Abstract
Despite its very narrow tropism for erythroid progenitor cells, human parvovirus B19 (B19V) has recently been shown to replicate and form infectious progeny virus in 293 cells in the presence of early adenoviral functions provided either by infection with adenovirus type 5 or by addition of the pHelper plasmid encoding the E2a, E4orf6, and VA RNA functions. In the present study we dissected the individual influence of these functions on B19V genome replication and expression of structural proteins VP1 and VP2. We show that, in the presence of the constitutively expressed E1A and E1B, E4orf6 alone is able to promote B19V DNA replication, resulting in a concomitant increase in VP expression levels. The stimulatory effects of E4orf6 require the integrity of the BC box motifs, which target cellular proteins such as p53 and the Mre11 DNA repair complex for proteosomal degradation through formation of an E3 ubiquitin ligase complex with E1B. VA RNA also strongly induces VP expression but, in contrast to E4orf6, in a replication-independent manner. This stimulation could be attributed exclusively to the VA I RNA transcript and does not involve major activating effects at the level of the B19V p6 promoter, but the nucleotide residues required for the well-defined pathway of VA I RNA mediated stimulation of translation through functional inactivation of protein kinase R. These data show that the cellular pathways regulating B19V replication may be very similar to those governing the productive cycle of the helper-dependent parvoviruses, the adeno-associated viruses.
INTRODUCTION
Human parvovirus B19 (B19V), a member of the genus Erythrovirus in the family Parvoviridae, shows a distinct tropism for erythroid progenitor cells (EPCs) in the human bone marrow, with a high level of progeny virus production in burst-forming unit erythroid cells (BFU-E) and colony-forming erythroid cells (CFU-E) cells (39, 51). Infection with B19V can result in various clinical manifestations, such as erythema infectiosum (also known as fifth disease) in children, hydrops fetalis after transplacental transmission, arthropathies (particularly observed in women), transient aplastic crisis in patients with chronic hemolytic anemia, and pure red cell aplasia due to persistent infection in immunocompromised patients (2). At least the latter two manifestations are thought to be a direct consequence of the cytopathic effects of B19V on EPCs. B19V has also been associated with acute and chronic forms of myocarditis (6). In myocarditis, endothelial cells of small intramyocardial arterioles and postcapillary venules have been identified as potential B19V target cells (25).
B19V virions with a diameter of about 25 nm are of icosahedral shape and composed of the two capsid proteins, VP1 and VP2, enclosing a single-stranded DNA genome of 5.6 kb (23). Within the genome, two major open reading frames (ORFs) are flanked by two identical inverted terminal repeats (ITRs) of 383 nucleotides (nt) each. Through palindromic sequences, the ITRs can adopt a hairpin configuration and thereby serve as origins of DNA replication (11). In addition, they also contain cis-acting sequences involved in the regulation of B19V gene expression (19, 34). B19V has only one functional promoter located at map unit 6 (p6). It initiates the transcription of a pre-mRNA, which leads to the generation of at least 12 different transcripts through alternative splicing and the use of two different polyadenylation sites (38, 59). Nonstructural protein 1 (NS1), encoded by the ORF in the left half of the genome, is translated from unspliced transcripts terminating at the proximal polyadenylation site in the central part of the genome. NS1 is essential for B19V DNA replication and, largely based on data from other parvoviruses, seems to possess site-specific DNA-binding, DNA-nicking, ATPase, and helicase activities implicated in its replicative functions (23). The ORF in the right part of the genome encodes two capsid proteins, VP1 and VP2, which are translated from spliced transcripts terminating at the distal polyadenylation site adjacent to the right ITR. VP2 is the major structural protein and accounts for ca. 95% of the total capsids (23), while the minor capsid protein VP1 is identical to VP2 except for a 227-amino-acid unique region at its amino terminus. In addition to NS1 and the two capsid proteins, additional transcripts encode at least two smaller proteins of 7.5 and 11 kDa, whose functions have only poorly been defined thus far. However, at least the 11-kDa protein seems to be critical for B19V infectivity (60).
The cellular tropism of B19V is not only controlled at the level of virus uptake through selective distribution of its primary receptor, the blood group P antigen (8), and the recently reported coreceptors integrin α5β1 (56) and KU80 antigen (35) but also by intracellular factors. In particular, low capsid protein concentrations in B19V nonpermissive cells due to a block in full-length transcription maturation (27) may severely hamper the formation of progeny virus. In nonpermissive cells, the polyadenylation of p6-initiated transcripts preferentially takes place at the proximal site, which generates unspliced transcripts encoding the NS1 protein and single-spliced transcripts encoding the 7.5-kDa protein. In contrast, efficient readthrough at the proximal polyadenylation site in favor of the distal site takes place in permissive cells, which results in high amounts of spliced mRNAs encoding the capsid proteins and the essential 11-kDa nonstructural protein. Replication of the B19V genome, either in the natural course of infection in permissive cells or forced by heterologous replication systems in nonpermissive cells, has been linked to enhanced readthrough at the proximal polyadenylation site (21) and may therefore represent a decisive factor for achieving a threshold level of capsid protein expression sufficient to promote assembly of progeny virus.
A few specialized cell lines, such as the megakaryoblastoid cell lines MB-02 (36) and UT7/Epo (49), and the human erythroid leukemia cell lines JK-1 (53) and KU812Ep6 (49) have been established for the investigation of the B19V replication cycle in vitro. They all support a low-level B19V replication in the presence of erythropoietin (Epo). Whereas in some of these semipermissive cell lines (e.g., UT7/Epo cells), conversion of the single-stranded B19V DNA genome into duplex replicative forms (RFs) has been observed at least in a subpopulation of cells, it has not been observed in others (16, 50). These findings point to the existence of additional, not-yet-characterized intracellular mechanisms of B19V restriction.
With the help of a full-length infectious B19V plasmid clone pB19-M20 (60), B19V genome replication has recently been demonstrated in the nonpermissive 293 cell line by complementation of the constitutively expressed E1A and E1B genes with the E2a, E4orf6, and VA-RNA adenoviral functions, which were expressed from the so-called pHelper plasmid (22). Furthermore, infectious B19V particles were produced in the presence of these five adenoviral functions at a level comparable to that in the semipermissive UT7/Epo-S1 cell line, as demonstrated by reinfection of ex vivo-expanded EPCs highly permissive for B19V replication. Intriguingly, the adenoviral functions required for B19V DNA replication and the production of infectious viral particles in 293 cells correspond to those identified as helper function for the productive infection cycle of the adeno-associated viruses (AAVs) belonging to the Dependovirus group of parvoviruses (4). The roles of these individual adenoviral functions in supporting AAV replication have been thoroughly characterized, and they were found not only to promote AAV DNA genome replication but also regulate AAV gene expression in a replication-independent manner at the transcriptional and posttranscriptional levels (4, 7).
The aim of our study was to elucidate the cellular mechanisms involved in adenovirus-mediated induction of productive B19V replication in otherwise nonpermissive 293 cells and to reveal possible similarities to the helper effect of adenovirus for the AAV dependoviruses. We therefore performed a detailed analysis of the influence of the individual functions encoded by the pHelper plasmid on B19V genome replication and on expression of the B19V structural proteins, since these represent the two most important determinants for the production of infectious B19V progeny virus. E4orf6 alone was found to strongly stimulate VP expression levels in a matter obviously due mainly to the induction of B19V DNA synthesis. In contrast, the effects of the VA RNA I, which was identified as a second major pHelper component capable of inducing B19V VP protein synthesis, were strictly replication independent and even partly suppressed by expression of the B19V NS1 protein. Analysis of individual E4orf6 and VA RNA I domains required for B19V transactivation revealed that the cellular pathways involved very closely resemble those, which participate in the induction of the productive cycle of helper-dependent AAV.
MATERIALS AND METHODS
Cell culture and transfection.
HEK293 (human embryonal kidney) cells were propagated in Dulbecco modified Eagle medium with GlutaMAX (Gibco-BRL, Karlsruhe, Germany) supplemented with 10% fetal calf serum (Gibco-BRL) and 100 μg of penicillin and streptomycin (Sigma, Munich, Germany)/ml at 37°C with 5% CO2. Transfections were performed by the calcium phosphate precipitate technique in 6-cm dishes with 5 × 105 cells at the day of transfection. A total of 4 μg of DNA was mixed with 150 μl of 260 mM CaCl2 and 150 μl of 2× BBS buffer (50 mM N,N-bis[2-hydroxyethyl]-2-aminoethanesulfonic acid, 280 mM NaCl, 1.5 mM Na2HPO4 [pH 6.98]), followed by incubation with the cells in 3 ml of medium for an initial incubation period of 16 h at 37°C and 5% CO2. The medium was the then replaced by fresh medium, and the cells were incubated for a further 24 h at 37°C in 5% CO2.
Plasmids.
The infectious B19V clone pB19-M20 was kindly provided by K. Brown (Centre for Infections, Health Protection Agency, London, United Kingdom) (61). The plasmid DNA was digested with SalI to obtain the linearized double-stranded B19V DNA full-length genomic fragment used for transfections. The NS1-negative B19V clone pB19-M20dNS1 has been described in detail before (41). It contains an insertion of 4 bp in the amino-terminal part of the NS1 gene, leading to a frameshift and a truncated nonfunctional protein of 213 amino acids. For transfections, it was also used in a SalI-linearized form.
Plasmid pTR-p6-Luc harboring the luciferase reporter gene under the control of the B19V p6 promoter has been described elsewhere (41). The pCATCH-NS1 plasmid for expression of an amino-terminally Flag-tagged NS1 protein under the control of the human cytomegalovirus (HCMV) early promoter/enhancer was cloned by amplification of the complete NS1 ORF from pB19-M20 with oligonucleotides generating EcoRI and XbaI restriction sites, respectively, and insertion via these sites into eukaryotic expression pCATCH (20). For the construction of plasmid pTR-p6-NS1 expressing the NS1 gene under the control of the B19V p6 promoter, the NS1 ORF was first amplified from pB19-M20 with oligonucleotides generating NcoI and XbaI restriction sites and ligated with plasmid pUC131 via these sites. The NS1 insert was then excised from the intermediate plasmid with BglII/SacI and cloned into BamHI/SacI-restricted pTR-p6-Luc to obtain the final ligation product.
The pHelper plasmid (31) containing the E2a, the E4, and the VA RNA region from adenovirus type 5 (Ad5) was obtained from Stratagene (Heidelberg, Germany). The CMV Flag-E2a plasmid for expression of a Flag-tagged Ad 5E2a protein, the CMV HA-E4orf6 wild-type plasmid for expression of HA-tagged Ad5 E4orf6, and the corresponding point mutants CMV HA-E4orf6-L122S, HA-E4orf6-L122S/C126M, HA-E4orf6-L122S/L47G, HA-E4orf6-L122S/L47G/C51V, and HA-E4orf6-R240E/R241E were all kindly provided by David Pintel (University of Missouri, Columbia, MO). The VA RNA construct with both VA RNAs I and II was constructed by cloning the corresponding SalI/ClaI fragment from pHelper into SalI/ClaI-digested Bluescript SK2 (Stratagene) vector. For the construct expressing the isolated VA RNA I, the respective SalI/NheI from pHelper was cloned into SalI/XbaI-digested Bluescript SK2 vector. The VA RNA II, contained within the neighbored NheI/ClaI fragment, was cloned into Bluescript SK2 in a similar fashion using the XbaI/ClaI restriction sites of the vector. Plasmids VA A119G, VA C120A, VA TT101/102CA, VA ΔIV, and VA RNA NES KO expressing VA RNA I mutant transcripts as described previously (37, 45) were generated by standard site-directed mutagenesis and verified by sequencing.
Extraction of viral DNA and Southern hybridization.
Extraction of viral DNA by a modified Hirt procedure was performed as described previously (55). Briefly, medium was removed from transfected cells, and the cells were washed twice with phosphate-buffered saline (PBS) in culture dishes. The cells were scraped into 2 ml of lysis buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 1% sodium dodecyl sulfate [SDS], and 200 μg of proteinase K per ml) and then incubated for 2 h at 37°C. The lysed cells were cooled on ice for 5 min, and 500 μl of 5 M NaCl was added slowly. After the precipitation of cellular DNA overnight at 4°C, samples were centrifuged at 10,000 × g for 15 min at 4°C, and the supernatant was extracted with chloroform and precipitated with ethanol in the presence of 0.3 M sodium acetate. After a washing step with 70% ethanol, the vacuum-dried DNA was resuspended in 50 μl of Tris-EDTA buffer (pH 7.6). For the selective separation of bacterial input DNA, a 4-μg sample of each DNA was digested with an excess amount of DpnI (40 U of enzyme for 2 h) and electrophoresed on a 0.8% agarose gel. Just prior to electrophoresis, the samples were digested with pancreatic RNase A (100 μg/ml for 15 min at room temperature). After depurinization, denaturation, and neutralization, the DNAs were transferred to a nylon membrane (Hybond-N; GE Healthcare) by capillary blotting in 25 mM phosphate buffer (pH 6.8) overnight. For the detection of B19 replicative DNA intermediates, a 0.94 HindIII fragment from the NS1 region encompassing B19V nt 775 to 1713 (numbering according to the B19V-J35 isolate) was used. The probe was either labeled with [32P]dCTP or, alternatively, with biotin-11-dUTP for nonradioactive labeling and detection. The nylon filters were prehybridized for 1 h at 42°C and then hybridized with 32P-labeled or biotin-11-dUTP-labeled probe in hybridization solution (7% [wt/vol] SDS, 0.125 M sodium phosphate buffer [pH 7.2], 0.25 M NaCl, 1 mM EDTA, 45% [vol/vol] formamide) at 42°C for 16 to 30 h. The filters were washed four times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 42°C for 5 min and subsequently twice with 0.1% SSC–0.1% SDS at 55°C for 30 min. Filters hybridized with the 32P-labeled probe were air dried and autoradiographed on Fuji RX films. Alternatively, the bound nonradioactive biotinylated probe was detected with Streptavidin horseradish peroxidase conjugate (High-Sensitivity HRP Conjugate; Thermo Scientific), followed by enhanced chemiluminescence (ECL) detection.
Western analysis.
For immunoblot analysis, cells from 6-cm dishes were washed twic with PBS, lysed directly in the culture dishes by the addition of 250 μl of protein sample buffer (1 mM EDTA, 50 mM Tris [pH 7.5], 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue), and boiled for 5 min. Then, 15-μl samples were loaded onto 8% polyacrylamide gels for SDS-PAGE. Separated proteins were transferred to nitrocellulose membranes by electroblotting, and membranes were blocked with 6% milk-PBS. Subsequently, the membranes were incubated with anti-B19V-VP1/2 mouse monoclonal antibody (MAB8293; Millipore, Schwalbach/Ts, Germany) diluted 1:2,000 or anti-B19V-NS1 (MAB1412; kindly provided by Susanne Modrow, Institute of Medical Microbiology Hygiene, University of Regensburg, Regensburg, Germany) diluted 1:1,000 and horseradish peroxidase-conjugated secondary antibodies, followed by ECL detection.
Quantitative B19V VP2 Western analysis corrected for β-actin expression as a loading control was performed with the Odyssey infrared imaging system (LI-COR Biosciences). As primary antibodies, a mixture of anti-B19V-VP1/2 MAB8293 (dilution, 1:2,000) and rabbit antibody ab8227 from Abcam (dilution, 1:5,000) recognizing β-actin was used. Secondary infrared fluorescence-labeled antibodies were anti-mouse IRDye680LT and anti-rabbit IRDye800CW (both at a dilution of 1:20,000). The ratios of the signal intensities for the B19V VP2 band and the β-actin band of the individual transfections are represented as relative values, with those of the control in the absence of pHelper functions usually set as 1.
Luciferase assays.
For determination of firefly luciferase activities, the cells were washed twice in PBS and scraped into 400 μl of Triton X-100 lysis buffer (1% [vol/vol] Triton X-100, 25 mM glycyl glycine [pH 7.8], 15 mM MgSo4, 4 mM EGTA, 1 mM dithiothreitol, 10% [vol/vol] glycerin), and the cell debris was removed by centrifugation. Quantization was performed with 50 μl of supernatant and 10 μl of Bright-Glo luciferase substrate (Promega GmbH, Mannheim, Germany) using a Centro LB960 luminometer (Berthold Technologies, Bad Wildbad, Germany).
RESULTS
DNA replication-dependent and -independent induction of B19V structural proteins by adenoviral functions in 293 cells.
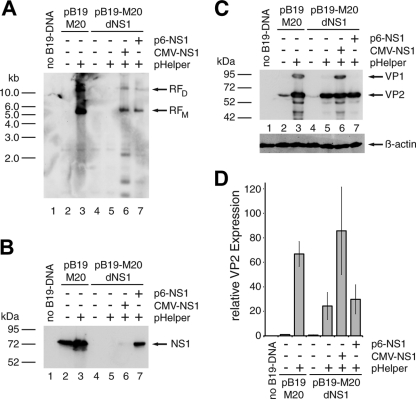
It has recently been demonstrated that the otherwise B19V nonpermissive 293 cell line, which constitutively expresses the Ad5 E1A and E1B genes, can support B19V DNA replication and the formation of infectious progeny virus in the presence of the additional Ad5 early functions E2a, E4orf6, and VA-RNA expressed from the pHelper plasmid (22). Intriguingly, this set of Ad5 functions, including E1A and E1B, also induces the productive replication cycle of the helper-dependent AAVs. In confirmation of the recent data of Guan et al., we observed large amounts of replicative B19V DNA intermediates in 293 cells after cotransfection of wild-type B19V DNA excised from the pB19-M20 infectious clone by SalI digestion, together with the pHelper plasmid (Fig. 1A, lane 3). In contrast, no DpnI-resistant B19V DNA replication intermediates were observed in the absence of the pHelper plasmid, (Fig. 1A, lane 2). In parallel, we also monitored the influence of pHelper cotransfection on abundance of the nonstructural (NS1) and structural (VP) proteins of B19V by Western blot analysis. Only a modest pHelper-induced increase in NS1 levels was observed (Fig. 1B, lanes 2 and 3). However, the abundance of B19V structural proteins was strongly stimulated by cotransfection of the pHelper plasmid, which led to the detection of large amounts of VP2 and distinct amounts of the minor capsid protein VP1, which was not detectable in the absence of pHelper (Fig. 1C, compare lane 3 to lane 2). Induction levels obtained with the pHelper plasmid were in the range of 50- to 100-fold, as determined by quantitative Western analysis through infrared fluorescence-labeled secondary antibodies (Fig. 1D). To monitor to what extent the increase in VP expression was dependent on B19V genome replication, we cotransfected the pHelper plasmid with an NS1-negative genome. As expected, no replicative DpnI-resistant DNA intermediates were formed in the absence of NS1 (Fig. 1A, lanes 4 and 5). Nevertheless, we could monitor a strong pHelper-dependent increase in VP expression levels (Fig. 1C, lanes 4 and 5, and Fig. 1D). Formation of replicative intermediates from the NS1-negative genome could be restored by complementation with NS1 expression constructs, driven either by the heterologous HCMV promoter (Fig. 1A, lane 6) or the isolated B19V p6 promoter (Fig. 1A, lane 7). Both NS1 constructs showed lower NS1 levels resulting in less induction of B19V DNA replication compared to the complete wild-type genome. Interestingly, NS1 expression from the isolated p6 promoter was higher than from the strong HCMV promoter, arguing for a high p6 promoter activity in 293 cells. At least the HCMV-driven construct also further enhanced VP protein levels (Fig. 1C, lane 6, and Fig. 1D). The lack of a positive effect for the p6-driven NS1 construct, which stimulated DNA replication to a similar extent as the HCMV-p6 construct, may be due to competition for transcription factors binding to the p6 promoter in pB19-M20 (i.e., a “squelching effect”). These initial results strongly suggested that functions of the pHelper plasmid were able to stimulate the expression of the B19V structural proteins in 293 cells both in a replication-dependent and a replication-independent manner.
Fig 1.
Adenoviral functions induce both B19V genome replication and structural protein expression in 293 cells. 293 cells were transfected with 1 μg of SalI-linearized wild-type (pB19-M20) or NS1-negative (pB19-M20dNS1) B19V plasmid DNA. Early adenoviral functions and the B19V NS1 protein were supplied by cotransfection of 1 μg of pHelper plasmid and 0.5 μg of HCMV- or p6-driven NS1 constructs as indicated. The total amount of DNA in each transfection was adjusted to 4 μg with plasmid pCATCH containing the HCMV promoter without any downstream coding sequences. As a negative control and for monitoring the transfection efficiency, cells were transfected with 4 μg of the yellow fluorescent protein (YFP) expressing plasmid pYFP-C2 (Clontech). (A) Southern blot hybridization of DpnI-digested Hirt DNA with a biotin-11-dUTP-labeled hybridization probe from the B19V NS1 region. Arrows indicate putative monomeric (RFM) and dimeric (RFD) replicative B19V intermediates. (B and C) Western blot analysis of whole-cell extracts with the monoclonal antibodies MAB1424 recognizing the NS1 protein (B) and MAB8293 (both from Millipore) recognizing the B19V capsid proteins (C). Arrows indicate the positions of the NS1 protein and the VP1 and VP2 capsid proteins, respectively. (D) Quantitative B19V VP Western analysis with infrared fluorescence-labeled secondary antibodies as described in Materials and Methods. The VP2 expression data were normalized for the β-actin loading control and are represented as relative values, with the wild-type B19V DNA in the absence of pHelper set as 1. The bars represent arithmetic means ± the standard deviations (SD) of two independent experiments with the corresponding B19V VP ECL analysis and the β-actin control for one of these experiments shown in panel C.
E4orf6 is the decisive function required for induction of B19V DNA synthesis in 293 cells.
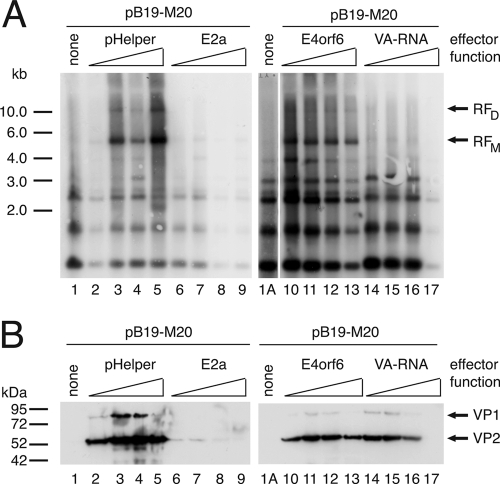
To more closely examine the mechanisms involved in the induction of B19V DNA replication and structural protein expression by adenoviral functions, we first monitored the influence of the individual gene products contained within the pHelper plasmid. Ad5 E2a or E4orf6 expressed as Flag- or HA-tagged proteins, respectively, under the control of the HCMV promoter or a VA RNA construct encoding both the VA RNA I and VA RNA II transcript in the context of their authentic promoters were individually cotransfected with SalI-linearized pB19-M20 wild-type DNA into 293 cells as described above. Since the individual adenoviral functions may possibly exhibit both positive and negative regulatory effects depending on their exact expression level, a wide range of effector concentrations ranging from 25 ng to 1 μg of cotransfected plasmid per 6-cm dish were used. For all transfections, the total amount of DNA was adjusted with a plasmid harboring the HCMV promoter without any downstream coding sequences. The Ad5 single-stranded DNA-binding protein E2a, which is essential for adenoviral genome replication, had no stimulatory effect on B19V DNA replication or gene expression at any concentration (Fig. 2A and B, lanes 6 to 9). In contrast, the E4orf6 protein strongly stimulated the appearance of B19V RFs already at the lowest effector concentration (Fig. 2A, lane 10), with a slightly reduced stimulation reproducibly observed at the highest concentrations used (Fig. 2A, lane 13). The increase in B19V DNA replication was accompanied by a marked increase in VP expression levels, which exhibited a maximum at intermediary E4orf6 concentrations (Fig. 2B, lanes 10 to 13). The isolated E4orf6 function showed similar maximum levels of induction of B19V DNA replication as the complete pHelper plasmid (Fig. 1A and B, lanes 2 to 5), although E4orf6 was more effective at lower plasmid concentrations. This may be due to expression of E4orf6 from the very strong HCMV promoter versus the authentic adenoviral promoter in the pHelper plasmid. With regard to VP expression levels, the complete pHelper plasmid showed a clearly stronger induction than E4orf6 alone (Fig. 2B, compare lanes 2 to 5 to lanes 10 to 13). These observations immediately suggested that, although E4orf6 may be sufficient to promote B19V replication in concert with the constitutively expressed E1A and E1B genes, additional pHelper functions are involved in the induction of capsid protein expression. In line with these considerations, the VA RNA, despite its inability to promote B19V DNA replication (Fig. 2A, lanes 14 to 17), showed a strong induction of VP expression (Fig. 2B, lanes 14 to 17). This induction was especially evident at low concentrations and sharply declined with higher VA RNA concentrations (Fig. 2B, lane 17). Thus, depending on its abundance, the VA RNA may exhibit both positive and negative effects on B19V VP expression levels.
Fig 2.
E4orf6 is required for induction of B19V DNA synthesis in 293 cells. 293 cells were cotransfected with 1 μg of SalI-linearized pB19-M20 DNA and increasing amounts (25 ng, 100 ng, 250 ng, and 1 μg) of either pHelper or the isolated pHelper functions E2a, E4orf6, or VA RNA as indicated. Adjustment of the total amount of DNA to 4 μg was performed with the pCATCH plasmid. (A) Southern blot detection of DpnI-resistant B19V replicative intermediates essentially as described in the legend to Fig. 1A, except that a [32P]dCTP-labeled hybridization probe was used. (B) Immunoblot analysis for B19V VP expression with the monoclonal antibody MAB8293, as described for Fig. 1B.
VA RNA-mediated induction of B19V structural protein expression is strictly replication independent.
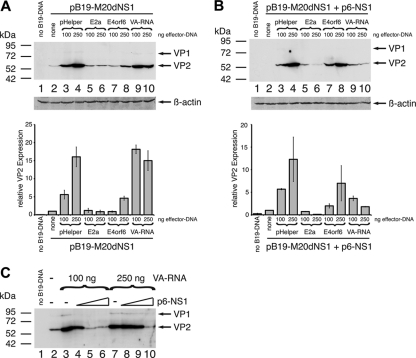
To more directly assess the dependency of E4orf6 and VA RNA-mediated induction of B19V VP expression on genome replication, the individual pHelper function were cotransfected with the SalI-linearized B19V NS1-negative genome in the absence or presence of complementing NS1 protein. Based on the results obtained with the NS1-positive genome, two intermediary effector concentrations of 100 and 250 ng of cotransfected plasmid DNA were chosen for the analysis. In agreement with the results of the initial experiments (compare with Fig. 1), the pHelper construct was able to stimulate B19V VP expression from the NS1-negative genome at both concentrations (Fig. 3A, lanes 3 and 4). E4orf6, in contrast, induced a minor increase in VP levels in the absence of NS1 only at the higher effector concentration (Fig. 3A, lanes 7 and 8). In accordance with a replication-independent mode of action, the VA RNA stimulated the NS1-negative B19V genome already in the absence of complementing NS1 protein with activation levels comparable to those of the complete pHelper plasmid (Fig. 3A, compare lanes 9 and 10 to lanes 3 and 4, respectively). For E4orf6, activation of VP expression could be restored by complementation with the p6-NS1 expression construct (Fig. 3B, lanes 7 and 8), which further corroborates a replication-dependent mechanism of E4orf6-mediated VP induction. A rather unexpected finding was that in, case of the VA RNA, complementation with NS1 not only failed to further stimulate B19V structural protein expression but contrarily even reduced VA RNA-mediated activation (compare Fig. 1A and B, lanes 9 and 10). These results identified the VA RNA as a strong activator of B19V VP expression and additionally implied the existence of a negative functional cross talk between the VA RNA and the NS1 protein. To more closely examine a possible interference of NS1 with the VA-RNA-mediated stimulation of VP expression, increasing amounts of the p6-driven NS1 construct were cotransfected with the NS1-negative B19V genome in the presence of two constant amounts of the VA RNA construct, 100 and 250 ng. As observed previously, the cotransfection of VA RNA alone strongly increased VP expression (Fig. 3C, compare lanes 3 and 7, respectively, to lane 2). However, activation was abrogated with increasing amounts of NS1 at both VA RNA concentrations.
Fig 3.
VA RNA activates B19V capsid protein expression independent of DNA replication. 293 cells were cotransfected with 1 μg of SalI-linearized NS1-negative B19V plasmid pB19-M20dNS1 and then evaluated for different early adenoviral functions and a p6-driven NS1 expression construct as indicated. An adjustment of the total amount of DNA to 4 μg for each transfection was performed with pCATCH plasmid DNA. Plasmid pYFP-C2 expressing the YFP was transfected as a negative control. Whole-cell extracts were subjected to B19V capsid protein immunoblot analysis with MAB8293 as described for Fig. 1B. (A and B) Two different concentrations (100 and 250 ng) of either pHelper or the isolated pHelper functions E2a, E4orf6, or VA RNA were cotransfected either in the absence (A) or presence (B) of 1 μg of complementing p6-NS1 construct. The lower panels show the results of the quantitative B19V VP Western analysis performed in parallel, which was normalized for the β-actin expression shown at the bottom of the upper panels. The bars represent arithmetic means ± the SD of two independent experiments. (C) Cells were cotransfected with increasing amounts of p6-NS1 construct (0.5, 1.0, and 2.0 μg) in the presence of two constant amounts (100 and 250 ng) of VA RNA.
E4orf6-mediated induction of B19V DNA synthesis in 293 cells requires functional domains involved in the formation of a cullin 5-containing E3 ligase complex.
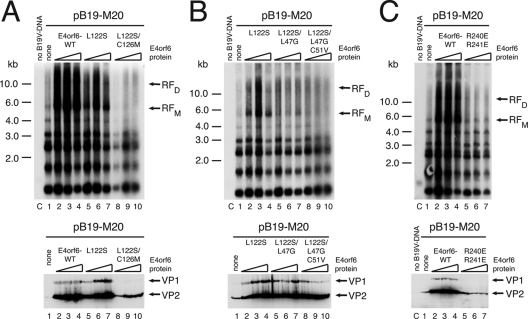
In the course of the adenoviral life cycle, the E4orf6 protein plays important roles in the regulation of cellular and viral RNA export and the modification of cellular DNA repair pathways. An important mechanism involved in several of these E4orf6 functions is the formation of an E3 ubiquitin ligase complex, together with the adenoviral E1B-55k protein and cellular components, which include cullin 5 and elongins B and C (42). The Mre11 DNA repair complex (52) and p53 (43) have been identified as cellular targets for the E3 ligase complex. Formation of the functional complex requires direct interaction of E4orf6 with elongins B and C via two so-called BC box motifs within E4orf6. The BC box motifs have been extensively characterized by a number of point mutants (5), and it has been shown that single-amino-acid mutations in either BC box 1 (L47G) or BC box 2 (L122S) lead to only a very modest impairment of p53 and Mre11 degradation. Degradation of Mre11, however, is almost completely abrogated when double mutations are introduced into BC box 1 (L47G/C51V) or 2 (L122S/C126M), whereas p53 degradation can be abolished by combined point mutations in both BC boxes (5). We used an analogous set of E4orf6 point mutants to investigate a possible involvement of the E3 ligase activity of E4orf6 in stimulating B19V DNA replication. Whereas only a very modest reduction of E4orf6-induced B19V DNA replication was observed for the L122S single mutant in BC box 2, we found the corresponding double mutant (L122S/C126M) to be completely deficient for induction of B19V DNA synthesis (Fig. 4A). The replication-inducing properties of the L122S single mutant could also be reduced by additional point mutations in BC box 1. Changing leucine to glycine at position 47 strongly reduced, and an additional point mutation of cysteine residue 51 to valine completely abolished the appearance of B19V RFs (Fig. 4B). As could be expected from the preceding experiments, B19V VP expression levels mostly paralleled the amount of B19V replication intermediates (Fig. 4A and B, lower panels) with the minor exception of the L122S/L47G/C51V triple mutant, which still showed clearly detectable levels of VP protein induction despite no effects on DNA replication. In addition to the BC box mutants, we also examined an E4orf6 mutant shown to lack the nuclear retention signal due to substitution of two essential arginine residues by glutamate (R240E/R241E). This mutant is unable to induce the degradation of p53 (44). As demonstrated in Fig. 4C, retention of E4orf6 in the nucleus was required for both the induction of B19V DNA synthesis and the activation of B19V VP expression. In summary, nuclear degradation of one or several cellular target proteins for the E4orf6/E1B-55k E3 ligase complex seems to comprise an important aspect of the stimulatory effects of E4orf6 on B19V DNA replication in 293 cells.
Fig 4.
E4orf6-mediated induction of B19V DNA synthesis requires the BC box motifs involved in formation of a cullin 5-based ubiquitin E3 ligase complex. 293 cells were cotransfected with 1 μg of SalI-linearized pB19-M20 DNA and increasing concentrations (25, 100, and 250 ng) of wild-type E4orf6 or constructs with various point mutations in the E4orf6 BC box motifs, which are required for the formation of a functional E3 ligase complex. The upper panels show the Southern blot detection of DpnI-resistant B19V replicative intermediates after isolation of Hirt DNA's as described for Fig. 2A, whereas the immunoblot analysis for B19V VP expression with MAB8293 as described for Fig. 1B is displayed in the lower panels. For the lanes marked “no B19V-DNA,” 4 μg of pYFP-C2 was transfected as a negative control.
Activation of B19V VP expression by VA RNA mainly involves VA RNA I and is probably mediated by its interaction with protein kinase R (PKR).
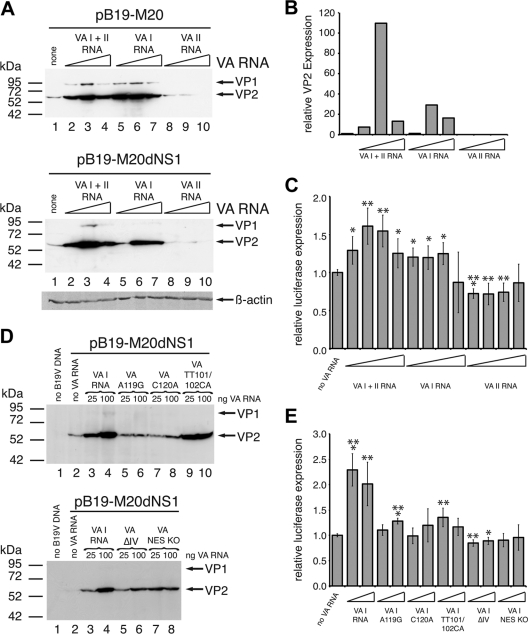
For a more detailed characterization of the VA RNA functions involved in activation of B19V gene expression, we first examined the effect of increasing concentrations of the individual VA I and II RNA genes on capsid protein levels from both a wild-type and an NS1-negative B19V genome. For the wild-type genome, the VA I RNA alone displayed very similar stimulatory effects as the construct containing the complete VA RNA region (Fig. 5A, upper panel, compare lanes 5 to 7 to lanes 2 to 4), whereas the VA II RNA was not able to upregulate VP expression (Fig. 5A, upper panel, lanes 8 to 10). It rather resulted in a further reduction of the already low levels of capsid proteins obtained in the absence of VA RNA. An almost identical result was obtained with the NS1-negative B19V genome, except that the VA I RNA displayed slightly reduced induction levels compared to joint expression of VA I and II RNAs (Fig. 5A lower panel, compare lanes 5 to 7 with lanes 2 to 4, a quantitative Western analysis for the NS1-negative B19V genome is shown in Fig. 5B). Thus, the VA I RNA accounts for most, if not all, of the stimulatory effects of the complete VA RNA region. To monitor possible effects of VA RNA on B19V gene expression at the transcriptional level, a reporter construct harboring the only functional B19V promoter p6 in front of the luciferase gene was cotransfected with increasing amounts of the individual VA RNA constructs. Considerable p6 promoter activities almost in the range of very strong constitutive viral promoters such as the HCMV early promoter/enhancer were observed in 293 cells already in the absence of VA RNA or other pHelper functions (data not shown). This high basal activity could only be stimulated to a minor extent by addition of either the complete VA RNA construct or the isolated VA I RNA with maximum activation levels in the range of 1.7-fold (Fig. 5C). The VA II RNA showed no stimulatory effect. These data clearly indicate that the overwhelming part of VA I RNA-induced activation of B19V VP protein expression is not mediated at the level of promoter activity. In the course of adenovirus infection, VA I RNA has been shown to be required for efficient translation of late mRNAs (54) by inhibiting the double-stranded RNA-mediated activation and autophosphorylation of the PKR (58). To examine the possible involvement of the PKR pathway in the VA I RNA-mediated activation of B19V VP expression, a set of well-characterized VA I RNA point mutants first described by Rahman et al. (45) was generated, and the individual constructs were cotransfected in two different concentrations with the NS1-negative B19V genome. The single nucleotide exchanges A119G and C120A in the PKR binding domain of VA I RNA, shown previously to largely abrogate the inhibition of PKR activation and autophosphorylation (45), also strongly compromised B19V VP activation (Fig. 5D, compare lanes 5 to 8 to lanes 3 and 4). VA TT101/102CA represents a double point mutant, which despite changes of two nucleotide residues in the central VA RNA domain retains the ability to inhibit PKR activation. Intriguingly, this point mutant showed a similar activation of VP expression as the wild-type VA RNA (Fig. 5D, lanes 9 and 10). Other VA RNA mutants, either unable to bind PKR because of deletion of parts of the central domain (VA ΔI) or defective for transport to the cytoplasm (VA NES KO), also showed clearly reduced levels of VP induction (Fig. 5D, lower panel). As expected from the results with the wild-type VA I RNA, the VA I RNA point mutants were also unable to efficiently stimulate the isolated B19V p6 promoter (Fig. 5E). In summary, VA I RNA-mediated induction of B19V VP expression seems to critically depend on nucleotide residues that have been shown to be required for the binding and functional inactivation of PKR. The minor transcriptional effects observed might be caused indirectly by alterations in the synthesis of transcription factors required for p6 promoter activity.
Fig 5.
VA RNA-mediated stimulation of B19V capsid protein depends on the interaction of VA I RNA with protein kinase R (PKR). (A and D) 293 cells were cotransfected with 1 μg of either pB19-M20 plasmid DNA or pB19-M20dNS1 plasmid DNA in SalI-linearized form as indicated and either increasing concentrations (25, 100, and 250 ng) of constructs encoding either both VA I and VA II RNA or the individual VA RNAs (A) or two different concentrations (25 and 100 ng) of constructs encoding wild-type VA I RNA or various VA I RNA mutants as indicated (D). Plasmid pYFP-C2 expressing the YFP was transfected as a negative control in panel D. (B) Quantitative B19V VP Western analysis of the protein extracts from the experiment with the NS1-negative B19V genome shown in the lower part of panel A with the β-actin loading control shown below the B19V VP ECL detection. (C and E) 293 cells were cotransfected with low amounts (20 ng) of a p6-luciferase reporter construct and either four different concentrations (25 ng, 100 ng, 250 ng, and 1 μg) of constructs encoding either both VA I and VA II RNA or the individual VA RNAs as indicated (C) or two different concentrations (100 and 250 ng) of wild-type VA I RNA or various VA I RNA mutants (E). Total DNA amounts were adjusted to 4 μg with the Bluescript SK2 vector. At 40 h posttransfection, the luciferase activities were determined, and these are represented as relative values with the respective activities in the absence of VA RNA set as 1. The data are presented as means ± the SD. P values of the statistical significances for the changes in luciferase activity compared to those in the absence of VA RNA are indicated above individual bars (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In the present study we dissected the roles of individual adenoviral functions in promoting B19V genome replication and structural gene expression in the otherwise nonpermissive human embryonal kidney cell line 293, which constitutively expresses the Ad5 E1A and E1B genes. E4orf6 is the sole additional component necessary to enable robust B19V DNA replication, which also results in a markedly enhanced VP expression levels. In contrast to E4orf6, the VA RNA stimulates VP expression in a replication-independent manner. For both E4orf6 and the VA RNA, the cellular pathways likely to be involved in the activation could be identified by point mutants. No role in stimulation of B19V DNA replication or structural gene expression could be assigned to the E2a function.
Already at low concentrations the E4orf6 function could account for the full magnitude of B19V genome replication observed in the presence of pHelper. In contrast, neither the Ad5 single-stranded DNA-binding protein E2a nor the VA RNA had a detectable positive effect on B19V replication. Several lines of evidence strongly indicate that the increased VP expression in the presence of E4orf6 is a direct consequence of genome replication. Major positive effects of E4orf6 on structural protein expression were only observed with an NS1-positive B19V genome or after complementation of the NS1-negative genome with NS1 expression plasmids. Furthermore, E4orf6 mutants unable to stimulate genome replication also failed to induce VP expression. The finding that, in comparison to VP, the expression of the B19V NS1 protein was induced to a much lesser by the pHelper plasmid argue against template amplification as the main reason for the induction but favor a specific link between B19V genome replication and structural gene expression. It has already been demonstrated that B19V genome replication is the key determinant for the production of full-length B19V transcripts encoding the structural proteins by favoring transcript polyadenylation at the distal site in the B19V genome in both permissive and nonpermissive cells (21). It has not been determined, however, whether B19V genome replication increases B19V RNA splicing, which in turn precludes polyadenylation at the internal site, or whether the enhanced readthrough at the internal polyadenylation site is a direct consequence of DNA replication. As a third possibility, splicing and polyadenylation may be modulated by a general increase in the abundance of primary B19V transcripts, since both processes can be stimulated by the carboxy-terminal domain of RNA polymerase II (9, 14).
The experiments performed with a series of well-defined E4orf6 point mutants strongly suggest that the main mechanism for E4orf6-mediated induction of B19V genome replication is the formation of a cullin 5-based E3 ubiquitin ligase complex (42). Further components of this complex are the adenoviral E1B-55k protein constitutively expressed in 293 cells, the cellular adapter proteins elongins B and C, and the ring finger protein Rbx1. The two BC box motifs of E4orf6 centered around amino acids 47 and 122, respectively, are both required for efficient assembly of the E3 ligase complex and the degradation of two of its cellular substrates, the p53 tumor suppressor protein and the Mre11 DNA repair complex (MRN) (5). A more recently identified potential third substrate for the E3 ligase complex is the DNA ligase IV (3). In particular, the point mutations in the BC box motifs, which had been shown to abrogate MRN degradation, also failed to induce B19V replication. MRN is composed of Mre11, Rad50, and Nbs1 and is one of the major players in sensing, signaling, and repair of cellular DNA damage (12). Inactivation of MRN by E1B-55k/E4orf6 is an important step in productive adenovirus infection (52). Intriguingly, degradation of MRN also promotes the genome replication of the helper-dependent AAV-2 parvovirus and the transduction with AAV-2-based recombinant vectors by enhancing second-strand-synthesis (47). MRN recruits ATM (for ataxia telangiectasia mutated), one of the three phosphatidylinositol 3-kinase-like kinases involved in DNA damage response (DDR), to double-stranded DNA breaks (10, 26). A very recent study addressed the involvement of the DDR in B19V replication in ex vivo-expanded erythroid progenitor cells (29). It was found that B19V infection induced phosphorylation of all upstream kinases of DNA repair pathways, namely, ATM, ATR (ATM and Rad3 related), and DNA-PKcs (DNA-dependent protein kinase catalytic subunit), which also localized to B19V replication centers. In contrast to the situation observed for other members of the autonomous parvoviruses (1, 28, 46), however, treatment with kinase inhibitors revealed requirements for ATR and DNA-PKcs, but not for activated ATM, in the promotion of viral replication and the phosphorylation of downstream substrates such as H2AX and RPA32 (29). The ATM/MRN pathway may therefore be either constitutively inactive in EPCs or inactivated by B19V without the requirement for further exogenous factors. The precise role of the ATM-dependent DDR in parvoviral replication may critically depend not only on the specific cell system under investigation but also on the exact nature of the viral DNA evoking the cellular response. Interestingly, within the Parvoviridae, only B19V and the AAV contain identical ITRs at both ends of their genome, and the B19V genome replication in 293 cells seems to proceed by very similar molecular mechanisms as those described for AAV (22). In summary, one can hypothesize that, similar to AAV, B19V actually relies on inactivation of the ATM-MRN pathway for its genome replication, a function fulfilled in 293 cells by the E4orf6/E1B-55k E3 ligase complex through the degradation of the MRN complex. Studies planned for the future will address this hypothesis.
The adenoviral VA RNA was able to stimulate B19V capsid protein expression levels in a replication-independent manner, and this effect could be clearly attributed to VA I RNA. One has to keep in mind, however, that these studies were performed with infectious B19V double-stranded DNA and not with B19V virions harboring the genome in a single-stranded DNA form. Thus, with B19V virions the VA I RNA may well depend on the initial DNA replication step of converting the incoming single-stranded virion DNA to a transcription-competent double-stranded form to exert its positive effect on VP expression levels. We also tried to determine whether the positive effects of VA I RNA on B19V VP expression require the E1A and E1B function constitutively expressed in 293 cells. However, in other B19V nonpermissive cell lines such as HeLa cells, the B19V VP expression level was generally below the Western blot detection limit, even after adenovirus infection or cotransfection of the E1A/E1B/pHelper set of adenoviral helper functions (data not shown). In immunofluorescence analysis, cotransfection of E1A led to the appearance of very occasional VP-positive cells, in line with data from the endothelial EA.hy926 cell line (41), but VA RNA did not further increase the number of positive cells (data also not shown). Therefore, a high p6 promoter activity, as present in 293 cells (40), may be prerequisite for the detection of the posttranscriptional effects of VA RNA. The known property of VA I RNA to stimulate translation by inhibiting the double-stranded RNA-induced activation of PKR through competition for the RNA-binding domain of PKR (32) seems to be absolutely necessary to promote B19V VP expression, as we demonstrated with a set of previously characterized point mutants (45). The PKR pathway represents an important cellular defense mechanism to inhibit productive replication of a variety of different viruses (17, 18). Double-stranded RNA formed in the early phase of the viral replication cycle induces autophosphorylation of PKR, leading to subsequent phosphorylation of the translation initiation factor eukaryotic translation initiation factor 2α (eIF2α) (58). The phosphorylated form of eIF2α is not able to participate in successive rounds of translation, since it blocks the function of the GDP-GPT exchange factor by formation of a stable complex (30). The requirement for the PKR binding properties of VA I RNA implicates that activation of B19V VP expression is mainly mediated at the level of translation. This conclusion is further supported by the results obtained in the reporter gene assays, where VA I RNA did not significantly stimulate transcription from the p6. An enhancement of viral protein accumulation at the translational level by VA RNA I has also been demonstrated for the helper-dependent AAV-5 (37). Furthermore, it has been shown that a short region of the AAV-5 capsid RNA leader sequence can induce the phosphorylation of eIF2α, and similar observations were made for AAV-2 and goat-derived AAV capsid expression units (37). The low levels of capsid proteins in the absence of additional adenoviral functions, despite the considerable activity of the B19V p6 promoter in 293 cells in the reporter gene assay, suggest that similar negative autoregulatory mechanisms as for AAV may govern B19V capsid levels. Not much is known yet about the PKR phosphorylation status in EPCs, the natural host cells for B19V replication. However, a recent study showed that gamma interferon and tumor necrosis factor alpha, which are potent inhibitors of hematopoiesis, induce a rapid PKR phosphorylation in erythroid CFU-E progenitor cells and that inhibition of PKR activity increases hematopoietic colony formation (48). Based on our results, such conditions may also support VP expression after B19V infection.
By including an NS1-negative B19V genome in our analysis, we were not only able to separate replication-dependent and replication-independent effects of the different pHelper functions but also assessed the role of NS1 in the regulation of B19V structural gene expression in 293 cells. NS1 is known to be essential for B19V genome replication, and several potential NS1 binding sites in the B19V-ITRs involved in this activity have been identified (22). These NS1 binding sites have also been implicated in replication-independent stimulation of the p6 promoter in stable HeLa cells after inducible expression of the NS1 protein (19). An NS1-mediated transactivation of the p6 has also been found in earlier studies in other B19V nonpermissive cells (13). However, our results rather argue against a major positive influence of NS1 on B19V capsid gene expression separate from its essential function in the promotion of B19V DNA replication. First, we found no dramatic difference between the capsid levels from the NS1-negative and the wild-type B19V genome in the absence of pHelper functions despite a robust NS1 expression from the wild-type genome. Second, complementing the NS1-negative genome with an NS1 protein expressed from its physiological promoter rather reduced capsid protein levels in the presence of the VA RNA. In contrast to the studies performed in other nonpermissive cells, we did not observe a clear NS1-mediated transactivation of the p6 promoter in 293 cells (data not shown). Although this may due to the high basal activity of the p6 promoter caused by constitutive expression of the E1A protein in 293 cells (40), we have also been unable to score a significant NS1-induced transactivation of the p6 promoter in endothelial cells, despite a low basal p6 activity (41). Thus, NS1 may not generally act as a positive regulator of B19V structural gene expression but may also exhibit negative effects under conditions, where B19V genome replication is blocked. Interestingly, activation of capsid gene expression from the p40 promoter of the helper-dependent AAV-2 by the corresponding nonstructural proteins Rep78 and Rep68 in the presence of helper virus also depends to a large degree on cis-acting sequences required for AAV genome replication (55).
In summary, B19V genome replication, high-level capsid protein expression, and the formation of substantial amounts of progeny virus (22) in normally nonpermissive cells is not only supported by the same set of adenoviral helper functions as the productive infection cycle of the AAV dependoviruses, but these individual functions also seem to fulfill similar roles and act by similar mechanisms for both genera. In the case of AAV, the roles of the individual adenoviral gene products in productive infection have been extensively characterized, and they have been shown to mainly function in an indirect matter by regulating the abundance, the modification state, and the subcellular localization of cellular factors important for AAV propagation (4). For example, it has been shown that AAV-2 replicates in the nuclear replication compartments established by infection with either adenovirus (57) or the second very effective AAV helper virus, herpes simplex virus 1 (24). AAV-2 then uses the cellular replication factors and factors involved in RNA transcription and processing recruited to these centers for its own propagation. Interestingly, the genome replication of the autonomous minute virus of mice (MVM) is increased strongly by Ad2 infection in HeLa cells (15) accompanied by a recompartmentalization of the MVM DNA from the nucleolus to the adenoviral replication centers (15, 33). In contrast to the situation with B19V, however, 293 cells are able to replicate MVM DNA quite efficiently in the absence of adenovirus, and no extra stimulation is observed by providing additional adenoviral functions, although recompartmentalization of the MVM DNA is also observed (15). Whether B19V genome replication in EPCs takes place in specialized replication centers remains to be determined, but it seems possible that in the presence of adenovirus the genome is recruited to the corresponding adenoviral replication centers.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft within the framework of the Sonderforschungsbereich/Transregio 19.
We thank Susanne Modrow (University of Regensburg, Regensburg, Germany), Kevin Brown (Health Protection Agency, United Kingdom), and David Pintel (University of Missouri-Columbia) for providing antibodies and plasmids used in this study. We also thank Eva Hammer for expert technical support.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. 2010. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog. 6:e1001147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson MJ, et al. 1985. Experimental parvoviral infection in humans. J. Infect. Dis. 152:257–265 [DOI] [PubMed] [Google Scholar]
- 3. Baker A, Rohleder KJ, Hanakahi LA, Ketner G. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berns KI, Giraud C. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1–23 [DOI] [PubMed] [Google Scholar]
- 5. Blanchette P, et al. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bock CT, Klingel K, Kandolf R. 2010. Human parvovirus B19-associated myocarditis. N. Engl. J. Med. 362:1248–1249 [DOI] [PubMed] [Google Scholar]
- 7. Bowles DJ, Rabinowitz JE, Samulski RJ. 2006. The genus Dependovirus, p 15–24 In Kerr J, et al. (ed), Parvoviruses. Hodder Arnold, London, United Kingdom [Google Scholar]
- 8. Brown KE, Anderson SM, Young NS. 1993. Erythrocyte P antigen: cellular receptor for B19 parvovirus. Science 262:114–117 [DOI] [PubMed] [Google Scholar]
- 9. Calvo O, Manley JL. 2003. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 17:1321–1327 [DOI] [PubMed] [Google Scholar]
- 10. Carson CT, et al. 2003. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 22:6610–6620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cotmore SF, Tattersall P. 2005. Structure and organization of the viral genome, p 73–94 In Kerr J, et al. (ed), Parvoviruses. Hodder Arnold, London, United Kingdom [Google Scholar]
- 12. D'Amours D, Jackson SP. 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell. Biol. 3:317–327 [DOI] [PubMed] [Google Scholar]
- 13. Doerig C, Hirt B, Antonietti JP, Beard P. 1990. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J. Virol. 64:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fong N, Bentley DL. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fox E, Moen PT, Jr, Bodnar JW. 1990. Replication of minute virus of mice DNA in adenovirus-infected or adenovirus-transformed cells. Virology 176:403–412 [DOI] [PubMed] [Google Scholar]
- 16. Gallinella G, et al. 2000. Different patterns of restriction to B19 parvovirus replication in human blast cell lines. Virology 278:361–367 [DOI] [PubMed] [Google Scholar]
- 17. Garcia MA, et al. 2006. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol. Mol. Biol. Rev. 70:1032–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811 [DOI] [PubMed] [Google Scholar]
- 19. Gareus R, et al. 1998. Characterization of cis-acting and NS1 protein-responsive elements in the p6 promoter of parvovirus B19. J. Virol. 72:609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Georgiev O, et al. 1996. Two versatile eukaryotic vectors permitting epitope tagging, radiolabeling, and nuclear localization of expressed proteins. Gene 168:165–167 [DOI] [PubMed] [Google Scholar]
- 21. Guan W, et al. 2008. Block to the production of full-length B19 virus transcripts by internal polyadenylation is overcome by replication of the viral genome. J. Virol. 82:9951–9963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus B19 can replicate in nonpermissive cells with the help of adenovirus genes and produces infectious virus. J. Virol. 83:9541–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heegaard ED, Brown KE. 2002. Human parvovirus B19. Clin. Microbiol. Rev. 15:485–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heilbronn R, et al. 2003. ssDNA-dependent colocalization of adeno-associated virus Rep. and herpes simplex virus ICP8 in nuclear replication domains. Nucleic Acids Res. 31:6206–6213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klingel K, et al. 2004. Molecular pathology of inflammatory cardiomyopathy. Med. Microbiol. Immunol. 193:101–107 [DOI] [PubMed] [Google Scholar]
- 26. Lee JH, Paull TT. 2004. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science 304:93–96 [DOI] [PubMed] [Google Scholar]
- 27. Liu JM, Green SW, Shimada T, Young NS. 1992. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J. Virol. 66:4686–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luo Y, Chen AY, Qiu J. 2011. Bocavirus infection induces a DNA damage response that facilitates viral DNA replication and mediates cell death. J. Virol. 85:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo Y, et al. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J. Virol. 85:8046–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsushita T, et al. 1998. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 5:938–945 [DOI] [PubMed] [Google Scholar]
- 32. McKenna SA, Kim I, Liu CW, Puglisi JD. 2006. Uncoupling of RNA binding and PKR kinase activation by viral inhibitor RNAs. J. Mol. Biol. 358:1270–1285 [DOI] [PubMed] [Google Scholar]
- 33. Moen PT, Jr, Fox E, Bodnar JW. 1990. Adenovirus and minute virus of mice DNAs are localized at the nuclear periphery. Nucleic Acids Res. 18:513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Momoeda M, et al. 1994. The transcriptional regulator YY1 binds to the 5′-terminal region of B19 parvovirus and regulates P6 promoter activity. J. Virol. 68:7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munakata Y, et al. 2005. Ku80 autoantigen as a cellular coreceptor for human parvovirus B19 infection. Blood 106:3449–3456 [DOI] [PubMed] [Google Scholar]
- 36. Munshi NC, Zhou S, Woody MJ, Morgan DA, Srivastava A. 1993. Successful replication of parvovirus B19 in the human megakaryocytic leukemia cell line MB-02. J. Virol. 67:562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nayak R, Pintel DJ. 2007. Adeno-associated viruses can induce phosphorylation of eIF2α via PKR activation, which can be overcome by helper adenovirus type 5 virus-associated RNA. J. Virol. 81:11908–11916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ozawa K, et al. 1987. Novel transcription map for the B19 (human) pathogenic parvovirus. J. Virol. 61:2395–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozawa K, Kurtzman G, Young N. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233:883–886 [DOI] [PubMed] [Google Scholar]
- 40. Ponnazhagan S, Woody MJ, Wang XS, Zhou SZ, Srivastava A. 1995. Transcriptional transactivation of parvovirus B19 promoters in nonpermissive human cells by adenovirus type 2. J. Virol. 69:8096–8101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pozzuto T, et al. 2011. Transactivation of human parvovirus B19 gene expression in endothelial cells by adenoviral helper functions. Virology 411:50–64 [DOI] [PubMed] [Google Scholar]
- 42. Querido E, et al. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Querido E, et al. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Querido E, et al. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rahman A, Malhotra P, Dhar R, Kewalramani T, Thimmapaya B. 1995. Effect of single-base substitutions in the central domain of virus-associated RNA I on its function. J. Virol. 69:4299–4307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruiz Z, Mihaylov IS, Cotmore SF, Tattersall P. 2011. Recruitment of DNA replication and damage response proteins to viral replication centers during infection with NS2 mutants of minute virus of mice (MVM). Virology 410:375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schwartz RA, et al. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 81:12936–12945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharma B, Altman JK, Goussetis DJ, Verma AK, Platanias LC. 2011. Protein kinase R as mediator of the effects of interferon (IFN) gamma and tumor necrosis factor (TNF) alpha on normal and dysplastic hematopoiesis. J. Biol. Chem. 286:27506–27514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimomura S, et al. 1992. First continuous propagation of B19 parvovirus in a cell line. Blood 79:18–24 [PubMed] [Google Scholar]
- 50. Srivastava A, et al. 1990. Parvovirus B19-induced perturbation of human megakaryocytopoiesis in vitro. Blood 76:1997–2004 [PubMed] [Google Scholar]
- 51. Srivastava A, Lu L. 1988. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J. Virol. 62:3059–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 53. Takahashi T, et al. 1993. DNA replication of parvovirus B 19 in a human erythroid leukemia cell line (JK-1) in vitro. Arch. Virol. 131:201–208 [DOI] [PubMed] [Google Scholar]
- 54. Thimmappaya B, Weinberger C, Schneider RJ, Shenk T. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31:543–551 [DOI] [PubMed] [Google Scholar]
- 55. Weger S, Wistuba A, Grimm D, Kleinschmidt JA. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep. proteins. J. Virol. 71:8437–8447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Weigel-Kelley KA, Yoder MC, Srivastava A. 2003. Alpha5beta1 integrin as a cellular coreceptor for human parvovirus B19: requirement of functional activation of beta1 integrin for viral entry. Blood 102:3927–3933 [DOI] [PubMed] [Google Scholar]
- 57. Weitzman MD, Fisher KJ, Wilson JM. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Williams BR. 1999. PKR; a sentinel kinase for cellular stress. Oncogene 18:6112–6120 [DOI] [PubMed] [Google Scholar]
- 59. Yoto Y, Qiu J, Pintel DJ. 2006. Identification and characterization of two internal cleavage and polyadenylation sites of parvovirus B19 RNA. J. Virol. 80:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhi N, et al. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J. Virol. 80:5941–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhi N, Zadori Z, Brown KE, Tijssen P. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142–152 [DOI] [PubMed] [Google Scholar]