Abstract
Cyclophilin A (CyPA) and its peptidyl-prolyl isomerase (PPIase) activity play an essential role in hepatitis C virus (HCV) replication, and mounting evidence indicates that nonstructural protein 5A (NS5A) is the major target of CyPA. However, neither a consensus CyPA-binding motif nor specific proline substrates that regulate CyPA dependence and sensitivity to cyclophilin inhibitors (CPIs) have been defined to date. We systematically characterized all proline residues in NS5A domain II, low-complexity sequence II (LCS-II), and domain III with both biochemical binding and functional replication assays. A tandem cyclophilin-binding site spanning domain II and LCS-II was identified. The first site contains a consensus sequence motif of AØPXW (where Ø is a hydrophobic residue) that is highly conserved in the majority of the genotypes of HCV (six of seven; the remaining genotype has VØPXW). The second tandem site contains a similar motif, and the ØP sequence is again conserved in six of the seven genotypes. Consistent with the similarity of their sequences, peptides representing the two binding motifs competed for CyPA binding in a spot-binding assay and induced similar chemical shifts when bound to the active site of CyPA. The two prolines (P310 and P341 of Japanese fulminant hepatitis 1 [JFH-1]) contained in these motifs, as well as a conserved tryptophan in the spacer region, were required for CyPA binding, HCV replication, and CPI resistance. Together, these data provide a high-resolution mapping of proline residues important for CyPA binding and identify critical amino acids modulating HCV susceptibility to the clinical CPI Alisporivir.
INTRODUCTION
Hepatitis C virus (HCV) is a member of the plus-strand RNA virus family Flaviviridae, which includes several additional significant human pathogens, such as West Nile virus, dengue virus, and yellow fever virus. HCV infection alone affects more than 130 million people worldwide and imposes a significant toll on the global health care system, because the majority of patients are chronically infected and in need of a more effective therapy.
Replication of the HCV genome occurs on intracellular membranes and requires the participation of multiple nonstructural proteins, including the nonstructural protein 3 (NS3)/NS4A protease, the NS3 helicase, membranous web-forming protein NS4B, the RNA-dependent RNA polymerase NS5B, and the replicase cofactor NS5A. In addition, the viral replication complex contains cellular proteins that are also essential for HCV replication (46). One of these cellular cofactors is human cyclophilin A (CyPA) (64), a cytosolic protein originally identified as the protein target of the immunosuppressive drug cyclosporine (CsA) (20). The targets of CsA and another immunosuppressive compound, FK506, are CyPs and FK506-binding proteins (FKBPs), respectively. The CsA-CyPA and the FK506-FKBP complexes bind to a common target, calcineurin, and block its phosphatase activity and T-cell activation (32). FK506 does not inhibit HCV in vitro (60), whereas derivatives of CsA, including the clinical-stage compounds DEB-025 (Alisporivir) and NIM811, inhibited HCV replication efficiently despite lacking any immunosuppressive function (39, 42). These results strongly indicate that the anti-HCV mechanism is independent of the calcineurin pathway. In addition, the mechanism of cyclophilin inhibitors (CPIs) is distinct from that of interferon (IFN) (47, 60), suggesting potential benefits of combining CPI treatment with IFN/ribavirin therapy.
Both CyPA and FKBPs are members of the superfamily of peptidyl-prolyl isomerases (PPIases) that catalyze the cis-trans isomerization of peptidyl-proline bonds (15, 49). The importance of CyPA's PPIase activity in HCV replication has been confirmed by several independent labs using both chemical and RNA interference approaches (5, 27, 34), but the relevant substrates of the PPIases are much less defined. At least three HCV proteins (NS2, NS5A, and NS5B) have been implicated in the action of CyPA as an HCV cofactor (7, 21, 33), and increasing evidence supports a direct interaction between CyPA and NS5A (8, 14, 21, 58, 63). Recombinant CyPA protein interacts with both full-length NS5A from infected cell lysates and purified NS5A domains. Importantly, mutations in the active site of CyPA or CPI treatment readily abolish the CyPA-NS5A interaction.
Like all NS proteins, NS5A is associated with intracellular membranes (3, 12, 43). It is strictly required for RNA replication (2, 36) and assembly of viral particles (1, 52), but little is known about its precise function or mechanisms of action, and no enzymatic activity has been assigned to it. The NS5A gene is a hot spot for cell culture adaptive mutations in the viral genome (2, 35), and hyperphosphorylation likely plays an important role in regulating NS5A function (13, 40). In addition to the N-terminal membrane anchor, NS5A contains three discrete domains (I, II, and III) connected by two solvent-exposed low-complexity sequences (LCS-I and LCS-II) (54). Domain I has been crystalized and may adopt two different dimer forms, with the monomers having the same structures (37, 55). One of the dimeric conformations reveals a large groove facing away from the membrane and suggests an RNA-binding function for domain I (55). In contrast to the highly structured domain I, domains II (30) and III (22) are intrinsically disordered and have resisted crystallization efforts so far. Consistent with the proline-rich nature of these domains, both domains II and III have been shown to be substrates of CyPA in vitro, and various proline residues showed chemical shift perturbation in nuclear magnetic resonance (NMR) studies when incubated with recombinant CyPA (21, 57); many of these residues, however, do not appear to contribute to HCV replication or CyPA dependence when examined in functional assays (53, 63).
Recently, an unbiased genetic approach, termed cofactor-independent mutant selection (CoFIM), that combines RNA interference and viral mutant selection (63) was developed to identify a dipeptide motif (Asp-Tyr) within domain II of NS5A that regulates HCV's dependence on CyPA. Mutations of this motif, either individually (D to E; Y to N) or in combination (DEYN), reduced viral dependence on CyPA and conferred resistance to CPIs such as cyclosporine. Here, we report a systematic characterization of proline residues within the NS5A domains II through III that revealed a tandem motif that constitutes the principal CyPA-binding site of NS5A and regulates HCV's susceptibility to CPIs.
MATERIALS AND METHODS
Compounds, cell lines, and antibodies.
CsA was purchased from Alexis Corporation (San Diego, CA), and DEB-025/Alisporivir was provided by Debiopharm (Lausanne, Switzerland) and Novartis Institutes for Biomedical Research (Cambridge, MA). Huh-7.5 cells were provided by Charles Rice (Rockefeller University) and Apath LLC, (St. Louis, MO). Anti-NS5A antibodies included 7B5 (BioFront Technologies, Tallahassee, FL) and 9E10 (provided by Timothy Tellinghuisen, Scripps Florida). The CyPA-KD cell line (sh-A161) has been described previously (64).
Overlapping peptide array spanning domain II through domain III.
A series of 18 peptides, each consisting of 18 amino acids, was designed to span the entire length of domain II through domain III of JFH-1 NS5A. Each peptide had an overlap with the previous peptide of five to seven residues. All peptides were acetylated at their N termini. Peptides were immobilized by their C termini on a cellulose membrane during synthesis (JPT Peptide Technologies GmbH, Berlin, Germany) and contained spacers of two alanine residues between each NS5A sequence and the membrane. Similarly, a set of soluble peptides was synthesized that covered the same region of NS5A (Sigma-Genosys, The Woodlands, TX). The soluble peptides were acetylated and amidated at their N termini and C termini, respectively.
CyPA-peptide spot-binding assay.
A CyPA-binding detection reagent (CyPA-horseradish peroxidase [HRP]) was generated with the Lightning-Link horseradish peroxidase kit (Innova Biosciences, Cambridge, United Kingdom) by following the manufacturer's procedures. Cellulose membranes containing the NS5A peptides (synthesized on the membrane) were first soaked in methanol for 5 min at room temperature. The membranes were then washed in Tris-buffered saline (TBS) buffer (50 mM Tris [pH 8.0], 137 mM NaCl, 2.7 mM KCl,) three times for 10 min each. The membrane was blocked with 5% nonfat milk in PBST (11.9 mM sodium phosphate [pH 7.4], 137 mM NaCl, 2.7 mM KCl) for 1 h at room temperature. The CyPA-HRP conjugate was diluted in blocking buffer to a final concentration of 0.5 nM and incubated with the membrane for 1 h at room temperature. The membrane was washed three times (10 min each) with PBST buffer and then briefly dried on filter paper and incubated with Immobilon HRP substrate (Millipore, Billerica, MA) for 5 min. The HRP substrate was removed, and the membrane was exposed to autoradiography film. For competition experiments, CyPA-HRP was first incubated with inhibitors (DEB-025, CsA, and various peptides), with a CyPA-to-inhibitor molar ratio of 1:1,000, for 1 h at room temperature while the membrane was being blocked. Spots on X-ray films were scanned and converted to JPEG images, which were then quantified with ImageJ software (NIH, MD).
Regeneration of peptide array membranes.
Spot membranes were regenerated by a series of washing steps, each lasting 10 min at room temperature. The membrane was first washed twice in Nanopure water and then washed sequentially in regeneration buffer A (8 M urea, 1% SDS, 0.1% β-mercaptoethanol) and regeneration buffer B (10% acetic acid, 50% ethanol), three times in each buffer. After that, the membrane was rinsed once in Nanopure water and then washed three times in T-TBS (TBS buffer with 0.05% Tween 20). Regenerated membranes were kept hydrated at 4°C for short-term storage or dried on filter paper for long-term storage at −20°C.
Recombinant vaccinia virus-mediated expression of HCV proteins.
Transfection of HCV constructs was conducted with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Four hours after transfection, a stock of recombinant vaccinia virus (vTF7-3; 2 × 109 PFU/ml; courtesy of Bernard Moss, NIH, Bethesda, MD) was diluted 1:125 in complete medium and passed through a 22-gauge needle 10 times. To each well, 167 μl of the diluted virus was added (multiplicity of infection [MOI] of ∼100). The vaccinia virus was removed after 2 h, and cells were incubated for an additional 36 h before lysates were harvested for the pulldown experiments.
Preparation of cell lysates from JFH-1 or vaccinia virus-infected cells.
Cell lysates were prepared by aspiration of culture media and direct addition of 600 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT], 1× Complete-Mini Roche protease cocktail) per 8 × 105 cells. The culture vessel was placed at 4°C and rocked gently for 1 h. Cell lysates were clarified by centrifugation at 18,000 × g for 10 min.
Pulldown assays.
In vitro binding of CyPA to NS5A-containing lysates was performed as follows: 100 μl of lysate was precleared with 75 μl of 50% Ni-nitrilotriacetic acid (NTA) agarose (Sigma-Aldrich, St. Louis, MO) in low-imidazole buffer (10 mM imidazole, 250 mM NaCl, 50 mM sodium phosphate, pH 8.0) for 1 h at 4°C. The lysate was then centrifuged at 1000 × g for 1 min, and the supernatant was saved as precleared lysate. A total of 300 μg of CyPA protein was added to cleared lysates and incubated for 1 h at 4°C. A total of 75 μl of 50% Ni-NTA agarose was added to the mixture, which was then incubated for 1 h at 4°C before centrifugation was used to separate the unbound from the bound proteins. For the competition experiments, the competitors (DEB-025 and soluble peptides) were first mixed with CyPA prior to its addition to the lysate. The molar ratio of competitor to CyPA was approximately 3.3 to 1. Protein bands on X-ray films were scanned and converted to JPEG images, which were then quantified with ImageJ software (NIH, MD).
NMR analysis.
Production and purification of 15N-labeled CyPA has been described previously (63). Purified 15N-labeled CyPA protein was dialyzed against NMR buffer (50 mM Na2HPO4, 20 mM NaCl, 2 mM EDTA, 1 mM DTT, pH 6.3). Synthetic peptides corresponding to P6 (residues 305 to 322 of JFH-1 NS5A) and P8 (residues 329 to 346 of JFH-1 NS5A) were obtained from Sigma-Genosys (The Woodlands, TX). Peptides were dissolved in dimethyl sulfoxide (DMSO) at 10 mM and added to 15N-labeled CyPA (200 μM) at the desired molar equivalents. Two-dimensional 1H,15N-heteronuclear single-quantum coherence buffer (HSQC) NMR spectra (28) were acquired at 298 K on a Bruker Avance spectrometer operating at 700 MHz (1H frequency). Spectra were collected as 1,024 by 200 complex point matrices with spectral widths of 11,261.3 and 2,838.5 Hz in the 1H and 15N dimensions, respectively, by means of 16 scans per complex t1 (time increment) point. Spectra were processed with nmrPipe (10) and analyzed with NMRViewJ (26). Chemical shift changes were calculated as described previously (63).
Mutagenesis, in vitro transcription, electroporation, and luciferase assays.
Mutagenesis, in vitro transcription, electroporation, and luciferase assay protocols have been described previously (63). Primer sequences for all the mutagenesis reactions are available upon request.
RESULTS
Scanning of NS5A domain II through domain III to identify proline-containing peptides that bind to CyPA in vitro.
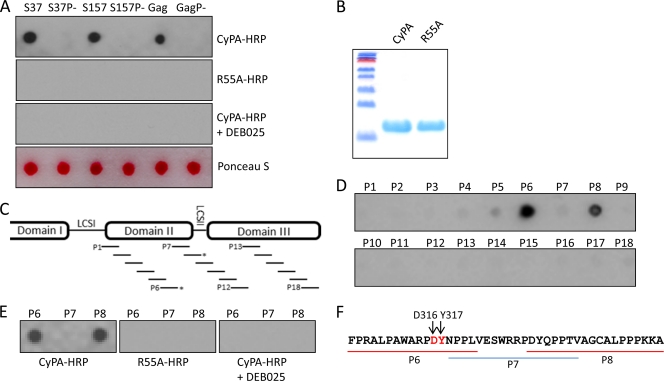
Both domains II and III of HCV NS5A are rich in prolines and implicated in CyPA's function as a cofactor for HCV replication (21, 57, 63). To identify proline substrates of CyPA in these domains using an unbiased approach, we screened a peptide array that consisted of 18 overlapping peptides spanning NS5A domain II, LCS-II, and domain III of JFH-1 with a spot-binding assay previously used to select consensus peptides from the phage display library (45). A slight modification to the published protocol was that a CyPA-HRP conjugate was used directly, as opposed to the glutathione S-transferase (GST)-CyPA that was then detected by anti-GST-HRP. To first validate the modified assay, we tested several peptides that bound to CyPA in this format (45) and an HIV Gag peptide (19, 56, 65), a known substrate for the PPIase activity of CyPA (see Table 1 for sequences). These positive-control peptides bound to CyPA in the spot assay, while corresponding mutant peptides devoid of proline residues failed to do so (Fig. 1A, top). The R55A mutant of CyPA, which is defective in PPIase activity (66) and deficient in binding to full-length NS5A (63), also failed to bind any of the spots even though equivalent amounts of the wild-type (wt) and mutant proteins were used (Fig. 1A and B). Finally, the spot signals were readily abolished by pretreatment of CyPA with DEB-025/Alisporivir (Fig. 1A). Taken together, these data indicated that the spot assay format was suitable for detecting biologically relevant interactions of CyPA in a PPIase-dependent manner. When the overlapping peptides from NS5A (Fig. 1C and Table 2) were assayed for CyPA-HRP binding, only two peptides, P6 and P8, bound strongly to CyPA (Fig. 1D). The interactions were sensitive to DEB-025 treatment, and the R55A mutant did not bind to either peptide under the same conditions (Fig. 1E). The P7 peptide, which overlapped with segments of both P6 and P8, did not bind CyPA (Fig. 1F), suggesting that CyPA binding by P6 and P8 was mediated by residues other than the two diproline motifs present in the P7 peptide. Finally, two proline-containing peptides from domain I and low-complexity sequence I of NS5A also failed to bind to CyPA in this assay (Table 3 and data not shown).
Table 1.
Sequences of CyPA-binding peptides and their proline-deficient mutantsa
| Spot name | Sequence | Reference |
|---|---|---|
| S37 | PRKGPPLPNGDTEVHGLA | 45 |
| S37P− | ARKGAALANGDTEVHGLA | This study |
| S157 | VIFDPYAPKLTSSVAEHK | 45 |
| S157P− | VIFDAYAAKLTSSVAEHK | This study |
| HIV Gag | VHAGPIAPGQMREPRGSD | 65 |
| HIV GagP− | VHAGAIAAGQMREARGSD | This study |
Positive-control peptides that have been described previously to bind CyPA and their derivatives devoid of proline residues were used to validate the spot-binding assay (see Fig. 1A). Alanine residues in bold indicate substitutions.
Fig 1.
CyPA interacts with JFH-1 NS5A peptides in a spot-binding assay. (A) Validation of a spot-binding assay using reported CyPA-binding peptides. Peptides (∼5 nmol) synthesized on a cellulose membrane filter were incubated with HRP-conjugated CyPA (top row), HRP-conjugated R55A mutant CyPA (second row), and CyPA-HRP in the presence of DEB-025 (450 nM) (third row). CyPA-bound spots were identified by exposure to an X-ray film after chemiluminescent substrate incubation. Ponceau S staining was used to demonstrate that the spots for both the wt and the proline mutant (P−) of each sequence contained comparable amounts of peptides. Note that each experiment was done at least twice and wt-CyPA experiments were carried out using regenerated membrane after the R55A and DEB-025 experiments, indicating that the negative results of the R55A and DEB-025 were not because of membrane deterioration. (B) SDS-PAGE image of CyPA and R55A recombinant proteins used in this study to make the HRP conjugates; (C) schematic representation of the 18 overlapping peptides (designated P1 through P18) that span domains II and III of JFH-1 NS5A; (D) CyPA-HRP interacts with immobilized P6 (residues 305 to 322) and P8 (residues 329 to 346) peptides; (E) CyPA binding by P6 and P8 peptides was sensitive to CPI DEB-025 and PPIase active-site mutation R55A; (F) NS5A sequence encompassing P6, P7, and P8 peptides. The D316 Y317 motif that regulates CsA sensitivity is shown in red.
Table 2.
Peptides used for probing CyPA bindinga
| Peptide | Sequence |
|---|---|
| P1 | THSNTYDVDMVDANLLME |
| P2 | ANLLMEGGVAQTEPESRV |
| P3 | EPESRVPVLDFLEPMAEE |
| P4 | EPMAEEESDLEPSIPSEC |
| P5 | SIPSECMLPRSGFPRALP |
| P6 | FPRALPAWARPDYNPPLV |
| P7 | NPPLVESWRRPDYQPPTV* |
| P8 | DYQPPTVAGCALPPPKKA |
| P9 | PPPKKAPTPPPRRRRTVG |
| P10 | RRRTVGLSESTISEALQQ |
| P11 | SEALQQLAIKTFGQPPSS |
| P12 | GQPPSSGDAGSSTGAGAA |
| P13 | TGAGAAESGGPTSPGEPA |
| P14 | SPGEPAPSETGSASSMPP |
| P15 | ASSMPPLEGEPGDPDLES |
| P16 | DPDLESDQVELQPPPQGG |
| P17 | PPPQGGGVAPGSGSGSWS |
| P18 | GSGSWSTCSEEDDTTVCC |
Sequences of 18 peptides (P1 through P18) that span the distance from low-complexity sequence I (LCS-I) to the C terminus of domain III of JFH-1 NS5A (residues 245 through 466) are shown. Each is 18 residues in length and includes a six-amino-acid overlap with the preceding peptide.
, the P7 peptide was shifted one residue to avoid an N-terminal tyrosine, which interfered with the HRP-based spot assay used in this study.
Table 3.
Proline-containing sequences from viruses that failed to bind CyPA-HRPa
| Viral peptide | Sequence |
|---|---|
| HCV NS5A-domain I | TEGQCAPKPPTNYKTAIW |
| HCV NS5A-LCS-I | ARRLARGSPPSEASSSVS |
| HCV NS2 | LRDLAVAVEPIIFSPMEK |
| HCV NS5B | KTKLKLTPLPEARLLDLS |
| GBV-B NS5A | RTGSLTLPPPPRSVPGVS |
| DEN-2 NS5 | SGVDVFFIPPEKCDTLLC |
| BVDV 1a NS5 | LFEELLRCPPATKSNKGH |
| WNV NS5 | KVDTKAPEPPEGVKYVLN |
Control peptides 18 residues in length that contain single or multiple prolines were not able to bind to CyPA in the spot-binding assay. Sequences were selected from HCV NS5A domain I and low-complexity sequence I, hepatitis C virus (HCV) nonstructural protein 2 (NS2; containing P164), HCV NS5B (containing P540), and the NS5 proteins of GBV-B, dengue virus 2, bovine viral diarrheal virus, and West Nile virus (WNV). Proline residues are highlighted in bold.
Residues in P6 that influence CyPA binding.
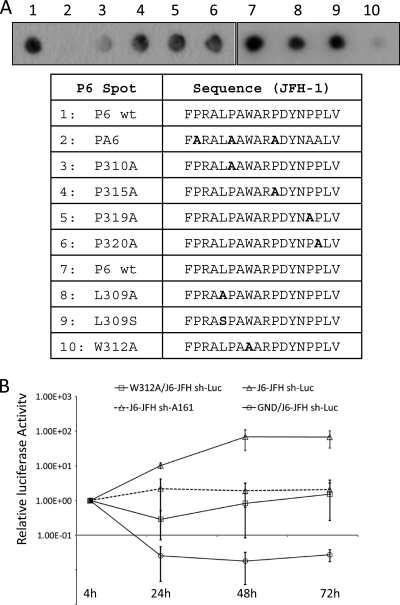
The P6 peptide of JFH-1 resides within a linear sequence in NS5A domain II that had been implicated in NS5A-CyPA interaction by both biophysics (21) and functional approaches (63). However, this peptide contains multiple prolines, and the relative contribution of the multiple prolines to CyPA binding had not been resolved. We tested a series of mutant P6 peptides with a single proline replaced by an alanine in the spot assay. As expected for a PPIase substrate, the mutant P6 peptide with all prolines replaced by alanines did not show any binding to CyPA (Fig. 2A). Of the individual single proline mutants, the P310A peptide exhibited the most significant decrease in CyPA binding, consistent with our recent finding that P310 had the most dramatic effect on regulating CsA sensitivity in a CsA-resistant virus (63). Because hydrophobic residues in the vicinity of proline substrates have been reported to influence CyPA binding (45), we next determined whether the two hydrophobic residues flanking P310 had any effect. We replaced the leucine residue at position 309 with either a polar residue (L309S) or a smaller hydrophobic alanine (L309A) and the tryptophan residue at position 312 with an alanine. Although the leucine substitutions only had a small (10 to 20%) reduction in CyPA binding, the W312A mutation reduced P6 binding to CyPA by up to 90% (Fig. 2A). We then engineered the W312A mutation into a full-length HCV genome based on J6-JFH and determined the replication kinetics of this mutant virus. The W312A mutant virus replicated 10- to 100-fold less than the wt virus (Fig. 2B), resembling a phenotype caused by the P310A mutation (63). Of note, mutations of these residues important for CyPA binding had an inhibitory effect on HCV produced in cell culture (HCVcc) replication similar to that of the depletion of CyPA from the cells (Fig. 2B) (63), suggesting a role of these amino acids in mediating CyPA's function as a cofactor of HCV replication.
Fig 2.
Amino acid residues within the P6 peptide influence CyPA binding. (A) Wild-type (wt), proline-deficient (PA), and single substitutions were evaluated for their ability to bind CyPA; (B) the W312A mutation resulted in reduced replication of a full-length J6-JFH genome. HCV RNAs of wt, W312A, and a polymerase-deficient mutant (GND) were electroporated into Huh-7.5 sh-Luc or sh-A161 stable cell lines. Luciferase assays were performed 4, 24, 48, and 72 h after electroporation. Values were normalized to the 4-h reading, and error bars represent standard deviations from two independent experiments.
The P6 CyPA-binding site of NS5A is conserved across all genotypes.
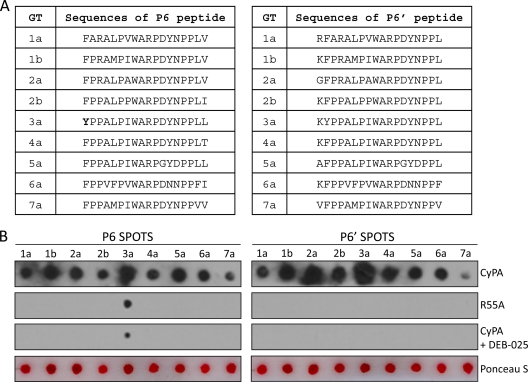
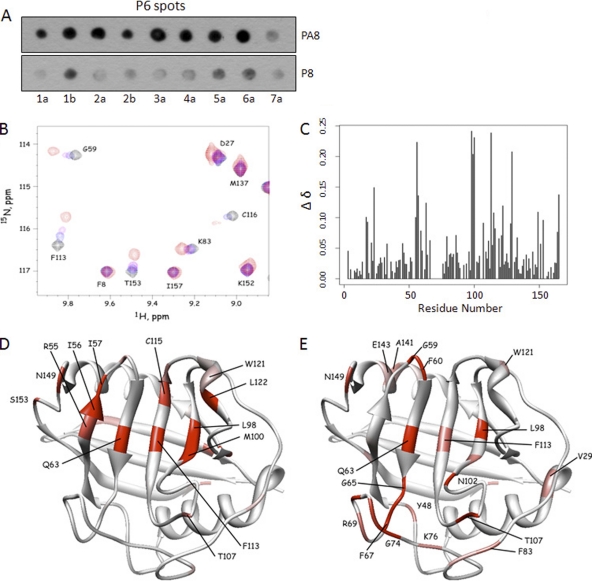
The peptide region represented by P6 is largely conserved in all the major genotypes of HCV (Fig. 3A). We next determined whether the corresponding P6 peptides from other genotypes could also bind to CyPA. P6 peptides from all seven genotypes (48) tested were capable of binding CyPA in the spot assay (Fig. 3B), although there were clear differences in the binding strengths among the different genotypes. For example, the P6 peptide from GT3a consistently generated the strongest spot signal, while the GT7a and GT1a spots were much weaker. All the interactions were sensitive to DEB-025 treatment and abolished by R55A mutation in CyPA with the exception of the GT3a P6 peptide, which still produced a spot signal, albeit much reduced, with R55A-HRP or when the binding was performed in the presence of DEB-025 (Fig. 3B, left). Of note, this peptide uniquely possesses an N-terminal tyrosine residue, and because we observed that an N-terminal tyrosine present in the immobilized peptides tends to generate a nonspecific spot signal in past experiments (H. Grise and H. Tang, unpublished data), we tested a modified version of the P6 peptides. The sequences of this set of peptides (P6′) were shifted upstream by one amino acid so that none of the new peptides had any tyrosine as the first residue at the N terminus (Fig. 3A, right). All P6′ peptides again bound to CyPA, and this time the signals from all the genotypes were abrogated by DEB-025 treatment and were also negative for R55A binding (Fig. 3B, right).
Fig 3.
CyPA binding to P6 peptide is conserved across all seven genotypes. (A) Amino acid sequences of the P6 and P6′ (sequence shifted one amino acid upstream of P6) peptides of the indicated genotypes (the NCBI accession numbers for the sequences selected are 1a, AAB67036.1; 1b, AJ238799.1; 2a, AB047639.1; 2b, D10988.1; 3a, ADF97231.1; 4a, CAA72338.1; 5a, AAC61696.1; 6a, CAA72801.1; 7a, ABN05226.1). Note the unique N-terminal tyrosine residue in the GT3a P6 sequence. (B) CyPA bound to P6/P6′ of different genotypes in a PPIase-dependent and CPI-sensitive manner. Ponceau S staining revealed that the differences in binding strengths of the different P6/P6′ sequences were not due to the amount of peptides spotted.
P8 represents the second part of a tandem CyPA-binding site of HCV NS5A.
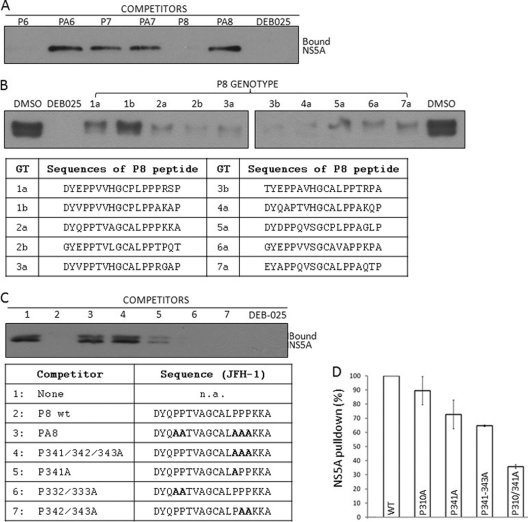
In addition to the P6 sequence, we also identified a second, novel CyPA-binding sequence (P8) that is located at the junction of NS5A domain II and LCS-II. To confirm the P8-CyPA interaction with an independent approach, we characterized the P8 peptide using a competition assay in which soluble peptides were incubated with histidine-tagged CyPA (His-CyPA) before the latter was used to affinity purify full-length NS5A from infected cell lysates. Consistent with our previous report that the NS5A pulldown by His-CyPA was dependent upon the active site of CyPA and sensitive to CsA (63), DEB-025 efficiently disrupted NS5A pulldown (Fig. 4A). Both P6 and P8 peptides competed with full-length NS5A protein for CyPA binding, whereas the proline-deficient control peptides (PA6 and PA8) failed to do so (Fig. 4A). The P7 peptide again did not show significant competition, suggesting that the major interacting prolines lie outside the P7 peptide (see below). P8 peptides from all seven genotypes were capable of binding to CyPA in the soluble format, although the P8 peptide from GT1b appeared to be less potent in competing with the full-length NS5A, which was from GT2a/JFH-1 (Fig. 4B). When proline residues within P8 were mutated to determine their relative importance for CyPA binding, residue P341 was found to be the major proline that contributed to CyPA binding, as any mutant P8 peptides with this proline mutated exhibited reduced ability to compete for CyPA binding (Fig. 4C, lanes 3 to 5). Double mutations of the other prolines (P332A/P333A and P342A/P343A) did not affect P8's ability to compete for CyPA binding (Fig. 4C, lanes 6 and 7).
Fig 4.
P6 and P8 represent a tandem CyPA-binding site of HCV NS5A. (A) Soluble P6, P7, and P8 peptides were evaluated for their ability to compete with NS5A from JFH-1-infected cell lysates for CyPA binding. The indicated peptides and DEB-025 were added to His-CyPA and incubated before NS5A-containing lysate was added to the pulldown assay. (B) Soluble P8 peptides from all seven genotypes were capable of binding to CyPA in the competition assay. Peptides were used at a 3.3-fold molar excess of the His-CyPA. (C) Proline at position 341 is the major contributor to CyPA binding by the JFH-1 P8 peptide. (D) P310 and P341 are the principal CyPA-binding sites on NS5A. HCV RNA containing NS5A of wt, P310A, P341A, P341-343A, or P310A P341A were expressed in Huh-7.5 cells using a recombinant vaccinia virus system and tested in the His-CyPA pulldown assay. The image shown is representative of two independent experiments. Note that in experiments where the two forms of phospho-NS5A (p56 and p58) were resolved by SDS-PAGE, CyPA binding was not biased toward either form, in agreement with previous results (63).
To determine the contribution of P310 and P341 to the interaction between CyPA and full-length NS5A, we generated NS5A mutant viral constructs where these proline residues were mutated to alanine, both individually and in combination. The NS5A proteins with these mutations were expressed in Huh-7.5 cells using a vaccinia-T7 expression system (18) and then tested in the His-CyPA pulldown assay. Wild-type NS5A bound efficiently to CyPA, and removing P310 or P341 individually slightly reduced CyPA binding. Importantly, mutating P310 and P341together was sufficient to eliminate close to 70% of CyPA binding by the full-length NS5A (Fig. 4D), indicating that these two prolines comprise the main binding sites of NS5A-CyPA interaction.
We next tested if P6 and P8 bind to a similar site on CyPA by using soluble P8 peptide to compete for CyPA binding to immobilized P6 spots. The P6 spots from various genotypes were incubated with CyPA in the presence of either P8 or PA8 soluble peptides. Compared to the PA8 peptide, the wt P8 peptide (based on JFH-1) reduced CyPA binding to P6 spots from all genotypes (Fig. 5A), although clear differences in relative decreases were again observed among the different genotypes. Nevertheless, these data suggest that P6 and P8 bind to the same site on CyPA. We next performed NMR experiments to identify amino acids of CyPA that undergo chemical shift perturbation upon binding to P6 or P8 peptide. We first obtained chemical shift assignments of CyPA without peptides by using 13C- and 15N-labeled CyPA by following published guidelines (41, 61) and then determined the perturbation of chemical shifts by titration of the P8 peptide (Fig. 5B and C). Residues exhibiting the largest chemical shift perturbations in the presence of P8 were generally localized to the known cyclophilin-active site (Fig. 5D). When P6 peptide was studied in the same experiment, a similar chemical shift perturbation profile was observed (Fig. 5E), again indicating a tandem arrangement of duplicate CyPA-binding sites.
Fig 5.
P6 and P8 bind to the ligand-binding pocket of CyPA with similar binding strengths. (A) Soluble P8 peptide competed with P6 for CyPA binding in a proline-dependent manner. Soluble P8/PA8 peptides were included in approximately equal amounts as the total P6 peptides on the membrane (∼5 nmol per spot) in the spot-binding assay that measured P6-CyPA interaction. CyPA binding to P6 spots was reduced by the inclusion of the P8 but not the PA8 peptide. (B) NMR spectra showing the effect of P8 titration on the chemical shifts. Some resonances are insensitive to P8, while others show high sensitivity in both the 1H and 15N chemical shifts. (C) Chemical shift changes (Δδ) were quantified and plotted for each residue of CyPA. (D) Residues that exhibited chemical shift perturbations were highlighted in a structure of CyPA complexed with a fragment of the HIV-1 Gag protein (PDB access code 1FGL). Normalized Δδ values for P8 titration (65) were used to generate the color scheme: red, 0.75 < Δδ < 1; pink, 0.5 < Δδ < 0.74; white, Δδ < 0.50. (E) Residues with normalized Δδ values for P6 titration are shown in the same structure of CyPA. The color scheme is the same as that described for panel C.
P310 and P341 regulate HCV's CyPA dependence and susceptibility to CsA.
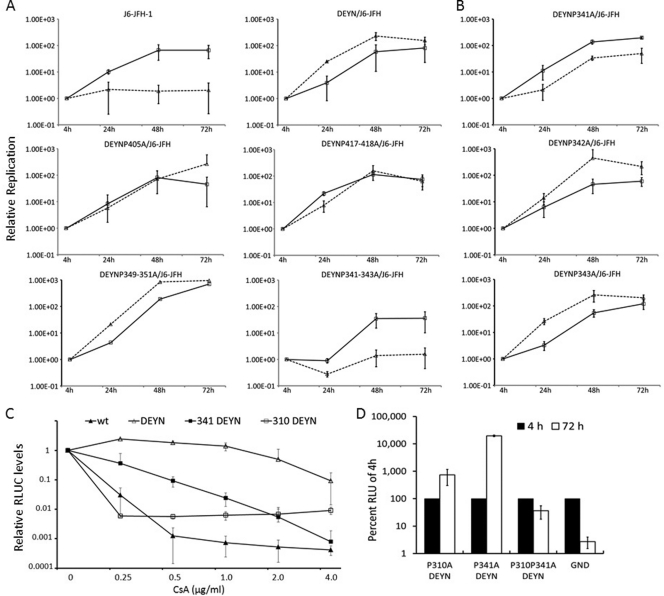
HCVcc was not able to replicate in Huh-7.5 cells expressing a small hairpin RNA (shRNA) directed at CyPA mRNA (64), but point mutations D316E and Y317N in NS5A enabled the mutant virus (DEYN) to efficiently replicate in CyPA-KD cells (63). Mutations of prolines relevant for CyPA function are expected to abolish DEYN′s ability to replicate in CyPA-KD cells, because neither the aspartate nor the tyrosine is expected to serve as the substrate for a PPIase. Indeed, we previously found the P310A mutation to be lethal in the wt JFH-1 background, and it severely reduces the ability of DEYN virus to infect CyPA-KD cells (63). To functionally characterize such prolines in LCS-II and domain III, we engineered a panel of HCV constructs that contain proline-to-alanine mutations in the DEYN background and then determined their ability to replicate in control (expressing an shRNA against firefly luciferase; sh-Luc) and in CyPA-KD (sh-A161) cells. As shown in Fig. 6A and Table 4, most of the mutants (single, double, and triple prolines) retained their ability to replicate in both cell lines. Like the DEYN virus, these mutants replicated to higher levels in the CyPA-KD cells than in the control cells, indicating that they retained the adaptation to the low levels of CyPA for optimum replication. Mutation of one specific stretch of prolines, P341 through P343, however, abrogated replication in CyPA-KD cells and reversed the relative efficiencies in the two cell lines (Fig. 6A). Further parsing of the three prolines revealed that P341 is the principal mediator of the phenotype (Fig. 6B), as P341A was the only mutant that could reverse the replication phenotype of the DEYN virus. These data independently identify P341 as an important residue involved in CyPA dependence.
Fig 6.
Alanine scanning of NS5A LCS-II and domain III in the background of DEYN mutations identify P310 and P341 as important for CyPA dependence. Prolines from P341 to P446 were mutated in singlets, doublets, or triplets within the DEYN/J6-JFH construct. Mutant RNAs were electroporated into Huh-7.5 sh-Luc and sh-A161 cell lines. Luciferase assays were performed 4, 24, 48, and 72 h after electroporation. Values were normalized to the 4-h reading in panels A, B, and D, and error bars represent standard deviations from two independent experiments. Solid lines, sh-Luc cells; dashed lines, sh-A161 cells. (A) Replication profiles of proline mutants of LCS-II and domain III. Top, the wt phenotype compared to the DEYN phenotype demonstrates rescue in the sh-A161 cell line; middle left, representative replication kinetics of single mutations (e.g., DEYN P405A); middle right, those of double mutations (e.g., DEYN P417-418A); bottom, those of triple mutations. Whereas the DEYN P349-351A mutant exhibited a phenotype similar to that of the DEYN virus, the DEYN P341-343A mutant uniquely abrogates the ability of DEYN mutations to rescue replication in the sh-A161 cell line (please see the summary of all mutants in Table 4). (B) The individual DEYN P341A, P342A, and P343A mutations highlight the importance of P341 in CyPA dependence. (C) P310A and P341A increase sensitivity to CsA treatment. J6-JFH RNAs with indicated mutations were electroporated into the Huh-7.5 sh-Luc cell line and treated with CsA 24 h after electroporation. Cells were collected 72 h after treatment and assayed for luciferase activity. Fold changes in response to treatment were normalized to no treatment (set at 1) and plotted. (D) DEYN mutations were unable to rescue the lethal phenotype of P310A P341A.
Table 4.
Alanine-scanning mutagenesis of proline residues in NS5A domain II
| Proline mutation (in DEYN) | Replication capacitya |
sh-A161/sh-Luc ratio of replication capacityb | |
|---|---|---|---|
| sh-Luc | sh-A161 | ||
| None/D316/Y317 | +++ | − | 0.04 ± 0.03 |
| None | ++++ | ++++ | 2.50 ± 0.32 |
| P310A | + | − | 0.05 ± 0.01 |
| P341-343A | ++ | − | 0.07 ± 0.06 |
| P310A/P341A | − | − | NA |
| P341A | ++++ | ++ | 0.24 ± 0.02 |
| P342A | +++ | ++++ | 1.94 ± 0.21 |
| P343A | +++ | ++++ | 2.71 ± 0.09 |
| P347A | + | ++++ | 4.28 ± 0.10 |
| P349-351A | ++++ | ++++ | 1.36 ± 0.08 |
| P379-380A | ++ | +++ | 1.49 ± 0.04 |
| P399A | +++ | ++++ | 2.85 ± 1.35 |
| P402A | ++++ | ++++ | 1.42 ± 0.38 |
| P405A | +++ | ++++ | 1.79 ± 0.50 |
| P407A | ++++ | ++++ | 1.08 ± 0.28 |
| P417-418A | ++++ | ++++ | 1.37 ± 0.75 |
| P423A | ++++ | ++++ | 1.11 ± 0.14 |
| P426A | ++++ | ++++ | 1.89 ± 0.09 |
| P437-439A | ++ | ++++ | 3.01 ± 1.20 |
| P446A | ++ | +++ | 2.92 ± 1.37 |
Replication capacity of the WT (fold increase from 4 h to day 3) was set at 100%, and the following scales were used: −, 0 to 10%; +, 11 to 50%; ++, 51 to 75%; +++, 76 to 100%; ++++, >100%.
The ratio of replication capacity in sh-A161 cells to that in sh-Luc cells was calculated and presented for all the D316E/Y317N mutation-based genomes. NA, not assayed.
We also determined whether the regulation of CyPA dependence by P310A and P341A was correlated with HCV's sensitivity to CsA. The wt J6-JFH was highly sensitive to and effectively inhibited by CsA at a concentration between 0.25 and 0.5 μg/ml. The DEYN mutant, however, remained largely resistant to up to 2 μg/ml of CsA. Both P310A and P341A reduced the CsA-resistant levels of the DEYN mutant virus (Fig. 6C). The double proline mutant, even in the DEYN background, was nonreplicative (Fig. 6D). These results suggest that DEYN mutations regulate CsA resistance and CyPA independence through the action of CyPA on both P310 and P341.
DISCUSSION
An essential role of CyPA-NS5A interaction in HCV replication is supported by increasingly consistent results (8, 14, 58, 63). The work described here identifies a tandem CyPA-binding site within NS5A domain II and LCS-II by means of an integrated approach of biochemical and functional assays. Two specific proline residues relevant for CyPA function were mapped to P310 and P341 in JFH-1 NS5A, and the two binding peptides (P6 and P8) centered on these prolines are well conserved in all genotypes. A cross-competition assay and NMR chemical shift experiments revealed that P6 and P8 bind to CyPA similarly, by interacting with its ligand-binding site. Interestingly, although each region of domain II was capable of binding to CyPA individually, both prolines were required for optimal replication of HCV. These results suggest that binding to both sites by CyPA molecules, either simultaneously or in an ordered sequence, is required for NS5A to function properly in the HCV life cycle. The involvement of multiple prolines in CyPA binding has been observed for another viral protein, the HIV Vpr (50). In contrast to the well-characterized Gag-CyPA interaction, which involved a single proline (38), Vpr interacts with CyPA with up to three proline residues, each with weak binding.
Results from previous NMR experiments suggested that many more residues in NS5A domains II and III can be isomerized by CyPA in vitro when purified domains II or III were analyzed (21, 57). For full-length JFH-1 NS5A expressed in Huh-7.5 cells, however, CyPA binding is mediated mostly by P310 and P341, because mutations of just these two prolines eliminated the majority of CyPA binding. The binding affinity between CyPA and NS5A domain II is already quite low (∼90 uM) (21), so any additional interactions outside the P6 and P8 peptides, which bound to CyPA with similar strengths, are expected to be even weaker. Indeed, any residual interaction between CyPA and the P310A P341A mutant was insufficient to sustain NS5A function in viral replication, as the double proline mutant was not viable, even in wild-type cells with normal levels of endogenous CyPA. These results do not rule out the possibility that the additional parts of domains II and III exhibiting low CyPA-binding affinity can contribute to NS5A's function beyond replication, such as signaling and/or pathogenesis. Recently, a distinct PPIase, the phosphorylation-dependent isomerase Pin1, was shown to be required for HCV replication in vitro (31). Although Pin1 also interacted with NS5A and NS5B, the Pin1-NS5A interaction appeared to be distinct from the CyPA-NS5A interaction (31), and the ALP motif identified in this study does not conform to the consensus sequence (Ser/Thr-Pro) required for Pin1 binding (62). The putative proline substrates of Pin1 relevant for HCV replication remain to be identified.
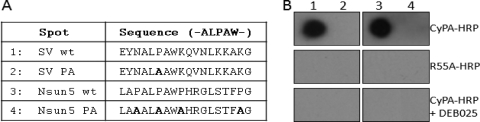
In GT2a, represented by the JFH-1 isolate, the tandem site contains a perfectly repeated motif of ALP in the P6 and P8 peptides. The AØPXW motif (Ø represents a hydrophobic residue) of P6 is highly conserved in all the HCV sequences (over 97% of the 4,307 sequences found in the European HCV database; Table 5). A homology search of GenBank with the JFH-1 P6 motif, ALPAW, identified two cellular proteins, supervillin (44) and Nsun5/WBSCR20 (11), each with an exact match of the linear peptide sequence. The corresponding peptides from these two proteins were also able to bind to CyPA in a PPIase-dependent and DEB-025-sensitive manner (Fig. 7). Similar to the reported interaction between CyPA and neuronal Wiskott-Aldrich syndrome protein (N-WASP) (4), an interaction between CyPA and supervillin, which is a major F-actin-binding protein, would be consistent with the observed effects of CsA treatment on altered cytoskeleton and contractile functions (23, 29).
Table 5.
Amino acids found at CBS-1 (P6) and CBS-2 (P8), showing that sequences of the tandem CBSs are highly conserveda
| Site | aa position in NS5A of JFH-1/H77 | aa frequency | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CBS-1 (P6) | 308/312 | A: 97.63* | G: 0.02 | P: 0.11 | S: 2.14 | T: 0.02 | V: 0.09 | |||
| 309/313 | F: 0.04* | I: 2.00* | L: 63.78* | M: 33.90* | P: 0.07 | S: 0.02 | T 0.02 | V: 0.14* | W: 0.02* | |
| 310/314 | A: 0.02 | L: 0.07 | P: 99.77* | S: 0.14 | ||||||
| 312/316 | G: 0.02 | L: 0.02 | R: 0.16 | W: 99.80* | ||||||
| CBS-2 (P8) | 340/344 | C: 0.02 | F: 0.07* | H: 0.02 | I: 0.12* | L: 99.37* | M: 0.04* | V: 0.35* | ||
| 341/345 | A: 0.39 | G: 0.02 | H: 0.02 | P: 99.37* | S: 0.14 | |||||
A total of 4,307 NS5A sequences obtained from the European HCV database (EuHCVdb) were analyzed, and the numberings are of JFH-1 NS5A.
, residues that conform to A∅PXW of CBS-1 and ∅P of CBS-2.
Fig 7.
ALPAW-containing peptides from cellular proteins interact with CyPA in a spot assay. A BLAST search of the GenBank database identified two cellular proteins, Supervillin and Nsun5, that contain a linear peptide with the ALPAW motif. The wt peptides, but not the proline-deficient mutants (sequences shown in panel A), bound to CyPA in a PPIase-dependent and DEB-025-sensitive manner (B).
The second ALP motif in the JFH-1 P8 site is less conserved in the different genotypes of HCV, although the ØP motif again appears to be the dominant sequence in most (over 99%) of the sequences found in the database (Table 5). P341 of this ØP motif locates in the LCS-II region, which connects domains II and III and is sensitive to protease digestion in vitro (54). Surface-exposed, proline-containing loops are often found in proteins that are substrates of PPIases (50, 65), and isomerization of P341 in LCS-II can conceivably regulate NS5A function by adjusting the relative positioning of domains II and III. The importance of this LCS-II region is underscored by the observation that P342 is critical for the replication of a genotype 1b replicon and the P341-to-P343 proline stretch can influence HCV assembly (24). Proline residues in proximity to position 341 may mediate CyPA binding by the P8 peptide of genotype 6a, as this peptide contains an alanine, rather than a proline, at this location. Interestingly, the P6 peptide of genotype 6a is also the only genotype that does not conform to the ALP motif. Residues other than the proline substrates themselves can also regulate CyPA dependence and replication efficiency. Two distinct mechanisms of modulation have been identified. The previously identified DEYN mutations do not affect CyPA binding; rather, they create a local conformation change that likely mimics the action of the PPIase (63). Similarly, a corresponding mutation in genotype 1b, D320E, similarly did not affect CyPA binding (6). The tryptophan residue in the AØPXW motif, on the other hand, appears to be directly required for CyPA binding and robust replication. Mutating tryptophan to alanine alters the hydrophobicity of the local environment that might be important for optimal CyPA binding (45). Similarly, the ALP sequence of the P6 peptide was present in the immobilized peptide P5 used in this study, which did not show significant binding in the spot assay, again suggesting that proper context of the proline-containing sequence is important for CyPA binding. Key residues in JFH-1 NS5A identified in the present study have also been characterized in the genotype 1b replicon (53). P314 and W316 of Con1, which correspond to P310 and W312 of JFH-1, were both required for efficient replication of a genotype replicon.
The specific aspect(s) of the NS5A function that depends upon isomerization is not clear, partly because the precise roles that NS5A plays in HCV replication and virion assembly (1, 52) are not fully understood. The C-terminal half of NS5A that contains the tandem CyPA-binding site belongs to a class of proteins that are designated intrinsically disordered proteins (IDPs), also known as natively unfolded proteins or natively disordered proteins. In contrast to a classical globular protein that exists in a single stable structural state (i.e., all the molecules in a sample of the native state have nearly the same structure), a sample of an IDP consists of a broad ensemble of molecules, each having a different conformation. The diversity of structures gives an IDP or a protein having IDP domains (e.g., HCV NS5A) the capacity to interact with multiple partners and serve as a hub for protein-protein interaction networks. It also raises the question of how the multiple interactions with a single IDP are regulated, both temporally and spatially. Proline isomerization, individually or collectively, may trigger subtle, local changes in structural conformers of an ensemble that would favor certain structures over others, thus providing an excellent mechanism for modulating IDP-partner interactions in response to the biological needs of the cell or the virus. Isomerization of multiple prolines would provide an even higher regulatory capacity by providing a larger number of possible peptide backbone conformations. Consistent with this idea, NS5A has been reported to interact with a myriad of proteins, including other components of the HCV replication complex such as NS5B and NS3 (9). We had initially hypothesized that disruption of NS5A-NS5B interaction may account for the observed effect of CPIs on the integrity of the HCV replication complex (34); preliminary data, however, indicated that neither the NS5A-NS5B nor the NS5A-NS3 interaction was sensitive to CsA treatment (A. Nag and H. Tang, unpublished data). It would be interesting to systematically determine whether any additional NS5A interactions, especially the ones that involve domains II and III, are dependent on CyPA's PPIase activity and thus sensitive to CPIs. CyPA has also been proposed to regulate NS5A/5B cleavage kinetics (27) and/or RNA binding by HCV replicase (17, 33). Although domain I is likely the major RNA-binding domain of NS5A and some controversies currently surround whether domains II and III are directly involved in NS5A's RNA-binding ability or not (16, 25), it is theoretically possible for structural changes in domain II and LCS-II to influence the configuration of domain I dimers. Alternatively, modulation of protein-protein interaction can indirectly contribute to RNA-binding regulation.
The remarkably consistent results obtained in this study regarding the role of P310 and P341 in CyPA's action highlight the power of the CoFIM method that originally identified the CPI resistance motif (DY) (63). Delineating the molecular interface of the NS5A-CyPA interaction represents an important step toward the elucidation of the structural and functional consequences of CyPA's isomerase activity, which is the target of a new class of compounds currently being tested in clinical trials as a candidate HCV therapy (51, 59).
ACKNOWLEDGMENTS
This study was supported by NIH grant R01 AI079150 to H.T.
We thank Takaji Wakita, Charles Rice, Timothy Tellinghuisen, Jean-Maurice Dumont, and Kai Lin for reagents, Margaret Seavy for help with protein purification, and Anne B. Thistle for proofreading the manuscript.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Appel N, et al. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 3. Brass V, et al. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130–8139 [DOI] [PubMed] [Google Scholar]
- 4. Calhoun CC, Lu YC, Song J, Chiu R. 2009. Knockdown endogenous CypA with siRNA in U2OS cells results in disruption of F-actin structure and alters tumor phenotype. Mol. Cell. Biochem. 320:35–43 [DOI] [PubMed] [Google Scholar]
- 5. Chatterji U, et al. 2009. The isomerase active site of cyclophilin A is critical for hepatitis C virus replication. J. Biol. Chem. 284:16998–17005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterji U, et al. 2010. HCV resistance to cyclosporin A does not correlate with a resistance of the NS5A-cyclophilin A interaction to cyclophilin inhibitors. J. Hepatol. 53:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ciesek S, et al. 2009. Cyclosporine A inhibits hepatitis C virus nonstructural protein 2 through cyclophilin A. Hepatology 50:1638–1645 [DOI] [PubMed] [Google Scholar]
- 8. Coelmont L, et al. 2010. DEB025 (Alisporivir) inhibits hepatitis C virus replication by preventing a cyclophilin A induced cis-trans isomerisation in domain II of NS5A. PLoS One 5:e13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Chassey B, et al. 2008. Hepatitis C virus infection protein network. Mol. Syst. Biol. 4:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delaglio F, et al. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 11. Doll A, Grzeschik KH. 2001. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet. Cell Genet. 95:20–27 [DOI] [PubMed] [Google Scholar]
- 12. Elazar M, et al. 2003. Amphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replication. J. Virol. 77:6055–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Evans MJ, Rice CM, Goff SP. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:13038–13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fernandes F, Ansari IU, Striker R. 2010. Cyclosporine inhibits a direct interaction between cyclophilins and hepatitis C NS5A. PLoS One 5:e9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fischer G, Wittmann-Liebold B, Lang K, Kiefhaber T, Schmid FX. 1989. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337:476–478 [DOI] [PubMed] [Google Scholar]
- 16. Foster TL, Belyaeva T, Stonehouse NJ, Pearson AR, Harris M. 2010. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 84:9267–9277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Foster TL, Gallay P, Stonehouse NJ, Harris M. 2011. Cyclophilin A interacts with domain II of hepatitis C virus NS5A and stimulates RNA binding in an isomerase-dependent manner. J. Virol. 85:7460–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fuerst TR, Earl PL, Moss B. 1987. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol. Cell. Biol. 7:2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gamble TR, et al. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285–1294 [DOI] [PubMed] [Google Scholar]
- 20. Handschumacher RE, Harding MW, Rice J, Drugge RJ, Speicher DW. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544–547 [DOI] [PubMed] [Google Scholar]
- 21. Hanoulle X, et al. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589–13601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanoulle X, et al. 2009. Domain 3 of non-structural protein 5A from hepatitis C virus is natively unfolded. Biochem. Biophys. Res. Commun. 381:634–638 [DOI] [PubMed] [Google Scholar]
- 23. Haskova V, Rozprimova L, Hasek J, Jelinkova M. 1994. Immunolocalization of cyclophilin in normal and cyclosporin A-treated human lymphocytes. Immunol. Lett. 41:267–272 [DOI] [PubMed] [Google Scholar]
- 24. Hughes M, et al. 2009. A conserved proline between domains II and III of hepatitis C virus NS5A influences both RNA replication and virus assembly. J. Virol. 83:10788–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hwang J, et al. 2010. Hepatitis C virus nonstructural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteins. J. Virol. 84:12480–12491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Johnson BA, Blevins RA. 1994. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4:603–614 [DOI] [PubMed] [Google Scholar]
- 27. Kaul A, et al. 2009. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 5:e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kay LE, Keifer P, Saarinen T. 1992. Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114:10663–10665 [Google Scholar]
- 29. Kolcz J, et al. 1999. Effects of cyclosporin A on contractile activity and cytoskeleton in chick embryo cardiomyocytes. Biochem. Cell Biol. 77:133–140 [PubMed] [Google Scholar]
- 30. Liang Y, Ye H, Kang CB, Yoon HS. 2007. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46:11550–11558 [DOI] [PubMed] [Google Scholar]
- 31. Lim YS, Tran HT, Park SJ, Yim SA, Hwang SB. 2011. Peptidyl-prolyl isomerase Pin1 is a cellular factor required for hepatitis C virus propagation. J. Virol. 85:8777–8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu J, et al. 1991. Calcineurin is a common target of cyclophilin-cyclosporin A and FKBP-FK506 complexes. Cell 66:807–815 [DOI] [PubMed] [Google Scholar]
- 33. Liu Z, et al. 2009. Mutations in the hepatitis C virus polymerase that increase RNA binding can confer resistance to cyclosporine A. Hepatology 50:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Z, Yang F, Robotham JM, Tang H. 2009. Critical role of cyclophilin A and its prolyl-peptidyl isomerase activity in the structure and function of the hepatitis C virus replication complex. J. Virol. 83:6554–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lohmann V, et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 37. Love RA, Brodsky O, Hickey MJ, Wells PA, Cronin CN. 2009. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83:4395–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luban J, Bossolt KL, Franke EK, Kalpana GV, Goff SP. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067–1078 [DOI] [PubMed] [Google Scholar]
- 39. Ma S, et al. 2006. NIM811, a cyclophilin inhibitor, exhibits potent in vitro activity against hepatitis C virus alone or in combination with alpha interferon. Antimicrob. Agents Chemother. 50:2976–2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Neddermann P, et al. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78:13306–13314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Neri P, et al. 1991. 1H, 13C and 15N backbone assignments of cyclophilin when bound to cyclosporin A (CsA) and preliminary structural characterization of the CsA binding site. FEBS Lett. 294:81–88 [DOI] [PubMed] [Google Scholar]
- 42. Paeshuyse J, et al. 2006. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43:761–770 [DOI] [PubMed] [Google Scholar]
- 43. Penin F, et al. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279:40835–40843 [DOI] [PubMed] [Google Scholar]
- 44. Pestonjamasp KN, Pope RK, Wulfkuhle JD, Luna EJ. 1997. Supervillin (p205): a novel membrane-associated, F-actin-binding protein in the villin/gelsolin superfamily. J. Cell Biol. 139:1255–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piotukh K, et al. 2005. Cyclophilin A binds to linear peptide motifs containing a consensus that is present in many human proteins. J. Biol. Chem. 280:23668–23674 [DOI] [PubMed] [Google Scholar]
- 46. Ploss A, Rice CM. 2009. Towards a small animal model for hepatitis C. EMBO Rep. 10:1220–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robida JM, Nelson HB, Liu Z, Tang H. 2007. Characterization of hepatitis C virus subgenomic replicon resistance to cyclosporine in vitro. J. Virol. 81:5829–5840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. 2011. Recombinant HCV variants with NS5A from genotypes 1–7 have different sensitivities to an NS5A inhibitor but not alpha interferon. Gastroenterology 140:1032–1042 [DOI] [PubMed] [Google Scholar]
- 49. Siekierka JJ, Hung SH, Poe M, Lin CS, Sigal NH. 1989. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature 341:755–757 [DOI] [PubMed] [Google Scholar]
- 50. Solbak SM, et al. 2010. The intriguing cyclophilin A-HIV-1 Vpr interaction: prolyl cis/trans isomerisation catalysis and specific binding. BMC Struct. Biol. 10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang H. 2010. Cyclophilin inhibitors as a novel HCV therapy. Viruses 2:1621–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 55. Tellinghuisen TL, Marcotrigiano J, Rice CM. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435:374–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vajdos FF, Yoo S, Houseweart M, Sundquist WI, Hill CP. 1997. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 6:2297–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Verdegem D, et al. 2011. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 286:20441–20454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Waller H, Chatterji U, Gallay P, Parkinson T, Targett-Adams P. 2010. The use of AlphaLISA technology to detect interaction between hepatitis C virus-encoded NS5A and cyclophilin A. J. Virol. Methods 165:202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Watashi K. 2010. Alisporivir, a cyclosporin derivative that selectively inhibits cyclophilin, for the treatment of HCV infection. Curr. Opin. Investig. Drugs 11:213–224 [PubMed] [Google Scholar]
- 60. Watashi K, Hijikata M, Hosaka M, Yamaji M, Shimotohno K. 2003. Cyclosporin A suppresses replication of hepatitis C virus genome in cultured hepatocytes. Hepatology 38:1282–1288 [DOI] [PubMed] [Google Scholar]
- 61. Weber C, et al. 1991. The NMR structure of cyclosporin A bound to cyclophilin in aqueous solution. Biochemistry 30:6563–6574 [DOI] [PubMed] [Google Scholar]
- 62. Yaffe MB, et al. 1997. Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278:1957–1960 [DOI] [PubMed] [Google Scholar]
- 63. Yang F, et al. 2010. A major determinant of cyclophilin dependence and cyclosporine susceptibility of hepatitis C virus identified by a genetic approach. PLoS Pathog. 6:e1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang F, et al. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82:5269–5278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhao Y, Chen Y, Schutkowski M, Fischer G, Ke H. 1997. Cyclophilin A complexed with a fragment of HIV-1 Gag protein: insights into HIV-1 infectious activity. Structure 5:139–146 [DOI] [PubMed] [Google Scholar]
- 66. Zydowsky LD, et al. 1992. Active site mutants of human cyclophilin A separate peptidyl-prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci. 1:1092–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]