Abstract
The Epstein-Barr virus (EBV) lytic transactivator Rta activates promoters through direct binding to cognate DNA sites termed Rta response elements (RREs). Rta also activates promoters that apparently lack Rta binding sites, notably Zp and Rp. Chromatin immunoprecipitation (ChIP) of endogenous Rta expressed during early replication in B95-8 cells was performed to identify Rta binding sites in the EBV genome. Quantitative PCR (qPCR) analysis showed strong enrichment for known RREs but little or no enrichment for Rp or Zp, suggesting that the Rta ChIP approach enriches for direct Rta binding sites. Rta ChIP combined with deep sequencing (ChIP-seq) identified most known RREs and several novel Rta binding sites. Rta ChIP-seq peaks were frequently upstream of Rta-responsive genes, indicating that these Rta binding sites are likely functioning as RREs. Unexpectedly, the BALF5 promoter contained an Rta binding peak. To assess whether BALF5 might be activated by an RRE-dependent mechanism, an Rta mutant (Rta K156A), deficient for DNA binding and RRE activation but competent for Zp/Rp activation, was used. Rta K156A failed to activate BALF5p, suggesting this promoter can be activated by an RRE-dependent mechanism. Rta binding to late gene promoters was not seen at early time points but was specifically detected at later times within the Rta-responsive BLRF2 and BFRF3 promoters, even when DNA replication was inhibited. Our results represent the first characterization of Rta binding to the EBV genome during replication, identify previously unknown RREs, such as one in BALF5p, and highlight the complexity of EBV late gene promoter activation by Rta.
INTRODUCTION
Epstein-Barr virus (EBV) infection is associated with infectious mononucleosis and, rarely, B-cell lymphoproliferative disease, B-cell lymphomas, Hodgkin lymphoma, and nasopharyngeal carcinoma (51, 67). Although EBV malignancies are linked to latent infection, recent evidence suggests replication may play a more direct role in EBV pathogenesis (39, 58). Moreover, lytic replication is essential for the continued high prevalence of EBV infection in the human population. During EBV replication, most viral genes are expressed, the EBV genome is replicated by viral enzymes, and infectious virions are produced. This cascade of EBV lytic gene expression is controlled by two viral transcription factors, Zta (Z, ZEBRA) and Rta (R), encoded by the BZLF1 and BRLF1 genes, respectively (50, 61). Deletion of either BZLF1 or BRLF1 renders the virus incompetent for DNA replication and virion production (21).
Zta and Rta are transcription factors that activate lytic gene promoters in a temporally controlled manner (47). Initial Zta or Rta expression activates the Zta (Zp) and Rta (Rp) promoters (1, 23, 64), as well as those of early genes, which encode proteins involved in viral DNA replication (17, 22). Late transcription follows DNA replication and can be blocked by inhibitors of DNA replication, such as phosphonoacetic acid (PAA) (37, 38). Lytic gene promoters vary in their responsiveness to Zta and Rta (61). Some genes are activated primarily by Rta and others primarily by Zta, and some are synergistically activated by Zta and Rta (15, 21, 25, 26, 41, 46, 57, 61, 65).
Zta is a bZIP DNA binding protein that activates gene expression by binding to Zta response elements within promoters (6, 20, 24, 44, 54, 72). Multiple mechanisms have been proposed for Rta promoter activation. Rta consists of an amino-terminal dimerization (amino acids 1 to 231) and DNA binding (amino acids 1 to 280) domain (60) fused to a carboxy-terminal activation domain (amino acids 416 to 605) that interacts with basal cellular transcription factors (32, 59, 60) and the EBV Rta regulator LF2 (8, 9, 33). Rta activates several promoters through direct binding to cognate DNA at sites called Rta response elements (RREs). In vitro, Rta binds preferentially to DNA containing the sequence GNCCN9GGNG, where N can be any base, but the 9-nucleotide spacing appears to be critical (13, 29). Although this consensus sequence occurs 354 times in the B95-8 genome, only a limited number of RREs have been characterized in the EBV genome. These include Rta binding sites in the BMLF1, BMRF1, BALF2, BaRF1, and BLRF2 promoters (13, 27, 41, 62, 65), as well as in the BHLF1/BHRF1 promoter (also called the BHLF1/LF3 promoter or the DL/DR enhancer), which is part of the viral origin of replication (oriLyt) (28, 30, 66). Other Rta-activated promoters apparently lack Rta binding sites. The best-studied examples of these so-called non-RRE promoters are Zp and Rp, the immediate-early gene promoters. Rta activates its own promoter via Sp1 sites, potentially via interactions with the Sp1 binding protein MCAF1 (10, 64). However, it is not clear whether nuclear Rta is required for Rp activation, and thus the Sp1 sites may instead be downstream of cytoplasmic signaling cascades triggered by Rta expression (10, 33, 56, 64). Rta activation of Zp requires the Zp promoter element ZII and is thought to be mediated by Rta-dependent activation of cellular signaling pathways in the cytoplasm (1, 16, 71). Recently, Rta has been reported to bind Zp through the cellular transcription factor Oct-1, which acts as a positive regulator of EBV lytic gene expression (68). A third example of a promoter that may be activated without direct Rta binding is that of the EBV DNA polymerase (BALF5), which has been reported to require USF and E2F binding sites for activation (55).
Rta activation of lytic genes is critical for EBV to enter and progress through the viral replication cycle. As described above, Rta appears to activate its target genes through at least two distinct mechanisms. Our previous work suggests that LF2-mediated relocalization of nuclear Rta provides a switch between Rta activation modes, since LF2 specifically inhibits RRE-dependent promoter activation but allows activation of Zp or Rp. This highlights the importance of characterizing which Rta-responsive promoters are downstream of the different activation modes. The fact that the BALF2 RRE does not conform to the GNCCN9GGNG consensus, while Rp, which does not require direct Rta binding for activation, contains a GNCCN9GGNG consensus site, underscores the limitations of predicting functional RREs and highlights the need for an empirical assessment of Rta binding to DNA. Ideally this method should capture interactions of endogenous Rta with the EBV episome during replication to account for modifications of Rta and the chromatin state of the viral DNA. Therefore, we established a chromatin immunoprecipitation (ChIP) protocol for endogenous Rta in B95-8 cells that were induced for replication, followed by deep sequencing (ChIP-seq) that allowed the genome-wide analysis of Rta DNA binding. Our study represents the most detailed characterization of Rta binding to the EBV genome during replication reported to date, confirms that many previously characterized RREs are bound in vivo, and indicates several more Rta binding sites that are likely RREs.
MATERIALS AND METHODS
Cell culture.
293T is a human embryonic kidney cell line and was cultured in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) supplemented with l-glutamine (Gibco), penicillin-streptomycin (Gibco), and 10% FetalPlex (Gemini Bio-Products, West Sacramento, CA). The B95-8 Z-HT cell line was derived by stable expression of Z-HT in the EBV-transformed cotton-top tamarin cell line B95-8 as previously described (42) and was cultured in RPMI 1640 medium (Gibco) supplemented with l-glutamine, penicillin-streptomycin, and 10% FetalPlex. B95-8 Z-HT cells were induced for lytic replication at a density of 3 × 105 cells/ml by addition of 300 nM 4-hydroxytamoxifen (4HT) (Sigma, St. Louis, MO). When indicated, viral DNA replication was blocked with 1.6 mM PAA (225 μg/ml; Sigma) that was present in the B95-8 Z-HT cell culture medium from the start of 4HT treatment until harvest.
Plasmids.
pcDNA3-Rta, pGL3-BALF2p, pGL3-BMLF1p, pGL3-BMRF1p, pGL3-Zp, and pGL3-Rp have been described previously (8, 33). The BLRF2 (−349/+28; B95-8 nucleotides 88548 to 88924), BFRF3 (−727/+128; B95-8 nucleotides 60585 to 61439), and BALF5 (−376/+16; B95-8 nucleotides 156859 to 157250) promoters were amplified from B95-8 genomic DNA and cloned into the pGL3 promoter (Promega, Madison, WI). pRL-EF1 was constructed by cloning the human elongation factor 1 alpha promoter into the pRL-null reporter control vector (Promega).
Antibodies.
The following antibodies were used for Western blotting: mouse monoclonal antibodies against EBV Rta (8C12, 1:500; Argene, Varilhes, France), EBV Zta (AZ-69; 1:500; Argene), EBV BMRF1 (R3, 1:1,000; Millipore, Bedford, MA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (6C5, 1:12,500; Millipore, Bedford, MA), rabbit polyclonal serum against EBV BLRF2 (SLO25-1, 1:200; generous gift from George Miller, Yale University School of Medicine, New Haven, CT), and rat monoclonal antibody against EBV BFRF3 (VCA-p18, 1:250; generous gift from Jaap Middledorp, VU University Medical Center, Amsterdam, The Netherlands).
Western blot analysis.
Total cell lysates were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, blotted onto a nitrocellulose membrane, and probed with appropriate antibodies. After extensive washing, horseradish peroxidase-conjugated secondary antibodies (Jackson Immuno Research, West Grove, PA) were applied, and the membrane was washed again, developed with a chemiluminescence reagent (Perkin Elmer, Waltham, MA) and visualized on a Kodak Image Station 4000R system (Kodak Molecular Imaging Systems, Rochester, NY).
Reporter assays.
Up to 0.75 μg of total DNA was transfected with Effectene (Qiagen, Valencia, CA) into 293T cells grown in 6-well plates. After 48 h, cells were lysed in reporter lysis buffer (Promega) and clarified by centrifugation. Firefly luciferase and renilla luciferase (Dual-Luciferase reporter assay system; Promega) values were measured using an LMax II384 luminescence microplate reader (Molecular Devices, Sunnyvale, CA). Firefly luciferase reporter values were controlled for experimental variability by correction with the renilla luciferase values.
ChIP assays.
B95-8 Z-HT cells were treated with 4HT or PAA as indicated. ChIP assays were performed following the Millipore chromatin immunoprecipitation (ChIP) assay kit protocol (Millipore). Briefly, 1 × 106 cells per ChIP were fixed in 1% (wt/vol) formaldehyde (Sigma) for 10 min at 37°C, followed by two washes with PBS. Cells were lysed in 1 ml of lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% [wt/vol] SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml leupeptin, and 20 μg/ml aprotinin) for 10 min on ice before extensive sonication using a 550 sonic dismembrator (Fisher Scientific, Pittsburgh, PA). After extract clearing by centrifugation, supernatants were diluted 1:10 in dilution buffer (16.7 mM Tris-HCl [pH 8.1], 1.2 mM EDTA, 167 mM NaCl, 1.1% [vol/vol] Triton X-100, 0.01% [wt/vol] SDS, 1 mM PMSF, 1 μg/ml leupeptin, and 20 μg/ml aprotinin) and incubated with protein A-Sepharose 4B (Sigma) for 1 h with rotation at 4°C. Protein A-Sepharose was pelleted, and supernatants were used in ChIP experiments. Input DNA (25%) was stored at −20°C until elution of samples. Four micrograms of monoclonal Rta antibody (8C12) was added per 1 × 106 cells, followed by incubation overnight at 4°C with rotation. After incubation, 65 μl of salmon sperm DNA-protein A agarose (Millipore) was added to each sample and incubated for 2 h at 4°C with rotation to capture the antibody. Samples were then centrifuged, and protein A beads were washed once with cold low-salt wash buffer (20 mM Tris-HCl [pH 8.1], 2 mM EDTA, 150 mM NaCl, 1% [vol/vol] Triton X-100, and 0.1% [wt/vol] SDS), twice with high-salt wash buffer (identical to low-salt wash buffer, except with 500 mM NaCl), twice with LiCl wash buffer (10 mM Tris-HCl [pH 8.1], 1 mM EDTA, 0.25 M LiCl, 1% [vol/vol] NP-40, and 1% deoxycholic acid), and finally twice with TE buffer (10 mM Tris-HCl [pH 8.1], 1 mM EDTA). Beads were then resuspended in 250 μl of elution buffer (0.1 M NaHCO3, 1% [wt/vol] SDS) and rotated for 15 min at room temperature. Two rounds of elution of protein-DNA complexes were pooled, and NaCl was added to the samples and inputs to a final concentration of 200 mM NaCl and heated at 65°C for at least 4 h. DNA was purified using the QIAquick PCR purification kit and quantified with the Power SYBR green PCR master mix (Applied Biosystems, Warrington, United Kingdom) using a 7300 real-time PCR system (Applied Biosystems). Purified inputs were serially diluted to 5, 1, and 0.2% and used in real-time PCRs for standardization. The sequences of primers used for real-time PCR are listed in Table 1.
Table 1.
Sequences of primers used for real-time PCR
| Site | Forward/reverse | Sequence (5′–3′) |
|---|---|---|
| BALF2p RRE | Forward | TGTGGTCATCCAGGTAGTTTCGCA |
| Reverse | ACTACGACTACTGGTCGCGGCT | |
| BALF2p CTL | Forward | CAACGCCCGGGTCCTTGCTA |
| Reverse | AAAGGAGGACATGCGAGAATTGGC | |
| BMLF1p RRE | Forward | GAGGGCCAGATGCAGGAGCTGA |
| Reverse | GTGGAGAATGTCTGCGCCATGATA | |
| BMLF1p CTL | Forward | TAGTCACTGGTGAGGTGGAGC |
| Reverse | GGTGACACCAAGTCCATCTCCAT | |
| BMRF1p RRE | Forward | ATCGCATCCTGGGTGACATTGGT |
| Reverse | TGATGCTAGTCATGTAGGTGAGCG | |
| BMRF1p CTL | Forward | AGCATGGCAACGACCAGTCATGT |
| Reverse | GGTCTGAGTTGGCCTTGACCTTTA | |
| Zp-3 | Forward | CATGCAGCAGACATTCATCATTTAGA |
| Reverse | CCATTTGGACGAACTGACCACAAC | |
| Zp-2 | Forward | CCATGCATATTTCAACTGGGCTGTC |
| Reverse | GGTGTGTCTATGAGGTACATTAGCA | |
| Zp-1 | Forward | CACGTCCCAAACCATGACATCACA |
| Reverse | GGCAAGGTGCAATGTTTAGTGAGT | |
| Rp-3 | Forward | TCCAGGGACGATTTAATCCCAGC |
| Reverse | AGGTCTCACCTGGAATAACACCCA | |
| Rp-2 | Forward | TCTACCAGATGTTGGGAGGGCATT |
| Reverse | TTCGCGATGCTATAAACCAGACCC | |
| Rp-1 | Forward | CCCATGTGATGGTCAGGGTTT |
| Reverse | AAAGGCCGGCTGACATGGATTACT | |
| BALF5p-1 | Forward | ATGGGATTAATGCCTGGACCCTCA |
| Reverse | AGCAGCCGGTGCAGATGCTCTA | |
| BLRF2p-2 | Forward | GGGAAGGTCCTAAGAAAGCCGTTT |
| Reverse | CCGCGTTTGTGGGACTGGAA | |
| BLRF2p-1 | Forward | ACTGAAGCCCAGGACCAGTTCTA |
| Reverse | TAAGACAAGCGTCAGAAGTGCCCA | |
| BFRF3p-2 | Forward | TGGACTGCGGTTACTGCTTGAACT |
| Reverse | AGGTCAGCACATGCTTCTCCTTCT | |
| BFRF3p-1 | Forward | TCCCGCCTCTTGGATGCCATCAT |
| Reverse | CTGGCCTCTGTCCGCAAAGTTAAA | |
| CTLsite | Forward | ATCTACCTCGGTTGTGCAGGAA |
| Reverse | AGGTGGGCATCTTCTGCTTCCTTT |
ChIP-seq.
The ChIP protocol described above was scaled up for subsequent Illumina sequencing (ChIP-seq). The large-scale ChIP deviated from the small-scale ChIP as follows. Cells (450 × 106) were cross-linked, washed, and lysed at a density of 25 × 106 cells/ml. Immunoprecipitation was carried out with 160 μg of monoclonal Rta antibody (8C12). The immunocomplexes were captured with a total of 900 μl of salmon sperm DNA-protein A agarose, which was washed once with each wash buffer and twice with the TE buffer. After elution and DNA purification, libraries were built from the chromatin-immunoprecipitated DNA and from an input sample (0.2%) using the ChIP-Seq DNA sample prep kit (Illumina, San Diego, CA). Single-end reads of 50 bp in length were sequenced on an Illumina HiSeq2000 platform (Illumina). Library preparations and Illumina sequencing were performed by The Center for Cancer Computational Biology, Dana-Farber Cancer Institute, Boston, MA. Sequence reads were mapped to the B95-8 genome using the aligner Mapping and Assembly with Qualities (MAQ) (53) and displayed with a local installation of the UCSC genome browser. Position-specific read depth was calculated by summing the first 36 bp of all forward and reverse reads at each location in the EBV genome. Rta ChIP-enriched regions were identified using the SPP software package (49) with the foreground window size set to 500 bp and a P value threshold of 0.01 (enrichment score). High-confidence peaks were defined as Rta ChIP-seq peaks with a read depth of at least 600 and an enrichment score of higher than 0.1. Three hundred fifty-four consensus Rta binding sites were identified using the Vector NTI software program (Invitrogen) to search the B95-8 genome (GenBank V01555.2) for the GNCCN9GGNG sequence. Rta binding sites were also predicted using a 17-bp position-specific scoring matrix based on oligonucleotides bound by Rta (29). Matrix scores were calculated as log odds relative to the background, and the higher score (sense or antisense) was assigned for each position. A threshold value of 0.5 predicting 480 Rta binding sites in the B95-8 genome was used for Tables 2 and 3 and for Fig. 4A.
Table 2.
High-confidence Rta ChIP-seq peaks (read depth of at least 600 and enrichment score higher than 0.1)
| Position (B95-8 nucleotide) | Read depth | Enrichment score | Distance (bp) to nearest predicted Rta binding site | Downstream gene(s) (nearest TSS within 2 kb) | Gene classa | Rta-responsive gene (reference) | Known RRE |
|---|---|---|---|---|---|---|---|
| 53550 | 11,891 | 3.96 | 6 | BHRF1, BHLF1 | E, E | Yes (31), yes (31) | Yes |
| 56800 | 774 | 0.87 | 20 | No | |||
| 57300 | 1,168 | 0.79 | 240 | BFLF2 | E | No (57) | No |
| 58400 | 961 | 0.96 | 49 | BFRF0.5 | E | unknown | No |
| 62050 | 970 | 0.93 | 3 | No | |||
| 76050 | 1,063 | 0.95 | 85 | BORF2 | E | Yes (57) | No |
| 79550 | 1,037 | 0.66 | 10 | BMRF1 | E | Yes (35) | Yes |
| 81050 | 679 | 0.42 | 94 | No | |||
| 84675 | 2,753 | 1.85 | 33 | SM (BMLF1) | E | Yes (7), (46) | Yes |
| 100375 | 734 | 0.65 | 64 | No | |||
| 119175 | 724 | 0.56 | 248 | BBLF2/3 | E | Yes (57) | No |
| 124350 | 642 | 0.13 | 46 | BGLF4 | E | Yes (73) | No |
| 125175 | 918 | 0.81 | 25 | BGLF3 | Unknown | No (57) | No |
| 157010 | 714 | 0.47 | 26 | BALF5 | E | Yes (55) | No |
| 165050 | 1,092 | 0.77 | 36 | BALF2, BARF1 | E, E | Yes (41), unknown | Yes |
E, early.
Table 3.
Rta ChIP-seq peaks that met only one of the two criteria applied to select the high-confidence peaks
| Position (B95-8 nucleotide) | Read depth | Enrichment score | Distance (bp) to nearest predicted Rta binding site | Downstream gene(s) (nearest TSS within 2 kb) | Gene classa | Rta responsiveness (reference) | Noteb | Known RRE |
|---|---|---|---|---|---|---|---|---|
| 13300 | 250 | 0.41 | 12 | Wp | No | |||
| 16425 | 237 | 0.58 | 41 | Wp | No | |||
| 19425 | 289 | 0.71 | 31 | Wp | No | |||
| 22325 | 294 | 0.80 | 120 | Wp | No | |||
| 25625 | 289 | 0.80 | 25 | Wp | No | |||
| 28525 | 282 | 0.76 | 147 | Wp | No | |||
| 31725 | 329 | 0.84 | 19 | Wp | No | |||
| 34800 | 226 | 0.82 | 16 | Wp | No | |||
| 37800 | 315 | 0.74 | 88 | Wp | No | |||
| 41000 | 260 | 0.72 | 40 | Wp | No | |||
| 44100 | 254 | 0.56 | 68 | Wp | No | |||
| 47125 | 229 | 0.41 | 21 | Wp | No | |||
| 48425 | 633 | 0.00 | 352 | No | ||||
| 50550 | 526 | 0.71 | 31 | No | ||||
| 52100 | 552 | 0.71 | 365 | BHRF1 | E | Yes (57) | No | |
| 59800 | 525 | 0.45 | 58 | BFLF1, BFRF3 | E (BFLF1), L (BFRF3) | Yes (57), yes (69) | No | |
| 64400 | 359 | 0.20 | 204 | No | ||||
| 66350 | 412 | 0.30 | 320 | No | ||||
| 71450 | 313 | 0.11 | 290 | No | ||||
| 75300 | 374 | 0.24 | 6 | BOLF1 | L | No (57) | No | |
| 91350 | 646 | 0.03 | 232 | BLLF2 | E | Yes (57) | No | |
| 110025 | 600 | 0.00 | 162 | BKRF3 | E | Yes (57) | No | |
| 160500 | 311 | 0.17 | 10 | BALF4 | L | No (57) | No | |
| 161300 | 351 | 0.13 | 36 | No |
E, early; L, late.
Wp, W repeat promoter.
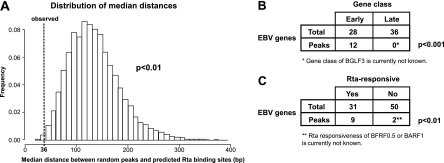
Fig 4.
The high-confidence Rta ChIP-seq peaks are within significant proximity of predicted Rta binding sites, and the genes downstream of the peaks are significantly enriched for early genes and for Rta-responsive genes. (A) Histogram of median distance between randomized peaks and predicted Rta-binding sites. Rta binding sites in the EBV B95-8 genome were predicted using a position-weight matrix, and the distances between these sites and 15 randomly distributed peaks were simulated (sample size, 1,000; repeated for 10 times). The empirical P value for 15 peaks to have the observed median distance of 36 bp, which is indicated by the dashed line, is 0.0021 (standard deviation, 0.0012). (B) Contingency table showing the distribution of early and late EBV genes within the 64 early and late EBV genes (Total) profiled by Yuan et al. (77) and within the downstream genes (Peaks) assigned to the high-confidence Rta ChIP-seq peaks (for genes, see Table 2). Among the 13 downstream genes, BGLF3 was exempt from the analysis because its gene class has not been determined yet. The P value was calculated using Fisher's exact test. (C) Contingency table showing the distribution of Rta-responsive genes within the 81 EBV genes (Total) profiled by Lu et al. (57) and within the downstream genes (Peaks) assigned to the high-confidence Rta ChIP-seq peaks (for genes and references, see Table 2). Among the 13 downstream genes, BARF1 and BFRF0.5 were exempt from the analysis because their Rta responsiveness has not been determined yet. The P value was calculated using Fisher's exact test.
Quantification of viral DNA amplification by real-time PCR.
Cells were resuspended in 75 μl of HE buffer (10 mM HEPES [pH 7.9] and 1 mM EDTA) and boiled at 95°C for 5 min. Lysates were digested with 130 μg/ml of proteinase K (Sigma) at 37°C for 12 h, followed by proteinase K inactivation at 95°C for 20 min. EBV DNA was quantified with the 7300 real-time PCR system and the Power SYBR green PCR master mix using primers specific for the BMLF1 RRE and corrected with the relative amount of cellular DNA as quantified by real-time PCR.
EMSA.
The electrophoretic mobility shift assay (EMSA) was performed as previously described using the BMLF1 RRE probe (8). EMSA buffer [10 mM HEPES (pH 7.9), 5% glycerol, 0.5 mM EDTA, 1 mM dithiothreitol, 125 μM PMSF, 100 μg/ml bovine serum albumin (BSA), 40 μg/ml poly(dI-dC), 50 mM KCl] was mixed with 1 to 2 ng of probe and 2 μg of nuclear extract from transfected 293T cells and incubated for 15 min at room temperature. For supershift assays, 8C12 anti-Rta antibody was preincubated with nuclear extract in reaction buffer on ice for 10 min. Samples were separated on a 5% polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE). The gel was dried and visualized on a PhosphorImager (Molecular Dynamics).
RESULTS
Rta binds to known RREs within early promoters during EBV replication.
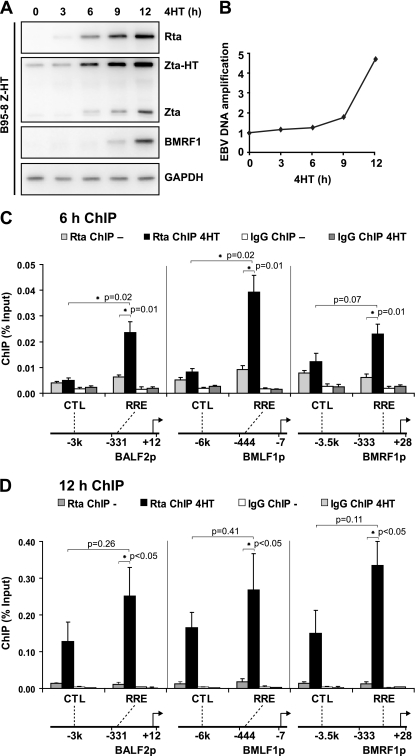
In orderto identify sites in the EBV genome bound by Rta during replication, an Rta chromatin immunoprecipitation (ChIP) assay was established. B95-8 Z-HT cells were used for these experiments, since the stable expression of Zta fused to the hydroxytamoxifen-responsive estrogen receptor hormone binding domain (Z-HT) allowed for efficient and synchronous induction of virus replication. Furthermore, this approach ensured that Rta was expressed under the control of its own promoter at endogenous levels. After the addition of 4HT, Rta protein expression was first detected at 3 h, endogenous Zta was observed at 6 h, and the BMRF1 protein, an early gene product, was detected at 9 h (Fig. 1A). To further assess the activation of EBV replication, viral DNA was quantified by real-time PCR. Significant EBV DNA amplification was not observed until at least 9 h after 4HT addition (Fig. 1B). At later time points, after DNA replication occurred, we observed an increased background signal in our quantitative PCR (qPCR) assays at the non-RRE control sites (Fig. 1D). This is not directly attributable to increased DNA levels in the cell, since all ChIP results are expressed relative to input. Instead, this may be caused by the complex branching structures that form during herpesviral DNA replication (70), which, in combination with the cross-linking used in the ChIP assay, increases the precipitation of EBV DNA distant from Rta binding sites. In order to avoid this effect, we elected to restrict our analysis to early time points. This also ensured that all Rta-DNA interactions characterized were with parental EBV episomes as opposed to interactions with newly synthesized DNA.
Fig 1.
Rta interacts specifically with known RREs during lytic replication. (A) Western blot results for the indicated EBV proteins from B95-8 Z-HT cells treated with 4HT for 0, 3, 6, 9,or 12 h and lysed in SDS loading buffer. Detection of GAPDH served as a loading control. (B) Relative EBV DNA levels in B95-8 Z-HT cells treated with 4HT for 0, 3, 6, 9, and 12 h are shown. EBV DNA in PCR lysates was quantified by real-time PCR for a site in the viral genome (BMLF1 RRE) and corrected for cellular DNA input by real-time PCR for a site in the cellular genome (CD21 exon 5). (C) Chromatin immunoprecipitation assay for Rta (Rta ChIP) and with IgG control (IgG ChIP). B95-8 Z-HT cells were induced for viral replication by treatment with 4HT for 6 h (black and dark-gray bars) or left untreated (gray and white bars). Amounts of precipitated viral DNA, as measured by real-time PCR using primers specific to the RRE in the BALF2, BMLF1, or BMRF1 promoter (RRE) and control (CTL) sites 3 to 6 kb upstream of the corresponding RRE, are shown on the y axis. Locations of RRE and CTL sites are indicated relative to the transcriptional start site and the core promoter used in reporter gene assays (BALF2p, BMLF1p, BMRF1p); schematic drawings on the bottom are not to scale. The bar graph represents the amount of DNA precipitated relative to the amount of DNA in the corresponding input sample. Data are arithmetic means from four independent ChIP experiments. Error bars indicate standard errors of the means. Asterisks mark statistically significant changes in the Rta ChIP from induced cells (Rta ChIP 4HT) relative to the Rta ChIP from untreated cells (Rta ChIP −) or in the Rta ChIP 4HT results at RREs relative to the results at the corresponding CTL sites (one-tailed t test for unequal variances). (D) Rta ChIP and IgG ChIP from B95-8 Z-HT cells that were induced for viral replication by treatment with 4HT for 12 h (black and dark-gray bars) or left untreated (gray and white bars). The results were analyzed and are presented as in Fig. 1C. Data are arithmetic means from three independent ChIP experiments.
Rta ChIPs were performed with B95-8 Z-HT cells that had been treated with 4HT for 6 h (Fig. 1C, black bars) or left untreated (gray bars), and chromatin-immunoprecipitated DNA was quantified relative to input DNA using real-time PCR with primer sets covering the BALF2p, BMLF1p, or BMRF1p RRE (RRE) or control (CTL) sites that were chosen to be 3 to 6 kb upstream of the corresponding RRE and distant from any GNCCN9GGNG consensus site. Rta occupancy of all three RREs significantly increased over that of the untreated control, while there was no significant increase of Rta occupancy at any of the control sites (Fig. 1C). ChIPs with control IgG resulted in consistently low signals (Fig. 1C, white and dark-gray bars). These results indicate that the Rta ChIP can specifically detect endogenous Rta interactions with known RREs in the context of the viral episome during EBV replication.
Rta occupancy of the Rp and Zp promoters is low.
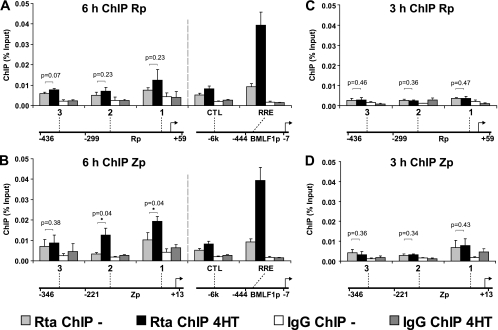
Although the immediate-early gene promoters Rp and Zp are widely acknowledged to be Rta responsive despite lacking functional Rta binding sites, there is controversy as to whether Rta localization to these promoters is required for their activation. To assess Rta occupancy of these promoters under endogenous conditions, Rta binding to the core promoter sequences (Rp, −299/+59; Zp, −221/+13), the minimal regions reported to confer Rta responsiveness, was examined by ChIP assay. This was quantified by real-time PCR for two sites at each promoter (Rp-1, −86/+3; Rp-2, −345/−251; Zp-1, −80/+13; Zp-2, −221/−91) and an additional site, located just upstream of the corresponding core promoter (Rp-3, −436/−352; Zp-3, −346/−222) (Fig. 2). Quantitative PCR signals for each of the three Rp sites did not increase significantly during replication (Fig. 2A). While there was no significant enrichment at the Zp-3 site, Zp core (Zp-1 and Zp-2) enrichment in Rta ChIPs increased slightly with induction of viral replication (Fig. 2B). This increase was statistically significant (Zp-1, P = 0.044; Zp-2, P = 0.038) but low compared to the increase at BMLF1p RRE, where the largest increase in Rta occupancy had been observed. Enrichment for Rp and Zp DNA in Rta ChIPs at 3 h (Fig. 2C and D), was even lower than observed at 6 h, suggesting that the low Rta binding observed at 6 h was not due to a decline from higher levels at earlier times. The Rta occupancy observed at Zp may reflect low-affinity interaction with nonconsensus Rta binding sites or an indirect association of Rta with the promoter. Overall these data suggest that the ChIP assay preferentially detects direct Rta binding to cognate DNA.
Fig 2.
Rta occupancy of Rp and Zp is low. Chromatin immunoprecipitation of the Rta protein (Rta ChIP) from B95-8 Z-HT cells induced for viral replication by treatment with 4HT for 6 h (A and B) or 3 h (C and D) (4HT, black bars) or left untreated (light-gray bars). Amounts of precipitated viral DNA were measured by real-time PCR using primers specific to three sites in the Rp promoter region (Rp-1, -2, and -3) (A and C) or three sites in the Zp promoter region (Zp-1, -2, and -3) (B and D). Locations of these sites are indicated relative to the transcriptional start site and the core promoter used in the reporter gene assays (Rp or Zp); schematic drawings on the bottom are not to scale. The bar graph represents the amount of DNA precipitated relative to the amount of DNA in the corresponding input sample. Arithmetic means from three independent Rta ChIP experiments and error bars indicating standard errors of the means are shown. Asterisks mark statistically significant (P < 0.05) Rta ChIP changes postinduction (Rta ChIP 4HT) relative to results for the uninduced control (Rta ChIP −) (one-tailed t test for unequal variances). For comparison, the 6-h ChIP results for the BMLF1p RRE and control site (CTL) and the corresponding locations are shown in panels A and B (results taken from Fig. 1C). Control IgG ChIPs (4HT, dark-gray bars; untreated, white bars) were performed with the same cell lysates used for the Rta ChIPs and analyzed in parallel.
Rta ChIP-seq identifies known and novel Rta binding sites in the EBV genome.
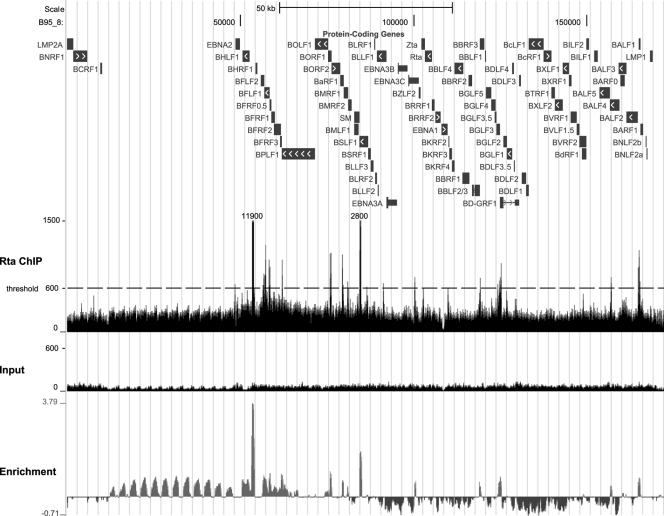
In order to identify Rta interactions with the EBV genome, a large-scale Rta ChIP was performed, and ChIP DNA as well as a DNA input sample (0.2%) were analyzed by high-throughput sequencing. Approximately 100 × 106 and 120 × 106 sequence reads were obtained from the Rta ChIP sample and the input sample, respectively; 1.14 × 106 ChIP reads (1.1%) and 0.21 × 106 of the input reads (0.18%) were mapped to the EBV B95-8 genome and displayed using the UCSC genome browser (http://genome.ucsc.edu/FAQ/FAQlicense.html) (5, 19, 48). The combined forward and reverse sequence reads mapping to the EBV genome from the Rta ChIP or the input sample are depicted in Fig. 3. Enrichment or depletion for the Rta ChIP sequences relative to the input was calculated and is displayed as the lower bound of the 95% confidence interval for enrichment using a log base 2 scale (Fig. 3). Fifteen high-confidence peaks were identified based on a read depth of at least 600 and an enrichment score relative to input higher than 0.1 (Table 2). Consistent with the observation that Rta ChIP enriches for Rta cognate DNA, 4 of the 6 tallest peaks corresponded to well-described RREs within the BHLF1/BHRF1, BMLF1, BMRF1, and BALF2 promoters. Further, the relative height of the peaks corresponded to the degree of Rta responsiveness, with BHLF1/BHRF1 being the strongest, followed by the BMLF1 RRE. All but two peaks were within 100 bp of a predicted Rta binding site. The median distance between the peaks and the nearest Rta binding site predicted by a position-specific weight matrix (27) was only 36 nucleotides (nt), which is highly significant (P = 0.0021) based on the null hypothesis that the peaks are randomly distributed over the EBV genome (Fig. 4A). Eleven of the 15 high-confidence peaks were tentatively associated with a regulated gene(s) based on being 2 kb or less upstream of its transcriptional start site. Two peaks were located between divergent transcriptional start sites. These included the peak within the known bidirectional BHLF1/BHRF1 promoter as well as a potential bidirectional promoter between BALF2 and BARF1. In agreement with the early time point chosen for the Rta ChIP-seq, EBV genes downstream of peaks were significantly enriched for early lytic genes (12 out of 13; P < 0.001) (Fig. 4B). Except for BGLF3, all genes listed had been previously classified as early genes (77). Furthermore, there was significant enrichment for genes previously demonstrated to exhibit Rta responsiveness (9 out of 13; P < 0.01) (Fig. 4C) (7, 31, 35, 41, 46, 55, 57, 73), suggesting that the observed Rta binding sites are upstream of Rta target genes.
Fig 3.
Rta ChIP-seq identifies sites in the EBV genome bound by Rta during replication. The UCSC genome browser window shows Rta ChIP and input sequence reads mapped to the B95-8 genome from ChIP-seq experiments (middle and lower tracks, respectively). The upper track displays protein-coding genes in the B95-8 genome, drawn schematically as blocks, with white arrows indicating open reading frame orientations. The Rta ChIP track represents the combined tracks of mapped forward and reverse Rta ChIP reads. Two Rta ChIP peaks are approximately 8- and 2-fold taller than the displayed scale and are therefore labeled with their approximate read depths (11,900 and 2,800). The dashed line indicates the read depth threshold of 600, one of two criteria applied for selection of high-confidence peaks (Table 2). The input track represents the combined tracks of mapped forward and reverse input reads. Sharp peaks were called using the SPP software package (49): the enrichment track shows conservative fold enrichment or depletion of Rta ChIP over the input profile on a log base 2 scale. The conservative fold enrichment reported by SPP is the lower bound of the 95% confidence interval; the depletion is the upper bound of the 95% confidence interval.
An additional set of 24 peaks met only one of the two selection criteria (read depth of at least 600 or an enrichment score higher than 0.1) and are listed in Table 3. These may represent lower-affinity Rta binding sites or sites where the chromatin state was not favorable for stable Rta binding. Alternatively, the weaker signal or lack of significant enrichment relative to background may indicate that there is not significant Rta binding at these sites.
An Rta DNA binding domain mutant fails to activate RRE promoters.
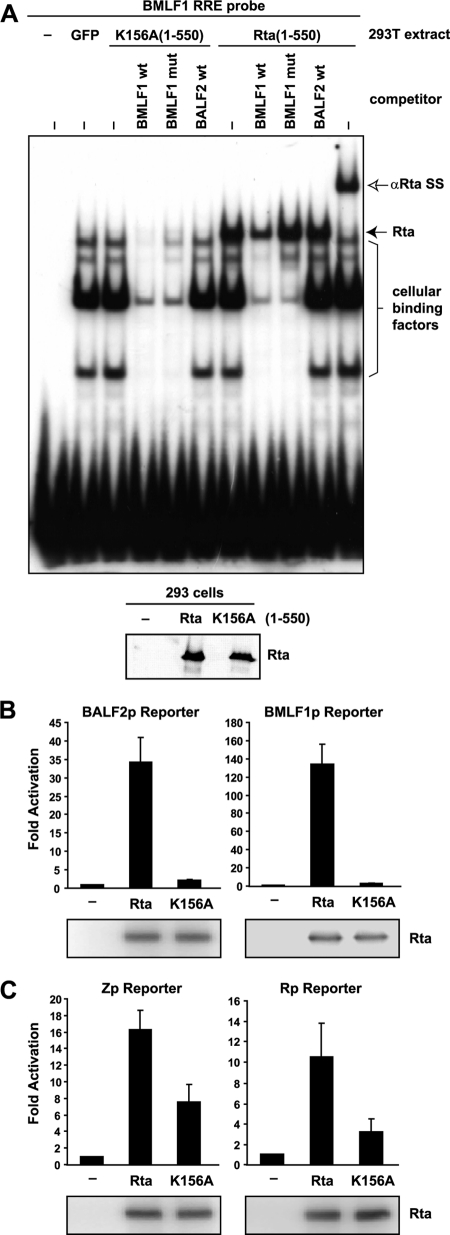
In order to develop a reagent that could discern whether promoters depend on RRE binding for their activation by Rta, we constructed an Rta DNA binding mutant that fails to bind RREs (Rta K156A), based on a DNA binding mutant described for its Kaposi's sarcoma-associated herpesvirus (KSHV) homologue, ORF50 (12). As expected, the K156A mutation abolished Rta DNA binding to the BMLF1 RRE in electrophoretic mobility shift assays, and Western blotting confirmed that Rta K156A was expressed at levels similar to those for wild-type Rta (Fig. 5A). In reporter assays, Rta K156A failed to activate the RRE-dependent BALF2p and BMLF1p constructs (Fig. 5B) but still stimulated Zp or Rp, albeit less efficiently than wild-type Rta (Fig. 5C). These results indicate that the Rta K156A mutant is completely defective for activation of RRE promoters and somewhat impaired for activation of Rp and Zp, which are upregulated independent of RRE binding.
Fig 5.
An Rta DNA binding domain mutant fails to activate RRE promoters. (A) Electrophoretic mobility shift assay (EMSA) comparing binding of Rta(1–550) K156A and Rta(1–550) to the BMLF1 RRE probe. EMSA was performed as previously described (8). Rta binding activity is indicated (filled arrow; Rta) and confirmed by supershifting (nonfilled arrow; SS). This activity was competed with excess cold probe (BMLF1wt) but not by the probe with the RRE site mutated (BMLF1mut) or by an oligonucleotide derived from the BALF2 promoter lacking a consensus RRE (BALF2wt). Cellular, Rta-independent binding activities are marked with a bracket on the right side. Nuclear extracts for the EMSA were prepared from 293T cells transiently transfected with plasmids expressing Rta 1–550 wt or K156A. Deletion of the C-terminal 55 residues of Rta has been shown to enhance RRE binding in EMSAs (13). Rta expression levels were confirmed by Western blotting of nuclear extracts (bottom panel). (B and C) Reporter assay results from 293T cells transfected with firefly luciferase reporter constructs from the BMLF1 (BMLF1p) or BALF2 (BALF2p) promoter (B) or BZLF1 (Zp) or BRLF1 (Rp) promoter (C) with or without Rta or Rta K156A. Firefly luciferase activities are shown as fold activation over reporter alone and were normalized with an Rta-independent renilla luciferase control. Data are averages for six transfections from three independent experiments, with error bars indicating standard deviations. Western blots of the cell lysates with anti-Rta antibody demonstrate Rta protein expression.
Rta DNA binding contributes to BALF5 promoter activation.
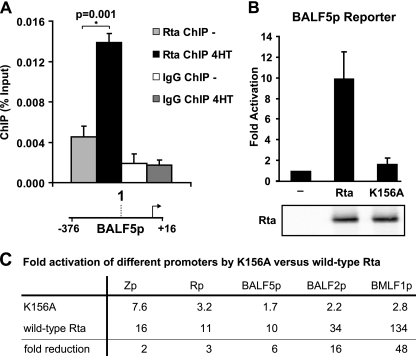
The significant Rta occupancy observed at the BALF5 promoter (read depth, 714; 0.47 log enrichment score) was unexpected, since this promoter had been previously reported to be activated indirectly by Rta (25, 55). Interestingly, the observed Rta binding peak corresponds to a region of the promoter previously demonstrated to confer Rta responsiveness (55). Rta binding to the BALF5 promoter was confirmed by Rta ChIP followed by quantitative PCR for a BALF5 promoter site (BALF5p-1, −166/−82), which is located within the Rta ChIP-seq peak. Rta occupancy was significantly increased at the BALF5 promoter 6 h after 4HT addition, confirming that Rta is present at this promoter (Fig. 6A). A portion of the BALF5p promoter (−376/+16), containing the putative RRE (located around −250 relative to the transcription start site), was cloned into the pGL3 reporter plasmid and tested for Rta activation in reporter gene assays (Fig. 6B). While wild-type Rta activated the BALF5p reporter 9.9-fold, the Rta K156A mutant activated only 1.7-fold. This fold activation ranks lowest among those of the lytic gene promoters tested for activation by Rta K156A (Fig. 6C) but corresponds to only a 6-fold reduction in activation, since the BALF5 promoter is activated less strongly by wild-type Rta than either the BALF2 or BMLF1 promoter. Overall, the finding of an Rta binding peak within a known Rta-responsive DNA element and the effect of the K156A mutation are most consistent with BALF5 promoter activation depending at least partially on Rta DNA binding.
Fig 6.
Rta DNA binding contributes to BALF5 promoter activation. (A) Chromatin immunoprecipitation assay for Rta (Rta ChIP) and with IgG control (IgG ChIP). B95-8 Z-HT cells were induced for replication by treatment with 4HT for 6 h (black and dark gray bars) or left untreated (gray and white bars). Amounts of precipitated viral DNA, as measured by real-time PCR using primers specific for a site within the BALF5 promoter (BALF5p-1), are shown on the y axis. The location of this site is indicated relative to the transcriptional start site and the promoter used in the reporter assays (BALF5p); the schematic drawing is not to scale. The bar graph represents the amount of DNA precipitated relative to the amount of DNA in the corresponding input sample. Arithmetic means from three independent ChIP experiments and error bars indicating standard errors of the means are shown. The asterisk marks the statistically significant (P < 0.05) Rta ChIP change relative to results for the uninduced control (one-tailed t test for unequal variances). (B) Reporter assay results from 293T cells transfected with firefly luciferase reporter constructs from the BALF5 (BALF5p) promoter with or without Rta or Rta K156A. Firefly luciferase activities are shown as fold activation over results with the reporter alone and were normalized with an Rta-independent renilla luciferase control. Data are averages for six transfections from three independent experiments, and error bars indicate standard deviations. Western blots of the cell lysates with anti-Rta antibody demonstrate Rta protein expression. (C) Summary of fold activations of different promoters by Rta K156 and wild-type Rta that were observed in reporter assays presented in Fig. 5 and 6B. Promoters were ranked according to fold reduction (ratio of activation by wild-type Rta over activation by Rta K156A) in ascending order.
Rta interacts with late gene promoters even when DNA replication is inhibited.
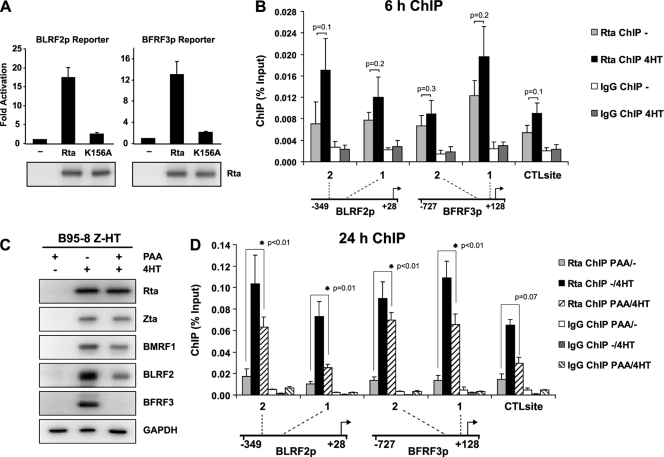
None of the 15 high-confidence Rta ChIP-Seq peaks were located upstream of a late gene, suggesting that Rta may not localize to late gene promoters at early time points in replication. To test whether the activation of late gene promoters requires Rta DNA binding, the responsiveness of BLRF2p and BFRF3p (13, 18, 69) to Rta and Rta K156A was compared in reporter assays (Fig. 7A). Wild-type Rta activated both reporters robustly (17.5-fold and 13.1-fold, respectively), but activation by Rta K156A was as low as that for RRE-dependent promoters (compare Fig. 5 and 6C), indicating that these late gene promoters are activated through direct Rta DNA binding. Given this, our failure to observe Rta binding to these promoters at early time points suggests that temporal regulation of Rta may be a mechanism governing sequential gene expression during replication. To further evaluate the binding of Rta to these promoters, Rta ChIP DNA from different time points was quantified by real-time PCR for BLRF2p and BFRF3p. Quantitative PCR signals for two sites in both BLRF2p and BFRF3p did not increase significantly 6 h post-induction of viral replication (BLRF2p-1, P = 0.2; BLRF2p-2, P = 0.1; BFRF3p-1, P = 0.2; BFRF3p-2, P = 0.3), corroborating the Rta ChIP-seq data (Fig. 7B) that indicate these promoters are not bound by Rta at early time points.
Fig 7.
Rta interacts with late gene promoters even when DNA replication is inhibited. (A) Reporter assay results from 293T cells transfected with firefly luciferase reporter constructs from the BLRF2 (BLRF2p) or BFRF3 (BFRF3p) promoter with or without Rta or Rta K156A. Firefly luciferase activities are shown as fold activation over reporter alone and were normalized with an Rta-independent renilla luciferase control. Data are averages for six transfections from three independent experiments, and error bars indicate standard deviations. Western blots of the cell lysates with anti-Rta antibody demonstrate Rta protein expression. (B) Rta ChIP and control IgG ChIP DNA from B95-8 Z-HT cells, which had been induced with 4HT for 6 h (black and light gray bars) or left untreated (gray and white bars), was analyzed by real-time PCR using primers specific for two sites each in the BLRF2 and the BFRF3 promoter (BLRF2p-1, BLRF2p-2, BFRF3p-1, and BFRF3p-2). The locations of these sites are indicated relative to the TSSs and the promoters used in the reporter assays (BLRF2p, BFRF3p); schematic drawings are not to scale. The bar graph represents the amount of DNA precipitated relative to the amount of DNA in the corresponding input sample. Arithmetic means from three independent Rta ChIP experiments and error bars indicating standard errors of the means are shown. Rta ChIP changes were tested for statistical significance using the one-tailed t test for unequal variances, and corresponding P values are depicted. (C) Western blot results from B95-8 Z-HT cells treated with PAA, 4HT, or both for 24 h and lysed in SDS loading buffer. Lysates were analyzed by Western blotting for the indicated EBV proteins. Detection of GAPDH served as a loading control. (D) Chromatin immunoprecipitation of the Rta protein (Rta ChIP). B95-8 Z-HT cells were induced for viral replication by treatment with 4HT for 24 h. PAA was added to 4HT-treated (dashed bars) and untreated (gray bars) cells in order to block viral replication. As positive controls, ChIPs were performed with cells that were induced for viral replication but not treated with PAA (Rta ChIP -/4HT, black bars). Amounts of precipitated viral DNA, as measured by real-time PCR using primers specific to two sites each in the BLRF2 promoter (BLRF2p-1 and BLRF2p-2) or in the BFRF3 promoter (BFRF3p-1 and BFRF3p-2), are shown on the y axis. Locations of these four sites are indicated relative to the TSSs and the promoters; schematic drawings are not to scale. Real-time PCR was additionally performed with primers specific to a control site (CTLsite) distant from any GNCCN9GGNG consensus site. The bar graph represents the amount of DNA precipitated relative to the amount of DNA in the corresponding input sample. Arithmetic means from three independent Rta ChIP experiments and error bars indicating standard errors of the means are shown. Asterisks mark statistically significant (P < 0.05) Rta ChIP changes (Rta ChIP PAA/4HT) relative to the uninduced control (Rta ChIP PAA/-) (one-tailed t test for unequal variances). Control IgG ChIPs were performed in parallel with cells that had been treated with PAA and/or 4HT.
Thus, Rta interactions with late gene promoters at 12 or 24 h were examined; however, a general increase in DNA precipitated with Rta was observed (Fig. 1D) at these times, likely due to the amplification and branching of viral DNA. Therefore, Rta ChIPs were performed with B95-8 Z-HT cells that had been induced for lytic replication for 24 h in the presence of 1.6 mM PAA, which efficiently abrogated viral DNA replication as observed in the ChIP input DNA quantification (data not shown). PAA treatment did not affect Rta or Zta protein expression but slightly reduced BMRF1 protein levels compared to those seen in the absence of PAA (Fig. 7C). In the presence of PAA, BLRF2 expression was dramatically decreased and BFRF3 expression was abrogated, confirming that expression of these late genes depends on DNA replication (Fig. 7C). Twenty-four hours after induction of replication with 4HT in the presence of PAA (Rta ChIP PAA/4HT), we detected a significant increase of Rta occupancy at the two sites within both BLRF2p (BLRF2p-1 and BLRF2p-2) (P ≤ 0.01) and BFRF3p (BFRF3p-1 and BFRF3p-2) (P < 0.01) compared to results for uninduced cells treated with PAA only (Rta ChIP PAA/−) (Fig. 7D). In contrast, Rta occupancy at a site (CTLsite) distant from any GNCCN9GGNG consensus sites did not increase significantly (P = 0.07) (Fig. 7D). These data suggest that Rta interaction with late gene promoters is delayed compared to its binding of early promoters. Since BFRF3 was not expressed in the presence of PAA despite significant Rta recruitment to its promoter, we conclude that Rta interaction with this late gene promoter is not sufficient for its activation.
DISCUSSION
In this article, we present a detailed characterization of Rta binding to the EBV genome during viral replication. The Rta ChIP-seq during early replication identified 15 high-confidence binding sites within the B95-8 genome (Table 2). Four of these peaks correspond to the BHLF1/BHRF1, BMLF1, BMRF1, and BALF2 promoter RREs (13, 41) and represent the first demonstration that these sites are occupied by Rta during early EBV replication (Fig. 1C and 3). The lone early gene RRE for which we did not observe a peak was the BaRF1 promoter. This may be due to the weak Rta responsiveness of this element, since we observed a correlation between responsiveness and read depth in our ChIP-seq. Alternatively, this element may not be bound by Rta during EBV replication. The status of the BaRF1 element as an RRE rests on its ability to bind Rta in EMSAs and to confer Rta responsiveness to the heterologous E4 promoter, but this does not guarantee that the chromatin state of this element in the replicating EBV genome is conducive to Rta binding (13). Seven of the remaining 11 Rta ChIP-seq peaks were near the potentially regulated genes BFLF2, BFRF0.5, BORF2, BBLF2/3, BGLF4, BGLF3, and BALF5. The following observations support the assertion that the Rta binding sites upstream of these genes are strong RRE candidates. First, the ChIP technique applied here enriches for direct Rta DNA-binding events. While known Rta-binding sites were reliably detected, the unbiased Rta ChIP-seq did not detect Rta interactions with Rp or Zp. Small-scale ChIPs confirmed the limited Rta presence at these promoters. These results do not exclude indirect Rta interactions with Zp or Rp (10, 68) but suggest that our Rta ChIP assay preferentially identifies direct Rta-DNA interactions. Additionally, the Rta ChIP-seq peak centers are significantly closer to in vitro Rta-binding consensus sites than expected for a random distribution of peaks, providing in vivo evidence for the GNCCN9GGNG consensus (29). Finally, our data establish a correlation between Rta binding and transactivation, because genes downstream of the Rta ChIP-seq peaks are significantly enriched for known Rta-responsive genes.
To test the hypothesis that the novel Rta binding sites in the EBV genome are RREs, we focused on the DNA polymerase promoter (BALF5p). Two Rta-responsive regions of BALF5p had been previously reported: an upstream element in which USF sites at −795/−786 appear to be critical and a downstream element at −239/−138, in which E2F sites at −186/−170 are important (25, 55). The peak of Rta binding within the downstream (−239/−138) element seen in our ChIP-seq analysis was unexpected and suggested that Rta may directly activate this promoter. To further explore this possibility, we first confirmed Rta occupancy of BALF5p at the −239/−138 site using local ChIP assays (Fig. 6A). Analysis of this BALF5p region (−376/+16) demonstrated that its Rta responsiveness is abrogated by the K156A mutation, implying that activation of this element is dependent on Rta binding directly to DNA (Fig. 6B). The inability to observe direct Rta binding to this element previously may be the result of an incompletely understood, but well-documented, inhibitory effect of the C-terminal 55 amino acids of Rta in EMSAs (8, 13, 14). While Rta may also activate BALF5p through cellular transcription factors, our observations suggest that the proximal part of BALF5p (−376/+16) in fact contains an RRE.
Our study represents the most complete characterization of early gene promoter RREs undertaken to date. Based on our capture of 4 of the 5 early gene RREs and a large number of peaks upstream of Rta-responsive early genes, we conservatively estimate that we have identified at least half of all early promoter RREs. This supports the concept that direct DNA binding is the most important mode of Rta early promoter activation in early replication. Since LF2 sequesters Rta outside the nucleus (33) and prevents Rta from binding to cognate DNA (8), we predict that LF2 decreases activation of most of the Rta-dependent early genes. In contrast, LF2 does not inhibit Rta-mediated activation of Rp or Zp (33), where we detected little or no Rta binding (Fig. 2). Since LF2 might have a strong impact on Rta binding to early gene promoters, future studies will need to contrast Rta binding observed in LF2-positive genomes with that observed here. In Akata cells, the LF2 transcript becomes detectible at 8 h postinduction (77). Thus, it is unlikely, for the 6-h time point studied here, that Rta binding to wild-type EBV genomes differs significantly from that observed in B95-8 EBV, used for our experiment (63).
Although the primary goal of this study was to characterize Rta binding to the EBV genome during early replication, our results yielded important insights regarding late gene promoter regulation. Previous work has shown that EBV late gene regulation, like that of alphaherpesviruses, is determined primarily by sequences near the TATA box and appears to require an origin of lytic replication in cis (2, 36, 74). Our Rta ChIP-seq did not identify peaks at known Rta-responsive late gene promoters, such as the BLRF2 promoter (2, 18, 65, 69). At late time points, however, Rta binding to the BRLF2 and BFRF3 promoters could be observed by ChIP-qPCR, but this binding was not dependent on new DNA replication. This suggests, in addition to proximal promoter elements responsible for late gene regulation, that Rta binding to late gene RREs is temporally regulated. There must, however, be events downstream of Rta recruitment to promoters that determine late gene expression. These events are possibly most important for true-late genes, such as BFRF3, that absolutely require viral DNA replication in order to be expressed, since Rta recruitment to the BFRF3 promoter was not sufficient for BFRF3 expression (Fig. 7). The BLRF2 gene belongs to the subclass of late genes, sometimes referred to as delayed-early genes, that are dramatically upregulated after DNA replication but can be expressed even when DNA replication is blocked (34, 43, 77). In agreement with studies done in the replication-incompetent Raji cell line (65), the BLRF2 protein was detected in our system despite the block of viral DNA synthesis.
Data presented here suggest direct binding of Rta to DNA not only as the most important mode of early gene activation but also as the least likely mode of Rp and Zp activation. Cellular transcription factors that cooperate with Rta in order to activate immediate-early gene promoters have already been characterized (1, 10, 52, 68). These can either be activated by cellular signaling pathways in the cytoplasm or interact with Rta. While the regulation of Zp, Rp, and early genes has been extensively studied, relatively little is known about the regulation of late genes. This study suggests that Rta binding to late promoters is temporally regulated and specific for the late replication cycle, raising the question of how the delayed recruitment of Rta to late promoters is regulated. That synthesis of new viral DNA is not required for Rta interactions with late promoters narrows down the possibilities for cis or trans signals that direct Rta to late promoters (2, 69). Chromatin changes in the parental episome, viral or cellular gene expression independent of viral DNA replication, Rta modifications, such as sumoylation (11, 33), or translocation of the episome to nuclear sites, such as PML bodies (3), might play a role in the recruitment of Rta to late gene promoters. Recent studies in murine gammaherpesvirus 68 (MHV68) have demonstrated that late gene expression depends not only on DNA replication but also on a subset of genes present in beta- and gammaherpesviruses but absent in alphaherpesviruses. These viral proteins appear to be essential for RNA polymerase II recruitment to late promoters, and their expression may be required for Rta (ORF50) recruitment to these promoters at late times as well (4, 45, 75, 76) (40). While different herpesviruses seem to share the cis requirements for late gene timing, the trans signals that contribute to late gene expression appear to differ between the subfamilies, though the mechanisms and advantages of these differences remain to be discovered.
ACKNOWLEDGMENTS
We thank Amy Holthaus and Melissa Duarte for excellent technical support, Elliott Kieff, Bo Zhao, and Hongfang Wang for helpful discussions and sharing laboratory equipment, and The Center for Cancer Computational Biology, Dana-Farber Cancer Institute, Boston, MA, for next-generation DNA sequencing services.
This work was supported by a Howard Hughes Medical Institute Physician-Scientist Early Career Award (to E.J.), by National Institutes of Health Public Health Service grants CA47006 and CA085180 (to Elliott Kieff), by National Human Genome Research Institute grant P50HG004233, and by the exchange program between Harvard Medical School and the graduate training program 1071 (DFG-GK 1071) at the Friedrich-Alexander University Erlangen-Nuremberg, Germany (to A.M.F.H.).
Footnotes
Published ahead of print 29 February 2012
REFERENCES
- 1. Adamson AL, et al. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amon W, et al. 2004. Lytic cycle gene regulation of Epstein-Barr virus. J. Virol. 78:13460–13469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amon W, White RE, Farrell PJ. 2006. Epstein-Barr virus origin of lytic replication mediates association of replicating episomes with promyelocytic leukaemia protein nuclear bodies and replication compartments. J. Gen. Virol. 87:1133–1137 [DOI] [PubMed] [Google Scholar]
- 4. Arumugaswami V, et al. 2006. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J. Virol. 80:9730–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baer R, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 [DOI] [PubMed] [Google Scholar]
- 6. Bhende PM, Seaman WT, Delecluse HJ, Kenney SC. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099–1104 [DOI] [PubMed] [Google Scholar]
- 7. Buisson M, et al. 1989. The Epstein-Barr virus (EBV) early protein EB2 is a posttranscriptional activator expressed under the control of EBV transcription factors EB1 and R. J. Virol. 63:5276–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calderwood MA, Holthaus AM, Johannsen E. 2008. The Epstein-Barr virus LF2 protein inhibits viral replication. J. Virol. 82:8509–8519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calderwood MA, et al. 2007. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U. S. A. 104:7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang LK, et al. 2005. Activation of Sp1-mediated transcription by Rta of Epstein-Barr virus via an interaction with MCAF1. Nucleic Acids Res. 33:6528–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang LK, et al. 2004. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 279:38803–38812 [DOI] [PubMed] [Google Scholar]
- 12. Chang PJ, Shedd D, Miller G. 2005. Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 79:8750–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen LW, Chang PJ, Delecluse HJ, Miller G. 2005. Marked variation in response of consensus binding elements for the Rta protein of Epstein-Barr virus. J. Virol. 79:9635–9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen LW, et al. 2009. Two phenylalanines in the C-terminus of Epstein-Barr virus Rta protein reciprocally modulate its DNA binding and transactivation function. Virology 386:448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox MA, Leahy J, Hardwick JM. 1990. An enhancer within the divergent promoter of Epstein-Barr virus responds synergistically to the R and Z transactivators. J. Virol. 64:313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Darr CD, Mauser A, Kenney S. 2001. Epstein-Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J. Virol. 75:6135–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. El-Guindy A, Heston L, Miller G. 2010. A subset of replication proteins enhances origin recognition and lytic replication by the Epstein-Barr virus ZEBRA protein. PLoS Pathog. 6:e1001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El-Guindy AS, Miller G. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 78:7634–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farrell PJ. 2001. Epstein-Barr virus. The B95-8 strain map. Methods Mol. Biol. 174:3–12 [DOI] [PubMed] [Google Scholar]
- 20. Farrell PJ, Rowe DT, Rooney CM, Kouzarides T. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feederle R, et al. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flemington E, Speck SH. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flemington EK, Borras AM, Lytle JP, Speck SH. 1992. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J. Virol. 66:922–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furnari FB, Adams MD, Pagano JS. 1992. Regulation of the Epstein-Barr virus DNA polymerase gene. J. Virol. 66:2837–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Granato M, et al. 2006. Regulation of the expression of the Epstein-Barr virus early gene BFRF1. Virology 347:109–116 [DOI] [PubMed] [Google Scholar]
- 27. Gruffat H, et al. 1992. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J. Virol. 66:46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gruffat H, Manet E, Rigolet A, Sergeant A. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gruffat H, Sergeant A. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 22:1172–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hammerschmidt W, Sugden B. 1988. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr virus. Cell 55:427–433 [DOI] [PubMed] [Google Scholar]
- 31. Hardwick JM, Lieberman PM, Hayward SD. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hardwick JM, Tse L, Applegren N, Nicholas J, Veliuona MA. 1992. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J. Virol. 66:5500–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heilmann AM, Calderwood MA, Johannsen E. 2010. Epstein-Barr virus LF2 protein regulates viral replication by altering Rta subcellular localization. J. Virol. 84:9920–9931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holland LE, Anderson KP, Shipman C, Jr, Wagner EK. 1980. Viral DNA synthesis is required for the efficient expression of specific herpes simplex virus type 1 mRNA species. Virology 101:10–24 [DOI] [PubMed] [Google Scholar]
- 35. Holley-Guthrie EA, Quinlivan EB, Mar EC, Kenney S. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Homa FL, Otal TM, Glorioso JC, Levine M. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 6:3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Honess RW, Roizman B. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. U. S. A. 72:1276–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Honess RW, Watson DH. 1977. Herpes simplex virus resistance and sensitivity to phosphonoacetic acid. J. Virol. 21:584–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong GK, et al. 2005. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 79:13993–14003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hong Y, Qi J, Gong D, Han C, Deng H. 2011. Replication and transcription activator (RTA) of murine gammaherpesvirus 68 binds to an RTA-responsive element and activates the expression of ORF18. J. Virol. 85:11338–11350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hung CH, Liu ST. 1999. Characterization of the Epstein-Barr virus BALF2 promoter. J. Gen. Virol. 80:2747–2750 [DOI] [PubMed] [Google Scholar]
- 42. Johannsen E, et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones PC, Roizman B. 1979. Regulation of herpesvirus macromolecular synthesis. VIII. The transcription program consists of three phases during which both extent of transcription and accumulation of RNA in the cytoplasm are regulated. J. Virol. 31:299–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalla M, Schmeinck A, Bergbauer M, Pich D, Hammerschmidt W. 2010. AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc. Natl. Acad. Sci. U. S. A. 107:850–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kayhan B, et al. 2007. A replication-deficient murine gamma-herpesvirus blocked in late viral gene expression can establish latency and elicit protective cellular immunity. J. Immunol. 179:8392–8402 [DOI] [PubMed] [Google Scholar]
- 46. Kenney S, Holley-Guthrie E, Mar EC, Smith M. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kenney SC. 2007. Reactivation and lytic replication of EBV, p 403–433 In Arvin A, et al. (ed), Human herpesviruses. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 48. Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kharchenko PV, Tolstorukov MY, Park PJ. 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 26:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kieff E, Rickinson AB. 2007. Epstein-Barr virus and its replication, In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2, p 2602–2654 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 51. Kutok JL, Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu. Rev. Pathol. 1:375–404 [DOI] [PubMed] [Google Scholar]
- 52. Lee YH, Chiu YF, Wang WH, Chang LK, Liu ST. 2008. Activation of the ERK signal transduction pathway by Epstein-Barr virus immediate-early protein Rta. J. Gen. Virol. 89:2437–2446 [DOI] [PubMed] [Google Scholar]
- 53. Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lieberman PM, Hardwick JM, Sample J, Hayward GS, Hayward SD. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu C, Sista ND, Pagano JS. 1996. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J. Virol. 70:2545–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu P, Speck SH. 2003. Synergistic autoactivation of the Epstein-Barr virus immediate-early BRLF1 promoter by Rta and Zta. Virology 310:199–206 [DOI] [PubMed] [Google Scholar]
- 57. Lu CC, et al. 2006. Genome-wide transcription program and expression of the Rta responsive gene of Epstein-Barr virus. Virology 345:358–372 [DOI] [PubMed] [Google Scholar]
- 58. Ma SD, et al. 2011. A new model of Epstein-Barr virus infection reveals an important role for early lytic viral protein expression in the development of lymphomas. J. Virol. 85:165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manet E, et al. 1993. The acidic activation domain of the Epstein-Barr virus transcription factor R interacts in vitro with both TBP and TFIIB and is cell-specifically potentiated by a proline-rich region. Gene Expr. 3:49–59 [PMC free article] [PubMed] [Google Scholar]
- 60. Manet E, Rigolet A, Gruffat H, Giot JF, Sergeant A. 1991. Domains of the Epstein-Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 19:2661–2667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miller IG, Jr, El-Guindy A. 2002. Regulation of Epstein-Barr virus lytic cycle activation in malignant and nonmalignant disease. J. Natl. Cancer Inst. 94:1733–1735 [DOI] [PubMed] [Google Scholar]
- 62. Quinlivan EB, et al. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus early promoter, BMRF1. Nucleic Acids Res. 21:1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Raab-Traub N, Dambaugh T, Kieff E. 1980. DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell 22:257–267 [DOI] [PubMed] [Google Scholar]
- 64. Ragoczy T, Miller G. 2001. Autostimulation of the Epstein-Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J. Virol. 75:5240–5251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ragoczy T, Miller G. 1999. Role of the Epstein-Barr virus RTA protein in activation of distinct classes of viral lytic cycle genes. J. Virol. 73:9858–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rennekamp AJ, Lieberman PM. 2011. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J. Virol. 85:2837–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rickinson AB, Kieff E. 2007. Epstein-Barr virus. In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2, p 2655–2700 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 68. Robinson AR, Kwek SS, Hagemeier SR, Wille CK, Kenney SC. 2011. Cellular transcription factor Oct-1 interacts with the Epstein-Barr virus BRLF1 protein to promote disruption of viral latency. J. Virol. 85:8940–8953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Serio TR, Kolman JL, Miller G. 1997. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J. Virol. 71:8726–8734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Severini A, Scraba DG, Tyrrell DL. 1996. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 70:3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Speck SH, Chatila T, Flemington E. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399–405 [DOI] [PubMed] [Google Scholar]
- 72. Urier G, Buisson M, Chambard P, Sergeant A. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang JT, et al. 2010. Characterization of Epstein-Barr virus BGLF4 kinase expression control at the transcriptional and translational levels. J. Gen. Virol. 91:2186–2196 [DOI] [PubMed] [Google Scholar]
- 74. Weir JP. 2001. Regulation of herpes simplex virus gene expression. Gene 271:117–130 [DOI] [PubMed] [Google Scholar]
- 75. Wong E, Wu TT, Reyes N, Deng H, Sun R. 2007. Murine gammaherpesvirus 68 open reading frame 24 is required for late gene expression after DNA replication. J. Virol. 81:6761–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wu TT, et al. 2009. ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. J. Virol. 83:2265–2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yuan J, Cahir-McFarland E, Zhao B, Kieff E. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]