Abstract
Maintenance of HIV latency in vitro has been linked to methylation of HIV DNA. However, examinations of the degree of methylation of HIV DNA in the latently infected, resting CD4+ T cells of infected individuals receiving antiretroviral therapy have been limited. Here, we show that methylation of the HIV 5′ long terminal repeat (LTR) in the latent viral reservoir of HIV-infected aviremic individuals receiving therapy is rare, suggesting that other mechanisms are likely involved in the persistence of viral latency.
TEXT
Although antiretroviral therapy (ART) can lower the level of plasma viremia in HIV-infected individuals to below the limit of detection (<50 copies/ml) (12), it does not eradicate the virus. In this regard, it has been demonstrated that a pool of latently infected CD4+ T cells persists in virtually all infected individuals receiving ART for prolonged durations, and the existence of such infected cells has been suggested as one of the major obstacles to achieving eradication of the virus (4, 5, 13). Therefore, understanding the mechanisms contributing to the maintenance of HIV latency is essential for the development of therapeutic strategies aimed at eradicating the virus in infected individuals receiving ART.
CpG methylation, an epigenetic transcriptional silencing mechanism critical for organismal development and cell differentiation, has been implicated in suppressing the expression of various endogenous and/or infectious retroviruses, such as human T-cell leukemia virus type 1 (8), Moloney murine leukemia virus (11), Rous sarcoma virus (6), and human endogenous retroviruses of the H, K, and W families (9, 10). Recently, CpG methylation has been proposed as an important restriction factor contributing to the maintenance and stability of HIV latency (1, 7). It has been suggested that the degree of methylation in the promoter/enhancer region, the 5′ long terminal repeat (LTR), of HIV DNA negatively correlates with the level of viral expression following stimulation of chronically infected Jurkat cell lines and in vitro infection models (1, 7). However, studies carefully evaluating the level of methylation in the 5′ LTR of HIV DNA in resting CD4+ T cells from aviremic individuals receiving effective ART have been limited thus far. We conducted the present study to address this issue.
To study the level of CpG methylation in the HIV promoter/enhancer region, we isolated resting CD4+ T cells from peripheral blood mononuclear cells of 11 infected individuals receiving ART. Study participants were receiving various antiretroviral regimens at the time of study, and all maintained undetectable levels of plasma viremia (<50 copies/ml) (Table 1). CD4+ T cells were isolated using an automated cell separation system (StemCell Technologies) followed by isolation of resting cells by depletion of CD25-, HLA-DR-, and CD69-expressing cells with phycoerythrin (PE)-conjugated antibodies (BD Biosciences) and anti-PE microbeads (Miltenyi Biotec). Genomic DNA was isolated from 2 × 106 resting CD4+ T cells (Qiagen). The purity of resting CD4+ T cells was greater than 98.5%.
Table 1.
Profile of 11 HIV-infected study participants receiving ART
| Subject identifier | Plasma viremia (copies/ml) | % CD4 | CD4 count/mm3 | % CD4, nadir | CD4 count/mm3, nadir | % CD8 | CD8 count/mm3 | Duration of ART (yr) | Antiretroviral therapy at time of studya | No. of HIV copies/106 cellsb | mCpGc (%) (5′ LTR) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | <50 | 27 | 412 | 14 | 260 | 48 | 733 | 6.6 | 3TC/AZT, TDF, LPV/r | 2,921 | 5.1 |
| 2 | <50 | 31 | 806 | 20 | 448 | 53 | 1,379 | 1.8 | ABC, 3TC, ATV | 1,139 | 4.4 |
| 3 | <50 | 30 | 499 | 14 | 294 | 38 | 632 | 4.2 | FTC, TDF, EFV | 2,419 | 10.0 |
| 4 | <50 | 27 | 742 | 9 | 318 | 54 | 1,484 | 3.6 | AZT, FTC, TDF, ATV/r | 3,946 | 1.7 |
| 5 | <50 | 28 | 639 | 26 | 559 | 57 | 1,300 | 2.1 | FTC, TDF, EFV, RAL | 1,166 | 2.2 |
| 6 | <50 | 33 | 540 | 22 | 314 | 43 | 704 | 0.9 | FTC, TDF, ATV/r | 3,196 | 0.0 |
| 7 | <50 | 37 | 823 | 34 | 916 | 50 | 1,112 | 3.9 | 3TC, AZT, FPV/r | 931 | 1.9 |
| 8 | <50 | 40 | 412 | 22 | 236 | 39 | 402 | 2.9 | ABC, 3TC, DRV/r | 905 | 2.4 |
| 9 | <50 | 41 | 500 | 22 | 264 | 22 | 268 | 5.1 | FTC, TDF, EFV | 1,930 | 3.0 |
| 10 | <50 | 18 | 274 | 8 | 86 | 40 | 609 | 2.5 | FTC, TDF, EFV | 1,146 | 1.3 |
| 11 | <50 | 53 | 1,468 | 36 | 873 | 36 | 997 | 1.9 | ABC, 3TC, ATV, RAL, MVC | 958 | 2.5 |
| Median | 31 | 540 | 22 | 314 | 43 | 733 | 2.9 | 1,166 | 2.4 |
Nucleoside reverse transcriptase inhibitors, abacavir (ABC), emtricitabine (FTC), lamivudine (3TC), tenofovir (TDF), and zidovudine (AZT). Nonnucleoside reverse transcriptase inhibitor, efavirenz (EFV). Protease inhibitors, atazanavir (ATV), atazanavir/ritonavir (ATV/r), darunavir/ritonavir (DRV/r), fosamprenavir/ritonavir (FPV/r), and lopinavir/ritonavir (LPV/r). Integrase inhibitor, raltegravir (RAL). CCR5 inhibitor, maraviroc (MVC).
Determined by real-time PCR.
mCpG, methylated CpG dinucleotide.
First, we used real-time PCR, as described previously (3), to determine the frequency of resting CD4+ T cells carrying HIV DNA in the study subjects. The median copy number of HIV DNA was 1,166 (range, 905 to 3,946) per 106 resting CD4+ T cells (Table 1).
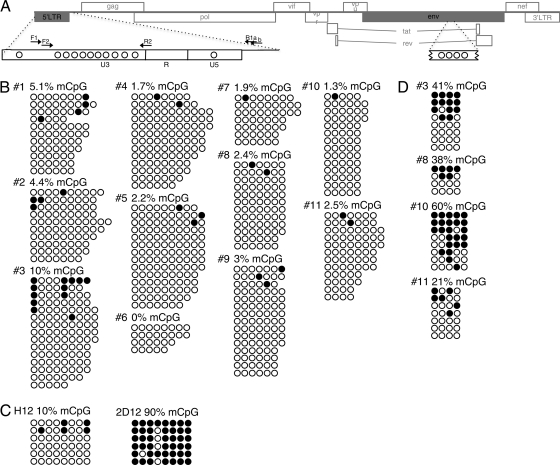
Next, we measured the level of DNA methylation within the HIV 5′ LTR (Fig. 1A) of the resting CD4+ T cells isolated from the study subjects. To quantify the frequency of methylated CpG dinucleotides, 1 μg of genomic DNA was subjected to sodium bisulfite treatment (EpiTect Plus DNA bisulfite kit; Qiagen), according to the manufacturer's instructions. Single-genome PCR amplification was conducted using primers specific for the bisulfite-converted HIV 5′ LTR: primers for sequencing F1 (5′-TAGATATTTATTGATTTTTGGATGGTG-3′) and R1a (5′-CACCCATCTCTCTCCTTCTAACCTC-3′) or R1b (5′-AAAAAACTCCTCTAATTTYHCTTTC-3′) and plasmid M13-tagged primers for sequencing F2 (5′-GTAAAACGACGGCCAGTAGTGTTAGTGTGGAGGTTTGATA-3′) and R2 (5′-GGAAACAGCTATGACCATGCAAAAAAACCCAATACAAACAAAAAAC-3′). An average of 12 nested-PCR products per study subject were sequenced, and the content of methylated CpG dinucleotides was determined. The percentage of methylated CpGs within the 217-bp region of the HIV 5′ LTR was calculated by enumerating the CpGs that were methylated and dividing this number by the number of CpG sites that could potentially be methylated, based on the consensus HIV sequence. As shown in Fig. 1B, the median frequency of methylated CpG dinucleotides within the HIV 5′ LTR was 2.4% (range, 0 to 10%). These data suggest that methylation of the HIV 5′ LTR may not play a prominent role in the maintenance of viral persistence in infected individuals receiving ART. Of note, the level of HIV 5′ LTR methylation in activated CD4+ T cells was comparable (median, 2.5%; range, 0 to 2.7%) to that of resting CD4+ T cells. To verify the ability of our methodology to detect methylated CpG dinucleotides, we used DNA from previously described Jurkat cell lines, H12 and 2D12, harboring differentially methylated HIV DNA (1). In accordance with previous data (1), we could detect hypo- and hypermethylated HIV 5′ LTR in H12 and 2D12 cell lines, respectively (Fig. 1C).
Fig 1.
Level of HIV DNA methylation in latently infected, resting CD4+ T cells of infected individuals receiving ART and Jurkat cell lines. (A) Scheme of the HIV genome detailing where CpG methylation (mCpG) was analyzed within the 5′ LTR and env regions. The circles depict the approximate distribution of CpG dinucleotides, and the arrows show the positions of primers used for amplification/sequencing. (B) Levels of CpG methylation in the bisulfite-treated HIV 5′ LTR of resting CD4+ T cells isolated from the study subjects. (C) Examination of CpG methylation in the HIV 5′ LTR of Jurkat cell lines (H12 and 2D12). (D) Levels of CpG methylation in the HIV env region of resting CD4+ T cells isolated from the study subjects. Open and closed circles represent nonmethylated and methylated CpG residues, respectively. Each line of circles represents one copy of the HIV genome.
In addition to the analysis of DNA methylation in the HIV 5′ LTR, we examined HIV envelope sequences (135 bp within the envelope coding region) comprising a CpG island (2) that does not contribute to the regulation of viral transcription. We found 21 to 60% of methylated CpG dinucleotides in HIV env from the resting CD4+ T cells of four study subjects (Fig. 1D). These data indicate that HIV DNA in resting CD4+ T cells of aviremic individuals is capable of being CpG methylated.
In the present study, we have demonstrated very low levels of methylated CpG dinucleotides within the HIV 5′ LTR of resting CD4+ T cells isolated from 11 infected individuals receiving ART. These results are in contrast with those of previous studies, in which DNA methylation analyses were performed using chronically infected Jurkat cell lines, in vitro-infected primary CD4+ T cells, and/or cells from a small number of infected individuals receiving ART (1, 7). In this regard, it is unlikely that chronically infected cell lines or in vitro-infected primary T cells could recapitulate physiologically relevant features of latently infected, resting CD4+ T cells in individuals receiving ART. Nonetheless, one study involving a small number of infected individuals receiving ART has shown relatively high levels of methylation in the HIV 5′ LTR of primary CD4+ T cells (1). It is possible that several factors, such as subtypes of CD4+ T cells, history of infection, duration and types of therapy, degrees of turnover of latently infected CD4+ T cells, and inclusion of unintegrated HIV DNA could have contributed to the contrasting results. In addition, the vast majority of infected resting CD4+ T cells carrying HIV DNA are replication defective. Although it is not technically feasible to enrich for cells carrying infectious virus, it is possible that such cells carry higher degrees of methylated HIV 5′ LTR. It is important to point out that our study utilized highly purified resting CD4+ T cells isolated from well-characterized infected individuals receiving ART. Moreover, we performed single-genome sequencing to measure the frequency of methylated CpG dinucleotides in the HIV 5′ LTR in order to eliminate any bias associated with amplifying more than one copy of HIV DNA per reaction tube.
In summary, our data demonstrate that HIV DNA carrying high levels of 5′ LTR methylation is rare in latently infected, resting CD4+ T cells of aviremic infected individuals. Thus, the latent viral reservoir is likely maintained predominantly by mechanisms other than methylation of HIV 5′ LTR.
ACKNOWLEDGMENTS
J.B. and T.-W.C. designed the research; J.B., D.M., J.S.J., and T.-W.C. performed the research; E.K.F. and A.N. recruited and monitored the study subjects; J.B., S.M., T.-W.C., and A.S.F. analyzed the data; and J.B., T.-W.C., and A.S.F. wrote the paper.
We declare no potential conflicts of interest.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Blazkova J, et al. 2009. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 5:e1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chavez L, Kauder S, Verdin E. 2011. In vivo, in vitro, and in silico analysis of methylation of the HIV-1 provirus. Methods 53:47–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chun TW, et al. 2011. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J. Infect. Dis. 204:135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chun TW, et al. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finzi D, et al. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300 [DOI] [PubMed] [Google Scholar]
- 6. Hejnar J, et al. 2001. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl. Acad. Sci. U. S. A. 98:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koiwa T, et al. 2002. 5′-Long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 76:9389–9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lavie L, Kitova M, Maldener E, Meese E, Mayer J. 2005. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2). J. Virol. 79:876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matouskova M, Blazkova J, Pajer P, Pavlicek A, Hejnar J. 2006. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell Res. 312:1011–1020 [DOI] [PubMed] [Google Scholar]
- 11. Robbins PB, et al. 1998. Consistent, persistent expression from modified retroviral vectors in murine hematopoietic stem cells. Proc. Natl. Acad. Sci. U. S. A. 95:10182–10187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Walensky RP, et al. 2006. The survival benefits of AIDS treatment in the United States. J. Infect. Dis. 194:11–19 [DOI] [PubMed] [Google Scholar]
- 13. Wong JK, et al. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295 [DOI] [PubMed] [Google Scholar]