Abstract
Our understanding of the third domain of life, Archaea, has greatly increased since its establishment some 20 years ago. The increasing information on archaea has also brought their viruses into the limelight. Today, about 100 archaeal viruses are known, which is a low number compared to the numbers of characterized bacterial or eukaryotic viruses. Here, we have performed a comparative biological and structural study of seven pleomorphic viruses infecting extremely halophilic archaea. The pleomorphic nature of this novel virion type was established by sedimentation analysis and cryo-electron microscopy. These nonlytic viruses form virions characterized by a lipid vesicle enclosing the genome, without any nucleoproteins. The viral lipids are unselectively acquired from host cell membranes. The virions contain two to three major structural proteins, which either are embedded in the membrane or form spikes distributed randomly on the external membrane surface. Thus, the most important step during virion assembly is most likely the interaction of the membrane proteins with the genome. The interaction can be driven by single-stranded or double-stranded DNA, resulting in the virions having similar architectures but different genome types. Based on our comparative study, these viruses probably form a novel group, which we define as pleolipoviruses.
INTRODUCTION
Extremophilic archaea inhabit ecological niches pushing the limits of life, such as hypersaline or hyperthermic environments. Archaea are also abundant in nonextreme environments affecting global biogeochemical cycles (16, 20, 52). To date, about 100 archaeal viruses have been described, and the virus host range is currently limited only to extremophilic archaea: hyperthermophiles, halophiles, and methanogens (4, 7, 29, 50, 51). The domain Archaea is divided into two major phyla, Euryarchaeota and Crenarchaeota (62, 63), and several novel morphologies have been described for crenarchaeal viruses (37, 42, 50). In contrast, the majority of the described euryarchaeal viruses has icosahedrally symmetric heads with tails attached and are classified into the order Caudovirales, originally containing only head-tailed bacteriophages (37, 50). However, electron microscopy (EM) studies have indicated that head-tailed viruses represent only a minor fraction of the viruslike particles found in hypersaline habitats (30, 46, 59). Other morphotypes described for euryarchaeal viruses are spindle shaped, icosahedral, and pleomorphic (12, 33, 49). Except for a few pleomorphic viruses, which have a single-stranded DNA (ssDNA) genome, all of the archaeal viruses studied so far have a double-stranded DNA (dsDNA) genome (49, 50, 58).
In our recent study, 40 viruses infecting halophilic archaea were isolated from five globally distinct locations (7). Based on virion morphology and the structural protein profile, three isolates, HRPV-2 (Halorubrum pleomorphic virus 2), HRPV-3 (Halorubrum pleomorphic virus 3), and HGPV-1 (Halogeometricum pleomorphic virus 1), are reminiscent of two previously described, closely related pleomorphic viruses, HRPV-1 (Halorubrum pleomorphic virus 1) and HHPV-1 (Haloarcula hispanica pleomorphic virus 1), infecting haloarchaea as well (7, 49, 53).
HRPV-1 and HHPV-1 are composed of a membrane vesicle containing two major structural proteins (48, 49, 53). Although HRPV-1 and HHPV-1 have different genome types (ssDNA versus dsDNA), their circular genomes share the same gene order, and they have homologous structural proteins (49, 53). The nine (HRPV-1) and eight (HHPV-1) open reading frames (ORF) encode a putative replication initiation protein; two major structural proteins, VP3 and VP4 (VP for virion protein); and a postulated ATPase (HRPV-1 VP8) (49, 53). Biochemical analyses of HRPV-1 have shown that VP4 is a spike protein anchored to the membrane and that VP3 is a membrane protein facing the particle interior where the genome resides, without associated nucleoproteins (48). The genome synteny of HRPV-1 and HHPV-1 (from the VP4-encoding gene to the ATPase-encoding gene) is also shared with the linear genome of the haloarchaeal virus His2, which was proposed previously to be spindle shaped (11, 49, 53).
Due to the rapid evolution of viral genome sequences (28, 60), additional approaches complementing genomic studies are needed to establish relationships between viruses. Structural studies of icosahedrally symmetric viruses infecting hosts from different domains of life have shown that they are related, and a large number of them can be classified into four structure-based viral lineages (2, 3, 8, 9, 13, 39). Similarities between HRPV-1 and the bacterial virus L172, which infects Acholeplasma laidlawii, have also led to the proposal of a viral lineage containing pleomorphic viruses from bacteria and archaea (48, 49).
Here, we have isolated a sixth member of the pleomorphic virus type infecting haloarchaea, and we report a comparative analysis of all known pleomorphic archaeal viruses isolated so far (Tables 1 and 2). Biochemical dissociation analyses of the virions combined with cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) reveal the structural organization of this virion type. The unique but common properties of these pleomorphic viruses support their classification as a novel group of pleolipoviruses having the same virion architecture but different genome types.
Table 1.
Viruses and their host strains used in this study
| Virus | Origin of virus | Reference for virus | Host archaeon | Reference for host archaeon |
|---|---|---|---|---|
| HRPV-1 | Trapani, Italy | 49 | Halorubrum sp. PV6 | 49 |
| HRPV-2 | Samut Sakhon, Thailand | 7 | Halorubrum sp. SS5-4 | 7 |
| HRPV-3 | Sedom ponds, Israel | 7 | Halorubrum sp. SP3-3 | 7 |
| HRPV-6 | Samut Sakhon, Thailand | This study | Halorubrum sp. SS7-4 | This study |
| HGPV-1 | Cabo de Gata, Spain | 7 | Halogeometricum sp. CG-9 | 7 |
| HHPV-1 | Margherita di Savoia, Italy | 53 | Haloarcula hispanica ATCC 33960 | 34 |
| His2 | Victoria, Australia | 11 | Haloarcula hispanica ATCC 33960 | 34 |
Table 2.
Experiments performed here and in previous studies
| Virus | Reference(s) |
GenBank accession no. of genome sequence (reference) | |||||
|---|---|---|---|---|---|---|---|
| Life cycle | Buffer optimization | Production and purification optimization | TLC analysis of lipids | Cryo-EM | Dissociation studies | ||
| HRPV-1 | 49 | 48 | 49 | 48 | This study | 48 | FJ685651 (49) |
| HRPV-2 | This study | This study | This study | This study | This study | This study | JN882264 (58) |
| HRPV-3 | This study | This study | This study | This study | This study | This study | JN882265 (58) |
| HRPV-6 | This study | This study | This study | This study | This study | This study | JN882266 (58) |
| HGPV-1 | This study | This study | This study | This study | This study | This study | JN882267 (58) |
| HHPV-1 | 53 | This study | 53 | 53 | This study | This study | GU321093 (53) |
| His2 | This study, 11 | This study | This study, 11 | This study | This study | This study | AF191797 (11) |
MATERIALS AND METHODS
Viruses, archaeal strains, and media.
The viruses and archaeal strains used in this study are listed in Table 1. Cultures were grown aerobically at 37°C, unless stated otherwise, using modified growth medium (MGM) containing 5 g peptone (Oxoid) and 1 g Bacto yeast extract (Difco Laboratories) per liter (44). Solid and top-layer media contained 14 and 4 g of Bacto agar (Difco Laboratories) per liter, respectively. A 30% (wt/vol) stock solution of artificial salt water (SW) was added to broth, solid, and top-layer media to give final concentrations of 23%, 20%, and 18% (wt/vol), respectively. The preparation of the artificial SW is described in the Halohandbook (http://www.haloarchaea.com/resources/halohandbook/Halohandbook_2008_v7.pdf).
For the isolation of halophilic archaeal and bacterial strains, salt crystals collected from a solar saltern in Samut Sakhon, Thailand (13°32′N, 100°17′E), during the year 2009 were mixed with 18% SW and plated and incubated as described previously by Atanasova et al. (7). Colonies were picked and colony purified three consecutive times. A partial 16S rRNA gene sequence of isolate SS7-4 was determined as described previously (7). PCR was performed by using primers D30 and D56 (6). The PCR product was sequenced with primers D30, D56 (6), and pD (24) at the Institute of Biotechnology, DNA Sequencing and Genomics Laboratory (University of Helsinki). The assembled sequence was analyzed by using BLASTN (64), and the 16S rRNA gene similarity was calculated by using the EzTaxon server (17). Virus isolation was performed by the direct plating of salt crystals dissolved in 18% SW as described previously (7).
Life cycle studies.
Host cells in the middle exponential growth phase were collected by centrifugation (12,000 × g for 10 min at 22°C [Sorvall SA600 rotor]), resuspended in the same volume of fresh growth medium, and infected with a virus at a multiplicity of infection (MOI) of 15. After 2 h of growth, the cells were washed to remove unbound viruses and resuspended in the same volume of fresh growth medium. The turbidity (A550) of the infected and uninfected cultures was measured at suitable intervals (3 to 29 h postinfection [p.i.]). The number of free viruses was enumerated by a plaque assay concurrently with the turbidity measurements (3 to 10 and 24 h p.i.). The number of viable cells was determined at 29 h p.i. HRPV-2 life cycle studies were carried out at 34°C.
Stability of the viruses.
The sensitivity of the viruses to a lowered ionic strength was studied by diluting the virus stocks (see below) in 50 mM Tris-HCl (pH 7.2) using appropriate volume ratios. The salinity of the dilutions decreased from 2.698 to 0.034 M, corresponding to 18% and 0.23% SW, respectively. The dilutions were incubated at 4°C, and the infectivity was determined after 1 day by using the plaque assay. Stocks were also diluted 1,000-fold in 23% SW and in the optimal SW buffer of the virus (Table 3) and incubated at 4°C. The infectivity was determined after 1, 3, 7, and 14 days.
Table 3.
Virion parameters
| Virus | Optimal buffer, salinity (M) | Density in CsCl (g ml−1) | Mean virion diam (nm) ± SD | Genome packaging density (nucleotides nm−3) | Genome type (reference) |
|---|---|---|---|---|---|
| HRPV-1 | HRPV-1 buffer, 1.6a | 1.29b | 41.1 ± 2.2 | 0.192 | Circular ssDNA (49) |
| HRPV-2 | 9% SW, 1.3 | 1.28 | 54.0 ± 4.3 | 0.127 | Circular ssDNA (58) |
| HRPV-3 | 15% SW, 2.2 | 1.27 | 67.2 ± 5.2 | 0.133 | Circular dsDNA (58) |
| HRPV-6 | 12% SW, 1.8 | 1.26 | 48.5 ± 2.7 | 0.153 | Circular ssDNA (58) |
| HGPV-1 | 15% SW, 2.2 | 1.34 | 55.5 ± 5.2 | 0.211 | Circular dsDNA (58) |
| HHPV-1 | 18% SW, 2.7 | 1.27c | 51.7 ± 4.0 | 0.227 | Circular dsDNA (53) |
| His2 | 18% SW, 2.7 | 1.30d | 70.6 ± 3.6 | 0.173 | Linear dsDNA (11) |
Growth and purification of the virions.
Virus stocks were prepared by using semiconfluent or confluent top-layer agar plates incubated at 34°C or 37°C for 3 to 5 days (see Table S1 in the supplemental material), as previously described (49). Prior to virus purification, HRPV-3, HGPV-1, and His2 stocks were treated with DNase I (0.1 mg ml−1; Sigma-Aldrich) for 1 h at 37°C.
Liquid culture propagation and virus purification of HRPV-1 and HHPV-1 were previously described (49, 53). Other viruses were purified as described previously for HRPV-1 and HHPV-1, using virus stocks. In brief, impurities were precipitated from HRPV-1 and HRPV-3 preparations with 6% (wt/vol) polyethylene glycol 6000 (PEG 6000), and virus particles were precipitated with 11% (wt/vol) PEG. Other viruses were precipitated by using 10% (wt/vol) PEG without pre-precipitation. Aggregates were removed from the virus concentrate, which was then purified with a linear 5 to 20% (wt/vol) sucrose gradient by rate-zonal centrifugation (104,000 × g at 15°C for 1 h 30 min for HGPV-1, 2 h for HRPV-6, 2 h 20 min for His2, 2 h 30 min for HRPV-3, 3 h 30 min for HRPV-2, and 4 h for HRPV-1 and HHPV-1 [Sorvall AH629]), yielding “1×-purified” virus. The “1× virus” was further purified with a CsCl gradient (mean densities of 1.4 g ml−1 for HGPV-1 and 1.3 g ml−1 for the other viruses) by equilibrium centrifugation (79,000 × g for 20 h at 15°C [Sorvall AH629]), yielding “2×-purified” virus. The density of the viruses in CsCl was determined after equilibrium centrifugation (83,000 × g for 20 h at 15°C [Sorvall TH641]) by fractionating the gradients and determining the density and A260 of the fractions. The “2× virus” was diluted 2-fold with 18% SW without NaCl and concentrated by differential centrifugation (114,000 × g for 3 h at 15°C [Sorvall T647.5]).
HRPV-1-buffer (48) (1.5 M NaCl, 100 mM MgCl2, 2 mM CaCl2, and 20 mM Tris-HCl [pH 7.5]) was used for the purification of HRPV-1. Other viruses were purified in 18% SW, except that the 2×-purified virus pellets were resuspended in 9% SW (HRPV-2), 12% SW (HRPV-6), 15% SW (HRPV-3 and HGPV-1), or 18% SW (HHPV-1 and His2). The compositions of the different salt waters are listed in Table S2 in the supplemental material.
A modified purification method was used for electron microscopy samples. After purification in a linear 5 to 20% sucrose gradient (228,000 × g at 15°C for 30 min for HGPV-1 and His2; 45 min for HRPV-6; 60 min for HRPV-1, HRPV-2, and HRPV-3; and 75 min for HHPV-1 [Sorvall TH660]), the 1× virus was concentrated and washed with the buffers indicated above by using ultrafiltration (with Amicon Ultra Centrifugal filter devices [Millipore], with a 100,000 nominal molecular weight limit; an Eppendorf 5810R centrifuge; and an A-4-62 rotor at 3,220 × g at 15°C). For HRPV-1, HRPV-6, and HHPV-1, the 1× virus was further purified with a CsCl gradient (83,000 × g for 20 h at 15°C [Sorvall TH641]), and the 2× virus was concentrated as described above for the 1× virus. For HRPV-1, the virus particles were also concentrated from the liquid culture and washed with HRPV-1 buffer by using ultrafiltration (centrifugation as described above).
For the HRPV-1 sedimentation analysis, PEG-precipitated virus particles were purified with a linear 5 to 20% sucrose gradient by rate-zonal centrifugation (210,000 × g for 2 h 5 min at 15°C [Sorvall TH641]). After centrifugation, gradients were fractionated. The protein, DNA, and lipid compositions; appearance by cryo-EM; and virus titers of the fractions were determined.
Virion dissociation.
The 2×-purified virus particles (10 μg of protein) were treated with proteinase K (10 μg; Finnzymes) at different salinities for 3 h at 37°C. Nine salinities were tested, and the salinity decreased linearly from 2.698 M to 82 mM, corresponding to a decrease of SW from 18 to 0.55%. The minimum concentration of Tris-HCl (pH 7.2) was 20 mM.
Detailed dissociation experiments and their analyses were carried out as described previously for HRPV-1 (48), using freshly made 2×-purified virus preparations (84 μg of protein per reaction). The following dissociations were performed: (i) proteinase K (0.1 mg/ml) treatment at high salinity for 3 h at 37°C, (ii) proteinase K (0.2 mg/ml) treatment at low salinity for 1 h at 37°C, (iii) low-salinity dissociation for 1 h at 37°C, and (iv) 0.1% (vol/vol) Nonidet P-40 (NP-40) treatment at high salinity for 1 h at 22°C. High-salinity incubations were performed with the optimal buffers of the viruses (Table 3). For low-salinity dissociations, a buffer containing 75 mM NaCl, 2.7 mM MgCl2, 2.6 mM MgSO4, 1.7 mM KCl, 0.1 mM CaCl2, and 20 mM Tris-HCl (pH 7.2) was used. After incubation, dissociation products were analyzed with a linear 5 to 20% sucrose gradient in 18% SW by rate-zonal centrifugation (210,000 × g for 1 h 45 min or 4 h at 15°C [Sorvall TH641]). The shorter centrifugation time was used for the first and third dissociations of His2. After centrifugation, gradients were fractionated, and protein, lipid, and DNA contents of the fractions were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Dissociation of HRPV-1 for cryo-EM.
The 1×-purified virus preparations were used. After proteinase K treatment (0.1 mg/ml) in HRPV-1 buffer (48), the particles were purified with a linear 5 to 20% sucrose gradient in HRPV-1 buffer (222,000 × g for 1 h 30 min at 15°C [Beckman SW50.1]) and concentrated and washed with the same buffer by using ultrafiltration (50,000 nominal molecular weight limit, with centrifugation as described above). Proteinase K-treated particles were further dissociated at low salinity (75 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris-HCl [pH 7.5]) for 1 h at 37°C. Particles were purified with a linear 5 to 20% sucrose gradient in a buffer containing 50 mM NaCl and 20 mM Tris-HCl (pH 7.5) (192,000 × g for 2 h 5 min at 15°C [Beckman SW50.1]) and concentrated and washed with the same buffer using ultrafiltration as described above. A protease inhibitor cocktail (Complete EDTA free; Roche) was used throughout further dissociations.
Proteinase K treatment (0.2 mg/ml) at low salinity at 37°C and dissociation at low salinity at 60°C were described previously (48). Particles produced at low salinity at 60°C were further treated with proteinase K (0.2 mg/ml) at low salinity (75 mM NaCl, 5 mM MgCl2, 0.1 mM CaCl2, and 20 mM Tris-HCl [pH 7.5]) for 1 h at 37°C. After treatments, particles were purified as described above for the low-salinity treatment. In addition to cryo-EM, DNA, proteins, and lipids of the dissociation products were analyzed.
Protein, DNA, and lipid analyses.
The protein concentrations were measured by the Coomassie blue method (14), using bovine serum albumin as a standard. Virus samples were analyzed by using either modified Tricine-SDS-PAGE with 4% and 14% (wt/vol) acrylamide concentrations or SDS-PAGE with 4% and 16% (wt/vol) acrylamide concentrations in the stacking and separation gels, respectively (45, 56). Tricine-SDS-polyacrylamide gels were stained with Coomassie blue to show proteins. After the Coomassie blue staining, gels were stained with Sudan Black B (Sigma) to detect lipids, according to the manufacturer's instructions. The intensities of the protein and lipid bands were analyzed by using the 1D Evaluation mode of the AIDA Image Analyser, version 3.44, software (Rayest Isotopenmeßgeräte GmbH, Germany). SDS-polyacrylamide gels were stained with ethidium bromide (EtBr) to detect DNA. N-terminal sequences were determined at the Protein Chemistry Core Facility of the Institute of Biotechnology, University of Helsinki, as described previously (48).
Lipids from the host cells and 2×-purified viruses were extracted according to a method described previously by Folch et al. (26), as modified by Kates (35) for halophilic organisms. Lipid extracts in chloroform-methanol (9:1, vol/vol) were subjected to thin-layer chromatography (TLC) on silica gel 60 plates (Merck). Polar lipids were separated by using chloroform–methanol–90% (vol/vol) acetic acid as the solvent (65:4:35, vol/vol/vol) and visualized with iodine vapor.
Electron microscopy.
For negative-stain transmission electron microscopy (TEM), the 1×-purified virus particles were adsorbed on grids for 1 min prior to negative staining with 1% (wt/vol) ammonium molybdate (pH 7.0) for 2 min or 3% (wt/vol) uranyl acetate (pH 4.5) for 1 min. The micrographs were taken with a Jeol 1200EX electron microscope operating at 80 kV at the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki. For titration, virus particles were mixed with a negative stain at a volume ratio of 1:1 and incubated for the staining time described above, and infectivity was determined by using a plaque assay.
For cryo-electron microscopy, 3 μl of a freshly purified virus suspension in the optimal buffers was pipetted onto Quantifoil R 2/2 grids and vitrified in liquid ethane (5). The sample was transferred into a Gatan 626 Cryo-Holder and observed at liquid nitrogen temperature with an FEI Tecnai F20 field emission gun transmission electron microscope operating at 200 kV. Images were collected under low-dose conditions either on Kodak SO163 film at a nominal magnification of ×50,000 or with a Gatan Ultrascan 4000 charge-coupled-device (CCD) camera at a nominal magnification of ×68,000. Images were recorded far from focus (typically 4 to 8 μm underfocus) to increase the contrast. For the tomography of HRPV-1, 10-nm colloidal gold was added prior to vitrification as a fiducial marker. Tilt series of untreated HRPV-1 and proteinase K-treated HRPV-1 further dissociated at low salinity at 37°C were collected, covering an angular range of −50° to +50° in 2° increments, using serialEM (41) under low-dose conditions. The images were recorded with a CCD camera at an underfocus of 6 μm and a nominal magnification of ×26,000, which gave a sampling of 0.57 nm per pixel. Average diameters and their standard deviations were calculated from 100 digital particle images. The genome packaging densities were calculated by first subtracting 7 nm from the average radius of each virus, and this value was then used to calculate the volume of a sphere of that radius, assuming that only one genome copy was present per virion.
Image processing.
The data were binned once, resulting in a final sampling of 1.15 nm per pixel. A low-pass filter to a 1/4-nm spatial frequency was applied to the raw data set to exclude all information beyond the first zero point of the contrast transfer function. Gold particles were used to align the tilt images, and weighted back-projection was used to calculate the three-dimensional (3D) reconstructions with IMOD (38). Further image processing was carried out with Jsubtomo (32), unless stated otherwise. A total of 78 smaller volumes (60 by 60 by 60 pixels), each containing a single virus particle, were extracted from one of the 17 tomograms for further analysis. Particles were iteratively centered, first against a spherical model of a 17.3-nm radius and then against their own 3D radial density profile, without any symmetry imposed. These particles were all roughly spherical and were assumed to have the membrane at a similar radius in subsequent steps. This enabled the automatic selection of 3,588 membrane subvolumes from all over the surface of the centered particles, on a regular grid, irrespective of whether or not the volume contained spikes, in a box of 30 by 30 by 30 pixels. A surface normal in each location defined the orientation of each subvolume relative to the viral surface. A radial density profile was calculated with Bsoft (31) for the average of the membrane subvolumes. To locate the VP4 spikes in the membrane of HRPV-1 and refine their positions, particle subtomograms were analyzed with Jsubtomo, using the averaged membrane subvolumes as a search template. A cross-correlation coefficient cutoff and a distance filter were applied to identify matches, which were then visualized with UCSF Chimera (47). The spike subvolumes were iteratively aligned and averaged with Jsubtomo, allowing limited angular and translational refinements, using only the information for which there was strong Fourier terms in order to account for the missing wedge (32). The final spike average contained information from 1,019 subvolumes.
Tomograms of the proteinase K-treated HRPV-1 particles, further dissociated at low salinity at 37°C, were treated in a similar fashion. Briefly, 89 particles were extracted and centered, 989 membrane subtomograms were aligned and averaged, and a radial profile of the resulting average was calculated with Bsoft (31, 32).
Nucleotide sequence accession number.
The sequence data were submitted to GenBank under accession number JN971009.
RESULTS
In order to judge the relationship between different pleomorphic archaeal viruses and to see how general their characteristics are, we compared three isolates described previously by Atanasova et al. (7) and one isolate from the present report with three previously described isolates, HRPV-1, HHPV-1, and His2. The viruses and hosts are listed in Table 1, and the experiments carried out with these viruses are described in Table 2.
The sixth pleomorphic virus, HRPV-6, infects a Halorubrum strain.
A salt crystal sample collected from Samut Sakhon, Thailand, was screened for halophilic strains and their viruses. One of the halophilic strains, SS7-4, supported the plaque formation of a virus from the same sample. The partial 16S rRNA gene sequence of SS7-4 (1,333 bp) (GenBank accession number JN971009) revealed that the closest relatives were Halorubrum chaoviator (accession number AM048786) and Halorubrum sodomense (accession number D13379), with sequence similarities of 99.4 and 99.2%, respectively. Accordingly, this new archaeal strain was designated Halorubrum sp. SS7-4. The obtained plaques were hazy, with a small clear center. The virus was named Halorubrum pleomorphic virus 6 (HRPV-6) (see the virion description below).
Pleomorphic viruses share nonlytic life cycles.
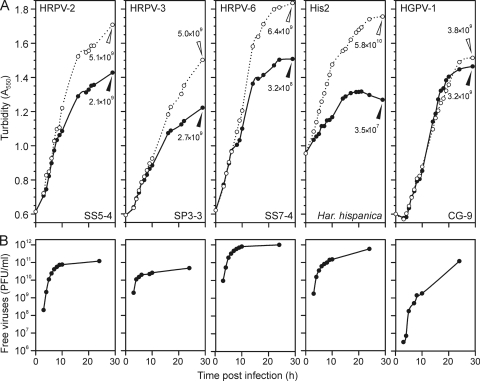
The plaque morphologies of all seven viruses were similar, characterized by a hazy appearance indicating an inhibition of host growth. The life cycles of HRPV-2, HRPV-3, HRPV-6, HGPV-1, and His2 were studied by infecting logarithmically growing cells at an MOI of 15 (Fig. 1). In all cases, the major difference between the infected and uninfected cultures was the lower growth rate of the infected cultures (Fig. 1A). The production of progeny viruses proceeded continuously during logarithmic growth, and the virus titers in the growth medium reached high levels, from 1010 up to 1012 PFU ml−1 (Fig. 1B). No decrease in the turbidity was observed during virus production (Fig. 1).
Fig 1.
Virus life cycles. (A) Turbidities of infected (closed circles) and uninfected (open circles) cultures monitored up to 29 h p.i. Cell densities (CFU ml−1) of the infected (closed triangles) and uninfected (open triangles) cultures were measured at 29 h p.i. The viruses and their hosts are indicated in Table 1. (B) Amount of free viruses enumerated at 3 to 10 and 24 h p.i. The time scales in panels A and B are the same.
There were subtle differences between the virus life cycles. HGPV-1 virus production started later than the release of the other viruses (Fig. 1B). The release of HRPV-2, HRPV-3, HRPV-6, and His2 viruses started at 3 h p.i. at the latest, and the greatest increase in virus production was observed at 3 to 4 h p.i. (Fig. 1B). HGPV-1 also had the most modest effect on host growth, emphasized by the equal numbers of viable cells in the infected and uninfected cultures at the stationary growth phase (Fig. 1A). HRPV-2 and HRPV-3 slowed host growth more than did HGPV-1, and the greatest growth retardation with the highest level of virus production was observed for HRPV-6 and His2 (Fig. 1).
Stability, morphology, and structural components of the virions.
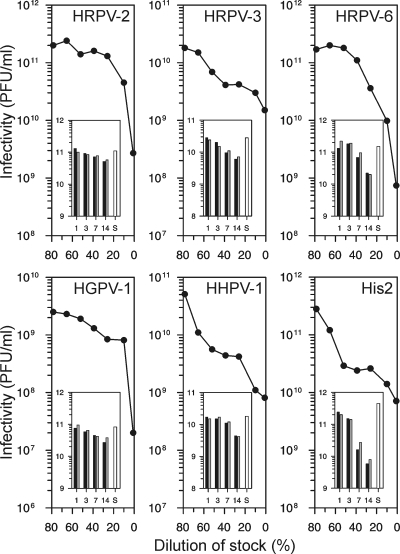
Virus stocks, with titers from 109 to 1012 PFU ml−1, were stable for several months. Stocks contained 23% (wt/vol) SW, corresponding to a 3.4 M salt concentration (see Table S2 in the supplemental material). All viruses were sensitive to low salt concentrations (Fig. 2), and optimal SW concentrations with the minimal salinity preserving infectivity were defined for each virus (Table 3). The infectivity was maintained for at least 3 days, and there was no major difference between virus stabilities in the optimal buffer and 23% SW (Fig. 2, insets). The optimized virus purification methods (see Materials and Methods) gave, on average, a recovery of about 20% of infectious virions, with a specific infectivity of 3 × 1013 PFU mg−1 of protein for the 2×-purified viruses. The low buoyant density of the virions indicated the presence of lipids (Table 3).
Fig 2.
Salt dependence of viral infectivity. The virus stocks were diluted in 50 mM Tris-HCl (pH 7.2), and infectivity was determined after 1 day of incubation at 4°C. The x axis indicates the proportion of the stock in the dilution mixture. The insets show the stability of the viruses in optimal SW buffer (black bars) and in 23% SW (gray bars). The virus stocks were diluted 1:1,000 in the buffers, and infectivity was determined after 1, 3, 7, and 14 days of incubation at 4°C. The white bars indicate the titers of the starting material (S). The scale of the y axis in the insets is the logarithm of infectivity (PFU ml−1).
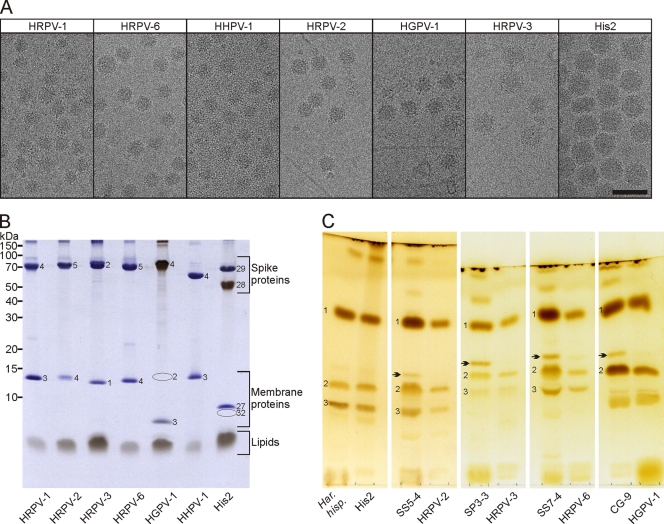
The sensitivity of the viruses to two negative stains commonly used for TEM was determined. Based on infectivity, viruses were more sensitive to uranyl acetate than to ammonium molybdate (see Fig. S1A in the supplemental material). In addition, the stains affected virion morphology (see Fig. S1B in the supplemental material). To avoid negative-staining artifacts, we employed cryo-EM to characterize the virus particles. HRPV-1 particles were imaged by cryo-EM at each step of the production and purification process to ensure that the apparent pleomorphicity of the viral particles (see below) was not induced by the purification steps (data not shown). The cryo-electron micrographs of the seven viruses showed roughly spherical, intact particles, with mean diameters and standard deviations varying from 41.1 ± 2.2 nm to 70.6 ± 3.6 nm (Fig. 3A and Table 3). Distinctively, all the viral particles showed spikes decorating the surface of the viral membrane (Fig. 3A). The genome packaging densities (Table 3) did not correlate with the genome type (ssDNA versus dsDNA).
Fig 3.
Virion morphology and composition. (A) Cryo-EM of the virions. The viruses are shown in order of increasing particle size. Either the 1×-purified virions (HRPV-1, HRPV-2, HGPV-1, HRPV-3, and His2) or the 2×-purified virions (HRPV-6 and HHPV-1) are shown in the cryo-electron micrographs. For HRPV-6 and HHPV-1, the 2×-purified virions were used because of ribosome impurities in the 1×-purified material. The micrographs of HRPV-2 and HGPV-1 contain some host cell flagella. Scale bar, 100 nm. (B) Protein and lipid profiles of the 2×-purified virions in a Tricine-SDS-polyacrylamide gel stained with Coomassie blue to stain proteins and Sudan Black B to stain lipids. Numbers on the left indicate the molecular masses of the markers. Numbers on the gel indicate the virion proteins (VPs). The positions of HGPV-1 VP2 and His2 VP32 are marked by ovals. (C) Lipids of host cells and 2×-purified virions analyzed by TLC and stained with iodine vapor. Viruses and their hosts are shown next to each other. The host strains are Haloarcula hispanica (Har. hisp.); Halorubrum sp. strains SS5-4, SP3-3, and SS7-4; and Halogeometricum sp. CG-9. The polar lipids which migrated to the same positions as those of Haloarcula hispanica (10) are indicated by the numbers 1 to 3 and correspond to PG, PGP-Me, and PGS, respectively.
Analyses of highly purified virions by Tricine-SDS-PAGE revealed almost identical protein patterns. All viruses had one large major structural protein of about 50 to 70 kDa, except for His2, which had two of these (Fig. 3B). In addition, all viruses had one small major structural protein of about 7 to 14 kDa (Fig. 3B), except for HGPV-1, which had two such proteins (see below). The large major structural proteins were identified to be spikes on the virion surface, and the small proteins were identified as membrane-associated ones (see below). HGPV-1 VP2 and VP3 and His2 proteins were identified by N-terminal sequencing (see Table S3 in the supplemental material). The other proteins were identified elsewhere previously (49, 53, 58).
Three major proteins and one minor protein of His2 gave N-terminal sequences matching the proteins encoded by genes 27, 28, 29, and 32, respectively (Fig. 3B). Previously, VP29 was identified by the sequencing of tryptic peptides, and the protein was named VP1 (11). However, here, the proteins are named according to the genes encoding them. The N-terminal sequences of His2 VP28 and VP29 started from amino acids 41 and 38, respectively, indicating that the proteins have a signal peptide. The Coomassie blue stain bound weakly to VP28, but when the gel was stained with Sudan Black B, a lipid stain, the VP28 band gave a strong signal (Fig. 3B). In addition, the apparent (about 50 kDa) and calculated (23.5 kDa) molecular masses of VP28 differ considerably. These observations indicated that VP28 is a lipid-modified protein.
One small major structural protein of HGPV-1 gave an N-terminal sequence matching the product of gene 3 (VP3 in Fig. 3B). The other small major structural protein of HGPV-1 was identified from the virions treated with proteinase K at low salinity as a C-terminal digestion product encoded by gene 2 and was thus named VP2 (see Fig. S2 in the supplemental material). This protein did not appear in the gel when intact virions were analyzed (Fig. 3B). Similarly to the His2 VP28 protein, HGPV-1 VP4 had a low affinity for the Coomassie blue stain, but it bound strongly to Sudan Black B, and the apparent (about 70 kDa) and calculated (48.5 kDa) molecular masses differed, indicating that VP4 is also a lipid-modified protein. The N termini of HGPV-1 VP4 and His2 VP28 did not contain a consensus sequence for myristoylation or a lipobox motif, which can typically be found in N-terminally modified lipoproteins (21, 25).
Sudan Black B staining of the Tricine-SDS-polyacrylamide gel showed that all viruses contained lipids (Fig. 3B). The lipid extracts of His2, HRPV-2, HRPV-3, HRPV-6, and HGPV-1 as well as their host cells were examined by thin-layer chromatography (TLC) (Fig. 3C). Based on the known lipid species of Haloarcula hispanica (10), three major polar lipids were identified: phosphatidylglycerol (PG), phosphatidylglycerophosphate methyl ester (PGP-Me), and phosphatidylglycerosulfate (PGS). PGS was not detected in Halogeometricum sp. CG-9 or its virus, HGPV-1 (Fig. 3C). The lipid compositions of His2 and its host, Haloarcula hispanica, were identical. All other virus-host pairs contained minor differences in lipid patterns, with at least one of the host lipids being missing from the viral membrane (Fig. 3C, arrows).
Pleomorphicity of the virions.
All studied viruses formed wide, diffusive, light-scattering zones by rate-zonal centrifugation, suggesting particle variability. For HRPV-1, this phenomenon was studied in more detail. After centrifugation, infectivity was high (above 1011 PFU ml−1) from the middle to the bottom of the gradient (Fig. 4A). The light-scattering virus zone was analyzed by Tricine-SDS-PAGE (Fig. 4B), and the protein band intensity analysis showed that the ratio of the major structural proteins, VP3 and VP4, remained almost identical throughout the zone (see Fig. S3A in the supplemental material). There was no major change in the ratio of proteins to lipids (see Fig. S3B in the supplemental material). We also established by cryo-EM that the average particle diameter did not vary significantly in different parts of the virus zone, increasing from 40.7 ± 2.2 nm to 41.7 ± 2.9 nm (see Fig. S3C in the supplemental material). The specific infectivity remained approximately the same across the zone, varying between 5.3 × 1012 and 7.7 × 1012 PFU mg−1 of protein. Hence, the wide particle band most probably reflects that the virions do not have identical structures, as seen for icosahedrally symmetric viruses, but instead display pleomorphicity.
Fig 4.
Sedimentation analysis of HRPV-1 virions. (A) HRPV-1 particles were analyzed by rate-zonal centrifugation, and the infectivity (open circles) and absorbance at 260 nm (closed circles) of the gradient fractions were determined. (B) The light-scattering virus zone was fractionated. The fractions were analyzed by SDS-PAGE and staining with EtBr (for nucleic acids) and with Coomassie blue and Sudan Black B (for proteins and lipids, respectively). Numbers on the left indicate the molecular masses of the markers. The protein and lipid band intensities are shown on the right.
Cryo-EM studies of HRPV-1 show randomly distributed spikes on the virion surface.
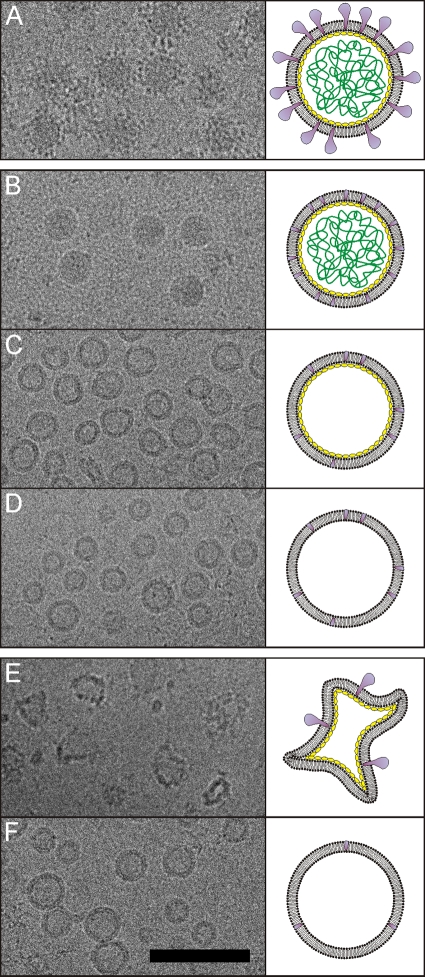
The virion structure of HRPV-1 was previously described by quantitative dissociation experiments (48). Here, we determined the HRPV-1 structure using cryo-EM. HRPV-1 subviral particles of known compositions were compared to identify densities belonging to VP3, VP4, the DNA, and the membrane. Cryo-EM images of the viral samples along with schematic diagrams are illustrated in Fig. 5. Two further dissociation methods (Fig. 5C and F) were developed, in addition to those described previously, and the protein and DNA compositions of the subviral particles were confirmed by SDS-PAGE analysis (data not shown).
Fig 5.
Dissociation products of HRPV-1 analyzed by cryo-EM. Cryo-electron micrographs of the particles are shown on the left, and schematic representations are shown on the right. Colors indicate the VP4 protein (purple), the VP3 protein (yellow), and the genome (green). The two membrane-associated domains of VP3 are not indicated in the schematic representations. (A) Untreated 1×-purified virions. (B) Virions treated with proteinase K at high salinity at 37°C (48). (C) Proteinase K-treated particles (see panel B) further dissociated at low salinity at 37°C. (D) Virions treated with proteinase K at low salinity at 37°C (48). (E) Virions treated at low salinity at 60°C (48). (F) Particles produced at low salinity at 60°C (see panel E) and further treated with proteinase K at low salinity at 37°C. Scale bar, 100 nm.
It is evident that VP4 forms the surface spikes (all but the C-terminal VP4 membrane anchor is removed by proteinase K treatment under conditions of high salinity) (Fig. 5A and B). The release of DNA from the proteinase K-treated particles at low salinity localizes the genome inside the membrane vesicle (Fig. 5B and C). The additional removal of VP3 (all but the N- and C-terminal VP3 membrane-associated domains) at low salinity by proteinase K treatment did not change the vesicle morphology; hence, the density from VP3 does not significantly contribute to the membrane density (Fig. 5C and D). Particles are angular after treatment at 60°C under conditions of low salinity, which releases the genome and most of the spikes (Fig. 5E). This could be due to phase changes of the lipids or the denaturation of the proteins. Further treatment of the angular particles with proteinase K restored the round morphology of the vesicles (Fig. 5F). This finding implied that the high temperature caused the denaturation of VP3, leading to the distorted shape of the vesicle.
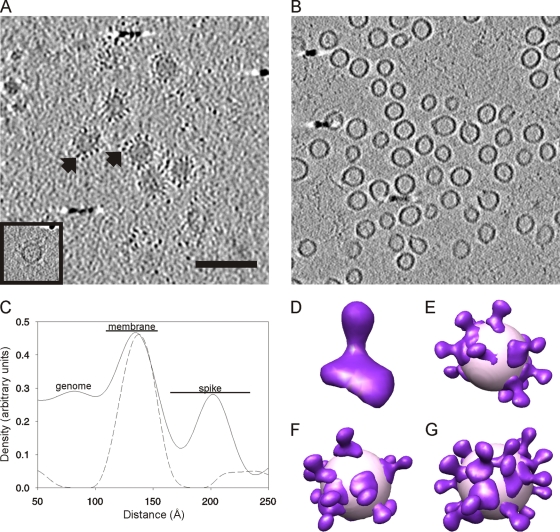
The HRPV-1 virion and subviral particle containing VP3 and the membrane (Fig. 5C) were analyzed in more detail by cryo-electron tomography and subtomographic averaging. Initially, we attempted single-particle averaging of the HRPV-1 images, as the particles had similar diameters, and some particles appeared to be ordered. However, the models did not converge to a stable answer (data not shown), and so we used cryo-ET, which accommodates the apparent heterogeneity in the particles and revealed the reasons for their pleomorphicity (Fig. 4 and see Fig. S3 in the supplemental material). Seventeen tomograms were computed, with angular ranges from ±48° to ±60° at 1° to 4° increments. The best three-dimensional tomographic reconstruction of the intact HRPV-1 virions clearly revealed approximately spherically shaped viruses not flattened by the vitrification process. The surface of each particle was covered by spikes, which were sitting on top of a continuous membrane layer (Fig. 6A).
Fig 6.
Structural organization of HRPV-1. (A) Tomographic slice of HRPV-1 showing intact spherically shaped virions containing the genome and spikes projecting out from the surface of the membrane (black arrows). An image of an empty particle is shown in the inset. Scale bar, 100 nm (same for panel B). (B) Tomographic slice of HRPV-1 subviral particles composed of the lipid membrane and VP3 protein (see also Fig. 5C). The slices are 5.7 nm thick in panels A and B. (C) Density distribution over the membrane, calculated from the aligned extracted subvolumes for HRPV-1 virions (solid line) and subviral particles containing the membrane and VP3 (dotted line). The membrane peaks were normalized. (D) Isosurface representation of a single VP4 spike. (E to G) The model of the VP4 spike protein (violet) was placed into individual virion tomograms based on the refined spike positions that contributed to the averaged VP4 model. The viral surface (light pink) was approximated by a sphere of 35 nm in diameter.
A detailed analysis using subtomographic processing showed the radial distribution of the membrane and the spike protein VP4 and led to an average structure of VP4 (Fig. 6). The densities of VP3 and the membrane were defined by averaging a large number of subtomograms from the virion or the VP3-containing membrane vesicle (Fig. 6A and B). This allowed the membranes of particles with slightly different membrane shapes and diameters to be aligned and averaged together. The resulting two average subtomograms were then compared by calculating radial profiles from each one, resulting in one profile from the virions and one from the VP3-containing membrane vesicles (Fig. 6C). The membrane, containing VP3, was clearly visible in the subviral particles as an approximately 4.2-nm-thick continuous layer (Fig. 6B and C). The profile also revealed the average length of the VP4 spike structure embedded in the membrane to be about 7 nm, measured from the center of the membrane peak to the center of the spike peak.
The subtomograms from the HRPV-1 virion were extensively refined in order to determine the structure of VP4. VP4 was club shaped, with the distal end being thicker than the proximal end with respect to the viral membrane (Fig. 6D). Cross-correlating this averaged spike structure with the virions in the tomograms revealed the organization of the VP4 spikes on the membrane and showed that they form irregular arrays on the surface of the particles (Fig. 6E to G). The relatively low density of the HRPV-1 VP4 spike protein in the radial profile compared to the membrane is explained by the occupancy of the spike on the surface of the membrane, where it is relatively loosely packed (Fig. 6C to G). The virion reconstructions are affected by the missing wedge of information in the tomograms, and as a result, the models of intact virions have low spike densities in two opposing areas on the volume (Fig. 6D to G).
Spike proteins have different sensitivities to protease digestion.
Using HRPV-1 dissociation (48) as a model, we carried out similar analyses for the other six viruses studied here. The virions were treated with proteinase K or dissociated at low salinity, or a combination of these treatments was used (see Materials and Methods). After each treatment, dissociation products were analyzed by rate-zonal centrifugation, and the protein, lipid, and DNA contents of the fractionated gradients were determined. Based on the results summarized in Table 4, HRPV-6 was the most different isolate compared to HRPV-1, and thus, the gradient analyses of this virus are presented as an example (Fig. 7).
Table 4.
Summary of the virion dissociation experiments
| Virus | Result(s) of experiment |
|||||
|---|---|---|---|---|---|---|
| Proteinase K, high salinity, 3 h at 37°C |
Low salinity, 1 h at 37°C |
Proteinase K, low salinity, 1 h at 37°C |
||||
| Major soluble fragments of spikes | Membrane-associated domain of spikes | Dissociation of spikes | Genome release | Genome release | Membrane protein digestedb | |
| HRPV-1a | No | Yes | Partial | Yes | Yes | ++++ |
| His2 | No | Yes | Partial (VP28), no (VP29) | Yes | Yes | +++ |
| HHPV-1 | Yes | No | Yes | Yes | Yes | ++++ |
| HRPV-2 | Yes | No | Partial | No | Yes | ++ |
| HRPV-6 | Yes | No | Partial | No | Partial | + |
| HRPV-3 | Yes | Yes | No | Yes | Yes | + |
| HGPV-1 | Yes | Yes | Partial | Yes | Yes | +++ (VP2), + (VP3) |
See reference 48.
++++, all or almost all the membrane protein molecules were digested; +, hardly any membrane protein molecules were digested.
Fig 7.
Dissociation analysis of HRPV-6. The virions and subviral particles were separated by rate-zonal centrifugation. The gradient fractions were subjected to gel electrophoresis followed by staining of the SDS-PAGE gel with EtBr (for nucleic acids) and of the Tricine-SDS-PAGE gel with Coomassie blue and Sudan Black B (for proteins and lipids, respectively). Numbers at the top indicate gradient fractions 1 to 13 from top to bottom. Numbers on the left indicate the molecular mass markers. A minor protein of about 20 kDa, indicated by arrowheads, was identified as VP4 (about 12 kDa) by N-terminal sequencing (see Table S3 in the supplemental material) and thus most likely represents a dimeric form of VP4. (A) Untreated 2×-purified virions. (B) Proteinase K treatment at high salinity at 37°C. The arrows indicate soluble fragments of VP5, and asterisks indicate the proteinase K bands. (C) Dissociation at low salinity at 37°C. (D) Proteinase K treatment at low salinity at 37°C. The asterisks indicate the proteinase K bands.
As described previously for HRPV-1 (48), the proteinase K treatment of all six viruses at high salinity resulted in DNA-containing lipid vesicles sedimenting more slowly than the untreated virions (HRPV-6 in Fig. 7A and B). Only the large major structural proteins were digested, indicating that they are located on the virion surface and were thus designated spike proteins. We also tested the proteinase K sensitivities of all seven viruses during a stepwise decrease of the ionic strength (see Fig. S4 in the supplemental material). In all cases, the spike proteins were digested even at the highest salinity (see Fig. S4 in the supplemental material), and based on the digestion products (Table 4), the viruses could be divided into three categories.
In category I, which includes HRPV-6, HRPV-2, and HHPV-1, the spike proteins were digested, producing major soluble fragments (HRPV-6 in Fig. 7B, arrows, and see Fig. S4A to S4C in the supplemental material). For HRPV-6, all three fragments were identified as the N-terminal fragments of the VP5 spike protein (see Table S3 in the supplemental material). In category II, which includes HRPV-3 and HGPV-1, the digestion resulted in major soluble fragments and one membrane-associated one (see Fig. S4D and S4E, arrows and arrowheads, in the supplemental material). The membrane-associated fragment of HRPV-3 was identified as the C-terminal fragment of the VP2 spike protein (see Table S3 in the supplemental material). The major soluble fragments were digested when the salinity was further decreased (see Fig. S4 in the supplemental material), indicating that the salinity affected the conformation and, thus, the protease sensitivity of the spikes.
In category III, which includes His2 and HRPV-1, the proteinase K treatment produced no major soluble fragments but produced membrane-associated ones (see Fig. S4F and S4G in the supplemental material). The His2 spike proteins VP28 and VP29 were both digested, which was confirmed by Sudan Black B staining (data not shown), and two membrane-associated fragments were observed (see Fig. S4F, arrowheads, in the supplemental material). The N-terminal sequence of one of the bands corresponded to the C-terminal fragment of the VP28 spike protein (see Table S3 in the supplemental material). No protein sequence information was obtained from the other band.
Virion dissociation shows the diversity of the pleomorphic viruses.
The treatment of the HRPV-1 virions with the nonionic detergent NP-40 releases the genome without associated proteins (48). The other virions were also treated with NP-40, leading to the same result (data not shown). The low-salinity dissociation at 37°C also releases the HRPV-1 genome without attached proteins (48). Under the same conditions, the genomes of HRPV-3, HGPV-1, HHPV-1, and His2 were released, and the small major structural proteins remained associated with the empty vesicle (Table 4), indicating that they are membrane proteins. The small major structural proteins became proteinase K sensitive only when the salinity was decreased (see Fig. S4 in the supplemental material), most likely due to the vesicle opening. In the case of the most sensitive small major structural proteins, His2 VP27, HGPV-1 VP2, and HHPV-1 VP3 (Table 4), we can conclude that they are most likely internal membrane proteins.
HRPV-2 and HRPV-6 were the only viruses whose genomes remained associated within the membrane vesicle when exposed to low salinity at 37°C (HRPV-6 in Fig. 7C). Interestingly, the addition of proteinase K caused the genome release in HRPV-2 and a partial release in HRPV-6 (HRPV-6 in Fig. 7D). A minor digestion of the small major structural proteins of HRPV-2 and HRPV-6 was observed (HRPV-6 in Fig. 7D). The undigested portion remained associated with the vesicle, indicating that these are most probably membrane proteins interacting with the genome.
The HRPV-1 spike protein VP4 is partially removed by low-salinity incubation at 37°C (48). The spike proteins of HGPV-1, HRPV-2, and HRPV-6 behaved similarly (HRPV-6 in Fig. 7C). In contrast, the spike protein of HRPV-3 did not dissociate, and only one spike protein of His2, VP28, was partially released. The spike protein of HHPV-1 was completely dissociated (Table 4).
DISCUSSION
This study expands our understanding of archaeal viruses by describing a worldwide-distributed, unique group of seven pleomorphic, lipid-containing viruses infecting extremely halophilic archaea, which we refer to as pleolipoviruses. All seven pleolipoviruses obey the same simple structural organization, having a lipid vesicle with one spike protein and one, most likely internal, membrane protein (Fig. 3). Of the seven presented isolates, HGPV-1 and His2 are more elaborated, with two membrane and two spike proteins, respectively (Fig. 3B). As previously shown for HRPV-1 (48), no major structural proteins associated with the genome were detected, indicating that the pleolipoviruses have no nucleocapsid. A parallel comparative genomic study illustrated that the genomes of the pleolipoviruses have a colinear gene organization and show similarity to each other at the amino acid level (58). Altogether, this information implies that the pleolipoviruses form a novel group of related viruses.
Currently, His2 is a tentative species of the genus Salterprovirus, whose type species is the spindle-shaped virus His1 (11, 37). Both His1 and His2 infect the same host and have linear dsDNA genomes encoding a putative DNA polymerase. His1 and His2 were proposed to be distantly related, as they share no significant amino acid sequence similarity except for that of the putative polymerases (11). Consequently, the amino acid sequence similarity between His2 and the pleomorphic viruses HRPV-1 and HHPV-1 (49, 53) led us to investigate if His2 is a pleomorphic rather than a spindle-shaped virus. Noticeably, cryo-electron microscopy and lipid and protein analyses confirmed that His2 belongs to the pleolipoviruses established in this work (Fig. 3).
The pleolipoviruses are extremely host specific, as the host range is limited to a single known species (7). They do not cause cell lysis, but virions are released continuously, slightly retarding the growth of the hosts (Fig. 1) (49, 53). Even though His2-infected cells showed lysis-like behavior late in the growth phase, this did not coincide with virus production, contradicting the previously reported suggestion that His2 is lytic (11). The life cycle of the pleolipoviruses is most similar to those of nonlytic, spindle-shaped viruses infecting hyperthermophilic archaea, such as SSV1 (Sulfolobus spindle-shaped virus 1) and TPV1 (Thermococcus prieurii virus 1), which persist in a carrier state within the host cells, producing viruses in a continuous fashion (29, 57).
It was shown previously that HRPV-1 and HHPV-1 acquire lipids unselectively from the host cell membrane (48, 53). Our TLC analysis confirmed that this is also true for the other pleolipoviruses (Fig. 3C). Based on the unselective lipid acquisition, it is likely that all pleolipoviruses are released from the cell by budding, as suggested previously for HRPV-1 and HHPV-1 (49, 53). Except for HGPV-1, all pleolipoviruses infect either Haloarcula or Halorubrum strains and contain the same major phospholipids, PG, PGP-Me, and PGS (Fig. 3C) (48, 53). Unlike Halorubrum and Haloarcula cells, Halogeometricum cells lack PGS (19, 43), explaining the major lipid pattern differences between HGPV-1 and the others (Fig. 3C). In addition, the spike proteins His2 VP28 and HGPV-1 VP4 were observed to possess lipid modifications (Fig. 3B). Although N-terminal myristoylation of proteins is commonly used among membrane-containing viruses (21), we found no evidence for this. However, the cell wall protein of a haloarchaeon was shown previously to contain a covalently attached lipid moiety in the C-terminal region of the protein, possibly contributing to membrane anchoring (36), and the spike proteins of His2 and HGPV-1 might contain similar modifications. HRPV-1 is the only pleolipovirus reported to have glycosylated spikes (48).
Cryo-EM and cryo-ET analyses of the virions in their native hydrated state showed that the pleolipoviruses are more or less spherical, with spikes decorating the virion surface (Fig. 3A and 6). For HRPV-1, we found no evidence for an ordered protein capsid made of the internal membrane protein VP3; rather, it seems to reside mainly in the membrane (Fig. 5 and 6). Although mostly spherical, the pleomorphic nature of the virions is obvious. The apparently random distribution of the spikes in the membrane (Fig. 6) and the asymmetry observed by rate-zonal centrifugation (Fig. 4) support this notion.
The irregular organization of the VP4 spikes in HRPV-1 indicates that there are only weak lateral spike-to-spike interactions, giving rise to a “sloppy” assembly compared to what has been seen, for instance, for membrane-containing icosahedrally symmetric viruses such as Semliki forest virus, PRD1, and SH1 (1, 18, 27, 33). Limited lateral order has been seen for Tula virus (32), but HRPV-1 is most reminiscent of pleomorphic viruses such as influenza virus, where no long-range order in the glycoproteins has been detected, although there is a segregation of the glycoproteins in the membrane (15, 54). The sloppy assembly of the pleolipoviruses is also reflected in the heterogeneity of the genome packaging densities (Table 3). The structure of the pleolipoviruses is probably influenced most by the proposed budding mechanism, and it is highly likely that the interaction of the membrane proteins with the genome is important for assembly.
Based on the protease sensitivity of the spike proteins, the pleolipoviruses were divided into different categories (see Fig. S4 in the supplemental material). Interestingly, Halorubrum viruses are found in all three categories, and HHPV-1 and His2, which infect the same host, Haloarcula hispanica, are in different categories. This finding implies that the pleolipoviruses infecting related or the same hosts may employ different cell receptors. Moreover, spikes cannot be used as the only classification criterion because they most likely evolve rapidly due to their function in host recognition (55). When other dissociation data (Table 4) are taken into account, the pleolipoviruses assemble into the following three groups: (i) HRPV-1, His2, and HHPV-1; (ii) HRPV-2 and HRPV-6; and (iii) HRPV-3 and HGPV-1.
The pleomorphic, lipid-containing virion type with two major structural proteins has also been detected among bacterial viruses (23), and the pleolipoviruses may be extended to a structural viral lineage. Interestingly, the circular ssDNA genome of the bacterial virus L172 is almost as large as the linear dsDNA genome of His2 (11, 23), which is clearly larger than the genomes of the other pleolipoviruses. It can be speculated that His2 originated by the fusion of a fragment containing the DNA polymerase gene (11) responsible for replication and a “pleolipovirus-like” fragment determining virion morphotype and architecture.
The conserved virion architecture implies that the pleolipoviruses have a common origin. However, despite the shared virion architecture, these viruses have different genome types (11, 49, 53, 58). It has been shown that viral genes responsible for functions like replication are not inherited vertically within a viral lineage but are changed by horizontal gene transfer (2, 3, 8, 40). Recombination is most probably frequent in hypersaline habitats, where the viral particle concentration reaches high levels and viruses infect the same limited range of host organisms (30, 46, 61). Thus, the pleolipoviruses most likely employ different replication mechanisms, resulting in different genome types. HRPV-1 and HHPV-1 were proposed previously to use rolling-circle replication, but the ssDNA genome is packaged in the case of HRPV-1 and the dsDNA is packaged in the case of HHPV-1 (49, 53). His2 is predicted to encode a type B DNA polymerase and most likely uses protein-primed replication (11). Since the pleolipoviruses probably bud from infected cells, it does not make a difference whether the genome enclosed in the membrane vesicle is circular or linear, single or double stranded.
This study illustrates that the pleolipoviruses infecting haloarchaea are defined by similar life cycles, protein and lipid patterns, and virion organizations. The pleolipoviruses might also compose a novel structure-based lineage of viruses using host-derived lipids to envelope the genome and only two or three major proteins, the spike and internal membrane proteins, to interact with the host and to direct virion assembly. In addition to the seven virus isolates, there are also possible pleolipoproviruses in the genomes of at least six haloarchaea (22, 49, 53), and a viral metagenome obtained from a hypersaline lake contained a sequence similar to those of the pleolipovirus genomes (59). These observations emphasize the abundance of these novel viruses in hypersaline environments.
Supplementary Material
ACKNOWLEDGMENTS
We thank Päivi Hannuksela, Sari Korhonen, Soile Storman, Pasi Laurinmäki, Veli-Pekka Kestilä, and Eevakaisa Vesanen for excellent technical assistance. We acknowledge Gunilla Rönnholm for the protein sequencing and Mike Dyall-Smith for kindly providing His2. We also thank Ana Senčilo, Lars Paulin, Stefanie Kellner, Mark Helm, and Elina Roine for providing the genomic data. We thank the University of Helsinki for the support to the European Union ESFRI Instruct Associate Centre for Virus Production and Purification and the Biocenter Finland National Cryo Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki, and the CSC-IT Center for Science, Ltd., for providing computation facilities.
This research was supported by Academy of Finland Centre of Excellence Programme in Virus Research grant 1129684 (2006 to 2011) (D.H.B. and S.J.B.), Academy of Finland grant 1127665 (H.M.O.), and Academy Professor (Academy of Finland) funding grants 255342 and 256518 (D.H.B.). M.K.P., N.S.A., V.M., and L.L. are fellows of the Viikki Doctoral Programme in Molecular Biosciences.
Footnotes
Published ahead of print 22 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abrescia NG, et al. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68–74 [DOI] [PubMed] [Google Scholar]
- 2. Abrescia NGA, Bamford DH, Grimes JM, Stuart D. Structure unifies the viral universe. Annu. Rev. Biochem., in press [DOI] [PubMed] [Google Scholar]
- 3. Abrescia NGA, et al. 2010. What does it take to make a virus: the concept of the viral ‘self’, p 35–58 In Stockley PG, Twarock R. (ed), Emerging topics in physical virology. Imperial College Press, London, United Kingdom [Google Scholar]
- 4. Ackermann HW. 2007. 5500 phages examined in the electron microscope. Arch. Virol. 152:227–243 [DOI] [PubMed] [Google Scholar]
- 5. Adrian M, Dubochet J, Lepault J, McDowall AW. 1984. Cryo-electron microscopy of viruses. Nature 308:32–36 [DOI] [PubMed] [Google Scholar]
- 6. Arahal DR, Dewhirst FE, Paster BJ, Volcani BE, Ventosa A. 1996. Phylogenetic analyses of some extremely halophilic archaea isolated from Dead Sea water, determined on the basis of their 16S rRNA sequences. Appl. Environ. Microbiol. 62:3779–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM. 2012. Global network of specific virus-host interactions in hypersaline environments. Environ. Microbiol. 14:426–440 [DOI] [PubMed] [Google Scholar]
- 8. Bamford DH. 2003. Do viruses form lineages across different domains of life? Res. Microbiol. 154:231–236 [DOI] [PubMed] [Google Scholar]
- 9. Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15:655–663 [DOI] [PubMed] [Google Scholar]
- 10. Bamford DH, et al. 2005. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J. Virol. 79:9097–9107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bath C, Cukalac T, Porter K, Dyall-Smith ML. 2006. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology 350:228–239 [DOI] [PubMed] [Google Scholar]
- 12. Bath C, Dyall-Smith ML. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J. Virol. 72:9392–9395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Benson SD, Bamford JK, Bamford DH, Burnett RM. 2004. Does common architecture reveal a viral lineage spanning all three domains of life? Mol. Cell 16:673–685 [DOI] [PubMed] [Google Scholar]
- 14. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 15. Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. 2010. Structural organization of a filamentous influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 107:10685–10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chaban B, Ng SY, Jarrell KF. 2006. Archaeal habitats—from the extreme to the ordinary. Can. J. Microbiol. 52:73–116 [DOI] [PubMed] [Google Scholar]
- 17. Chun J, et al. 2007. EzTaxon: a Web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2259–2261 [DOI] [PubMed] [Google Scholar]
- 18. Cockburn JJ, et al. 2004. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432:122–125 [DOI] [PubMed] [Google Scholar]
- 19. Cui HL, et al. 2010. Halogeometricum rufum sp. nov., a halophilic archaeon from a marine solar saltern, and emended description of the genus Halogeometricum. Int. J. Syst. Evol. Microbiol. 60:2613–2617 [DOI] [PubMed] [Google Scholar]
- 20. DeLong EF, Pace NR. 2001. Environmental diversity of bacteria and archaea. Syst. Biol. 50:470–478 [PubMed] [Google Scholar]
- 21. Du Y, Zuckermann FA, Yoo D. 2010. Myristoylation of the small envelope protein of porcine reproductive and respiratory syndrome virus is non-essential for virus infectivity but promotes its growth. Virus Res. 147:294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dyall-Smith ML, et al. 2011. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6:e20968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dybvig K, Nowak JA, Sladek TL, Maniloff J. 1985. Identification of an enveloped phage, Mycoplasma virus L172, that contains a 14-kilobase single-stranded DNA genome. J. Virol. 53:384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edwards U, Rogall T, Blöcker H, Emde M, Böttger EC. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843–7853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eichler J, Adams MW. 2005. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 69:393–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Folch J, Lees M, Sloane Stanley GH. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226:497–509 [PubMed] [Google Scholar]
- 27. Fuller SD, Butcher SJ, Cheng RH, Baker TS. 1996. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol. 116:48–55 [DOI] [PubMed] [Google Scholar]
- 28. Glazko G, Makarenkov V, Liu J, Mushegian A. 2007. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. 2012. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ. Microbiol. 14:503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guixa-Boixareu N, Calderón-Paz JI, Heldal M, Bratbak G, Pedrós-Alió C. 1996. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat. Microb. Ecol. 11:215–227 [Google Scholar]
- 31. Heymann JB. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156–169 [DOI] [PubMed] [Google Scholar]
- 32. Huiskonen JT, et al. 2010. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 84:4889–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jäälinoja HT, et al. 2008. Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc. Natl. Acad. Sci. U. S. A. 105:8008–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Juez G, Rodriquez-Valera F, Ventosa A, Kushner DJ. 1986. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic archaebacteria. Syst. Appl. Microbiol. 8:75–79 [Google Scholar]
- 35. Kates M. 1972. Techniques of lipidology: isolation, analysis and identification of lipids. North-Holland Publishing Co, Amsterdam, Netherlands [Google Scholar]
- 36. Kikuchi A, Sagami H, Ogura K. 1999. Evidence for covalent attachment of diphytanylglyceryl phosphate to the cell-surface glycoprotein of Halobacterium halobium. J. Biol. Chem. 274:18011–18016 [DOI] [PubMed] [Google Scholar]
- 37. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. 2011. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 38. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 39. Krupovič M, Bamford DH. 2010. Order to the viral universe. J. Virol. 84:12476–12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krupovič M, Bamford DH. 2007. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics 8:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 [DOI] [PubMed] [Google Scholar]
- 42. Mochizuki T, et al. 2010. Diversity of viruses of the hyperthermophilic archaeal genus Aeropyrum, and isolation of the Aeropyrum pernix bacilliform virus 1, APBV1, the first representative of the family Clavaviridae. Virology 402:347–354 [DOI] [PubMed] [Google Scholar]
- 43. Montalvo-Rodríguez R, et al. 1998. Halogeometricum borinquense gen. nov., sp. nov., a novel halophilic archaeon from Puerto Rico. Int. J. Syst. Bacteriol. 48(Pt 4):1305–1312 [DOI] [PubMed] [Google Scholar]
- 44. Nuttall SD, Dyall-Smith ML. 1993. HF1 and HF2: novel bacteriophages of halophilic archaea. Virology 197:678–684 [DOI] [PubMed] [Google Scholar]
- 45. Olkkonen VM, Bamford DH. 1989. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology 171:229–238 [DOI] [PubMed] [Google Scholar]
- 46. Oren A, Bratbak G, Heldal M. 1997. Occurrence of virus-like particles in the Dead Sea. Extremophiles 1:143–149 [DOI] [PubMed] [Google Scholar]
- 47. Pettersen EF, et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Pietilä MK, Laurinavičius S, Sund J, Roine E, Bamford DH. 2010. The single-stranded DNA genome of novel archaeal virus Halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J. Virol. 84:788–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. 2009. An ssDNA virus infecting archaea: a new lineage of viruses with a membrane envelope. Mol. Microbiol. 72:307–319 [DOI] [PubMed] [Google Scholar]
- 50. Pina M, Bize A, Forterre P, Prangishvili D. 2011. The archeoviruses. FEMS Microbiol. Rev. 35:1035–1054 [DOI] [PubMed] [Google Scholar]
- 51. Prangishvili D, Garrett RA, Koonin EV. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52–67 [DOI] [PubMed] [Google Scholar]
- 52. Prosser JI, Nicol GW. 2008. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 10:2931–2941 [DOI] [PubMed] [Google Scholar]
- 53. Roine E, et al. 2010. New, closely related haloarchaeal viral elements with different nucleic acid types. J. Virol. 84:3682–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saren AM, et al. 2005. A snapshot of viral evolution from genome analysis of the Tectiviridae family. J. Mol. Biol. 350:427–440 [DOI] [PubMed] [Google Scholar]
- 56. Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 57. Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. U. S. A. 89:7645–7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Senčilo A, Paulin L, Kellner S, Helm M, Roine E. Related haloarchaeal pleomorphic viruses contain different genome types. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sime-Ngando T, et al. 2011. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ. Microbiol. 13:1956–1972 [DOI] [PubMed] [Google Scholar]
- 60. Steinhauer DA, Holland JJ. 1987. Rapid evolution of RNA viruses. Annu. Rev. Microbiol. 41:409–433 [DOI] [PubMed] [Google Scholar]
- 61. Tang SL, Nuttall S, Dyall-Smith M. 2004. Haloviruses HF1 and HF2: evidence for a recent and large recombination event. J. Bacteriol. 186:2810–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Winker S, Woese CR. 1991. A definition of the domains Archaea, Bacteria and Eucarya in terms of small subunit ribosomal RNA characteristics. Syst. Appl. Microbiol. 14:305–310 [DOI] [PubMed] [Google Scholar]
- 63. Woese CR, Kandler O, Wheelis ML. 1990. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 87:4576–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203–214 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.