Abstract
Despite overwhelming interest in the impact exerted by recombination during evolution of RNA viruses, the relative contribution of the polarity of inoculum templates remains poorly understood. Here, by agroinfiltrating Nicotiana benthamiana leaves, we show that brome mosaic virus (BMV) replicase is competent to initiate positive-strand [(+)-strand] synthesis on an ectopically expressed RNA3 negative strand [(−) strand] and faithfully complete the replication cycle. Consequently, we sought to examine the role of RNA polarity in BMV recombination by expressing a series of replication-defective mutants of BMV RNA3 in (+) or (−) polarity. Temporal analysis of progeny sequences revealed that the genetic makeup of the primary recombinant pool is determined by the polarity of the inoculum template. When the polarity of the inoculum template was (+), the recombinant pool that accumulated during early phases of replication was a mixture of nonhomologous recombinants. These are longer than the inoculum template length, and a nascent 3′ untranslated region (UTR) of wild-type (WT) RNA1 or RNA2 was added to the input mutant RNA3 3′ UTR due to end-to-end template switching by BMV replicase during (−)-strand synthesis. In contrast, when the polarity of the inoculum was (−), the progeny contained a pool of native-length homologous recombinants generated by template switching of BMV replicase with a nascent UTR from WT RNA1 or RNA2 during (+)-strand synthesis. Repair of a point mutation caused by polymerase error occurred only when the polarity of the inoculum template was (+). These results contribute to the explanation of the functional role of RNA polarity in recombination mediated by copy choice mechanisms.
INTRODUCTION
Among a wide range of events, genome replication is considered to be the fundamental aspect in the biology of positive-strand [(+)-strand] RNA viruses pathogenic to humans, animals, or plants. Production of copious amounts of infectious (+)-strand progeny occurs in two distinct phases that are preferentially modulated by virus-encoded RNA-dependent RNA polymerase (RdRp): initiation and synthesis of complementary negative-strand [(−)-strand] RNA using genomic (+)-strand RNA as a template, followed by the synthesis of progeny (+)-strand RNA using a (−)-strand RNA as the template (53). The accuracy with which the viral genomes are copied by viral RdRp plays an important role in establishing a successful infection in susceptible hosts (20). However, due to the lack of inherent proofreading activity during RNA synthesis by viral RdRp, polymerase errors result in the emergence of highly heterogeneous populations (quasispecies) held together in a dynamic equilibrium (20, 22, 56). In addition to polymerase error, RNA recombination has been shown to be one of the major driving forces toward genetic variation in RNA viruses (15, 20, 36, 37, 50, 56). Whether it is polymerase error or RNA recombination, natural selection is likely to maintain the most favorable consensus sequences for optimal replication of viral genomes (20). Ever since the first report of RNA recombination in poliovirus (38), it has been extensively studied in experimental and natural settings of animal viruses (37, 68), plant viruses (1, 2, 50, 56, 58, 60), and bacteriophages (18, 44). Literature is replete with documentation of different mechanisms (14, 15, 19, 31, 36, 39), sequences (46, 48), structures (19, 27, 45), host proteins and environmental factors (33), viral replicase (28, 29), RNA interference (RNAi) defense pathways (8, 26), and host transgenes (11) that regulate RNA recombination. However, despite these significant advances, the contribution of RNA strand polarity in recombination has not been discerned due to our inability to initiate in vivo replication on a (−)-strand RNA template except in a few cases (see Discussion).
Brome mosaic virus (BMV) is a (+)-strand, multicomponent RNA virus of plants, belonging to the family Bromoviridae (53). The genome of BMV is divided among three RNAs. Proteins encoded by monocistronic genomic RNA1 (B1) and RNA2 (B2) constitute the viral RdRp (53). Genomic RNA3 (B3) is dicistronic: the protein encoded by the 5′ cistron is the designated movement protein (MP), while the capsid protein (CP) encoded by the 3′ cistron is translated from a subgenomic RNA4 (sgB4) synthesized from the B3(−) progeny (43). RNA recombination in BMV was first observed more than 2 decades ago (13). Since then, it has been extensively used as a useful model for investigating various aspects associated with RNA recombination (15, 49, 51, 56–58, 66). Studies on RNA recombination in BMV involved mechanical inoculation of whole plants (49, 58, 66) with a mixture of (+)-strand RNA transcripts as the starting inoculum. Depending on the type of host used, progeny were analyzed at 7 to 15 days postinfection (dpi). Although these studies provided valuable information in the generation of recombinants, the following limitations are associated with the use of plants and (+)-strand RNA transcripts. First, establishment of BMV infection in whole plants requires the entry of all three components into the same cell, an essential prerequisite for multicomponent viruses such as BMV. Infection resulting from mechanical inoculation is synchronous but is less efficient. This inherent constraint often resulted in the inoculation of disproportionate doses of RNA transcripts (66) and imposed longer incubation periods of inoculated plants to maximize the yield of viral progeny RNA. In full-blown infections, occurring 7 to 10 dpi, postrecombination sequence rearrangements, high selection pressure, and packaging constraints dictated by the size of the viral capsid resulted in the survival and systemic movement of only those recombinants that were biologically fit and encapsidated (13, 58). Consequently, the genetic makeup of the primary recombinant pool generated during early phases of replication (e.g., 2 days) remained obscure. Second, as observed with other (+)-strand viruses (52), BMV replication is asymmetric, resulting in a (+)/(−)-strand ratio of 100:1 (30, 41). Despite the significance of (−)-strand RNA in replication, our inability to initiate replication from (−)-strand transcripts precluded the precise determination of the relative contribution of RNA polarity in recombination. These limitations can be circumvented by the application of Agrobacterium-mediated transient expression (agroinfiltration) (5). Agroinfiltration of viral components offers the following benefits: (i) synchronized delivery of multiple plasmids to the same cell (5); (ii) the steady-state supply of ectopically expressed RNA transcripts for up to 4 days (5, 7); (iii) unlike in mechanically inoculated plants, robust BMV replication can be initiated as early as 2 dpi (10); and (iv) BMV infection initiated by agroinfiltration recapitulates the scenario of mechanically inoculated plants (10). In addition, as demonstrated in this study using agroinfiltration (3, 5), we are able to initiate and complete replication on ectopically expressed B3 (−)-strand inoculum templates (Fig. 1B). Consequently, the major focus of this study is to exploit the agroinfiltration approach in dissecting and determining the relative contribution of RNA polarity in recombination.
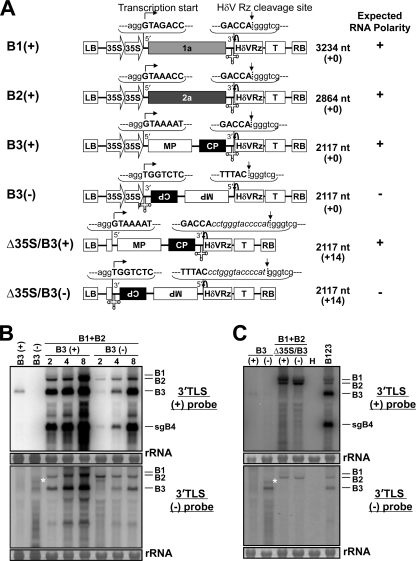
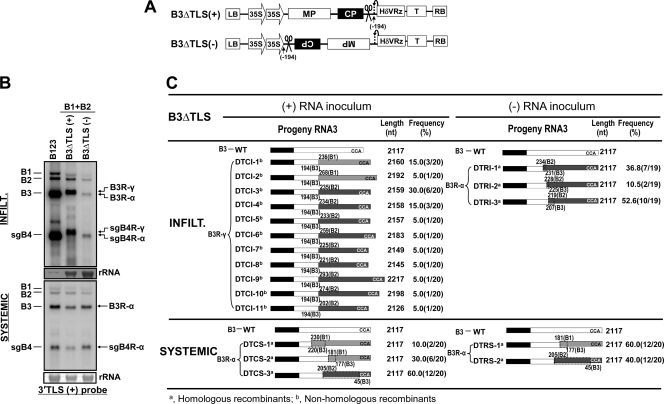
Fig 1.
Transfer DNA (T-DNA) constructs of BMV RNAs used for Agrobacterium-mediated transient expression in plants. (A) All cDNAs were subcloned into the binary vector pCassHδV. The vector contained a double cauliflower mosaic virus (CaMV) 35S promoter (35S), a ribozyme sequence derived from hepatitis delta virus (HδVRz), and a nos terminator (T). The agroconstructs B1(+), B2(+), and B3(+) contained full-length cDNA copies of BMV genomic RNAs 1 (B1), 2 (B2), and 3 (B3), respectively. In each agroconstruct, (+) and (−) represent the polarity of the ectopically produced transcripts. In agroconstruct Δ35S/B3, the double 35S promoter was deleted (see Materials and Methods). At the 5′ junction of (+) and the 3′ junction of (−) agroconstructs, the nucleotide sequences of nonviral and viral origin are shown in lowercase and uppercase, respectively. The bent arrows at the 5′ end of (+) and the 3′ end of (−) constructs represent transcription start sites. Cloverleaf-like structures at the 3′ end of (+) and the 5′ end of (−) constructs represent the tRNA-like structures. Arrows at the 3′ end of (+) and the 5′ end of (−) constructs indicate the predicted self-cleavage sites of the HδV Rz. The size in nucleotides (nt) of ectopically expressed RNA transcripts and their expected polarity are shown to the right of each construct. The numbers shown in parentheses are the numbers of nonviral RNA nucleotides left after self-cleavage by the ribozyme. (B) Progeny RNA analysis at various days postinfiltration (dpi). N. benthamiana leaves were infiltrated with either single or indicated mixtures of agroconstructs at an optical density at 600 nm (OD600) of 0.1. The total RNA recovered at 2, 4, and 8 dpi was analyzed on duplicate Northern blots and hybridized with 32P-labeled riboprobes of the indicated desired strand specificity. The positions of all four BMV RNAs are shown to the right of each panel. The strand specificity of riboprobes is exemplified by the detection of ectopically expressed B3(+) or B3(−) in N. benthamiana leaves infiltrated autonomously with agroconstructs of B3(+) or B3(−). rRNA was used as a loading control. (C) Ectopic expression of B3(+) or B3(−) agroconstructs without a 35S promoter (Δ35S). Following infiltration of the indicated agrocultures, total RNA was isolated at 4 dpi and analyzed on duplicate Northern blots using 32P-labeled riboprobes of indicated specificity. Total RNA recovered from leaves infiltrated with a mixture containing all three WT BMV agroconstructs (B123) served as a positive control, while that from healthy leaves (H) represents a negative control. Note that the slower-migrating band in B3(−), indicated with an asterisk in panels B and C, marks incompletely processed mRNA, perhaps due to the secondary structure of (−)-sense RNA transcripts that had rendered the self-cleavage activity of the HδV ribozyme inefficient.
MATERIALS AND METHODS
Construction of BMV plasmids for agroinfiltration.
Each of three full-length BMV genomic RNAs was amplified by PCR from 35S-B1, 35S-B2, and 35S-B3 (5). After treating with kinase, the resulting product was ligated into the binary vector pCassHδV, digested with StuI and NcoI, and blunt ended using mung bean nuclease. The B3 mutants, containing GUA, Δknob, or 5′Psk, were amplified by PCR using cDNA clones (21) with the same forward and reverse primers as those used for amplifying the wild-type B3 and were ligated into the binary vector pCassHδV as described above. To obtain B3ΔTLS, B3ΔUTR-297, and B3ΔUTR-250, RNA3 fragments were amplified by PCR with the same 5′ forward primer and reverse primers (TLS Rev [5′-ACAAGCTTTTAACCTTAGCCA-3′], 297 Rev [5′-CTACCTATAAACCGGGG-3′], and 250 Rev [5′-AGGGGCCTGTGACTTTA-3′], respectively) and were then ligated into the binary vector pCassHδV as described above. To generate (+)- and (−)-sense expressing vectors, we screened the orientation of inserts and obtained correct orientation of the inserts as B3(+), B3-GUA(+), B3-Δknob(+), B3-5′Psk(+), B3ΔTLS(+), B3ΔUTR-297(+), or B3ΔUTR-250(+) and reverse orientation of the inserts as B3(−), B3-GUA(−), B3-Δknob(−), B3-5′Psk(−), B3ΔTLS(−), B3ΔUTR-297(−), or B3ΔUTR-250(−). The sequences encompassing the BMV 1a and 2a genes were amplified by PCR using B1 and B2 as the template and forward primers 1a 5′Fw (5′-ATGTCAAGTTCTATCGATTTGCT-3′) and 2a 5′Fw (5′-ATGTCTTCGAAAACCTGGGA-3′) and reverse primers 1a 3′Rv-Spe-Stop (5′-GGACTAGTTACTTAACACAATTAAAGATCAAATC-3′, with the SpeI site underlined and the termination codon indicated in bold) and 2a 3′Rv-Xba-Stop (5′-GCTCTAGATTATCTCAGATCAGAGGGCTTA-3′, with the XbaI site, which can be useful for cloning to the SpeI site, underlined and the termination codon indicated in bold), respectively. The resulting PCR products were digested with SpeI or XbaI and subcloned into StuI- and SpeI-digested pZP vectors, respectively. The resulting final plasmids were referred to as pZP-1a and pZP-2a, respectively.
Construction of Δ35S plasmid for agroinfiltration.
To test whether there is cryptic promoter activity in our agroinfiltration system, we constructed a pCassHδV vector lacking a 35S promoter. To delete the double 35S promoter in the pCassHδV vector, we amplified the upstream sequence of the 35S promoter on pCassHδV and engineered a StuI site at the 3′ end of the amplified product for cloning. Briefly, the vector sequence was amplified by PCR using a forward primer (5′-TACGCCAAGCTTGCATGCCT-3′, with the HindIII site underlined), which corresponds to nucleotides (nt) 177 to 197 of the pCassHδV vector, and a reverse primer (5′-CCTAGATAGCTGGGCAATGGA-3′, with the CCT nucleotides which can be used as a StuI site after cloning underlined), which is complementary to nt 323 to 340 of pCassHδV. The resulting PCR product was digested with HindIII and ligated into the HindIII- and StuI-digested pCassHδV vector, yielding the Δ35S-pCassHδV vector. Then the BMV RNA3 was inserted into the StuI-digested Δ35S-pCassHδV vector.
Agroinfiltration, in vitro transcription, and inoculation of Chenopodium quinoa plants.
For agroinfiltration, following transformation of binary vectors into Agrobacterium sp. strain GV3101, infiltration into Nicotiana benthamiana leaves was performed as described previously (5). Capped BMV RNA transcripts were synthesized in vitro using T7 RNA polymerase (24) and mechanically inoculated into C. quinoa plants (55).
Progeny RNA analysis.
For analysis of the progeny RNA, total RNAs from leaves of either mechanically inoculated C. quinoa or agroinfiltrated N. benthamiana plants at various dpi were extracted using TRIzol reagent (Invitrogen), and the RNA pellet was resuspended in RNase-free water. Virions were purified from agroinfiltrated leaves as described previously (6). For Northern blot analysis, samples of virion RNA (0.2 μg) or total RNA (10 μg) were electrophoresed in 1.2% agarose-formaldehyde gels (5). After transfer to a nylon membrane, the blot was processed for prehybridization and hybridization using 32P-riboprobe corresponding to a 3′ conserved region.
Cloning and analysis of recombinants.
For analysis of recombinants, total RNAs were extracted from either C. quinoa leaves or agroinfiltrated leaves at 4 dpi or uninoculated systemic leaves at 15 dpi and subjected to electrophoresis on 1% agarose gel. After being stained with ethidium bromide, the region of gel corresponding to B3-α, -β, or -γ that compared to the size of the viral RNA as a marker was excised and the progeny RNA was extracted by a Qiagen gel extraction kit (Qiagen, Inc.). cDNA of progeny RNAs was produced using M-MLV reverse transcriptase (RT) (New England BioLabs) with a reverse primer (5′-TGGTCTCTTTTAGAGATTTAC-3′, complementary to nt 2097 to 2117 of RNA3). The 3′ end sequence of the progeny RNA3 was amplified by PCR using Crimson Taq DNA polymerase (New England BioLabs), a forward primer (5′-GTTGTACATCTAGAAGTTGAGCAC-3′, nt 1752 to 1775 of RNA3), and the same reverse primer used for RT. The PCR products were purified with a Qiagen gel extraction kit and ligated into pGEM-T Easy cloning vector (Promega). Crossover sites of recombinants were determined by sequencing.
RESULTS
Biological activity of ectopically expressed B3(−)-strand templates.
Agroinfiltration is a robust system for the production of steady-state levels of ectopic expression of viral RNAs to detectable levels in the absence of sustained replication (5, 32). To verify whether ectopically expressed (−)-strand RNA transcripts would accumulate to detectable levels and subsequently serve as templates for viral replicase to initiate (+)-strand synthesis, an agrotransformant of B3, referred to as B3(−), was constructed (Fig. 1A). Northern blot analysis of total RNA from N. benthamiana leaves infiltrated autonomously with the B3(−) agrotransformant resulted in the accumulation of detectable levels of (−)-strand RNA transcripts of the expected size (Fig. 1B, bottom panel). In addition, unlike homogeneous populations of native-length B3(+), synthesis of a slower-migrating population of B3(−) was also occasionally observed (indicated by an asterisk in Fig. 1B, bottom panel). We attribute the production of this slower-migrating B3(−) strand to inefficient self-cleavage activity of the HδV ribozyme resulting from the secondary structure of (−)-strand RNA. As observed previously (5), this assumption was confirmed when the ribozyme was deleted and infiltrated into N. benthamiana leaves (data not shown). To evaluate the biological activity of ectopically expressed transcripts of B3(−), Agrobacterium cultures transformed with B1(+) and B2(+) were mixed with B3(−) and infiltrated into N. benthamiana leaves. Plants infiltrated with B1(+), B2(+), and B3(+) served as controls. Total RNA isolated at various days postinfiltration (dpi) was analyzed by Northern blot hybridization. Results are shown in Fig. 1B. At 2 dpi, efficient replication and accumulation of progeny B3 and sgB4 were evident in control plants, and their accumulation increased with time (Fig. 1B, top panel). In contrast, in samples infiltrated with B3(−), at 2 dpi, the detection of the (+)-strand progeny for each BMV RNA was weak (Fig. 1B, top panel). However, from 4 dpi onward, efficient accumulation of all four BMV RNAs was evident (Fig. 1B, top panel). Despite this delayed onset of replication initiated with B3(−), the (+)/(−)-strand ratio was indistinguishable from that of control wild-type (WT) infections (data not shown). Possible reasons for this delayed initiation of (+)-strand synthesis on ectopically expressed B3(−) are considered in the Discussion. Taken together, the results indicate that ectopically expressed B3(−) is biologically active and that it served as a functional template for BMV RdRp to initiate and faithfully complete replication.
To further verify that initiation of (+)-strand synthesis on ectopically expressed B3(−) is not due to inconspicuous synthesis of (+)-strand transcripts by the action of a cryptic promoter present in the B3(−) agrotransformant, a region encompassing the double 35S promoter was deleted from the B3(−) agroconstruct (Fig. 1A) [Δ35S/B3(−)]. As a control, the double 35S promoter region was also deleted from the B3(+) agroconstruct (Fig. 1A) [Δ35S/B3(+)]. Agrocultures of B1(+) and B2(+) were mixed with either Δ35S/B3(+) or Δ35S/B3(−) and infiltrated into N. benthamiana leaves, and progeny isolated at 4 dpi were subjected to Northern blot hybridization. No accumulation of B3 or sgB4 occurred in the absence of the 35S promoter (Fig. 1C). These observations unequivocally established that initiation of (+)-strand synthesis on ectopically expressed B3(−) strands by viral RdRp is authentic and is not due to inadvertent synthesis of B3(+) transcripts by a cryptic promoter.
Recombinants generated from B3(+)- and B3(−)-strand inoculum templates are distinct.
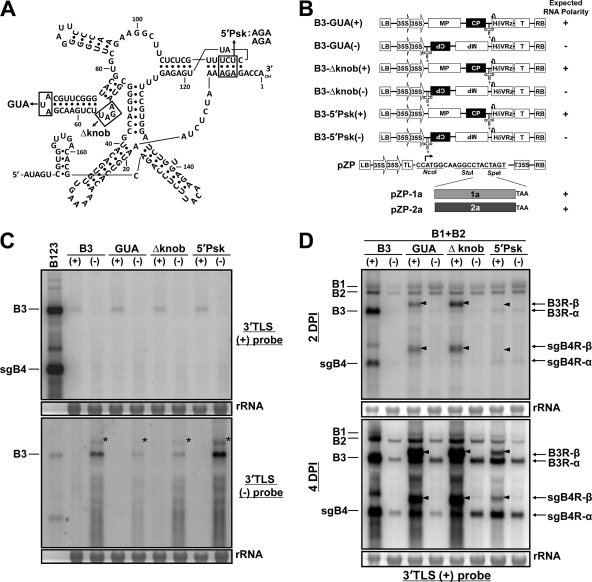
Previously, it has been shown that inoculation of Chenopodium hybridum plants with B3 mutants severely debilitated in (−)-strand synthesis are repaired by homologous recombination with the 3′ ends of either B1 or B2 (58). Since the mutant B3 inoculum used in that study was (+) in in vitro transcripts, it is unclear which RNA polarity was involved in recombination. Thus, to discriminate and evaluate the distinct contributions of (+)- and (−)-strand inoculum templates in RNA recombination, we opted to use the same set of B3 mutants that had previously been shown to severely affect (−)-strand synthesis in vitro (23, 24) and in vivo (58). These are B3-GUA, B3-Δknob, and B3-5′Psk (Fig. 2A), and the characteristic features of the (+) and (−) agrotransformants of each B3 mutant are shown in Fig. 2B. Autonomous expression of each B3 mutant (+) and (−) agrotransformant resulted in a steady-state level of RNA synthesis of the expected polarity (Fig. 2C).
Fig 2.
Characteristics of replication-deficient mutant sequences used in this study. (A) The 3′ 200-nt sequence with a tRNA-like structure (TLS) encompassing the minus-strand promoter of BMV RNA3 and engineered mutations (23) evaluated for recombination in this study. The nucleotides are numbered from the 3′ end. The locations of engineered mutations GUA (65AUA67→65GUA67), Δknob (deletion of 53AGAU56), and 5′Psk (113UCU115→113AGA115) are indicated by open boxes. (B) Agroconstructs of B3(+) or B3(−) harboring mutations GUA [B3-GUA(+)/B3-GUA(−)], ΔKnob [B3-Δknob(+)/B3-Δknob(−)], and 5′Psk [B3-5′Psk(+)/5′Psk(−)]. The expected polarity following agroinfiltration of each construct is shown to the right. Characteristics of each construct are identical to those of WT B3(+) or WT B3(−), shown in Fig. 1A. Schematic representation of the pZP binary vector used to subclone the open reading frames (ORFs) of BMV replicase proteins 1a and 2a. The basal pZP binary vector contains, in sequential order, a left border of T-DNA (LB), a double 35S promoter (35S), a multiple cloning site cassette, a 35S terminator (T35S), and a right border of T-DNA (RB). The bent arrow indicates the translation initiation site. cDNA copies corresponding to ORFs 1a and 2a were amplified by PCR and subcloned in frame into StuI and SpeI sites. (C) Autonomous ectopic expression of each mutant B3(+)- or B3(−)-strand polarity. The lane marked with B123 is a control marker lane, as described in the Fig. 1D legend. Following agroinfiltration, at 4 dpi, total RNA of 0.5 μg for the B123 and 10 μg for all other samples was analyzed on duplicate Northern blots, as described in the legend for Fig. 1B. The positions of B3 and sgB4 are shown to the left. The nature of a slower-migrating band (indicated by an asterisk) is as described in the Fig. 1B legend. (D) Progeny analysis of B3(+) or B3(−) mutants. N. benthamiana leaves were infiltrated with the indicated mixtures of agroconstructs. Total RNA preparations recovered from the infiltrated leaves at 2 dpi (the top panel) and 4 dpi (the bottom panel) were subjected to Northern blot hybridization with the indicated riboprobe. The positions of all BMV RNAs are shown to the left. The native-length recombinant progeny of B3 and sgB4 are labeled as B3R-α and sgB4R-α, respectively. Another set of slower-migrating nonhomologous recombinant B3 and sgB4 progeny are labeled B3R-β and sgB4R-β, respectively (indicated by arrowheads). rRNA was used as the loading control.
Agrobacterium cultures transformed with B1(+) and B2(+) were mixed with the mutant B3(+) or B3(−) agrotransformant and infiltrated into N. benthamiana leaves, and progeny RNA was analyzed at 2 and 4 dpi by Northern blot hybridization. A careful analysis of the electrophoretic mobility patterns of RNA progeny resulting from (+)-strand inocula revealed the accumulation of two distinct sets of B3 and sgB4 populations. The mobility phenotype of the first set, referred to as B3R-α and sgB4R-α (Fig. 2D), is indistinguishable from that of control WT B3 and its sgB4. At 2 dpi, the accumulation levels for B3R-α and sgB4R-α remained low due to the engineered mutation but increased to detectable levels by 4 dpi (Fig. 2D). A second set, referred to as B3R-β and sgB4R-β, exhibited a slower mobility phenotype than the WT control and accumulated to detectable levels at 2 and 4 dpi (Fig. 2D). In this case, the accumulation levels of B3R-β and sgB4R-β for mutants GUA and Δknob were higher than those for 5′Psk. It is important to note that the stronger hybridization signal seen for progeny B3R-β than for WT B3 (Fig. 2D) is not due to higher replication levels but to the binding of the 3′ probe to each of the two 3′ untranslated regions (UTRs) (see below). Since all three mutants are known to debilitate B3 replication in protoplasts (24), the replication profiles observed in this study for these mutants suggested that the engineered mutations have been restored, presumably by recombination. To verify this, cDNAs corresponding to B3R-α and B3R-β progeny of each mutant from the 4-dpi samples were amplified by RT-PCR, subcloned into pGEM-T vector, and subjected to sequencing.
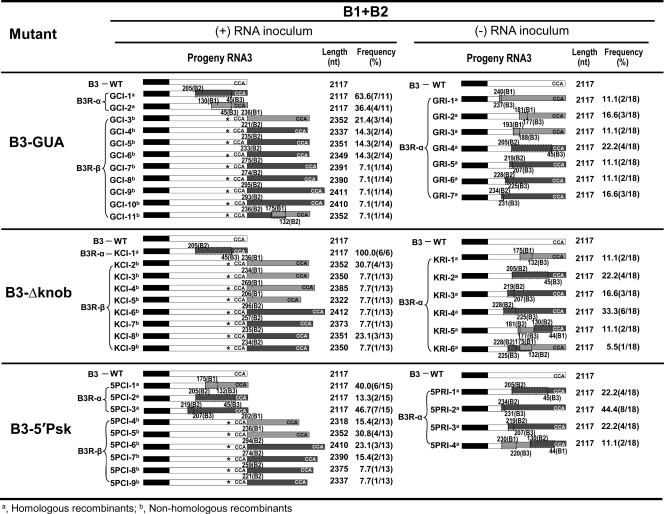
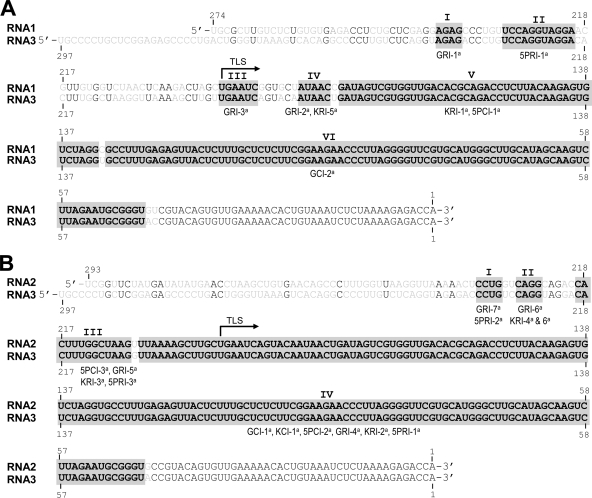
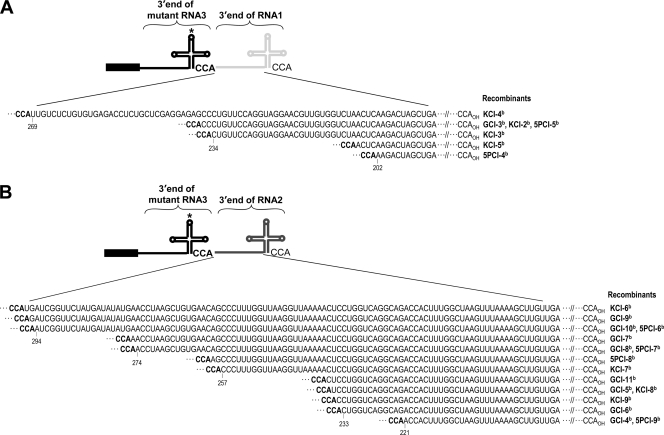
Based on the sequence analyses, as summarized in Fig. 3, progeny of B3R-α and B3R-β are classified as homologous and nonhomologous recombinants, respectively. Despite being identical in size to WT B3 (i.e., 2,117 nt), B3R-α is classified as recombinant since, in each case, the engineered mutation was replaced with that of the 3′ UTR from either B1 (Fig. 3) (36.4% and 40% for GUA and 5′Psk, respectively, and none for Δknob) or B2 (Fig. 3) (63.6%, 100%, and 60% for GUA, Δknob, and 5′Psk, respectively). The recombinant nature of B3R-α was confirmed by the presence of nucleotides diagnostic for the 3′ UTRs of B1 and B2 (the 3′ 200-nt region of B3 differs from that of B2 by a single A→G substitution at position 44 and from that of B1 at 10 additional positions) (Fig. 4). For example, recombinant GCI-2 (Fig. 3) was generated by recombination with the 3′ UTR of B1 due to the presence of U at position 43 and G at position 44 (Fig. 4). Since the GUA mutation involves changes at positions 65 to 67 (Fig. 2A), the absence of C at position 131 (the next diagnostic nucleotide for B1) suggested that template switching of RNA polymerase with the nascent strand of the B1 3′ UTR had occurred between nucleotides 68 and 130 (Fig. 4). Among B3R-α recombinants characterized, three clones, one derived from each mutant inoculum (Fig. 3) (GCI-1, KCI-1, and 5PCI-2), had identical sequences. The presence of the A→G substitution at position 44 and the absence of a single G→U substitution at position 206 (the next diagnostic nucleotide for B2) (Fig. 4) suggested that in each case, template switching of viral RdRp with the nascent WT B2 3′ UTR had occurred between nucleotides 45 and 205. Similar sequence comparisons allowed us to conclude that other B3R-α recombinants were generated by template switching of viral replicase with the 3′ UTR of B1 (e.g., GCI-2, 5PCI-1) or B2 (e.g., 5PCI-3) (Fig. 3).
Fig 3.
A summary of RNA recombinants generated from replication initiated on B3 mutants of (+)- and (−)-strand inocula. N. benthamiana plants were coinfiltrated with a mixture of agroconstructs containing B1(+) and B2(+) and each B3 mutant in either (+)- or (−)-strand orientation. Open boxes represent the 3′ untranslated region (UTR) of B3, and black boxes represent the coat protein ORF. Stippled and crosshatched boxes represent the 3′ UTRs of B1 and B2, respectively. An asterisk in the B3 3′ UTR represents the location of the engineered mutation. Numbers above each recombinant represent the 3′ nucleotides of B1 or B2 recombined with the 3′ end of B3. For example, in recombinant GCI-4, 221(B2) indicates that a 3′ 221-nt region from B2 was recombined with the 3′ end of B3. The dotted boxes in the columns labeled (+) RNA inoculum and (−) RNA inoculum show the overlapped and commonly shared homologous sequences between RNAs of B3 and B1 or B2. CCA denotes the last three nucleotides in the 3′ end of the BMV RNA. The length is the number of nucleotides (nt) in a given recombinant. The recombination frequency (%) was determined based on the total number of clones sequenced. B3R-α and B3R-β represent homologous and nonhomologous recombinants (see text for details). Naming of recombinants is as follows (for example). GCI, GUA mutant, correct orientation (i.e., plus strand), inoculated leaves; GRI GUA mutant, reverse orientation (i.e., minus strand), inoculated leaves; K, Δknob; P, 5′Psk. Recombinant progeny isolated at 4 dpi were amplified from total RNA using RT-PCR and sequenced after subcloning into pGEM-T vector (see Materials and Methods).
Fig 4.
Comparison of the entire 3′ UTR of B3 to those of B1 (A) and B2 (B). Nucleotides are numbered from the 3′ end. The diagnostic nucleotides (shown in gray font) used to distinguish the 3′ end of B3 (shown in black font) from those of B1 and B2 are shown. Roman numerals I to VI and I to IV indicate nucleotide domains of B1 and B2, respectively, that are involved in template switching of the BMV RdRp to generate recombinant sequences. Recombinant clones shown below each domain (e.g., GRI-1) correspond to those shown in Fig. 3. a, homologous recombinants; b, nonhomologous recombinants.
As schematically shown in Fig. 3 and 5, the genetic makeup of B3R-β progeny was clearly distinguishable from that of B3R-α by two characteristic features. (i) The primary characteristic feature was their distinct lengths. Progeny of B3R-β were longer than the mutant inoculum template (Fig. 3) (range between 2,337 and 2,411 nt for GUA, between 2,322 and 2,412 nt for Δknob, and between 2,318 and 2,410 nt for 5′Psk). (ii) A second unique characteristic feature is the duplication of the 3′ UTR: an internal 3′ UTR is the original inoculum sequence of the engineered mutation, while the terminal 3′ UTR of various lengths is derived from either WT B1 (202 to 269 nt) or WT B2 (221 to 296 nt) (Fig. 5). One pool of cDNA clones (e.g., GCI-11) (Fig. 3) generated from a B3-GUA (+)-strand mutant inoculum contained 3′ sequences from both B1 and B2. Based on these unique characteristic features, we conclude that progeny of B3R-β are nonhomologous recombinants generated by a copy choice mechanism for reasons given in the Discussion. The generation of B3R-α- and B3R-β-type recombinants was consistently reproduced in at least three additional independent experiments (data not shown).
Fig 5.
Junction sequences of B3R-β nonhomologous recombinants. The TLS of B3 is shown in bold, that of B1 (A) is in light gray, and that of B2 (B) is in dark gray. The junction sequence between the 3′ CCA of B3 (in bold font) and the various sizes of the recombined 3′ UTR of B1 or B2 is shown. Recombinant clones shown to the right correspond to those shown in Fig. 3.
Recovery of B3R-β-type recombinants from plants mechanically inoculated with in vitro transcripts.
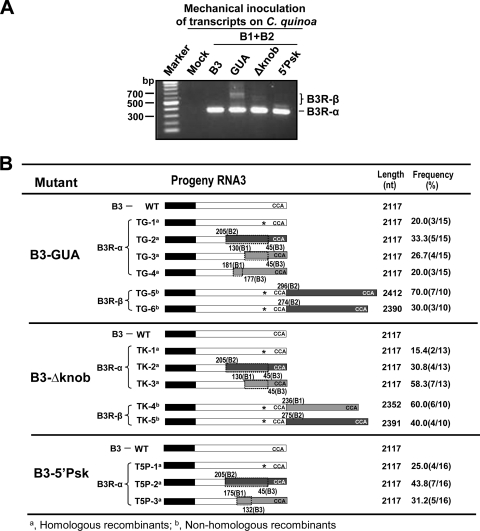
Although unlikely, a remote possibility exists that recombinants (specifically, B3R-β type) generated in agroinfiltrated leaves may have originated in the nucleus by RNA-based ligation between various ribozyme-cleaved 3′ ends. If this were to happen, recombinants of the B3R-β type would have also emerged from WT control infiltrations. But no such recombinants were seen in control infiltrations (Fig. 2D). However, as a precaution, to rule out that generation of B3R-β-type recombinants was an artifact of agroinfiltration, we opted to verify the products of recombination in plants mechanically inoculated with RNA transcripts. We preferred inoculating plants over protoplasts since detection of recombination is more difficult in protoplasts than in plants and often requires multiple passages due to the influence of natural selection (47). Furthermore, several RNA3 variants of BMV, including those examined in this study (12, 24), and cowpea chlorotic mottle virus (CCMV) (2) failed to recombine in protoplasts but they do so rapidly in whole plants (2, 13, 56, 58). Consequently, Chenopodium quinoa plants were mechanically inoculated with in vitro-synthesized, full-length RNA transcripts of WT B1 and B2 and each B3 mutant, and progeny from asymptomatic leaves were analyzed at 4 dpi for recombinants. Results shown in Fig. 6A and B clearly demonstrate that recombinants of the B3R-β type, similar to those generated in agroinfiltrated N. benthamiana plants, were successfully recovered from C. quinoa leaves inoculated with mutant GUA and Δknob, while inoculation with the mutant 5′Psk resulted in the recovery of only B3R-α-type recombinants (Fig. 6B). This failure to recover B3R-β type from leaves inoculated with mutant 5′Psk may be attributed to the inefficient synchronized infection resulting from mechanical inoculation. Taken together, these observations confirmed that B3R-β-type recombinants generated in agroinfiltrated N. benthamiana plants are authentic and are not the artifacts of agroinfiltration.
Fig 6.
Emergence of B3R-β-type recombinants in mechanically inoculated C. quinoa plants. (A) Agarose gel electrophoresis of RT-PCR products amplified from total RNA preparations of C. quinoa leaves mechanically inoculated with the indicated mixture of RNA transcripts. Conditions of RT-PCR are as described in the Fig. 3 legend. PCR products corresponding to B3R-α and B3R-β were excised and subjected to sequencing. (B). Summary and characteristic features of RNA recombinants generated in mechanically inoculated C. quinoa leaves as specified in panel A. Description of recombinants is similar to that in the Fig. 3 legend. Naming of the recombinants is as follows: TG-1, transcript, GUA mutant; TK-1, transcript, Δknob mutant; T5P, transcript, 5′Psk mutant.
Recombinants generated from B3(−)-strand inoculum templates are homologous in nature.
The newly proven ability of BMV RdRp to recognize the ectopically expressed B3(−)-strand template and initiate synthesis in vivo (Fig. 1B) has prompted us to find an answer to the following question: are recombinants generated using a (−) strand as an inoculum template different from those generated from (+)-strand inoculum templates? To address this question, Agrobacterium cultures transformed with B1(+) and B2(+) were mixed with either B3-5′Psk(−) or B3-Δknob(−) or B3-GUA(−) (Fig. 2B) and infiltrated into N. benthamiana leaves. Although replication initiated on ectopically expressed B3(−) strands is slower than that on B3(+) strands (Fig. 1B), to avoid variations due to sampling time, the progeny were analyzed at 2 and 4 dpi by Northern blot hybridization and subjected to sequencing. At 2 dpi, similar to control infiltrations performed with an agrotransformant of WT B3(−), replication of each B3 mutant was low and barely detectable (Fig. 2D, top panel). However, by 4 dpi, B3(+) and sgB4(+) progeny corresponding to each mutant accumulated to detectable levels (Fig. 2D, bottom panel). A most intriguing observation that emerged from these assays is that, in contrast to replication initiated on mutant B3(+)-strand inoculum templates that had resulted in the emergence of B3R-β recombinants, replication initiated on all three mutant B3(−)-strand inoculum templates had generated recombinant progeny identical in size to those of B3R-α recombinants (Fig. 2D). Longer exposure of the Northern blots failed to detect any slower-migrating progeny reminiscent of B3R-β recombinants (data not shown). Sequence analysis (Fig. 3) showing the presence of diagnostic nucleotides located in the 3′ UTRs of B1 and B2 revealed that in each case, the 3′ UTR of mutant B3 was restored to a functional sequence due to recombination with either B1 (38.8% for GUA involving 3′ nt 181 to 240, 11.1% for Δknob involving 3′ nt 175, and none for 5′Psk) or B2 (61.1% for GUA involving 3′ nt 205 to 234, 72.2% for Δknob involving 3′ nt 205 to 228, and 88.8% for 5′Psk involving nt 205 to 234). In addition, one pool of recombinants that emerged from B3-Δknob (KRI-5 and KRI-6) and B3-5′Psk (5PRI-4) (Fig. 3) was assembled from the 3′ UTRs of both B1 and B2. Based on the sequence composition and length, we conclude that the recombinants generated from each mutant B3(−) inoculum are homologous in nature, mediated by a copy choice mechanism. A collective compilation of homologous recombinant sequences assembled from the replication initiated on (+)- and (−)-strand inocula suggested that the generation of homologous recombinants with the 3′ UTR of WT B1 involved six domains while that of B2 involved four domains (Fig. 4).
Encapsidation competence, stability, and systemic movement of B3R-β recombinants.
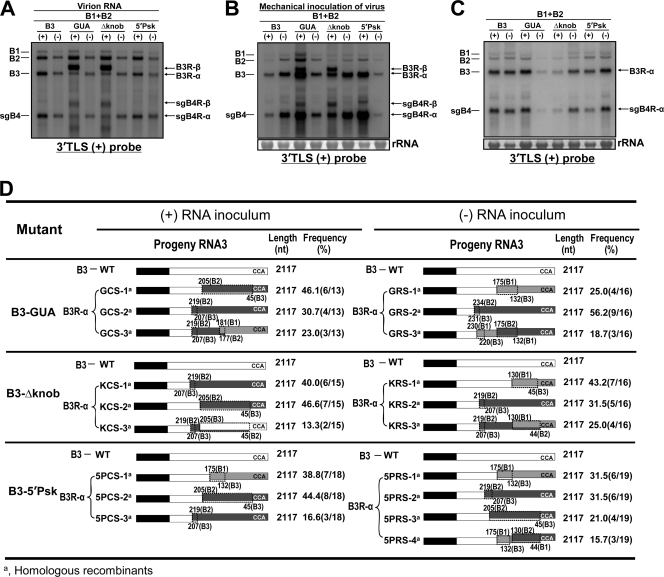
To verify the encapsidation competence and the ability to maintain the B3R-β recombinant nature during serial passage in N. benthamiana plants, the following experiments were performed. First, to verify the encapsidation phenotype, virions were purified at 4 dpi from leaves infiltrated with each (+)- or (−)-strand mutant inoculum and their physical morphology and nucleic acid composition were verified by electron microscopy (EM) and Northern blot hybridization, respectively. Results are shown in Fig. 7A. Irrespective of the polarity of the mutant inoculum, the morphology of virions recovered in each case was indistinguishable from that of control infiltrations (data not shown). Northern blot hybridization revealed that, in each case, the B3R-α/sgB4R-α recombinant progeny were encapsidated (Fig. 7A). Likewise, the B3R-β/sgB4R-β recombinant progeny were also encapsidated to detectable levels for mutants GUA and Δknob, while those corresponding to mutant 5′Psk were only weakly detectable (Fig. 7A). To examine the stability of these recombinant sequences and their competence for systemic movement, purified virions of each mutant were mechanically inoculated into N. benthamiana and the resulting progeny in inoculated (∼4 dpi) and uninoculated systemic (∼15 dpi) leaves were subjected to Northern blot hybridization followed by sequence analysis. Results are summarized in Fig. 7B and C. The data revealed that, although progeny of B3R-α and B3R-β recombinants (and their sgB4 counterparts) were accumulated to detectable levels in the inoculated leaves (Fig. 7B), the uninoculated systemic leaves contained only native-length progeny reminiscent of B3R-α recombinants (Fig. 7C). The sequence composition and the nature of the B3 recombinants recovered from uninoculated systemic leaves are summarized in Fig. 7D. The data suggested that in every case, the recombinant progeny had a single 3′ UTR from either B1 (e.g., 5PCS-1), B2 (e.g., KCS-1), or a mosaic of B1 and B2 (e.g., GCS-3) (Fig. 7D). Failure to detect slower-migrating B3 species reminiscent of B3R-β either in Northern blots exposed for prolonged periods of time or by PCR (data not shown) suggested that B3R-β recombinants were incompetent for systemic movement. A closer examination of the recombinant sequences revealed the following additional interesting features. First, a majority of the progeny derived from inoculating (+)-strand (85%) and (−)-strand (67%) inocula involved recombination with the 3′ UTR of B2 as a donor (Fig. 7D). Second, a selected recombinant pool was exactly identical in sequence to that present in the original inocula. For example, for the (+)-strand inoculum, compare sequences of recombinants GCS-1, KCS-2, and 5PCS-2 (Fig. 7D) to those of GCI-1, KCI-1, and 5PCI-2, respectively (Fig. 3); likewise, for the (−)-strand inoculum, compare sequences of recombinants GRS-2, KRS-2, and 5PRS-2 (Fig. 7D) to those of GRI-7, KRI-3, and 5PRI-3, respectively (Fig. 3). Recombinant sequences that are not found in the original inocula (e.g., GCS-3 or KCS-3) may have been derived from additional sequence rearrangement that occurred during subsequent replication of B3R-β recombinants. Possible reasons for this rearrangement of recombinant sequences are considered in the Discussion.
Fig 7.
Encapsidation profiles and stability of recombinants during passage in N. benthamiana. (A) Northern blot analysis of 200 ng of each virion RNA preparation recovered from leaves infiltrated with the indicated agrocultures. (B) Northern blot analysis of total RNA preparations recovered at 4 dpi from N. benthamiana leaves mechanically inoculated with virion RNA preparations shown in panel A. (C) Northern blot analysis of progeny RNA recovered from uninoculated systemic leaves of N. benthamiana at 15 dpi. Conditions of Northern blot hybridization with riboprobes are as described in the Fig. 1B legend. The positions of all BMV RNAs are shown to the left, and the recombination progeny of B3R-α and B3R-β are shown to the right. (D) Schematic representation of recombinants recovered from uninoculated systemic leaves. A representative example of the nomenclature used to describe each recombinant clone is as follows: GCS, GUA, correct orientation, systemic leaves. All other features of recombinant clones are as described in the legend of Fig. 3.
Repair of 3′ point mutations by polymerase error occurs with (+)-strand but not (−)-strand template inocula.
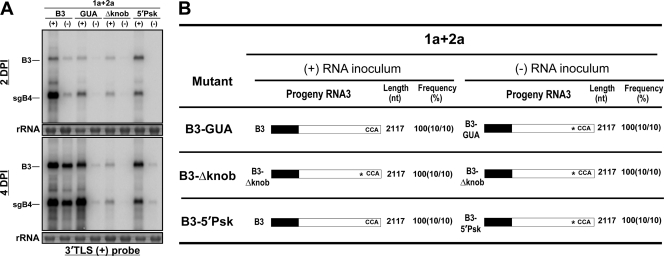
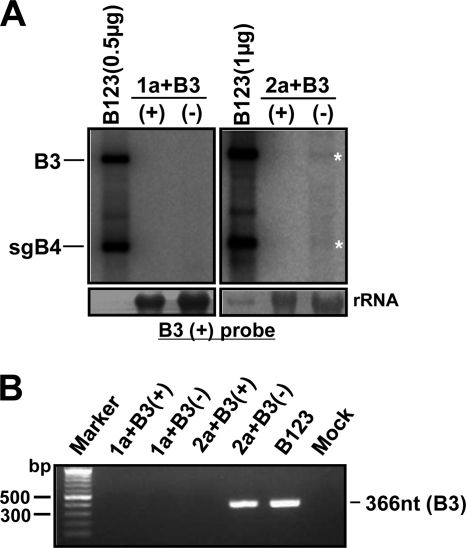
The data presented above clearly demonstrate that replication-defective B3 mutant templates of either (+) or (−) polarity are rapidly restored to replication-competent sequences due to recombination with the 3′ UTR from B1 and/or B2 by a copy choice mechanism. In addition to recombination, polymerase error has been implicated as a mechanism responsible for the repair of deleterious point mutations (21, 56). However, the role of polarity and the extent to which a given strand polarity influences polymerase error are not known. To investigate this, it is imperative to prevent recombination mediated by the 3′ UTR of biologically active WT B1 and WT B2. Therefore, the open reading frame (ORF) sequences encoding replicase proteins 1a and 2a were subcloned independently into a pZP binary vector (pZP-1a and pZP-2a) (Fig. 2B). Initially, to verify the biological activity of ectopically expressed 1a and 2a, Agrobacterium cultures transformed with pZP-1a and pZP-2a were coinfiltrated with either the WT B3(+) or WT B3(−) agroconstruct, and progeny at 2 and 4 dpi were analyzed by Northern blot hybridization. Results are summarized in Fig. 8. Failure to detect transcripts of B1 or B2 by the riboprobe complementary to the highly conserved 3′ end (Fig. 8A) confirmed that ectopically expressed mRNAs of 1a and 2a are devoid of the 3′ UTR. However, in leaves coinfiltrated with WT B3(+) or WT B3(−), their replicated progeny accumulated to detectable levels at 2 dpi and increased by 4 dpi (Fig. 8A). These results suggested that the ectopically expressed replicase proteins 1a and 2a are biologically active.
Fig 8.
Evidence for restoration of functional B3 sequences by polymerase error occurring on (+)- but not (−)-strand inoculum. (A) Northern blot analysis of progeny RNA isolated at 2 and 4 dpi from N. benthamiana leaves coinfiltrated with agrotransformants of pZP-1a and pZP-2a and the indicated mutant B3(+) or B3(−). Conditions of Northern blot hybridization with riboprobes are as described in the Fig. 1B legend. The positions of B3 and sgB4 are shown to the left. (B) Schematic representation of progeny of B3. Open boxes represent the 3′ UTR, and black boxes represent the CP ORF. An asterisk in the 3′ UTR represents the location of the engineered mutation. All other features of recombinant clones are as described in the legend of Fig. 3.
Analysis of progeny from leaves coinfiltrated with agroconstructs of 1a and 2a and each B3 variant in (+) or (−) polarity revealed a distinct RNA profile. At 2 dpi, replication initiated on each mutant B3(+) strand template resulted in the generation of detectable levels of sgB4. However, at 4 dpi, the accumulation of progeny B3(+) and its sgB4(+) for mutants GUA and 5′ Psk was increased (<5%), while that for mutant Δknob remained unchanged (Fig. 8A). In contrast, at 2 and 4 dpi, progeny accumulation resulting from replication initiated on all three mutant B3(−)-strand templates remained low (<2%) (Fig. 8A). Sequence analysis of B3 progeny recovered from plants infiltrated with mutant B3(+)-strand templates revealed that for mutants GUA and 5′Psk, the engineered mutation was restored to the WT, while that of Δknob remained unaltered (Fig. 8B). Absence of the diagnostic nucleotides characteristic for the 3′ UTR of B1 or B2 (Fig. 4) confirmed that restoration of the 3′ functional sequence in B3 progeny is not due to recombination. Therefore, we conclude that in the absence of a donor 3′ UTR from B1 or B2, the engineered point mutations (e.g., GUA and 5′Psk), but not the deletion (e.g., Δknob), are restored to the WT due to polymerase error. In contrast, sequence analysis of the B3 progeny recovered from plants infiltrated with agrotransformants of mutant B3(−)-strand templates revealed that in each case, the engineered mutation was conserved. Collectively, the data show that repair of deleterious point mutations by error-prone viral RdRp occurs on (+)-strand but not on (−)-strand template inocula.
Additional evidence showing that B3R-β and B3R-α recombinants emerge from (+)- and (−)-strand inoculum templates, respectively.
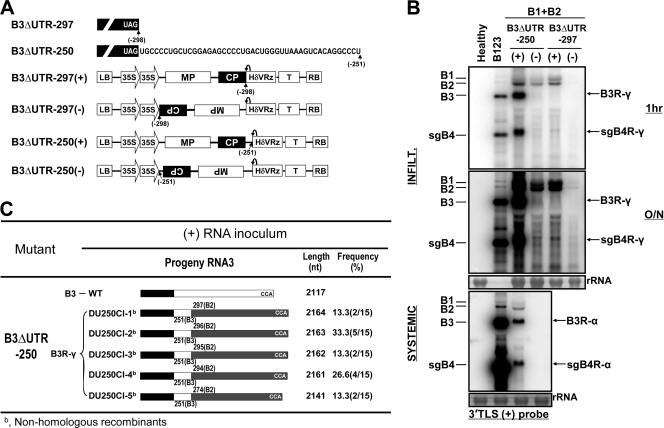
Based on the data presented above, we conclude that the recombinants that accumulate from mutant (+)-strand inocula are of B3R-α and B3R-β type (Fig. 3) whereas those generated from (−)-strand inocula are exclusively of B3R-α type (Fig. 3). However, a reverse scenario is also possible, since irrespective of the polarity of the RNA inoculum, replication of BMV RNA involves synthesis of both (+) and (−) strands. Therefore, to further clarify this issue, we performed the following two experiments. In the first experiment, a replication-defective B3 was constructed by deleting a 3′ 193-nt UTR encompassing the tRNA-like structure (TLS; the minus-strand promoter) and subcloning it into a binary vector amenable for ectopic expression of (+)- and (−)-strand RNA transcripts (B3ΔTLS) (Fig. 9A). Agrocultures of B3ΔTLS(+) or B3ΔTLS(−) were mixed with those of WT B1(+) and WT B2(+) and coinfiltrated into N. benthamiana leaves. Progeny were subjected to Northern blot hybridization and sequence analysis. Results are shown in Fig. 9B and C. Detection of progeny B3(+) and its sgB4(+), corresponding to B3ΔTLS(+) and B3ΔTLS(−), respectively, by a riboprobe complementary to a 3′ 200-nt sequence suggested that, in each case, the engineered deletion of the 3′ TLS had been restored (Fig. 9B). The electrophoretic mobility profiles suggested that the B3 recombinant progeny generated from B3ΔTLS(+) inoculum were longer in length than the inoculum template (range, 2,126 and 2,217 nt) (Fig. 9B and C). Based on the sequence composition of these recombinants, they are classified as nonhomologous recombinants and referred to as B3R-γ (Fig. 9B and C). B3R-γ recombinants are distinguished in sequence from B3R-β by having only one 3′UTR. However, similar to the generation of B3R-β-type recombinants, each B3R-γ recombinant retained the truncated 3′ end of the original inoculum, and either a 3′ UTR of 236 nt (DTCI-1 in Fig. 9C) or 268 nt (DTCI-2 in Fig. 9C) from B1 or a 3′ UTR of one of various lengths (202 to 293 nt) from B2 (Fig. 9C) was added. In contrast, as observed above (Fig. 3), recombinants generated from the B3ΔTLS(−) inoculum were of the same length as those generated from WT B3 (i.e., 2,117 nt, and hence referred to as B3R-α in Fig. 9B). Sequence analysis showed that in these recombinants, template switching of viral RdRp with a nascent B2 template in domain I (Fig. 4), encompassing nt 231 to 234 (indicated by a stippled box in Fig. 9C), had resulted in the generation of recombinant DTRI-1. Similarly, switching of the nascent B2 template in domain II (Fig. 4), encompassing nt 225 to 228 (indicated by a stippled box in Fig. 9C), and domain III, encompassing nucleotides 207 to 219, resulted in the generation of recombinant DTRI-2 and DTRI-3, respectively (Fig. 9C). Sequence analysis of B3 progeny recovered from systemic leaves of plants infiltrated with the B3ΔTLS(+) or B3ΔTLS(−) inoculum revealed that, as observed in the case of the above-described B3 mutants (Fig. 7C and D), all recombinants were of native WT B3 size (Fig. 9C). These data confirmed that a postrecombination process further rearranged sequences of B3R-γ recombinants to native-length molecules.
Fig 9.
Evidence for generation of nonhomologous recombinants exclusively from (+)-strand inoculum templates. (A) Features of B3ΔTLS agroconstructs designed to ectopically express (+)- or (−)-strand transcripts. Features of each construct are the same as those described in the legend of Fig. 1A. (B) Northern blot analysis of progeny RNA in infiltrated (top panel) and uninoculated systemic (bottom panel) leaves of N. benthamiana leaves infiltrated with the indicated mixture of agrocultures. Conditions of Northern blot hybridization with riboprobes are as described in the Fig. 1B legend. Approximately 10 μg of total RNA was subjected to Northern blot hybridization, and 1 μg of B123 was used as a marker for a positive control. The positions of the four WT BMV RNAs are shown to the left of each panel. Since nonhomologous recombinants generated from B3ΔTLS (+) strands are distinct in sequences from those shown in Fig. 3, they are referred to as B3R-γ. The positions of recombinant progeny generated from infiltrating B3ΔTLS (+) strands (B3R-γ and sgB4R-γ) and (−) strands (B3R-α and sgB4R-α) are shown to the right. (C) Profiles of recombinants generated during restoration of the deleted TLS region from B3. All features of recombinant clones are as described in the legend of Fig. 3.
The data shown above (Fig. 9) suggested that, despite the absence of the highly conserved (−)-strand promoter (i.e., TLS), the remaining sequence in the 3′ UTR (i.e., 3′ nt 194 to 297), despite having limited homology to corresponding B1 or B2 regions, functioned as a template for reinitiating RNA synthesis, generating B3R-β- and B3R-γ-type recombinants. To verify the minimal 3′ UTR sequence required to promote recombination, in the second experiment, either the 3′ 250-nt region (B3ΔUTR-250) (Fig. 10A) or a 3′ 297-nt region encompassing the entire 3′ UTR (B3ΔUTR-297) (Fig. 10A) was deleted and each deletion mutant was coexpressed as either a (+) or (−) strand along with WT B1(+) and WT B2(+). Northern blot hybridization (Fig. 10B) revealed that among four inocula, only the ectopically expressed templates of the B3ΔUTR-250(+) strand, and not others [i.e., B3ΔUTR-250(−), B3ΔUTR-297(+), B3ΔUTR-297(−)] (Fig. 10B), were able to replicate. Prolonged exposure of Northern blots confirmed that ectopically expressed templates of B3ΔUTR-250(−), B3ΔUTR-297(+), and B3ΔUTR-297(−) are deficient in replication and that no evidence for restoration of sequences by recombination was obtained. Once again, hybridization of the riboprobe specific for the 3′ 200-nt region to the progeny of the B3ΔUTR-250(+) strand suggested that the engineered deletion was restored by recombination. Furthermore, as observed above (Fig. 2D and 9B), the electrophoretic mobilities of B3 and its sgB4 progeny of B3ΔUTR-250(+) were slower than that of control samples (i.e., WT B3 and B4), suggesting that recombinant progeny were longer than the native B3 template. Sequence analysis (Fig. 10C) revealed the following. First, the recombinants are heterogeneous in size (between 2,411 nt and 2,164 nt) (Fig. 10C). Second, template switching of RdRp on the 3′ 250-nt deletion mutant of the (+)-strand inoculum had generated recombinants having a single 3′ UTR and are hence referred to as B3R-γ (Fig. 10C). Third, as observed in the case of the B3ΔTLS(+) inoculum, in each recombinant, viral RdRp with nascent strands of various lengths (274 to 297 nt) from the 3′ UTR of B2 was able to reinitiate synthesis at the truncated 3′ end of the original mutant inoculum template. Consistent with the above-mentioned results of progeny recovered from systemic leaves, sequences of B3R-γ recombinants generated from the B3ΔUTR-250(+) strand were rearranged to attain native-length molecules (Fig. 10B). Failure to restore functional sequences when the entire UTR was deleted [B3ΔUTR-297(+) or B3ΔUTR-297(−)] suggested that viral RdRp is unable to reinitiate synthesis in the absence of a 3′ UTR. Results from these two experiments clearly demonstrate that, despite the absence of the conserved 3′ TLS, viral RdRp can reinitiate synthesis of the UTR spanning the 3′ region of nt 251 to 297. Similar results were obtained when mutants lacking sequences between 3′ nt 251 and 291 were tested in planta (data not shown).
Fig 10.
Effect of B3 3′ UTR deletions on recombination. (A) Agroconstructs of B3 deletion mutants lacking either the entire 3′ UTR (B3ΔUTR-297) or 250 nt of the 3′ UTR (B3ΔUTR-250). Features of agroconstructs are the same as those described in the legend of Fig. 1A. (B) Northern blot analysis. Following agroinfiltration of N. benthamiana leaves with the indicated mixture of agroconstructs, approximately 10 μg of total RNA was recovered from either infiltrated leaves (the top and middle panels) at 4 dpi or uninoculated systemic leaves (the bottom panel) at 15 dpi and subjected to Northern blot hybridization as described in the legend of Fig. 1B. Total RNA preparations from healthy leaves (10 μg) and B123 (1 μg) were used as negative and positive controls, respectively. The positions of the four WT BMV RNAs and B3 recombinants are shown to the left and right, respectively, of each panel. (C) Schematic representation of the profiles of recombinants recovered from total RNA preparations. All features of recombinant clones are as described in the legend of Fig. 3.
DISCUSSION
In this study, we exploited the inherent advantages of an agroinfiltration method to specifically evaluate the relative contributions of (+)- and (−)-strand RNA inocula in recombination using BMV as a model system. Our study revealed two major phenomena that have not been observed before with respect to the generation of functional recombinants. First, the polarity of the inoculum RNA is the primary determinant of the type of recombinants to be generated, i.e., when viral RdRp reinitiates synthesis on a given replication-defective B3 mutant of (+)-strand polarity, nonhomologous recombinants longer than the inoculum template are generated. In contrast, reinitiation of synthesis from (−)-strand inoculum templates had generated homologous recombinants of native WT B3 size (Fig. 2, 3, and 9). Second, restoration of functional sequences due to repair of an engineered point mutation by polymerase error had occurred only when viral RdRp reinitiated synthesis on (+)-strand, and not (−)-strand, inoculum templates (Fig. 8). In addition, as discussed below, progeny analysis during early phases of replication (2 to 4 dpi) as well as in uninoculated systemic leaves (15 dpi) is imperative to get a better insight into the composition and genetic makeup of biologically active recombinants.
In vivo initiation of BMV replication on an ectopically expressed (−)-strand template.
In BMV and other (+)-strand RNA viruses, undoubtedly (−)-strand RNAs are the most efficient templates for (+)-strand synthesis, since each (−) strand serves as a template for 100-fold excess of (+) strands (41, 52). However, efforts to initiate BMV replication on (−)-strand templates either in vitro or in vivo have been unsuccessful (40, 42). Only in a few cases has either replication (9, 63, 64) or infectivity (67) been reported to occur from (−)-strand templates. Despite these few reports, for unknown reasons, no further advancement in using (−) strands as templates toward elucidation of viral RNA replication has been made. We provide the following explanations for unsuccessful attempts in previous studies to initiate replication on in vitro-synthesized (−)-strand templates. First, the lack of biological activity of (−)-strand templates may be attributed to extraneous nonviral nucleotides at the termini of (−) strands (42). Second, some inherent requirements of replication-derived RNA molecules may preclude replication, as in the case of poliovirus (59). Third, procedures used to deliver (−)-strand RNA templates, either by mechanical inoculation (as in the case of plant viruses) or by transfections, result in uncontrolled cellular partitioning and may either affect the stability (degraded by either RNases or the RNAi pathway) or prevent access to interactions with viral replicase. The precise reasons for successful initiation of replication by BMV replicase from (−)-strand templates delivered by agroinfiltration are currently obscure. It is likely that, compared to mechanical inoculation, agroinfiltration results in the accumulation of much higher concentrations of RNA transcripts of (−)-strand polarity. This would increase the chances for (−) strands to be “seen” and used by viral RdRp. Alternatively, synthesis of RNA transcripts of (−) polarity by the 35S promoter is an exclusively host-controlled process that directs RNA templates to subcellular compartments of the cytoplasm, providing access for viral RdRp to initiate replication. Regarding the delayed onset of replication with B3 (−)-strand templates (Fig. 1B), we provide the following explanation. BMV replication occurs in spherule-like invaginations of the endoplasmic reticulum (ER), induced by replicase protein 1a (61). Membrane fractionation experiments showed that although protein 1a is entirely membrane associated, protein 2a is present in two forms: one associated with 1a and the other with cytoplasm (17). Similar results were reported for a genetically related cucumber mosaic virus (CMV) (62). Interestingly, CMV 2a alone was shown to be responsible for the synthesis of (+) strands from ectopically expressed (−)-strand templates of RNA3 (62).
To verify whether similar activity exists for BMV 2a, leaves were infiltrated with agrocultures of 2a (Fig. 2B) along with either B3(−) or B3(+) and the progeny were subjected to Northern blot hybridization. Since B3(−) serves as the template for B3(+) and sgB4 (43), bands corresponding to full-length B3(+) and sgB4(+) were weakly detected in leaves infiltrated with agrocultures of 2a and B3(−) but not B3(+) (Fig. 11A). The identity of weakly detected bands was further confirmed by RT-PCR analysis (Fig. 11B). These results suggest that 2a alone is sufficient to initiate synthesis of (+) strands on (−)-strand templates. Similar observations were recently reported while examining the activity of BMV 1a and 2a in transiently transfected human cells (65). Keeping these results and the fact that viral RdRp recruits (+)-strand templates to spherules for replication in perspective, it is possible that synthesis of B3(+)-strand templates from ectopically expressed B3(−)-strand templates is initiated by the 2a protein associated with the cytoplasm prior to spherule recruitment. This process can likely delay the onset of replication from B3(−)-strand templates.
Fig 11.
Initiation of (+)-strand synthesis on a B3 (−)-strand template by ectopically expressed BMV replicase protein 2a. (A) Northern blot analysis of progeny RNA (10 μg) recovered from leaves infiltrated with the indicated mixture of agrotransformants. The blot was hybridized with a riboprobe complementary to the CP ORF detecting B3 and sgB4. The positions of B3 and sgB4 are shown to the left. The asterisks indicate B3(+) and sgB4(+) synthesized from B3(−) by 2a. (B) Agarose gel electrophoretic analysis of the RT-PCR products corresponding to the sample shown in panel A. Following reverse transcription, cDNA products are subjected to PCR as described in the Fig. 6A legend. The position of an expected 366-nt fragment is indicated.
Role of polarity of the inoculum RNA in sequence restoration by recombination and polymerase error.
Our study clearly showed that the polarity of the inoculum RNA determines the type of recombinants to be generated. Reinitiation of viral RdRp from (+)-strand inoculum templates had generated exclusively two groups of nonhomologous recombinants that were longer than the original inoculum template. The first group of recombinants are of B3R-β type, characterized by having two 3′ TLSs: an internal 3′ TLS with an engineered mutation and the terminal 3′ TLS from either WT B1 (e.g., GCI-3) (Fig. 3) or WT B2 (e.g., GCI-4). All these recombinants replicated and synthesized their respective sgB4 RNAs and accumulated to detectable levels (Fig. 2D). Since the 3′ TLS harboring either 5′Psk or Δknob or GUA is partially defective in (−)-strand synthesis (23), replication of these recombinants had occurred using the terminal WT 3′ TLS of either B1 or B2. Despite having the WT 3′ TLS, the replication of recombinants of 5′Psk was lower than that of either GUA or Δknob (Fig. 2D). This differential replication may perhaps be attributed to relative (−)-strand promoter strengths of each B3β-type recombinant. For example, the 3′ 200-nt regions of all three BMV RNAs are similar but not identical due to the conservation of certain nucleotide differences (Fig. 4) (B2 differs from B3 by a single base substitution, while B1 differs from B3 in 11 positions). These nucleotide differences are not only important in determining the relative strengths of the (−)-strand promoter of each BMV RNA (B3 TLS > B2 TLS > B1 TLS), but they play a critical role in the maintenance of overall replication of the BMV genome (25). The second group of nonhomologous recombinants of the B3R-γ type was generated when the highly conserved 3′ TLS or a portion of the 3′ UTR was deleted (Fig. 9 and 10). In contrast to the B3R-β type, B3R-γ-type nonhomologous recombinants are characterized by having only one 3′ TLS, derived from either WT B1 or WT B2.
In contrast to the reinitiation of synthesis on (+)-strand inoculum templates, reinitiation of synthesis on (−)-strand inoculum templates resulted in the generation of only homologous recombinants of native WT B3 size (e.g., GRI-1 or KRI-3 in Fig. 3 or DTRI-1 in Fig. 9C). This observation also suggests that homologous recombinants recovered from partially defective (+)-strand inoculum templates (e.g., GCI-1 and 5PCI-1 in Fig. 3) were actually generated from (−)-strand progeny resulting from a (+)-strand inoculum. This conjecture is further supported by the recovery of identical homologous recombinants from leaves infiltrated with each mutant of B3(+) and B3(−) (compare recombinant sequences of GCI-1 and GRI-4, KCI-1 and KRI-2, and 5PCI-2 and 5PRI-1 in Fig. 3). It is also significant to note that, irrespective of the polarity of the inoculum template, a majority of the recombinants had a 3′ TLS from B2. This may be due to the following two reasons. (i) In BMV, B3 is by far the most efficiently replicated and accumulated RNA component, followed by B2 and then B1 (B3 > B2 > B1) (54). Therefore, a majority of recombination events between the supplied mutant B3 template and WT B2 probably result from the abundant accumulation of nascent WT B2(−)-strands, since these represent the most-available donor templates for recombination. (ii) The sequence spanning the 3′ UTR of B2 is more similar to that of B3 than that of B1 (Fig. 4). Therefore, the 3′ UTR of B2 might participate more frequently in recombination than that of B1.
Another significant outcome of this study is the correction of lethal point mutations by polymerase error occurring on (+)-strand, but not (−)-strand, inoculum templates. Since a single (−) strand can serve as a template for ∼100 (+) strands (41), a correction mechanism during (−)-strand synthesis would be more advantageous than one during (+)-strand synthesis, which needs to undergo another round of (−)-strand synthesis followed by (+)-strand synthesis.
Mechanism regulating the generation of nonhomologous (B3R-β and B3R-γ) and homologous (B3R-α) recombinants.
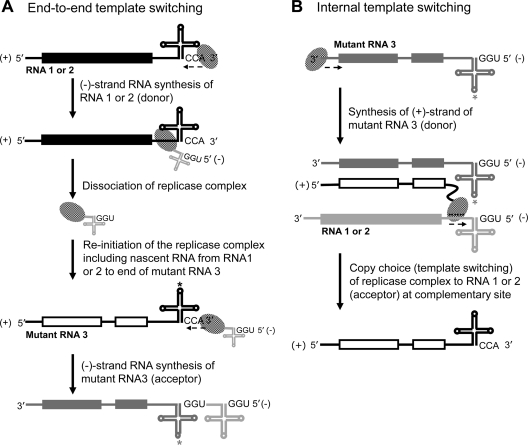
Consistent with previous reports on most RNA viruses (13, 36, 37, 48, 50), generation of recombinants in this study involved a copy choice mechanism. RNA recombination mediated by a copy choice mechanism is a widespread phenomenon reported to occur in RNA-containing viruses pathogenic to plants (46, 50), animals (37), and humans (36). Nagy and Simon (50) further classified the copy choice mechanism as similarity (in sequence) essential, similarity assisted, and similarity nonessential. In similarity essential and assisted mechanisms, the direction of the template switch is predominantly regulated by the template sequence and/or the structure of the acceptor and donor template molecules (35, 36, 48, 50, 56). Consequently, depending on the crossover sites, the resulting recombinants are either WT length (13, 56) or longer (13). In contrast, recombinants generated by a similarity nonessential mechanism involve complex RNA structures and either are nonfunctional or accumulate rarely in vivo due to selection pressure (27). In addition, evidence for the generation of products longer than the template length by end-to-end template switching is available from in vitro replicase assays for BMV (35), turnip crinkle virus (64), bovine viral diarrhea virus (35), hepatitis C (34), and poliovirus (69). Data shown in Fig. 2, 3, 9, and 10 clearly show that, as observed in vitro for BMV (35), end-to-end template switching can occur in vivo as well. As schematically shown in Fig. 12, recombination initiated using a (+)-strand inoculum involved the following two likely mechanisms. (i) Copy choice mediated by efficient end-to-end template switching during (−)-strand synthesis results in products longer than the template, as exemplified by recovery of B3R-β- and B3R-γ-type recombinants (Fig. 3 and 9). (ii) Since mutant (+)-strand inoculum templates are not completely defective in (−)-strand synthesis, the generation of B3R-α type recombinants may have mediated by using progeny (−) strands, as schematically shown in Fig. 12B. Whereas template switching during (+)-strand synthesis generates native-length homologous recombinants, exemplified by B3R-α type recombinants (Fig. 3), generation of B3R-β and B3R-γ recombinants occurs during the early phase of replication. For example, in a previous study, mechanical inoculation of whole plants with WT B1(+) and WT B2(+) and mutant B3(+) transcripts bearing the replication-defective mutations that were used in this study (i.e., GUA or Δknob or 5′Psk) (Fig. 2) had generated only homologous recombinants of WT length (56, 58). The question is why nonhomologous recombinants involving end-to-end template switching (i.e., B3R-β) were not recovered in previous studies using B3 harboring GUA or Δknob or 5′Psk mutations. We provide the following explanations.
Fig 12.
A copy choice model for the generation of B3R-β/B3R-γ- and B3R-α-type recombinants. (A) End-to-end template switching. According to this model, following minus-strand synthesis, viral RdRp with a nascent 3′ UTR from B1 or B2 switches templates and reinitiates (−)-strand synthesis at the very 3′ end of mutant B3, resulting in the generation of longer-than-template-size recombinants (i.e., B3R-β or B3R-γ type). (B) Internal template switching. In this model, following (+)-strand synthesis of mutant B3 from a (−)-strand template, viral RdRp with a nascent B3 (+)-strand switches templates and reinitiates (+)-strand synthesis on an internally located homologous region of the 3′ UTR of WT B1 or B2. This process generates exclusively native-template-length recombinants (i.e., B3R-α type).
Characterization of BMV RNA recombinants in planta involved the use of either local lesion hosts (e.g., C. quinoa or C. hybridum) or systemic hosts (barley or N. benthamiana). The onset of excisable discrete local lesions in C. quinoa or C. hybridum requires >7 days, while systemic infection in either barley or N. benthamiana requires >10 days. Furthermore, efficient cell-to-cell movement (required for local lesion development in C. quinoa or C. hybridum) and long-distance spread (in barley or N. benthamiana) requires assembly of infectious virions (55), a process which is functionally coupled to replication (4). To meet these functional constraints, recombinants recovered from either local lesions or systemically infected leaves must have endured a series of sequence rearrangements. As shown in this study, the primary pool of recombinants formed during the early phase of replication (e.g., 4 dpi) was a mixture of homologous and nonhomologous recombinants (Fig. 3). These results are in agreement with those obtained with turnip crinkle virus (16) and exemplify the importance of characterizing recombinants generated at various times postinfection, specifically those formed at the onset of replication (i.e., 2 to 4 dpi). For example, in every single instance when a B3(+)-strand mutant was coexpressed with WT B1(+) and WT B2(+), nonhomologous recombinants mediated by end-to-end template switching were consistently recovered (Fig. 2, 9, and 10). Subsequent host passage experiments (Fig. 7B and C) or recombinants recovered from uninoculated systemically infected leaves (Fig. 7C and D) revealed that nonhomologous recombinants that are longer than the inoculum template (e.g., B3R-β in Fig. 2 and 3 and B3R-γ in Fig. 9B and C) endured postrecombination events to generate WT-length homologous recombinants (Fig. 7D and 9C). This sequence rearrangement to WT length is perhaps required to attain optimal folding of the TLS for (−)-strand initiation and synthesis. Despite efficient encapsidation of nonhomologous recombinants (Fig. 7A), their inability to move to uninoculated systemic leaves may perhaps be due to a lack of optimal interaction between the host and the surface architecture of virions packaging nonhomologous recombinant RNAs. In conclusion, our results provide a new impetus for studying RNA recombination and, specifically, our ability to reinitiate (+)-strand synthesis from (−)-strand templates is likely to help in unraveling the precise role played by the (−)-strand template in recombination.
ACKNOWLEDGMENTS
We thank Deb Mathews and Jang K. Seo for critical readings of the manuscript.
Research in this laboratory was partly supported by a grant from the University of California, Riverside.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Aaziz R, Tepfer M. 1999. Recombination in RNA viruses and in virus-resistant transgenic plants. J. Gen. Virol. 80(Pt 6):1339–1346 [DOI] [PubMed] [Google Scholar]
- 2. Allison R, Thompson C, Ahlquist P. 1990. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc. Natl. Acad. Sci. U. S. A. 87:1820–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Annamalai P, Rao AL. 2006. Delivery and expression of functional viral RNA genomes in planta by agroinfiltration, p 2.1–2.15 In Downey T. (ed), Current protocols in microbiology, vol 16B John Wiley & Sons, New York, NY: [DOI] [PubMed] [Google Scholar]
- 4. Annamalai P, Rao AL. 2006. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 80:10096–10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Annamalai P, Rao AL. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96–111 [DOI] [PubMed] [Google Scholar]
- 6. Annamalai P, Rao AL. 2008. RNA encapsidation assay. Methods Mol. Biol. 451:251–264 [DOI] [PubMed] [Google Scholar]
- 7. Annamalai P, Rofail F, Demason DA, Rao AL. 2008. Replication-coupled packaging mechanism in positive-strand RNA viruses: synchronized coexpression of functional multigenome RNA components of an animal and a plant virus in Nicotiana benthamiana cells by agroinfiltration. J. Virol. 82:1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Asaoka R, Shimura H, Arai M, Masuta C. 2010. A progeny virus from a cucumovirus pseudorecombinant evolved to gain the ability to accumulate its RNA-silencing suppressor leading to systemic infection in tobacco. Mol. Plant Microbe Interact. 23:332–339 [DOI] [PubMed] [Google Scholar]
- 9. Ball LA. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. U. S. A. 91:12443–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bamunusinghe D, Seo JK, Rao AL. 2011. Subcellular localization and rearrangement of endoplasmic reticulum by brome mosaic virus capsid protein. J. Virol. 85:2953–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Borja M, Rubio T, Scholthof HB, Jackson AO. 1999. Restoration of wild-type virus by double recombination of tombusvirus mutants with a host transgene. Mol. Plant Microbe Interact. 12:153–162 [DOI] [PubMed] [Google Scholar]
- 12. Bujarski JJ, Ahlquist P, Hall TC, Dreher TW, Kaesberg P. 1986. Modulation of replication, aminoacylation and adenylation in vitro and infectivity in vivo of BMV RNAs containing deletions within the multifunctional 3′ end. EMBO J. 5:1769–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bujarski JJ, Kaesberg P. 1986. Genetic recombination between RNA components of a multipartite plant virus. Nature 321:528–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bujarski JJ, Nagy PD. 1994. Targeting of the site of nonhomologous genetic recombination in brome mosaic virus. Arch. Virol. Suppl. 9:231–238 [DOI] [PubMed] [Google Scholar]
- 15. Bujarski JJ, Nagy PD, Flasinski S. 1994. Molecular studies of genetic RNA-RNA recombination in brome mosaic virus. Adv. Virus Res. 43:275–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carpenter CD, Simon AE. 1996. Changes in locations of crossover sites over time in de novo generated RNA recombinants. Virology 223:165–173 [DOI] [PubMed] [Google Scholar]
- 17. Chen J, Ahlquist P. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chetverin AB. 1997. Recombination in bacteriophage Qb and its satellite RNAs: the in vivo and in vitro studies. Semin. Virol. 8:121–129 [Google Scholar]
- 19. Chetverin AB, Chetverina HV, Demidenko AA, Ugarov VI. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88:503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Domingo E, et al. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859–864 [DOI] [PubMed] [Google Scholar]
- 21. Domingo E, et al. 1985. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene 40:1–8 [DOI] [PubMed] [Google Scholar]
- 22. Drake JW, Holland JJ. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 96:13910–13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dreher TW, Hall TC. 1988. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J. Mol. Biol. 201:31–40 [DOI] [PubMed] [Google Scholar]
- 24. Dreher TW, Rao AL, Hall TC. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206:425–438 [DOI] [PubMed] [Google Scholar]
- 25. Duggal R, Rao AL, Hall TC. 1992. Unique nucleotide differences in the conserved 3′ termini of brome mosaic virus RNAs are maintained through their optimization of genome replication. Virology 187:261–270 [DOI] [PubMed] [Google Scholar]
- 26. Dzianott AM, Sztuba-Solinska JM, Bujarski JJ. 2012. Mutations in the antiviral RNAi defense pathway modify brome mosaic bromovirus RNA recombinant profiles. Mol. Plant Microbe Interact. 25:97–196 [DOI] [PubMed] [Google Scholar]
- 27. Figlerowicz M. 2000. Role of RNA structure in non-homologous recombination between genomic molecules of brome mosaic virus. Nucleic Acids Res. 28:1714–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Figlerowicz M, Nagy PD, Bujarski JJ. 1997. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl. Acad. Sci. U. S. A. 94:2073–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Figlerowicz M, Nagy PD, Tang N, Kao CC, Bujarski JJ. 1998. Mutations in the N terminus of the brome mosaic virus polymerase affect genetic RNA-RNA recombination. J. Virol. 72:9192–9200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. French R, Ahlquist P. 1987. Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J. Virol. 61:1457–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gmyl AP, Korshenko SA, Belousov EV, Khitrina EV, Agol VI. 2003. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA 9:1221–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gopinath K, Kao CC. 2007. Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 19:1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaag HM, Nagy PD. 2010. The combined effect of environmental and host factors on the emergence of viral RNA recombinants. PLoS Pathog. 6:e1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kao CC, et al. 2000. Template requirements for RNA synthesis by a recombinant hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 74:11121–11128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kim MJ, Kao C. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 98:4972–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kirkegaard K, Baltimore D. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lai MM. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:61–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ledinko N. 1963. Genetic recombination with poliovirus type 1. Studies of crosses between a normal horse serum-resistant mutant and several guanidine-resistant mutants of the same strain. Virology 20:107–119 [DOI] [PubMed] [Google Scholar]
- 39. Li Y, Ball LA. 1993. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J. Virol. 67:3854–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marsh LE, Dreher TW, Hall TC. 1988. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16:981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marsh LE, Huntley CC, Pogue GP, Connell JP, Hall TC. 1991. Regulation of (+):(−)-strand asymmetry in replication of brome mosaic virus RNA. Virology 182:76–83 [DOI] [PubMed] [Google Scholar]
- 42. Miller WA, Bujarski JJ, Dreher TW, Hall TC. 1986. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J. Mol. Biol. 187:537–546 [DOI] [PubMed] [Google Scholar]
- 43. Miller WA, Dreher TW, Hall TC. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68–70 [DOI] [PubMed] [Google Scholar]
- 44. Mindich L. 1996. Heterologous recombination in the segmented dsRNA genome of bacteriophage phi6. Semin. Virol. 7:389–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagy PD, Bujarski JJ. 1995. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 69:131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nagy PD, Bujarski JJ. 1997. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 71:3799–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nagy PD, Bujarski JJ. 1992. Genetic recombination in brome mosaic virus: effect of sequence and replication of RNA on accumulation of recombinants. J. Virol. 66:6824–6828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nagy PD, Bujarski JJ. 1996. Homologous RNA recombination in brome mosaic virus: AU-rich sequences decrease the accuracy of crossovers. J. Virol. 70:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nagy PD, Ogiela C, Bujarski JJ. 1999. Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots. Virology 254:92–104 [DOI] [PubMed] [Google Scholar]
- 50. Nagy PD, Simon AE. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1–9 [DOI] [PubMed] [Google Scholar]
- 51. Nagy PD, Zhang C, Simon AE. 1998. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 17:2392–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nassuth A, Bol JF. 1983. Altered balance of the synthesis of plus- and minus-strand RNAs induced by RNAs 1 and 2 of alfalfa mosaic virus in the absence of RNA 3. Virology 124:75–85 [DOI] [PubMed] [Google Scholar]
- 53. Noueiry AO, Ahlquist P. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77–98 [DOI] [PubMed] [Google Scholar]
- 54. Pogue GP, Marsh LE, Connell JP, Hall TC. 1992. Requirement for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology 188:742–753 [DOI] [PubMed] [Google Scholar]
- 55. Rao AL, Grantham GL. 1995. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 211:42–52 [DOI] [PubMed] [Google Scholar]
- 56. Rao AL, Hall TC. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67:969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rao AL, Hall TC. 1990. Requirement for a viral trans-acting factor encoded by brome mosaic virus RNA-2 provides strong selection in vivo for functional recombinants. J. Virol. 64:2437–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rao AL, Sullivan BP, Hall TC. 1990. Use of Chenopodium hybridum facilitates isolation of brome mosaic virus RNA recombinants. J. Gen. Virol. 71(Pt 6):1403–1407 [DOI] [PubMed] [Google Scholar]
- 59. Richards OC, Ehrenfeld E. 1990. Poliovirus RNA replication. Curr. Top. Microbiol. Immunol. 161:89–119 [DOI] [PubMed] [Google Scholar]
- 60. Roossinck MJ. 1997. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 35:191–209 [DOI] [PubMed] [Google Scholar]
- 61. Schwartz M, et al. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505–514 [DOI] [PubMed] [Google Scholar]
- 62. Seo JK, Kwon SJ, Choi HS, Kim KH. 2009. Evidence for alternate states of Cucumber mosaic virus replicase assembly in positive- and negative-strand RNA synthesis. Virology 383:248–260 [DOI] [PubMed] [Google Scholar]
- 63. Shaklee PN. 1990. Negative-strand RNA replication by Q beta and MS2 positive-strand RNA phage replicases. Virology 178:340–343 [DOI] [PubMed] [Google Scholar]
- 64. Song C, Simon AE. 1994. RNA-dependent RNA polymerase from plants infected with turnip crinkle virus can transcribe (+)- and (−)-strands of virus-associated RNAs. Proc. Natl. Acad. Sci. U. S. A. 91:8792–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Subba-Reddy C, et al. February 2012. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase in human cells reveals requirements for de novo initiation and protein-protein interaction. J. Virol. doi:10.1128/JVI.00069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sztuba-Solinska J, Dzianott A, Bujarski JJ. 2011. Recombination of 5′ subgenomic RNA3a with genomic RNA3 of brome mosaic bromovirus in vitro and in vivo. Virology 410:129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tousch D, Jacquemond M, Tepfer M. 1994. Replication of cucumber mosaic virus satellite RNA from negative-sense transcripts produced either in vitro or in transgenic plants. J. Gen. Virol. 75(Pt 5):1009–1014 [DOI] [PubMed] [Google Scholar]
- 68. Worobey M, Holmes EC. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80(Pt 10):2535–2543 [DOI] [PubMed] [Google Scholar]
- 69. Young DC, Tuschall DM, Flanegan JB. 1985. Poliovirus RNA-dependent RNA polymerase and host cell protein synthesize product RNA twice the size of poliovirion RNA in vitro. J. Virol. 54:256–264 [DOI] [PMC free article] [PubMed] [Google Scholar]