Abstract
Nonstructural protein 4 (NSP4) viroporin activity is critical for the replication and assembly of serogroup A rotavirus (RVA); however, the dramatic primary sequence divergence of NSP4s across serogroups raises the possibility that viroporin activity is not a common feature among RVs. We tested for NSP4 viroporin activity from divergent strains, including RVA (EC and Ty-1), RVB (IDIR), and RVC (Cowden). Canonical viroporin motifs were identified in RVA, RVB, and RVC NSP4s, but the arrangement of basic residues and the amphipathic α-helices was substantially different between serogroups. Using Escherichia coli and mammalian cell expression, we showed that each NSP4 tested had viroporin activity, but serogroup-specific viroporin phenotypes were identified. Only mammalian RVA and RVC NSP4s induced BL21-pLysS E. coli cell lysis, a classical viroporin activity assay. In contrast, RVA, RVB, and RVC NSP4 expression was universally cytotoxic to E. coli and disrupted reduction-oxidation activities, as measured by a new redox dye assay. In mammalian cells, RVB and RVC NSP4s were initially localized in the endoplasmic reticulum (ER) and trafficked into punctate structures that were mutually exclusive with RVA NSP4. The punctate structures partially localized to the ER-Golgi intermediate compartment (ERGIC) but primarily colocalized with punctate LC3, a marker for autophagosomes. Similar to RVA NSP4, expression of RVB and RVC NSP4s significantly elevated cytosolic calcium levels, demonstrating that despite strong primary sequence divergence, RV NSP4 has maintained viroporin activity across serogroups A to C. These data suggest that elevated cytosolic calcium is a common critical process for all rotavirus strains.
INTRODUCTION
Rotaviruses (RVs) are the leading cause of infantile gastroenteritis, which is marked by life-threatening dehydration due to vomiting and diarrhea (17, 24). Virus isolates are classified into distinct groups based on the serological response to the inner capsid protein VP6, with seven groups (RVA through RVG) currently being recognized. RVA, RVB, and RVC strains have been isolated from humans, other mammals, and birds, whereas RVD, RVE, RVF, and RVG strains have been isolated from animals but not humans (39). RVA strains cause the vast majority of RV-associated morbidity and mortality in humans, but RVB and RVC also can cause severe gastroenteritis in children and adults (30, 48, 50). The lack of robust culture systems for non-RVA strains has limited our understanding of these viruses, but many studies have compared the conservation and divergence of homologous genes across groups (27, 32, 39–41).
RV gene 10, which encodes nonstructural protein 4 (NSP4), has substantially diverged based on the primary sequence but has retained similar biochemical and functional characteristics (27). NSP4 genes cluster according to RV group and share more than 30% amino acid sequence identity within each group. In contrast, RVA NSP4s share only approximately 10% and 20% sequence identity with RVB and RVC NSP4s, respectively. Similarly, RVB and RVC NSP4s share less than 10% identity (23, 37). Despite this lack of primary sequence conservation, NSP4s from different groups appear to have well-conserved biochemical properties, including two or three prominent N-terminal hydrophobic domains, at least one predicted transmembrane domain, and a C-terminal hydrophilic domain (23, 27, 34). This observation has been confirmed with one RVD NSP4 gene and NSP4 genes from two unclassified novel adult diarrhea RV strains (B219 and J19), which cluster with RVB NSP4 genes (45, 59).
In addition to shared biochemical properties, the expectation that NSP4s from different groups carry out similar functions is supported by several observations. RVB (in infected rats) and RVC (in infected ST-1 cells) undergo final morphogenesis by budding through endoplasmic reticulum (ER) membranes to assemble mature infectious particles, similar to RVA (28, 46). The C-terminal tail of RVA NSP4 facilitates this budding-and-assembly process by acting as an intracellular receptor in ER membranes (1, 2, 42). Therefore, it is reasonable to predict that non-RVA NSP4 also serves as a receptor for particle maturation. Additionally, since the discovery that RVA NSP4 has enterotoxin activity in neonatal mice (3), numerous studies have shown that NSP4s from divergent RVA murine and avian strains, as well as from RVB and RVC human strains, all exhibit enterotoxin activity in the neonatal mouse model, suggesting a possible common mode of action (26, 32, 43, 52). Thus, divergent NSP4s have shared functions in RV morphogenesis and pathogenesis.
Nevertheless, little experimental data regarding the intracellular functions of non-RVA NSP4s have been generated. Early studies demonstrated that RVB NSP4 increases cytoplasmic calcium levels ([Ca2+]cyto) of Sf9 insect cells similarly to RVA NSP4. However, the functional importance of elevated [Ca2+]cyto has been investigated only for RVA, not RVB or RVC, strains (49, 58). Recently, we showed that NSP4 from the RVA strain SA11 has a viroporin domain that is functionally responsible for the elevation in [Ca2+]cyto (29). Viroporins are a family of diverse, virus-encoded pore-forming proteins that fulfill a variety of functions important for the replication of their cognate virus. Viroporins have several characteristic structural motifs, including a cluster of basic residues (Lys or Arg) and an amphipathic α-helix, such that oligomerization forms an aqueous pore through the membrane; however, it is currently unknown which oligomeric state(s) of NSP4 is the functional form (9, 20). Viroporin-mediated increases in [Ca2+]cyto induce formation of NSP4 vesicular structures (puncta), which have been shown to colocalize with the autophagy marker microtubule-associated protein light chain 3 (LC3), bypassing normal Golgi-dependent trafficking pathways (4, 29). Since RVB NSP4 also elevates [Ca2+]cyto in Sf9 cells, we predicted that NSP4 viroporin activity is conserved across RV groups despite the negligible primary sequence identity. To test this hypothesis, we examined NSP4s from RVA, RVB, and RVC strains for canonical viroporin structural motifs and assessed their viroporin activity using Escherichia coli and mammalian epithelial cell expression systems.
MATERIALS AND METHODS
Expression constructs and cell lines.
All bacterial expression constructs were generated using the pET46 Ek/LIC vector (EMD Biosciences, Gibbstown, NJ) according to the manufacturer's instructions. The pET46 expression vector encodes an N-terminal hexahistidine tag for affinity chromatography purification. The mammalian red fluorescent protein (RFP)-tagged NSP4 expression constructs were generated using the TagRFP-N1 vector (Evrogen, Moscow, Russia). Each NSP4 gene was amplified with appropriate primers that incorporated a 5′ EcoRI and 3′ AgeI restriction enzyme site for insertion into the TagRFP-N1 vector, resulting in a C-terminal RFP tag with a linker amino acid sequence of GGGPVAT, similar to the previously described NSP4-EGFP construct (4).
Based on a new nomenclature system, NSP4s from RVA strains have been classified into 14 enterotoxin (E) genotypes (39). Genes used in these studies are from RV strains SA11 clone 3 (RVA genotype E2; accession number AAC61867), OSU (RVA genotype E1; accession number BAA13728) and IDIR (RVB; accession number U03557) and were previously cloned (39, 58, 61, 62). Constructs reported here were subcloned from these cDNA clones. NSP4 genes from RRV (RVA genotype E3; accession number AAA64924) and Cowden (RVC; accession number AF093202, originally from Linda Saif) were amplified from virus stock using a Superscript one-step reverse transcription-PCR (RT-PCR) kit (Life Technologies, Carlsbad, CA) (61). The attenuated Cowden virus stock was a kind gift from Frank Ramig at Baylor College of Medicine. NSP4 gene from murine EC RV (RVA genotype E7; accession number AAB58700) was amplified by RT-PCR from virus-infected mouse stool samples. Finally, Ty-1 NSP4 (RVA genotype E4; accession number AB065285) was generated by primer-based gene synthesis (Epoch Bioscience, Missouri City, TX) and cloned into the NcoI and XhoI sites of pET46. The sequences of the constructs were determined by DNA sequence analysis and were identical to known sequences in GenBank.
The bacterial expression constructs were used to transform BL21(DE3) and BL21(DE3)pLysS E. coli (Promega, Madison, WI), which express the T7 RNA polymerase under the control of the LacI operator and in which recombinant protein expression is therefore induced by isopropyl β-d-1-thiogalactopyranoside (IPTG). In all cases, E. coli transformants carrying a NSP4 expression vector were grown in LB medium supplemented with 1% (vol/vol) glucose and appropriate antibiotics for selection. Due to the toxic nature of NSP4, the addition of 1% glucose is essential to reduce leaky expression of NSP4.
The African green monkey kidney cell line MA104 was used for mammalian expression of NSP4-RFP, NSP4-EGFP, EGFP-LC3, and Sec61β-EGFP, which was a gift from Tom Rapoport (Addgene plasmid 15108) (4, 60). MA104 cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum under standard conditions.
Lysis assay and partial purification of NSP4.
Starter cultures from well-isolated BL21(DE3)pLysS colonies were diluted 1:100 into fresh medium (LB, 1% glucose, 100 μg/ml ampicillin, 35 μg/ml chloramphenicol) and then plated into a 96-well culture plate. The plates were incubated at 37°C until the cultures reached mid-log phase. NSP4 expression was induced with 1 mM IPTG and the plates were incubated for 10 h. Optical density at 600 nm (OD600) readings were taken at 15 min intervals. Data are reported as the mean OD600 from three independent experiments. A similar procedure was used for immunoblot analysis of protein expression, except that a 1-liter total culture volume was used and cells were harvested at 3 h postinduction. Cells and membrane fragments were pelleted by centrifugation (22,000 × g, 30 min) and solubilized in lysis buffer (50 mM Tris, 500 mM NaCl, 10 mM imidazole, and 0.5% N-laurylsarcosine) overnight at 4°C with stirring. Lysates were clarified by centrifugation (22,000 × g, 30 min) and incubated with Ni2+-nitriloacetic acid (NTA) beads (Qiagen, Valencia, CA) to concentrate the His-tagged proteins. The beads were washed and eluted in lysis buffer containing 40 mM and 500 mM imidazole, respectively.
Immunoblot analysis.
Immunoblot analysis was performed as previously described (31). Protein samples were mixed with sodium dodecyl sulfate (SDS) sample buffer, heated at 100°C for 10 min, and separated by polyacrylamide gel electrophoresis (PAGE). Resolved proteins were transferred onto nitrocellulose membranes, blocked with 5% nonfat dry milk dissolved in phosphate-buffered saline (PBS) (BLOTTO) for 10 min at room temperature, and then incubated overnight with the NSP4-specific monoclonal antibody B4-2/55 (31) at 1:1,000, antisera specific for the NSP4 peptide from amino acid (aa) 114 to 135 at 1:1,000, His probe antibody [EMD Biosciences (Calbiochem)] at 1:500, or TagRFP-specific antibody at 1:1,000 diluted in 0.5% BLOTTO. Membranes were washed three times in 0.5% BLOTTO, incubated with the appropriate alkaline phosphatase-conjugated secondary antibody for ∼2 h, and washed again. Blots were developed with a colorimetric substrate (50 mM Tris base, 3 mM MgCl2, 0.1 mg/ml nitroblue tetrazolium chloride, and 0.5 mg/ml 5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt).
Cytotoxicity assay.
Overnight cultures of BL21(DE3) E. coli were serially diluted 10-fold, and 100 μl of each dilution was plated on agar medium (LB, 1% glucose, ampicillin) in the absence or presence of 1 mM IPTG. The plates were then incubated overnight at 37°C, and the CFU/ml was calculated. The experiment was performed in triplicate, and the mean values are reported.
Redox Sensor Green assay.
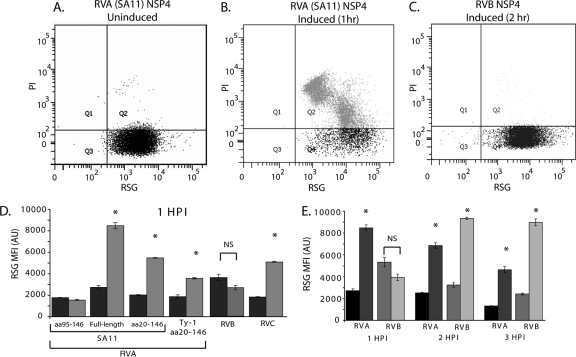
Redox Sensor Green (RSG; Life Technologies) is a novel redox indicator that becomes fluorescent green upon being modified by bacterial reductases. Thus, this dye is a sensitive and direct measure of cell viability and vitality. We chose this assay to measure NSP4 viroporin activity because disruption of the bacterial inner membrane would neutralize the proton gradient similarly to proton ionophores, which increase RSG fluorescence (21). To measure RSG fluorescence, mid-log-phase uninduced or IPTG-induced BL21(DE3) cultures were incubated at 37°C for the times indicated in Fig. 5. Aliquots were removed from the culture and diluted to ∼106 cells/ml in 0.22-μm-filtered PBS containing 1 μM RSG and 20 μM propidium iodide (PI) for 10 min at 37°C. Cells were then analyzed by flow cytometry on an LSR Fortessa cytometer. RSG and PI fluorescence was excited with an argon laser (488 nm), and emission was detected using 520/20 and 630/22 band pass filters, respectively. Gates were set using unstained cells and cells stained with RSG or PI alone. The assay was performed in triplicate, and the results are presented as the mean fluorescence intensity.
Fig 5.
NSP4 viroporin activity disrupts E. coli redox homeostasis. (A to C) Flow cytometry scatter plots of BL21(DE3) E. coli stained with Redox Sensor Green (RSG) and propidium iodide (PI). Gates were set using unstained and singly RSG- or PI-stained cells. (A and B) Uninduced (A) and IPTG-induced (B) cells expressing full-length RVA NSP4 at 1 h postinduction. (C) IPTG-induced cells expressing RVB NSP4 at 2 h postinduction. (D) RSG mean fluorescent intensity (MFI) of uninduced (dark gray) and IPTG-induced cells (light gray) from three independent experiments. (E) The RSG MFI of full-length RVA (uninduced [black] and induced [dark gray]) and RVB (uninduced [gray] and induced [light gray]) was measured at 1-h intervals for 3 h. Error bars (D and E) represent standard deviations. *, P < 0.01; NS, nonsignificant difference.
Calcium measurements.
Vectors encoding TagRFP NSP4s from SA11, Ty-1, IDIR, and Cowden rotavirus strains were used to transfect MA104 cells using the transfection reagent Lipofectamine 2000. At ∼16 h posttransfection, the cells were washed with trypsin diluent (137 mM NaCl, 5.4 mM KCl, 423 μM Na2HPO4, 441 μM KH2PO4, 5.6 mM dextrose, 4.5 mM NaHCO3) and trypsinized with 0.25% trypsin-EDTA (Life Technologies) to release them from the well, and then DMEM–10% fetal bovine serum (FBS) was added to neutralize the trypsin. The cells were pelleted by centrifugation at ∼71 × g for 5 min, the supernatant was removed, and then loading buffer [alpha MEM supplemented with 10 mM HEPES, 1× PowerLoad, 2.5 mM probenecid (Life Technologies), and 2 μM Fluo-2-AM (TEFLabs, Austin, TX)] was added. Cells were incubated for 30 min at 37°C to load them with Fluo-2 and then analyzed by flow cytometry on an LSR Fortessa cytometer. Fluo-2 fluorescence was excited by the argon laser (488 nm), and emission was detected with a 520/20 band pass filter. TagRFP was excited by the yellow/green laser (561 nm), and emission was detected with a 610/20 band pass filter. Gates were set using unstained cells, cells expressing TagRFP alone, and cells loaded with Fluo-2-AM alone.
Confocal microscopy.
Cells were grown on glass coverslips (12-mm circular, no. 1.5; Fisherbrand) and transfected with NSP4-RFP alone or cotransfected with RVA SA11 NSP4-EGFP, EGFP-LC3, or Sec61β-EGFP (4, 60). At 24 h posttransfection, cells were fixed with 4% paraformaldehyde in PBS for 30 min and blocked in 5% bovine serum albumin (BSA) for 30 min. Indirect immunofluorescence detection for the ER [rabbit anti-calnexin (1:300; Santa Cruz Biotechnology, Inc., Santa Cruz, CA)], ER-Golgi intermediate compartment [rabbit anti-ERGIC-53 (1:250; Sigma-Aldrich)], or Golgi apparatus [rabbit anti-giantin (1:500; Abcam, Cambridge, MA)] was then performed as previously described (4). Alexa Fluor 488 conjugates (1:500) were used for detection of these markers. Finally, cells were washed three times with PBS, and quinolinium {4-[3-(3-methyl-2(3H)-benzothiazolylidene)-1-propenyl]-1-[3-(trimethylammonio)propyl]diiodide (TO-PRO-3; Life Technologies)} was added for nucleus staining. The cells were then washed extensively with PBS and mounted on slides using Pro-Long Gold antifade mounting reagent. Mounted slides were observed with a Zeiss Axiovert 100 M microscope equipped with a Carl Zeiss LSM 510 META confocal system and Zeiss 63× PlanAPO/1.4NA oil immersion objective (Carl Zeiss, Germany) in multitrack mode. For EGFP imaging, cells were excited using a 488-nm laser line, and fluorescence emission spectra were collected between 500 and 530 nm. For excitation of RFP, a 543-nm laser line was used, and fluorescence emission spectra were collected between 570 and 620 nm. For excitation of TO-PRO-3, a 633-nm laser line was used, and emission spectra were collected between 640 and 680 nm. The pinhole aperture was set to obtain an equal focal-plane thickness for all three colors and as close as possible to 1 Airy unit. Pixel time was set at 3.20 μs, and the results of four scans were averaged for each slice. z stack slices were set to 0.25 μm. Collected images were processed using LSM 510 Image v3.2 SP2 (Carl Zeiss, Inc., Thornwood, NY) and saved as 12-bit tagged TIFF images in RGB-format MDB files. All observations were carried out in the Integrated Microscopy Core Laboratory, Baylor College of Medicine, Houston, TX.
Membrane protein fractionation and Endo H assay.
MA104 cells expressing RFP-tagged NSP4 from RVA strains SA11 and Ty-1, RVB, and RVC were harvested by treatment with PBS plus 5 mM ethylene glycol tetraacetic acid (EGTA) on ice for 10 min. Cells were collected and lysed by sonication for 30 s. Membranes were pelleted by centrifugation (100,000 × g, 2 h). The supernatant containing soluble proteins (S) was collected, and the membrane pellet (M) was solubilized in PBS plus 1% SDS. Aliquots of M were treated with endoglycosidase H (Endo H) for 2 h at 37°C to remove N-linked glycans. Protein samples were then analyzed by SDS-PAGE followed by immunoblotting.
Computational and statistical analysis.
Full-length NSP4 amino acid sequences from 80 RVA, 10 RVB, 14 RVC, 1 RVD, and 2 ungrouped RV strains (data not shown) were aligned using the ClustalW software program (56). Helical wheel diagrams and amphipathic-moment calculations were generated using the WinPep software program (25). The amphipathic moment is a vector calculation that represents the mean hydrophobicity of side chains of a helix (16). All experiments were performed in triplicate. Statistical significance was determined by the t test, with a P value of <0.05 being considered significant.
RESULTS
Lack of viroporin domain homology among non-RVA NSP4s.
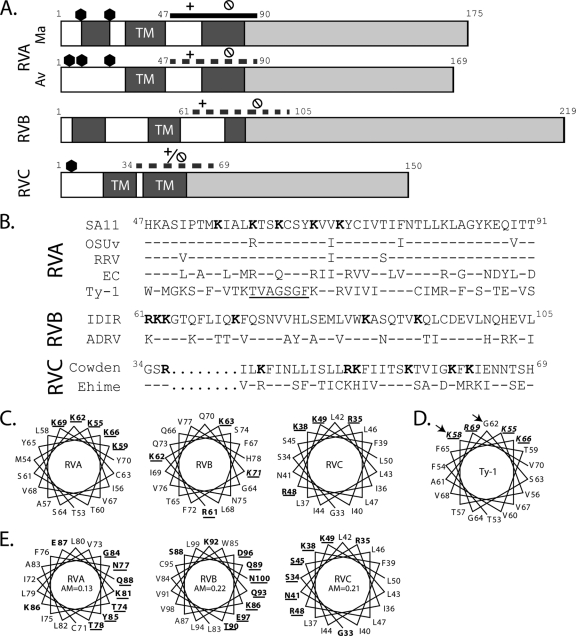
We recently showed that RVA NSP4 has a viroporin domain (aa 47 to 90) that elevates [Ca2+]cyto (29). Since NSP4-mediated elevation of [Ca2+]cyto is necessary for RV replication and assembly, and since non-RVA strains appear to utilize a generally similar morphological assembly pathway, we predicted that non-RVA NSP4s also have a viroporin domain. We first performed a ClustalW alignment of NSP4 sequences from 107 different RV strains (data not shown). NSP4 genes clustered with respect to their serogroup, though RVA avian NSP4s diverged substantially from mammalian RVA NSP4s, and the ungrouped J19 and B219 RV strains clustered most closely with RVB strains, as previously observed (37, 45). Based on the ClustalW alignment, the RVA viroporin domain (aa 47 to 90) aligned with the C-terminal-most hydrophobic domains of RVB and RVC NSP4s (Fig. 1A). We then analyzed the amino acid sequences of these domains for the characteristic viroporin motifs (Table 1).
Fig 1.
Identification of putative NSP4 viroporin domains. (A) Linear schematic of full-length mammalian (Ma) and avian (Av) RVA NSP4, RVB NSP4, and RVC NSP4. Numbers represent amino acid positions. Predicted glycosylation sites are indicated with hexagons, hydrophobic domains are in dark gray with predicted transmembrane domains indicated (TM), and the hydrophilic C-terminal tail is shown in light gray. The previously mapped viroporin domain of Ma RVA NSP4 (aa 47 to 90; solid line) shows the relative location of the polybasic domain (+) and the amphipathic domain ( ). Putative viroporin domains for Av RVA NSP4 (aa 47 to 90), RVB NSP4 (aa 61 to 105), and RVC NSP4 (aa 34 to 69) are shown with dashed lines with the identified polybasic (+) and amphipathic (
). Putative viroporin domains for Av RVA NSP4 (aa 47 to 90), RVB NSP4 (aa 61 to 105), and RVC NSP4 (aa 34 to 69) are shown with dashed lines with the identified polybasic (+) and amphipathic ( ) domains indicated. (B) ClustalW alignment of the rotavirus NSP4 viroporin domain from RVA strains SA11 (genotype E2), OSUv (genotype E1), RRV (genotype E3), EC (genotype E7), and Ty-1 (genotype E4); RVB strains IDIR and ADRV; and RVC strains Cowden and Ehime. Superscript numerals are the amino acid numbers in the full-length sequence. Conserved lysine (K) and arginine (R) residues are in bold in the SA11 sequence. The underlined segment of Ty-1 highlights the disruption in the helical arrangement of basic residues. Conservation of residues is based on SA11 for RVA, IDIR for RVB, and Cowden for RVC. Conserved residues are represented by dashes, and gaps are represented by dots. (C) Helical wheel diagrams of highly basic domains of RVA (SA11), RVB (IDIR), and RVC (Cowden). Conserved lysine (K) or arginine (R) residues are in bold and underlined. (D) Helical wheel diagram of Ty-1 NSP4 aa 53 to 70. The amino acids in the basic residue cluster that differ from SA11 (aa 58 and 62) are indicated by arrows. (E) Helical wheel diagrams of the RVA (SA11), RVB (IDIR), and RVC (Cowden) amphipathic α-helices. Charged and polar residues are in bold, and polar face residues are underlined. AM, amphipathic moment.
) domains indicated. (B) ClustalW alignment of the rotavirus NSP4 viroporin domain from RVA strains SA11 (genotype E2), OSUv (genotype E1), RRV (genotype E3), EC (genotype E7), and Ty-1 (genotype E4); RVB strains IDIR and ADRV; and RVC strains Cowden and Ehime. Superscript numerals are the amino acid numbers in the full-length sequence. Conserved lysine (K) and arginine (R) residues are in bold in the SA11 sequence. The underlined segment of Ty-1 highlights the disruption in the helical arrangement of basic residues. Conservation of residues is based on SA11 for RVA, IDIR for RVB, and Cowden for RVC. Conserved residues are represented by dashes, and gaps are represented by dots. (C) Helical wheel diagrams of highly basic domains of RVA (SA11), RVB (IDIR), and RVC (Cowden). Conserved lysine (K) or arginine (R) residues are in bold and underlined. (D) Helical wheel diagram of Ty-1 NSP4 aa 53 to 70. The amino acids in the basic residue cluster that differ from SA11 (aa 58 and 62) are indicated by arrows. (E) Helical wheel diagrams of the RVA (SA11), RVB (IDIR), and RVC (Cowden) amphipathic α-helices. Charged and polar residues are in bold, and polar face residues are underlined. AM, amphipathic moment.
Table 1.
Characteristics of RV NSP4 proteinsa
| Group | Length (aa) of: |
|||
|---|---|---|---|---|
| Full protein | Hydrophobic domainsb | Putative viroporin domain | Hydrophilic tail | |
| RVA | 175 | 3 (9–21; 29–46; 66–83) | 47–90 | 84–175 |
| RVA (avian) | 169 | 2 (29–46; 66–83) | 47–90 | 84–169 |
| RVB | 218–219 | 3 (6–20; 41–55; 76–84) | 61–105 | 106–219 |
| RVC | 150 | 2 (19–33; 36–55) | 33–69 | 70–150 |
| RVD | 127 | 3 (9–19; 29–46; 52–70) | 42–85 | 71–127 |
| Ungrouped | 213 | 3 (33–41; 51–72; 74–98) | 66–106 | 99–213 |
Summary of NSP4 domain organization of known serogroups.
Underlining indicates predicted transmembrane domains.
Among the representative RVA sequences selected, there was more than 90% identity between SA11, OSU, and RRV, and the two critical viroporin motifs [the clustered basic residues from aa 55 to 70 (pentalysine domain) and the amphipathic α-helix from aa 71 to 88 (amphipathic domain)] identified in SA11 NSP4 were fully conserved (29). In contrast, NSP4s from the murine strain EC and avian strain Ty-1 were further divergent, having only 53% and 29% identity to SA11, respectively (Table 2). Despite the greater divergence, the EC viroporin motifs were essentially conserved. For Ty-1, the amphipathic domain was conserved, but a number of changes in the pentalysine domain (relative to SA11 NSP4) were noted. The conserved K58 is shifted N-terminally by one residue relative to SA11, and there is a K-to-G substitution at aa 62. These two changes create a gap between conserved K residues that disrupts the helical organization of the positive charge (Fig. 1B and D). Since the SA11 pentalysine domain (PD) is critical for viroporin activity, these mutations might affect viroporin activity (29).
Table 2.
Sequence identity of RVA viroporin domains
| Strain | % identity with: |
||||
|---|---|---|---|---|---|
| SA11 | OSU | RRV | EC | Ty-1 | |
| SA11 | 100 | ||||
| OSU | 91 | 100 | |||
| RRV | 93 | 89 | 100 | ||
| EC | 53 | 58 | 53 | 100 | |
| Ty-1 | 29 | 22 | 24 | 18 | 100 |
RVB and RVC NSP4s also have motifs characteristic of viroporins. We used helical wheel diagrams to look at the orientation of the basic residues within the RVB and RVC putative viroporin domains. In RVB NSP4, the basic residues are linearly arranged rather than being aligned on one face of the α-helix, as in RVA NSP4 (Fig. 1B and C). In RVC NSP4, the basic residues are dispersed throughout the putative viroporin domain, though Arg35 and Lys38 are adjacent on the α-helix (Fig. 1C). While the organization of the basic residues differs substantially across the different groups, all NSP4 sequences examined had a strong amphipathic α-helical domain. NSP4s from all RVA strains had comparable amphipathic domains, with amphipathic moments of ∼0.13 (Fig. 1E). The RVB NSP4 amphipathic α-helix extended from aa 83 to 100 and had an amphipathic moment of 0.22. In contrast, the RVC amphipathic α-helix, which had an amphipathic moment of 0.21, was considerably longer (extending from aa 33 to 63) and contained several basic residues (Fig. 1E). Finally, we analyzed the RVB and RVC sequences for polybasic and amphipathic domains outside the putative viroporin domains identified through the ClustalW alignment, but no other domains of either protein satisfied these criteria (data not shown). Thus, despite substantial divergence in the primary sequence, characteristic viroporin structural motifs were found in the C-terminal-most hydrophobic domain of non-RVA NSP4s. However, the organization of the viroporin motifs, particularly the arrangement of basic residues, was not conserved. Since the polybasic motif was previously shown to be critical for RVA NSP4 viroporin activity, these dramatic differences raised the possibility that viroporin activity was not a feature of all NSP4s. So, using both E. coli and mammalian cell model systems, we looked for NSP4 viroporin activity in representative RVA, RVB, and RVC NSP4s [RVA (SA11), RVB (IDIR), and RVC (Cowden)]. Additionally, we tested for differences in NSP4 viroporin activity between mammalian RVA NSP4 (SA11) and the divergent avian RVA NSP4 from Ty-1 RV.
E. coli lysis assay.
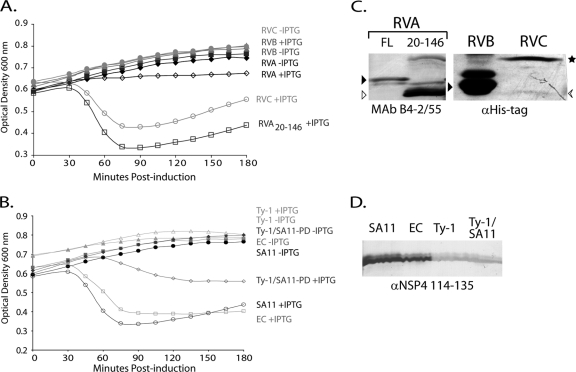
We recently showed that SA11 NSP4 causes viroporin-domain dependent lysis of BL21(DE3)pLysS cells. Previous studies showed that expression of full-length NSP4 or truncated forms that include the N-terminal hydrophobic domains results in no or low protein yield (6, 33), and this is likely due to rapid viroporin-induced death of NSP4-expressing cells (29). Thus, we first compared the lysis activity of full-length RVA, RVB, and RVC NSP4s. In the absence of IPTG, the NSP4 gene-containing E. coli cells grow normally (Fig. 2A). In contrast, expression of full-length RVA NSP4 (aa 1 to 175) appeared to kill the cells in the absence of lysis, as the optical density remained at the preinduced level (Fig. 2A). The lack of cell lysis upon expression of full-length RVA NSP4 was puzzling, because we previously observed rapid cell lysis using RVA NSP4 truncations. We then tested forms of RVA with both N- and C-terminal truncations for E. coli lysis. Truncation of the C terminus, RVA NSP4 aa 1 to 146, killed the cells in the absence of cell lysis, similar to full-length RVA (data not show). Truncation of both the N and C termini, RVA NSP4 aa 20 to 146, supported rapid E. coli lysis, as was seen in previous reports (Fig. 2A) (6, 29). We harvested cells at 3 h postinduction to assess protein expression, and immunoblot analysis of both RVA NSP4 full-length and the aa 20-146 truncation product showed very low protein expression levels for full-length NSP4 and substantially more expression of the truncated protein (Fig. 2C, left).
Fig 2.
NSP4 E. coli lysis activity is not conserved across rotavirus groups. (A) Full-length NSP4 from RVA, RVB, and RVC rotaviruses were tested for activity in an E. coli lysis assay. The optical densities at 600 nm of uninduced (closed symbols) or NSP4-expressing (open symbols) cultures were determined at 15-min intervals for 180 min. Black diamonds, RVA (SA11) NSP4; gray triangles, RVB (IDIR); light gray circles, RVC (Cowden); and dark gray squares, RVA aa 20-146 truncation. (B) E. coli lysis activity of SA11 NSP4 aa 20-146 (black circles) was compared to that of murine rotavirus EC (gray squares) and the avian rotavirus Ty-1 (light gray triangles). A chimeric Ty-1/SA11 construct was also tested (Ty-1/SA11-PD, dark gray diamonds) that replaced the Ty-1 NSP4 pentalysine domain (aa 47 to 72) with the SA11 NSP4 sequence. The optical densities at 600 nm of uninduced (closed symbols) and NSP4-expressing (open symbols) cultures were measured as described above. In all cases, the values are the mean from three independent experiments. (C and D) Immunoblots of NSP4 expressed in BL21(DE3)pLysS cells. (C, left) Full-length RVA NSP4 (FL; black arrowhead) and the aa 20-146 truncated NSP4 (white arrowhead) were detected with NSP4 MAb B4-2/55. (C, right) RVB (black arrowhead) and RVC NSP4 [arrow (monomer) and star (oligomer)] were detected anti-His-tag antisera. (D) NSP4s with aa 20-146 truncations from SA11, EC, and Ty-1 and the Ty-1/SA11 pentalysine domain (PD) chimera were detected with anti-NSP4 114-135 antisera.
Expression of full-length RVB NSP4 had no effect on the E. coli growth kinetics, did not induce cell lysis (Fig. 2A), and occurred at very high levels (Fig. 2C, right). In contrast, expression of full-length RVC NSP4 caused rapid cell lysis, similar to expression of the RVA with the aa 20-146 truncation (Fig. 2A). RVC NSP4 was expressed at very low levels and was difficult to detect using the anti-His-tag antibody, with the SDS-resistant dimer being more stable than the monomer (Fig. 2C, right).
We next sought to determine whether NSP4 viroporin activity was conserved among different RVA strains. Because the aa 20-146 truncation supported cell lysis in this assay, we generated similar truncations for several different RVA strains. We found that OSU and RRV NSP4s both induced lysis similarly to SA11 (data not shown). This result was expected, given the high degree of sequence conservation between NSP4 genotypes E1, E2, and E3. EC NSP4 also induced cell lysis similarly to SA11, but Ty-1 NSP4 did not induce cell lysis (Fig. 2B). We hypothesized this was due to the disrupted helical orientation of conserved lysine and/or arginine residues in Ty-1 NSP4 between aa 55 and 69 (Fig. 1B and D), which are important for lysis activity (29). To test this, we made a chimeric Ty-1 NSP4, in which we inserted the SA11 NSP4 pentalysine domain (PD; aa 47 to 72) in place of the Ty-1 sequence. The Ty-1/SA11-PD chimera induced lysis, albeit to a lesser extent than SA11 (Fig. 2B), indicating that the cluster of conserved lysine residues on a single helical face is important for NSP4-mediated cell lysis via membrane permeabilization. Immunoblot analysis showed good expression levels for all of the truncation constructs. Detection of Ty-1 NSP4 by the anti-NSP4 aa 114-135 peptide antibody was lower than that of EC NSP4, due to lack of conservation of this epitope between murine and avian NSP4s (Fig. 2D) (31). The anti-His-tag antibody detected Ty-1 NSP4 better than either SA11 or EC NSP4 (data not shown). Using the lysis assay, we demonstrated that NSP4s from mammalian RVA strains all have similar viroporin activity, and therefore we subsequently used SA11 NSP4 as the representative of these proteins. Additionally, RVC NSP4 had viroporin activity that was similar to that of RVA NSP4, but avian RVA and RVB NSP4s did not support E. coli lysis, initially suggesting that these NSP4s may lack viroporin activity.
E. coli cytotoxicity of NSP4.
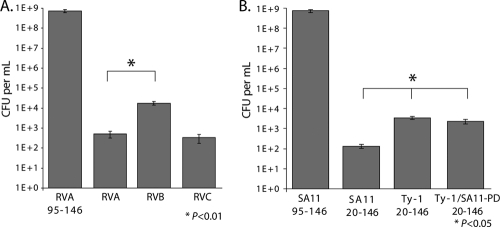
Lysis of BL21(DE3)pLysS cells is mediated by T7 phage lysozyme and not by NSP4 itself; however, expression of NSP4 is cytotoxic to E. coli in a viroporin domain-dependent manner. Viroporin mutants that fail to cause lysis of BL21(DE3)pLysS cells remain significantly cytotoxic even in the absence of lysozyme, which indicates that E. coli cell viability is a more sensitive measure of intermediate viroporin activity than the lysis assay (29). Thus, we determined the number of viable BL21(DE3) E. coli cells after plating cultures in the absence or presence of IPTG (Fig. 3). In the absence of IPTG, all cultures had 108 to 109 CFU/ml (data not shown). In the presence of IPTG, cells with the negative-control RVA NSP4 containing aa 95 to 146 had high viability, with 7.3 × 108 CFU per ml. Expression of full-length NSP4s from RVA, RVB, and RVC strains significantly reduced cell viability (Fig. 3A); thus, the RVB NSP4 phenotype was consistent with possible viroporin activity even though it did not induce cell lysis (Fig. 2). Nevertheless, cells expressing RVB NSP4 had significantly greater viability than cells expressing either RVA or RVC NSP4, which may account for the greater expression of RVB NSP4 (Fig. 2C). We also compared the truncated mammalian RVA NSP4 (SA11) to both the wild-type avian RVA NSP4 (Ty-1) and the Ty-1/SA11-PD chimera described above (Fig. 3B). Expression of all of the NSP4s caused a significant decrease in cell viability; however, cells expressing the wild-type or chimeric Ty-1 NSP4 had significantly higher viability than cells expressing SA11 NSP4. These data indicated that all of the NSP4s tested significantly reduced E. coli viability (P < 0.01), which is consistent with their having viroporin-like activity, as previously demonstrated with SA11 NSP4 (29). Since RVB and Ty-1 NSP4s were expressed at a higher level than SA11 RVA and RVC NSP4s but lacked lysis activity and were the least cytotoxic in E. coli, the data suggest a weaker viroporin activity of these NSP4s in bacteria. Thus, while no lysis activity was detected (Fig. 2), the nearly 5-log decrease in cell viability demonstrated that NSP4s that do not promote lysozyme-mediated cell lysis are still extremely cytotoxic to E. coli, and that, as seen previously, the E. coli cytotoxicity assay is a more sensitive measure of NSP4 viroporin activity than the T7 lysozyme-mediated-lysis assay (29).
Fig 3.
NSP4 Cytotoxicity in E. coli is conserved across rotavirus groups. Tenfold serial dilutions of BL21(DE3) E. coli bearing expression plasmids for the indicated NSP4 were plated on LB-Amp supplemented with 1 mM IPTG and incubated overnight at 37°C. Error bars represent standard deviations. (A) Full-length RVA, RVB, and RVC NSP4s expressed in E. coli compared to the noncytotoxic RVA NSP4 with the aa 95–146 truncation. (B) SA11 and Ty-1 aa 20–146 truncation constructs and the Ty-1/SA11-PD chimera, expressed in E. coli. *, P < 0.05.
Redox Sensor Green assay in E. coli.
The results of the lysis and cytotoxicity assays suggested that NSP4-mediated E. coli cytotoxicity may be due to disruptions of ion gradients across the inner membrane. E. coli maintains a proton gradient across the inner membrane, and this electrochemical gradient is used to generate ATP and is critical for cell viability and vitality. Establishment of the proton gradient is directly linked to cellular oxidation and reduction activity, as many oxidases and reductases are inner membrane proton pumps (53). Therefore, the redox state of cells is an informative measure of general cell vitality and whether the proton gradient is intact. Chemicals that uncouple the proton gradient, such as the proton ionophore carbonyl cyanide 3-chlorophenylhydrazone (CCCP), increase the reductive activity of E. coli cells (21, 53).
To determine whether NSP4 viroporin activity alters the redox state of bacteria, we stained uninduced cells and cells induced to express the different NSP4s with Redox Sensor Green (RSG), a new redox-sensitive dye used to measure bacterial vitality (21). In these experiments, we used E. coli BL21(DE3), which does not express the T7 phage lysozyme gene and does not lyse in response to NSP4 viroporin activity (29). Mid-log-phase cultures were split evenly; one half was untreated, and the other half was treated with IPTG to induce NSP4 expression. At various times postinduction, aliquots of each culture were stained with RSG and PI, a red fluorescent DNA binding dye used as a general indicator of membrane integrity. The stained cells were analyzed by flow cytometry for PI staining and the level of RSG fluorescence. First, we showed that expression of SA11 NSP4 aa 47 to 146, but not the viroporin-deficient ASDASA mutant, increased RSG fluorescence, indicating that NSP4 increased RSG fluorescence in a viroporin activity-dependent manner (Fig. 4) (29). For all of the tested NSP4s, uninduced cells were all RSG positive and essentially PI negative (Fig. 5A). Expression of the negative control RVA NSP4 aa 95 to 146, which lacks the viroporin domain, did not significantly alter RSG fluorescence (Fig. 5D) or increase PI staining (data not shown) at 1 h postinduction (HPI).
Fig 4.
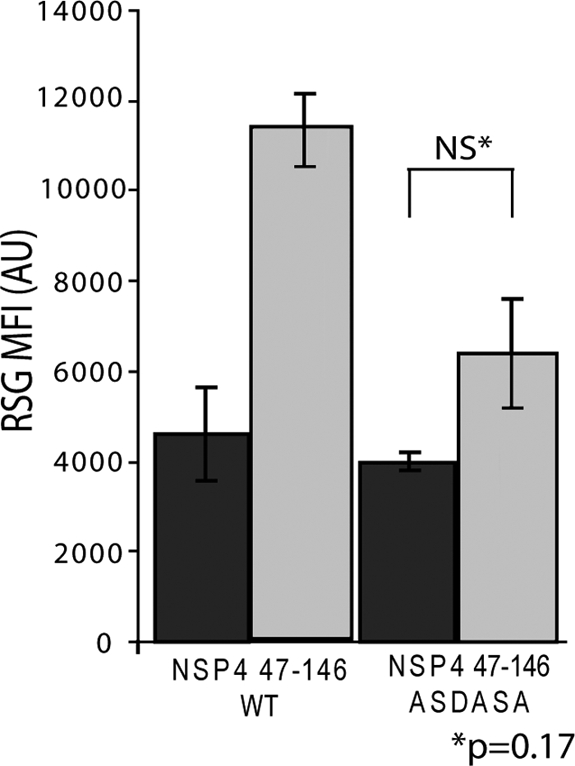
Enhanced Redox Sensor Green staining is dependent on viroporin activity. Redox Sensor Green (RSG) staining of BL21(DE3) E. coli bearing expression vectors encoding either wild-type NSP4 aa 47–146 or the viroporin-deficient mutant NSP4 aa 47–146 ASDASA, in which residues 75 to 80 are mutated from IFNTLL to ASDASA. Cells were left untreated (dark gray), or NSP4 expression was induced with 1 mM IPTG (light gray). At 1 h postinduction, cells were stained with RSG for 10 min at 37°C and then analyzed by flow cytometry for the RSG mean fluorescence intensity (MFI). Samples were tested in triplicate, and error bars represent standard deviations. There was no statistically significant difference in RSG staining between uninduced and induced cells expressing the ASDASA viroporin mutant.
We next tested the redox state of cells expressing NSP4 from the different RV groups. We found that expression of RVA, RVB, and RVC NSP4s significantly increased RSG fluorescence (Fig. 5B to E), suggesting that all the NSP4s uncoupled the inner membrane proton gradient similarly to CCCP. We then compared RSG staining between mammalian RVA and avian RVA NSP4. Both the full-length protein and the aa 20-146 truncation of mammalian RVA NSP4 significantly increased RSG fluorescence; however, full-length RVA stimulated a much greater increase in RSG signal than the truncation construct (Fig. 5D), despite the expression level being substantially lower (Fig. 2C). Finally, avian RVA NSP4 also significantly elevated RSG fluorescence, but to a lesser extent than mammalian RVA NSP4 (Fig. 5D).
RVA and RVC NSP4 expression also increased PI staining (Fig. 5B and data not shown), but RVB NSP4-expressing cells remained PI negative (Fig. 5C). There were also differences in the kinetics of the increase in RSG fluorescence between groups. RVA and RVC NSP4 stimulated the increase in RSG by 1 HPI (Fig. 5D), and the increase continued until 3 HPI for RVA NSP4 (Fig. 5E). In contrast, there was no difference in RSG staining of RVB NSP4-expressing cells at 1 HPI (Fig. 5D), but by 2 to 3 HPI, there was a significant increase in RSG staining (Fig. 5E). These data indicate that regardless of their genetic divergence, NSP4s from the different groups all collapsed the E. coli proton gradient and significantly disrupted the cellular redox state. As with the other bacterium-based viroporin assays, Ty-1 and RVB NSP4s showed a more attenuated phenotype than RVA or RVC NSP4; however, both Ty-1 and RVB NSP4s had significant viroporin-like activity. Together, the results of the bacterial assays support our prediction that viroporin activity is a common NSP4 function.
Molecular characterization of RVA, RVB and RVC NSP4 in mammalian cells.
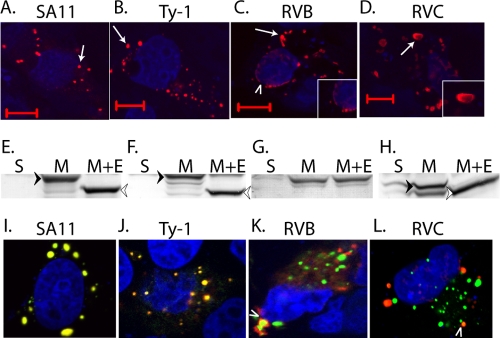
The E. coli-based assays represent classical and rapid tests for viroporin-like activity of proteins, but ultimately viroporin activity has to be shown by expression of the protein in a relevant mammalian cell. We have shown that mammalian RVA (SA11) NSP4 localizes to the ER, the primary Ca2+ storage organelle, and a viroporin-mediated elevation in [Ca2+]cyto triggers the formation of punctate structures (29). Formation of punctate structures by mammalian RVA NSP4 is seen both in RV-infected cells and in vector-based expression of recombinant NSP4 (4, 12). Therefore, we next examined avian RVA, RVB, and RVC NSP4s for (i) their ability to form punctate structures, (ii) membrane localization, and (iii) the presence of N-linked glycosylation. We C-terminally tagged full-length mammalian (SA11) and avian (Ty-1) RVA, RVB, and RVC NSP4s with TagRFP and expressed these proteins in MA104 cells by transient transfection. We first examined the subcellular distribution of the different NSP4s for the formation of punctate structures. Two pools of NSP4 were observed in the cells: a diffuse reticular pattern consistent with ER localization and punctate structures (Fig. 6). RVA NSP4 from SA11 and Ty-1 NSP4 formed discrete round puncta (Fig. 6A and B), but the puncta from both RVB and RVC NSP4 had slightly different morphologies. RVB NSP4 showed stronger perinuclear localization (arrowhead) and elongated puncta (Fig. 6C), whereas RVC NSP4 exhibited a patchy distribution with large disc-like puncta (Fig. 6D). We confirmed membrane localization of all of the NSP4s by preparing membrane protein fractions and treating them with Endo H, which removes ER-derived N-linked glycans. These fractions were analyzed by immunoblotting for RFP-tagged NSP4 (Fig. 6E to H). RVA SA11 NSP4 was found completely in the membrane protein fraction and Endo H treatment resulted in a faster-migrating, unglycosylated species (Fig. 6E). A similar pattern was observed for RVA Ty-1 and RVC NSP4, with three glycosylated species being observed for Ty-1 NSP4 (Fig. 6F) and only one being observed for RVC NSP4 (Fig. 6H). The observed glycosylation patterns are consistent with the N-linked glycosylation motifs present in these proteins (27, 44). RVB was also exclusively membrane localized, but Endo H treatment did not shift the electrophoretic mobility (Fig. 6G), indicating that RVB NSP4 was not modified by N-linked glycans, as previously predicted (23).
Fig 6.
NSP4 forms serogroup-specific puncta that colocalize with EGFP-LC3. RFP-tagged NSP4s from RVA SA11 (A), RVA Ty-1 (B), RVB (C), and RVC (D) strains were expressed in MA104 cells and examined by confocal microscopy for their ability to form punctate structures (arrows). RVB NSP4 also showed prominent perinuclear localization (C, arrowhead), which is shown in detail in the inset. (D) An RVC NSP4 disc-like punctum is shown in detail in the inset. Bars, 10 μm. (E to H) Soluble (S) and membrane (M) protein fractions or membrane proteins treated with Endo H (M+E) were prepared from cells expressing RVA SA11 (E), RVA Ty-1 (F), RVB (G), and RVC NSP4-RFP (H). Samples were analyzed by immunoblotting using an RFP-specific antibody. Fully glycosylated (black arrowhead) and nonglycosylated (white arrowhead) species are indicated. (I to L) Coexpression of RVA SA11 NSP4-EGFP and RFP-tagged NSP4 from RVA SA11 (I), RVA Ty-1 (J), RVB (K), and RVC (L). Only a small subset of RVA and RVC puncta colocalize (L, arrowhead). For confocal microscopy, TO-PRO-3 was used to stain nuclei, which is pseudocolored blue.
Since the punctum morphologies were different, we coexpressed RVA NSP4 with either RVB or RVC NSP4 to determine whether the puncta would colocalize or form distinct structures. EGFP-tagged RVA SA11 NSP4 was transfected into MA104 cells with RFP-tagged Ty-1 RVA, RVB, or RVC NSP4, and confocal microscopy was performed 24 h posttransfection. Coexpressed SA11 NSP4-EGFP and SA11 NSP4-RFP completely colocalized (Fig. 6I), as did coexpressed SA11 NSP4-EGFP and Ty-1 NSP4-RFP (Fig. 6J). In contrast, most RVB and RVC NSP4-RFP did not colocalize with SA11 NSP4-EGFP but formed distinct punctate structures (Fig. 6K and L), with very few puncta showing colocalization (Fig. 6K and L).
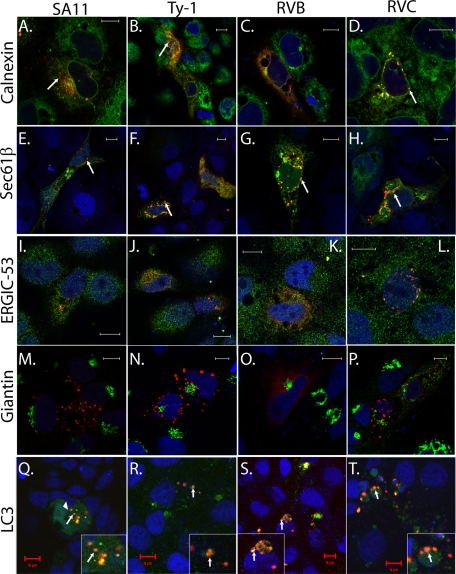
The above data suggest that RVB and RVC NSP4 may traffic differently than RVA NSP4. Studies have shown that mammalian RVA NSP4 (including SA11) localizes to the ER and the ER-Golgi intermediate compartment (ERGIC) but does not transverse the Golgi apparatus (4). RVA NSP4 punctate structures colocalize with the autophagy protein LC3, and in RV-infected cells these structures appear to cap viroplasms (4). Using markers for ER, ERGIC, Golgi, and autophagosomes, we examined the subcellular localization of each different NSP4 protein (Fig. 7). We first looked at ER localization by immunofluorescence for calnexin and EGFP-tagged Sec61β, two ER transmembrane proteins that detect the reticular cisternae of the ER (60). Both SA11 (Fig. 7A and E) and Ty-1 NSP4s (Fig. 7B and F) partially localized to the reticular ER; however, the punctate structures showed much less colocalization with either calnexin or Sec61β (arrows). RVB (Fig. 7C and G) and RVC (Fig. 7D and H) NSP4 also colocalized with the calnexin and Sec61β ER markers in the reticular pools but showed greater colocalization with these ER markers in the punctate structures as well. We previously reported that a distinct pool of SA11 NSP4 localizes to ERGIC in human embryonic kidney (HEK) 293 cells. We confirmed that observation here and showed that Ty-1, RVB, and RVC NSP4s also show some limited colocalization with ERGIC-53, a marker for ERGIC (Fig. 7I to L). Finally, we looked for the NSP4 in the Golgi apparatus, using giantin as a marker (4). Neither the reticular nor the punctate pools of SA11 RVA (Fig. 7M), Ty-1 RVA (Fig. 7N), RVB (Fig. 7O), or RVC (Fig. 7P) localized to the Golgi. This is consistent with the carbohydrate moieties on RVA and RVC NSP4 remaining sensitive to Endo H (Fig. 6E, F, and H).
Fig 7.
Subcellular localization of NSP4 using organelle-specific markers. Markers for the endoplasmic reticulum [calnexin (A to D) and Sec61β (E to H)], ER-Golgi intermediate compartment [ERGIC-53 (I to L)], Golgi [giantin (M to P)], and autophagosomes [LC3 (Q to T)] were used to examine the subcellular localization of RFP-tagged SA11 (A, E, I, M, and Q), Ty-1 (B, F, J, N, and R), RVB (C, G, K, O, and S), or RVC NSP4 (D, H, L, P, and T). Calnexin, ERGIC-53, and giantin were detected by indirect immunofluorescence as described in Materials and Methods. Sec61β and LC3 were tagged with EGFP. For confocal microscopy, TO-PRO-3 was used to stain nuclei, which is pseudocolored blue. Bars, 10 μm.
The punctate structures formed by SA11 NSP4 are induced by elevated [Ca2+]cyto and colocalize with the autophagy marker LC3, a soluble cytosolic protein that forms punctate structures upon induction of autophagy. To determine whether the different NSP4s also induced and colocalized with LC3 puncta, we coexpressed EGFP-LC3 with NSP4-RFP. Much EGFP-LC3 remained cytoplasmic and did not colocalize with NSP4 (Fig. 7Q). As previously reported, SA11 RVA NSP4 induced formation of LC3 puncta that colocalized with NSP4 puncta (Fig. 7Q). Ty-1 RVA NSP4s also induced LC3 puncta that colocalized with NSP4 (Fig. 7R). RVB and RVC NSP4s both strongly colocalized with EGFP-LC3 punctate structures (Fig. 7S and T). Although the morphologies of the NSP4/LC3 puncta were different among RVA, RVB, and RVC NSP4s, they were consistent with the differences in the NSP4 puncta described above (Fig. 6A to D).
Elevation of cytoplasmic calcium by NSP4 in mammalian cells.
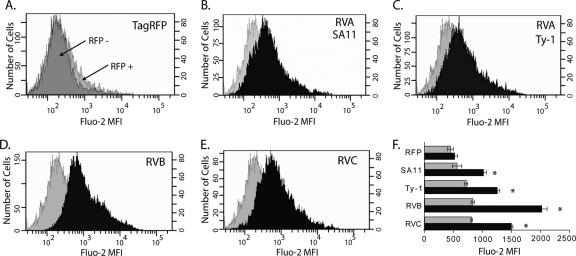
For SA11 NSP4, viroporin-mediated elevation of [Ca2+]cyto triggers the movement of SA11 NSP4 from the ER into punctate structures (4, 29). Therefore, the localization of each of the NSP4s to the ER and their ability to form punctate structures suggested that they all would disrupt in Ca2+ homeostasis. While early studies using Sf9 insect cells showed that both RVA SA11 and RVB IDIR NSP4 both elevate [Ca2+]cyto levels, there has been little molecular characterization of the intracellular functions of non-serogroup A rotavirus NSP4 in mammalian cells (58). Since NSP4s from RVA, RVB, and RVC strains all showed evidence of viroporin activity in E. coli, we predicted that all of the NSP4s would elevate [Ca2+]cyto. To detect the levels of free Ca2+ in the cytoplasm of mammalian cells expressing these NSP4s, we expressed the different RFP-tagged NSP4s in MA104 cells by transient transfection and then loaded the cells with the green fluorescent Ca2+ indicator Fluo-2. We then used flow cytometry to measure Ca2+ in NSP4-expressing and nonexpressing cells. The RFP-positive population was gated, and the mean fluorescence intensity (MFI) of Fluo-2 was determined. Expression of the TagRFP protein alone was used as the negative control, and the RFP-positive and -negative populations had similar distributions of Fluo-2 fluorescence (Fig. 8A).
Fig 8.
Elevation of cytoplasmic calcium by NSP4 from RVA, RVB, and RVC. (A) Population histograms for TagRFP-negative (dark gray) and TagRFP-positive (light gray) cells. (B to E) Representative population histogram of TagRFP-positive cells (gray) compared to a representative population histogram of NSP4-RFP-positive cells (black) for NSP4 from the indicated rotavirus. The x axis shows the mean fluorescent intensity of the Ca2+ indicator Fluo-2. The left and right y axes show the number of cells for the NSP4-RFP-positive and TagRFP-positive populations, respectively. (F) The Fluo-2 MFIs of RFP-negative (gray) and RFP-positive (black) populations from three independent experiments are graphed. Error bars represent standard deviations. *, P < 0.01 for TagRFP-positive versus individual NSP4-RFP-positive populations.
SA11 NSP4 is known to significantly elevate [Ca2+]cyto, and SA11 NSP4-RFP was therefore used as a positive control (5, 29). The SA11 NSP4-RFP-positive population had a higher Fluo-2 MFI than the negative-control TagRFP-positive population, as indicated by the right-shifted histogram (Fig. 8B). Similar increases in the Fluo-2 MFI were observed for the RFP-tagged avian Ty-1 RVA NSP4 (Fig. 8C). Further, RVB and RVC NSP4 also increased the intracellular Ca2+ level (Fig. 8D and E). Quantitation of the Fluo-2 MFI for each construct showed that TagRFP alone did not cause a significant difference in the Fluo-2 MFI between the RFP-positive and -negative populations (P > 0.4). In contrast, all of the NSP4-RFP constructs significantly elevated [Ca2+]cyto (P < 0.01). Curiously, RVB NSP4 stimulated the greatest increase in [Ca2+]cyto, which was significantly greater than RVA and RVC NSP4 (P < 0.05), even though in bacteria RVB NSP4 appeared to have a less robust viroporin activity than RVA NSP4. Elevation of [Ca2+]cyto by RVC NSP4 was also significantly higher than that caused by both RVA NSP4s (P < 0.05). Finally, the RFP-negative population of NSP4-transfected samples exhibited a significantly higher Fluo-2 MFI than the RFP-negative population in the TagRFP negative-control-transfected cells (P < 0.05). This was likely because cells had NSP4-RFP expression that was below the level of detection but had elevated [Ca2+]cyto. Together, these data showed that NSP4s from RVA, RVB, and RVC all function to elevate [Ca2+]cyto, which is consistent with the observed viroporin activity in E. coli. However, the extent of NSP4-mediated [Ca2+]cyto elevation differed significantly between rotaviruses from different serogroups, underlining possible functional differences as a result of genetic divergence.
DISCUSSION
The goal of this study was to determine whether divergent NSP4 genes from RV serogroups A, B, and C retained viroporin-like activity despite dramatic sequence divergence. Of rotavirus proteins, NSP4 shows the least sequence homology across serogroups, and in most cases the amino acid sequence identity between groups falls well below 20% (23, 41, 55, 59). For RVA strains, the NSP4-mediated elevation in [Ca2+]cyto is caused by viroporin activity of the N-terminal amphipathic domain (aa 47 to 90). The importance of high [Ca2+]cyto is well established for RVA RV, but this has not been shown for RVB or RVC strains (49); therefore, it was unclear whether NSP4 viroporin activity would be conserved across groups. Similar analyses of picornavirus 2B viroporin demonstrated that closely related 2B homologs that had highly similar viroporin motifs also elevated [Ca2+]cyto, but distantly related 2B homologs that had less than 20% sequence identity and mutations in the viroporin motifs did not (11). The RVA NSP4 viroporin domain aligned with the C-terminal-most hydrophobic domain of NSP4 from the other serogroups, and while viroporin-like motifs were found within each of the NSP4s, there was no sequence identity, and the arrangements of these motifs were strikingly different. Thus, the sequence divergence and differences in the organization of the putative viroporin domains across serogroups made it possible that NSP4 viroporin activity was not conserved.
We tested for potential viroporin activity in mammalian and avian RVA, RVB, and RVC NSP4s using E. coli and mammalian cell systems. Though these are mammalian virus proteins, E. coli-based assays, which were first developed by Lama and Carrasco in 1992 to examine viroporin activity of eukaryotic viral proteins (35), have been used extensively to identify and characterize viroporins for many different viruses, including poliovirus, Sindbis virus, influenza A virus, respiratory syncytial virus, HIV, coronavirus, and simian virus 40 (SV40) (10, 19, 22, 36, 38, 47, 51). Each of the NSP4s tested exhibited viroporin activity both in E. coli and in mammalian cells; however, there was considerable variability in the phenotype between assays and for different NSP4s. A summary of the viroporin activity for each of the tested NSP4 is given in Table 3. Surprisingly, only full-length RVC NSP4 supported lysis of BL21(DE3)pLysS cells, but lysis was not observed for full-length RVA or RVB NSP4. So while the lysozyme-mediated lysis assay is a classical test for viroporin activity, it was the least sensitive test used in this study.
Table 3.
Summary of NSP4 viroporin activitya
| Group | Construct | E. coli lysis | Cytotoxicity | Altered redox activity (RSG) | Membrane perm. (PI) | Localization | Elevation of [Ca]cyto |
|---|---|---|---|---|---|---|---|
| RVA (E2) | SA11 1-175 (FL) | No | Yes | + | + | LC3 puncta | Yes |
| SA11 20-146 | Yes | Yes | + | + | NA | NA | |
| RVA (murine) (E7) | EC 20-146 | Yes | Yes | + | + | NA | NA |
| RVA (avian) (E4) | Ty-1 20-146 | No | Yes | + | + | LC3 puncta | Yes |
| RVA chimera | Ty-1/SA11-PD 20-146 | Yes | Yes | ND | ND | NA | NA |
| RVB (rat) | IDIR 1-218 (FL) | No | Yes | + | − | LC3 puncta | Yes |
| RVC (porcine) | Cowden 1-150 (FL) | Yes | Yes | + | + | LC3 puncta | Yes |
The viroporin activities observed with each NSP4 in the different assays are summarized here. Boldface type indicates data not shown. FL, full-length constructs; ND, not done; NA, not applicable; RSG, Redox Sensor Green; perm, permeabilization; PI, propidium iodide.
Although full-length RVA NSP4 was cytotoxic to E. coli in the absence of cell lysis, the mammalian RVA NSP4s with the aa 20-146 truncation induced BL21(DE3)pLysS lysis (6, 29). It is interesting that the primary difference between RVC NSP4 and RVA NSP4 is that RVC NSP4 lacks the first N-terminal hydrophobic domain that is present in RVA NSP4 (Fig. 1A) (27). Thus, the RVA NSP4 aa 20-146 truncation construct closely mimics the RVC N terminus and suggests that the RVA NSP4 N-terminal domain regulates the viroporin domain in some way. The RVA avian NSP4 aa 20-146 truncation construct failed to induce E. coli lysis, but activity was partially rescued in the Ty-1/SA11 chimera, confirming our previous results indicating that changes in the basic residues of the pentalysine domain disrupt RVA NSP4 viroporin activity (29). These data further underline the importance of the clustered basic residues for viroporin activity, by facilitating membrane penetration, which has been noted for several viroporins, including hepatitis C virus p7 and picornavirus 2B (11, 54).
We also used a bacterial cytotoxicity assay and a redox assay, which assess E. coli viability and vitality, to test for NSP4 viroporin activity. Both of these assays detected viroporin-like activity with greater sensitivity than the lysis assay, likely because they measure parameters of cell health and not the release of lysozyme into the periplasmic space (35). In particular, the RSG redox assay represents a compelling new and highly sensitive screen for unidentified viroporins. Fluorescence of the RSG reagent requires it to be reduced in the E. coli cytoplasm, which is enhanced upon uncoupling of the inner membrane proton gradient, as is seen with the proton ionophore CCCP (21, 53). All of the NSP4s enhanced RSG reduction, though it is not clear whether this was due to direct conduction of protons, permeability of the membrane due to NSP4 overexpression, or an indirect effect on proton pumps due to NSP4-mediated disruption in Ca2+ levels in E. coli (14). Indirect permeability seems unlikely, because the reduced (fluorescent) dye would also be able to diffuse out of cells rather than accumulating and increasing the green staining.
In both the cytotoxicity and RSG E. coli assays, Ty-1 and RVB NSP4 both showed viroporin-like activity but were less cytotoxic and showed a lower and/or delayed RSG response compared to SA11 RVA NSP4. RVB NSP4 reportedly forms inclusion bodies when expressed in E. coli, so it is possible that these could sequester some of the protein in an inactive form, resulting in an apparently weak viroporin phenotype (23). The blunted phenotypes are interesting, because Ty-1 and RVB NSP4 were expressed at higher levels than mammalian RVA and RVC NSP4, suggesting that viroporin-mediated cytotoxicity is the primary reason for the low expression level of full-length RVA and RVC NSP4 in E. coli. We were able to demonstrate recombinant expression of full-length RVA NSP4 in E. coli for the first time, though this has been attempted by several groups (6, 33). Detection by immunoblot required affinity isolation of NSP4, but these results suggest that overexpression of full-length RVA NSP4, which would facilitate structural studies, may be possible if the cytotoxicity of viroporin activity can be reduced.
The E. coli assays are useful means of identifying and mapping putative viroporin domains, but the results must be confirmed by validation of viroporin-like activity in mammalian cells. For example, the rotavirus spike protein VP4 contains a membrane fusion peptide within the VP5* subdomain. Expression of VP5* permeabilizes E. coli to the small β-galactosidase substrate o-nitrophenyl-β-d-galactopyranoside but does not cause cell lysis or reduce cell viability (15). Further, in mammalian cells, expression of VP4 does not elevate [Ca2+]cyto, so VP4 would not make a strong viroporin candidate (12). Thus, the E. coli lysis activity of RVC NSP4 and the observed cytotoxicity of avian RVA, RVB, and RVC NSP4 help build the case for the conservation of viroporin activity among these proteins.
RVA, RVB, and RVC NSP4 were likely synthesized in the ER, because they were all found in the membrane-associated cellular fraction, colocalized with two ER markers (calnexin and Sec61β), and RVA and RVC NSP4 were modified by the addition of N-linked glycans, with mammalian RVA NSP4 having two glycosylation sites, avian RVA NSP4 having three sites, and RVC NSP4 having only one site (43, 44, 46). In addition to their ER localization, each NSP4 tested formed punctate structures. The shape and distribution of the punctate structures associated with NSP4 was unique for each serogroup, underlining the presence of serogroup-specific characteristics that are likely related to the high sequence divergence observed between groups. To test this experimentally, we confirmed that NSP4 from different serogroups formed distinct punctate structures that did not colocalize with one another.
Using several organelle-specific antibodies, we showed that the different NSP4s localized to the ER and ERGIC compartments but did not enter the Golgi apparatus. Further, as seen with SA11 NSP4, the punctate structures all colocalized with the autophagy marker LC3. We previously connected RVA NSP4 viroporin activity and elevation of [Ca2+]cyto with induction of the formation of puncta, which appear to vesiculate from ER-like cellular structures (4). To extend this observation, we measured the [Ca2+]cyto in MA104 cells expressing RFP-tagged avian RVA, RVB, and RVC NSP4s. Each of the NSP4s significantly elevated [Ca2+]cyto, confirming that disruption of Ca2+ homeostasis, likely by viroporin activity, is a common property of NSP4s among serogroups A, B, and C. The RFP tag would not be expected to perturb NSP4, as it was previously shown that EGFP-tagged NSP4 functioned identically to untagged NSP4 and that both fully recapitulate alterations in Ca2+ homeostasis observed in RV-infected cells (12). Interestingly, despite an apparently blunted viroporin phenotype in bacteria, RVB NSP4 showed the greatest [Ca2+]cyto elevation at ∼16 h posttransfection. The significance of this observation requires further experimentation, because RVA and RVB NSP4 disrupted [Ca2+]cyto with different kinetics in Sf9 insect cells, and differences in the rate of expression could affect the timing of peak Ca2+ levels (58).
While previous mutational analyses showed that viroporin activity is the mechanism by which NSP4 ultimately elevates [Ca2+]cyto, the detailed pathways involved for RVA NSP4 remain unknown. Two possible models have been proposed. In the first, NSP4 viroporin activity in the ER releases stored Ca2+ into the cytosol, which subsequently stimulates store-operated Ca2+ entry (SOCE) through plasma membrane-resident host Ca2+ channels (8, 29). Alternatively, the NSP4 viroporin remains inactive in the ER but forms an active channel in the plasma membrane, directly conducting extracellular Ca2+ into the cytoplasm (12). Our data indicate that NSP4 likely activates SOCE (57). While recent reports show cell surface targeting of RVA NSP4 in infected human epithelial cells (Caco-2 and HT-29) and canine kidney cells (MDCK), recombinant EGFP- or RFP-tagged NSP4 does not traffic to the cell surface but does recapitulate the disruption in Ca2+ homeostasis observed in RV-infected cells (4, 7, 13, 18). In contrast, a substantial pool of NSP4, including non-RVA NSP4, localizes to the ER. In Sf9 cells, RVA NSP4 increases the Ca2+ leakiness of the ER, causing store depletion in the absence of extracellular Ca2+ (57). Ongoing studies aim to directly measure ER luminal Ca2+ levels upon NSP4 expression. Nevertheless, a possible contribution of NSP4 viroporin activity at the plasma membrane in RV-infected cells cannot be ruled out, and NSP4-induced [Ca2+]cyto elevation may involve both activation of cellular Ca2+ uptake channels via ER store depletion and direct Ca2+ entry through plasma membrane-localized NSP4.
Together, these data show that despite significant sequence divergence, NSP4s from RVA, RVB, and RVC all have viroporin activity in both E. coli and mammalian cells. This suggests that many of the Ca2+-dependent and -triggered processes observed in RVA-infected cells are likely to occur during the replication of RVB and RVC RV as well. The dependence of RVA RV replication on elevated [Ca2+]cyto is well characterized, and NSP4 viroporin activity is known to be the mechanism by which RV accesses the Ca2+ needed for replication (29, 49). Thus, it is amazing that a protein with such a critical function shows so much sequence divergence, and the differences may aid the protein in carrying out undiscovered functions that are unique to the different RV serogroups. The amino acid differences between these three ER-localized Ca2+ viroporins should facilitate a better understanding of NSP4 channel properties.
ACKNOWLEDGMENTS
We thank Miguel Saldana for his assistance with these studies. We acknowledge the assistance of the Integrated Microscopy Core, which is supported by SCPPR U54 HD-007495, DDC P30 DK-56338 (to M. K. Estes), DLDCC P30 CA-125123, and the Dan L. Duncan Cancer Center of Baylor College of Medicine. We also acknowledge the Baylor College of Medicine Cytometry and Cell Sorting Core, which is supported by National Institutes of Health grants NCRR S10RR024574, NIAID AI036211, and NCI P30CA125123.
This work was supported in part by NIH grant R01AI080656, NIH Research Training in Pediatric Gastroenterology grant T32DK007664, Public Health Service grant P30DK56338, which funds the Texas Medical Center Digestive Diseases Center (TMCDDC), and a TMCDDC Pilot & Feasibility grant.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Au KS, Chan WK, Burns JW, Estes MK. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553–4562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Au KS, Mattion NM, Estes MK. 1993. A subviral particle binding domain on the rotavirus nonstructural glycoprotein NS28. Virology 194:665–673 [DOI] [PubMed] [Google Scholar]
- 3. Ball JM, Tian P, Zeng CQ, Morris AP, Estes MK. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101–104 [DOI] [PubMed] [Google Scholar]
- 4. Berkova Z, et al. 2006. Rotavirus NSP4 induces a novel vesicular compartment regulated by calcium and associated with viroplasms. J. Virol. 80:6061–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berkova Z, Morris AP, Estes MK. 2003. Cytoplasmic calcium measurement in rotavirus enterotoxin-enhanced green fluorescent protein (NSP4-EGFP) expressing cells loaded with Fura-2. Cell Calcium 34:55–68 [DOI] [PubMed] [Google Scholar]
- 6. Browne EP, Bellamy AR, Taylor JA. 2000. Membrane-destabilizing activity of rotavirus NSP4 is mediated by a membrane-proximal amphipathic domain. J. Gen. Virol. 81:1955–1959 [DOI] [PubMed] [Google Scholar]
- 7. Bugarcic A, Taylor JA. 2006. Rotavirus nonstructural glycoprotein NSP4 is secreted from the apical surfaces of polarized epithelial cells. J. Virol. 80:12343–12349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cahalan MD. 2009. STIMulating store-operated Ca2+ entry. Nat. Cell Biol. 11:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chacko AR, et al. 2011. Novel pentameric structure of the diarrhea-inducing region of the rotavirus enterotoxigenic protein NSP4. J. Virol. 85:12721–12732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daniels R, Sadowicz D, Hebert DN. 2007. A very late viral protein triggers the lytic release of SV40. PLoS Pathog. 3:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. de Jong AS, et al. 2008. Functional analysis of picornavirus 2B proteins: effects on calcium homeostasis and intracellular protein trafficking. J. Virol. 82:3782–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Diaz Y, et al. 2008. Expression of nonstructural rotavirus protein NSP4 mimics Ca2+ homeostasis changes induced by rotavirus infection in cultured cells. J. Virol. 82:11331–11343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Didsbury A, et al. 2011. Rotavirus NSP4 is secreted from infected cells as an oligomeric lipoprotein and binds to glycosaminoglycans on the surface of non-infected cells. Virol. J. 8:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dominguez DC. 2004. Calcium signalling in bacteria. Mol. Microbiol. 54:291–297 [DOI] [PubMed] [Google Scholar]
- 15. Dowling W, Denisova E, LaMonica R, Mackow ER. 2000. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 74:6368–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eisenberg D, Weiss RM, Terwilliger TC. 1982. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 299:371–374 [DOI] [PubMed] [Google Scholar]
- 17. Estes MK. 2001. Rotaviruses and their replication, p 1747–1785 In Knipe DM, Howley PM. (ed), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18. Gibbons TF, et al. 2011. Rotavirus NSP4: cell type-dependent transport kinetics to the exofacial plasma membrane and release from intact infected cells. Virol. J. 8:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez ME, Carrasco L. 1998. The human immunodeficiency virus type 1 Vpu protein enhances membrane permeability. Biochemistry 37:13710–13719 [DOI] [PubMed] [Google Scholar]
- 20. Gonzalez ME, Carrasco L. 2003. Viroporins. FEBS Lett. 552:28–34 [DOI] [PubMed] [Google Scholar]
- 21. Gray DR, Yue S, Chueng C, Godfrey W. 2005. Bacterial vitality detected by a novel fluorogenic redox dye using flow cytometry, p. 331, abstr. I-111. Abstr. 105th Gen. Meet. Am. Soc. Microbiol American Society for Microbiology, Washington, DC [Google Scholar]
- 22. Guinea R, Carrasco L. 1994. Influenza virus M2 protein modifies membrane permeability in E. coli cells. FEBS Lett. 343:242–246 [DOI] [PubMed] [Google Scholar]
- 23. Guzman E, McCrae MA. 2005. Molecular characterization of the rotavirus NSP4 enterotoxin homologue from group B rotavirus. Virus Res. 110:151–160 [DOI] [PubMed] [Google Scholar]
- 24. Hagbom M, et al. 2011. Rotavirus stimulates release of serotonin (5-HT) from human enterochromaffin cells and activates brain structures involved in nausea and vomiting. PLoS Pathog. 7:e1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hennig L. 2001. WinPep 2.11: novel software for PC-based analyses of amino acid sequences. Prep. Biochem. Biotechnol. 31:201–207 [PubMed] [Google Scholar]
- 26. Horie Y, et al. 1999. Diarrhea induction by rotavirus NSP4 in the homologous mouse model system. Virology 262:398–407 [DOI] [PubMed] [Google Scholar]
- 27. Horie Y, Nakagomi T, Oseto M, Masamune O, Nakagomi O. 1997. Conserved structural features of nonstructural glycoprotein NSP4 between group A and group C rotaviruses. Arch. Virol. 142:1865–1872 [DOI] [PubMed] [Google Scholar]
- 28. Huber AC, Yolken RH, Mader LC, Strandberg JD, Vonderfecht SL. 1989. Pathology of infectious diarrhea of infant rats (IDIR) induced by an antigenically distinct rotavirus. Vet. Pathol. 26:376–385 [DOI] [PubMed] [Google Scholar]
- 29. Hyser JM, Collinson-Pautz MR, Utama B, Estes MK. 2010. Rotavirus disrupts calcium homeostasis by NSP4 viroporin activity. mBio 1:e00265–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hyser JM, Estes MK. 2009. Rotavirus vaccines and pathogenesis: 2008. Curr. Opin. Gastroenterol. 25:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hyser JM, Zeng CQ, Beharry Z, Palzkill T, Estes MK. 2008. Epitope mapping and use of epitope-specific antisera to characterize the VP5* binding site in rotavirus SA11 NSP4. Virology 373:211–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishino M, et al. 2006. Comparison of NSP4 protein between group A and B human rotaviruses: detection of novel diarrhea-causing sequences in group B NSP4. Arch. Virol. 151:173–182 [DOI] [PubMed] [Google Scholar]
- 33. Jagannath MR, et al. 2006. N- and C-terminal cooperation in rotavirus enterotoxin: novel mechanism of modulation of the properties of a multifunctional protein by a structurally and functionally overlapping conformational domain. J. Virol. 80:412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]
- 35. Lama J, Carrasco L. 1992. Expression of poliovirus nonstructural proteins in Escherichia coli cells. Modification of membrane permeability induced by 2B and 3A. J. Biol. Chem. 267:15932–15937 [PubMed] [Google Scholar]
- 36. Lama J, Carrasco L. 1992. Inducible expression of a toxic poliovirus membrane protein in Escherichia coli: comparative studies using different expression systems based on T7 promoters. Biochem. Biophys. Res. Commun. 188:972–981 [DOI] [PubMed] [Google Scholar]
- 37. Lin SL, Tian P. 2003. Detailed computational analysis of a comprehensive set of group A rotavirus NSP4. Virus Genes 26:271–282 [DOI] [PubMed] [Google Scholar]
- 38. Madan V, Garcia MJ, Sanz MA, Carrasco L. 2005. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 579:3607–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matthijnssens J, et al. 2011. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 156:1397–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McDonald SM, Aguayo D, Gonzalez-Nilo FD, Patton JT. 2009. Shared and group-specific features of the rotavirus RNA polymerase reveal potential determinants of gene reassortment restriction. J. Virol. 83:6135–6148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDonald SM, Patton JT. 2011. Rotavirus VP2 core shell regions critical for viral polymerase activation. J. Virol. 85:3095–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meyer JC, Bergmann CC, Bellamy AR. 1989. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology 171:98–107 [DOI] [PubMed] [Google Scholar]
- 43. Mori Y, Borgan MA, Ito N, Sugiyama M, Minamoto N. 2002. Diarrhea-inducing activity of avian rotavirus NSP4 glycoproteins, which differ greatly from mammalian rotavirus NSP4 glycoproteins in deduced amino acid sequence in suckling mice. J. Virol. 76:5829–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mori Y, Borgan MA, Ito N, Sugiyama M, Minamoto N. 2002. Sequential analysis of nonstructural protein NSP4 derived from group A avian rotaviruses. Virus Res. 89:145–151 [DOI] [PubMed] [Google Scholar]
- 45. Nagashima S, et al. 2008. Whole genomic characterization of a human rotavirus strain B219 belonging to a novel group of the genus Rotavirus. J. Med. Virol. 80:2023–2033 [DOI] [PubMed] [Google Scholar]
- 46. Nilsson M, von Bonsdorff CH, Svensson L. 1993. Biosynthesis and morphogenesis of group C rotavirus in swine testicular cells. Arch. Virol. 133:21–37 [DOI] [PubMed] [Google Scholar]
- 47. Perez M, Garcia-Barreno B, Melero JA, Carrasco L, Guinea R. 1997. Membrane permeability changes induced in Escherichia coli by the SH protein of human respiratory syncytial virus. Virology 235:342–351 [DOI] [PubMed] [Google Scholar]
- 48. Phan TG, et al. 2004. Virus diversity and an outbreak of group C rotavirus among infants and children with diarrhea in Maizuru city, Japan during 2002–2003. J. Med. Virol. 74:173–179 [DOI] [PubMed] [Google Scholar]
- 49. Ruiz MC, Cohen J, Michelangeli F. 2000. Role of Ca2+ in the replication and pathogenesis of rotavirus and other viral infections. Cell Calcium 28:137–149 [DOI] [PubMed] [Google Scholar]
- 50. Sanekata T, Ahmed MU, Kader A, Taniguchi K, Kobayashi N. 2003. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J. Clin. Microbiol. 41:2187–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sanz MA, Perez L, Carrasco L. 1994. Semliki Forest virus 6K protein modifies membrane permeability after inducible expression in Escherichia coli cells. J. Biol. Chem. 269:12106–12110 [PubMed] [Google Scholar]
- 52. Sasaki S, Horie Y, Nakagomi T, Oseto M, Nakagomi O. 2001. Group C rotavirus NSP4 induces diarrhea in neonatal mice. Arch. Virol. 146:801–806 [DOI] [PubMed] [Google Scholar]
- 53. Smith JJ, McFeters GA. 1996. Effects of substrates and phosphate on INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride) and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli. J. Appl. Bacteriol. 80:209–215 [DOI] [PubMed] [Google Scholar]
- 54. StGelais C, et al. 2009. Determinants of hepatitis C virus p7 ion channel function and drug sensitivity identified in vitro. J. Virol. 83:7970–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Taraporewala ZF, et al. 2006. Structure-function analysis of rotavirus NSP2 octamer by using a novel complementation system. J. Virol. 80:7984–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tian P, et al. 1995. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J. Virol. 69:5763–5772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tian P, et al. 1994. The nonstructural glycoprotein of rotavirus affects intracellular calcium levels. J. Virol. 68:251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Trojnar E, Otto P, Roth B, Reetz J, Johne R. 2010. The genome segments of a group D rotavirus possess group A-like conserved termini but encode group-specific proteins. J. Virol. 84:10254–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. 2006. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124:573–586 [DOI] [PubMed] [Google Scholar]
- 61. Zhang M, et al. 1998. Mutations in rotavirus nonstructural glycoprotein NSP4 are associated with altered virus virulence. J. Virol. 72:3666–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang M, Zeng CQ, Morris AP, Estes MK. 2000. A functional NSP4 enterotoxin peptide secreted from rotavirus-infected cells. J. Virol. 74:11663–11670 [DOI] [PMC free article] [PubMed] [Google Scholar]