Abstract
A crucial step in the life cycle of arenaviruses is the biosynthesis of the mature fusion-active viral envelope glycoprotein (GP) that is essential for virus-host cell attachment and entry. The maturation of the arenavirus GP precursor (GPC) critically depends on proteolytic processing by the cellular proprotein convertase (PC) subtilisin kexin isozyme-1 (SKI-1)/site-1 protease (S1P). Here we undertook a molecular characterization of the SKI-1/S1P processing of the GPCs of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) and the pathogenic Lassa virus (LASV). Previous studies showed that the GPC of LASV undergoes processing in the endoplasmic reticulum (ER)/cis-Golgi compartment, whereas the LCMV GPC is cleaved in a late Golgi compartment. Herein we confirm these findings and provide evidence that the SKI-1/S1P recognition site RRLL, present in the SKI-1/S1P prodomain and LASV GPC, but not in the LCMV GPC, is crucial for the processing of the LASV GPC in the ER/cis-Golgi compartment. Our structure-function analysis revealed that the cleavage of arenavirus GPCs, but not cellular substrates, critically depends on the autoprocessing of SKI-1/S1P, suggesting differences in the processing of cellular and viral substrates. Deletion mutagenesis showed that the transmembrane and intracellular domains of SKI-1/S1P are dispensable for arenavirus GPC processing. The expression of a soluble form of the protease in SKI-I/S1P-deficient cells resulted in the efficient processing of arenavirus GPCs and rescued productive virus infection. However, exogenous soluble SKI-1/S1P was unable to process LCMV and LASV GPCs displayed at the surface of SKI-I/S1P-deficient cells, indicating that GPC processing occurs in an intracellular compartment. In sum, our study reveals important differences in the SKI-1/S1P processing of viral and cellular substrates.
INTRODUCTION
Arenaviruses are single-strand, enveloped, negative-strand RNA viruses with a worldwide distribution that are of significance as experimental models and important human pathogens (7). The prototypic arenavirus lymphocytic choriomeningitis virus (LCMV), a neglected pathogen in humans (4), has been widely used as an experimental model and provided key concepts in virus-host cell interactions and viral pathogenesis (25). In immunocompetent individuals, LCMV causes a self-limiting febrile illness frequently associated with aseptic meningitis. In contrast, LCMV infection of the fetus and newborn has been linked to spontaneous abortion and severe central nervous system malformations, respectively (1). Immunocompromised patients are particularly vulnerable to LCMV, as tragically demonstrated by several fatal cases of LCMV infection in transplanted patients (13, 26). Lassa virus (LASV) causes severe hemorrhagic fever in humans and is the most prevalent pathogen among the arenaviruses, with over 300,000 infections and 5,000 deaths yearly in Western Africa (22). In South America, the New World arenaviruses Junin, Machupo, Guanarito, and Sabiá viruses are causative agents of severe hemorrhagic fevers in humans (29). To date, no licensed vaccine is available, and the only antiarenavirus treatment is restricted to the antiviral agent ribavirin, which is efficacious only when administered early after infection but also results in several side effects.
The genome of arenaviruses is comprised of two RNA segments that each encode two proteins in the ambisense orientation, separated by an intergenic region (9). The RNA-dependent RNA polymerase (L) and the small RING finger protein (Z) are encoded by the large segment (L), whereas the nucleoprotein (NP) and the viral envelope glycoprotein (GP) precursor (GPC) are encoded by the small segment (S). The envelope GP mediates cell surface attachment, entry, and fusion (8). The GPC polypeptide undergoes proteolytic processing that is essential for its maturation into a fusion-competent envelope GP. The arenavirus GPC is cleaved by the cellular proprotein convertase (PC) subtilisin kexin isozyme-1 (SKI-1)/site-1 protease (S1P) into GP1 and GP2 (3, 16, 18, 33). Upon cleavage, GP1 remains attached to GP2 by noncovalent interactions, and homotrimers of GP1/GP2 form the viral spikes that decorate the viral surface (12). GP1 constitutes the distal part of the spike and mediates the attachment of the virus to the cellular receptor(s). Upon attachment, the viral particle is internalized and delivered to late endosomes. An acidic pH triggers the dissociation of GP1 from GP2, which undergoes conformational changes, allowing the exposure of a fusogenic peptide that initiates the fusion between viral and host cell endosomal membranes. The processing of the GPC by SKI-1/S1P is required for the cell-to-cell propagation of infection and the release of infectious virion progeny from infected cells (3, 16, 18, 33).
Mammalian PCs are a family of serine proteases closely related to bacterial subtilisin and yeast kexin (38). PCs are involved in the limited proteolysis of polypeptides in the secretory pathway, in a time- and tissue-specific manner, generating a large diversity of bioactive molecules. PCs are synthesized as inactive precursors whose activation requires the autocatalytic removal of a conserved N-terminal prosegment that assists the correct folding of the protease. To date, nine PC members have been described. Among them, seven (PC1/3, PC2, furin, PC4, PC5/6, PACE4, and PC7) are basic PCs that cleave after the consensus sequence (R/K)-(X)n-(R/K)↓, where n is 0, 2, 4, or 6 and X is any amino acid except Cys. The ninth member is subtilisin/kexin type 9 (PCSK9), whose activity is restricted to its own prosegment processing (2). SKI-1/S1P is the eighth member of the PC family. It cleaves after the consensus sequence (R)-(X)-(hydrophobic)-(X)↓, where X is any amino acid except Cys (32, 37). As for other PCs, SKI-1/S1P is synthesized as an inactive zymogen (pre-pro-SKI-1/S1P) that undergoes three sequential cleavages to reach its mature and active form (39). The first cleavage occurs during translation in the endoplasmic reticulum (ER), where the signal peptide is excised at site A by a cellular signal peptidase. Pro-SKI-1/S1P then autocatalytically cleaves its prosegment at sites B′ (RKVF↓133), B (RSLK↓137), and C (RRLL↓186) in cis and trans, respectively. After the processing of its prosegment, SKI-1/S1P is translocated in the cis-Golgi/medial Golgi compartment, where it exerts its cellular functions. Some of the critical cellular substrates of SKI-1/S1P include the transcription factors SREBP1/2 and ATF6 (5, 46). Specifically, SKI-1/S1P plays a crucial role in lipid homeostasis and the ER stress response, via sterol regulatory element binding proteins (SREBPs) and ATF6 processing, respectively. Upon stimulation, the uncleaved, inactive form of the transcription factor dissociates from its ER-sequestrating protein to translocate into the Golgi compartment, where it undergoes two consecutive cleavages, first by SKI-1/S1P and then by site-2 protease (S2P). The processed active cytosolic fragment then translocates to the nucleus, where it activates the synthesis of target genes. Recently, N-acetylglucosamine-1-phosphotransferase, a Golgi-resident enzyme involved in the sorting of proteins to lysosomes, was described as a novel substrate of SKI-1/S1P (21).
Previous proof-of-concept studies using specific inhibitors of SKI-1/S1P demonstrated that the SKI-1/S1P-mediated processing of arenavirus GPCs is a promising antiviral strategy (20, 34, 40). Considering the important function of SKI-1/S1P in the host cell, the possible identification of novel inhibitors that specifically block the recognition and/or processing of arenavirus GPCs, but not endogenous substrates, is of high priority. In an attempt to achieve such a challenging goal, in the present study, we undertook a structure-function analysis of the molecular interaction of SKI-1/S1P with the GPC of the prototypic arenaviruses LCMV and LASV.
MATERIALS AND METHODS
Cell lines and viruses.
Human embryonic kidney (HEK293T) cells and human alveolar epithelial (A549) cells were cultivated in Dulbecco's modified Eagle medium (DMEM) (Gibco BRL, NY) supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Chinese hamster ovary (CHO-K1) and FD11 CHO cells deficient in furin (15) were cultivated in DMEM–Ham's F12 medium (1:1; Biochrom AG, Berlin, Germany) supplemented with 10% FBS and penicillin-streptomycin. SKI-1/S1P-deficient CHOK1 cells (SRD12B) were supplemented with 5 μg/ml cholesterol (Sigma), 20 μM sodium oleate (Sigma), and 1 mM sodium mevalonate (Sigma). All the cell lines were grown at 37°C in 5% CO2. LCMV ARM53b was produced and titers were determined as previously described (10). The following recombinant viruses were described elsewhere previously: rLCMV-LASVGP (35) and rLCMV-RRRR (34).
Antibodies.
Monoclonal antibodies (MAbs) 113 (anti-LCMV NP) and 83.6 (anti-LCMV GP) were described previously (8, 42). Anti-V5 MAb was purchased from Invitrogen, and anti-α-tubulin MAb was obtained from Sigma. Anti-5×His MAb was used to detect SKI-1/S1P-BTMD (Qiagen, Chatsworth, CA). Polyclonal rabbit anti-mouse secondary antibodies conjugated to horseradish peroxidase (HRP) (Dako, Glostrup, Denmark) and goat anti-mouse antibody conjugated with rhodamine RedX (Jackson ImmunoResearch Laboratories) were also used.
Plasmids and transfections.
Plasmids coding for wild-type (WT) SKI-1/S1P and SKI-1/S1P mutants (pIR SKI-1/S1P FL WT, pIR SKI-1/S1P H249A, pIR SKI-1/S1P-BTMD, pIR SKI-1/S1P-KDEL, pIR SKI-1/S1P K948A,L952A,L953A [KLL], pIR SKI-1/S1P R130E,R134E [RR], pIR SKI-1/S1P S1026A,T1049A,S1051A [Pmut], and pIR SKI-1/S1P ΔCT) were provided by Nabil G. Seidah (30). All the constructs were cloned into a pIRES-EGFP vector (Invitrogen), and a V5 tag was added to the C terminus, except for SKI-1/S1P-BTMD (6×His tag) and KDEL (no tag). Expression plasmids for LCMV GP, LCMV GP-RRRR, and LASV GP were described elsewhere previously (16, 17, 34). Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Briefly, 2 × 105 cells were seeded into a 24-well plate 24 h prior transfection. Various amounts of DNA (0.4 to 0.8 μg) were dissolved in OPTI-MEM medium (Gibco BRL, NY), mixed with OPTI-MEM–Lipofectamine 2000 (2 μl/transfection), incubated for 15 min at room temperature, and added to the cells. After 4 h, transfection solutions were replaced with fresh medium. The efficiency of transfection was evaluated after 48 h by the detection of the enhanced green fluorescent protein (EGFP) reporter.
Western blotting.
Cells were washed twice with cold phosphate-buffered saline (PBS) and lysed in SDS-PAGE sample buffer (62.5 mM Tris-HCl [pH 6.8], 20% [wt/vol] glycerol, 2% [wt/vol] SDS, 100 mM dithiothreitol [DTT]) supplemented with Complete protease inhibitors (Roche). Samples were boiled for 10 min at 95°C and centrifuged for 5 min at 13,000 rpm prior to gel loading. Proteins were separated on 10% polyacrylamide gels and blotted onto a nitrocellulose membrane. Membranes were blocked in blocking solution (PBS, 0.2% [wt/vol] Tween 20, 3% [wt/vol] nonfat milk) and then incubated with primary antibody (MAb 83.3 to LCMV GP2 at 1:1,000, MAb to V5 at 1:5,000, and MAb to α-tubulin at 1:10,000) at 4°C. After overnight incubation, membranes were washed thoroughly with PBS–0.2% (wt/vol) Tween 20 and incubated for 1 h at room temperature with the HRP-conjugated secondary antibodies (1:3,000). After three washes in PBS–0.2% (wt/vol) Tween 20, membranes were developed with the LiteABlot kit (EuroClone, Pero, Italy).
Real-time quantitative PCR.
Total RNA was isolated by using an RNeasy minikit (Qiagen). The lysate was homogenized with a Qiashredder kit (Qiagen). The real-time (RT) reaction was performed by using QuantiTect a reverse transcription kit (Qiagen), using 1 μg of template RNA per reaction. Real-time quantitative PCR (RT-qPCR) was performed by using SYBR green PCR Master mix (Applied Biosystems) on a StepOnePlus real-time PCR system (Applied Biosystems). CHO-specific primers for low-density lipoprotein (LDL) receptor (LDLR) and hydroxymethylbilane synthase (HMBS) cDNA were described previously (43). Primers for heat shock 70-kDa protein 5 (HSPA5) (forward primer CAC TTG GAA TGA CCC TTC AG and reverse primer GTT TGC CCA CCT CCA ATA TC) were designed based on data reported under GenBank accession no. M17169.1, while primers for 3-hydroxy-3-methylglutaryl-coenzyme A synthase (HMGCS1) (forward primer GCC CTT GAG ATC TAC TTT CC and reverse primer CCA GGC CAA TGG TAT ACT TC) were designed based on a cDNA sequence reported previously (14). Primer pair specificity was assessed by melting-curve analysis and by the sequencing of amplification products. Amplification reactions were performed in triplicate by using the following protocol: a 10-min hold at 95°C and 40 cycles of 15 s at 95°C, followed by 1 min at 60°C. Gene expression levels relative to those of HMBS were determined according to the 2−ΔΔCT method (19).
Pharmacological treatment.
The induction of genes regulated by SREBP was triggered by CHOK1 treatment with a solution containing Ham's F-12 medium–DMEM (1:1), 5% delipidated FBS, 50 μM sodium mevalonate, and 50 μM mevastatin (Enzo Lifescience) for 18 h, as reported previously (31). The induction of genes downstream of ATF6 was triggered by the addition of 5 μg/ml tunicamycin (Sigma-Aldrich) for 4 h before lysis, as described previously (46). Dimethyl sulfoxide (DMSO) (Sigma-Aldrich) was used as a negative control.
Viral infection assay.
CHOK1 or SRD12B cells were seeded into a Lab-Tek II chamber slide (Nunc, Rochester, NY). On the next day, monolayers were transfected with expression plasmids containing the cDNA of the indicated SKI-1/S1P variants. The transfection efficiencies of the different SKI-1/S1P constructs were comparable (>60%) based on EGFP expression. At 24 h posttransfection, cells were infected with viruses (LCMV, rLCMV-RRRR, and rLCMV-LASVGP) at a low multiplicity of infection (MOI) (0.01) at 37°C in 5% CO2. After 1 h, the inoculums were removed, cells were washed with PBS, and fresh medium was added. At the indicated time points, cells were fixed, and intracellular NP staining was performed. Briefly, cells were fixed in CellFix solution (BD Biosciences, Franklin Lakes, NJ) for 10 min at room temperature, permeabilized for 10 min in PBS–1% (vol/vol) fetal calf serum (FCS)–0.2% (wt/vol) saponin, and incubated for 1 h at room temperature with primary MAb 113 to LCMV NP (10 μg/ml). Monolayers were washed three times in PBS–1% (vol/vol) FCS and incubated with secondary goat anti-mouse IgG conjugated with rhodamine red (Jackson ImmunoResearch Laboratories) at a dilution of 1:200 for 45 min in the dark. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) solution (Invitrogen) for 5 min at room temperature. Slides were mounted in Mowiol solution and analyzed under an Axioplan 2 fluorescence microscope (Carl Zeiss Inc.).
Determination of enzymatic activities of SKI-1/S1P and furin in vitro.
The enzymatic activities of commercially available purified furin (New England BioLabs, Ipswich, MA) diluted in DMEM (Gibco BRL, NY) and conditioned SKI-1/S1P-BTMD medium (31) were measured as described previously (28), using either the furin substrate pyroglutamic acid-RTKR-methyl coumaryl-7-amide (Pyr-RTKR-MCA) (Peptide Institute Inc., Osaka, Japan) or the SKI-1/S1P substrate acetyl-IYISRRLL-MCA (Ac-IYSRRLL-MCA) (custom synthesis; GenScript). The reactions were carried out at room temperature with a buffer solution containing 25 mM Tris-HCl (pH 7.5), 25 mM morpholineethanesulfonic acid (MES), and 1 mM CaCl2, and fluorescence was detected with a TriStar LB 941 multimode microplate spectrofluorometer (Berthold Technologies, Bad Wildbad, Germany).
Medium swap experiments.
SKI-1/S1P-BTMD conditioned medium was obtained from HEK293T cells transfected with SKI-1/S1P-BTMD and centrifuged for 15 min at 6,000 × g at 4°C. For the enrichment of SKI-I/S1P activity, conditioned medium was concentrated by using Centricon filter devices with a 10-kDa cutoff. The furin active medium was obtained by the supplementation of HEK293T medium with 1 to 20 U of purified recombinant furin. Fresh medium and conditioned medium from mock-transfected HEK293T cells were used as controls. SKI-1/S1P and furin enzymatic activities were assessed in vitro as described above, and medium that contained comparable amounts of enzymatic activity was prepared. SRD12B cells and FD11 cells were infected with LCMV and LCMV-RRRR, respectively, at a low MOI (0.1). After 12 h, cells were then exposed to the medium containing SKI-1/S1P or furin for 36 h in the presence of lipid supplements. The cells were then lysed and analyzed by Western blotting, or the number of infected cells was determined by the immunofluorescence (IF) detection of LCMV NP, as described above.
RESULTS
The SKI-1/S1P recognition motif RRLL is critical for the processing of the LASV GPC in the ER/cis-Golgi compartment.
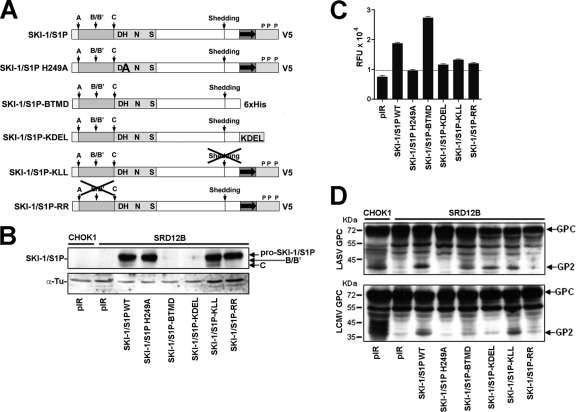
Previous studies provided evidence that the GPCs of LCMV and LASV undergo proteolytic processing in different intracellular compartments. Previous pulse-chase experiments, combined with a 15°C temperature block, which prevents ER exit, demonstrated that the LCMV GPC undergoes processing in a late Golgi compartment (44). In contrast, similar pulse-chase experiments, combined with the detection of endo-β-N-acetylglucosaminidase H (endo H) resistance, mapped the processing of the LASV GPC to the ER/cis-Golgi compartment (18). Accordingly, the treatment of cells with brefeldin A, which disassembles the cis- and medial Golgi compartments and inhibits the anterograde transport of secreted proteins, blocked the processing of the LCMV GPC but not the LASV GPC (3). Moreover, the addition of monensin, an ionophore that blocks transport from the cis-Golgi to the medial/late Golgi compartment, also affected the cleavage of the LCMV GPC (3). To confirm and extend data from these studies, we addressed the question of whether the transport of SKI-1/S1P to the late Golgi compartment is required for the processing of the LCMV GPC. For this purpose, we expressed a soluble SKI-1/S1P variant containing a C-terminal KDEL sequence (24) (Fig. 1A) that is largely retained in the ER/cis-Golgi compartment and cannot process SREBPs (11). To test the ability of SKI-1/S1P-KDEL to process the GPCs of LCMV and LASV, we made use of the SKI-1/S1P-deficent CHOK1-derived cell line SRD12B (32). The expression of recombinant SKI-1/S1P-KDEL in SKI-1/S1P-deficient cells resulted in only a minimal shedding of catalytically active protease compared to the wild type (Fig. 1C), indicating efficient ER retention and an impaired release of SKI-1/S1P-KDEL. To assess GPC processing, SRD12B and wild-type CHOK1 cells were cotransfected with the recombinant LCMV GPC and LASV GPC together with either wild-type SKI-1/S1P or SKI-1/S1P-KDEL, using Lipofectamine. In both cell lines, transfection efficiencies were consistently 50 to 60%, as assessed by a control plasmid expressing green fluorescent protein (GFP). As controls, we used a catalytically inactive SKI-1/S1P variant bearing the active-site H249A mutation in its catalytic domain (11) and an empty vector. After 48 h, total protein was extracted and separated by SDS-PAGE, and GPC processing was detected by Western blotting using MAb 83.6, which recognizes a conserved epitope present in the GPC and mature GP2 (42). The expression of SKI-1/S1P-KDEL in SKI-1/S1P-deficient cells partially rescued the processing of the LASV GPC but not of the LCMV GPC (Fig. 1D), suggesting that the translocation of SKI-1/S1P beyond the ER/cis-Golgi compartment is required for the processing of the LCMV GPC.
Fig 1.
Processing of recombinant LASV and LCMV GPCs by wild-type and mutant SKI-1/S1P. (A) Recombinant SKI-1/S1P variants used in our studies. The prodomain is shown in gray; the catalytic domain is shown in light gray, with critical residues of the catalytic triad highlighted; and the transmembrane domain is represented by a gray box with an arrow, followed by the cytoplasmic tail. The processing site for signal peptidase (site A) and autoprocessing sites B/B′ and C are marked. Putative phosphorylation sites within the cytoplasmic tail (P) and the C-terminal V5 tag are indicated. The SKI-1/S1P H249A mutant bears a mutation in the catalytic center and is enzymatically inactive. The soluble variant SKI-1/S1P-BTMD consists of the ectodomain of SKI-1/S1P and contains a six-histidine (6×His) tag. SKI-1/S1P-KLL is deficient in shedding, whereas SKI-1/S1P-RR is unable to undergo autoprocessing at the B/B′ site. (B) Expression of recombinant SKI-1/S1P variants in SKI-1/S1P-deficient SRD12B cells. SRD12B cells were transfected with the indicated SKI-1/S1P variants or the empty vector (pIR) by use of Lipofectamine. After 48 h, cells were lysed, and proteins were separated by SDS-PAGE and blotted onto nitrocellulose. Recombinant SKI-1/S1P variants were detected with an antibody to the V5 tag combined with an HRP-conjugated secondary antibody, and blots were developed with enhanced chemiluminescence (ECL). Blots were stripped and incubated with an antibody to α-tubulin (α-Tu), which was used for normalization. The positions of pro-SKI-1/S1P, the form processed at the B/B′ site, and the fully processed form (C site) are indicated. Please note the absence of processed (lower-molecular-mass) forms of SKI-1/S1P in cells transfected with the SKI-1/S1P H249A and SKI-1/S1P-RR variants. (C) Detection of SKI-1/S1P enzymatic activity in the cell culture supernatants of SRD12B cells transfected with recombinant SKI-1/S1P variants or an empty vector (pIR). SKI-1/S1P-deficient SRD12B cells were transfected with recombinant SKI-1/S1P as described above for panel B. After 48 h, conditioned cell culture supernatants were harvested, cleared, and assayed for the enzymatic activity of SKI-1/S1P, as reported previously (28). Briefly, conditioned medium samples containing soluble SKI-1/S1P were incubated with 10 μM Ac-YISRRLL-MCA in pH 7.5 buffer solution, and the released fluorescent 7-amino-4-methyl-coumaride (AMC) (relative fluorescence units [RFU]) was monitored over 6 h. Data shown are mean relative fluorescence units obtained from three independent experiments ± standard deviations (SD). (D) Processing of recombinant LCMV and LASV GPCs by SKI-1/S1P variants. SKI-1/S1P-deficient SRD12B cells were cotransfected with LCMV and LASV GPCs and the indicated SKI-1/S1P variants or an empty vector (pIR). As a positive control, wild-type CHOK1 cells cotransfected with LASV and LCMV GPCs and an empty vector (pIR) were used. The processing of the GPC was assessed by Western blotting using MAb 83.6 to LCMV GP, whose epitope is conserved and present in the GPC and mature GP2 of LCMV and LASV (42). The positions of the GPC and mature GP2, as well as apparent molecular masses, are indicated. The prominent band at 55 kDa was consistently detected and likely represents an underglycosylated GPC precursor.
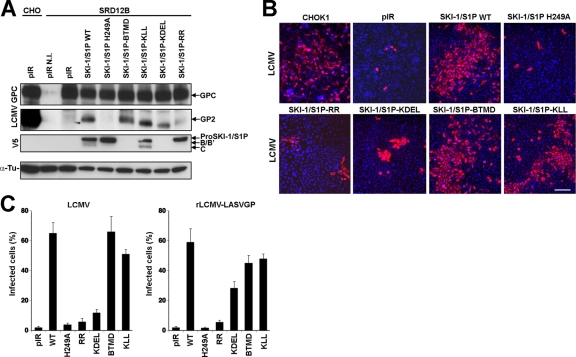
To confirm our findings in the context of viral infection, SRD12B cells were transfected with SKI-1/S1P-KDEL, wild-type SKI-1/S1P, as well as the catalytically inactive SKI-1/S1P H249A mutant. At 48 h posttransfection, cells were infected with LCMV at an MOI of 1. To detect GPC processing, infected cells were lysed at 48 h postinfection (p.i.), followed by the detection of the GPC and GP2 by Western blotting. In line with our findings obtained with recombinant proteins, the expression of SKI-1/S1P-KDEL inefficiently rescued the processing of the GPC in cells infected with LCMV (Fig. 2A). Compared to wild-type GP2, the residual GP2 detected in cells expressing SKI-1/S1P-KDEL seemed to be of a lower apparent molecular mass. The exact reason for this is currently unclear, but the altered migration of GP2 may be due to a different glycosylation caused by the processing of the GPC in the ER. Since arenavirus GPC processing by SKI-1/S1P is essential for the cell-to-cell propagation of the virus (3, 16, 18, 33), we studied the ability of SKI-1/S1P-KDEL to rescue viral spread in SKI-1/S1P-deficient cells. SRD12B cells were transfected with SKI-1/S1P-KDEL, wild-type SKI-1/S1P, and the SKI-1/S1P H249A mutant, followed by infection with LCMV and a recombinant LCMV expressing the GPC of LASV, rLCMV-LASVGP (35), at a low MOI (0.01). After 48 h, cultures were fixed, and infected cells were detected by immunofluorescence (IF) staining for LCMV NP. None of the viruses spread significantly in SRD12B cells transfected with the empty vector (Fig. 2B and C). As expected, transfection with wild-type SKI-1/S1P, but not the SKI-1/S1P H249A mutant, efficiently rescued the spread of both LCMV and rLCMV-LASVGP (Fig. 2B and C). Upon transfection with SKI-1/S1P-KDEL, only a limited spread of LCMV was observed, whereas rLCMV-LASVGP propagated more efficiently (Fig. 2B and C). Compared to wild-type SKI-1/S1P, the SKI-1/S1P-KDEL construct resulted in only a partial rescue of the cell-to-cell spread of rLCMV-LASVGP (Fig. 2C), which may be due to some catalytic impairment of SKI-1/S1P-KDEL. On the other hand, we observed a residual spread of LCMV in SRD12B cells transfected with SKI-1/S1P-KDEL. This finding suggests that some processing of the LCMV GPC can occur, likely due to the “leakiness” of the overexpressed SKI-1/S1P-KDEL mutant. This leakiness may be caused by the saturation of the ER retention receptors (6), likely allowing the translocation of a fraction of SKI-I/S1P-KDEL into late Golgi compartments. In sum, previously reported results and our data demonstrate the effective processing of the closely related GPCs of LASV and LCMV in distinct intracellular compartments, the ER/cis-Golgi and late Golgi compartments, respectively.
Fig 2.
Rescue of LCMV infection in SKI-1/S1P-deficient SRD12B cells. (A) Processing of the LCMV GPC in infected cells by SKI-1/S1P mutants. SRD12B cells were transfected with the indicated SKI-1/S1P variants, followed by infection with LCMV at an MOI of 1. After 48 h, cells were lysed, and total protein was extracted, separated by SDS-PAGE, and blotted onto nitrocellulose. Blots were probed with MAb 83.6 to LCMV GP2 (LCMV GPC), the V5 tag (V5), and α-tubulin (α-Tu). The positions of the LCMV GPC and GP2 as well as pro-SKI-1/S1P, SKI-1/S1P processed at the B and B′ sites (B, B′), and fully processed SKI-1/S1P (C site) are indicated. N.I., noninfected. (B) Rescue of LCMV cell-to-cell propagation in SKI-1/S1P-deficient SRD12B cells by recombinant SKI-1/S1P variants. CHOK1 cells and SRD12B cells were transfected with either an empty vector (pIR) or the indicated SKI-1/S1P variants. After 48 h, cells were infected with LCMV at an MOI of 0.01. After 48 h, cells were fixed, and infected cells were detected by IF using MAb 113 to LCMV NP, combined with a rhodamine RedX-labeled secondary antibody (red). Cell nuclei were counterstained with DAPI (blue). Bar, 50 μm. (C) Infection of LCMV and rLCMV-LASVGP in SRD12B cells transfected with recombinant SKI-1/S1P variants. SRD12B cells were transfected with the indicated SKI-1/S1P variants and infected with LCMV and rLCMV-LASVGP as described above for panel B. After 48 h, cells were fixed and subjected to IF staining for LCMV NP. One hundred cells were examined, and NP-positive cells were scored. Data shown are means ± SD (n = 3).
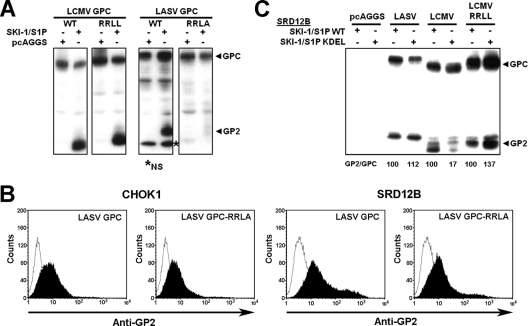
Our recent studies showed that the GPCs of some arenaviruses mimicking the autoprocessing sites of SKI-1/S1P are preferred substrates for this protease (28). Intriguingly, the cleavage sites of African arenaviruses, like LASV, mimic the C-autoprocessing site (RRLL↓), and the SKI-1/S1P recognition site of the GPC of the prototypic LCMV (RRLA↓) differs in sequence at the P1 position. Could this sequence difference account for the subcellular localization of the processing event of GPCs? Accordingly, we assessed the importance of the integrity of the RRLL motif for the ER/cis-Golgi processing of the LASV GPC. To this end, we used a two-pronged approach. First, we mutated the SKI-1/S1P recognition site RRLL of the LASV GPC into RRLA, the corresponding sequence found in the LCMV GPC, resulting in the LASV GPC-RRLA mutant, and second, we used an LCMV GPC variant bearing the sequence RRLL of the LASV GPC (LCMV GPC-RRLL). We previously showed that LCMV GPC-RRLL is expressed at levels similar to those of the wild type, is efficiently processed, and retains its functions in host cell attachment and viral entry (34). The coexpression of wild-type and mutant GPCs together with wild-type SKI-1/S1P in SRD12B cells resulted in the efficient processing of wild-type GPCs and LCMV GPC-RRLL but not LASV GPC-RRLA (Fig. 3A). Despite the apparent lack of processing, LASV GPC-RRLA was transported to the cell surface similarly to the wild type, as assessed by flow cytometry on live nonpermeabilized cells (Fig. 3B), which is in line with previous studies that showed the cell surface transport of arenavirus GPCs in the absence of cleavage (3, 16). To get first hints on the subcellular location of the processing of LCMV GPC-RRLL, SRD12B cells were cotransfected with either wild-type SKI-1/S1P or the SKI-1/S1P-KDEL mutant. As shown in Fig. 3C, SKI-1/S1P-KDEL efficiently processed LCMV GPC-RRLL, similarly to the LASV GPC, whereas, as expected, the cleavage of the wild-type LCMV GPC was impaired (Fig. 3C). Together, the data indicate that the RRLL motif allows the early processing of the LASV GPC in the ER/cis-Golgi compartment.
Fig 3.
The SKI-1/S1P recognition site RRLL is crucial for the processing of the LASV GPC in the ER/cis-Golgi compartment. (A) Processing of LCMV GPC-RRLL and LASV GPC-RRLA. SRD12B cells were cotransfected with the indicated wild-type (WT) and mutant LCMV and LASV GPCs together with an empty vector (pcAGGS) and SKI-1/S1P. After 48 h, GPC processing was assessed by Western blotting as described in the legend of Fig. 1D. The positions of the GPC and GP2 are indicated. The lower-molecular-mass band was occasionally detected but was found to be nonspecific (NS). (B) Detection of mutant and wild-type GPCs at the cell surface. CHOK1 and SRD12B cells were transfected with the indicated wild-type and mutant GPCs. After 48 h, cell surface staining was performed with MAb 83.6 combined with Alexa 594 secondary antibody. Data were acquired with a FACSCalibur flow cytometer and analyzed by using Cell Quest software. In histograms, the y axis represents cell counts, and the x axis represents the Alexa 594 fluorescence intensity. Dark areas indicate primary and secondary antibodies, and empty areas indicate secondary antibody only. (C) Efficient processing of LCMV GPC-RRLL by SKI-1/S1P-KDEL. SRD12B cells were cotransfected with the indicated GPCs and WT SKI-1/S1P or SKI-1/S1P-RR. After 48 h, GPC processing was examined by Western blotting as described above for panel A. Signals for the GPC and GP2 were quantified by densitometric analysis, and the GP2/GPC ratio was calculated. The GP2/GPC ratios in samples from cells transfected with WT SKI-1/S1P were set at 100%.
Autoprocessing of SKI-1/S1P is required for efficient cleavage of arenavirus GPCs but not cellular substrates.
During biosynthesis, SKI-1/S1P undergoes maturation that involves the autocatalytic cleavage of the proenzyme at three processing sites, sites B, B′, and C, yielding the mature protease. An SKI-1/S1P variant bearing the point mutations R130E and R134E (SKI-1/S1P-RR) (Fig. 1A) lacks B/B′ autoprocessing and subsequent autocatalytic cleavage at the C site. However, previous reports showed that this mutant is still able to partially process cellular SKI-1/S1P substrates like ATF6 and SREBP2, likely due to some low-extent processing at the C site (11). To study the role of SKI-1/S1P autoprocessing and maturation in the context of arenavirus GPC cleavage, the SKI-1/S1P-RR mutant was expressed in SKI-1/S1P-deficent SRD12B cells. The detection of SKI-1/S1P-RR by Western blotting revealed that the mutant reached expression levels comparable to those of wild-type SKI-1/S1P (Fig. 1B). Culture supernatants of cells transfected with SKI-1/S1P-RR contained only small amounts of catalytically active SKI-1/S1P, compared to levels of the wild type, indicating an impaired but still detectable shedding of the mutant (Fig. 1C). To test the ability of SKI-1/S1P-RR to process arenavirus GPCs, the mutant was coexpressed with LCMV and LASV GPCs in SRD12B cells as described above. Western blot analysis revealed a markedly reduced processing of both the LCMV GPC and LASV GPC in cells transfected with SKI-1/S1P-RR (Fig. 1D). To confirm our results in the context of productive virus infection, SRD12B cells were transfected with SKI-1/S1P-RR, followed by infection with LCMV, and GPC processing was assessed after 48 h. Similar to the situation with recombinant GPCs (Fig. 1D), an inefficient processing of the GPC was detected in infected cells (Fig. 2A). When SRD12B cells expressing SKI-1/S1P-RR were infected with LCMV and rLCMV-LASVGP at a low MOI, we observed only limited viral spread. Infected cells localized in compact foci (Fig. 2B), suggesting an impaired release of infectious progeny from infected cells. Altogether, our data indicated that the effective maturation of SKI-1/S1P by autoprocessing is required for the efficient cleavage of both viral GPCs and the cell-to-cell spread of infection.
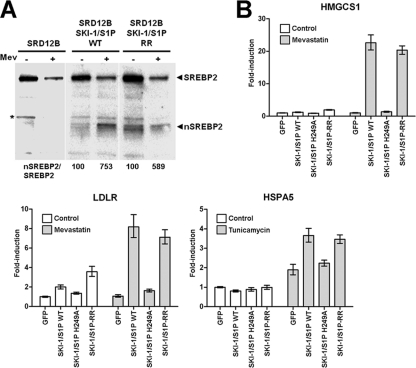
Previous studies showed the activity of SKI-1/S1P-RR in the context of the processing of the cellular substrates, particularly the transcription factors SREBP2 and ATF6 (11). In the context of our findings with the processing of viral GPCs, we sought to validate the activity of the SKI-1/S1P-RR mutant against cellular substrates in our system. First, we tested the ability of the SKI-1/S1P variants to process the endogenous transcription factor SREBP2 in response to cholesterol depletion. For this purpose, SKI-1/S1P-deficient SRD12B cells were transfected with SKI-1/S1P-RR and wild-type SKI-1/S1P. For sterol depletion, cells were treated with the drug mevastatin, an inhibitor of cholesterol biosynthesis, resulting in enhanced SREBP2 cleavage conditions by SKI-1/S1P. After 18 h of treatment, cells were lysed, and the SKI-1/S1P-dependent processing of SREBP2 was examined by Western blotting. As shown in Fig. 4A, upon cholesterol depletion, SKI-1/S1P-RR was able to process the SREBP protein nearly as efficiently as the wild type, consistent with previously reported data that a small amount of active SKI-1/S1P is enough to fully process SREBP2 (11).
Fig 4.
Normal processing of the cellular substrates SREBP2 and ATF6 by the SKI-1/S1P-RR mutant. (A) Processing of the SREBP2 protein by SKI-1/S1P-RR. SRD12B cells were transfected with wild-type (WT) SKI-1/S1P and SKI-1/S1P-RR or mock transfected. After 48 h, cells were subjected to cholesterol depletion by the addition of 50 μM mevastatin (Mev). As a control, cells were treated with vehicle only (−). After 18 h, cells were lysed, and the processing of SREBP2 was examined by Western blot analysis. The positions of full-length SREBP2 and the proteolytically cleaved nuclear form (nSREBP2) are indicated. To determine the extent of processing, blots were subjected to densitometric analysis. The signals for nSREBP were normalized to the signals for full-length SREBP2 (nSREBP2/SREBP2), and the ratios obtained for untreated cells were set at 100%. (B) Activation of cellular genes downstream of SREBP2 and ATF6. SRD12B cells were transfected with WT SKI-1/S1P, the enzymatically inactive SKI-1/S1P H249A mutant, SKI-1/S1P-RR, and GFP, which was used as a negative control. After 48 h, cells were subjected to ER stress to assess the ATF6-mediated induction of heat shock 70-kDa protein 5 (HSPA5) and cholesterol depletion to monitor the SREBP2-mediated upregulation of the genes for 3-hydroxy-3-methylglutaryl-coenzyme A synthase (HMGCS1) and LDL receptor (LDLR). Genes downstream of SREBP2 were induced by the treatment of cells with 50 μM mevastatin as described above for panel A. To induce genes downstream of ATF6, cells were treated with 5 μg/ml tunicamycin for 4 h. After treatment, cells were washed twice with PBS, and total RNA was isolated to perform RT-qPCR analyses as described in Materials and Methods. Data were normalized by using the calibrator gene hydroxymethylbilane synthase (HMBS). Data are presented as fold induction above levels for mock (DMSO)-treated cells (means ± SD; n = 3). The differences between the signals obtained with WT SKI-1/S1P and SKI-1/S1P-RR in cells induced with mevastatin and tunicamycin were not statistically significant.
In a complementary approach, we tested the ability of SKI-1/S1P-RR to process SREBP2 and ATF6 in response to cholesterol depletion and ER stress, using the regulation of downstream genes as a readout. The SKI-1/S1P-mediated activation of SREBP2 in response to cholesterol depletion was assessed by looking at the upregulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase (HMGCS1) and the LDL receptor (LDLR). The activation of ATF6 was detected by monitoring the mRNA levels of ATF6-regulated heat shock 70-kDa protein 5 (HSPA5), triggered by ER stress. Briefly, SRD12B cells were transfected with wild-type SKI-1/S1P, the SKI-1/S1P H249A mutant, SKI-1/S1P-RR, or GFP, which was used as a negative control. To induce ER stress, cells were treated with tunicamycin, an inhibitor of protein N-glycosylation, leading to the accumulation of underglycosylated and misfolded proteins in the ER. Cholesterol depletion was performed as described above. Following sterol depletion or ER stress induction, cells were lysed, total RNA was extracted, and mRNA levels for HMGCS1, LDLR, and HSPA5 were determined by real-time quantitative PCR (RT-qPCR), as described in Materials and Methods. As shown in Fig. 4B, the expression of SKI-1/S1P-RR in SRD12B cells fully rescued the induction of SREBP2- and ATF6-regulated genes, whereas the catalytically inactive SKI-1/S1P H249A mutant was unable to do so, confirming the functional activity of the SKI-1/S1P-RR mutant toward cellular substrates.
Membrane anchorage and the cytoplasmic domain of SKI-1/S1P are dispensable for viral GPC processing.
Full-length SKI-1/S1P is a type I membrane-anchored protein that contains a transmembrane domain and a cytoplasmic tail. To assess the role of the membrane anchorage in the SKI-1/S1P-mediated processing of viral GPCs, we used a truncated version of SKI-1/S1P that lacks the transmembrane and cytoplasmic tail (SKI-1/S1P-BTMD) (Fig. 1A) but retains catalytic activity. Upon transfection in SKI-1/S1P-deficient cells, SKI-1/S1P-BTMD was efficiently secreted, as evidenced by the enhanced SKI-1/S1P activity in the cell culture supernatant compared to that of the wild type (Fig. 1C). The cotransfection of SKI-1/S1P-BTMD with the GPCs of LASV and LCMV resulted in significant GPC processing (Fig. 1D). In line with these findings, the expression of SKI-1/S1P-BTMD in SKI-1/S1P-deficient cells rescued GPC processing in infected cells and the cell-to-cell propagation of virus (Fig. 2B and C). The efficient processing of both the LASV and LCMV GPCs by soluble SKI-1/S1P-BTMD indicated that the membrane anchorage of the protease is not required for the processing of viral GPCs.
Membrane-bound SKI-1/S1P is subject to the autocatalytic shedding of its ectodomain at the KHQKLL953 site close to the transmembrane domain, thereby releasing a soluble protein (amino acids [aa] 188 to 953) and leaving behind a membrane-bound stump consisting of aa 954 to 1052 (11). To address the role of ectodomain shedding in the ability of SKI-1/S1P to process arenaviral GPCs, we employed the SKI-1/S1P-KLL mutant, which bears the mutations K948A, L952A, L953A and which is deficient in shedding (11). Upon transfection into SRD12B cells, SKI-1/S1P-KLL was expressed at levels similar to those of the wild type and underwent complete autoprocessing (Fig. 1B), indicating full catalytic activity. Compared to the wild type, SKI-1/S1P-KLL showed significantly reduced shedding, detected by the diminished SKI-1/S1P activity in cell culture supernatants (Fig. 1C). When expressed in SRD12B cells, SKI-1/S1P-KLL mediated a normal processing of the recombinant LCMV GPC and LASV GPC (Fig. 1D). The mutant further rescued GPC processing and viral spread in SKI-1/S1P-deficient cells (Fig. 2), indicating that SKI-1/S1P shedding is not required for GPC processing.
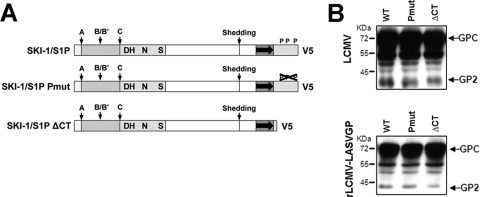
To address a possible role for the C-terminal domain of SKI-1/S1P in arenavirus GPC processing by the membrane-anchored enzyme, we tested two SKI-1/S1P mutants (displayed in Fig. 5A). The SKI-1/S1PΔCT mutant lacks the cytoplasmic domain, whereas SKI-1/S1P-Pmut contains mutations in the putative phosphorylation sites of the cytosolic tail, namely, S1026A, T1049A, and S1051A (30). When expressed in SRD12B cells, SKI-1/S1P-Pmut and SKI-1/S1PΔCT rescued the processing of the LCMV and LASV GPCs to a degree comparable to that of the wild-type enzyme (Fig. 5B). This finding indicates that the cytoplasmic domain is dispensable for GPC processing by the membrane-anchored SKI-1/S1P and further excludes a possible role of the phosphorylation of S1026, T1049, and S1051 in the SKI-1/S1P–GPC interaction.
Fig 5.
The cytoplasmic domain of SKI-1/S1P is dispensable for the processing of LASV and LCMV GPCs. (A) Schematic of the recombinant SKI-1/S1P variants. The different domains are indicated as described in the legend of Fig. 1A. SKI-1/S1P-Pmut is deficient in the phosphorylation of the intracellular domain. (B) Processing of recombinant LCMV and LASV GPCs by SKI-1/S1P variants. SKI-1/S1P-deficient SRD12B cells were cotransfected with LCMV and LASV GPCs and the indicated SKI-1/S1P variants. The processing of the GPC was assessed by Western blotting using MAb 83.6 as described in the legend of Fig. 1D. Relative molecular masses and the positions of the GPC and mature GP2 are indicated.
Soluble SKI-1/S1P is unable to process arenavirus GPCs at the cell surface.
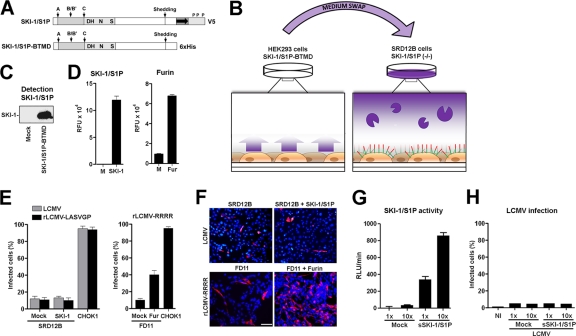
SKI-1/S1P undergoes shedding into the extracellular space, and our present study showed that soluble SKI-1/S1P can efficiently rescue the processing of the LASV GPC and LCMV GPC in SKI-1/S1P-deficient cells (Fig. 1D and 2). To further investigate this issue, we assessed whether soluble SKI-1/S1P was also able to process LCMV and LASV GPCs present at the surface of infected SKI-1/S1P-deficient cells. For this purpose, we used soluble SKI-1/S1P-BTMD (Fig. 6A) and performed medium swap experiments (shown schematically in Fig. 6B). Briefly, SKI-1/S1P-deficient SRD12B cells were infected with wild-type LCMV and rLCMV-LASVGP. Upon infection, their media were removed, and the cells were exposed to a medium containing recombinant soluble SKI-1/S1P. As a readout, we assessed the cell-to-cell propagation of the virus, which is highly dependent on GPC processing. As a control for our infection and medium swap approach (Fig. 6B), we used a recombinant LCMV variant bearing a canonical furin recognition site (RRRR) in its GPC (rLCMV-RRRR). This LCMV variant grows independently of SKI-1/S1P and critically depends on cellular furin for GPC processing and cell-to-cell propagation (34).
Fig 6.
Soluble SKI-1/S1P is unable to process arenavirus GPCs at the cell surface. (A) Schematic representation of the soluble SKI-1/S1P variant SKI-1/S1P-BTMD with the different domains indicated in the legend of Fig. 1A. (B) Schematic of the medium-swapping experiment. HEK293 cells stably transfected with SKI-1/S1P-BTMD were cultured for 48 h to produce soluble SKI-1/S1P. After 48 h, conditioned cell culture supernatants from SKI-1/S1P-BTMD-producing HEK293 cells were harvested, cleared by centrifugation, supplemented with lipids, and added to LCMV-infected SRD12B cells that displayed the unprocessed GPC at their cell surface. The GP1 moiety is in red, and the GP2 part is in green. (C) Detection of SKI-1/S1P-BTMD. Conditioned supernatants of stably transfected HEK293 and mock-transfected cells were harvested after 48 h. Total protein was extracted, and the soluble recombinant SKI-1/S1P-BTMD was detected by Western blotting using an antibody to the His tag, an HRP-conjugated secondary antibody, and ECL for detection. The position of SKI-1/S1P-BTMD (SKI-1) is indicated. (D) Detection of enzymatic activity in medium containing SKI-1/S1P-BTMD or recombinant furin. The activity of SKI-1/S1P in the conditioned supernatant (SKI-1) and control medium (M) was determined as described in the legend of Fig. 1C. Recombinant furin diluted in medium (Fur) and control medium (M) were incubated with 10 μM Pyr-RTKR-MCA in pH 7.5 buffer solution, and the released fluorescent AMC was monitored over 6 h. Data shown are mean relative fluorescence units (RFU) over 6 h obtained from three independent experiments ± SD. (E) Exogenous soluble SKI-1/S1P is unable to rescue the spread of LCMV in SKI-1/S1P-deficient cells. SRD12B cells were infected with LCMV and rLCMV-LASVGP at an MOI of 0.1. For comparison, furin-deficient FD11 cells were infected with rLCMV-RRRR. After 12 h, the medium was removed, cells were washed, and medium containing soluble SKI-1/S1P (SKI-1), furin, or control medium (Mock) was added. After 36 h, cells were fixed, and infection was detected by IF staining for LCMV NP as described in the legend of Fig. 2B. As a control, CHOK1 cells were infected with the viruses at an MOI of 0.1 and fixed after a total of 48 h. For quantification, 100 cells were examined, and NP-positive cells were scored. Data shown are means ± SD (n = 3). (F) Representative specimens from panel E. LCMV NP appears in red, and cell nuclei appear in blue. Please note the lack of viral spread in SRD12B cells treated with soluble exogenous SKI-1/S1P. Bar, 50 μm. (G) Conditioned supernatants containing soluble SKI-1/S1P were concentrated 10-fold, and enzymatic activity was determined as described above for panel D. RLU, relative light units. (H) SRD12B cells were infected with LCMV, as described above for panel E, and exposed to increasing amounts of SKI-1/S1P activity, as described above for panel G. The cell-to-cell propagation of virus was assessed as described above for panels E and F. None of the conditions applied resulted in a significant propagation of virus infection in treated cultures. NI, noninfected.
In a first step, we produced recombinant SKI-1/S1P-BTMD in stably transfected HEK293 cells, as described previously (28). Soluble SKI-1/S1P-BTMD was detected in the cell culture supernatants from transfected cells but not in the media of untransfected controls (Fig. 6C). Recombinant soluble furin was obtained from a commercial source. The enzymatic activity of soluble SKI-1/S1P was determined by a sensitive and specific in vitro assay based on fluorogenic peptides derived from the recognition site of the LASV GPC (Ac-IYSRRLL-MCA) (28) (for details, please see Materials and Methods). The activity of the recombinant furin preparation was determined by use of the same assay format, using the substrate Pyr-RTKR-MCA (47). Catalytic activity was detected in our preparation of soluble recombinant SKI-1/S1P and furin (Fig. 6D). For the medium swap experiments, we infected SKI-1/S1P-deficient SRD12B cells with LCMV and rLCMV-LASVGP at an MOI of 0.1. Furin-deficient CHO-FD11 cells were infected with rLCMV-RRRR, and wild-type CHOK1 cells were used as controls. After 12 h, the cell culture media were replaced by those containing either exogenous recombinant SKI-1/S1P or furin. Thirty-six hours later, cells were fixed, and infection was detected by IF staining for LCMV NP. The addition of exogenous soluble SKI-1/S1P to infected SRD12B cells did not increase the spread of LCMV or rLCMV-LASVGP above the low background levels observed in mock-treated SDR12B cells (Fig. 6E and F). In contrast, the addition of soluble furin to CHO-FD11 cells infected with rLCMV-RRRR resulted in a significant spread of the virus (Fig. 6E and F).
Since the processing of peptide substrates in vitro may not accurately reflect the activity of an enzyme toward its full-length protein substrate in the cellular context, our detection of SKI-1/S1P activity shown in Fig. 6D may have been an overestimation. To address this issue, we produced and concentrated soluble SKI-1/S1P by 10-fold, which resulted in markedly increased catalytic activity (Fig. 6G). When applied in our medium swap assay, concentrated SKI-1/S1P did not result in a significant detection of viral spread (Fig. 6H). Altogether, the data suggest that neither the LCMV GPC nor the LASV GPC can be significantly processed by soluble SKI-1/S1P at the cell surface. Although to some extent artificial, the efficient processing of the furin-dependent LCMV GPC by recombinant soluble furin excluded the gross misfolding of the unprocessed GPC displayed at the cell surface. In sum, the above-described data suggested that soluble SKI-1/S1P can process viral GPCs in an intracellular compartment but not at the plasma membrane.
DISCUSSION
In the present study, we undertook a molecular characterization of the processing of arenavirus GPCs by SKI-1/S1P and reached the following conclusions. (i) We confirmed that the GPCs of LCMV and LASV and cellular substrates undergo processing in different intracellular compartments and found a determining role for the RRLL motif in the processing of the LASV GPC in the ER/cis-Golgi compartment. (ii) Our structure-function analysis revealed that the efficient cleavage of arenavirus GPCs, but not cellular substrates, requires efficient SKI-1/S1P autoprocessing. (iii) Membrane anchorage and the cytosolic domain of SKI-I/S1P were dispensable for arenavirus GPC processing. (iv) However, exogenous soluble SKI-1/S1P was unable to process LCMV and LASV GPCs present at the surface of SKI-I/S1P-deficient cells.
In the host cell, SKI-1/S1P is implicated in the proteolytic processing of a defined set of substrates, including the membrane-bound SREBPs and the ATF6 family of transcription factors (27). The SKI-1/S1P-mediated processing of these cellular substrates is under tight spatiotemporal control, as illustrated by the activation of ATF6 upon ER stress. In the resting cell, ATF6 is retained in the ER by the interaction of its luminal domain with the chaperone GRP78/BiP. The engagement of GRP78/BiP by unfolded proteins upon ER stress liberates ATF6, allowing its translocation to the Golgi compartment, where processing and activation by SKI-1/S1P occur in a tightly controlled manner in the early Golgi compartment (36). In contrast to cellular substrates whose processing seems to be confined to the Golgi compartment, a significant fraction of the LASV GPC undergoes processing already in the ER (18). Here we confirmed these findings by showing that the soluble SKI-1/S1P-KDEL variant retained in the ER/cis-Golgi compartment efficiently processed the LASV GPC. In contrast, the GPC of LCMV, which is structurally closely related to LASV, is processed in a late Golgi compartment (3, 44) and, accordingly, was poorly processed by the ER-retained SKI-1/S1P-KDEL construct.
Interestingly, the SKI-1/S1P processing site of LCMV, RRLA, differs by only one amino acid at P1 from the one found in LASV GPC (RRLL). The GPC of the bunyavirus Crimean-Congo hemorrhagic fever (CCHF) virus also contains an RRLL SKI-1/S1P recognition site and undergoes cleavage in the ER/cis-Golgi compartment (41), suggesting that the RRLL motif allows the processing of substrates early along the secretory pathway, likely in the ER/cis-Golgi compartment. To address this issue, we used an LCMV GPC variant containing the RRLL motif (LCMV GPC-RRLL) and, conversely, an LASV GPC mutant bearing the LCMV recognition site RRLA (LASV GPC-RRLA). LCMV GPC-RRLL was efficiently processed by SKI-1/S1P-KDEL in the ER/cis-Golgi compartment. In contrast, a mutation of the RRLL motif in the LASV GPC to RRLA abrogated its early processing. This finding suggests that the RRLL motif is a discriminatory determinant for the selection of the SKI-1/S1P substrates that would be processed in the ER/cis-Golgi compartment. Altogether, the data at hand indicate that SKI-1/S1P processes cellular and viral substrates in at least three different subcellular compartments: (i) GPCs of LASV, CCHF virus, and likely other arenaviruses in the ER/cis-Golgi compartment; (ii) cellular substrates in the medial Golgi compartment; and (iii) the LCMV GPC in late Golgi compartments. It is conceivable that the processing of viral GPCs in distinct compartments may prevent a perturbation of the SKI-1/S1P-mediated processing of cellular substrates. Indeed, our recent studies revealed that arenavirus infection results in a transient induction of ATF6 but does not interfere with ATF6-regulated gene expression upon the induction of ER stress (27). This ability of arenaviruses to utilize a cellular protease without interfering with its function in the host cell may provide a strategy for the virus to limit cellular damage during its nonlytic infection.
Cellular PCs are synthesized as inactive zymogens that undergo autocatalytic processing and activation. In most PCs, including furin, the cleaved prosegment remains associated with the enzyme and inhibits is activity, thereby generating a latent form of the protease (23). Among the PCs, SKI-1/S1P seems unusual, as a mutant impaired in primary B/B′ autoprocessing is still functional in the cleavage of cellular substrates, likely due to its ability to autoprocess itself to a small extent at its secondary C site, even in the absence of primary cleavage at the B/B′ site (11). Our present study indicates that a variant of SKI-1/S1P deficient in B/B′ processing (SKI-1/S1P-RR) cleaved the GPCs of LCMV and LASV inefficiently and was unable to rescue productive virus infection and cell-to-cell spread in SKI-1/S1P-deficient cells. In line with previously reported work, we were able to show that SKI-1/S1P-RR processed the SREBP2 protein normally and was fully functional in the activation of SREBP2 and ATF6 downstream genes (11). Thus, contrary to cellular substrates, the SKI-1/S1P processing of viral GPCs seems to critically depend on the efficient autocatalytic processing of the inhibitory prosegment. The underlying reasons for this are currently unclear. One possible explanation could be the markedly different affinities of cellular and viral substrates for SKI-1/S1P. In this scenario, residual amounts of activity in SKI-1/S1P-RR may be sufficient to fully process endogenous cellular transcription factors, whereas the higher load of viral GPCs may overwhelm the system, resulting in the accumulation of the unprocessed GPC, with a negative impact on the formation of infectious virus particles. It is also possible that the intracellular SREBPs and ATF6 are much better SKI-1/S1P substrates than the viral GPCs, hence requiring much less SKI-1/S1P activity for the processing of the former. Alternatively, the distinct activity of SKI-1/S1P-RR toward the SREBP2 protein and viral GPCs may be due to as-yet-unknown differences in the molecular recognition of cellular and viral substrates by SKI-1/S1P. Interestingly, SKI-1/S1P-RR was impaired in the processing of the GPCs of LCMV and LASV that undergo processing in distinct subcellular compartments. This finding indicates that the dependence on SKI-1/S1P autoprocessing is not related to cleavage in a particular compartment but may be a common feature of arenaviral GPCs, which may require high levels of active SKI-1/S1P for their efficient cleavage. This suggests that even a partial inhibition of the cellular SKI-1/S1P activity may be enough to block viral infection without significantly affecting its ability to activate its cognate cellular substrates. This is supported by the observation that the liver-specific knockout of SKI-1/S1P in mice revealed that a more than 95% loss of SKI-1/S1P activity was necessary to observe a loss of SREBP processing and, hence, sterol regulation (45). Thus, SKI-1/S1P inhibitors may be effective agents against arenavirus infection, since for a lowering of lipid levels, a nearly complete inhibition of SKI-1/S1P is required.
Previous studies identified the SKI-1/S1P-mediated processing of arenavirus GPCs as a promising novel target for the development of antiviral therapeutics (20, 34, 40). Considering the important roles of SKI-I/S1P in cell biology, the development of SKI-I/S1P inhibitors that specifically block the processing of arenaviral GPCs without affecting cellular substrates remains a major unaddressed and challenging issue. Specific inhibitors of SKI-I/S1P B/B′ prodomain processing are expected to predominantly affect virus infection, with lesser effects on the cellular function of SKI-1/S1P. Our finding that SKI-I/S1P B/B′ prodomain processing is specifically required for the enzyme's ability to efficiently process arenavirus GPCs, but not cellular substrates, may thus have revealed an “Achilles' heel” of the virus to be exploited for the development of novel antiviral strategies.
In the host cell, SKI-1/S1P exists in two active forms: a membrane-associated form that localizes predominantly in the Golgi compartment and, to some extent, endosomal compartments, but not at the cell surface (30), as well as a soluble secreted form that arises from its autocatalytic shedding (11). To address the role of the membrane anchorage of SKI-1/S1P in arenavirus GPC processing, we expressed a soluble recombinant form comprising the entire ectodomain in SKI-1/S1P-deficient cells. The efficient rescue of GPC processing and productive virus infection by soluble SKI-1/S1P indicated that membrane anchorage was dispensable for GPC processing. To address a possible specific role for soluble SKI-1/S1P in GPC processing, we employed an SKI-1/S1P form deficient in shedding and found that the lack of shedding had no significant effect on GPC processing. In sum, the data suggest that a critical concentration of enzymatically active SKI-1/S1P in the respective compartment of the secretory pathway is a necessary and sufficient condition for GPC processing.
The efficient processing of the LASV GPC and LCMV GPC by soluble SKI-1/S1P raised the question of whether shed SKI-1/S1P was able to process the viral GPCs at the cell surface. Previous studies showed a normal cell surface transport of unprocessed LCMV GPC (3, 16) as well as the presence of detectable amounts of uncleaved GPC at the surface of infected cells (16). By performing medium-swapping experiments, we found no evidence for the processing of the LCMV and LASV GPCs by soluble SKI-1/S1P. These data suggest that arenavirus GPC processing by SKI-1/S1P can occur exclusively in intracellular compartments, a finding that is consistent with the absence of membrane-anchored SKI-1/S1P from the cell surface (30). In sum, our study revealed the first significant differences in the processing of cellular and viral substrates by SKI-1/S1P. Considering the importance of SKI-1/S1P for normal cell function, and its promise as an antiviral drug target, these findings will help the development of novel therapeutic strategies that specifically target the role of SKI-1/S1P in virus infection.
ACKNOWLEDGMENTS
We thank Juan Carlos de la Torre (Scripps Research Institute, La Jolla, CA) for valuable reagents and helpful discussions. We also thank Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern Medical Center, Dallas, TX) for the S1P-deficient SRD12B cells.
This research was supported by Swiss National Science Foundation grants FN 31003A-120250 and FN 31003A-135536 (Stefan Kunz), funds from the University of Lausanne (Stefan Kunz), CIHR grant MOP 93792, and Canada Research Chair grant 216684 (Nabil G. Seidah).
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Barton LL, Mets MB, Beauchamp CL. 2002. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am. J. Obstet. Gynecol. 187:1715–1716 [DOI] [PubMed] [Google Scholar]
- 2. Benjannet S, et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279:48865–48875 [DOI] [PubMed] [Google Scholar]
- 3. Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. 2003. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J. Virol. 77:2866–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonthius DJ. 2009. Lymphocytic choriomeningitis virus: a prenatal and postnatal threat. Adv. Pediatr. 56:75–86 [DOI] [PubMed] [Google Scholar]
- 5. Brown MS, Goldstein JL. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 [DOI] [PubMed] [Google Scholar]
- 6. Bu G, Rennke S, Geuze HJ. 1997. ERD2 proteins mediate ER retention of the HNEL signal of LRP's receptor-associated protein (RAP). J. Cell Sci. 110(Pt 1):65–73 [DOI] [PubMed] [Google Scholar]
- 7. Buchmeier MJ, de la Torre JC, Peters CJ. 2007. Arenaviridae: the viruses and their replication, p 1791–1828 In Knipe DL, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 8. Buchmeier MJ, Lewicki HA, Tomori O, Oldstone MB. 1981. Monoclonal antibodies to lymphocytic choriomeningitis and pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology 113:73–85 [DOI] [PubMed] [Google Scholar]
- 9. de la Torre JC. 2009. Molecular and cell biology of the prototypic arenavirus LCMV: implications for understanding and combating hemorrhagic fever arenaviruses. Ann. N. Y. Acad. Sci. 1171(Suppl 1):E57–E64 [DOI] [PubMed] [Google Scholar]
- 10. Dutko FJ, Oldstone MB. 1983. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J. Gen. Virol. 64:1689–1698 [DOI] [PubMed] [Google Scholar]
- 11. Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG. 2002. Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J. Biol. Chem. 277:11265–11275 [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 12. Eschli B, et al. 2006. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J. Virol. 80:5897–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fischer SA, et al. 2006. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N. Engl. J. Med. 354:2235–2249 [DOI] [PubMed] [Google Scholar]
- 14. Gil G, Goldstein JL, Slaughter CA, Brown MS. 1986. Cytoplasmic 3-hydroxy-3-methylglutaryl coenzyme A synthase from the hamster. I. Isolation and sequencing of a full-length cDNA. J. Biol. Chem. 261:3710–3716 [PubMed] [Google Scholar]
- 15. Gordon VM, Klimpel KR, Arora N, Henderson MA, Leppla SH. 1995. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect. Immun. 63:82–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kunz S, Edelmann KH, de la Torre J-C, Gorney R, Oldstone MBA. 2003. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology 314:168–178 [DOI] [PubMed] [Google Scholar]
- 17. Kunz S, Rojek J, Perez M, Spiropoulou C, Oldstone MB. 2005. Characterization of the interaction of Lassa fever virus with its cellular receptor α-dystroglycan. J. Virol. 79:5979–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. U. S. A. 98:12701–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 20. Maisa A, Stroher U, Klenk HD, Garten W, Strecker T. 2009. Inhibition of Lassa virus glycoprotein cleavage and multicycle replication by site 1 protease-adapted alpha(1)-antitrypsin variants. PLoS Negl. Trop. Dis. 3:e446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marschner K, Kollmann K, Schweizer M, Braulke T, Pohl S. 2011. A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science 333:87–90 [DOI] [PubMed] [Google Scholar]
- 22. McCormick JB, Fisher-Hoch SP. 2002. Lassa fever. Curr. Top. Microbiol. Immunol. 262:75–109 [DOI] [PubMed] [Google Scholar]
- 23. Muller JM, et al. 2002. Sequential SNARE disassembly and GATE-16-GOS-28 complex assembly mediated by distinct NSF activities drives Golgi membrane fusion. J. Cell Biol. 157:1161–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munro S, Pelham HR. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899–907 [DOI] [PubMed] [Google Scholar]
- 25. Oldstone MBA. 2002. Biology and pathogenesis of lymphocytic choriomeningitis virus infection, p 83–118 In Oldstone MBA. (ed), Arenaviruses I: the epidemiology, molecular and cell biology of arenaviruses. Springer-Verlag, New York, NY: [DOI] [PubMed] [Google Scholar]
- 26. Palacios G, et al. 2008. A new arenavirus in a cluster of fatal transplant-associated diseases. N. Engl. J. Med. 358:991–998 [DOI] [PubMed] [Google Scholar]
- 27. Pasqual G, Burri DJ, Pasquato A, de la Torre JC, Kunz S. 2011. Role of the host cell's unfolded protein response in arenavirus infection. J. Virol. 85:1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pasquato A, et al. 2011. Arenavirus envelope glycoproteins mimic autoprocessing sites of the cellular proprotein convertase subtilisin kexin isozyme-1/site-1 protease. Virology 417:18–26 [DOI] [PubMed] [Google Scholar]
- 29. Peters CJ. 2002. Human infection with arenaviruses in the Americas. Curr. Top. Microbiol. Immunol. 262:65–74 [DOI] [PubMed] [Google Scholar]
- 30. Pullikotil P, Benjannet S, Mayne J, Seidah NG. 2007. The proprotein convertase SKI-1/S1P: alternate translation and subcellular localization. J. Biol. Chem. 282:27402–27413 [DOI] [PubMed] [Google Scholar]
- 31. Pullikotil P, Vincent M, Nichol ST, Seidah NG. 2004. Development of protein-based inhibitors of the proprotein of convertase SKI-1/S1P: processing of SREBP-2, ATF6, and a viral glycoprotein. J. Biol. Chem. 279:17338–17347 [DOI] [PubMed] [Google Scholar]
- 32. Rawson RB, Cheng D, Brown MS, Goldstein JL. 1998. Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J. Biol. Chem. 273:28261–28269 [DOI] [PubMed] [Google Scholar]
- 33. Rojek JM, Lee AM, Nguyen N, Spiropoulou CF, Kunz S. 2008. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J. Virol. 82:6045–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rojek JM, et al. 2010. Targeting the proteolytic processing of the viral glycoprotein precursor is a promising novel antiviral strategy against arenaviruses. J. Virol. 84:573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rojek JM, Sanchez AB, Nguyen NT, de la Torre JC, Kunz S. 2008. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J. Virol. 82:7677–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schroder M, Kaufman RJ. 2005. The mammalian unfolded protein response. Annu. Rev. Biochem. 74:739–789 [DOI] [PubMed] [Google Scholar]
- 37. Seidah NG, Chretien M. 1999. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 848:45–62 [DOI] [PubMed] [Google Scholar]
- 38. Seidah NG, Day R, Marcinkiewicz M, Chretien M. 1998. Precursor convertases: an evolutionary ancient, cell-specific, combinatorial mechanism yielding diverse bioactive peptides and proteins. Ann. N. Y. Acad. Sci. 839:9–24 [DOI] [PubMed] [Google Scholar]
- 39. Toure BB, et al. 2000. Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J. Biol. Chem. 275:2349–2358 [DOI] [PubMed] [Google Scholar]
- 40. Urata S, et al. 2011. Antiviral activity of a small-molecule inhibitor of arenavirus glycoprotein processing by the cellular site 1 protease. J. Virol. 85:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vincent MJ, et al. 2003. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J. Virol. 77:8640–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weber EL, Buchmeier MJ. 1988. Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology 164:30–38 [DOI] [PubMed] [Google Scholar]
- 43. Wong J, Quinn CM, Brown AJ. 2006. SREBP-2 positively regulates transcription of the cholesterol efflux gene, ABCA1, by generating oxysterol ligands for LXR. Biochem. J. 400:485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wright KE, Spiro RC, Burns JW, Buchmeier MJ. 1990. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology 177:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang J, et al. 2001. Decreased lipid synthesis in livers of mice with disrupted site-1 protease gene. Proc. Natl. Acad. Sci. U. S. A. 98:13607–13612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye J, et al. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6:1355–1364 [DOI] [PubMed] [Google Scholar]
- 47. Zhong M, et al. 1999. The prosegments of furin and PC7 as potent inhibitors of proprotein convertases. In vitro and ex vivo assessment of their efficacy and selectivity. J. Biol. Chem. 274:33913–33920 [DOI] [PubMed] [Google Scholar]