Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is closely associated with several malignancies, including Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. KSHV can establish lifelong latency in the host, but the mechanism is not fully understood. Previous studies have proposed a feedback model in which the viral replication and transcription activator (RTA) can induce the expression of the latency-associated nuclear antigen (LANA) during early infection. LANA, in turn, represses transcription and RTA function to establish and maintain KSHV latency. The interaction between LANA and the recombination signal sequence binding protein Jκ (RBP-Jκ, also called CSL), a major transcriptional repressor of the Notch signaling pathway, is essential for RTA repression. In the present study, we show that the LANA carboxyl-terminal amino acids 1052 to 1082 are responsible for the LANA interaction with RBP-Jκ. The secondary structure of the LANA carboxyl terminus resembles the RBP-Jκ-associated module (RAM) of Notch receptor. Furthermore, deletion of the region of LANA residues 1052 to 1082 resulted in aberrant expression of RTA, leading to elevated viral lytic replication. For the first time, we dissected a conserved RBP-Jκ binding domain in LANA and demonstrated that this domain was indispensable for LANA-mediated repression of KSHV lytic genes, thus helping the virus maintain latency and control viral reactivation.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, was identified in Kaposi's sarcoma (KS) lesions in 1994 and is classified into the gammaherpesvirus family, which also includes Epstein-Barr virus (EBV) and herpesvirus saimiri (6). In addition to KS, which is a common neoplasm in HIV-1-infected individuals, KSHV is also associated with primary effusion lymphoma (PEL) and multicentric Castleman's disease, causing significant morbidity and mortality (4, 39, 41, 44). KSHV is a large, double-stranded DNA virus whose genome is a 140-kb unique coding region flanked by multiple GC-rich terminal repeats. More than 90 open reading frames (ORFs) are present in the long unique region (38).
Like other herpesviruses, KSHV has two alternative life cycle programs: latent and lytic infection. During latency, only a small number of viral genes are expressed, and no infectious progeny are produced. Under specific conditions, KSHV can undergo lytic replication in which most viral genes are expressed in an ordered cascade, infectious virions are released, and host cells die. Both latent and lytic infections are important to KS pathogenesis (30, 34, 40). Following primary infection, the default pathway of KSHV infection is latency. However, the mechanism by which KSHV establishes latency is not fully understood. Previous studies have shown that during early infection, the viral replication and transcription activator (RTA) can induce latency-associated nuclear antigen (LANA) expression by activating the LANA promoter. LANA represses the RTA promoter to inhibit its expression and interacts with RTA to antagonize its function. During de novo infection, RTA transcription correlates with LANA expression (21, 22). These findings suggest that RTA contributes to the establishment of KSHV latency by activating LANA expression in the early stages of infection. This describes a feedback mechanism by which LANA and RTA regulate each other in the establishment of KSHV latency.
The recombination signal sequence binding protein Jκ (RBP-Jκ) is the major downstream transcription factor of the Notch signaling pathway and acts as either a transcriptional repressor or an activator in different situations (16, 20, 31, 33). RBP-Jκ plays important roles in KSHV life cycle control. RBP-Jκ is critical to viral lytic replication since in RBP-Jκ-null murine fibroblasts, KSHV reactivation is dramatically inhibited (25). Extensive studies have shown that the induction of numerous viral lytic promoters such as ORF57, ORF59, and ORF47 by RTA depends on RBP-Jκ binding (5, 24, 27). RBP-Jκ is also involved in the establishment of KSHV latency. The activation of the LANA promoter by RTA is RBP-Jκ dependent (21). A recent study by Lu, et al. shows that genetic disruption of the RBP-Jκ binding site within the LANA promoter leads to enhanced lytic replication during de novo infection of human peripheral blood mononuclear cells (28). In addition, LANA physically interacts with RBP-Jκ to repress the RTA promoter and competes with RTA for binding to the RBP-Jκ cognate sequence, which is believed to be important for maintaining KSHV latency (21).
In addition to KSHV, RBP-Jκ is also involved in transcriptional regulation of other tumor viruses. In EBV, Epstein-Barr virus nuclear antigen 2 (EBNA2) interacts with RBP-Jκ to transcriptionally activate responsive elements on viral promoters, which is essential for EBV transformation (12, 46). EBNA2 uses a highly conserved PPWWPP motif to interact with RBP-Jκ (15). This feature is also observed in the murine protein KyoT2, for negative regulation of transcription in mammalian systems (46). The related sequence WXP is found in the RBP-Jκ-binding region of all Notch family members. This region, termed the RBP-Jκ-associated module (RAM), is less than 50 amino acids (aa) in length and is immediately downstream of the Notch transmembrane domain (45). Unlike EBNA2, KyoT2, and Notch, the KSHV lytic protein RTA interacts with RBP-Jκ but lacks this conserved motif. In RTA, the smallest fragment capable of binding RBP-Jκ is a centrally located region of amino acids 170 to 270, which contains a leucine-rich repeat region (24).
Previous studies showed that the LANA carboxyl terminus was important for interaction with RBP-Jκ, but the precise amino acid sequence of LANA responsible for RBP-Jκ binding was not fully characterized (21). Thus, we wanted to identify the key amino acids of LANA that interact with RBP-Jκ. Since interaction between LANA and RBP-Jκ is essential for LANA-mediated RTA repression (21), we hypothesize that disruption of the interaction between LANA and RBP-Jκ may destroy KSHV latency. We used a KSHV bacterial artificial chromosome that does not express LANA (BAC36ΔLANA) and LANA constructs to reconstitute a recombinant virus in human 293T cells and found that LANA with a deletion of amino acids 1052 to 1082 [LANAΔ(1052-1082)], which constitute the RBP-Jκ binding region, was defective in repressing RTA expression, leading to elevated viral lytic replication and virion production. These results further confirmed the important role of RBP-Jκ in maintaining KSHV latency. Our study also implied a potential target for disruption of KSHV latency which could be used for oncolytic viral therapy.
MATERIALS AND METHODS
Constructs, antibodies, and cell lines.
The KSHV LANA expression plasmid pA3M-LANA, described previously (47), was used as a PCR template to construct a series of C-terminal LANA truncations. PCR primers are listed in Table 1. The PCR products were subcloned into vector pGEX4T-1 (GE Health Care, Buckinghamshire, United Kingdom) at the EcoRI and XhoI sites. The LANA truncated constructs, schematically shown in Fig. 1A, were as follows: glutathione S-transferase fused to a fragment of LANA consisting of amino acids 1022 to 1162 [GST-LANA(1022–1162)], GST-LANA(1052–1162), GST-LANA(1082–1162), GST-LANA(1112–1162), and GST-LANA(1142–1162). These constructs were used in GST pulldown assays.
Table 1.
Primers for PCR amplification and analysis
| Primer name | Sequence |
|---|---|
| GST-LANA1022aa-1162aa F | CGGAATCAGACAGATAGATGATTGTCCT |
| GST-LANA1052aa-1162aa F | CGGAATCTGTCAATGGAAGTTTGCAGTG |
| GST-LANA1082aa-1162aa F | CGGAATCGCAGGCCCCGTGTCCTGCTTG |
| GST-LANA1112aa-1162aa F | CGGAATCACAAGTAAGAAAGTACAAATG |
| GST-LANA1142aa-1162aa F | CGGAATCAAGCCCCTGCCATTAACCCAG |
| GST-LANA1142aa-1162aa R | CGCTCGAGTTATGTCATTTCCTGTGGAG |
| GST-LANAW1054AW1056A F | AAGACGAGATCCAAAGTGTCAAGCGAAGTTTGCAGTGATTTTTTGG |
| GST-LANAW1054AW1056A R | TGGAAGACGAGATCCAAAGTGTCAATGGAAGTTTGCAGTGATTTTTTGGGGC |
| pCAGGS-LANA F | CGGAATTCATGGCGCCCCCGGGAATGCGC |
| pCAGGS-LANA R | CGGAATTCTTAAGCGTAATCTGGAACATCGTATGGGTATGTCATTTCCTGTGGAGAGTC |
| pCAGGS-LANAΔ1052-1082aa F | AAGACGAGATCCAAAGTGTCAAGCGAAGTTTGCAGTGATTTTTTGG |
| pCAGGS-LANAΔ1052-1082aa R | CCAAAAAATCACTGCAAACTTCGCTTGACACTTTGGATCTCGTCTT |
| Mut1 F | GGGAAAGGATGGAAGACGAGATCCAAAGGCTGCAGCGAAGTTTGCAGTGATTTTTTGGGGCAAT |
| Mut1 R | ATTGCCCCAAAAAATCACTGCAAACTTCGCTGCAGCCTTTGGATCTCGTCTTCCATCCTTTCCC |
| Mut2 F | GACGAGATCCAAAGTGTCAATGGGCGGCTGCAGTGATTTTTTGGGGCAATG |
| Mut2 R | CATTGCCCCAAAAAATCACTGCAGCCGCCCATTGACACTTTGGATCTCGTC |
| Mut3 F | CAAAGTGTCAATGGAAGTTTGCAGCGGCTGCTTGGGGCAATGACCCATACGGACT |
| Mut3 R | AGTCCGTATGGGTCATTGCCCCAAGCAGCCGCTGCAAACTTCCATTGACACTTTG |
| Mut4 F | ATCCAAAGTGTCAATGGAAGTTTGCAGTGATTTTTGCGGCCGCTGACCCATACGGACTTAA |
| Mut4 R | TTAAGTCCGTATGGGTCAGCGGCCGCAAAAATCACTGCAAACTTCCATTGACACTTTGGAT |
| Mut5 F | TTGCAGTGATTTTTTGGGGCAATGCCGCAGCCGGACTTAAAAAATTATCTCAGGC |
| Mut5 R | GCCTGAGATAATTTTTTAAGTCCGGCTGCGGCATTGCCCCAAAAAATCACTGCAA |
| Mut6 F | GATTTTTTGGGGCAATGACCCATACGCAGCTGCAAAATTATCTCAGGCCTTCCAGTTTG |
| Mut6 R | CAAACTGGAAGGCCTGAGATAATTTTGCAGCTGCGTATGGGTCATTGCCCCAAAAAATC |
| Mut7 F | TTGGGGCAATGACCCATACGGACTTAAAGCAGCAGCTCAGGCCTTCCAGTTTGGA |
| Mut7 R | TCCAAACTGGAAGGCCTGAGCTGCTGCTTTAAGTCCGTATGGGTCATTGCCCCAA |
| Mut8 F | CAATGACCCATACGGACTTAAAAAATTATCTGCGGCCGCCCAGTTTGGAGGAGTA |
| Mut8 R | TACTCCTCCAAACTGGGCGGCCGCAGATAATTTTTTAAGTCCGTATGGGTCATTG |
| Mut9 F | CTTAAAAAATTATCTCAGGCCTTCGCGGCTGCAGGAGTAAAGGCAGGCCCCGTGTC |
| Mut9 R | GACACGGGGCCTGCCTTTACTCCTGCAGCCGCGAAGGCCTGAGATAATTTTTTAA |
| Mut10 F | GCCTTCCAGTTTGGAGCAGCAGCGGCAGGCCCCGTGTCC |
| Mut10 R | GGACACGGGGCCTGCCGCTGCTGCTCCAAACTGGAAGGC |
| qRT-RTA-F | CACAAAAATGGCGCAAGATGA |
| qRT-RTA-R | TGGTAGAGTTGGGCCTTCAGTT |
| qRT-vCyclin-F | GCTGATAATAGAGGCGGGCAATGAG |
| qRT-vCyclin-R | GTTGGCGTGGCGAACAGAGGCAGTC |
| qRT-TK-F | CGTAGCCGACGCGGATAA |
| qRT-TK-R | TGCCTGTAGATTTCGGTCCAC |
| qRT-PAN F | GCCGCTTCTGGTTTTCATTG |
| qRT-PAN R | TTGCCAAAAGCGACGCA |
| qRT-ORFK9 F | GTCTCTGCGCCATTCAAAAC |
| qRT-ORFK9 R | CCGGACACGACAACTAAGAA |
| RpChIP F | GGCACCCGTGGGAAGGAGTACTGAAA |
| RpChIP R | TCGGGAACTTAGGCTAACCACCACAGTGT |
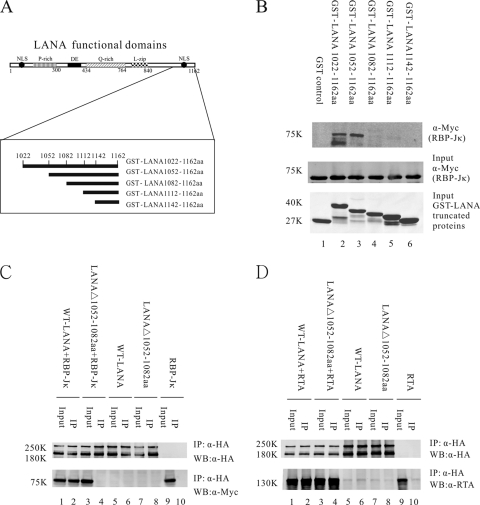
Fig 1.
Deletion of LANA C-terminal aa 1052 to 1082 disrupts LANA interaction with RBP-Jκ but not LANA interaction with RTA. (A) Schematic diagram of LANA C-terminal mutant proteins fused with GST. Amino acids in LANA C-terminal region of aa 1022 to 1162 were deleted sequentially. (B) GST pulldown assays to detect Myc-tagged RBP-Jκ protein bound to GST-LANA mutant proteins. Equivalent amounts of GST-LANA proteins and RBP-Jκ nuclear extract were used for in vitro binding assays (middle and lower panels). Two constructs, GST-LANA(1022–1162) and GST-LANA(1052–1162), interacted with RBP-Jκ in vitro, while other constructs did not (upper panel). (C) Coimmunoprecipitation assay indicated that LANAΔ(1052–1082), with a deletion of C-terminal aa 1052 to 1082, did not interact with RBP-Jκ in 293T cells (lanes 3 and 4). Nonspecific binding was not detected (lanes 5 to 10). (D) Coimmunoprecipitation assays showed that LANAΔ(1052–1082) still interacted with RTA (lanes 3 and 4). Control panels showed no binding affinity (lanes 5 to 10). IP, immunoprecipitation; WB, Western blotting.
LANA C-terminal mutations designated Mut1 to Mut10, schematically shown in Fig. 2A, were constructed using site-directed mutagenesis following the manufacturer's protocol (QuikChange Mutagenesis Kit; Stratagene, Santa Clara, CA). PCR primers were designed using a QuikChange Primer Design tool (Agilent Technology, Santa Clara, CA). Every three amino acids in the region of residues 1052 to 1082 were mutated to alanines (A) on GST-LANA(1022–1162). Using the same method, GST-LANA(W1054AW1056A) was constructed to substitute alanines for tryptophans with primers containing W1054A and W1056A mutations. Theses constructs were also used in GST pulldown assays. All primers are listed in Table 1.
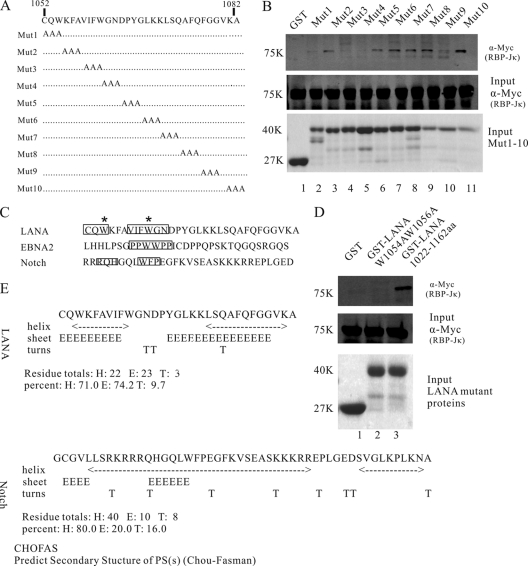
Fig 2.
Multiple amino acids in the LANA C-terminal region of aa 1052 to 1082 contribute to the LANA interaction with RBP-Jκ. (A) Schematic diagram of LANA mutations. Amino acids from LANA 1052 to 1082 were mutated to alanines (A) sequentially to determine the contribution of each site in the interaction. (B) GST-pulldown assays to detect Myc-tagged RBP-Jκ protein bound to Mut1 to Mut10. Mut1, Mut3, Mut4, and Mut9 failed to interact with RBP-Jκ, while other mutations interacted with RBP-Jκ normally. (C) Comparisons of LANA C-terminal aa 1052 to 1082 to the EBNA2 PPWWPP and Notch RAM domains. Important amino acids are boxed. Two conserved Ws are marked with asterisks. (D) GST pulldown assays to detect the interaction between GST-LANA(W1054AW1056A) and RBP-Jκ. (E) Predicted secondary structure of the LANA C-terminal aa 1052 to 1082 and the Notch RAM domain using the CHOFAS method.
The plasmid pCAGGS-LANA expressing wild-type LANA (WT-LANA) with a C-terminal hemagglutinin (HA) tag, was amplified by PCR and subcloned into the pCAGGS vector at the EcoRI site. pCAGGS-LANAΔ(1052–1082), which encodes LANAΔ(1052–1082), was obtained by performing a one-step mutation PCR on pA3M-LANA, using primers containing the deletion of aa 1052 to 1082, and then subcloned into the pCAGGS vector at the EcoRI site. The pCAGGS vector was kindly provided by Jun-Ichi Miyazaki (Osaka University, Osaka, Japan) (32). The BAC36ΔLANA plasmid, which contains the entire KSHV genome but lacks LANA expression and uses a green fluorescent protein (GFP) cassette as a tracking marker for the KSHV episome, was a kind gift from S. J. Gao (University of Southern California, CA). The Myc-tagged RBP-Jκ expression plasmid pA3M-RBP-Jκ, reporter plasmids pRpluc, pRplucΔ1327, p57pluc, and p59pluc and the RTA expression plasmid pCR3.1-RTA were described previously (14, 22, 24). All primers are listed in Table 1.
The 9E10 hybridoma, which produces c-Myc monoclonal antibodies (MAbs), was obtained from the University of Michigan Hybridoma Core Facilities. An anti-HA MAb was purchased from Sigma. A KSHV RTA mouse MAb was a kind gift from Koichi Yamanishi (Osaka University, Osaka, Japan). A KSHV ORF45 mouse MAb was purchased from Abmart. A mouse MAb against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Abcam.
293T and 293.219 cells (a 293 cell line that harbors the KSHV genome inserted into a bacterial artificial chromosome) were grown in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine growth serum (BGS; HyClone, MA), 2 mM l-glutamine, 25 U/ml penicillin, and 25 μg/ml streptomycin. DG75 (KSHV-negative B cell line) was grown in RPMI 1640 medium supplemented with 10% BGS, 2 mM l-glutamine, 25 U/ml penicillin, and 25 μg/ml streptomycin. All cells were cultured at 37°C in the presence of 5% CO2.
Protein expression, purification, and GST pulldown assay.
To purify glutathione S-transferase (GST) or GST-LANA mutant proteins, Escherichia coli strain BL21 was transformed with pGEX4T-1 or GST-LANA mutant plasmids, and expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested after 4 h of induction at 37°C, resuspended in ice-cold phosphate-buffered saline (PBS), and homogenized by sonication. The lysates were cleared by centrifugation, and the supernatant was subjected to Sepharose 4B-glutathione resin (GE Health Care, Buckinghamshire, United Kingdom) for affinity purification. The pA3M-RBP-Jκ plasmid was transfected into 293T cells, and at 48 h posttransfection, nuclear extract was prepared. For pulldown assays, equal amounts of purified GST or GST-LANA proteins bound to Sepharose 4B-glutathione were mixed for 4 h at 4°C with 30 μl of RBP-Jκ nuclear extract expressing Myc-tagged RBP-Jκ proteins. The beads were washed three times with PBS. Bound proteins were analyzed by SDS-PAGE and Western blotting.
Coimmunoprecipitation (co-IP) and Western blotting.
Five micrograms of pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) plasmid was transfected along with 10 μg of pA3M-RBP-Jκ into 10 million 293T cells, and at 24 h posttransfection, each cell was lysed in 1 ml of radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1% Nonidet P-40 [NP-40], 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 1 h on ice, with brief vortexing every 15 min. A portion of the lysate was removed for use as a control. Lysates were precleared by incubation with protein A-Sepharose beads for 1 h. One microgram of anti-HA antibody was incubated with each lysate overnight at 4°C. Immunoprecipitates were collected by rotation with 30 μl of protein A-Sepharose beads for 1 h and five washes in RIPA buffer. Proteins were heated in SDS–β-mercaptoethanol lysis buffer (SDS loading buffer) and analyzed by 8% SDS-PAGE. Western blot analysis was performed using 9E10 to detect Myc-tagged RBP-Jκ protein and an anti-HA MAb to detect WT-LANA or LANAΔ(1052–1082) protein. Using the same method, pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) plasmid was transfected along with pCR3.1-RTA into 293T cells. Anti-HA MAb was used to immunoprecipitate LANA proteins. Western blot analysis was performed using KSHV RTA MAb to detect bound RTA protein and anti-HA MAb to detect WT-LANA or LANAΔ(1052–1082) protein.
To determine the protein expression levels of the sample, 2 million cells were lysed in 200 μl of 1× SDS-loading buffer. After being heated at 100°C for 15 min, the cells were briefly sonicated and spun at 10,000 × g for 10 min. An appropriate amount of supernatant was subjected to Western blotting.
Immunofluorescence assay (IFA).
One microgram of pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) was transfected into 293T cells. At 24 h posttransfection, the cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.1% Triton X-100 for 15 min, blocked with 5% normal goat serum (Invitrogen, Carlsbad, CA), and then incubated with anti-HA MAb at a 1:500 dilution for 1 h. Cells were washed with PBS three times and further stained with goat anti-mouse IgG fused with Alexa Fluor 488 (Invitrogen, Carlsbad, CA) at a 1:1,000 dilution for 1 h. Cell nuclei were indicated by staining with 4′,6′-diamidino-2-phenylindole ([DAPI] Sigma, St. Louis, MO). Following staining, coverslips were mounted onto an inverted fluorescence microscope (DM IRB; Leica, Wetzlar, Germany) and photographed by using a digital camera and software (Leica, Wetzlar, Germany).
ChIP assay.
Fifteen micrograms of pCAGGS-LANA, pCAGGS-LANAΔ(1052–1082), or pCAGGS was transfected into 10 million KSHV-positive 293.219 cells. After 24 h, cells were washed with PBS and cross-linked with 1% formaldehyde at 37°C for 10 min. Every 10 million cross-linked cells were washed with PBS twice and resuspended in 1 ml of buffer A (10 mM Tris [pH 7.5], 10 mM NaCl, 3 mM MgCl2, 0.2% Triton X-100, 1 mM dithiothreitol [DTT], 0.5 mM EDTA, 0.2 mM PMSF). The extracted nuclei were pelleted by low-speed centrifugation and resuspended in 800 μl of buffer B (10 mM Tris [pH 7.5], 10 mM NaCl, 3 mM MgCl2, and 1 mM CaCl2), and 50 U of micrococcal endonuclease (Takara, Japan) was added. After the incubation, 3 mM EGTA (pH 7.8) (final concentration) was added to terminate the reaction, and the solution was spun at 10,000 × g for 5 min at 4°C. The supernatants (SP1) were collected, and the pellet was resuspended in 800 μl of LS buffer (10 mM Tris [pH 7.5], 10 mM NaCl, and 1 mM EDTA) and spun again. The supernatants from this spin were mixed with SP1 and divided into two aliquots for immunoprecipitation. The 800-μl supernatant samples were subjected to immunoprecipitation with either 1 μg of mouse anti-HA antibody or 1 μg of control mouse immunoglobulin and 30 μl of protein A-Sepharose, which was preincubated in a binding buffer containing 0.2 mg of salmon sperm DNA per ml for 30 min. After an overnight incubation at 4°C, the precipitates were washed five times with immunoprecipitation assay buffer (50 mM Tris [pH 7.6], 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 0.5% deoxycholate [DOC], and 1% NP-40), four times with LiCl buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.1% SDS, 0.5% DOC, 1% NP-40, and 150 mM LiCl), and once with TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA). To extract the DNA fragment, TE buffer with 1% SDS, 0.2 M NaCl, and proteinase K (Roche) was added to the washed precipitates. After incubation at 65°C for 8 h, the eluted solution was subjected to phenol-CHCl3 extraction and ethanol precipitation. Specific primers used for chromatin immunoprecipitation (ChIP) DNA amplification matched the RTA promoter region and were set around an RBP-Jκ consensus sequence located at nucleotides (nt) 68821 to 69065 within the RTA promoter (GQ 994935). The amplified fragment was 245 bp. The procedures for quantitative PCR are described below. The primers are listed in Table 1.
Dual luciferase reporter assays.
A dual luciferase reporter assay system (E1910) was purchased from Promega and used according to the manufacturer's protocol. Two million 293T cells or 5 million DG75 cells were transfected with 1 μg of pRpluc, 0.02 μg of pRL-SV40 (where SV40 is simian virus 40), and 2 μg of pCR3.1-RTA and 1 μg, 2 μg, or 3 μg of pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) plasmid. The total amount of DNA was normalized with empty vector during transfection. The pRL-SV40 plasmid, which constitutively expresses Renilla luciferase in 293T cells, was used to normalize firefly luciferase activity. After 24 h, cells were harvested and washed once with PBS. Cells were treated with 500 μl of reporter lysis buffer, and 20 μl of lysate was mixed with 20 μl of luciferase assay reagent II. Light emission of each sample was quantified in a luminometer (Veritas 209; Turner Biosystems, PA). Stop and Glo buffer was added to stop firefly luciferase activity, and Renilla luciferase was measured. Firefly luciferase was normalized using Renilla luciferase, and results are shown as the fold change relative to WT-LANA and LANAΔ(1052–1082) cells. The results shown represent experiments performed in triplicate. Similarly, luciferase assays were conducted to determine the LANA repression effect on pRplucΔ1327, a mutant RTA promoter with a deletion of all RBP-Jκ binding sites, the ORF57 promoter p57pluc, and the ORF59 promoter p59pluc.
Transfection of episome DNA and monitoring episome persistence.
Cells from the 293T line were seeded in a six-well plate before transfection and grown to 90% confluence. Purified BAC36ΔLANA episomes were transfected into 293T cells at 4 μg/well with Targefect reagent (Targefect System, CA). pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) plasmid was delivered into 293T cells by transfection to complement LANA expression. As a control, BAC36ΔLANA was cotransfected with the vector pCAGGS into 293T cells. Each transfection assay was repeated three times. Transfected cells were visualized with a DM6000B fluorescence microscope (Leica, Wetzlar, Germany) and photographed by using a digital camera and software (Leica, Wetzlar, Germany). A portion of cells were washed twice with PBS buffer and fixed with 1% formaldehyde, and the percentage of GFP-positive cells was counted using fluorescence-activated cell sorting (FACS) with a FACScan (Becton Dickinson, NJ).
Reconstitution of recombinant viruses in mammalian cells and induction of viral lytic replication.
Cells from the 293T line were plated into T-25 flasks 1 day before transfection. Cells at 90% confluence were transfected with 10 μg of a fresh DNA preparation of BAC36ΔLANA with 5 μg of pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) plasmid using Targefect transfection reagent. At 24 h posttransfection, one T-25 flask of transfected cells was split into 3- by 60-mm dishes at 60% confluence. After 12 h, recombinant viruses were treated with 25 ng/ml of 12-O-tetradecanoylphorbol-13-acetate (TPA) and 1.5 mM valproate (VPA) to induce viral lytic replication. To examine the expression of viral genes, cells were collected at various times postinduction and examined for the expression of viral transcripts.
Reverse transcription real-time PCR.
Quantitative real-time PCR (qRT-PCR) was used for relative quantitative comparison of KSHV gene expression before and after TPA/VPA induction for BAC36ΔLANA cells cotransfected with pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) plasmid. At 0, 24, and 48 h postinduction, cells were harvested, and total RNA was collected using TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. cDNA (20 μl) for real-time PCR was generated from 1 μg of total RNA with a First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) according to the manufacturer's instructions. Primer sequences for qRT-PCR are shown in Table 1. Briefly, qRT-PCR was performed with a SYBR green Real-Time PCR Master Mix kit (Toyobo, Japan). Reaction mixtures of 20 μl contained 10 μl of Master Mix, 1 mM each primer, and 5 μl of diluted cDNA product. Following 2 min at 50°C, DNA polymerase was activated at 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and at 60°C for 1 min. Values for the relative quantification were calculated by the ΔΔCT method (where CT is threshold cycle), and melting curve analysis was performed to verify the specificity of the products. As a negative control, each plate contained a minimum of three wells without template. Relative mRNA levels were normalized to GAPDH and then to percentages of GFP-positive cells; values are reported as the increase in mRNA accumulation. All reactions were carried out three times with a 7900HT sequence detection system (Applied Biosystems, CA).
Quantification of virion DNAs.
At 96 h after TPA/VPA induction, cell suspensions were centrifuged at 3,000 rpm for 20 min, and the supernatant was filtered through a 0.45-μm-pore-size filter. Viral particles were concentrated by ultracentrifugation at 20,000 × g at 4°C for 3 h. The pellet was resuspended in 50 μl of PBS, heated to 95°C for 15 min, and switched to 56°C for 1 h with proteinase K treatment (10 mg/ml). The enzyme was then destroyed by treatment at 95°C for 30 min. A 5-μl portion of virus lysate was used for PCR amplification of the KSHV-specific region of ORFK9. The primers used are listed in Table 1. A standard curve was made using a serial dilution of the BAC36 DNA plasmid. KSHV virions were normalized to GFP-positive cells, and fold change relative to BAC36ΔLANA cells cotransfected with pCAGGS was calculated.
Statistical analysis.
Data are shown as mean values with standard errors of the means (SEM). P values were determined by a Student's t test with GraphPad Prism, version 5, software (n = 3 for each group).
RESULTS
LANA carboxyl-terminal amino acids 1052 to 1082 are essential for interaction with RBP-Jκ in vitro.
To identify the RBP-Jκ binding motif of LANA, we performed in vitro binding assays. Amino acids from the LANA C-terminal region of residues 1022 to 1162 were deleted sequentially (Fig. 1A). GST-fused LANA truncations were purified using GST-Sepharose, and RBP-Jκ nuclear extracts were prepared from 293T cells transfected with a Myc-tagged RBP-Jκ plasmid, pA3M-RBP-Jκ. Equivalent amounts of GST-LANA mutant proteins were incubated with RBP-Jκ nuclear extracts for binding assays (Fig. 1B, middle and lower panels). Binding assay results indicated that a C-terminal truncated protein of LANA consisting of amino acids 1022 to 1162 bound strongly to RBP-Jκ (Fig. 1B, lane 2, upper panel). In addition, GST-LANA(1052–1162) also bound to RBP-Jκ (Fig. 1B, lane 3, upper panel). However, after deletion of amino acids 1052 to 1082, GST-LANA(1082–1162) showed little or no specific binding affinity to RBP-Jκ (Fig. 1B, lane 4, upper panel). Two other proteins, GST-LANA(1112–1162) and GST-LANA(1142–1162), also lost the ability to interact with RBP-Jκ (Fig. 1B, lanes 5 and 6, upper panel). The GST control, alone, did not interact with RBP-Jκ (Fig. 1B, lane 1, upper panel). These data suggested that the primary region responsible for binding to RBP-Jκ in vitro was the LANA carboxyl-terminal aa 1052 to 1082.
Coimmunoprecipitation (co-IP) experiments were carried out to determine if the region identified by in vitro binding was also responsible for LANA and RBP-Jκ interaction in mammalian cells. We constructed a full-length LANA expression plasmid, pCAGGS-LANA, expressing WT-LANA with the HA tag at the C terminus, and a LANA mutant plasmid, pCAGGS-LANAΔ(1052–1082), expressing LANAΔ(1052–1082), which had a deletion of C-terminal residues 1052 to 1082. pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) was transfected with pA3M-RBP-Jκ into 293T cells. At 24 h posttransfection, cells were harvested and lysed. WT-LANA or LANAΔ(1052–1082) protein was immunoprecipitated with anti-HA monoclonal antibody (MAb). Bound complexes coimmunoprecipitating with WT-LANA or LANAΔ(1052–1082) were probed for RBP-Jκ with mouse MAb 9E10 against the Myc epitope. Consistent with the previous experiments, WT-LANA coimmunoprecipitated with RBP-Jκ in 293T cells (Fig. 1C, lanes 1 and 2, lower panel), but LANAΔ(1052–1082) did not show a signal for RBP-Jκ (Fig. 1C, lanes 3 and 4, lower panel), indicating that amino acids 1052 to 1082 were essential for LANA to interact with RBP-Jκ in vitro. Control blots to determine if WT-LANA and LANAΔ(1052–1082) were expressed and immunoprecipitated equally showed that both proteins were expressed to similar levels and were efficiently immunoprecipitated by the antibody (Fig. 1C, upper panel). When the plasmids were transfected alone, we did not detect bound RBP-Jκ (Fig. 1C, lanes 5 to 10, lower panel). Taken together, GST pulldown assays demonstrated that LANA carboxyl-terminal amino acids 1052 to 1082 were responsible for direct binding of LANA and RBP-Jκ. Co-IP assays confirmed the importance of this region for LANA interaction with RBP-Jκ in mammalian cells.
Since previous reports have indicated that LANA interacted with RTA and that C-terminal amino acids 990 to 1162 of LANA were responsible for direct interaction with RTA, we therefore wanted to determine whether the deletion of LANA C-terminal amino acids 1052 to 1082 also influenced LANA interaction with RTA (22). We performed co-IP experiments as described above. pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) was transfected with pCR3.1-RTA into 293T cells. Then WT-LANA or mutant LANA proteins were immunoprecipitated with anti-HA MAb. The results indicated that WT-LANA bound to RTA (Fig. 1D, lanes 1 and 2, lower panel) and that LANAΔ(1052–1082) also bound to RTA (Fig. 1D, lanes 3 and 4, lower panel), while control panels showed no binding affinity (Fig. 1D, lanes 5 to 10). These results therefore showed that deletion of LANA C-terminal amino acids 1052 to 1082 did not influence the ability of LANA to interact with RTA; thus, this region of LANA was not responsible for direct interaction with RTA.
Multiple amino acids are required for LANA to interact with RBP-Jκ.
To further verify the sites of RBP-Jκ binding to LANA, we generated a panel of constructs in which every three amino acids in the C-terminal region of residues 1052 to 1082 were mutated to alanines (A) sequentially (Fig. 2A). These constructs, termed Mut1 to Mut10, were generated using site-directed mutagenesis methods and GST-LANA(1022–1162) as a template. GST pulldown assays were performed as above. LANA mutant proteins Mut1, Mut3, and Mut4 showed no detectable binding to RBP-Jκ, while Mut9 showed only weak interaction with RBP-Jκ (Fig. 2B, lanes 2, 4, 5, and 10, upper panel). The other mutant proteins bound to RBP-Jκ normally (Fig. 2B, lanes 3, 6 to 9, and 11, upper panel). Interestingly, mutations in Mut1, Mut3, and Mut4 were adjoining, indicating that the first 12 amino acids in the C-terminal region of residues 1052 to 1082 were critical for the interaction between LANA and RBP-Jκ.
To further analyze the composition of these amino acids, we compared the LANA C-terminal residues 1052 to 1082 with the EBNA2 PPWWPP motif and the Notch RAM domain (15, 45). We found two tryptophans (W) close to each other but separated by several amino acids (Fig. 2C). This feature might indicate a conserved motif although the two Ws were not adjacent, as they are in the conserved EBNA2 PPWWPP motif (Fig. 2C). As anticipated, when the two Ws were replaced with As, GST-LANA(W1054AW1056A) failed to interact with RBP-Jκ (Fig. 2D, lane 2), suggesting that these two Ws were essential for LANA interaction with RBP-Jκ.
LANA carboxyl-terminal secondary structure might contribute to RBP-Jκ interaction.
Structural studies revealed a striking conformational change in RBP-Jκ upon RAM binding, creating a docking site for the transcriptional coactivator Mastermind to bind (9). We postulated that the secondary structure of the LANA carboxyl terminus was also important for successful interaction with RBP-Jκ. The secondary structures of the LANA C-terminal amino acids 1052 to 1082 and the Notch RAM domain were predicted using the CHOFAS program (Fig. 2E) (7, 35). Interestingly, the predicted secondary structure of the 12 LANA C-terminal amino acids, corresponding to mutations from Mut1 to Mut4, contained a helix and a sheet that were also seen in the Notch RAM domain (Fig. 2E). From these experiments, we proposed that the secondary structure of the LANA C terminus may contribute to its interaction with RBP-Jκ.
LANAΔ(1052 to 1082) fails to form a complex with the RBP-Jκ consensus sequence within the RTA promoter.
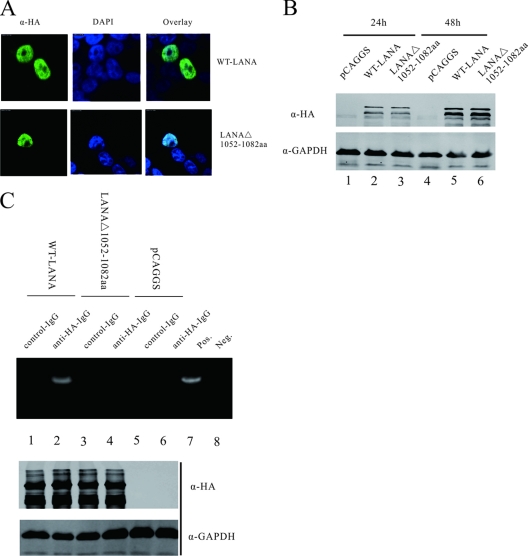
To exclude the possibility that the differences observed with LANAΔ(1052–1082) were due to the change of mutant protein expression or location, we conducted IFA and Western blot assays. As indicated in Fig. 3A, both WT-LANA and LANAΔ(1052–1082) were localized to the nucleus. And in Fig. 3B, Western blot assays showed that WT-LANA and LANAΔ(1052–1082) were expressed to similar levels at 24 h and 48 h after transfection. Thus, deletion of LANA C-terminal residues 1052 to 1082 did not change LANA expression and localization.
Fig 3.
LANAΔ(1052–1082) fails to form a complex with the RBP-Jκ consensus sequence within the RTA promoter. (A) IFAs showed that WT-LANA and LANAΔ(1052–1082) localized to the nucleus area. Green indicates the expression of LANA proteins; blue indicates the nucleus stained with DAPI. (B) The protein levels of WT-LANA and LANAΔ(1052–1082) were detected 24 h and 48 h after transfection. Both proteins were expressed in cells to similar levels. (C) ChIP assays indicated that WT-LANA associated with the RBP-Jκ consensus sequence within the RTA promoter (lanes 1 and 2, upper panel). However, LANAΔ(1052–1082) lacking the C-terminal aa 1052 to 1082 failed to form a complex with the RBP-Jκ consensus sequence (lane 3 and 4, upper panel). Protein lysates were analyzed by Western blotting for transfected protein expression (middle and lower panels).
Previous reports using electrophoretic mobility shift assays (EMSAs) suggested that RBP-Jκ can bind to the consensus binding site within the RTA promoter and that LANA is able to form a complex with RBP-Jκ bound to its specific sequence (21). To determine whether deletion of LANA C-terminal amino acids 1052 to 1082 influenced the complex formation between LANA and the RTA promoter, we performed a chromatin immunoprecipitation assay (ChIP). pCAGGS-LANA or pCAGGS-LANAΔ(1052-1082) was transfected into 293.219 (KSHV-positive) cells, and Western blot analysis indicated that the protein expression levels were similar (Fig. 3C, middle and lower panels). Then, WT-LANA and mutant LANA proteins were immunoprecipitated with anti-HA MAb. The immunoprecipitates were amplified by quantitative PCR using specific primers set around a RBP-Jκ consensus sequence which was located at nt 68821 to 69056 within the RTA promoter. The specificity was evaluated by comparing the amplification levels to control immunoprecipitates (Fig. 3C, lanes 5 to 8). The results suggested that WT-LANA was involved in binding the RBP-Jκ consensus sequence (Fig. 3C, lanes 1 and 2). But after deletion of LANA C-terminal amino acids 1052 to 1082, the complex formation between LANA and the RBP-Jκ consensus sequence within the RTA promoter was disrupted (Fig. 3C, lanes 3 and 4). Taken together, deletion of LANA C-terminal amino acids 1052 to 1082 disrupted the interaction between LANA and RBP-Jκ and destroyed the complex of LANA on the RTA promoter but did not influence LANA interaction with RTA.
LANAΔ(1052–1082) fails to repress RTA autoactivation in vitro.
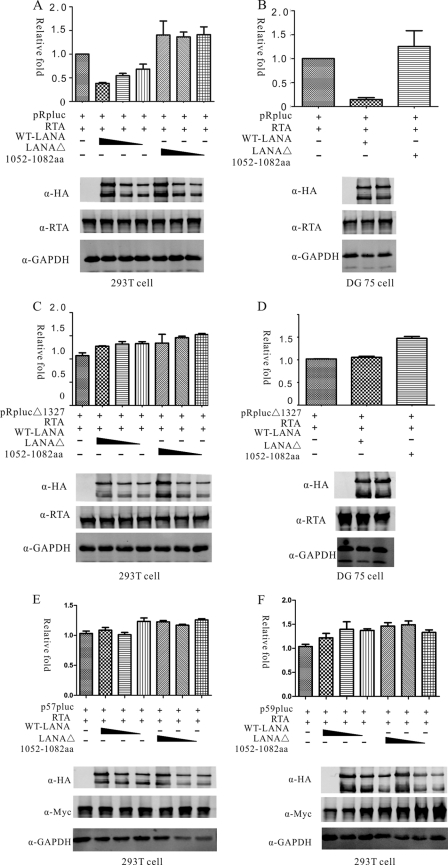
Previous research showed that LANA can replace RTA from RBP-Jκ to inhibit transcription mediated by RTA/RBP-Jκ (21). Thus, LANAΔ(1052–1082), which was deficient in interacting with RBP-Jκ, might not be able to repress the RTA promoter. We conducted luciferase assays to test this postulate. pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) was transfected into 293T cells with the reporter plasmid pRpluc containing the RTA promoter, RTA expression plasmid pCR3.1-RTA, and an endogenous control plasmid, pRL-SV40, expressing Renilla luciferase. In 293T cells, WT-LANA protein repressed pRpluc transactivation mediated by RTA in a dose-dependent manner. More than 50% of activation was repressed at most (Fig. 4A). However, LANAΔ(1052–1082) failed to repress the RTA promoter in 293T cells. When LANAΔ(1052–1082) expression was increased, the induction levels were still similar to the levels in the absence of LANA (Fig. 4A). This observed increase in the induction levels indicated that aa 1052 to 1082 of LANA are important for mediating transcriptional repression of RTA. To determine whether this observation was cell type dependent, we performed luciferase reporter assays in a B cell line, DG75. Repression was observed with WT-LANA, with induction approximately 80% repressed. However, this repression was not observed with LANAΔ(1052–1082) (Fig. 4B). Meanwhile, the expression levels of RTA and LANA proteins were assayed by Western blotting (Fig. 4), which indicated that the observed differences of reporter assays were not due to the exogenous protein expression. Next, we conducted luciferase assays using pRplucΔ1327, which had a deletion of all RBP-Jκ binding sites and showed no repression by LANA (21). As expected, both WT-LANA and LANAΔ(1052–1082) were unable to repress pRplucΔ1327 in either 293T cells or DG75 cells (Fig. 4C and D). Taken together, these data suggested that the LANA C-terminal residues 1052 to 1082 contributed to regulation of the RTA promoter through interaction with RBP-Jκ. Therefore, loss of these sites might affect overall viral gene expression. Since RBP-Jκ binding sites were widely present in KSHV lytic genes and RTA mediated transcriptional activation were largely through interactions with RBP-Jκ (36). In this case, we would like to determine whether LANA could also repress other lytic promoters through RBP-Jκ. We chose ORF57 and ORF59 promoters which have been shown to be activated by RTA through RBP-Jκ (24, 27). Luciferase assays indicated that although these promoters contained active RBP-Jκ binding sites, they failed to be repressed by either wild-type LANA or mutant LANA proteins (Fig. 4E and F). It is possible that LANA-mediated repression could be promoter dependent or that the RBP-Jκ binding site alone might not be sufficient to mediate LANA repression as other flanking sequences may also be critical.
Fig 4.
LANAΔ(1052–1082) fails to repress RTA autoactivation in vitro. WT-LANA was able to repress RTA promoter, but LANAΔ(1052–1082) did not repress RTA induction in 293T cells (A) or in DG75 cells (B). In 293T cells, RTA repression by WT-LANA was dose dependent, while LANAΔ(1052–1082) did not repress RTA induction at three different doses (A). pRplucΔ1327 is a mutant RTA reporter with a deletion of all RBP-Jκ binding sites. Neither WT-LANA nor LANAΔ(1052–1082) repressed pRplucΔ1327 induction in 293T cells (C) or DG75 cells (D). The protein expression levels of each group were estimated by Western blotting and are shown in the lower panels. The expression of RTA, WT-LANA, or LANAΔ(1052–1082) and endogenous GAPDH were estimated. Neither WT-L ANA nor mutant LANA repressed ORF57 or ORF59 promoter autoactivation in 293T cells (E and F).
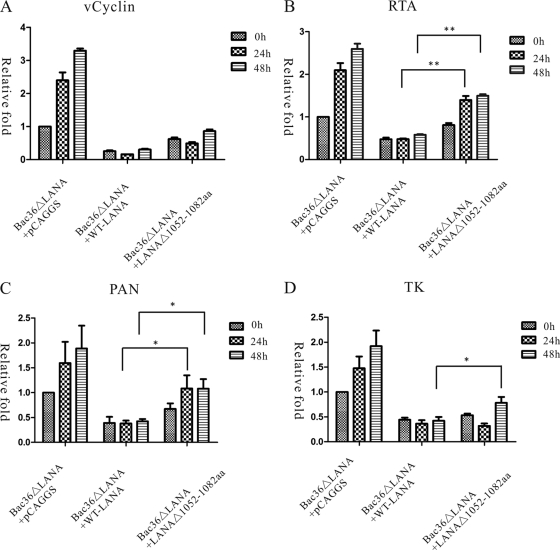
LANAΔ(1052–1082) is ineffective in repressing viral lytic genes in the full-length KSHV genome.
In vitro studies indicated that LANAΔ(1052–1082) was defective in repressing RTA transactivation. To further analyze whether this observed loss of function also occurred in the context of the entire viral genome, we reconstituted recombinant virus using BAC36ΔLANA in 293T cells and cotransfected pCAGGS-LANA or pCAGGS-LANAΔ(1052–1082) to recover LANA expression. After 12 h, recombinant viruses were treated with TPA/VPA to induce viral lytic replication. BAC36ΔLANA is a KSHV mutation with LANA disrupted. It is based on the KSHV BAC36 and uses a GFP cassette as a tracking marker for the KSHV episome. Previous study found that genetic disruption of LANA enhances the KSHV viral lytic transcriptional program without affecting expression of latently expressed genes (23). Thus, we hypothesized that WT-LANA would rescue this phenotype and repress viral lytic genes while LANAΔ(1052–1082), without RBP-Jκ binding sites, would be less efficient at rescue. To test this postulation, we examined the expression of a set of KSHV genes representing different classes of viral genes: a latent gene encoding viral cyclin D homolog (vCyclin); an immediate early (IE) gene, RTA; a delayed early (DE) gene, polyadenylated nuclear RNA (PAN); and a late gene encoding thymidine kinase (TK). qRT-PCR was used to determine viral gene expression levels before or after TPA/VPA chemical induction. Relative mRNA levels were normalized to an endogenous control gene, GAPDH, and then to the percentages of GFP-positive cells.
As shown in Fig. 5, in BAC36ΔLANA cells cotransfected with pCAGGS, the expression of viral lytic genes and latent genes increased after TPA/VPA induction. However, when BAC36ΔLANA was coexpressed with WT-LANA, the expression of viral genes was greatly reduced. Even after TPA/VPA induction, viral genes were expressed at a low level. Similarly, in the presence of LANAΔ(1052–1082), viral genes were also repressed.
Fig 5.
LANAΔ(1052–1082) is ineffective in repressing viral lytic genes in the context of the full-length KSHV genome. qRT-PCR was used to examine viral gene expression levels at 0, 24, and 48 h after chemical induction. The relative mRNA levels were first normalized to GAPDH and then to percentages of GFP-positive cells. Four viral genes were investigated: a latent gene, vCyclin (A); an immediate-early gene, RTA (B); a delayed-early gene, PAN (C); and a late gene, TK (D). Compared to WT-LANA, LANAΔ(1052–1082) was less efficient in repressing KSHV lytic genes RTA, PAN, and TK although the patterns of repression were slightly different. Statistical analysis was conducted using a Student's t test. *, P < 0.05; **, P < 0.01.
vCyclin is encoded by a viral latent gene and is not believed to be influenced by the latent LANA gene (23). In our experiments, with exogenous expression of LANA, the expression of vCyclin was also greatly repressed. This might be an effect of the large amount of exogenous LANA production. However, we showed that expression of vCyclin transcripts was not significantly different between LANAΔ(1052–1082) and WT-LANA as determined by statistical analysis (P > 0.05). Thus, deletion of the LANA C-terminal aa 1052 to 1082 had little effect on vCyclin transcription (Fig. 5A).
RTA is a critical reactivator which regulates the KSHV switch from latent to lytic replication. As indicated in Fig. 5B, expression of the RTA transcript in LANAΔ(1052–1082)was 1.7-fold greater than in WT-LANA cells. Upon induction with TPA/VPA, the expression of RTA at 24 h postinduction increased 1.8-fold in LANAΔ(1052–1082) but was almost unchanged in WT-LANA cells. Thus, the expression of RTA transcripts remained 2.9-fold higher in LANAΔ(1052–1082) than in WT-LANA following chemical induction. These results indicated that LANAΔ(1052–1082) is less efficient at complementing LANA function and leads to elevated RTA transcription.
We next examined the expression of other viral lytic genes. PAN is a DE gene, which is transcribed into a noncoding RNA. Similar to RTA, in uninduced cells, the expression level of PAN was 1.8-fold higher in LANAΔ(1052–1082) cells than in WT-LANA cells. At 24 h postinduction, PAN expression had increased by 1.6-fold in LANAΔ(1052–1082), making it 2.5-fold higher than in WT-LANA cells (Fig. 5C). The expression pattern of the late TK gene was slightly different. While we detected minimal differences in TK transcripts between LANAΔ(1052–1082) and WT-LANA cells at 24 h postinduction, TK transcript levels at 48 h postinduction were 1.9-fold higher in LANAΔ(1052–1082) cells than in WT-LANA cells (Fig. 5D). These results suggested that LANAΔ(1052–1082) is less efficient at repressing a DE and a late gene. The different patterns of these genes might reflect their different expression kinetics.
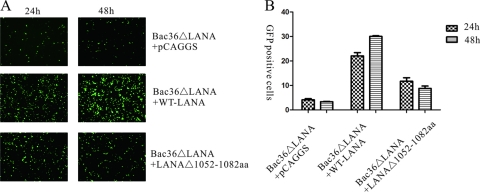
LANAΔ(1052–1082)is defective in episome persistence of BAC36ΔLANA.
Previous research found fewer GFP-positive cells in BAC36ΔLANA cultures than in WT BAC36 at the early stage following episome delivery by transfection. This could be partially rescued by cotransfection with a LANA expression plasmid, indicating that LANA is essential for KSHV episome persistence (48). Using the same method described previously, we delivered BAC36ΔLANA into 293T cells by transfection. WT-LANA or LANAΔ(1052–1082) was coexpressed, and GFP-positive cells were monitored. At 24 and 48 h posttransfection, cells were visualized and photographed with a fluorescence microscope, and the percentage of GFP-positive cells was counted using FACS (Fig. 6A and B). In the control group, the vector pCAGGS was cotransfected with BAC36ΔLANA, and the episome was present in the absence of LANA expression. As shown in Fig. 6, at 24 h posttransfection, approximately 3.7% of cells cotransfected with BAC36ΔLANA and the vector pCAGGS were GFP positive. The percentage of GFP-positive cells in cultures of BAC36ΔLANA coexpressed with WT-LANA was significantly higher at 23.4%. For BAC36ΔLANA cells coexpressed with LANAΔ(1052–1082) cells, approximately 10.3% were GFP positive, which was higher than coexpression with pCAGGS but lower than with WT-LANA. Similarly, at 48 h posttransfection, 30.3% of cells coexpressing BAC36ΔLANA with WT-LANA were GFP positive, which was much higher than for coexpression with pCAGGS (3.2%) or LANAΔ(1052–1082) (7.8%). These results indicated that LANAΔ(1052–1082) could not efficiently maintain the BAC36ΔLANA episome.
Fig 6.
LANAΔ(1052–1082) is inefficient in maintaining the BAC36ΔLANA episome in 293T cells. WT-LANA or LANAΔ(1052–1082) was coexpressed with BAC36ΔLANA to complement the expression of LANA. At 24 and 48 h posttransfection, cells were virtualized and photographed with a fluorescence microscope (A), and the percentage of GFP-positive cells was counted using FACS (B). BAC36ΔLANA transfected with vector pCAGGS was used as a control. Both photography and FACS data indicated that there were fewer GFP-positive cells in LANAΔ(1052–1082) cells than in WT-LANA.
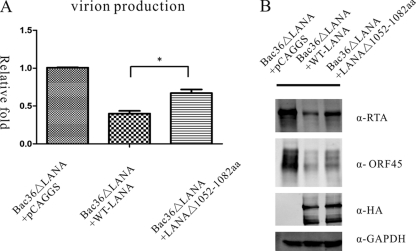
Deletion of LANA aa 1052 to 1082 leads to increased KSHV lytic replication.
The above-described experiments demonstrated that, compared to WT-LANA, LANAΔ(1052–1082) leads to increased expression of viral lytic genes. We determined whether this enhanced viral lytic replication resulted in increased production of KSHV virions. We induced cells with TPA/VPA, and at 96 h postinduction, cultured supernatants were collected and titrated for infectious virions by qRT-PCR to detect viral K9 DNA. KSHV virions were normalized to GFP-positive cells, and fold change relative to BAC36ΔLANA transfected with pCAGGS was determined. Compared to WT-LANA, LANAΔ(1052-1082) cells produced 1.7-fold more virions after TPA/VPA induction (Fig. 7A). To confirm this result, we conducted Western blotting to detect the endogenous KSHV protein expression. As shown in Fig. 7B, in the presence of LANAΔ(1052-1082), cells expressed more of the endogenous KSHV proteins RTA and ORF45 than WT-LANA, indicating increased lytic replication. This result suggested that disruption of LANA and RBP-Jκ interaction by deletion of the LANA C-terminal aa 1052 to 1082 leads to increased lytic replication and virion production.
Fig 7.
Deletion of LANA C-terminal aa 1052 to 1082 leads to elevated virion production. At 96 h postinduction, cultured supernatants were collected and titrated for infectious virions using qRT-PCR to detect viral K9 DNA. KSHV virions were normalized to GFP-positive cells, and fold change relative to BAC36ΔLANA cotransfected with pCAGGS is shown. LANAΔ(1052–1082) cells produced more virions than WT-LANA (A). Furthermore, the protein expression levels of KSHV endogenous RTA and ORF45 were higher in LANAΔ(1052–1082)-expressing cells than in WT-LANA-expressing cells (B).
DISCUSSION
Understanding the precise RBP-Jκ interaction domain in LANA is important for elucidating the function of LANA in latent infection and the establishment and maintenance of KSHV. Unlike our extensive knowledge about RBP-Jκ-mediated viral reactivation, the role of RBP-Jκ in KSHV latency is poorly understood. Previous reports showed that LANA physically associates with RBP-Jκ in vitro and in KSHV-infected cells, forming a complex capable of binding to RBP-Jκ cognate sequences (21). In this study, we confirmed the interaction between LANA and RBP-Jκ and discovered that LANA C-terminal aa 1052 to 1082 were responsible for LANA interaction with RBP-Jκ. Furthermore, we reconstituted the virus and analyzed the function of LANAΔ(1052–1082) in the context of the full-length KSHV genome and found that interactions between LANA and RBP-Jκ were important for repressing viral lytic replication and maintaining KSHV latency.
Among the limited number of latent genes, the ORF73 gene, which encodes LANA, is critical for establishing latent KSHV infection. LANA is a large, multifunctional protein (about 222 to 234 kDa) with three distinct domains: a proline-rich N-terminal region with a putative nuclear localization signal; a long, glutamic acid-rich internal repeat domain; and a carboxyl-terminal domain including a putative nuclear localization signal (17, 37). Extensive studies indicate that the LANA C terminus is involved in multiple biologic processes including transcription repression, DNA binding, protein interaction with RTA and CBP, and ubiquitin-mediated VHL and p53 degradation (3, 11, 22, 26). Here, we showed that the LANA C-terminal aa 1052 to 1082 were the primary region responsible for LANA interaction with RBP-Jκ. Deletion of LANA C-terminal amino acids 1052 to 1082 disrupted the interaction between LANA and RBP-Jκ and destroyed the complex of LANA on the RTA promoter but did not influence LANA interaction with RTA. Then we compared the amino acid sequences of this region to the EBNA2 PPWWPP and the Notch PXW motifs and found that two Ws were also present in the LANA region although they were not adjacent. We searched around the Ws and found a proline (P). However, GST pulldown assays indicated that when P was replaced with A, the mutant LANA could still interact with RBP-Jκ. Thus, whether this P also plays a role in the interaction with RBP-Jκ is yet to be resolved.
Although LANA shows no homology at the sequence level, its phenotype and structural features are reminiscent of the EBNA2 protein, and one such example is its extended central region of repeats (42). The predicted secondary structure of LANA's C terminus reveals striking similarity to the known structure of the DNA-binding domain of EBNA1 (13). Similarly, our data showed that the predicted secondary structure of the LANA C-terminal RBP-Jκ binding region resembled the Notch RAM domain, suggesting conservation of the key mechanistic features of RBP-Jκ binding proteins.
In a previous study, after deletion of RBP-Jκ binding sites within the RTA promoter, the transcriptional repression of RTA mediated by LANA was greatly reduced (21). In this study, when we repressed the interaction of LANA and RBP-Jκ by deleting the LANA C-terminal amino acids 1052 to 1082, LANA-mediated RTA repression was also decreased. These results indicated that RBP-Jκ played important roles in LANA-mediated RTA repression. Furthermore, we analyzed the contribution of RBP-Jκ in viral transcriptional regulation in the context of the full-length KSHV genome. With the C-terminal aa 1052 to 1082 were deleted, LANAΔ(1052-1082) was less effective in repressing transcripts of RTA and other viral lytic genes, indicating that RBP-Jκ was involved in viral transcriptional regulation. However, we also found that both WT-LANA and LANAΔ(1052-1082) repressed RTA transcription but to different degrees. This indicated that mechanisms other than RBP-Jκ interaction are also responsible for LANA-mediated RTA repression. The finding that acetylation of LANA regulates repression of RTA could explain this result (29). The observed elevated viral lytic gene transcription might be due to direct regulation by LANAΔ(1052-1082) or to indirect regulation by altered RTA levels. Since our in vitro studies indicated that LANA did not directly regulate many viral lytic genes, we speculated that different regulatory effects might reflect indirect regulation by RTA.
One of the most studied functions of LANA is KSHV episome persistence in the host. Indeed, LANA alone is sufficient for the maintenance, replication, and segregation of the KSHV episome via binding of its C-terminal domain to the terminal repeat sequence (TR), which is tethered to cellular chromosomes via its N-terminal domain (11). LANA regulates transcription by activating or repressing various cellular and viral promoters (1, 19). LANA is also reported to regulate oncogene and tumor suppressors at the posttranscriptional level (2, 10). Here, we showed that disruption of interaction between LANA and RBP-Jκ by deletion of the LANA C-terminal aa 1052 to 1082 eliminated the ability of LANA to maintain the KSHV episome, as shown by decreased GFP-positive cells. LANA tethered the viral episomal DNA to the host chromosomes by directly binding to its cognate binding sequence within the TR region of the genome through its C terminus and to the nucleosomes through the N terminus of the molecule (11, 43). This decreased episome persistence may be because deletion of LANA C-terminal aa 1052 to 1082 disrupted the interaction between LANA and the KSHV episome. Most importantly, viral lytic gene transcription levels were extensively altered, indicating that RBP-Jκ-mediated repression is important in KSHV gene regulation. We also constructed a plasmid which contains the LANA C-terminal aa 1052 to 1082 fused with GFP, termed pEGFP(1052-1082), and wanted to determine whether this small peptide had a dominant negative effect. Our results indicated that pEGFP(1052-1082) was expressed to high levels in 293T cells; however, in a luciferase reporter assay, it did not influence the ability of LANA to repress the RTA promoter and thus did not have a dominant negative effect (data not shown). This may indicate that the binding affinity of the small peptide alone was rather weak.
In summary, we dissected a conserved RBP-Jκ binding domain in LANA and showed that this region is required for LANA-mediated repression of RTA. This finding indicates the important and extensive role of RBP-Jκ, which is involved not only in viral gene activation but also in mediating transcriptional repression, thus playing important roles in maintaining KSHV latency and controlling reactivation. Our finding also provided valuable evidence for understanding the regulation of viral genes by a host transcription factor and its potential role in maintaining KSHV latency.
Conventional chemotherapeutic regimens provide no specific cure for PEL, but other approaches to develop anti-PEL therapies are under way. Most PEL and KS tumor cells are latently infected by KSHV and are thus resistant to antiherpesvirus drugs that are dependent on lytic replication (18). Alternatively, the replication and maintenance of the KSHV episome during latency can be disrupted by glycyrrhizic acid or hydroxyurea, so the virus no longer contributes to tumorigenesis (8). Our results also imply that the LANA C-terminal aa 1052 to 1082 could be a potential target for disruption of viral latency, which could be a strategy for oncolytic viral therapy, either by inducing KSHV to reenter the lytic cascade in the presence of antiherpesvirus drugs or by selectively inducing death of KSHV-infected cells, leading to virus eradication.
ACKNOWLEDGMENTS
We thank S. J. Gao at University of Southern California for his kind gift of the Bac36ΔLANA construct. We also thank Jun-ichi Miyazaki at Osaka University for providing the vector pCAGGS.
This work was supported by grants from the National Basic Research Program of China (2011CB504800 and 2012CB519002), the 100 Talent Program of the Chinese Academy of Sciences, and Natural Science Foundation of China (30970154) to K.L.
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. An F-Q, et al. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862–3874 [DOI] [PubMed] [Google Scholar]
- 2. Bubman D, Guasparri I, Cesarman E. 2007. Deregulation of c-Myc in primary effusion lymphoma by Kaposi's sarcoma herpesvirus latency-associated nuclear antigen. Oncogene 26:4979–4986 [DOI] [PubMed] [Google Scholar]
- 3. Cai Q-L, Knight JS, Verma SC, Zald P, Robertson ES. 2006. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the p53 tumor suppressors. PLoS Pathog. 2:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186–1191 [DOI] [PubMed] [Google Scholar]
- 5. Chang P-J, Boonsiri J, Wang S-S, Chen L-Y, Miller G. 2010. Binding of RBP-Jκ (CSL) protein to the promoter of the Kaposi's sarcoma-associated herpesvirus ORF47 (gL) gene is a critical but not sufficient determinant of transactivation by ORF50 protein. Virology 398:38–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang Y, et al. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869 [DOI] [PubMed] [Google Scholar]
- 7. Chou PY, F GD. 1978. Prediction of the secondary structure of proteins from their amino acid sequence. Adv. Enzymol. 47:45–148 [DOI] [PubMed] [Google Scholar]
- 8. Curreli F, Friedman-Kien AE, Flore O. 2005. Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes. J. Clin. Invest. 115:642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedmann DR, Wilson JJ, Kovall RA. 2008. RAM-induced allostery facilitates assembly of a Notch pathway active transcription complex. J. Biol. Chem. 283:14781–14791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujimuro M, et al. 2003. A novel viral mechanism for dysregulation of β-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300–306 [DOI] [PubMed] [Google Scholar]
- 11. Garber AC, Shu MA, Hu J, Renne R. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J. kappa recombination signal binding protein. Proc. Natl. Acad. Sci. U. S. A. 91:7568–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grundhoff A, Ganem D. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He Z, et al. 2010. Cellular corepressor TLE2 inhibits replication-and-transcription-activator-mediated transactivation and lytic reactivation of Kaposi's sarcoma-associated herpesvirus. J. Virol. 84:2047–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Henkel T, Ling P, Hayward S, Peterson M. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92–95 [DOI] [PubMed] [Google Scholar]
- 16. Hsieh JJ, Zhou S, Chen L, Young DB, Hayward SD. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. U. S. A. 96:23–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kedes DH, Lagunoff M, Renne R, Ganem D. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Invest. 100:2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klass CM, Offermann MK. 2005. Targeting human herpesvirus-8 for treatment of Kaposi's sarcoma and primary effusion lymphoma. Curr. Opin. Oncol. 17:447–455 [DOI] [PubMed] [Google Scholar]
- 19. Knight JS, Cotter MA, II, Robertson ES. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971–22978 [DOI] [PubMed] [Google Scholar]
- 20. Kovall RA, Hendrickson WA. 2004. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23:3441–3451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lan K, Kuppers DA, Robertson ES. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jκ, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lan K, Kuppers DA, Verma SC, Robertson ES. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Q, Zhou F, Ye F, Gao S-J. 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liang YY, Chang J, Lynch SJ, Lukac DM, Ganem D. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jκ (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liang YY, Ganem D. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 100:8490–8495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lim C, Gwack Y, Hwang S, Kim S, Choe J. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016–31022 [DOI] [PubMed] [Google Scholar]
- 27. Liu YH, et al. 2008. Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jκ and a cis-acting RTA responsive element. Virology 380:264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu J, Verma SC, Cai Q, Robertson ES. 2011. The Single RBP-Jκ site within the LANA promoter is crucial for establishing Kaposi's sarcoma-associated herpesvirus latency during primary infection. J. Virol. 85:6148–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu J, et al. 2009. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (KSHV) upregulates survivin expression in KSHV-associated B-lymphoma cells and contributes to their proliferation. J. Virol. 83:7129–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura H, et al. 2003. Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator. J. Virol. 77:4205–4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124:973–983 [DOI] [PubMed] [Google Scholar]
- 32. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–200 [DOI] [PubMed] [Google Scholar]
- 33. Oswald F, et al. 2005. RBP-Jκ/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 25:10379–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Paulose-Murphy M, et al. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pearson WR. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63–98 [DOI] [PubMed] [Google Scholar]
- 36. Persson LM, Wilson AC. 2010. February Wide-scale use of Notch signaling factor CSL/RBP-Jκ in RTA-mediated activation of Kaposi's sarcoma-associated herpesvirus lytic genes. J. Virol. 84:1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rainbow L, et al. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Russo JJ, et al. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Said W, et al. 1996. Kaposi's sarcoma-associated herpesvirus (KSHV or HHV8) in primary effusion lymphoma: ultrastructural demonstration of herpesvirus in lymphoma cells. Blood 87:4937–4943 [PubMed] [Google Scholar]
- 40. Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sarid R, Klepfish A, Schattner A. 2002. Virology, pathogenetic mechanisms, and associated diseases of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8). Mayo Clin. Proc. 77:941–949 [DOI] [PubMed] [Google Scholar]
- 42. Schwam DR, Luciano RL, Mahajan SS, Wong L, Wilson AC. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532–8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shinohara H, et al. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917–12924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Soulier J, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276–1280 [PubMed] [Google Scholar]
- 45. Tamura K, et al. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-Jκ/Su(H). Curr. Biol. 5:1416–1423 [DOI] [PubMed] [Google Scholar]
- 46. Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. 1998. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 18:644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verma SC, Borah S, Robertson ES. 2004. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78:10348–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ye F-C, et al. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78:11121–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]