.5 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml aprotinin, and 1 μg/μl leupeptin]), sonicated for 10 s, and cleared by centrifugation. Extracts were precleared using protein-A/G PLUS-agarose (Santa Cruz Biotechnology, Inc.) and immunoprecipitated with anti-Flag M2 agarose (Sigma) or anti-HA agarose (Sigma). Beads were washed 6 times with the same buffer, and bound proteins were detected by Western blotting. The antibodies used for Western blot analysis included mouse anti-HA (Sigma); rabbit anti-Flag (Sigma); mouse anti-Myc (Millipore); rabbit anti-PP2A A and anti-PP2A B (Cell Signaling Technology); mouse anti-PP2A C (Millipore); mouse anti-LANA (Novocastra); rabbit anti-XPA and anti-RPA2, mouse anti-RPA1, and goat anti-TRF1 (Santa Cruz Biotechnology, Inc.); and mouse anti-beta actin (Sigma) antibodies.
Chromatin immunoprecipitation assays.
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (90). ChIP assays on HEK 293 cells were performed on cells transfected with a LANA expression vector using Lipofectamine 2000 (Invitrogen). Antibodies against RPA1 were used to immunoprecipitate endogenously expressed proteins. U2OS-TRE and U2OS-LANA cells were treated with 2 μg/ml doxycycline for 48 h before ChIP analysis. Primers for the KSHV terminal repeats were CCTCTCTCTACTGTGCGAGGA (forward) and CTCCACGTAGCAAGCACTGA (reverse) and for telomeres were as published (72).
Telomere length.
Southern blotting for analysis of telomere length was performed as described previously (50). Briefly, genomic DNA was isolated using proteinase K digestion followed by phenol-chloroform extraction. Terminal DNA fragments were generated by HinfI and RsaI restriction enzyme digestion, electrophoresed through a 0.5% agarose gel, and transferred to a membrane. DNA bands were detected by hybridization with a 3′ biotin-labeled probe (CCCTAACCCTAACCCTAA) and visualized using a chemiluminescent nucleic acid detection kit (Thermo Scientific). Band intensity was measured using the ImageJ program (http://rsb.info.nih.gov).
RESULTS
Protein array screen for LANA-interacting cell proteins.
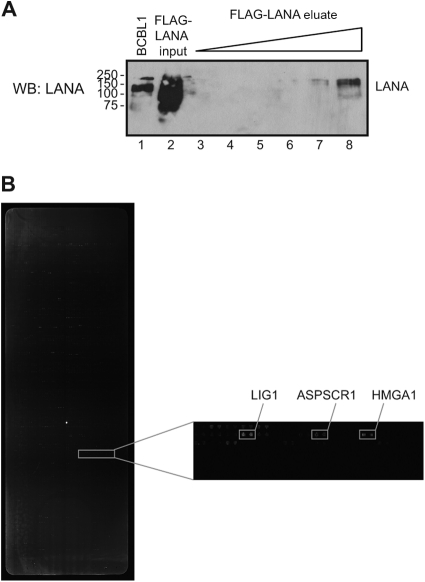
LANA was purified from HEK 293 cells infected with a recombinant adenovirus expressing 3× Flag-LANA. Cell extract was immunoprecipitated with anti-Flag M2 affinity beads, and 3× Flag-LANA was eluted from the column with 3× Flag peptide (Fig. 1A). The human protein array, displaying a total of 4,191 nonredundant human proteins comprising known and predicted transcription factors as well as representative proteins from other functional classes, has been described (46, 59). The human proteins were expressed in yeast as N-terminal GST fusions, and purified proteins were printed onto nitrocellulose-coated slides (FAST). The purified 3× Flag-LANA protein was incubated with the human protein arrays, and interactions were detected using mouse anti-LANA antibody followed by Cy3-conjugated anti-mouse IgG. The arrays were scanned and analyzed using Genepix software (Fig. 1B). Signals detected on arrays incubated with the anti-LANA mouse monoclonal antibody and Cy3-anti-mouse IgG in the absence of LANA were eliminated from further analysis. Three independent array incubations were performed, and each protein on the array was printed in duplicate. A protein was identified as a positive interactor in this screen if the signal on both paired protein spots was greater than 4 SD above the background signal and the protein was positive in at least two out of the three assays. The 61 cell proteins identified in this way are listed in Table 1 along with their functional categories. As might be expected given the known properties of LANA, the majority of the proteins identified were in the categories of replication, DNA damage/repair, transcription regulation, signal transduction, and cell cycle. There were also multiple proteins in the category of mRNA processing, an activity that does not have a documented association with LANA.
Fig 1.
Expression of adenovirus 3× Flag-LANA and detection of LANA-interacting proteins on the protein array. (A) Expression of adenovirus 3× Flag-LANA. HEK 293 cells were infected with adenovirus Ad-1-LANA, and after 42 h cells were lysed, immunoprecipitated with Flag beads, and eluted with 3× Flag peptide. Cell extract from BCBL1 cells served as the LANA control (lane 1). Input (lane 2) and increasing amounts of eluted 3× Flag-LANA, dilution 1:1,000 (lane 3), 1:250 (lane 4), 1:100 (lane 5), 1:25 (lane 6), 1:10 (lane 7), and 1:5 (lane 8) were blotted with anti-LANA antibody. (B) Detection of LANA-interacting proteins on the protein array. The protein arrays were incubated with 3× Flag-LANA, followed by mouse anti-LANA antibody and Cy3-labeled goat anti-mouse antibody. One section of the protein array is presented with the gene name for some of the positive spots.
Table 1.
LANA-interacting proteins identified in a protein array screen
| Protein | Full name | Functional annotations for proteins identified in protein array screen by categorya |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Replication (n = 10 proteins) | DNA damage/repair (n = 13 proteins) | Transcription regulation (n = 24 proteins) | Signal transduction (n = 13 proteins) | Cell cycle (n = 6 proteins) | Antiviral (n = 2 proteins) | Metabolism (n = 2 proteins) | Ribosome/mRNA processing (n = 12 proteins) | ||
| APEX1 | APEX nuclease 1 | ||||||||
| ASPSCR1 | Alveolar soft part sarcoma chromosome region, candidate 1 | ||||||||
| CDK2AP1 | Cyclin-dependent kinase 2-associated protein 1 | ||||||||
| CENTB2 | ArfGAP with coiled-coil, ankyrin repeat, and PH domains 2 | ||||||||
| CIAO1 | Cytosolic iron-sulfur protein assembly 1 | ||||||||
| CREM | cAMP-responsive element modulator | ||||||||
| ERI1/THEX1 | Exoribonuclease 1 | ||||||||
| FHL1 | Four-and-a-half LIM domains 1 | ||||||||
| GSPT1 | G-to-S phase transition 1 | ||||||||
| GTF3C2 | General transcription factor IIIC | ||||||||
| H3 | Histone 3 | ||||||||
| H4 | Histone 4 | ||||||||
| HCFC2 | Host cell factor C2 | ||||||||
| HMGA1 | High-mobility group AT-hook 1 | ||||||||
| HNRPK | Heterogeneous nuclear ribonucleoprotein K | ||||||||
| HNRPUL1 | Heterogeneous nuclear ribonucleoprotein U-like 1, E1B 55-kDa-associated protein | ||||||||
| HSFY1 | Heat shock transcription factor, Y-linked 1 | ||||||||
| ID1 | Inhibitor of DNA binding 1, dominant negative helix-loop-helix protein | ||||||||
| IL17B | Interleukin 17B | ||||||||
| IRF6 | Interferon regulatory factor 6 | ||||||||
| KAT5 | K(lysine) acetyltransferase 5, TIP60 | ||||||||
| LIG1 | Ligase I, DNA, ATP dependent | ||||||||
| NRF1 | Nuclear respiratory factor 1 | ||||||||
| NUDT21 | Nudix (nucleoside diphosphate-linked moiety X)-type motif 21 | ||||||||
| NXF3 | Nuclear RNA export factor 3 | ||||||||
| OLIG3 | Oligodendrocyte transcription factor 3 | ||||||||
| PCBP2 | Poly(rC) binding protein 2 | ||||||||
| POLB | Polymerase (DNA directed), beta | ||||||||
| PPP2R5E | Protein phosphatase 2, regulatory subunit B′, epsilon isoform | ||||||||
| PPP5C | Protein phosphatase 5, catalytic subunit | ||||||||
| PYGO2 | Pygopus homolog 2 (Drosophila) | ||||||||
| RAD51C | RAD51 homolog C (Saccharomyces cerevisiae) | ||||||||
| RAD51L3 | RAD51-like 3 (S. cerevisiae) | ||||||||
| RBPJ | Recombination signal binding protein for immunoglobulin kappa J region | ||||||||
| REEP2 | Receptor accessory protein 2 | ||||||||
| REEP5 | Receptor accessory protein 5 | ||||||||
| RGS14 | Regulator of G-protein signaling 14 | ||||||||
| RLIM | Ring finger protein, LIM domain interacting | ||||||||
| RPA1 | Replication protein A1, 70 kDa | ||||||||
| RPA2 | Replication protein A2, 32 kDa | ||||||||
| SERBP1 | SERPINE1 mRNA binding protein 1 | ||||||||
| SFRS11 | Serine/arginine-rich splicing factor 11 | ||||||||
| SMARCC2 | SWI/SNF related, matrix associated | ||||||||
| SND1 | Staphylococcal nuclease and tudor domain containing, EBNA2 coactivator p100 | ||||||||
| SNRP70 | Small nuclear ribonucleoprotein, 70 kDa | ||||||||
| SUMO1 | SMT3 suppressor of mif two 3 homolog 1 | ||||||||
| TCEAL6 | Transcription elongation factor A (SII)-like 6 | ||||||||
| TERF1 | Telomeric repeat binding factor 1 | ||||||||
| TFAP2C | Transcription factor AP-2 gamma | ||||||||
| TTF2 | Transcription termination factor, RNA polymerase II | ||||||||
| WWP1 | WW domain containing E3 ubiquitin protein ligase 1 | ||||||||
| XPA | Xeroderma pigmentosum, complementation group A | ||||||||
| ZRANB2 | Zinc finger, RAN-binding domain containing 2 | ||||||||
| DPPA2 | Developmental pluripotency associated 2 | Pluripotency | |||||||
| PPIE | Peptidylprolyl isomerase E (cyclophilin E) | PPIase | |||||||
| C5orf17 | Chromosome 5 open reading frame 17 | Unknown | |||||||
| CCDC25 | Coiled-coil domain containing 25 | Unknown | |||||||
| RBM19 | RNA binding motif protein 19 | Unknown | |||||||
| RELL2 | RELT-like 2 | Unknown | |||||||
| SLC4A1AP | Solute carrier family 4 (anion exchanger), member 1, adaptor protein | Unknown | |||||||
| ZMAT2 | Zinc finger, matrin-type 2 | Unknown | |||||||
Functional annotations were generated using the GeneCards Human Gene Database (www.genecards.org). Shading indicates functional annotations.
LANA interaction with HMGA1, HMGB1, PYGO2, and TRF1.
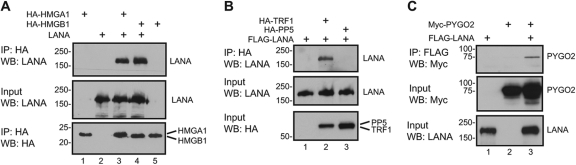
As an initial validation of selected interactions detected on the human protein array, the high mobility group protein HMGA1, the related HMGB1, the Wnt signal transducer pygopus homolog 2 (PYGO2), the telomeric repeat binding factor 1 (TRF1), and the protein phosphatase PP5 were coexpressed with LANA in cotransfected 293T cells. Coimmunoprecipitation assays were performed followed by Western blotting to detect coprecipitating proteins. This approach validated interaction between LANA and HA-HMGA1 (Fig. 2A, top, lane 3), LANA and HA-HMGB1 (Fig. 2A, top, lane 4), LANA and HA-TRF1 (Fig. 2B, top, lane 2), and LANA and Myc-PYGO2 (Fig. 2C, top, lane 3). No interaction was detected in the coprecipitation assay between LANA and HA-PP5 (Fig. 2C, lane 3, top).
Fig 2.
Validation of LANA interaction with HMGA1, HMGB1, TRF1, and PYGO2. (A) LANA interacts with HMGA1 and HMGB1. HEK 293T cells were transfected with expression vectors for HA-HMGA1 or HA-HMGB1 and LANA. Cell extracts were immunoprecipitated with anti-HA-conjugated beads and subjected to SDS-PAGE and Western blot analysis. (B) LANA interacts with TRF1. HEK 293T cells were transfected with expression vectors for Flag-LANA and HA-TRF1 or HA-PP5. Cell extracts were immunoprecipitated with anti-HA-conjugated beads. (C) LANA interacts with PYGO2. Cell extract from HEK 293T cells transfected with expression vectors for Flag-LANA and Myc-PYGO2 were immunoprecipitated with anti-Flag-conjugated beads.
LANA interaction with TIP60.
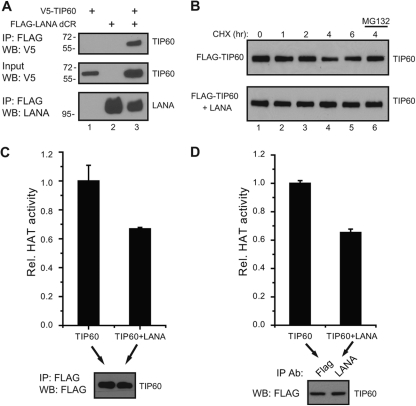
LANA interaction with the histone acetyltransferase TIP60 (also called KAT5) was validated using extracts from 293T cells cotransfected with V5-TIP60 and a Flag-LANA construction deleted of the LANA central repeats (Flag-LANA dCR). TIP60 coprecipitated with LANA in this assay (Fig. 3A, top, lane 3). Human papillomavirus (HPV) E6, HIV-1 TAT, and human cytomegalovirus (HCMV) pUL27 interact with TIP60 and induce TIP60 degradation (21, 47, 82). The effect of LANA on TIP60 turnover was examined in cells transfected with Flag-TIP60 in the presence or absence of LANA and treated with cycloheximide. Cotransfection of LANA did not decrease TIP60 stability but rather resulted in a slight stabilization (Fig. 3B).
Fig 3.
LANA interacts with TIP60 but does not decrease its stability or abolish TIP60 acetyltransferase activity. (A) LANA interacts with TIP60. Cell extract from HEK 293T cells transfected with expression vectors for Flag-LANA dCR and V5-TIP60 were immunoprecipitated with anti-Flag-conjugated beads. (B) LANA does not reduce TIP60 stability. Flag-TIP60 was transfected alone or with a LANA expression vector, and protein extracts were prepared from cells treated with 200 μg/ml cycloheximide for the indicated times or cycloheximide plus 10 μM MG132 for 4 h. (C and D) LANA does not ablate TIP60 acetyltransferase activity. Cells were transfected with Flag-TIP60 alone or Flag-TIP60 and LANA. Cell extracts were immunoprecipitated with anti-Flag antibody to directly precipitate Flag-TIP60 (C) or with anti-LANA antibody to obtain LANA-bound Flag-TIP60 (D), and Flag-TIP60 was assayed for acetyltransferase activity. The presence of equal amounts of TIP60 in the immunoprecipitates was determined by Western blotting (bottom).
HIV-1 TAT has also been reported to interfere with TIP60 acetyltransferase activity (22). To test whether LANA affected TIP60 acetyltransferase activity, an in vitro acetyltransferase assay was performed comparing Flag-TIP60 precipitated from transfected 293T cells with Flag-TIP60 precipitated from cells cotransfected with LANA (Fig. 3C). Equal amounts of Flag-TIP60 were added to the assay as determined by Western blotting (Fig. 3C, bottom). TIP60 acetyltransferase activity was only modestly decreased (68% of activity) in LANA-expressing cells. This decrease may represent an indirect effect of LANA on the cell environment. The activity of LANA-associated TIP60 was also examined by comparing Flag-TIP60 directly precipitated from singly transfected cells with the Flag-TIP60 coprecipitating with LANA in a LANA immunoprecipitate derived from dually transfected cells (Fig. 3D). Equal amounts of Flag-TIP60 were used in the in vitro assay (Fig. 3D, bottom). The Flag-TIP60 coprecipitating with LANA also retained 65% of the activity seen in the direct Flag-TIP60 immunoprecipitates. Thus, LANA does not destabilize TIP60 function and is able to recruit enzymatically active TIP60.
LANA interaction with the phosphatase PP2A.
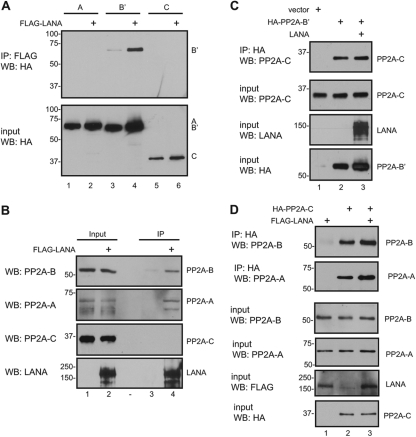
One of the novel LANA-interacting proteins identified in the protein array screen was the B′ subunit of protein phosphatase 2A (PP2A). PP2A accounts for most of the serine/threonine phosphatase activity in the cell and therefore regulates many cellular processes, including regulation of signal transduction pathways, cell cycle progression, DNA replication, gene transcription, and protein translation. PP2A exists as a core enzyme consisting of the A or scaffold subunit and the C or catalytic subunit and as a holoenzyme which contains the A and C subunits plus the B or regulatory subunit. The A subunit has two isoforms, while the B subunit comprises four families of proteins, B, B′, B′′, and B′′′, each of which has multiple isoforms (92). In the protein array screen, LANA interacted with the B′epsilon form of the B subunit. A coimmunoprecipitation assay was performed on extracts from 293T cells cotransfected with LANA and the HA-tagged A, B′, or C subunits of PP2A. Binding to LANA was seen with the B′ subunit (Fig. 4A, top, lane 4) but not with the A or C subunits (Fig. 4A, top, lanes 2 and 6). In this case, the transfected subunit was the B′ alpha form, suggesting that LANA interaction with B′ subunits is not restricted to a specific B′ variant.
Fig 4.
LANA interacts with PP2A. (A) LANA interacts with the B′ subunit of PP2A. Cell extract from HEK 293T cells transfected with expression vectors for Flag-LANA and HA-PP2A A, B′, or C subunits were immunoprecipitated with anti-Flag-conjugated beads. (B) LANA interacts with endogenous A and B but not C PP2A subunits. Extracts of Flag-LANA-transfected HEK 293T cells were immunoprecipitated with anti-Flag-conjugated beads and subjected to SDS-PAGE and Western blot analysis. Specific antibodies against PP2A B (top), PP2A A (second from top), and PP2A C (second from bottom) were used to detect endogenous PP2A proteins. Expression of LANA was detected with anti-LANA antibody (bottom). (C) LANA does not globally disrupt the PP2A complex. HEK 293T cells were transfected with expression vectors for HA-PP2A B′ and LANA. Total cell extracts were immunoprecipitated with anti-HA-conjugated beads and subjected to SDS-PAGE and Western blot analysis. Coimmunoprecipitated (top) and input (second from top) endogenous PP2A C were detected with anti-PP2A C antibody. (D) LANA does not globally disrupt the nuclear PP2A complex. HEK 293T cells were transfected with expression vectors for HA-PP2A C and LANA. Nuclear extracts were immunoprecipitated with anti-HA-conjugated beads and subjected to SDS-PAGE and Western blot analysis. Coimmunoprecipitated endogenous PP2A A and B were detected with anti-PP2A A and PP2A B antibodies, respectively.
Simian virus 40 (SV40) small t antigen binds to the A subunit of PP2A at a site that overlaps with the B subunit binding site. Small t antigen reduces B subunit binding affinity as well as inhibits the activity of the catalytic C subunit (17, 19), with the outcome being a disruption of PP2A function (18). We examined the composition of the PP2A complex associated with LANA in Flag-LANA-transfected cells. Flag-LANA coprecipitated the endogenous B and A subunits of PP2A but not the catalytic C subunit (Fig. 4B). This result suggests that LANA interaction with the B subunit prevents stable PP2A holoenzyme formation. The antibody used to detect the endogenous PP2A B subunit recognizes the B alpha form of PP2A, further suggesting that LANA binds to sequences common to the B and B′ families.
To further strengthen the model for loss of PP2A holoenzyme formation through LANA interaction with the B or B′ PP2A subunit, we examined the overall effect of LANA expression on the association of the catalytic C subunit with the other PP2A subunits. Cells were transfected with an HA-PP2A B′ subunit in the presence or absence of cotransfected LANA. The ability of the HA-PP2A B′ subunit to interact with the endogenous PP2A C subunit was not affected by expression of LANA in the cells, nor was there any change in the levels of the PP2A C subunit in the cells (Fig. 4C, lanes 2 and 3). PP2A is found in both the nucleus and the cytoplasm, while LANA is present only in the nucleus. To specifically examine the impact of LANA on PP2A holoenzyme formation in the nucleus, the transfection experiment was repeated using nuclear extracts of the transfected cells to examine the association of a transfected HA-PP2A C subunit with endogenous nuclear PP2A A and B subunits. No difference was seen in the nuclear levels of the endogenous PP2A A or B subunit in LANA-expressing cells (Fig. 4D, lanes 1 and 2), and there was no negative effect of LANA expression on the amount of the PP2A A or B subunits coprecipitating with the HA-PP2A C subunit (Fig. 4D, lanes 2 and 3). These observations are consistent with a localized LANA impairment of PP2A holoenzyme formation being mediated through binding of LANA to the PP2A B subunit rather than through any indirect or global effect on overall PP2A subunit expression or stability.
LANA interaction with XPA and RPA.
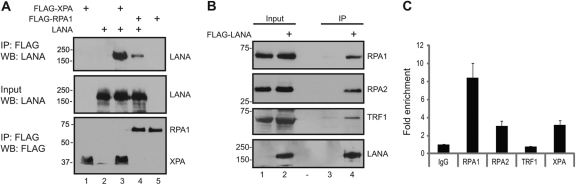
XPA functions in DNA damage recognition, where it is involved in the recruitment of nucleotide excision repair factors to sites of DNA damage. RPA is a heterotrimer of RPA1 (70 kd), RPA2 (34 kd), and a 14-kd subunit that binds single-stranded DNA. RPA is essential for homologous recombination repair of double-stranded DNA breaks and plays a supporting role in nonhomologous end joining (119). XPA interacts with RPA, and both proteins transmit DNA damage signals to cell cycle checkpoints: XPA to the G2/M checkpoint and RPA to the spindle assembly checkpoint (2, 64). RPA1 also has a critical role in telomere length maintenance (51). Interactions between LANA and XPA and between LANA and RPA1 were demonstrated by coimmunoprecipitation from extracts of cotransfected 293T cells (Fig. 5A, top, lanes 3 and 4). Coimmunoprecipitation of endogenous RPA1, RPA2, and TRF1 with transfected Flag-LANA was also observed (Fig. 5B, lane 4).
Fig 5.
RPA1 and RPA2 interact with LANA and associate with the KSHV terminal repeats. (A) LANA interacts with XPA and RPA1. Cell extract from HEK 293T cells transfected with expression vectors for Flag-XPA, Flag-RPA1, and LANA were immunoprecipitated with anti-Flag-conjugated beads. (B) LANA interacts with endogenous RPA1, RPA2, and TRF1. Extracts of Flag-LANA-transfected HEK 293T cells were immunoprecipitated with anti-Flag-conjugated beads and subjected to SDS-PAGE and Western blot analysis. Coimmunoprecipitation of endogenous RPA1 (top) RPA2 (second from top) and TRF1 (second from bottom) were detected with the respective specific antibodies. Expression of LANA was detected with anti-LANA antibody (bottom). (C) RPA1, RPA2, and XPA associate with the terminal repeats. Chromatin immunoprecipitation (ChIP) assays were performed on chromatin from BC3 cells that were immunoprecipitated with anti-RPA1, anti-RPA2, anti-TRF1, or anti-XPA antibody or control IgG. The immunoprecipitated DNA was subjected to real-time PCR using primers for the KSHV terminal repeat region. The results are presented as fold enrichment relative to control IgG. TRF1 was not associated with the terminal repeats.
Cell proteins binding to the KSHV terminal repeats that serve as the KSHV latent origin of replication have been identified individually and in screening assays (44, 73, 94, 97). However, RPA is not among those reported. To examine whether RPA binds to the terminal repeats, ChIP assays were performed using chromatin from BC3 and BCBL1 PEL cells. Binding of RPA1 and RPA2 to the terminal repeats was detected in both cell lines, and data from BC3 cells are shown in Fig. 5C. TRF1 can bind to the Epstein-Barr virus (EBV) latency origin of replication, OriP, but does not significantly affect OriP-dependent DNA replication (4, 25). TRF1 was not detected as a terminal repeat binding protein in our ChIP analysis (Fig. 5C).
LANA impairs binding of RPA1 and RPA2 to cellular telomeres.
DNA repair proteins are also essential participants in replication of the cell telomeric DNA. The initial phase of telomere replication produces single-stranded DNA that is bound by RPA and triggers an ataxia telangiectasia mutated (ATM)/ATM- and Rad3-related (ATR)-dependent DNA damage response that recruits the machinery necessary for completion of replication at the chromosome ends (102). The presence of endogenous RPA1 at the telomeres of 293 cells in the presence or absence of transfected LANA was examined by ChIP. RPA1 was readily detected on the telomeric DNA in the absence of LANA, but the ability to detect telomere-bound RPA1 was drastically decreased in LANA-transfected cells (Fig. 6A). We then examined the association of RPA1 and RPA2 with telomeric DNA in paired tet-U2OS cell lines that were stably transfected with an empty vector or with a LANA-expressing derivative and selected in puromycin. The binding of RPA1 and RPA2 to the telomeres of the two resulting cell lines, U2OS-TRE and U2OS-LANA, was compared using ChIP. As expected, RPA1 and RPA2 were readily detected binding to the telomeres of the U2OS-TRE cell line. However, there was a dramatic decrease in the ability to detect RPA1 and RPA2 binding in the U2OS-LANA cell line (Fig. 6B). The loss of telomeric binding of RPA1 and RPA2 was not due to a decrease in endogenous RPA protein levels, which were, if anything, slightly elevated in the LANA-expressing cell line (Fig. 6C).
Fig 6.
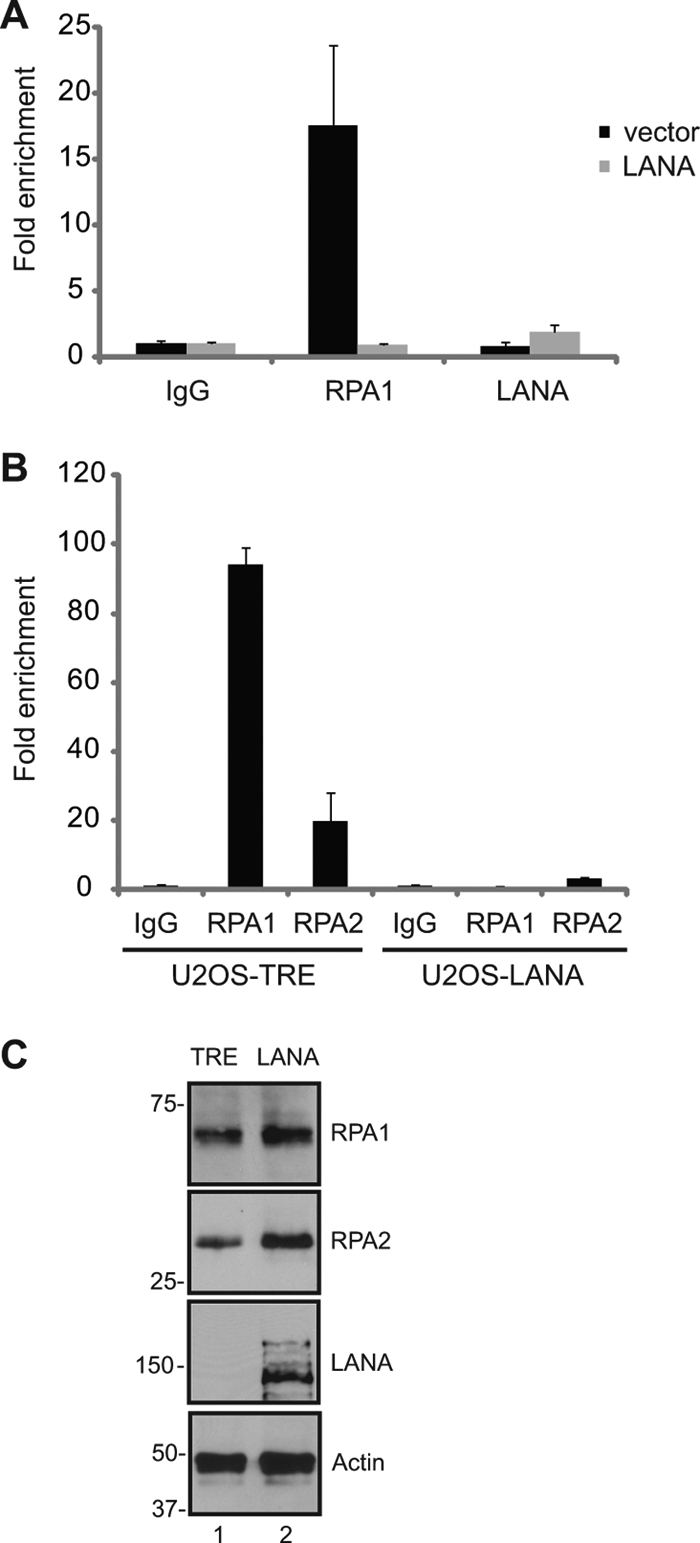
LANA disrupts the association of RPA1 and RPA2 with cellular telomeres. (A) LANA disrupts the association of RPA1 with telomeres in transfected cells. ChIP assays were performed on HEK 293 cells that were transfected with a LANA expression vector. Endogenous RPA1 was immunoprecipitated and detected by real-time PCR. Fold enrichment is relative to the IgG control. (B) LANA disrupts the association of RPA1 and RPA2 with telomeres in LANA-expressing cells. ChIP assays were performed on U2OS-TRE and U2OS-LANA cell lines. (C) Expression of RPA1, RPA2, LANA, and β-actin in U2OS-TRE and U2OS-LANA cell lines as determined by Western blotting.
LANA expression is associated with telomere shortening.
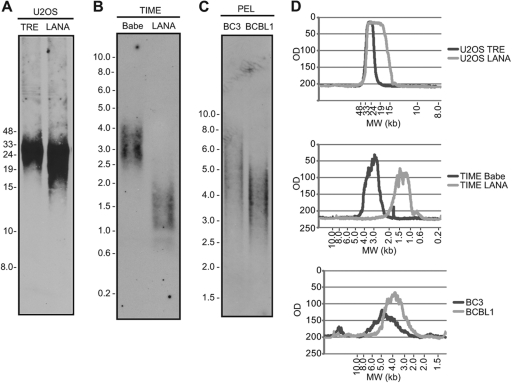
In budding yeast, cells carrying an N-terminal deletion in the yeast homolog of RPA2, Rfa2p, have impaired loading of the Est1p protein onto telomeres and have a defect in telomerase activation that results in severe telomere shortening (89). EST1A is conserved in humans (81). The diminished detection of RPA on the telomeres of cells expressing LANA raised the possibility that LANA might have an impact on telomere length. U2OS cells lack telomerase activity and maintain their telomeres by the alternative lengthening of telomeres (ALT) mechanism, which involves homologous recombination between telomere repeats (10). A Southern blot analysis was performed to compare the telomere length in vector-transformed U2OS cells and LANA-converted U2OS cells that had been passaged in parallel. The distribution of telomere length overlapped in the two cell lines, with a broadening at the lower end of the size range in the LANA-expressing cells (Fig. 7A and D). We had previously described the generation of paired telomerase immortalized microvascular endothelial (TIME) cell lines transformed with the pBabe retrovirus vector or with pBabe-LANA (91). These cell lines have also been passaged in parallel. Telomere length in the control cells (TIME-Babe) as determined by TRF Southern blot analysis is 3.0 kbp. This is in agreement with a previous report showing that telomere length in hTERT-expressing endothelial cells stabilizes at 3 to 4 kbp (114). Analysis of telomere length revealed a decrease in average telomere length from 3.0 to 1.25 kbp in LANA-expressing cells (Fig. 7B and D). Thus, LANA expression leads to a decrease in telomere length in cells dependent on telomerase activity for telomere maintenance. Analysis of telomere length in two KSHV-positive primary effusion lymphoma cell lines revealed short telomeres, 4.0 kbp in BCBL1 and 5.0 kbp in BC3 (Fig. 7C and D), compared to the published telomere length in normal B cells of 6.9 to 12.6 kbp (83). Thus, short telomeres are present in LANA-expressing endothelial cells and in KSHV-infected primary effusion lymphoma cells.
Fig 7.
LANA induces telomere shortening. Telomere length analysis of DNA from U2OS-TRE and U2OS-LANA cell lines (A), from TIME-Babe and TIME-LANA cell lines (B), and from BC3 and BCBL1 cell lines (C). Cell DNA was digested with HinfI and RsaI restriction enzymes and subjected to Southern blotting. (D) Quantitation of the size distribution of telomere DNA fragments.
DISCUSSION
Previously, we and others performed yeast two-hybrid screens to identify novel protein-protein interactions with KSHV LANA (34, 52, 53, 79, 107). The conventional two-hybrid screen, although very powerful, is based on a transcriptional readout and is therefore unsuited for identifying transcription repressors or proteins whose interaction results in degradation of the bait protein. A second approach using mass spectrometry has identified additional important LANA interactors (6, 15, 48, 103). This method preferentially identifies abundant cellular proteins, such as histones and ribosomal proteins. Here, we performed a protein array assay for LANA-interacting proteins that does not have the limitations of the previous assays but can itself lead to identification of false positives. The relatively high protein concentrations in the in vitro assay allow interactions mediated by conserved protein domains that may be restricted to specific individual proteins carrying these domains under in vivo conditions. The array assays identified 61 cell proteins, including the known LANA interactors histone H3, histone H4 (6), recombination signal binding protein for the immunoglobulin kappa J region (RBPJ)/CSL (55), EBNA2 coactivator p100 (SND1) (103), HNRPUL1 (15), and ID1, which was shown to be upregulated by LANA, although no interaction was found (101). Of the previously undocumented LANA interactors found in the screen that were selected for validation, only the serine/threonine protein phosphatase, PP5, was not validated in cotransfection assays. However, interaction with the protein phosphatase PP2A was validated. Interactions with HMGA1, HMGB1, and PYGO2 were validated but not further pursued. The high mobility group proteins HMGA1 and HMGB1 are chromatin binding proteins that change DNA conformation to allow formation of higher-order transcriptional complexes and promote transcription (84). HMGB1 can also function as an extracellular signaling molecule by binding to RAGE and Toll-like receptors to promote an inflammatory response (100). HMGB1 stimulates RTA transactivation of RTA-responsive promoters from KSHV and murine gamma herpesvirus 68 (MHV-68) (96). HMGB1 binds and synergistically upregulates the ORF50 promoter in collaboration with RTA (41). LANA interaction with HMG proteins may be relevant for both the DNA replication function and transcriptional reprogramming functions of LANA. PYGO2 is a component of the β-catenin/lymphoid-enhancing factor 1 (LEF) transcriptional activation complex required for the expression of canonical Wg/Wnt target genes. PYGO2 associates with histone-modifying methyltransferase and acetyltransferase complexes to facilitate their interaction with β-catenin (14). The interaction with PYGO2 represents a second LANA interaction with the Wnt/β-catenin pathway since LANA has been shown to increase nuclear β-catenin levels by interfering with glycogen synthase kinase 3 (GSK-3)-mediated turnover of β-catenin (33).
Several viruses encode proteins that target TIP60 for degradation. HIV-1 TAT and human papillomavirus (HPV) E6 induce TIP60 degradation to enable establishment of viral latency (21, 47). Knockdown of TIP60 also enhances establishment of an EBV latent infection (59). The HCMV UL27 protein degrades TIP60, and mutations that inhibit this activity allow the development of maribavir-resistant HCMV strains (20, 82). On the other hand, human T cell lymphotropic virus type 1 (HTLV-1) p30II stabilizes c-Myc-TIP60 containing chromatin remodeling complexes to enhance c-Myc-transforming activity (5). We found that LANA interaction with TIP60 was stabilizing rather than destabilizing. LANA was previously shown to stabilize and activate c-Myc by decreasing GSK-3-mediated phosphorylation of c-Myc Thr58 and increasing extracellular signal-regulated kinase (ERK)-mediated phosphorylation of Ser62 (8, 61). The association of LANA with enzymatically active TIP60 may be an additional mechanism enhancing c-Myc transcription function in KSHV-infected cells, since acetylation of c-Myc also increases c-Myc stability (77). TIP60 can act as a transcriptional activator or as a transcriptional repressor in different contexts. Knockdown studies suggested that TIP60 represses more genes than it activates (28). Repression is mediated through association with HDAC7 (113) and through acetylated histone H4 recruitment of bromodomain containing proteins such as Brd4 (47). Interestingly, LANA interactions with Brd4 have been previously described (74, 116). p53 is another protein that interacts with both LANA and TIP60 (30). TIP60 acetylation of p53 on lys120 selectively increases p53 transcription of proapoptotic target genes, such as BAX and PUMA (99). However, phosphorylation of TIP60 by GSK-3 is needed for this process (12), and again LANA-mediated inactivation of GSK-3 may allow interaction between TIP60 and p53 to occur without induction of proapoptotic gene expression.
The phosphatase PP2A regulates a variety of cellular processes (105). PP2A functions as a trimeric complex of the scaffold A subunit, the catalytic C subunit, and the regulatory B subunit. The B, B′, B′′, and B′′′ family of B subunits with their multiple members produce specificity for substrate recognition by the PP2A holoenzyme. The ability of LANA to interact in our assays with both B and B′ family members suggests that the LANA-mediated disruption of PP2A holoenzyme formation would impact PP2A dephosphorylation of a variety of substrates. PP2A dephosphorylates c-Myc at ser62, thus rendering c-Myc subject to degradation (3). Inactivation of PP2A by LANA would provide yet another mechanism to reinforce c-Myc stability. SV40 small tumor antigen and polyomavirus small and middle tumor antigens each associate with PP2A and displace B subunits, and this activity is essential for their cell transformation function (18). Cancer-associated mutations in the PP2A A subunit render the A subunit defective for binding B or C subunits, and expression of these mutant PP2A proteins in mice increases tumor formation, providing additional evidence that dysregulation of PP2A activity is cancer promoting (87).
The protein array screen also identified RPA1, RPA2, and XPA as LANA-interacting proteins. RPA was identified as a single-stranded DNA (ssDNA) binding protein required for replication of SV40 DNA in vitro (26, 109). RPA is a stable complex of three subunits, RPA1 (RPA70), RPA2 (RPA34), and RPA3 (RPA14), conserved in all eukaryotes (108). RPA is involved in replication, recombination, DNA damage response, and telomere maintenance. XPA interacts with the RPA1 and RPA2 subunits of the RPA complex. The association of XPA with RPA generates a double-check sensor that monitors DNA bending and unwinding to verify DNA integrity (57, 66). Coimmunoprecipitation assays confirmed the interaction of LANA with RPA1, RPA2, and XPA. Chromatin immunoprecipitation assays detected the association of RPA1, RPA2, and XPA with the KSHV terminal repeats. This is not surprising given the important role of RPA in DNA replication. In contrast, TRF1 was not detected as a terminal repeat binding protein in our ChIP analysis. LANA is essential for replication of KSHV during latency, through binding to the terminal repeats and recruitment of cellular replication proteins (37, 39, 43, 45, 97). RPA1 has been reported to interact with other viral proteins that support viral replication, such as SV40 large T antigen (76), papillomavirus E1 helicase (40), and Epstein-Barr virus (EBV) EBNA1 (118). Thus, similar to its functional homolog EBNA1 and other viral replication proteins, LANA interacts with RPA1.
The interaction of LANA with RPA and the binding of RPA to the terminal repeats prompted us to investigate the effect of LANA on telomeres. Telomeres are nucleoprotein complexes located at the extreme ends of eukaryotic chromosomes. They protect the chromosome ends from activating DNA damage signals and prevent chromosomal fusions. Expression of the aspartic acid 227 mutant of RPA1 (RPA70) leads to telomere shortening in human cells (51), similar to the phenotype of the corresponding mutation in yeast (95). Telomere shortening was observed also in Rfa2 (homolog of human RPA2) mutant yeast strains (63). As expected from a protein that regulates telomere length, both human RPA and yeast Rfa bind the telomeres. Chromatin immunoprecipitation assays detected tight association of RPA1 and RPA2 with the telomeres. This association was disrupted in the presence of LANA both in transfected HEK 293 and in LANA-expressing U2OS cells. The reduced detection of RPA on the telomeres was not due to a reduced RPA protein level in LANA-expressing cells as determined by Western blotting. Moreover, telomere length analyses in endothelial cells indicated that stable expression of LANA leads to shorter telomeres than in the matched control cells. The marked reduction of RPA binding to the telomeres in the presence of LANA may account for this telomere shortening.
Telomere length in cancer cells is shorter in high-grade tumors. Comparison of normal and tumor tissues has generally demonstrated shorter telomeres in the tumor samples (71). Small-cell lymphoma, large-cell lymphoma, and Hodgkin lymphoma exhibit significantly shortened telomeres (5.1 to 5.4 kbp) compared to those of the normal controls (6.9 to 12.6 kbp) (83). KSHV leads to the development of a rare lymphoma, primary effusion lymphoma, in immunocompromised patients. In agreement with the short telomeres in LANA-expressing cells, we found short telomeres in the two primary effusion lymphoma cell lines that we analyzed, 4.0 kbp in BCBL1 and 5.0 kbp in BC3. This finding supports the notion that the effect of LANA on telomere length is relevant to KSHV biology. In normal somatic cells, significant telomere shortening leads to p53-dependent senescence or apoptosis (23). In LANA-expressing cells, this obstacle may be overcome due to LANA's ability to block p53 function (30). Previously, it was shown that LANA can upregulate transcription from the telomerase promoter (104) and that telomerase activity is increased in KSHV-infected cells (29). A similar pattern of short telomeres despite the presence of increased telomerase activity is seen in other human tumor virus-associated cancers, such as those linked to infection with human papillomavirus (HPV), hepatitis B virus (HBV), hepatitis C virus (HCV), and human T-lymphotropic virus type 1 (HTLV-1) (67, 117).
Short telomeres become dysfunctional, and loss of telomere function can be a major mechanism for the generation of chromosomal abnormalities (38). The tight connection between telomere integrity and cancer is illustrated by the increase in tumor formation in mice with deficient telomerase activity that results in shorter telomeres (7). In yeast, the L221P missense mutation in one of the three DNA binding domains of RFA1 (homolog of human RPA1) leads to chromosomal rearrangements that resemble those frequently found in human cancer (13). In mice, introducing the corresponding L230P mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability, and cancer (106). Primary effusion lymphoma cells are prone to chromosomal instability (68, 70, 75). Chromosomal instability has also been observed upon KSHV infection of primary endothelial cells (75) and in LANA-expressing HeLa cells (93). Our finding that LANA inhibits telomere binding by RPA and enhances telomere shortening provides additional understanding of the contribution of LANA-to-chromosome instability and to the development of KSHV-associated malignancies.
ACKNOWLEDGMENTS
We thank Linda Resar for HMGA1 and HMGB1 plasmids, Victor Wong Hing Lok for generating U2OS-TRE and U2OS-LANA cell lines, Michal Zalzman for critical comments and suggestions, and Feng Chang for help with manuscript preparation.
This work was funded by NIH grant no. P01CA113239 and R21CA138163 to S.D.H., Cancer Center Core grant no. P30CA006973 to William Nelson, and grant no. GM076102 and RR020839 to H.Z.
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. An J, Sun Y, Rettig MB. 2004. Transcriptional coactivation of c-Jun by the KSHV-encoded LANA. Blood 103:222–228 [DOI] [PubMed] [Google Scholar]
- 2. Anantha RW, Borowiec JA. 2009. Mitotic crisis: the unmasking of a novel role for RPA. Cell Cycle 8:357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arnold HK, Sears RC. 2008. A tumor suppressor role for PP2A-B56alpha through negative regulation of c-Myc and other key oncoproteins. Cancer Metastasis Rev. 27:147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atanasiu C, Deng Z, Wiedmer A, Norseen J, Lieberman PM. 2006. ORC binding to TRF2 stimulates OriP replication. EMBO Rep. 7:716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Awasthi S, et al. 2005. A human T-cell lymphotropic virus type 1 enhancer of Myc transforming potential stabilizes Myc-TIP60 transcriptional interactions. Mol. Cell. Biol. 25:6178–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barbera AJ, et al. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 7. Blasco MA, et al. 1997. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91:25–34 [DOI] [PubMed] [Google Scholar]
- 8. Bubman D, Guasparri I, Cesarman E. 2007. Deregulation of c-Myc in primary effusion lymphoma by Kaposi's sarcoma herpesvirus latency-associated nuclear antigen. Oncogene 26:4979–4986 [DOI] [PubMed] [Google Scholar]
- 9. Cai Q, et al. 2006. Kaposi's sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1 alpha to upregulate RTA expression during hypoxia: latency control under low oxygen conditions. J. Virol. 80:7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cesare AJ, Reddel RR. 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 11:319–330 [DOI] [PubMed] [Google Scholar]
- 11. Cesarman E. 2011. Gammaherpesvirus and lymphoproliferative disorders in immunocompromised patients. Cancer Lett. 305:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Charvet C, et al. 2011. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol. Cell 42:584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen C, Umezu K, Kolodner RD. 1998. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol. Cell 2:9–22 [DOI] [PubMed] [Google Scholar]
- 14. Chen J, et al. 2010. Pygo2 associates with MLL2 histone methyltransferase and GCN5 histone acetyltransferase complexes to augment Wnt target gene expression and breast cancer stem-like cell expansion. Mol. Cell. Biol. 30:5621–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen W, Dittmer DP. 2011. Ribosomal protein S6 interacts with the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 85:9495–9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen W, Hilton IB, Staudt MR, Burd CE, Dittmer DP. 2010. Distinct p53, p53:LANA, and LANA complexes in Kaposi's sarcoma-associated herpesvirus lymphomas. J. Virol. 84:3898–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y, et al. 2007. Structural and biochemical insights into the regulation of protein phosphatase 2A by small T antigen of SV40. Nat. Struct. Mol. Biol. 14:527–534 [DOI] [PubMed] [Google Scholar]
- 18. Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. 2009. Cellular transformation by simian virus 40 and murine polyoma virus T antigens. Semin. Cancer Biol. 19:218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cho US, et al. 2007. Structural basis of PP2A inhibition by small T antigen. PLoS Biol. 5:e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chou S. 2009. Diverse cytomegalovirus UL27 mutations adapt to loss of viral UL97 kinase activity under maribavir. Antimicrob. Agents Chemother. 53:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Col E, et al. 2005. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 24:2634–2645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Creaven M, et al. 1999. Control of the histone-acetyltransferase activity of Tip60 by the HIV-1 transactivator protein, Tat. Biochemistry 38:8826–8830 [DOI] [PubMed] [Google Scholar]
- 23. d'Adda di Fagagna F, et al. 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426:194–198 [DOI] [PubMed] [Google Scholar]
- 24. De Leon Vazquez E, Kaye KM. 2011. The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence. J. Virol. 85:7622–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deng Z, Atanasiu C, Burg JS, Broccoli D, Lieberman PM. 2003. Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP. J. Virol. 77:11992–12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fairman MP, Stillman B. 1988. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 7:1211–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fakhari FD, Jeong JH, Kanan Y, Dittmer DP. 2006. The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J. Clin. Invest. 116:735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fazzio TG, Huff JT, Panning B. 2008. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell 134:162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Flore O, et al. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588–592 [DOI] [PubMed] [Google Scholar]
- 30. Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889–894 [DOI] [PubMed] [Google Scholar]
- 31. Fujimuro M, Hayward SD. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3beta. J. Virol. 77:8019–8030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fujimuro M, Liu J, Zhu J, Yokosawa H, Hayward SD. 2005. Regulation of the interaction between glycogen synthase kinase 3 and the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Virol. 79:10429–10441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujimuro M, et al. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300–306 [DOI] [PubMed] [Google Scholar]
- 34. Fukushi M, et al. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with human myeloid cell nuclear differentiation antigen induced by interferon alpha. Virus Genes 27:237–247 [DOI] [PubMed] [Google Scholar]
- 35. Ganem D. 2007. KSHV-induced oncogenesis. In Arvin A, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 36. Gantt S, Casper C. 2011. Human herpesvirus 8-associated neoplasms: the roles of viral replication and antiviral treatment. Curr. Opin. Infect. Dis. 24:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grundhoff A, Ganem D. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hackett JA, Feldser DM, Greider CW. 2001. Telomere dysfunction increases mutation rate and genomic instability. Cell 106:275–286 [DOI] [PubMed] [Google Scholar]
- 39. Han SJ, Hu J, Pierce B, Weng Z, Renne R. 2010. Mutational analysis of the latency-associated nuclear antigen DNA-binding domain of Kaposi's sarcoma-associated herpesvirus reveals structural conservation among gammaherpesvirus origin-binding proteins. J. Gen. Virol. 91:2203–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han Y, Loo YM, Militello KT, Melendy T. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harrison SM, Whitehouse A. 2008. Kaposi's sarcoma-associated herpesvirus (KSHV) Rta and cellular HMGB1 proteins synergistically transactivate the KSHV ORF50 promoter. FEBS Lett. 582:3080–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hillion J, et al. 2009. Upregulation of MMP-2 by HMGA1 promotes transformation in undifferentiated, large-cell lung cancer. Mol. Cancer Res. 7:1803–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu J, Garber AC, Renne R. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677–11687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu J, Liu E, Renne R. 2009. Involvement of SSRP1 in latent replication of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83:11051–11063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hu J, Renne R. 2005. Characterization of the minimal replicator of Kaposi's sarcoma-associated herpesvirus latent origin. J. Virol. 79:2637–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu S, et al. 2009. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139:610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jha S, et al. 2010. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol. Cell 38:700–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaul R, Verma SC, Robertson ES. 2007. Protein complexes associated with the Kaposi's sarcoma-associated herpesvirus-encoded LANA. Virology 364:317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kelley-Clarke B, De Leon-Vazquez E, Slain K, Barbera AJ, Kaye KM. 2009. Role of Kaposi's sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence. J. Virol. 83:4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kimura M, et al. 2010. Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 5:1596–1607 [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi Y, et al. 2010. Expression of mutant RPA in human cancer cells causes telomere shortening. Biosci. Biotechnol. Biochem. 74:382–385 [DOI] [PubMed] [Google Scholar]
- 52. Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596–11604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Krithivas A, Young DB, Liao G, Greene D, Hayward SD. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kusano S, Eizuru Y. 2010. Human I-mfa domain proteins specifically interact with KSHV LANA and affect its regulation of Wnt signaling-dependent transcription. Biochem. Biophys. Res. Commun. 396:608–613 [DOI] [PubMed] [Google Scholar]
- 55. Lan K, Kuppers DA, Robertson ES. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lan K, Kuppers DA, Verma SC, Robertson ES. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li L, Lu X, Peterson CA, Legerski RJ. 1995. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol. Cell. Biol. 15:5396–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Q, Zhou F, Ye F, Gao SJ. 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li R, et al. 2011. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe 10:390–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lim C, Gwack Y, Hwang S, Kim S, Choe J. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016–31022 [DOI] [PubMed] [Google Scholar]
- 61. Liu J, Martin HJ, Liao G, Hayward SD. 2007. The Kaposi's sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J. Virol. 81:10451–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu F, Day L, Gao SJ, Lieberman PM. 2006. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 80:5273–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mallory JC, et al. 2003. Amino acid changes in Xrs2p, Dun1p, and Rfa2p that remove the preferred targets of the ATM family of protein kinases do not affect DNA repair or telomere length in Saccharomyces cerevisiae. DNA Rep. (Amst.) 2:1041–1064 [DOI] [PubMed] [Google Scholar]
- 64. Marini F, et al. 2006. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc. Natl. Acad. Sci. U. S. A. 103:17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matsumura S, Persson LM, Wong L, Wilson AC. 2010. The latency-associated nuclear antigen interacts with MeCP2 and nucleosomes through separate domains. J. Virol. 84:2318–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Missura M, et al. 2001. Double-check probing of DNA bending and unwinding by XPA-RPA: an architectural function in DNA repair. EMBO J. 20:3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Miura N, et al. 1997. Progressive telomere shortening and telomerase reactivation during hepatocellular carcinogenesis. Cancer Genet. Cytogenet. 93:56–62 [DOI] [PubMed] [Google Scholar]
- 68. Mullaney BP, Ng VL, Herndier BG, McGrath MS, Pallavicini MG. 2000. Comparative genomic analyses of primary effusion lymphoma. Arch. Pathol. Lab. Med. 124:824–826 [DOI] [PubMed] [Google Scholar]
- 69. Muromoto R, et al. 2006. Physical and functional interactions between STAT3 and Kaposi's sarcoma-associated herpesvirus-encoded LANA. FEBS Lett. 580:93–98 [DOI] [PubMed] [Google Scholar]
- 70. Nair P, Pan H, Stallings RL, Gao SJ. 2006. Recurrent genomic imbalances in primary effusion lymphomas. Cancer Genet. Cytogenet. 171:119–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Norrback KF, Roos G. 1997. Telomeres and telomerase in normal and malignant haematopoietic cells. Eur. J. Cancer 33:774–780 [DOI] [PubMed] [Google Scholar]
- 72. O'Callaghan N, Dhillon V, Thomas P, Fenech M. 2008. A quantitative real-time PCR method for absolute telomere length. Biotechniques 44:807–809 [DOI] [PubMed] [Google Scholar]
- 73. Ohsaki E, et al. 2004. Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J. Virol. 78:9936–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ottinger M, et al. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80:10772–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pan H, Zhou F, Gao SJ. 2004. Kaposi's sarcoma-associated herpesvirus induction of chromosome instability in primary human endothelial cells. Cancer Res. 64:4064–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Park CJ, Lee JH, Choi BS. 2005. Solution structure of the DNA-binding domain of RPA from Saccharomyces cerevisiae and its interaction with single-stranded DNA and SV40 T antigen. Nucleic Acids Res. 33:4172–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Patel JH, et al. 2004. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol. Cell. Biol. 24:10826–10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Paudel N, Sadagopan S, Balasubramanian S, Chandran B. 2012. Kaposi's sarcoma associated herpesvirus (KSHV) latency associated nuclear antigen (LANA-1) and angiogenin interact with common host proteins including annexin A2 that is essential for the survival of latently infected cells. J. Virol. 86:1589–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Platt GM, Simpson GR, Mittnacht S, Schulz TF. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789–9795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Radkov SA, Kellam P, Boshoff C. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121–1127 [DOI] [PubMed] [Google Scholar]
- 81. Reichenbach P, et al. 2003. A human homolog of yeast Est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr. Biol. 13:568–574 [DOI] [PubMed] [Google Scholar]
- 82. Reitsma JM, et al. 2011. Antiviral inhibition targeting the HCMV kinase pUL97 requires pUL27-dependent degradation of Tip60 acetyltransferase and cell-cycle arrest. Cell Host Microbe 9:103–114 [DOI] [PubMed] [Google Scholar]
- 83. Remes K, et al. 2000. Telomere length and telomerase activity in malignant lymphomas at diagnosis and relapse. Br. J. Cancer 82:601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Resar LM. 2010. The high mobility group A1 gene: transforming inflammatory signals into cancer? Cancer Res. 70:436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rossetto C, Yamboliev I, Pari GS. 2009. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 K-bZIP modulates latency-associated nuclear protein-mediated suppression of lytic origin-dependent DNA synthesis. J. Virol. 83:8492–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Roupelieva M, et al. 2010. Kaposi's sarcoma-associated herpesvirus Lana-1 is a major activator of the serum response element and mitogen-activated protein kinase pathways via interactions with the Mediator complex. J. Gen. Virol. 91:1138–1149 [DOI] [PubMed] [Google Scholar]
- 87. Ruediger R, Ruiz J, Walter G. 2011. Human cancer-associated mutations in the Aα subunit of protein phosphatase 2A increase lung cancer incidence in Aα knock-in and knockout mice. Mol. Cell. Biol. 31:3832–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sarek G, et al. 2007. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J. Clin. Invest. 117:1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schramke V, et al. 2004. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat. Genet. 36:46–54 [DOI] [PubMed] [Google Scholar]
- 90. Shamay M, Greenway M, Liao G, Ambinder RF, Hayward SD. 2010. De novo DNA methyltransferase DNMT3b interacts with NEDD8-modified proteins. J. Biol. Chem. 285:36377–36386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shamay M, Krithivas A, Zhang J, Hayward SD. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554–14559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shi Y. 2009. Serine/threonine phosphatases: mechanism through structure. Cell 139:468–484 [DOI] [PubMed] [Google Scholar]
- 93. Si H, Robertson ES. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Si H, Verma SC, Robertson ES. 2006. Proteomic analysis of the Kaposi's sarcoma-associated herpesvirus terminal repeat element binding proteins. J. Virol. 80:9017–9030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Smith J, Zou H, Rothstein R. 2000. Characterization of genetic interactions with RFA1: the role of RPA in DNA replication and telomere maintenance. Biochimie 82:71–78 [DOI] [PubMed] [Google Scholar]
- 96. Song MJ, et al. 2004. The DNA architectural protein HMGB1 facilitates RTA-mediated viral gene expression in gamma-2 herpesviruses. J. Virol. 78:12940–12950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Stedman W, Deng Z, Lu F, Lieberman PM. 2004. ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78:12566–12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U. S. A. 102:13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sykes SM, et al. 2006. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 24:841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Tang D, Kang R, Zeh HJ, III, Lotze MT. 2010. High-mobility group box 1 and cancer. Biochim. Biophys. Acta 1799:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tang J, Gordon GM, Muller MG, Dahiya M, Foreman KE. 2003. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces expression of the helix-loop-helix protein Id-1 in human endothelial cells. J. Virol. 77:5975–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Verdun RE, Karlseder J. 2007. Replication and protection of telomeres. Nature 447:924–931 [DOI] [PubMed] [Google Scholar]
- 103. Verma SC, et al. 2006. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus recruits uracil DNA glycosylase 2 at the terminal repeats and is important for latent persistence of the virus. J. Virol. 80:11178–11190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Verma SC, Borah S, Robertson ES. 2004. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78:10348–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Virshup DM, Shenolikar S. 2009. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33:537–545 [DOI] [PubMed] [Google Scholar]
- 106. Wang Y, et al. 2005. Mutation in Rpa1 results in defective DNA double-strand break repair, chromosomal instability and cancer in mice. Nat. Genet. 37:750–755 [DOI] [PubMed] [Google Scholar]
- 107. Watanabe A, et al. 2007. A novel KRAB-Zinc finger protein interacts with latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus and activates transcription via terminal repeat sequences. Virus Genes 34:127–136 [DOI] [PubMed] [Google Scholar]
- 108. Wold MS. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61–92 [DOI] [PubMed] [Google Scholar]
- 109. Wold MS, Kelly T. 1988. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. U. S. A. 85:2523–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wong LY, Wilson AC. 2005. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces a strong bend on binding to terminal repeat DNA. J. Virol. 79:13829–13836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wood LJ, et al. 2000. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol. Cell. Biol. 20:5490–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Wu YH, et al. 2011. The manipulation of miRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood 118:2896–2905 [DOI] [PubMed] [Google Scholar]
- 113. Xiao H, Chung J, Kao HY, Yang YC. 2003. Tip60 is a co-repressor for STAT3. J. Biol. Chem. 278:11197–11204 [DOI] [PubMed] [Google Scholar]
- 114. Yang J, et al. 1999. Human endothelial cell life extension by telomerase expression. J. Biol. Chem. 274:26141–26148 [DOI] [PubMed] [Google Scholar]
- 115. Ye F, Lei X, Gao SJ. 2011. Mechanisms of Kaposi's sarcoma-associated herpesvirus latency and reactivation. Adv. Virol. 2011:pii=193860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. You J, et al. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zhang A, et al. 2004. Telomere attrition predominantly occurs in precursor lesions during in vivo carcinogenic process of the uterine cervix. Oncogene 23:7441–7447 [DOI] [PubMed] [Google Scholar]
- 118. Zhang D, Frappier L, Gibbs E, Hurwitz J, O'Donnell M. 1998. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 26:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zhang Y, Rohde LH, Wu H. 2009. Involvement of nucleotide excision and mismatch repair mechanisms in double-strand break repair. Curr. Genomics 10:250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]