Abstract
We generated influenza A viruses expressing mutant NS1 proteins unable to activate phosphoinositide 3-kinase (PI3K) in two mouse-lethal strains. The recombinant A/Puerto Rico/8/34 (rPR8) mutant virus strain was attenuated and caused reduced morbidity/mortality. For the recombinant A/WSN/33 (rWSN) virus strain, the inability to stimulate PI3K had minimal impact on replication or morbidity/mortality. Cell-based assays revealed subtly distinct intracellular sites of NS1 localization and PI3K activation between the strains. We hypothesize that specific spatially regulated NS1-activated PI3K signaling, rather than simply the total level of active PI3K, is important for virus replication and virulence.
TEXT
Influenza A virus NS1 protein is a multifunctional virulence factor with well-characterized abilities to counteract host antiviral defenses (10). NS1 also binds directly to the p85β regulatory subunit of the cellular lipid metabolism enzyme phosphoinositide 3-kinase (PI3K), thereby stimulating activation of PI3K/Akt signaling (2, 8, 17). A crystal structure of the NS1-p85β interaction interface, combined with mutagenesis and biochemical studies, has confirmed several key residues of NS1 that are essential for binding and activating PI3K (8, 9, 16). Among these, tyrosine 89 is at the very center of the interface, and its conservative substitution to phenylalanine (Y89F) completely abrogates the NS1-p85β interaction without detectably affecting its ability to antagonize host interferon production (7–9). The biological reasons for NS1 activation of this specific p85β-PI3K pathway are not understood, and there is debate in the literature as to whether this NS1 function acts to delay virus-induced apoptosis (2, 12, 18) or has another role, such as regulating lung epithelium cation currents (6).
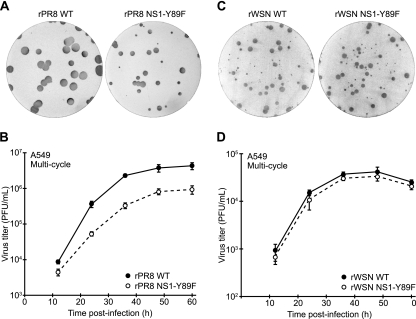
Using reverse genetics (3), we generated recombinant wild-type (WT) and NS1-Y89F viruses with the mouse-lethal A/Puerto Rico/8/34 (PR8) and A/WSN/33 (WSN) influenza virus strains. The four viruses (rPR8 WT, rPR8 NS1-Y89F, rWSN WT, and rWSN NS1-Y89F) were plaque purified, and virus stocks were grown and titrated in MDCK cells using serum-free medium. RNA was extracted from stock aliquots, and the eight genomic segments of each virus were fully sequenced after segment-specific reverse transcription-PCR (RT-PCR) to ensure the absence of undesired mutations. Compared to the rPR8 WT virus, the rPR8 NS1-Y89F virus was clearly attenuated, forming smaller plaques on MDCK cells (Fig. 1A) and growing to ∼10-fold-lower titers in the A549 human lung epithelial cell line during multicycle replication assays (Fig. 1B). In contrast, we were unable to discern any obvious difference between the rWSN WT virus and the rWSN NS1-Y89F virus; the two viruses formed similar plaque phenotypes on MDCK cells (Fig. 1C) and grew with identical replication kinetics in A549 cells (Fig. 1D).
Fig 1.
Replication of WT and NS1-Y89F viruses in tissue culture. Plaque phenotypes of rPR8 WT and rPR8 NS1-Y89F viruses (A) or rWSN WT and rWSN NS1-Y89F viruses (C) in MDCK cells. Multicycle growth analysis of rPR8 WT and rPR8 NS1-Y89F viruses (B) or rWSN WT and rWSN NS1-Y89F viruses (D) in a human lung epithelial cell line, A549 (infection at an MOI of 0.01 PFU/cell). Data points show mean values from three replicates, and error bars represent standard deviations (SD).
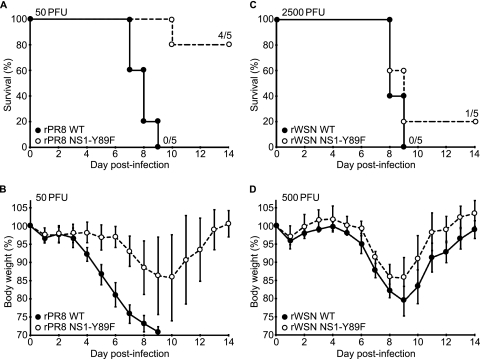
To assess the contribution of NS1-activated PI3K to influenza A virus replication and pathogenicity in vivo, we determined the 50% mouse lethal dose (MLD50) values for all four viruses by intranasally infecting ketamine-xylazine-anesthetized 6- to 8-week-old C57BL/6 mice (Jackson Laboratory, ME) with 5-fold serial dilutions of virus (5 mice/group) and monitoring body weight changes for 14 days. All procedures were performed in accordance with the Institutional Animal Care and Use Committee guidelines of Mount Sinai School of Medicine, and animals showing more than 25% weight loss were considered to have reached the experimental endpoint and were humanely euthanized. MLD50 values were subsequently calculated according to the method of Reed and Muench (Table 1). In the rPR8 virus background, the NS1-Y89F mutation led to a >8-fold increase in MLD50 (MLD50 values were 7.3 PFU for the rPR8 WT virus and 59.3 PFU for the rPR8 NS1-Y89F virus) (Table 1 and Fig. 2A), and mice infected with the rPR8 NS1-Y89F virus exhibited less morbidity (as determined by weight loss) than mice infected with the WT (Table 1 and Fig. 2B). Notably, even when comparable MLD50 values were used, there was an ∼2-day delay in the onset of disease symptoms and death with the rPR8 NS1-Y89F virus compared to results with the WT (Table 1 and data not shown). In stark contrast, the rWSN WT and rWSN NS1-Y89F viruses had similar MLD50 values (∼1.8-fold difference; MLD50 values were 593 PFU for the rWSN WT and 1,050 PFU for the rWSN NS1-Y89F virus) (Table 1 and Fig. 2C), and the weight loss profiles associated with the two viruses were almost identical (Fig. 2D).
Table 1.
Characterization of WT and NS1-Y89F virus pathogenicity in vivo
| Virus | Dose (PFU) | % Survival (no. of mice surviving/total no. tested) | Mean maximum wt loss (%) (range) | Median no. of days with indicated % wt loss (range) |
|
|---|---|---|---|---|---|
| >10% | >20% | ||||
| rPR8 WT | 10 | 60 (3/5) | 21.4 (15.6–>25) | 5 (3–9) | 2 (2–8) |
| 50 | 0 (0/5) | >25 (>25) | 10 (9–11) | 8 (8–9) | |
| 250 | 0 (0/5) | >25 (>25) | 11 (11) | 10 (9–10) | |
| 1,250 | 0 (0/5) | >25 (>25) | 12 (12) | 11 (10–11) | |
| rPR8 NS1-Y89F | 10 | 100 (5/5) | 10.1 (4.6–14.4) | 1 (0–2) | 0 (0) |
| 50 | 80 (4/5) | 16.4 (5.9–>25) | 4 (0–8) | 0 (2–7) | |
| 250 | 0 (0/4) | >25 (>25) | 9 (8–9) | 7 (7–8) | |
| 1,250 | 0 (0/5) | >25 (>25) | 10 (10–11) | 9 (8–9) | |
| rWSN WT | 20 | 100 (5/5) | 3.3 (0.8–5.4) | 0 (0) | 0 (0) |
| 100 | 100 (5/5) | 5.0 (2.5–7.8) | 0 (0) | 0 (0) | |
| 500 | 80 (4/5) | 21.2 (16.4–>25) | 4 (4–8) | 1 (0–7) | |
| 2,500 | 0 (0/5) | >25 (>25) | 9 (9–10) | 8 (7–8) | |
| rWSN NS1-Y89F | 20 | 100 (5/5) | 4.9 (0.3–11.6) | 0 (0–2) | 0 (0) |
| 100 | 100 (5/5) | 6.8 (1.2–19.8) | 0 (0–3) | 0 (0) | |
| 500 | 100 (5/5) | 15.6 (8.5–19.4) | 3 (0–3) | 0 (0) | |
| 2,500 | 20 (1/5) | >25 (20.8–>25) | 9 (5–9) | 7 (1–7) | |
Fig 2.
Characterization of WT and NS1-Y89F virus pathogenicity in vivo. (A and B) Survival data (A) and mean body weights (B) for 6- to 8-week-old C57/BL6 mice intranasally infected with the rPR8 WT and rPR8 NS1-Y89F viruses (50 PFU/mouse, n = 5). Error bars in panel B represent SD. (C and D) Survival data (C) and mean body weights (D) for 6- to 8-week-old C57/BL6 mice intranasally infected with the rWSN WT and rWSN NS1-Y89F viruses (2,500 or 500 PFU/mouse, n = 5). Error bars in panel D represent SD.
To assess viral replication in vivo, mice were intranasally infected with 1,250 PFU of each virus, and lungs were excised on days 2 and 4 postinfection. Following homogenization and centrifugation (10,000 × g, 5 min, 4°C) steps, the supernatants were used to determine viral titers. As shown in Fig. 3A and B, the rPR8 NS1-Y89F virus replicated to titers ∼10-fold lower than those of the rPR8 WT at both day 2 and day 4, while no differences in viral titer were observed between rWSN WT and rWSN NS1-Y89F viruses. The NS segments of all four viruses were fully sequenced after RT-PCR of homogenate samples to ensure the absence of revertants.
Fig 3.
Replication of WT and NS1-Y89F viruses in vivo. Six- to eight-week-old C57/BL6 mice were infected intranasally with 1,250 PFU of each virus. Lung titers were determined on days 2 and 4 postinfection from 3 or 4 mice per group. Bars represent mean values. (A) rPR8 WT and rPR8 NS1-Y89F viruses. (B) rWSN WT and rWSN NS1-Y89F viruses. Results shown were obtained in a single experiment and are representative of those of two similar experiments.
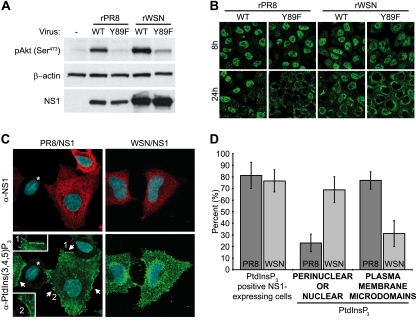
To confirm that the NS1-Y89F mutation in both the rPR8 and rWSN virus backgrounds is sufficient to abrogate activation of PI3K, individual A549 monolayers were infected at a multiplicity of infection (MOI) of 5 PFU/cell with each of the four viruses, and total cell lysates were harvested after 8 h. Following SDS-PAGE (4 to 12% NuPAGE gels; Invitrogen) and polypeptide transfer to polyvinylidene difluoride (PVDF) membranes, Western blotting was used to analyze the phosphorylation status of cellular Akt at serine 473 (mouse monoclonal antibody [MAb] 587F11; Cell Signaling Technology), a previously described marker for NS1-activated PI3K (8). As shown in Fig. 4A, infection with both rPR8 WT and rWSN WT viruses resulted in increased levels of pAkt compared to that in mock-infected cells. Furthermore, despite similar expression levels of NS1 between the WT and NS1-Y89F virus pairs, levels of pAkt in both NS1-Y89F virus-infected samples were comparable to that in the mock-infected sample, suggesting an inability of these mutants to activate PI3K signaling. This result correlates with previous data showing that both PR8/NS1 and WSN/NS1 proteins can bind p85β (8) and indicates that in both PR8 and WSN virus strains, the mutation of tyrosine 89 is sufficient to prevent this interaction.
Fig 4.
PI3K activation by influenza A virus NS1 proteins. (A) Phosphorylation of Akt during infection (or mock infection [-]) of A549 cells with rPR8 WT, rPR8 NS1-Y89F, rWSN WT, and rWSN NS1-Y89F viruses. Lysates were prepared 8 h postinfection (MOI, ∼5 PFU/cell) and subjected to SDS-PAGE followed by transfer to PVDF membranes. NS1 and phosphorylated Akt at serine 473 [pAkt (Ser473)] were detected using specific antibodies. β-Actin served as a loading control. (B) Indirect immunofluorescence of NS1 protein localization in A549 cells infected for the indicated times with rPR8 WT, rPR8 NS1-Y89F, rWSN WT, and rWSN NS1-Y89F viruses (MOI, ∼5 PFU/cell). Staining was performed using a polyclonal anti-NS1 serum (PAb 155). (C) Indirect immunofluorescence of HeLa cells cotransfected with plasmids encoding p85β/p110α and either PR8/NS1 or WSN/NS1. Costaining was performed using a monoclonal anti-PtdIns(3,4,5)P3 antibody (green) and a polyclonal anti-NS1 serum (PAb 155; red), together with 4′,6′-diamidino-2-phenylindole (DAPI) to stain nuclei (blue). Imaging and processing settings were kept identical for all fields to allow comparisons. Arrows indicate PtdIns(3,4,5)P3-enriched plasma membrane microdomains in PR8/NS1-transfected cells, which are not predominant in WSN/NS1-transfected cells. Magnified insets are numbered. An asterisk shows an untransfected cell with unstimulated levels of PtdIns(3,4,5)P3 and background levels of NS1 staining. Images are composite Z-stacks of multiple optical slices. α-, anti-. (D) Quantification of PtdIns(3,4,5)P3 staining in PR8/NS1- and WSN/NS1-transfected p85β/p110α-overexpressing HeLa cells (as in panel C). Bars represent the mean percentages of manually assigned individual cells from five randomly selected fields of view (∼20 cells per field; error bars represent SD).
It is intriguing that both PR8/NS1 and WSN/NS1 proteins are capable of binding PI3K and inducing Akt phosphorylation in a Y89-dependent manner, yet as viruses, the rPR8 and rWSN strains show differential sensitivities to abrogation of this signaling axis. We investigated possible phenotypic differences between PR8/NS1 and WSN/NS1 (which differ by only 7 amino acids) and observed that the intracellular localization of these two proteins was subtly distinct during infection (Fig. 4B). At 8 h postinfection, both NS1 proteins appeared predominantly nuclear, but at 24 h postinfection, PR8/NS1 was either mostly nuclear or cytoplasmic, and WSN/NS1 was mostly cytoplasmic with a concentrated perinuclear distribution. Notably, the Y89F mutation had little impact on the localization of either NS1 protein at 8 h postinfection, but it did exacerbate the cytoplasmic predominance of both PR8/NS1 and WSN/NS1 at 24 h postinfection (Fig. 4B). Thus, the Y89F mutation did not have a striking differential effect on the two virus strains that could correlate with the virus replication data.
We were intrigued by the subtle differences in WT PR8/NS1 and WT WSN/NS1 protein localizations and hypothesized that these could lead to differential spatial activation of PI3K. To characterize this, we established a cell-based assay to detect intracellular phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3], the membrane-bound lipid second-messenger product of PI3K that is responsible for recruiting Akt to membranes. Initial experiments using virus proved difficult to interpret, possibly because PI3K activation has multiple reported roles at different stages of the infection cycle (1, 5, 11, 14, 15). We also found that an enhanced signal-to-noise ratio could be achieved by providing catalytically active PI3K in trans. Thus, to characterize NS1-activated PI3K in the absence of other potentially confounding viral factors, HeLa cells were cotransfected for 16 h with plasmids encoding human PI3K (p85β/p110α subunits), as well as PR8/NS1 or WSN/NS1. Cells were then serum starved for 8 h, after which fixation and processing for indirect immunofluorescence was performed as described previously (13), except that permeabilization was done with 0.5% saponin in order to limit permeabilization of intracellular membranes. A monoclonal anti-PtdIns(3,4,5)P3 antibody (Echelon Biosciences) and a polyclonal anti-NS1 serum (polyclonal antibody [PAb] 155) were used as primary antibodies, and imaging was performed using a Zeiss LSM 510 Meta confocal microscope. In this system, PR8/NS1 and WSN/NS1 appeared to be expressed to similar levels, both were broadly detectable in the cytoplasm, and both induced production of PtdIns(3,4,5)P3 in a large proportion (∼80%) of transfected cells (Fig. 4C and D), findings in agreement with the observation that both NS1 proteins can interact with PI3K and promote Akt phosphorylation. However, using this assay, we observed a clear spatial difference in PtdIns(3,4,5)P3 accumulation between the two strains (Fig. 4C). PR8/NS1-induced PtdIns(3,4,5)P3 appeared to concentrate in distinct microdomains of the plasma membrane, possibly at the basal edges of the cell. In contrast, WSN/NS1-induced PtdIns(3,4,5)P3 was broadly distributed throughout the cell membrane, with an apparent concentration only where the membrane covered the perinuclear/nuclear area, something that was mostly absent for PR8/NS1. Quantification of PtdIns(3,4,5)P3 localization by scoring five randomly selected fields of view totaling >100 cells per condition confirmed these observations (Fig. 4D). Notably, we believe that the absence of both NS1 proteins from nuclei in this transfection-based assay is likely a consequence of permeabilization with saponin, which inefficiently permeabilizes nuclei (4) but is necessary to maintain membranes with PtdIns(3,4,5)P3 embedded.
Here, we used the mouse-adapted rPR8 and rWSN viruses to confirm the importance of tyrosine 89 for NS1-mediated PI3K activation and demonstrate a strain-specific contribution of NS1-activated PI3K to virus replication in tissue culture and in vivo: for rPR8, NS1-activated PI3K is important, while for rWSN, it is not. Our data, which suggest strain-specific differences in the abilities of the respective NS1 proteins to define intracellular sites of PI3K activation, provide a tempting correlation with the virus growth data, although further mapping and mutagenesis work is clearly necessary to confirm this. Nevertheless, we hypothesize that the biological function of NS1-activated PI3K during infection may not simply rely on total levels of PtdIns(3,4,5)P3 or phosphorylated Akt but may also depend on their correct intracellular redistribution by NS1 to certain signaling platform sites. It may therefore be that the particular PI3K phenotype activated by WT WSN/NS1 is not in itself beneficial to the virus, thus explaining why abrogation of such signaling in this viral strain does not significantly impact replication. Future studies in this area, including the identification and characterization of any NS1-specific signaling platform, may improve our understanding of both influenza virus biology and cellular PI3K signaling.
ACKNOWLEDGMENTS
We are grateful to Peter Palese and Elena Carnero (Mount Sinai School of Medicine [MSSM], New York, NY) for provision of reagents and Richard Cadagan and Osman Lizardo (MSSM) for excellent technical assistance.
This work was supported by NIH funding to A.G.-S. (Center for Research on Influenza Pathogenesis [CRIP], NIAID CEIRS contract HHSN266200700010C, and NIAID grant RO1AI046954). Confocal laser scanning microscopy was performed at the MSSM-Microscopy Shared Resource Facility, which is supported with funding from a National Institutes of Health-National Cancer Institute (NIH-NCI) shared resources grant (5R24 CA095823-04), an NSF Major Research Instrumentation grant (DBI-9724504), and an NIH shared instrumentation grant (1 S10 RR0 9145-01).
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Ehrhardt C, et al. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 8:1336–1348 [DOI] [PubMed] [Google Scholar]
- 2. Ehrhardt C, et al. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fodor E, et al. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679–9682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frisch S. 2004. Nuclear localization of FADD protein. Cell Death Differ. 11:1361–1362; author reply 1362–1364 [DOI] [PubMed] [Google Scholar]
- 5. Fujioka Y, et al. 2011. The Ras-PI3K signaling pathway is involved in clathrin-independent endocytosis and the internalization of influenza viruses. PLoS One 6:e16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gallacher M, et al. 2009. Cation currents in human airway epithelial cells induced by infection with influenza A virus. J. Physiol. 587:3159–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hale BG, Batty IH, Downes CP, Randall RE. 2008. Binding of influenza A virus NS1 protein to the inter-SH2 domain of p85 suggests a novel mechanism for phosphoinositide 3-kinase activation. J. Biol. Chem. 283:1372–1380 [DOI] [PubMed] [Google Scholar]
- 8. Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. 2006. Influenza A virus NS1 protein binds p85beta and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 103:14194–14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hale BG, et al. 2010. Structural insights into phosphoinositide 3-kinase activation by the influenza A virus NS1 protein. Proc. Natl. Acad. Sci. U. S. A. 107:1954–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 11. Hrincius ER, et al. 2011. Phosphatidylinositol-3-kinase (PI3K) is activated by influenza virus vRNA via the pathogen pattern receptor Rig-I to promote efficient type I interferon production. Cell. Microbiol. 13:1907–1919 [DOI] [PubMed] [Google Scholar]
- 12. Jackson D, Killip MJ, Galloway CS, Russell RJ, Randall RE. 2010. Loss of function of the influenza A virus NS1 protein promotes apoptosis but this is not due to a failure to activate phosphatidylinositol 3-kinase (PI3K). Virology 396:94–105 [DOI] [PubMed] [Google Scholar]
- 13. Kerry PS, et al. 2011. A transient homotypic interaction model for the influenza A virus NS1 protein effector domain. PLoS One 6:e17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu X, Masic A, Liu Q, Zhou Y. 2011. Regulation of influenza A virus induced CXCL-10 gene expression requires PI3K/Akt pathway and IRF3 transcription factor. Mol. Immunol. 48:1417–1423 [DOI] [PubMed] [Google Scholar]
- 15. Marjuki H, et al. 2011. Influenza A virus-induced early activation of ERK and PI3K mediates V-ATPase-dependent intracellular pH change required for fusion. Cell. Microbiol. 13:587–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shin YK, et al. 2007. SH3 binding motif 1 in influenza A virus NS1 protein is essential for PI3K/Akt signaling pathway activation. J. Virol. 81:12730–12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 88:13–18 [DOI] [PubMed] [Google Scholar]
- 18. Zhirnov OP, Klenk HD. 2007. Control of apoptosis in influenza virus-infected cells by up-regulation of Akt and p53 signaling. Apoptosis 12:1419–1432 [DOI] [PubMed] [Google Scholar]