Abstract
The role of the multifunctional accessory Nef protein in the immunopathogenesis of HIV-2 infection is currently poorly understood. Here, we performed comprehensive functional analyses of 50 nef genes from 21 viremic (plasma viral load, >500 copies/ml) and 16 nonviremic (<500) HIV-2-infected individuals. On average, nef alleles from both groups were equally active in modulating CD4, TCR-CD3, CD28, MHC-I, and Ii cell surface expression and in enhancing virion infectivity. Thus, many HIV-2-infected individuals efficiently control the virus in spite of efficient Nef function. However, the potency of nef alleles in downmodulating TCR-CD3 and CD28 to suppress the activation and apoptosis of T cells correlated with high numbers of CD4+ T cells in viremic patients. No such correlations were observed in HIV-2-infected individuals with undetectable viral load. Further functional analyses showed that the Nef-mediated downmodulation of TCR-CD3 suppressed the induction of Fas, Fas-L, PD-1, and CTLA-4 cell surface expression as well as the secretion of gamma interferon (IFN-γ) by primary CD4+ T cells. Moreover, we identified a single naturally occurring amino acid variation (I132T) in the core domain of HIV-2 Nef that selectively disrupts its ability to downmodulate TCR-CD3 and results in functional properties highly reminiscent of HIV-1 Nef proteins. Taken together, our data suggest that the efficient Nef-mediated downmodulation of TCR-CD3 and CD28 help viremic HIV-2-infected individuals to maintain normal CD4+ T cell homeostasis by preventing T cell activation and by suppressing the induction of death receptors that may affect the functionality and survival of both virally infected and uninfected bystander cells.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) has spread around the world and is the main causative agent of AIDS. HIV-2, on the other hand, shows a poor capacity for transmission in humans and is largely confined to West Africa (10). Furthermore, HIV-2 is significantly less virulent than HIV-1 (22, 32, 59). Both HIV-1 and HIV-2 are most likely the result of relatively recent cross-species transmissions of simian immunodeficiency viruses (SIVs) from nonhuman primates to humans in west-central Africa (20, 48). HIV-1 originated from SIVcpz (SIV infecting chimpanzees) and (in rare cases) from SIVgor, which is found in gorillas (20, 48). In contrast, HIV-2 is the result of several independent zoonotic transmissions of SIVsmm, which infects sooty mangabeys (Cercocebus atys atys), which are found in Senegal and the Ivory Coast (15, 16). Notably, it has recently been shown that SIVcpz causes AIDS in its chimpanzee host (24). In contrast, SIVsmm replicates with high efficiency in naturally infected sooty mangabeys (SMs) without causing disease but can induce simian AIDS in experimentally infected rhesus macaques (RMs) (6).
The finding that SIVsmm is usually nonpathogenic in SMs, moderately pathogenic in humans, and highly pathogenic in RMs illustrates that host factors play an important role in the clinical outcome of infection. However, viral properties also play a role, since HIV-2 is clearly less virulent in humans than HIV-1 and the majority of HIV-2-infected people become long-term nonprogressors (32, 59). HIV-2 differs in several aspects, including a broader coreceptor tropism, the presence of a vpx gene, and the absence of a vpu gene from HIV-1 (6, 34, 38). Furthermore, HIV-1 and HIV-2 show fundamental differences in Nef function. Nef alleles from HIV-2 and most SIVs block the activation and programmed death of virally infected T cells by the downmodulation of T cell receptor (TCR)-CD3 (36, 44). In contrast, HIV-1 and its closest SIV counterparts are unable to remove CD3 from the cell surface and enhance rather than suppress the responsiveness of virally infected T cells to stimulation (44). Furthermore, HIV-2 and many SIV Nefs are also substantially more active than those of HIV-1 in downmodulating the cell surface expression of CD28, a key costimulatory factor of T cell activation (36, 44, 55). As a consequence, HIV-2 and most SIVs disrupt the interaction between virally infected CD4+ helper T cells and antigen-presenting cells (APCs), whereas HIV-1 and its SIVcpz precursor just deregulate it (2, 44). In contrast, other Nef activities, such as the downmodulation of CD4 and major histocompatibility complex class I (MHC-I), the upmodulation of the MHC-II-associated invariant chain (Ii), as well as the enhancement of viral infectivity and replication, are generally conserved (29).
The importance of the lack of Nef-mediated TCR-CD3 downmodulation for the aberrant immune activation and decline of CD4+ T cells, which are hallmarks of disease progression in HIV-infected individuals, is currently unclear. On the one hand, it has been shown that the inefficient downmodulation of TCR-CD3 by Nef is associated with significantly reduced CD4+ T cell counts in naturally infected SMs (45). Thus, Nef may prevent a loss of CD4+ T cells in natural SIVsmm infection by suppressing their activation-induced cell death. On the other hand, CD3 downmodulation by Nef does not prevent the progression to AIDS in some HIV-2-infected individuals (13). Notably, the characteristics of nonprogressive SIVsmm and HIV-2 infections are different. SIVsmm-infected SMs do not develop disease despite high levels of viral replication (12, 40, 49). In contrast, most long-term nonprogressors of HIV-2 infection show very low viral loads (VLs) and thus usually do not progress to AIDS because they efficiently control the virus (3, 47).
A previous study failed to detect a protective effect of Nef-mediated TCR-CD3 downmodulation against disease progression in HIV-2-infected individuals (13). However, many HIV-2 nef alleles were derived from nonprogressors with undetectable viral loads. Furthermore, just a few Nef functions were examined and only using expression vectors, not in virally infected primary human T cells. We hypothesized that specific Nef functions have a significant effect on the clinical and immunological outcome of infection if the virus replicates to detectable levels. To challenge this possibility, we examined the effects of nef alleles from 21 viremic and 16 nonviremic HIV-2-infected individuals on the surface expression of CD4, CD3, CD28, MHC-I, and Ii (CD74), as well as the activation and apoptosis of virally infected primary T cells. We found that the downmodulation of TCR-CD3 and CD28 correlates with the suppression of T cell activation and apoptosis, as well as with the reduced expression of death and immunosuppressive receptors in virally infected T cells in vitro and with the preservation of CD4+ T cells in viremic but not in nonviremic HIV-2-infected individuals in vivo. Thus, although the Nef-mediated suppression of T cell activation by the downmodulation of TCR-CD3 and CD28 ultimately fails to prevent disease progression in viremic HIV-2-infected individuals, it most likely decelerates the loss of CD4+ T cells. We also identified a naturally occurring I132T mutation in the core domain of HIV-2 Nef that selectively disrupts its ability to interact with the CD3ζ chain and to downmodulate TCR-CD3. This was unexpected, because all previous studies have failed to map the exact interaction site between Nef and the CD3ζ chain. Notably, this single-amino-acid substitution results in functional properties that are highly reminiscent of Nef proteins from highly pathogenic HIV-1 strains. Thus, HIV-2 could evolve into a more virulent form by losing this protective Nef function. However, such loss-of-function mutations are very rare, suggesting a strong selective pressure for maintaining the TCR-CD3 downmodulation function in vivo.
MATERIALS AND METHODS
Patients and nef alleles.
Nef genes were derived from 26 HIV-2-infected individuals from the Caió cohort in Guinea Bissau (60) and from biological HIV-2 clones isolated from 11 individuals from the Rotterdam cohort (4, 5). The characteristics of these patients and the determination of CD4+ T cell numbers and plasma viral loads have been described previously (4, 5, 60) and are summarized in Table S1 in the supplemental material. Viremic HIV-2-infected individuals were defined as having a plasma viral load of >500 copies per ml, and nonviremic subjects were defined as having viral loads of <500 copies per ml. The cloning and partial functional characterization of patient-representative HIV-2 nef alleles from the Guinea Bissau cohort have been described elsewhere (13). All HIV-2-infected individuals examined were seronegative for HIV-1. With the exception of three individuals (RH2-8, RH2-22, and RH2-26), who were infected with HIV-2 subtype B, all individuals were infected with subtype A. For four nonviremic and five viremic individuals, more than one nef allele was examined (see Table S1). All study participants provided informed consent and the studies were approved by the local ethical committees.
Proviral constructs.
The generation of HIV-1 (NL4-3-based) proviral constructs carrying functional or disrupted nef genes followed by an internal ribosome entry site (IRES) and the enhanced green fluorescent protein (eGFP) gene has been described (43, 44). Splice-overlap-extension PCR was used to replace the NL4-3 nef gene with the HIV-2 nef genes. Briefly, PCR fragments containing the 3′ end of the NL4-3 env gene fused to the various HIV-2 nef genes were cloned into pBR-NL43-IRES-eGFP-nef+ using the unique HpaI and MluI sites. The accuracy of all PCR-derived inserts was confirmed by sequence analysis.
Cell culture.
Jurkat and 293T cells were cultured as described previously (44). 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum. Peripheral blood mononuclear cells (PBMCs) from healthy human donors were isolated using lymphocyte separation medium (Biocoll separating solution; Biochrom), stimulated for 3 days with phytohemagglutinin (PHA) (2 μg/ml), and cultured in RPMI 1640 medium with 10% fetal calf serum (FCS) and 10 ng/ml interleukin-2 (IL-2) prior to infection.
Virus stocks and transductions.
To generate viral stocks, 293T cells were cotransfected with the proviral HIV-1 constructs alone (for assays on infectivity and replication) or with a plasmid (pHIT-G) expressing the vesicular stomatitis virus G protein (VSG-G) (44) to achieve comparable high initial infection levels for flow-cytometric analyses. The medium was changed after overnight incubation, and virus was harvested 24 h later. Residual cells in the supernatants were pelleted, and the supernatants were stored at −80°C. Virus stocks were quantified using a p24 antigen capture assay provided by the NIH AIDS Research and Reference Reagent Program.
Infectivity assays.
Virus infectivity was determined using TZM-bl and P4-CCR5 cells as described previously (35). Briefly, the cells were sown out in 96-well dishes in a volume of 100 μl and infected after overnight incubation with virus stocks containing 1 μg of p24 antigen produced by transiently transfected 293T cells. Two days postinfection, viral infectivity was detected using the Gal screen kit from TROPIX as recommended by the manufacturer. β-Galactosidase activities were quantified as relative light units per second (RLU/s) using an Orion microplate luminometer.
Flow-cytometric analysis.
CD4, TCR-CD3, MHC-I, CXCR4, CD28, and eGFP reporter expression in Jurkat T cells or human PBMC and CD74 expression on THP-1 cells transduced with HIV-1 (NL4-3) constructs coexpressing Nef and eGFP were measured as described previously (44). CD69 and IL-2R expression was measured by standard fluorescence-activated cell sorter (FACS) staining using CD25 (clone M-A251; BD Pharmingen) and CD69 (clone FN50; BD Pharmingen) monoclonal antibodies (MAbs). For the quantification of the Nef-mediated modulation of specific surface molecules, the levels of receptor expression (red fluorescence) were determined for cells expressing a specific range of eGFP. The extent of downmodulation (n-fold) was calculated by dividing the mean fluorescence intensity (MFI) obtained for cells infected with the nef-defective NL4-3 control viruses by the corresponding values obtained for cells infected with viruses coexpressing Nef and eGFP.
PBMC activation and apoptosis.
Human PBMCs were stimulated with PHA (1 μg/ml) for 3 days and subsequently infected with various HIV-1 eGFP/Nef constructs and cultured in RPMI 1640 (10% FCS, 10 ng/ml IL-2) for another 2 days. At this time PBMC expressed very low levels of activation markers and hence had a resting phenotype. This treatment mimics the repeated stimulation of CD4+ CCR5+ T cells by antigen-presenting cells (APCs). Thereafter, the PBMCs were treated a second time with PHA, and CD69 and IL-2R expression levels were measured by FACS analysis 1 and 4 days later. The frequency of virally infected apoptotic cells was determined using the annexin V apoptosis detection kit (BD Bioscience) as recommended by the manufacturer.
NF-AT induction.
Jurkat cells stably transfected with an NF-AT-dependent reporter gene vector (19) were transduced with HIV-1 Nef/eGFP constructs expressing various nef alleles or were left uninfected and subsequently treated with PHA (1 μg/ml; Murex). Luciferase activity was measured and n-fold induction determined by calculating the ratio between measured relative light units of treated samples and untreated samples as described previously (13, 44).
Statistical analysis.
The activities of nef alleles derived from HIV-2-infected individuals with low (n = 16) or high (n = 20) VLs were compared using a two-tailed Student's t test. The PRISM package, version 5.0 (Abacus Concepts, Berkeley, CA), was used for all calculations.
Phylogenetic analysis.
Phylogenetic trees of Nef nucleotide sequences were inferred by the PHYML v2.4.4 software program (19) using an HKY model with a BIONJ initial tree.
Co-IP assay.
To analyze the interaction of Nef with the CD3ζ chain, 293T cells were cotransfected with a CD3ζ expression vector and C-terminally hemagglutinin (HA)-tagged Nef variants in pCGCG expression constructs coexpressing eGFP. To ensure the equal input of CD3ζ and Nef, Western blotting was performed after 40 h. Coimmunoprecipitation (co-IP) was performed with the ProFound mammalian HA tag IP/co-IP kit essentially as described by the manufacturer's protocol. Briefly, cell lysates were incubated with immobilized anti-HA antibody overnight and washed three times, and HA-tagged Nef and interacting proteins were eluted with nonreducing sample buffer. The eluate was reduced by the addition of ß-mercaptoethanol and analyzed by Western blotting. CD3ζ was detected by an anti-CD3ζ antibody from Abcam (ab11281).
Confocal microscopy.
To analyze the interaction of various Nef alleles with CD8-CD3ζ, HEK293T cells (20,000 per well) were cultivated on poly-lysine-coated 8-well Ibidi slides (Ibidi GmbH). The next day the cells were transfected with 130 ng (each) plasmid DNA expressing CD8-CD3 ζ-GFP and C-terminally HA-tagged Nefs. At 16 h posttransfection, cells were fixed with 4% paraformaldehyde for 30 min and stained by indirect immunofluorescence. Briefly, cells were treated with mouse MAb (1:500 dilution) against HA tag (Abcam) in a buffer containing 0.25% fish skin gelatin (Sigma), 0.2% bovine serum albumin (BSA), and 0.2% saponin for 2 h. Subsequently, the cells were washed with phosphate-buffered saline (PBS), incubated with Alexa Flour 568-conjugated goat anti-mouse IgG Ab (1:500 dilution; Invitrogen-Life Technologies) for another 2 h, washed again, and stained by Hoechst 33342. Images were acquired on an LSM 710 confocal microscope (Carl Zeiss, Germany) with Zeiss Zen 2010 software, and further analysis and brightness/contrast were performed using FIJI software (http://fiji.sc/wiki/index.php/Fiji). The mean fluorescence intensity was calculated using a region of interest (ROI), which was drawn across an entire cell so that it always included the membrane as well as intracellular compartments.
Specific Nef mutants.
To generate the N118D mutation in RH2-1_A8, we used a splice-overlap-extension PCR method; the first half of the nef gene was amplified with primers 5′-HpaI (5′-GCTGTTAACTTGCTCAATGCCACAGCC) and 3′-N118D (3′-GTGACATATCTACTGCCAAT), and the second half of the nef gene was amplified with 5′-N118D (5′-ATTGGCAGTAGATATGTCAC) and 3′-R2-MluI (5′-GCACGCGTTAACTGAACGGTAT) primers. In the second round of PCR, the first and second halves of the PCR products were used as the template in equimolar ratios, and the intact gene was amplified with 5′-HpaI and 3′R2-MluI outer primers. T132I mutation also was generated in a similar fashion using 5′-HpaI and 3′_T132I (5′-CACTGTAATATATCCCATCCA) primers for the first half of the gene and 5′-T132I (5′-TGGATGGGATATATTACAGTG) and 3′R2-MluI primers for the second half of the gene. Subsequently, double mutations were generated in the gene with same method. In RH2-1_D8, 3′-D118N (5′-GTGACATATTTACTGCCAAT) and 5′-D118N (5′-ATTGG-CAGTAAATATGTCAC) internal primers for the generation of D118N point mutation and 3′-I132T (5′-CACTGTAATATGTCCCATCCA) and 5′-I132T (5′-TGGATGGGACATATTACAGTG) internal primers for I132T point mutation were used with 5′-HpaI and 3′-R2-MluI outer primers. These mutations were confirmed by sequence analysis. The nef variants were subcloned into pBR-NL4-3-IRES-eGFP proviral vector using HpaI and MluI sites.
Western blotting.
To monitor Nef expression, 293T cells were transfected with 3 μg of vector DNA coexpressing HA-tagged Nefs and eGFP. The next day cells were harvested and lysed in radioimmunoprecipitation assay buffer (1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 0.15 M NaCl, 50 mM Tris-HCl [pH 7.4], and 5 mM EDTA), and cell lysates were separated in 12% SDS-polyacrylamide (PAA) gels in a Tris-Tricine buffer system. After gel electrophoresis, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes and probed with HA (18181-100; Abcam) and β-actin (8227-50; Abcam). For a transfection control, another gel was run with similar conditions, and after blotting the membrane was incubated with antibody specific for GFP (290-50, Abcam). Subsequently, blots were probed with anti-mouse or anti-rabbit IRDye Odyssey antibodies (926-32210 and 926-32221) and proteins revealed using a LI-COR Odyssey scanner.
Death receptors and IFN-γ.
Human PBMCs were stimulated and infected with HIV-1 NL4-3 eGFP constructs with various Nefs as described above. After the second stimulation of PHA, the expression of death receptors and ligand were measured by FACS staining using the FAS (clone DX2; eBioscience), FAS-L (clone NOK-1; BioLegend), programmed death-1 (PD-1) (clone MIH4; eBioscience), and cytotoxic T-lymphocyte antigen 4 (CTLA-4) (clone 14D3; eBioscience) MAbs. The expression levels of receptors and the frequency of ligand-expressing cells were calculated by dividing the MFI and percentage of cells infected with virus coexpressing different Nefs and eGFP by the corresponding values from uninfected cells. The supernatants from different infected cell cultures were collected and further used to detect the gamma interferon (IFN-γ) secretion using the RayBio human IFN-γ enzyme-linked immunosorbent assay (ELISA) kit (RayBio). ELISA was performed as recommended by the manufacturer.
Ethics statement.
Written informed consent was provided by all study participants or their legal guardians. Ethical approval was obtained from the Gambian Government/MRC Ethics Committee, from the Republic of Guinea Bissau Ministry of Health, from the Oxford Tropical Research Ethics Committee, Oxford, United Kingdom, and from the Ethics Committee of the University Hospital Centre Rotterdam, Rotterdam, The Netherlands.
Nucleotide sequence accession numbers.
HIV-2 nef sequences determined in the course of this work have been deposited in GenBank under accession numbers JQ746624 through JQ746644.
RESULTS
Patient characteristics and generation of proviral constructs expressing HIV-2 nef alleles.
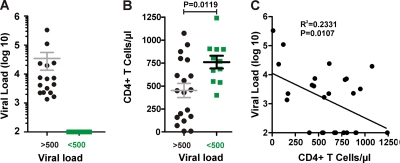
The nef alleles analyzed in the present study were derived from a total of 37 HIV-2-infected subjects; 26 of them (15 viremic, 11 nonviremic) were from the Guinea Bissau cohort (60) and 11 individuals (6 viremic, 5 nonviremic) were from the Rotterdam cohort (4, 5, 58). Some virological and immunological characteristics of these HIV-2-infected individuals have been described previously (4, 5, 13, 58, 60) and are summarized in Table S1 in the supplemental material. As shown in Fig. 1A, the viral loads (VLs) in the viremic group ranged from 3.13 to 5.52 (log10 copies/ml) with a mean of 3.87 ± 0.65. In the nonviremic group, the VLs were generally below the detection limit of about 100 viral RNA copies/ml. On average, the CD4+ T cell counts in the nonviremic individuals (760.5 ± 69.32; n = 11) was 1.6-fold higher than that in the viremic individuals (452 ± 77; n = 19) (Fig. 1B). The CD4+ T cell counts showed a significant but imperfect inverse correlation with the viral RNA loads (Fig. 1C). All but one of the nonviremic HIV-2-infected individuals had CD4 counts of >500/μl (see Table S1). By comparison, the CD4 counts in viremic subjects varied greatly from 10 to 1,075/μl, and 11 of 19 individuals had CD4 counts of <500/μl (Fig. 1B). Notably, the relatively old age of most individuals at the time of virus isolation (average age of 58.95 ± 16.1 years in the viremic group and 59.45 ± 17.0 years in the nonviremic group) most likely contributed to reduced levels of CD4+ T cells. In agreement with published data (12, 49), these findings suggest that a significant number of HIV-2-infected individuals do not develop immunodeficiency because they efficiently control the virus.
Fig 1.
Viral loads and CD4+ T cell counts of study subjects. Viral loads (A) and CD4+ T cell counts (B) in HIV-2-infected individuals at the time of PBMC or plasma sampling for nef analysis. Patient samples were grouped based on their VLs (above or below the detection limit of 500 viral RNA copies per ml plasma) and are color coded: black, viremic; green, nonviremic. The horizontal bars indicate average numbers per group. (C) Correlation between the VLs and the CD4+ T cell counts.
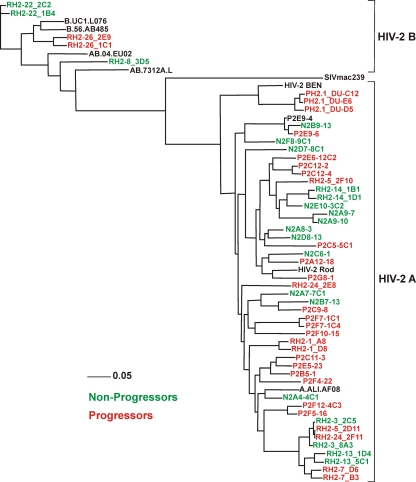
To study the role of Nef function in HIV-2 infection, we cloned a total of 50 nef alleles from the 16 nonviremic (<500 RNA copies/ml) and the 21 viremic (>500/ml) HIV-2-infected individuals into an HIV-1 NL4-3-based IRES-eGFP proviral vector coexpressing Nef and eGFP from bicistronic RNAs (43, 44). We used HIV-1-based constructs, because in HIV-2 nef overlaps env, which complicates meaningful analyses of Nef function. Phylogenetic analyses verified that the proviral constructs contained nef alleles from different HIV-2-infected individuals (Fig. 2). With the exception of three patients who were infected with group B viruses (RH2-8, RH2-22, and RH2-26), all individuals were infected with HIV-2 group A strains (Fig. 2; also see Table S1 in the supplemental material). The alignment of the deduced amino acid sequences showed that several domains previously described to be relevant for HIV-1 Nef function (17) were conserved in HIV-2 Nefs, including the N-terminal myristoylation signal, the acidic region, a diarginine motif, a C-proximal adaptor-protein interaction site, and a diacidic putative V1H binding site (see Fig. S1 in the supplemental material). By comparison, the P(xxP)3 motif, which is highly conserved in HIV-1 Nefs and interacts with cellular kinases, is not conserved in HIV-2 and SIVsmm Nef alleles, which usually contain only two proline residues at this location. Notably, all nef genes analyzed contained intact reading frames that predicted full-length Nef protein sequences.
Fig 2.
Evolutionary relationships among HIV-2 Nef sequences. A phylogenetic tree was constructed using the minimum evolution criterion as implemented in PHYML v2.4.4 (60). Bootstrap node support was calculated from 10,000 replicates.
Nef alleles from viremic and nonviremic HIV-2-infected individuals modulate cellular receptors.
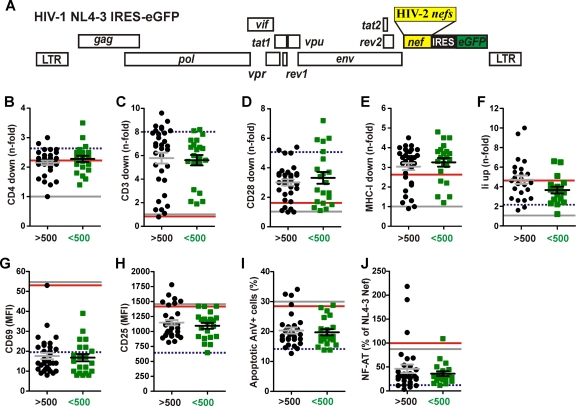
To determine the potency of HIV-2 nef alleles from individuals with detectable or undetectable VLs in downmodulating CD3, CD4, CD28, and MHC-I, we transduced human PBMCs with the proviral NL4-3-based IRES-eGFP constructs (Fig. 3A) and analyzed them by flow cytometry. HIV-1 virions pseudotyped with the VSV-G envelope protein were used to transduce the cells with comparable efficiency independently of their nef coding regions, and primary human cells were used to avoid in vitro artifacts. Our results showed that the vast majority of HIV-2 nef alleles from both viremic and nonviremic individuals modulated the various receptors analyzed, albeit with variable efficiency (Fig. 3B to E). No broadly reactive HIV-2 Nef-specific antibody is available, patient-derived sera varied substantially in their reactivity to the highly divergent HIV-2 Nef proteins, and tagging may alter their stability and functionality. Thus, the contribution of expression levels to differences in Nef function was not examined. However, the fact that the vast majority of nef alleles were functionally active implies that they are expressed. The detectable effect of Nef on CD4 (viremic, 2.5 ± 0.5; nonviremic, 2.6 ± 0.6; values give average n-fold downmodulation ± standard deviations [SD]) was relatively modest because the proviral HIV-1 constructs express Vpu and Env, which also reduce CD4 cell surface expression (30). Most HIV-2 nef alleles efficiently downmodulated TCR-CD3 (viremic, 6.1 ± 2.5; nonviremic, 5.8 ± 1.9) from the cell surface (Fig. 3C), whereas the effects on CD28 (viremic, 3.6 ± 1.4; nonviremic, 3.8 ± 2.0) and MHC-I (viremic, 3.0 ± 0.8; nonviremic, 3.1 ± 0.8) were more modest. Notably, for the subset of nef alleles that have been previously examined (13), we found a significant correlation (R2 = 0.35; P = 0.0006) between the published values on their efficiency of CD3 downmodulation in Jurkat T cells transfected with Nef expression constructs and in primary T cells infected with viral constructs expressing these nef alleles in the present study (data not shown). Several HIV-2 Nefs were fully active in downmodulating CD4, CD3, CD28, and MHC-I, although they contained deletions or mutations in domains proposed to be important for HIV-1 Nef function. By comparison, two HIV-2 Nefs (RH2-5 2D11 and RH2-26 2E9), which contain Y-to-C mutations in an otherwise highly conserved C-terminal Tyr residue that is critical for MHC-I downmodulation by the SIVmac239 Nef (37, 54), were inactive in modulating MHC-I (data not shown). Most notably, nef alleles from viremic and nonviremic HIV-2-infected individuals did not differ significantly in their ability to downmodulate CD4, CD3, CD28, and MHC-I.
Fig 3.
Functional characterization of HIV-2 nef alleles. (A) Schematic presentation of proviral HIV-1 IRES-eGFP constructs carrying HIV-2 nef alleles. (B to E) Quantitation of Nef-mediated downmodulation of CD4 (A), CD3 (C), CD28 (D), and MHC-I (E) on PBMCs infected with HIV-1 Nef/eGFP constructs. HIV-2 nef genes were grouped by VLs and are color coded as described for Fig. 1. Each symbol indicates the n-fold downmodulation of the indicated receptor molecule by one of the 45 different NL4-3 proviral recombinants expressing an HIV-2 nef allele. (F) Functional activity of HIV-2 nef alleles in up-modulating Ii (CD74) surface expression on THP-1 cells. (G to I) Expression of CD69 (G) and CD25 (H) and levels of apoptosis (I) in PBMCs infected with HIV-1 Nef/eGFP constructs. (J) Suppression of induction of NFAT in T-cell cultures infected with viral constructs expressing HIV-2 nef alleles. Values in panels B to K represent averages from triplicate experiments, and the horizontal bars indicate average activities per group. The results obtained for the HIV-1 Nef/eGFP control constructs are indicated by lines: red, NL4-3 nef; broken blue, HIV-2 BEN nef; and gray, disrupted nef gene. LTR, long terminal repeat.
Nef also may interfere with MHC-II antigen presentation by antigen-presenting cells by upregulating the cell surface expression of the invariant chain (Ii) (46, 52). To determine the effect of HIV-2 Nefs on Ii, we used the human monocytic leukemia THP-1 cell line, which shares many properties with monocyte-derived macrophages and expresses high levels of MHC-II (57). Our data showed that HIV-2 Nef proteins upmodulated Ii surface expression with efficiencies ranging from 1.2- to 10.0-fold (Fig. 3F). On average, nef alleles from viremic HIV-2-infected individuals were slightly more active (4.7 ± 0.4) than those from subjects with undetectable VLs (3.7 ± 0.4). However, this difference was mainly due to two unusually active nef alleles in the high VL (hVL) group and failed to reach significance (P = 0.08).
Most HIV-2 nef alleles suppress T cell activation and apoptosis.
It has previously been shown that HIV-2 Nefs prevent the responsiveness of virally infected T cells to activation (36, 44). To assess whether different viral loads in HIV-2-infected individuals are associated with differential responsiveness of infected T cells to stimulation, we determined the surface expression levels of CD69 and of the IL-2 receptor-α chain (IL-2Rα and CD25), well-known markers for early and late T cell activation, in HIV-1-infected, PHA-treated PBMC cultures. We found that all but 1 of the 50 HIV-2 nef alleles suppressed the induction of CD69 expression (Fig. 3G). Similarly, the great majority of HIV-2 nef alleles also inhibited the induction of IL-2R expression, NF-AT activation, and activation-induced cell death (Fig. 3H to J). Exceptions were a few nef alleles from viremic HIV-2-infected individuals. Altogether, however, nef alleles from viremic and nonviremic individuals did not differ significantly in their capability to suppress T-cell activation and apoptosis. In agreement with previous data (2, 36, 44), the suppression of CD69, IL-2Rα, and NF-AT expression and apoptosis correlated significantly with one another and with the efficiency of TCR-CD3 downmodulation (data not shown). Thus, nef alleles from most HIV-2-infected individuals suppress the responsiveness of virally infected T cells to the stimulation and induction of programmed cell death by the downmodulation of TCR-CD3 and CD28, the key ligand and costimulatory factor of T cell activation, irrespective of their viral loads.
nef alleles from viremic HIV-2-infected individuals are more effective in promoting viral spread in PBMCs.
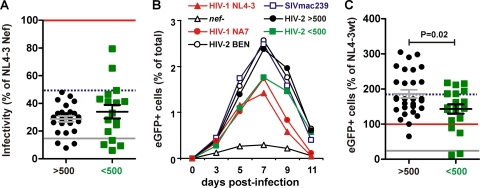
Nef facilitates viral spread not only indirectly by promoting viral immune evasion but also directly by enhancing virion infectivity and promoting viral replication in primary cells (7, 33, 51, 35). To examine whether the efficient replication of HIV-2 in vivo is associated with an increased capability of Nef to increase virion infectivity, we infected P4-CCR5 indicator cells with 293T cell-derived virus stocks normalized for their p24 content and determined the β-galactosidase activities 2 days later. Most HIV-2 nef alleles enhanced virion infectivity, albeit with various efficiencies and with lower potency than HIV-1 nef alleles (Fig. 4A and data not shown). On average, nef genes from viremic and nonviremic HIV-2-infected individuals promoted virion infectivity with similar efficiency (Fig. 4A).
Fig 4.
HIV-2 Nef-mediated enhancement of viral infectivity and spread in PBMCs. (A) Enhancement of viral infectivity by primary HIV-2 nef alleles. TZM-bl indicator cells were infected with HIV-1 NL4-3 IRES-eGFP constructs in triplicate with virus stocks containing 1 ng p24 antigen. Shown are average values for each HIV-2 nef allele compared to the infectivity of the virus expressing the NL4-3 Nef (100%). (B) Percentages of virally infected GFP+ cell levels detected in PBMCs derived from three donors after infection with proviral constructs expressing nef alleles from the indicated groups of HIV-2-infected individuals or control nef alleles. Shown are average values for the entire group of HIV-2 nef alleles (viremic, n = 23; nonviremic, n = 22). (C) The average cumulative numbers of HIV-1-infected GFP+ cells were measured for each of the 45 proviral constructs investigated. Values are shown relative to the proviral HIV-1 NL4-3 Nef (wt+) construct (100%).
We next examined whether nef alleles from HIV-2-infected subjects with high or undetectable VLs differ in their ability to promote viral spread in infected PBMC cultures. As reported previously (44), the HIV-1 NL4-3 Nef/eGFP constructs are replication competent and have an advantage in that the numbers of virally infected eGFP+ cells can readily be determined by flow-cytometric analysis. We found that all virus constructs containing intact nef alleles spread with substantially higher efficiency in the PBMC cultures than the control virus containing a disrupted nef gene (Fig. 4B). On average, nef alleles derived from viremic individuals were slightly more active in promoting viral spread in PBMCs than those derived from nonviremic subjects (Fig. 4B). For further analysis, we determined the average cumulative numbers of HIV-1-infected GFP+ cells for each of the 50 proviral constructs. We found that, on average, nef alleles from HIV-2-infected individuals with high VLs were significantly more active in promoting viral spread in PBMCs than those derived from the nonviremic subjects (186.1 ± 11.7 versus 143.1 ± 13.3; P = 0.02) (Fig. 4C). Although HIV-2 Nefs were generally less effective than the control HIV-1 Nef in enhancing virion infectivity in TZM-bl indicator cells, they were usually more efficient in increasing the number of virally infected GFP+ cells in PBMC cultures (Fig. 4). One possible reason for this is that T cells expressing HIV-1 Nefs show higher levels of apoptosis than those expressing HIV-2 Nefs (36, 44). Thus, the time span of eGFP and viral gene expression should be reduced in HIV-1-infected cells.
Efficient Nef-mediated downmodulation of TCR-CD3 and CD28 correlates with high CD4+ T cell counts in viremic HIV-2-infected individuals.
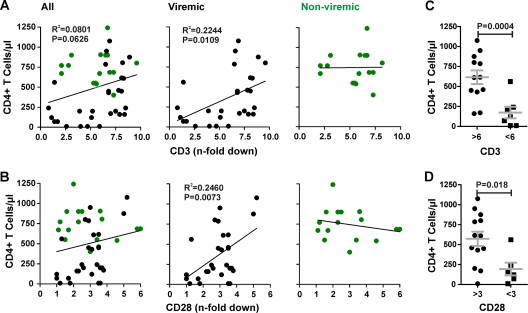
We next examined whether specific Nef functions correlate with the CD4+ T cell counts in HIV-2-infected individuals. We found that the efficiency of the Nef-mediated downmodulation of TCR-CD3 correlated significantly with higher numbers of CD4+ T cells in viremic (R2 = 0.2244; P = 0.0109) but not in nonviremic HIV-2-infected individuals (Fig. 5A). The same correlation was observed for the modulation of CD28 by Nef and CD4+ T cell counts in viremic individuals (R2 = 0.2460; P = 0.0073) but not for any of the other Nef functions investigated (Fig. 5B and data not shown).
Fig 5.
Correlation between Nef-mediated TCR-CD3 and CD28 downmodulation in virally infected PBMCs and the CD4+ T cell counts in HIV-2-infected individuals. Correlation between the absolute numbers of CD4+ T cells in HIV-2-infected individuals and n-fold downmodulation of CD3 (A) and CD28 (B). Assays were performed in PBMCs, and statistical analysis was performed using the PRISM 4.0 package. Data derived from viremic individuals are shown in black, and those derived from nonviremic individuals are in green. (C and D) Comparison of the CD4+ T cell counts in viremic individuals infected with HIV-2 strains expressing nef alleles showing high or low activity in the downmodulation of TCR-CD3 (C) and CD28 (D). Average values ± SD and P values of <0.05 are indicated.
To further assess the possible relevance of the Nef-mediated modulation of TCR-CD3 and CD28 for the immunological outcome of viremic HIV-2 infection, we compared the CD4+ T cell counts in individuals infected with HIV-2 strains expressing nef alleles showing high or low activities in these two functions. This analysis showed that viremic individuals infected with HIV-2 strains that efficiently downmodulate TCR-CD3 (>6-fold) had 3.6-fold higher CD4+ T cell counts than those infected with viruses that showed only modest (<6-fold) activity in this Nef function (615 ± 85 [n = 12] versus 173 ± 73 [n = 7]; P = 0.0004) (Fig. 5C). Similarly, the levels of CD4+ T cells were significantly higher in individuals infected with HIV-2 strains that effectively (>3-fold) downregulated CD28 (542 ± 88 [n = 14] and 193 ± 80 [n = 6], respectively; P = 0.018) (Fig. 5D). No such differences were observed in nonviremic individuals or for other Nef functions (data not shown). Taken together, these data demonstrate that the effective downmodulation of TCR-CD3 and CD28 by Nef is a strong correlate of preserved CD4+ T cell counts in viremic HIV-2 infection. In contrast, CD4+ T cells in nonviremic individuals are usually preserved irrespective of Nef function.
A single I132T substitution selectively disrupts HIV-2 Nef-mediated downmodulation of TCR-CD3 and interaction with the CD3ζ chain.
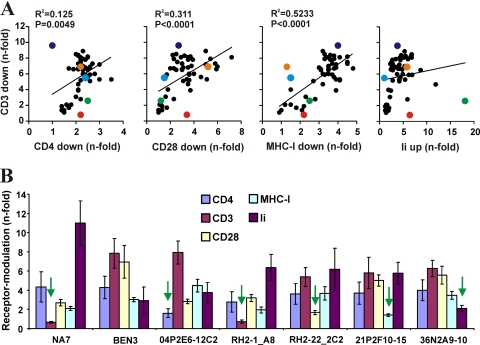
We next examined whether some HIV-2 nef alleles are selectively defective in specific functions. Correlation analyses showed that the efficiency of Nef-mediated TCR-CD3 downmodulation correlated significantly but imperfectly with the downmodulation of CD4, CD28, and MHC-I but not with the upmodulation of Ii or the enhancement of viral infectivity or replication (Fig. 6A and data not shown). Notably, some nef alleles were selectively impaired in modulating CD3, CD4, CD28, MHC-I, or Ii (Fig. 6B), which is in agreement with the results of previous studies showing that most or all Nef functions are genetically separable but mediated by overlapping domains (1, 18, 21, 42, 53, 55).
Fig 6.
Identification of HIV-2 Nefs impaired in specific functions. (A) Correlation between n-fold downmodulation of TCR-CD3 by HIV-2 Nefs and modulation of CD4, CD28, MHC-I, and Ii. Nef alleles selectively impaired in specific functions are highlighted by enlarged colored symbols: CD3 (red), CD28 (green), CD4 (purple), MHC-I (orange), and Ii (light blue). (B) Examples of HIV-2 Nef selectively impaired in specific functions. The HIV-1 NA7 Nef is shown for comparison. Quantitative assessment of Nef-mediated modulation of the indicated cellular receptors on PBMCs or THP-1 cells (Ii). Shown are average values ± SD obtained from four independent experiments.
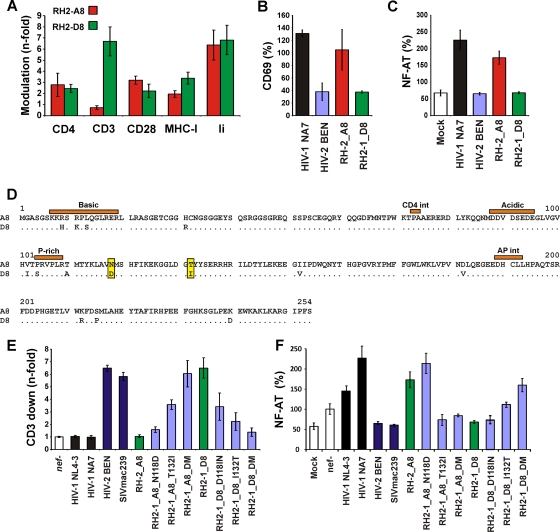
One pair of nef alleles from patient RH2-1, who was viremic and showed reduced numbers of CD4+ T cells (240/μl) at the time of virus isolation (see Table S1 in the supplemental material), was particularly interesting because both differed specifically in their capability to downmodulate TCR-CD3 (Fig. 7A) and thus to modulate the responsiveness of T cells to stimulation (Fig. 7B and C). The examination of the deduced amino acid sequences of the RH2-1_A8 and RH2-1_D8 Nefs revealed a total of 14 differences, including 5 in the conserved core domain previously reported to be important for the Nef-mediated downmodulation of TCR-CD3 (26, 53) (Fig. 7D). Mutational analyses revealed that the two amino acid changes N118D and T132I were sufficient to confer CD3 downmodulation activity to the inactive RH2-1_A8 Nef, and that the reverse changes disrupted this activity in the RH2-1_D8 Nef (Fig. 7E). Importantly, these two changes also affected the impact of Nef on the responsiveness of virally infected cells to stimulation (Fig. 7F) but had little, if any, effect on the levels of Nef expression and other activities, such as the modulation of CD4, CD28, and MHC-I (Fig. 8 and data not shown).
Fig 7.
I132T mutation in the core domain of HIV-2 Nef selectively disrupts downmodulation of TCR-CD3. (A to C) Comparison of the functional activity of the RH2-1_A8 and RH2-1_D8 Nef proteins in modulating various receptors (A), early T cell activation (B), and NF-AT activity (C). (D) Alignment of the RH2-1_A8 and RH2-1_D8 amino acid sequences. (E) The RH2-1_A8 Nef is selectively impaired in TCR-CD3 downmodulation and does not suppress the responsiveness of virally infected T cells to stimulation. A single-amino-acid variation of I132T disrupts the effect of HIV-2 Nefs on TCR-CD3. (F) Analysis of Jurkat cells stably transfected with an NFAT-dependent reporter gene (56) following transduction with the indicated HIV-1 Nef/eGFP constructs and subsequent stimulation with PHA. Levels of NFAT-dependent luciferase reporter activity are the averages (± SD) from four experiments.
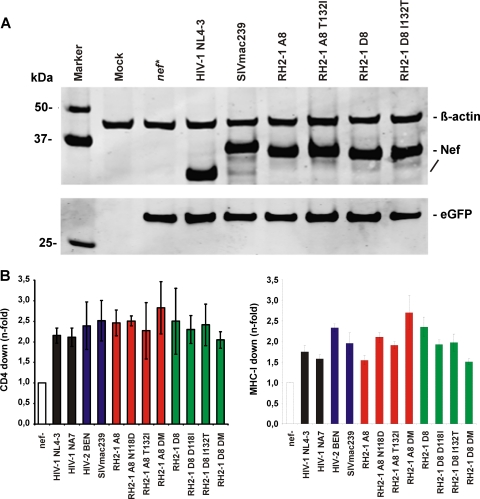
Fig 8.
T132I changes does not impair Nef expression and downmodulation of CD4 and MHC-I. (A) Western blot analysis of 293T cells transfected with pCGCG vectors encoding HA-tagged nef alleles followed by an IRES element and GFP. The Nef variants were probed with an anti-HA monoclonal antibody. β-Actin and GFP were measured to control for protein quantity and transfection efficiencies, respectively. (B) The N118D and T132I mutations in RH2-1 A8 Nef and the reverse changes in the RH2-1 D8 Nef do not impair the downmodulation of CD4 and MHC-1 in PBMCs transduced with HIV-1 NL4-3 constructs expressing the indicated nef alleles.
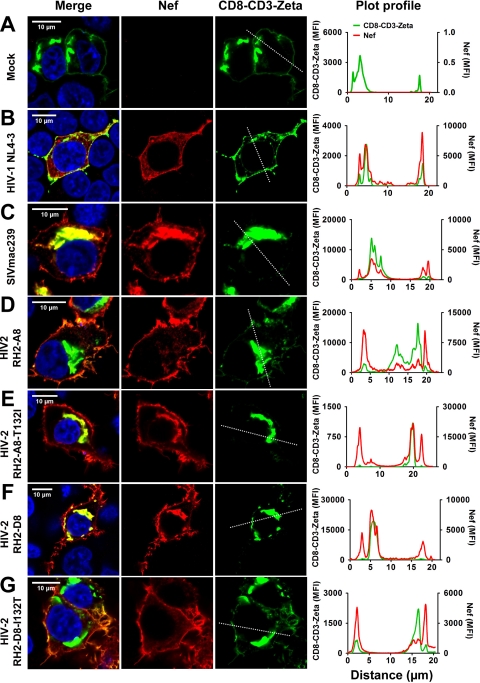
Both the D118N and I132T amino acid substitutions affected the Nef-mediated downmodulation of TCR-CD3, but the latter had a more prominent effect and thus was analyzed in more detail. We examined the effect of the I132T substitution on the ability of Nef to modulate the subcellular localization of a fusion between the cytoplasmic tail of TCR-CD3 and CD8 (39). We found that a major fraction of CD8-CD3ζ-GFP fluorescence was generally detected in perinuclear regions of the cells (Fig. 9). However, in the absence of Nef and in the presence of the HIV-1 NL4-3, HIV-2 RH2-A8, or HIV-2 RH2-D8 I132T Nef proteins, the CD8-CD3 fusion was also detected at the plasma membrane (Fig. 9A, B, D, and G). Thus, both Nef and the CD8-CD3 fusion usually showed three peaks in a fluorescence intensity profile spanning one cell: two indicative of membrane and a third of perinuclear localization. CD8-CD3ζ-GFP fluorescence was detected exclusively in perinuclear compartments in the presence of the SIVmac239, HIV-2 RH2-A8 T132I, or HIV-2 RH2-D8 Nef (Fig. 9C, E, and F). Consequently, the cells showed just a single peak of fluorescence for CD8-CD3ζ-GFP but still showed three peaks for Nef. These results confirmed that the I132T substitution is sufficient to disrupt the ability of HIV-2 RH-2_D8 Nef to remove the CD8-CD3 fusion from the surface, whereas the reciprocal change in the RH-2_A8 Nef restored this activity.
Fig 9.
Subcellular distribution of CD8-CD3ζ in the presence of wild-type and mutant HIV-2 Nef proteins. HEK293T cells cotransfected with equal amounts of plasmid DNA encoding a CD8-CD3ζ-GFP fusion and the indicated nef alleles were analyzed by confocal microscopy. CD8-CD3ζ was detected by GFP, whereas HA-tagged Nef proteins were detected by indirect immunofluorescence (anti-HA mouse monoclonal antibody followed by secondary Alexa Flour 568-conjugated goat anti-mouse IgG antibody). Merged and single images for various Nef alleles and CD8-CD3ζ-GFP are shown for mock (A), (B) HIV-1 NL4-3 Nef, (C) SIVmac239 Nef, (D) HIV-2 RH2_A8 Nef, (E) HIV-2 RH2_A8-T132I Nef, (F) HIV-2 RH2_D8 Nef, and (G) HIV-2 RH2_D8-I132T samples. The last panel under each condition shows the mean fluorescence intensity at the ROI drawn across the cells.
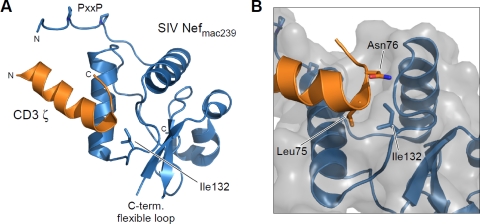
Recently, the crystal structure of the complex between the SIVmac239 core domain and the first ITAM motif of the CD3ζ chain has been reported (26). Isoleucine 132 is well conserved in SIV and HIV-2 Nef alleles and located at the surface of a hydrophobic crevice in Nef that is distal to the PxxP motif that is required for SH3 domain binding (Fig. 10A). Indeed, I132 performs tight interactions with leucine L75 of the YxxL ITAM motif in the ζ chain of CD3 and the following asparagine N76 (Fig. 10B). It is thus conceivable that these hydrophobic interactions of Nef are abrogated upon the mutation of I132 to the polar threonine residue, leading to a loss of CD3ζ binding and hence TCR-CD3 downmodulation.
Fig 10.
Structure of the complex formation between SIV Nefmac239 and CD3ζ. (A) Ribbon display of the core domain structure of SIV Nefmac239 (blue) with the first ITAM motif of CD3ζ indicated in orange (45). The hydrophobic residue Ile132 is located at the surface of Nef and is involved in tight interactions with the ζ chain of CD3. (B) Close-up view of CD3ζ-Nef binding. L75 and N76 of CD3ζ bind into a characteristic hydrophobic crevice formed by I132 of Nef. Similar interactions with receptor tail motifs are supposed for HIV-2 Nef. The mutation of I132T in HIV-2 Nef allele RH-2_A8 will disrupt the hydrophobic contacts formed between Nef and ζ, resulting in the impairment of CD3 downmodulation. Molecular diagrams were drawn using PyMOL (http://www.pymol.org/).
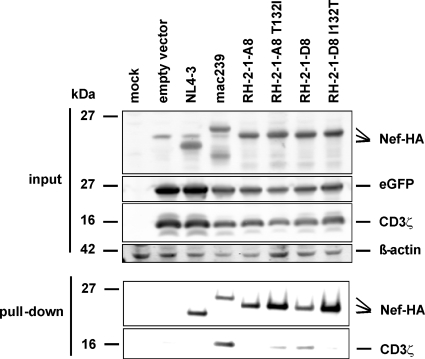
To directly examine whether residue I132 plays an important role in the interaction of the HIV-2 Nef with CD3ζ, we performed coimmunoprecipitation analyses. 293T cells were cotransfected with pCGCG constructs expressing HA-tagged Nef variants or CD3ζ together with eGFP. Western blot analysis showed that all wild-type and mutant Nef proteins as well as the CD3ζ chain were efficiently expressed (Fig. 11, upper). Coimmunoprecipitation experiments using immobilized anti-HA antibody demonstrated that the SIVmac239 Nef is highly effective in pulling down CD3ζ (Fig. 11, lower). As expected from the capability to downmodulate TCR-CD3, the HIV-2 RH2-1-A8 T132I and RH2-1-D8 Nefs also coprecipitated CD3ζ, whereas no interaction was detected for the RH-2-1A8 and RH2-1-D8 I132T Nef proteins (Fig. 11, lower). These data are further evidence that I132 is directly involved in the interaction of the HIV-2 Nef with the CD3ζ chain.
Fig 11.
Amino acid residue 132 is critical for interaction of HIV-2 Nef with CD3ζ. 293T cells were transfected with pCGCG vectors coexpressing GFP and CD3ζ or C-terminally HA-tagged Nef variants. At 40 h posttransfection, the cells were lysed and analyzed by Western blotting or coimmunoprecipitation analysis as described in Materials and Methods.
HIV-2 Nefs suppress death receptor expression.
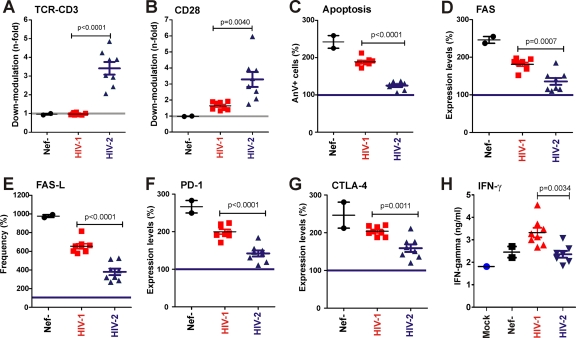
We have previously shown that the Nef-mediated downmodulation of TCR-CD3 protects infected T cells against programmed death (36, 44). To what extent the apoptosis of virally infected T cells contributes to the development of immunodeficiency in vivo in infected patients is still a matter of debate. It is generally accepted, however, that the programmed death of uninfected bystander cells, immune dysfunction, and high levels of inflammatory cytokine expression play a major role in the pathogenesis of AIDS. To assess whether HIV-1 and HIV-2 Nefs differentially affect these processes, we randomly selected a panel of 16 nef alleles from both viruses and examined their effect on the expression of death receptors and interferon production. We first confirmed that only HIV-2 but not HIV-1 nef alleles downmodulate TCR-CD3 (Fig. 12A), and that the former are also more active in modulating CD28 (Fig. 12B). In agreement with previous data (36, 44), HIV-2 Nefs suppressed the induction of apoptosis of virally infected T cells more efficiently than those of HIV-1 (Fig. 12C). It is well established that the Fas/Fas-L-mediated killing of both virally infected and uninfected bystander cells may play an important role in the pathogenesis of AIDS (23). We found that infection with nef-defective HIV-1 constructs induced both the levels of FAS expression (2.5-fold) and the frequency of cells expressing the Fas ligand (FAS-L+ cells; 9.8-fold) (Fig. 12D and E). The expression of HIV-1 nef alleles suppressed FAS/FAS-L induction only moderately, whereas HIV-2 Nefs reduced the levels of death receptor expression almost to those found in uninfected T cells (Fig. 12D and E). Similarly, HIV-2 Nefs also inhibited the induction of programmed death-1 (PD-1) and cytotoxic T-lymphocyte antigen 4 (CTLA-4) substantially more efficiently than HIV-1 Nefs (Fig. 12F and G). Both receptors are negative immune regulators that may downmodulate T cell immune responses (8, 11, 50). High levels of PD-1 on HIV-infected T cells correlates with viral load and with a state of immune exhaustion that results in decreased cellular proliferation and CTL activity (25, 56). Notably, blocking PD-1 restores HIV-specific T cell responses and thus has potential for immunotherapy (25, 56). The suppression of FAS, FAS-L, PD-1, and CTLA-4 expression correlated very well with the efficiency of the Nef-mediated downmodulation of TCR-CD3 (R2 > 0.6; P < 0.0001) (data not shown). Finally, lower levels of IFN-γ were detected in the supernatant of PBMC cultures infected with HIV-1 constructs expressing HIV-2 Nefs compared to those expressing HIV-1 Nef protein (Fig. 12H). Such differences in cytokine production could contribute to differences in the levels of chronic immune activation between HIV-1- and HIV-2-infected individuals.
Fig 12.
Functional differences between HIV-1 and HIV-2 Nefs. (A and B) Quantitative assessment of Nef-mediated downmodulation of TCR-CD3 (A) and CD28 (B) on virally infected PBMCs. (C to G) Frequency of apoptotic annexin V+ cells (C) and levels or percentages of FAS (D), FAS-L (E), PD-1 (F), and CTLA-4 (G) expression on PBMCs transduced with HIV-1 constructs expressing HIV-1 or HIV-2 nef alleles. (H) Levels of IFN-γ in the supernatant of PBMCs infected with viral constructs expressing HIV-1 or HIV-2 Nef proteins. Each symbol represents the value obtained for one of the 16 NL4-3 recombinants expressing HIV-1 or HIV-2 nef alleles or containing defective nef alleles. The results represent average values obtained from three independent experiments. The gray line indicates the nef-defective control, and the blue line indicates the values obtained for uninfected cells. P values of <0.05 are indicated.
DISCUSSION
In the present study, we examined the role of the accessory Nef protein in the virological and immunological outcome of HIV-2 infection. We show that the efficacy of the Nef-mediated downmodulation of TCR-CD3 and CD28 correlates with high CD4+ T cell counts in viremic HIV-2-infected subjects but not in individuals with undetectable viral loads. This result most likely explains the seeming discrepancy between two previous studies, which showed that the efficient Nef-mediated downmodulation of TCR-CD3 does not protect HIV-2-infected individuals against disease progression (13) but is associated with stable high levels of CD4+ T cells in SIVsmm-infected SMs, the original host of HIV-2 (45). Most long-term nonprogressors of HIV-2 infection have undetectable viral loads (12, 49), whereas SIVsmm-infected SMs do not develop disease despite high levels of viral replication (40). It is conceivable that viral properties only affect the clinical and immunological outcome of HIV-2 infection if the virus replicates to detectable levels. Taken together, our current knowledge supports that the ability of Nef to remove TCR-CD3 from the cell surface to suppress T cell activation helps both HIV-2-infected individuals and the natural hosts of SIV to maintain normal CD4+ T cell counts in the presence of significant levels of viral replication. It is also evident, however, that the Nef-mediated suppression of T cell activation by the efficient downmodulation of TCR-CD3 and CD28 may only decelerate and not entirely prevent the loss of CD4+ T cells and the development of immunodeficiency in the recent and poorly adapted human host.
It is well established that intact and functional nef genes play an important role in efficient viral replication in vivo, and that defective nef genes are associated with set point viral loads that were reduced about three to five orders of magnitude in SIVmac-infected macaques (25) and in HIV-1-infected human individuals (9, 28). Thus, it is unexpected that the only significant association between Nef function and detectable levels of viremia in HIV-2-infected individuals was a modestly increased activity in promoting viral spread in PBMC cultures. nef alleles from viremic and nonviremic individuals did not differ significantly in their capability to modulate CD4, TCR-CD3, CD28, MHC-I, and Ii cell surface expression, T cell activation, programmed cell death, and virion infectivity. Our finding that the great majority of HIV-2 nef alleles were intact and active in most assays for Nef function clearly supports that these activities are advantageous for virus replication in vivo. Thus, our results do not support previous data suggesting that defective nef alleles play a significant role in nonprogressive HIV-2 infection (17), but we suggest that many HIV-2 infected individuals can effectively control the virus despite the expression of fully functional nef alleles.
It is tempting to speculate that differences in the effect of Nef on the expression of PD-1 and CTLA-4 contribute to the different levels of immune control in HIV-1 and HIV-2 infection. Both receptors are negative immune regulators of the immune response, and their induction may contribute to the lack of immune control of HIV-1 replication and the impaired virus-specific CTL and helper T cell responses, which represent hallmarks of AIDS. It has been demonstrated that PD-1 and CTLA-4 are upregulated on CD4+ T cells in HIV-1-infected individuals and that the expression levels of these receptors correlate directly with viral load and inversely with the CD4+ T cell counts (9, 23). The blockade of the PD-1 and CTLA-4 pathways augmented HIV-specific CD4+ T cell proliferation (41, 56). We demonstrated that CD4+ T cells infected with viral constructs expressing HIV-2 Nef express significantly lower levels of PD-1 and CTLA-4 than those expressing HIV-1 Nef proteins. As a result, HIV-specific T cell dysfunction may be reduced in HIV-2-infected individuals, resulting in more effective helper CD4+ T cell and CD8+ CTL responses. In fact, it has been reported that these immune responses are maintained and are more effective in HIV-2 infection than in HIV-1 infection, and they are associated with the improved control of viral replication (27).
Further studies are necessary to challenge this possibility, but our data support that HIV-2 infection also is less pathogenic than HIV-1 infection at similar levels of virus replication. As mentioned above, HIV-2 and HIV-1 differ fundamentally in the two Nef functions that correlated with high numbers of CD4+ T cells in viremic HIV-2-infected individuals. In contrast to HIV-2 Nefs, those of HIV-1 generally lack the CD3 downmodulation activity and have substantially weaker effects on CD28 cell surface expression (36, 44). While the ability of primate lentiviruses to suppress the responsiveness of infected T cells to stimulation is not always sufficient to prevent disease progression, the Nef-mediated downmodulation of TCR-CD3 (and, to a lesser extent, CD28) may generally dampen damaging immune activation and decelerate the loss of CD4+ T cells and thus the development of immunodeficiency. In support of a role of Nef in the overall activation state of the immune system, it has been shown that the efficiency of CD3 downregulation by Nef, even in HIV-2-infected individuals with undetectable viral loads, correlates with reduced levels of immune activation (13).
The Nef proteins of primate lentiviruses can be divided into two groups based on their capability to downmodulate TCR-CD3 or lack thereof (44), and previous data suggested that the residues that govern this fundamental function are located in the conserved core domain of Nef (26, 53). Nonetheless, mapping the exact residues or domains in Nef that are critical for CD3 downmodulation has proven very difficult. Some changes that impair TCR-CD3 downmodulation by the SIVmac239 Nef have been described (26, 53). Similarly to the I132T change identified in the present study, these alterations were localized in the conserved core domain of Nef. However, all of them also impaired other Nef functions, suggesting that they have more general disruptive effects on the highly structured core region of Nef (26, 53). In contrast, the naturally occurring I132T change in the HIV-2 Nef identified in the present study was highly specific for the CD3 downmodulation function. Thus, together with the coimmunoprecipitation analyses, these results clearly support that residue I132 is directly involved in the interaction of Nef with the cytoplasmic domain of the CD3ζ chain.
It is noteworthy that the single I132T mutation in the RH2-1 HIV-2 Nef results in a phenotype highly reminiscent of HIV-1 Nefs, i.e., the selective loss of CD3 downmodulation and hence the lack of the capability to effectively suppress the responsiveness of virally infected T cells to stimulation (2, 44). It is well established that HIV-1 M strains are substantially more successful in spreading in the human population than HIV-2 (10). If it is relatively easy for HIV-2 to become more HIV-1-like by acquiring single-amino-acid changes in Nef, then why does this not happen more often? In fact, we found that none of the 122 HIV-2 Nef sequences present in the Los Alamos National Laboratory (LANL) database (http://www.hiv.lanl.gov/components/sequence/HIV/search/search.html) had a Thr at the amino acid position corresponding to I132 in the RH2 Nef. It seems that for HIV-2 it is not advantageous to lose the CD3 downmodulation function, possibly because it lacks a vpu gene and thus is less well equipped to counteract the host defense mechanisms induced by increased immune activation (14). Nonetheless, our result raises the possibility that HIV-2 can evolve into a more virulent form by losing a protective Nef function.
In summary, our data suggest that the Nef-mediated downmodulation of TCR-CD3 and CD28 protects viremic HIV-2-infected individuals against the loss of CD4+ T cells. Both of these functions presumably delay the apoptotic death of virally infected T cells by suppressing their activation-induced death. Furthermore, Nef may also help HIV-2 to maintain the functionality of uninfected bystander T cells by suppressing the induction of death (FAS/FAS-L) and immunosuppressive receptors (PD-1 and CTLA-4). Further studies are required to elucidate the importance of the differential effects of HIV-1 and HIV-2 Nefs on T cell activation for the overall systemic levels of immune activation, exhaustion, and control of virus replication. In support of a relevant role, however, it has been shown that the levels of the Nef-mediated downmodulation of TCR-CD3 correlate with the levels of immune activation in HIV-2-infected individuals with comparably low viral loads (13). Furthermore, CD4+ helper T cells that are infected and directly manipulated by HIV orchestrate the immune response, and it is their key role to manipulate many other cells in small numbers. Notably, the identification of naturally occurring nef alleles that are selectively impaired in specific functions also provides a means to study the role of these Nef activities in viral immune evasion and pathogenicity in appropriate animal models, and we have generated mutants of the pathogenic SIVmac239 molecular clone expressing Nef proteins selectively impaired in TCR-CD3 modulation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Martha Mayer and Susanne Engelhart for technical assistance and Dré van der Merwe, Christine Goffinet, and Hetty Blaak for comments and critical readings of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) to F.K. and M.G. (GE 976/6-1). R.A.G. is supported by Virgo FES.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Akari H, et al. 2000. Nef-induced major histocompatibility complex class I downregulation is functionally dissociated from its virion incorporation, enhancement of viral infectivity, and CD4 downregulation. J. Virol. 74:2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arhel N, et al. 2009. The inability to disrupt the immunological synapse between infected human T cells and APCs distinguishes HIV-1 from most other primate lentiviruses. J. Clin. Investig. 119:2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berry N, et al. 1998. Low peripheral blood viral HIV-2 RNA in individuals with high CD4 percentage differentiates HIV-2 from HIV-1 infection. J. Hum. Virol. 1:457–468 [PubMed] [Google Scholar]
- 4. Blaak H, et al. 2005. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. J. Virol. 79:1686–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blaak H, Boers PH, Schutten M, van der Endeñ ME, Osterhaus AD. 2004. HIV-2-infected individuals with undetectable plasma viremia carry replication-competent virus in peripheral blood lymphocytes. J. Acquir. Immune Defic. Syndr. 36:777–782 [DOI] [PubMed] [Google Scholar]
- 6. Brenchley JM, Silvestri G, Douek DC. 2010. Nonprogressive and progressive primate immunodeficiency lentivirus infections. Immunity 32:737–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chowers MY, et al. 1994. Optimal infectivity in vitro of HIV-1 requires an intact nef gene. J. Virol. 68:2906–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Day CL, et al. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350–354 [DOI] [PubMed] [Google Scholar]
- 9. Deacon NJ, et al. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988–991 [DOI] [PubMed] [Google Scholar]
- 10. de Silva TI, Cotton M, Rowland-Jones SL. 2008. HIV-2: the forgotten AIDS virus. Trends Microbiol. 16:588–595 [DOI] [PubMed] [Google Scholar]
- 11. D'Souza M, et al. 2007. Programmed death 1 expression on HIV-specific CD4+ T cells is driven by viral replication and associated with T cell dysfunction. J. Immunol. 179:1979–1987 [DOI] [PubMed] [Google Scholar]
- 12. Estes JD, et al. 2008. Early resolution of acute immune activation and induction of PD-1 in SIV-infected sooty mangabeys distinguishes nonpathogenic from pathogenic infection in rhesus macaques. J. Immunol. 180:6798–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feldmann J, et al. 2009. Downregulation of the T-cell receptor by human immunodeficiency virus type 2 Nef does not protect against disease progression. J. Virol. 83:12968–12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fortin JF, Barat C, Beausejour Y, Barbeau B, Tremblay MJ. 2004. Hyper-responsiveness to stimulation of human immunodeficiency virus-infected CD4+ T cells requires Nef and Tat virus gene products and results from higher NFAT, NF-kappaB, and AP-1 induction. J. Biol. Chem. 279:39520–39531 [DOI] [PubMed] [Google Scholar]
- 15. Gao F, et al. 1994. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J. Virol. 68:7433–7447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao F, et al. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 17. Geyer M, Fackler OT, Peterlin BM. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Greenberg ME, Iafrate AJ, Skowronski J. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 20. Hahn BH, Shaw GM, De Cock KM, Sharp PM. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607–614 [DOI] [PubMed] [Google Scholar]
- 21. Iafrate AJ, Bronson S, Skowronski J. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jaffar S, et al. 1997. Rate of decline of percentage CD4+ cells is faster in HIV-1 than in HIV-2 infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovir. 16:327–332 [DOI] [PubMed] [Google Scholar]
- 23. Kaufmann DE, Walker BD. 2009. PD-1 and CTLA-4 inhibitory cosignaling pathways in HIV infection and the potential for therapeutic intervention. J. Immunol. 182:5891–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keele BF, et al. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kestler HW, et al. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651–662 [DOI] [PubMed] [Google Scholar]
- 26. Kim WM, Sigalov AB, Stern LJ. 2010. Pseudo-merohedral twinning and noncrystallographic symmetry in orthorhombic crystals of SIVmac239 Nef core domain bound to different-length TCRzeta fragments. Acta Crystallogr. D Biol. Crystallogr. 66:163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kirchhoff F. 2009. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 7:467–476 [DOI] [PubMed] [Google Scholar]
- 28. Kirchhoff F, Greenough TC, Brettler DB, Sullivan JL, Desrosiers RC. 1995. Absence of intact nef sequences in a long-term, nonprogressing survivor of HIV-1 infection. N. Engl. J. Med. 332:228–232 [DOI] [PubMed] [Google Scholar]
- 29. Kirchhoff F, Schindler M, Specht A, Arhel N, Münch J. 2008. Role of Nef in primate lentiviral immunopathogenesis. Cell. Mol. Life Sci. 65:2621–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lama J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr. HIV Res. 1:167–184 [DOI] [PubMed] [Google Scholar]
- 31. Leonard JA, Filzen T, Carter CC, Schaefer M, Collins KL. 2011. HIV-1 Nef disrupts intracellular trafficking of major histocompatibility complex class I, CD4, CD8, and CD28 by distinct pathways that share common elements. J. Virol. 85:6867–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marlink R, et al. 1994. Reduced rate of disease development after HIV-2 infection compared to HIV-1. Science 265:1587–1590 [DOI] [PubMed] [Google Scholar]
- 33. Miller MD, Warmerdam MT, Gaston I, Greene WC, Feinberg MB. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 179:101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mörner A, et al. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Münch J, et al. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 81:13852–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Münch J, et al. 2005. Primary sooty mangabey simian immunodeficiency virus and human immunodeficiency virus type 2 nef alleles modulate cell surface expression of various human receptors and enhance viral infectivity and replication. J. Virol. 79:10547–10560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Münch J, Stolte N, Fuchs D, Stahl-Hennig C, Kirchhoff F. 2001. Efficient class I major histocompatibility complex down-regulation by simian immunodeficiency virus Nef is associated with a strong selective advantage in infected rhesus macaques. J. Virol. 75:10532–10536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Owen SM, et al. 1998. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 72:5425–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Poonia B, Pauza CD, Salvato MS. 2009. Role of the Fas/FasL pathway in HIV or SIV disease. Retrovirology 6:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rey-Cuillé MA, et al. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rowland-Jones SL, Whittle HC. 2007. Out of Africa: what can we learn from HIV-2 about protective immunity to HIV-1? Nat. Immunol. 8:329–331 [DOI] [PubMed] [Google Scholar]
- 42. Schaefer TM, et al. 2002. The conserved process of TCR/CD3 complex down-modulation by SIV Nef is mediated by the central core, not endocytic motifs. Virology 302:106–122 [DOI] [PubMed] [Google Scholar]
- 43. Schindler M, Münch JJ, Kirchhoff F. 2005. Human immunodeficiency virus type 1 inhibits DNA damage-triggered apoptosis by a Nef-independent mechanism. J. Virol. 79:5489–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schindler M, et al. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055–1067 [DOI] [PubMed] [Google Scholar]
- 45. Schindler M, et al. 2008. Inefficient Nef-mediated downmodulation of CD3 and MHC-I correlates with loss of CD4+ T cells in natural SIV infection. PLoS Pathog. 4:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schindler M, et al. 2003. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548–10556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shanmugam V, et al. 2000. Lower HIV-2 plasma viral loads may explain differences between the natural histories of HIV-1 and HIV-2 infections. J. Acquir. Immune Defic. Syndr. 24:257–263 [DOI] [PubMed] [Google Scholar]
- 48. Sharp PM, Hahn BH. 2010. The evolution of HIV-1 and the origin of AIDS. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365:2487–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Silvestri G, et al. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Simone R, Piatti G, Saverino D. 2009. The inhibitory co-receptors: a way to save from anergy the HIV-specific T cells. Curr. HIV Res. 7:266–272 [DOI] [PubMed] [Google Scholar]
- 51. Spina CA, Kwoh TJ, Chowers MY, Guatelli JC, Richman DD. 1994. The importance of nef in the induction of HIV-1 replication from primary quiescent CD4 lymphocytes. J. Exp. Med. 179:115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stumptner-Cuvelette P, et al. 2001. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. U. S. A. 98:12144–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Swigut T, Greenberg M, Skowronski J. 2003. Cooperative interactions of simian immunodeficiency virus Nef, AP-2, and CD3-zeta mediate the selective induction of T-cell receptor-CD3 endocytosis. J. Virol. 77:8116–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Swigut T, Iafrate AJ, Münch J, Kirchhoff F, Skowronski J. 2000. Simian and human immunodeficiency virus Nef proteins use different surfaces to down-regulate class I major histocompatibility antigen expression. J. Virol. 74:5691–5701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swigut T, Shohdy N, Skowronski J. 2001. Mechanism for down-regulation of CD28 by Nef. EMBO J. 20:1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Trautmann L, et al. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198–1202 [DOI] [PubMed] [Google Scholar]
- 57. Tsuchiya S, et al. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171–176 [DOI] [PubMed] [Google Scholar]
- 58. van der Ende ME, et al. 2000. Antiviral resistance of biologic HIV-2 clones obtained from individuals on nucleoside reverse transcriptase inhibitor therapy. J. Acquir. Immune Defic. Syndr. 25:11–18 [DOI] [PubMed] [Google Scholar]
- 59. Whittle H, et al. 1994. HIV-2-infected patients survive longer than HIV-1-infected patients. AIDS 8:1617–1620 [DOI] [PubMed] [Google Scholar]
- 60. Wilkins A, et al. 1993. The epidemiology of HIV infection in a rural area of Guinea-Bissau. AIDS 7:1119–1122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.