Abstract
Mycobacteriophages represent a genetically diverse group of viruses that infect mycobacterial hosts. Although more than 80 genomes have been sequenced, these still poorly represent the likely diversity of the broader population of phages that can infect the host, Mycobacterium smegmatis mc2155. We describe here a newly discovered phage, Marvin, which is a singleton phage, having no previously identified close relatives. The 65,100-bp genome contains 107 predicted protein-coding genes arranged in a noncanonical genomic architecture in which a subset of the minor tail protein genes are displaced about 20 kbp from their typical location, situated among nonstructural genes anticipated to be expressed early in lytic growth. Marvin is not temperate, and stable lysogens cannot be recovered from infections, although the presence of a putative xis gene suggests that Marvin could be a relatively recent derivative of a temperate parent. The Marvin genome is replete with novel genes not present in other mycobacteriophage genomes, and although most are of unknown function, the presence of amidoligase and glutamine amidotransferase genes suggests intriguing possibilities for the interactions of Marvin with its mycobacterial hosts.
INTRODUCTION
Bacteriophages are the most numerous biological entities in the biosphere, with an estimated global population of 1031 phage particles (56). Bacteriophages appear to have emerged early in evolutionary history and may have been evolving for more than three billion years (28, 29). The population is not only vast and old, but also dynamic, with an estimated 1023 phage infections per second on a global scale (53). It is, therefore, perhaps no great surprise that the limited genomic information to date reveals a highly diverse and complex population (5, 20, 24). This population is, however, dominated by viruses classified morphologically in the order Caudovirales, double-stranded DNA (dsDNA)-tailed phages whose genomes vary in size from 15 kbp to approximately 500 kbp (6, 20, 25).
Mycobacteriophages are a group of phages that infect mycobacterial hosts such as Mycobacterium tuberculosis and Mycobacterium smegmatis (19). To date, all characterized mycobacteriophages have either siphoviral or myoviral morphotypes (19). Currently, genomic characterization of 83 mycobacteriophages capable of infecting the nonpathogenic host M. smegmatis mc2155 has revealed a large degree of genetic diversity (19, 46, 47). When grouped by gross genomic nucleotide sequence comparisons, mycobacteriophages that infect the common host M. smegmatis mc2155 fall into 12 major groups (“clusters”) designated A to K (47); several of these clusters can be further divided into subclusters according to their gross nucleotide relationships (19, 47). Only the nine phages constituting cluster C have myoviral morphologies, and all of the others morphologically belong to the Siphoviridae. Five of the siphoviral mycobacteriophages (Giles, Corndog, Wildcat, Omega, and LeBron) were classified as singletons (47), although there have been recent findings of close relatives of Corndog, Omega, and LeBron (22).
The expanding collection of sequenced mycobacteriophage genomes continues to throw new light on mycobacteriophage diversity and the evolutionary processes that create these genomes (19, 21, 31, 47). For instance, the group of cluster A phages has increased significantly, representing a growing number of subclusters with information about superinfection immunity (47). Additionally, the presence of an A1 subcluster phage repressor gene in a cluster C phage, LRRHood, suggests that this gene has been recently acquired by LRRHood from a subcluster A1 phage (47). Mycobacteriophage genomes—like bacteriophages of other hosts—carry many genes that mediate their own mobility either within or between genomes, such as transposons (48), homing endonucleases (3, 21), and inteins (47, 54); although introns have been described in other phages (12), none have yet been identified in mycobacteriophages. Overall, the most striking observation to emerge from bacteriophage comparative genomics is that they are pervasively mosaic, with different segments of the genome—commonly containing just a single gene—having distinct evolutionary histories (20, 30).
The grouping of phages into clusters and subclusters is based on gross nucleotide sequence similarity and therefore reflects more recent evolutionary relationships. More distant relationships can be discerned by comparison of the predicted amino acid sequences of genes, and to facilitate this, a program, Phamerator, has been described that assorts genes sharing protein sequence similarity into “phamilies” (“phams”) (10). The 83 published genomes encode a total of 9,308 predicted genes, and these assemble into 2,367 phams, of which 1,120 (47.3%) are “orphams” (phams containing only a single gene member) (46, 47). Of these phams, about 80% have no significant database match to previously published sequences, and the functions of these large numbers of phage genes are unknown (46, 47). Notable exceptions to this are the virion structure and assembly genes that in the siphoviral phages are syntenically conserved, and thus gene location facilitates their functional assignments (19, 24).
Here, we report a novel siphoviral mycobacteriophage, Marvin, isolated and annotated by students at Cabrini College enrolled in an Honors Introductory Biology Laboratory course sponsored and funded by the Howard Hughes Medical Institute (HHMI) Science Education Alliance (SEA) program. Marvin is a new singleton mycobacteriophage with a 65,100-bp genome that is unrelated at the DNA level to any of the other 83 sequenced mycobacteriophages. Marvin has a mosaic genome, and over 70% of the genes have no homologues among known mycobacteriophages or other organisms. Of the 27 genes that are homologous to other mycobacteriophage genes, the matching genes are from genetically diverse mycobacteriophages, and the mosaic structure of the Marvin genome is clear. Surprisingly, a subset of the tail protein genes are displaced about 20 kbp away from their more typical location and are situated among the nonstructural genes in the right arm. The novelty of the Marvin genome and its large number of new genes support the hypothesis that in spite of the growing collection of mycobacteriophages, we are far from having a full understanding of this diverse population.
MATERIALS AND METHODS
Phage isolation and genomic DNA purification.
Mycobacteriophage Marvin was identified by direct plating on lawns of M. smegmatis mc2155 using an extract from soil on the campus of Cabrini College, located in Southeastern Pennsylvania. Phage isolation was accomplished by mixing approximately 1 g of a soil isolate with phage buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgSO4, 1 mM CaCl2, 68.5 mM NaCl) for a 30-min incubation period at room temperature. The extract was then filtered through a 0.22-μm-pore filter, and 50 μl of this sample was plated with 0.5 ml of late-log-phase M. smegmatis mc2155 and 4.5 ml of 7H9 agar (Middlebrook 7H9 broth base; Difco Laboratories, Detroit, MI) supplemented with 1 mM CaCl2. Following several rounds of plaque purification, a high-titer phage stock was prepared by treating 10 ml of a filtered phage crude lysate with RNase A and DNase I for 30 min at 37°C, followed by a 60-min incubation at room temperature. Intact particles were then precipitated with 30% polyethylene glycol 8000 (PEG 8000)–3.3 M NaCl overnight at 4°C and harvested by centrifugation at 10,000 × g for 20 min. DNA was extracted from the phage pellet using a Wizard DNA cleanup kit (Promega) as per the manufacturer's instructions. For other analyses, Marvin particles were purified by equilibrium-density CsCl centrifugation as described previously (23).
Digestion with DNA methylation-sensitive and DNA methylation-resistant enzymes.
One microgram of Marvin genomic DNA per reaction was digested overnight at 37°C with 1 U of restriction endonuclease. Products were separated by electrophoresis through a 1.2% agarose gel using Tris-acetate-EDTA buffer.
Phage genome sequencing and gene identification.
Purified phage genomic DNA was sequenced by the Joint Genome Institute (JGI) to a depth of ∼25-fold coverage using 454 sequencing and supplemented by an additional ∼60-fold coverage with SOLiD sequencing. Raw reads were assembled using 454's GS De Novo Assembler; assemblies were then quality controlled using Consed. Six Sanger reads were required to resolve weak areas in the assembly. Finished sequences were analyzed and annotated in genome editors, including DNAMaster (http://cobamide2.bio.pitt.edu), G Browse (52), Apollo (37), Glimmer (11), GeneMark (4), tRNA ScanSE (38), Aragorn (36), and Programmed Frameshift Finder (57) to identify genome features. Genes were assigned to phams, and genome maps and phamily circle diagrams were drawn using Phamerator, with the threshold parameters of 32.5% identity with ClustalW and a BlastP E value of 10−50, as described previously (10).
Electron microscopy.
A lysate of Marvin with a titer of approximately 1010 PFU/ml was serially diluted into phage buffer to approximately 104 PFU/ml, and 3 μl of each dilution was spotted onto a soft agar lawn seeded with M. smegmatis mc2155. After overnight incubation at 37°C, the spot that exhibited densely packed yet distinguishable plaques was gently washed with 10 μl of phage buffer by pipetting up and down several times. The 10 μl of buffer was diluted 1:2 in phage buffer, and 5 μl of that dilution was allowed to sit on freshly glow-discharged 400-mesh carbon-Formvar-coated copper grids for approximately 30 s. The grids were then rinsed with distilled water and stained with 1% uranyl acetate. Virus particles were imaged on an FEI Morgagni transmission electron microscope at 80 kV at a magnification of 56,000.
Identification of Marvin virion proteins.
Approximately 100 μl of CsCl-purifed Marvin particles (a total of 1012 PFU) was collected by centrifugation at 14,000 rpm for 30 min, and the pellet was resuspended in 75 μl of 20 mM dithiothreitol. Two microliters of 0.5 M EDTA was added, and the solution was heated to 65°C for several minutes, when it became viscous. The sample was sonicated on ice for 10 s and allowed to rest on ice for 1 min, and this cycle was repeated six times, at which point, the viscosity was greatly reduced. Finally, 4× SDS sample buffer was added, and the sample was boiled for 2.5 min. Several dilutions were loaded onto a 12% SDS–polyacrylamide gel and electrophoresed at 100 V until the dye front ran off the gel. The gel was stained with Coomassie blue and destained in 10% acetic acid. The visible bands were compared to a standard to determine the approximate molecular mass.
For protein identification by mass spectrometry (MS), 8 μl of sonicated Marvin particles was loaded into a single lane of a different 12% SDS–polyacrylamide gel and electrophoresed only until the sample was approximately 2 cm into the separating portion of the gel. The gel was stained with Coomassie blue and destained in H2O. The single visible band comprised of all particle proteins was excised, and the proteins were digested in situ with trypsin (at the University of Pittsburgh Genomics and Proteomics Core Labs), followed by peptide elution, chromatography, and tandem MS (MS/MS) on an LTQ Velos Orbitrap mass spectrometer. Peptides were matched against predicted Marvin proteins.
Analysis of the predicted secondary structure and coiled-coil propensity for selected protein sequences was carried out with the Psipred (http://bioinf.cs.ucl.ac.uk/psipred/) and Coils (http://www.ch.embnet.org/software/COILS_form.html) servers, respectively.
Nucleotide sequence accession number.
The GenBank accession number for mycobacteriophage Marvin is JF704100.
RESULTS
Phage isolation and morphological characteristics of mycobacteriophage Marvin.
Mycobacteriophage Marvin was isolated from soil on the campus of Cabrini College, Radnor, PA, by direct plating with M. smegmatis mc2155. Marvin is somewhat unusual among mycobacteriophages in that it propagates slowly and forms tiny barely identifiable plaques after 48 h of growth on a lawn of M. smegmatis at 37°C. The plaques are round and clear, suggesting that under standard growth conditions using M. smegmatis as the host, Marvin is either a lytic phage or a temperate phage that forms lysogens at only a low frequency.
To determine whether lysogens could be recovered from Marvin infections, cells from a spot where Marvin particles had infected a lawn of M. smegmatis were recovered and grown on solid media. Bacterial growth was observed, and two independent colonies were restreaked twice more and then patched onto M. smegmatis lawns to test for phage release; none of the colonies recovered showed phage release (data not shown). Thus, although bacterial survivors can be readily recovered, there is no evidence that Marvin is a temperate mycobacteriophage.
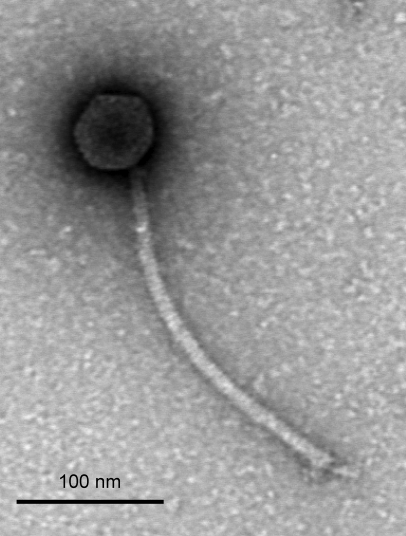
Electron microscopic images show that Marvin has a siphoviral morphotype with a long, flexible noncontractile tail and an isometric head (Fig. 1). The average tail length from several electron micrographic images of Marvin is 250 nm, and the head diameter is 58 nm. The tail length is longer than the average tail length of mycobacteriophages, but not as long as those of the cluster H phages Konstantine, Predator, and Barnyard (18a).
Fig 1.
Morphology of mycobacteriophage Marvin virions. An electron micrograph with uranyl acetate negative stain is shown. The scale bar corresponds to 100 nm.
Genome sequencing and classification.
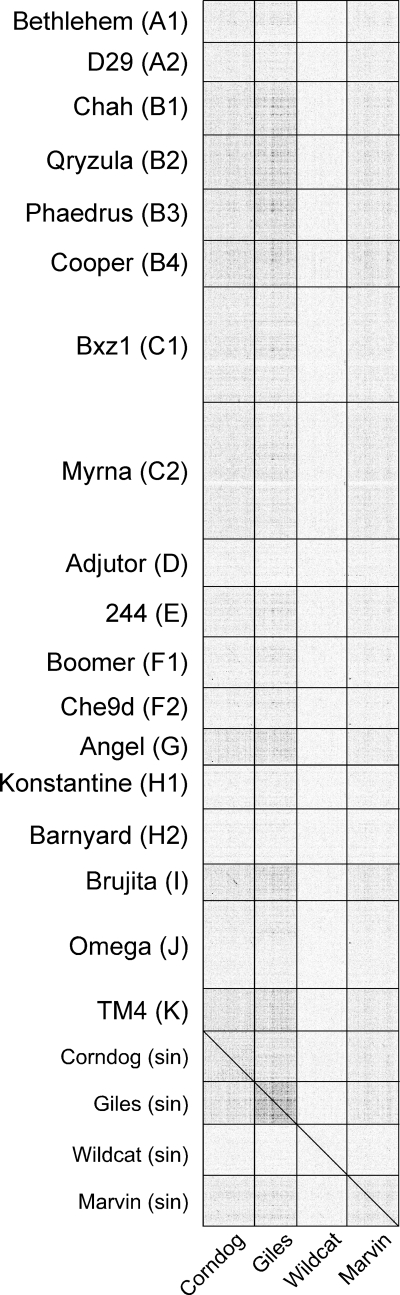
Marvin DNA was isolated and sequenced using a combination of 454 shotgun and SOLiD sequencing. The Marvin dsDNA genome is 65,100 bp in length, with 11-nucleotide 3′-terminal extensions. This genome length is near the average for the siphoviral mycobacteriophages. The GC% of the Marvin genome is 63.4%, close to both the mycobacteriophage average and to that of the host, M. smegmatis. Comparison of the Marvin genome with examples of each of the mycobacteriophage clusters shows little or no discernible DNA sequence similarity to any of them (Fig. 2), and Marvin has therefore been designated a new singleton phage.
Fig 2.
Dot plot comparison of mycobacteriophage Marvin with representative mycobacteriophages. A sequence file containing the four singleton (sin) phages Corndog, Giles, Wildcat, and Marvin was compared against a file containing a single representative of each cluster or subcluster (as indicated) using Gepard (35). Marvin is classified as a singleton phage because of its lack of identifiable sequence similarity to other known mycobacteriophages. Omega is not shown as a singleton phage here because it has recently been grouped with unpublished phages as cluster J.
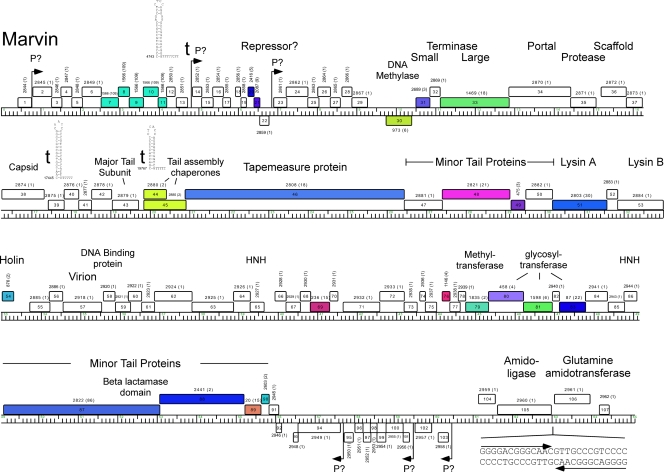
Analysis of the Marvin genome identified 107 putative open reading frames (ORFs), but no tRNA or other small RNA genes (Fig. 3 and Table 1). The ORF density is relatively high (92.85%), and there are only four noncoding intergenic gaps larger than 300 bp. Ninety-three of the ORFs are expressed from the top strand (shown rightwards in Fig. 3) spanning the leftmost 56 kbp of the genome. Twelve of the leftwards-transcribed ORFs (genes 92 to 103) are closely linked and situated about 10% of the genome length from the right end; the other two are interspersed with the rightwards-transcribed genes (Fig. 3). This overall organization is unlike any other mycobacteriophage genome (19), consistent with its assignment as a new singleton phage.
Fig 3.
Annotated genome map of mycobacteriophage Marvin. The viral 65,100-bp genome of Marvin is represented in four tiers with markers spaced at 1-kbp and 100-bp intervals. The predicted genes are shown as boxes either above or below the genome, depending on whether they are rightwards or leftwards transcribed, respectively. Gene numbers are shown within each box, and the phamily to which that gene belongs is shown above with the number of phamily members shown in parentheses; genes are color coordinated according to their phamily identity. Putative functions are shown above the genes. Other sequences, including putative promoters (P), a terminator (t), and a long palindromic sequence, are shown.
Table 1.
Mycobacteriophage Marvin predicted genes and gene products
| Gene | F/Ra | Position |
Product mass (kDa) | Pham no.b | Size (no. of members)c | Putative function | |
|---|---|---|---|---|---|---|---|
| Start | Stop | ||||||
| 1 | F | 424 | 768 | 12.98 | 2844 | 1 | |
| 2 | F | 802 | 1281 | 18.18 | 2845 | 1 | |
| 3 | F | 1278 | 1538 | 9.47 | 2846 | 1 | |
| 4 | F | 1511 | 1789 | 9.02 | 2847 | 1 | |
| 5 | F | 1789 | 2058 | 10.08 | 2848 | 1 | |
| 6 | F | 2048 | 2533 | 18.63 | 2849 | 1 | |
| 7 | F | 2530 | 2967 | 15.74 | 1566 | 109 | |
| 8 | F | 2964 | 3248 | 10.66 | 1566 | 109 | |
| 9 | F | 3241 | 3600 | 13.04 | 1566 | 109 | |
| 10 | F | 3597 | 3977 | 13.2 | 1566 | 109 | |
| 11 | F | 3974 | 4192 | 8.03 | 1566 | 109 | |
| 12 | F | 4189 | 4404 | 8.02 | 2850 | 1 | |
| 13 | F | 4433 | 4720 | 10.91 | 2851 | 1 | |
| 14 | F | 4843 | 5079 | 8.37 | 2852 | 1 | |
| 15 | F | 5076 | 5366 | 10.49 | 2853 | 1 | |
| 16 | F | 5359 | 5625 | 9.47 | 2854 | 1 | |
| 17 | F | 5612 | 5806 | 7.1 | 2855 | 1 | |
| 18 | F | 5935 | 6060 | 4.01 | 2856 | 1 | |
| 19 | F | 6072 | 6209 | 5.12 | 2858 | 1 | |
| 20 | F | 6250 | 6405 | 5.74 | 2416 | 5 | |
| 21 | F | 6402 | 6551 | 5.38 | 2087 | 9 | |
| 22 | R | 6781 | 6548 | 8.7 | 2859 | 1 | Repressor? |
| 23 | F | 6901 | 7233 | 11.75 | 2861 | 1 | |
| 24 | F | 7217 | 7768 | 19.89 | 2862 | 1 | |
| 25 | F | 7765 | 8046 | 11.04 | 2863 | 1 | |
| 26 | F | 8018 | 8338 | 11.36 | 2864 | 1 | |
| 27 | F | 8335 | 8568 | 9.46 | 2865 | 1 | |
| 28 | F | 8630 | 8884 | 8.91 | 2866 | 1 | |
| 29 | F | 8881 | 9276 | 15.34 | 2867 | 1 | |
| 30 | R | 10398 | 9748 | 24.6 | 973 | 6 | DNA methylase |
| 31 | F | 10504 | 10857 | 13.43 | 2689 | 3 | Terminase small subunit |
| 32 | F | 10858 | 11115 | 9.22 | 2869 | 1 | |
| 33 | F | 11120 | 12862 | 64.79 | 1469 | 18 | Terminase large subunit |
| 34 | F | 12859 | 14424 | 58.35 | 2870 | 1 | Portal |
| 35 | F | 14421 | 15065 | 24 | 2871 | 1 | Protease |
| 36 | F | 15195 | 15827 | 22 | 2872 | 1 | Scaffold |
| 37 | F | 15831 | 16235 | 14.49 | 2873 | 1 | Virion protein |
| 38 | F | 16247 | 17317 | 38.96 | 2874 | 1 | Capsid |
| 39 | F | 17422 | 17823 | 15.04 | 2875 | 1 | Virion protein |
| 40 | F | 17823 | 18194 | 14.21 | 2876 | 1 | Virion protein |
| 41 | F | 18194 | 18517 | 12.23 | 2877 | 1 | Virion protein |
| 42 | F | 18521 | 19027 | 19.54 | 2878 | 1 | Virion protein |
| 43 | F | 19043 | 19708 | 23.78 | 2879 | 1 | Major tail subunit |
| 44 | F | 19842 | 20414 | 20.93 | 2880 | 2 | Tail assembly |
| 45 | F | 19824 | 20911 | 40.42 | 2880 | 2 | Tail assembly |
| 46 | F | 20889 | 26438 | 195.74 | 2808 | 18 | Tapemeasure |
| 47 | F | 26438 | 27400 | 36.52 | 2881 | 1 | Minor tail protein |
| 48 | F | 27400 | 29112 | 63.28 | 2821 | 21 | Minor tail protein |
| 49 | F | 29144 | 29491 | 12.47 | 475 | 3 | Minor tail protein? |
| 50 | F | 29517 | 30194 | 25.41 | 2882 | 1 | Minor tail protein? |
| 51 | F | 30191 | 31570 | 50.63 | 2803 | 30 | Lysin A |
| 52 | F | 31567 | 31842 | 10.25 | 2883 | 1 | |
| 53 | F | 31842 | 32996 | 42.52 | 2884 | 1 | Lysin B |
| 54 | F | 33016 | 33306 | 10.26 | 876 | 2 | Holin |
| 55 | F | 33713 | 34222 | 19.5 | 2885 | 1 | |
| 56 | F | 34209 | 34547 | 12.07 | 2886 | 1 | |
| 57 | F | 34557 | 35534 | 35.63 | 2918 | 1 | Virion protein |
| 58 | F | 35578 | 35871 | 10.87 | 2920 | 1 | DNA-binding protein |
| 59 | F | 35885 | 36265 | 13.89 | 2921 | 1 | |
| 60 | F | 36210 | 36548 | 11.57 | 2922 | 1 | |
| 61 | F | 36548 | 36874 | 12.5 | 2923 | 1 | |
| 62 | F | 36874 | 37824 | 35.66 | 2924 | 1 | |
| 63 | F | 37821 | 38873 | 37.75 | 2925 | 1 | |
| 64 | F | 38873 | 39298 | 15.76 | 2926 | 1 | |
| 65 | F | 39295 | 39648 | 13.6 | 2927 | 1 | HNH protein |
| 66 | F | 39924 | 40205 | 10.34 | 2928 | 1 | |
| 67 | F | 40202 | 40567 | 13.6 | 2929 | 1 | |
| 68 | F | 40564 | 40827 | 10.12 | 2930 | 1 | |
| 69 | F | 40187 | 41308 | 18.4 | 236 | 15 | |
| 70 | F | 41313 | 41516 | 8 | 2931 | 1 | |
| 71 | F | 41650 | 42576 | 34.32 | 2932 | 1 | |
| 72 | F | 42578 | 43198 | 22.92 | 2933 | 1 | |
| 73 | F | 43195 | 43524 | 11.98 | 2935 | 1 | |
| 74 | F | 43591 | 43734 | 5.11 | 2936 | 1 | |
| 75 | F | 43731 | 44018 | 10.86 | 2937 | 1 | |
| 76 | F | 44158 | 44355 | 7.5 | 1146 | 4 | |
| 77 | F | 44352 | 44612 | 9.94 | 2938 | 1 | |
| 78 | F | 44566 | 44754 | 7.48 | 2939 | 1 | |
| 79 | F | 44750 | 45333 | 22.19 | 1835 | 2 | Methyltransferase |
| 80 | F | 45330 | 46217 | 33.77 | 458 | 4 | Glycosyltransferase |
| 81 | F | 46217 | 46945 | 27.77 | 1598 | 6 | Glycosyltransferase |
| 82 | F | 46955 | 47125 | 6.37 | 2940 | 1 | |
| 83 | F | 47128 | 47790 | 24.71 | 87 | 22 | Glycosyltransferase |
| 84 | F | 47787 | 48341 | 20.71 | 2941 | 1 | |
| 85 | F | 48363 | 48776 | 15.7 | 2943 | 1 | |
| 86 | F | 48763 | 49089 | 12.62 | 2944 | 1 | HNH protein |
| 87 | F | 49116 | 53072 | 140.89 | 2822 | 86 | Minor tail protein |
| 88 | F | 53069 | 55204 | 72.95 | 2441 | 2 | Minor tail protein |
| 89 | F | 55227 | 55634 | 14.19 | 20 | 15 | Minor tail protein |
| 90 | F | 55647 | 55841 | 7.08 | 2603 | 2 | Minor tail protein? |
| 91 | F | 55838 | 56083 | 9.23 | 2945 | 1 | |
| 92 | R | 56158 | 56018 | 5.27 | 2946 | 1 | |
| 93 | R | 56575 | 56453 | 4.55 | 2948 | 1 | |
| 94 | R | 57642 | 56572 | 40.15 | 2949 | 1 | |
| 95 | R | 57970 | 57740 | 8.29 | 2950 | 1 | |
| 96 | R | 58218 | 58015 | 7.55 | 2951 | 1 | |
| 97 | R | 58417 | 58226 | 7.89 | 2952 | 1 | |
| 98 | R | 58574 | 58404 | 6.46 | 2953 | 1 | |
| 99 | R | 58798 | 58571 | 8.45 | 2954 | 1 | |
| 100 | R | 59232 | 58801 | 15.68 | 2955 | 1 | |
| 101 | R | 59393 | 59235 | 6.27 | 2956 | 1 | |
| 102 | R | 59958 | 59536 | 15.4 | 2957 | 1 | |
| 103 | R | 60379 | 60122 | 10.13 | 2958 | 1 | |
| 104 | F | 61145 | 61567 | 15.37 | 2959 | 1 | |
| 105 | F | 61626 | 62987 | 51.84 | 2960 | 1 | Amidoligase |
| 106 | F | 63067 | 63966 | 33.65 | 2961 | 1 | Glutamine amidotransferase |
| 107 | F | 64205 | 64453 | 9.04 | 2962 | 1 | |
F/R, forward or reverse transcription.
Pham number derived using the Phamerator database “Marvin.”
Number of gene members of that pham.
Marvin genome architecture.
Each of the Marvin open reading frames was compared with all other mycobacteriophage genes using the program Phamerator (10) (using the database “Marvin”), and the ORFs were assorted into phamilies according to their amino acid sequence similarities. The “Marvin” Phamerator database contains 84 genomes, 9,415 genes, 2,446 phamilies, and 1,196 orphams. A striking outcome of this analysis is that 75 (70%) of the predicted Marvin protein coding genes are orphams, although this is not uncommon for a singleton phage for which there are no close relatives (19) (Fig. 3). Searching against the NCBI database revealed very few significant matches with any of these orphams, and only 19 of the predicted Marvin genes gave any informative matches (Table 1).
Although relatively few Marvin genes can be assigned putative functions, an overall architecture can be proposed. The virion structure and assembly genes likely span genes 33 to 50, deduced from the observations that the terminase genes are typically the leftmost of the operon, and the putative Marvin lysis genes lie to the right of gene 50 (Fig. 3). However, this segment spans only about 20 kbp, which would make this among the smallest of the virion structure and assembly operons of any of the mycobacteriophages. For example, although BPs and related cluster G phages have the smallest mycobacteriophage genomes (48), their 25 virion structural genes span more than 24 kbp of the genome. An explanation for this lies in the observation that Marvin's “missing” minor tail protein genes (87 to 90) are located elsewhere in the genome among nonstructural genes, displaced by more than 20 kbp from their typical position (see below). We also note that the terminase large subunit gene is separated from the physical end of the genome by more than 10.5 kbp. This is atypical but not unprecedented and is also seen in the cluster A phage genomes (14, 17, 23). However, in those examples, the lysis cassette also lies within this region, whereas in Marvin it is to the right of the structural operon (Fig. 3).
Temperate phages typically encode either a serine- or tyrosine-integrase that mediates prophage integration, and these genes are usually positioned near the center of their genomes (18). However, there are no Marvin ORFs with recognizable similarity to either type of integrase, and no apparent relatives of the ParAB functions that some mycobacteriophages use to stabilize extrachromosomally replicating prophages (47). This is consistent with the conclusion from the lysogen analysis described above that Marvin does not appear to be a temperate phage. The presence of putative transcriptional regulator genes and their potential roles are discussed below.
Nonstructural genes 1 to 30.
Marvin genes 1 to 30 occupy the space between the physical left end of the genome and the terminase genes. Twenty-two of these genes are orphams and have no close relatives in other mycobacteriophages; most also have no database matches, although protein gp2 (gene product 2′) has weak similarity (31% identity) to gp59 of Tsukamurella phage TPA2 (44). We note though that they are all small, and none is longer than 600 bp. Two of the 29 genes in this region have functionally informative database matches, and gene 22 encodes a 78-residue helix-turn-helix putative DNA-binding protein with similarity to putative repressors of the XRE class. Although some members of this family of proteins are predicted to be phage repressors, others are components of toxin-antitoxin systems. We note, for example, that Marvin gp22 shares 33% identity with the putative antitoxin component of Escherichia coli TA271. Because Marvin does not appear to form stable lysogens, gp22 seems unlikely to be a phage repressor, and an antitoxin component of a toxin-antitoxin system is an attractive role. It is possible that a closely linked gene, such as gene 23, encodes the toxin component, although gp23 has no close relatives. We note that toxin-antitoxin systems have been implemented in phage resistance mechanisms (13), and it is reasonable to expect these also to be carried by phage genomes. Indeed, the previously reported mycobacteriophage Fruitloop also encodes a putative toxin-antitoxin system (47).
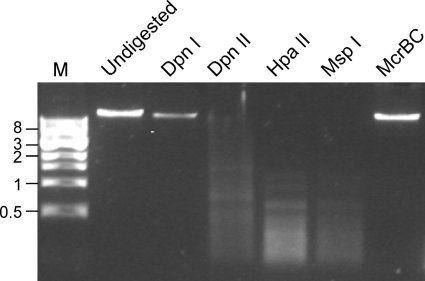
Marvin gp30 matches known proteins, suggesting that it functions as a cytosine-C5-specific DNA methylase. Related proteins are found in other mycobacteriophages, including U2, DD5, Jasper, Lockley, and Pukovnik (all cluster A phages). The specific role of this protein is not known, although it could act to modify Marvin DNA nonspecifically, or alternatively act as a component of a restriction modification system. Because Marvin DNA is readily digested by several restriction enzymes that are typically inhibited by cytosine methylation (Fig. 4), we favor the second explanation, although we have not been successful in identifying a restriction enzyme partner in the Marvin genome.
Fig 4.
Restriction enzyme sensitivity of Marvin DNA. Marvin DNA was digested with the enzymes DpnI, Dpn II, HpaII, MspI, and McRBC as indicated, and the products were separated by agarose gel electrophoresis. Lane 2 contains undigested genomic DNA; Lane M is a 1-kbp size marker. Note that DpnI and Dpn II are isoschizomers (recognizing 5′-GATC), and DpnI only cuts DNA if the recognition site is methlylated, whereas DpnII is blocked by dam methylation. Likewise, HpaII and MspI are isochizomers (recognizing 5′-CCGG), and HpaII is blocked by CpG methylation, whereas MspI is insensitive to site methylation. McrBC (lane 7) that recognizes 5′-PumC(N40–3000)PumC only cuts methylated DNA.
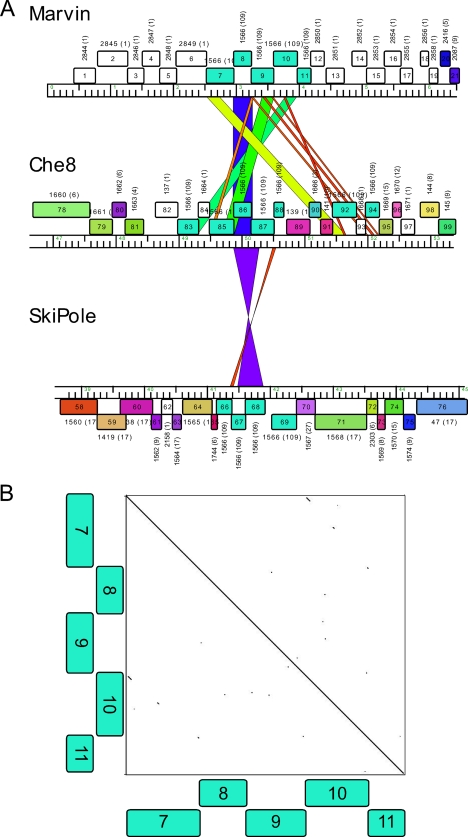
A striking feature of this region is the segment containing genes 7 to 11 (Fig. 5). These are all members of the same Pham (Pham1566), although distant relatives of each other. However, this is a large Pham, with 109 members in the current Phamerator database (database “Marvin”), and there are representatives in virtually every other mycobacteriophage cluster, the exceptions being clusters G, H, and K. Moreover, the sequence similarity extends to the nucleotide sequence level, with short but significant matches of similarity to many phages, including Che8 (subcluster F1) and SkiPole (subcluster A1) (Fig. 5A). For example, Marvin gene 8 has 95% nucleotide identity with Che8 gene 86, spanning a region of about 300 bp (Fig. 5A); 10 other mycobacteriophages contain genes with similar levels of sequence similarity. Within a genome, these related genes form short arrays, and in Che8, there are seven Pham1566 genes; however, the order of genes varies between genomes (Fig. 5). Although the genes within the array are related at the level of the protein sequences, there is little evidence of nucleotide sequence similarity between them (Fig. 5B), in sharp contrast to the intergenome relationships. Thus, while the arrays may have arisen through gene duplications, these must have been far distant evolutionary events, and individual members appear to have been exchanged between genomes during very recent evolutionary times. It is tempting to speculate that perhaps these represent novel mobile elements, although we have been unable to find any significant similarity to transposases or homing endonucleases using Psi-BLAST or HHPred.
Fig 5.
Marvin genes 7 to 11 and their homologues. Marvin genes 7 to 11 form a group of genes that are related to each other (Pham1566) and which are related to other similar groups in other mycobacteriophages. (A) Alignment of the Marvin genome with those of Che8 (subcluster F1) and SkiPole (subcluster A1) illustrates the nucleotide sequence similarities between the genomes. Genome representations are made in Phamerator, and the gene annotations are as described for Fig. 3. Segments of nucleotide sequence similarity are shown by colored regions between pairs of genomes and spectrum colored, with violet being the most similar and red the most dissimilar. (B) Although gp7 to gp11 are related at the amino acid sequence level, they are not related at the DNA sequence level as shown by a dot plot of genes 7 to 11 against themselves.
Within the region from genes 1 to 30, there are three plausible promoters, each of which contains a canonical −35 sequence (5′-TTGACA) of the σ70 class of promoters; promoters of this class have previously been described in mycobacteriophage L5 (41). These are located between genes 1 and 2, between genes 13 and 14, and between genes 22 and 23 (Fig. 3). Between genes 13 and 14—but located upstream of the putative promoter—there is a putative rightwards-facing stem-loop transcription terminator (Fig. 3). The activity and role of these putative transcription signals remain unclear.
Marvin virion structural and assembly genes.
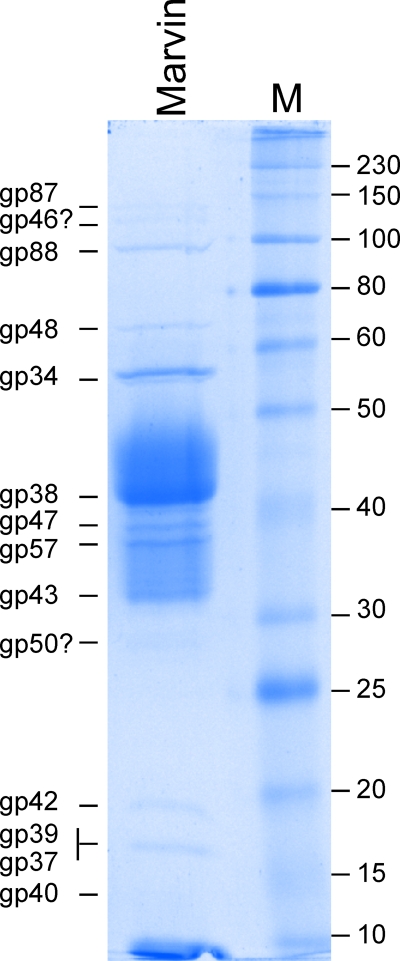
A putative operon of virion structure and assembly genes (31 to 50) shares the canonical organization and common synteny seen in phages with siphoviral morphologies: terminase, portal, protease, scaffold, capsid, head and tail completion proteins, major tail subunit, tail assembly chaperones, tapemeasure protein, and minor tail proteins (Fig. 3). The genes are generally tightly packed, with overlapping or minimal gaps between start and stop codons, with the exceptions of three larger gaps (100 to 130 bp) between genes 35 and 36, 38 and 39, and 43 and 44. The latter two contain putative transcriptional terminators that presumably modulate transcription levels throughout the operon (Fig. 3); there is little space also to accommodate promoters between these putative terminators and the downstream genes. The gene assignments within the operon correlate well with proteins present in intact virions, as determined by SDS-PAGE separation of virion proteins (Fig. 6) and identification of virion proteins by mass spectrometry (Table 2). These gene assignments are discussed in further detail below.
Fig 6.
SDS-PAGE analysis of Marvin virion proteins. Illustrated is SDS gel electrophoresis of Marvin virion proteins, showing the predicted gene products. Molecular mass markers (M) are shown in kDa.
Table 2.
Identification of virion-associated proteins
| No. of PSMsa | Coverage (%)b | No. of peptidesc | Productd | Mol mass (kDa) | Scoree |
|---|---|---|---|---|---|
| 197 | 71.91 | 19 | gp38 | 39.0 | 767.70 |
| 101 | 53.39 | 8 | gp43 | 23.8 | 474.64 |
| 120 | 40.29 | 56 | gp46 | 195.6 | 470.89 |
| 81 | 43.57 | 19 | gp34 | 58.3 | 270.10 |
| 52 | 73.85 | 17 | gp57 | 35.6 | 200.46 |
| 63 | 64.06 | 13 | gp47 | 36.5 | 183.93 |
| 41 | 55.70 | 19 | gp88 | 72.9 | 175.92 |
| 29 | 44.21 | 16 | gp48 | 63.3 | 116.50 |
| 18 | 15.86 | 14 | gp87 | 140.9 | 60.93 |
| 16 | 66.42 | 9 | gp37 | 14.5 | 56.33 |
| 4 | 33.08 | 3 | gp39 | 15.0 | 16.13 |
| 5 | 34.29 | 5 | gp36 | 22.0 | 15.74 |
| 5 | 27.98 | 4 | gp42 | 19.5 | 15.36 |
| 5 | 19.11 | 4 | gp50 | 25.4 | 15.29 |
| 3 | 20.09 | 3 | gp35 | 24.0 | 10.87 |
| 2 | 19.51 | 2 | gp40 | 14.2 | 9.98 |
PSMs, peptide spectrum matches.
Percentage of predicted protein sequence identified in peptides.
Number of different peptides identified corresponding to the protein.
Predicted gene product of mycobacteriophage Marvin.
The sum of the matching scores of individual peptides to the predicted sequence.
In Marvin, gene 33 encodes the terminase large subunit with relatives in other mycobacteriophages (Fig. 3), the closest being Bxz2 gp13 (32% identity). However, there are closer relatives in nonmycobacteriophage phage genomes, and the closest match is to the terminase of a putative prophage in Corynebacterium kroppenstedtii (47% identity). Curiously, Marvin gp33 has a short (32 residues) but significant (E value, 5 × 10−3) match to a conserved domain (pfam02459) in the adenoviral terminal protein, which is of interest given the related functionalities of these proteins. Marvin gp31 is a strong candidate for a terminase small subunit with homologues in the subcluster I1 phages (e.g., Brujita gp1 and Island3 gp1), where it is positioned close to the genome physical end and immediately upstream of the terminase large subunit gene. Marvin gp32 is of unknown function and has no database matches.
Marvin gp34 is a strong candidate for the portal protein and contains a DUF1484 domain common to phage portal proteins. It is well represented in the peptides identified by mass spectrometry (Table 2), and a product of the expected size is seen by SDS-PAGE (Fig. 6).
Gene 35, for which only a small number of peptides are represented in the mass spectrometry data, likely codes for a protease, with weak sequence matches to other putative mycobacteriophage proteases, including gp5 of both phages Ramsey and Boomer. A similar small number of peptides are found corresponding to the putative scaffolding protein, gp36. Marvin gp36 has poor (29% identity) but significant similarity to a putative scaffolding protein encoded in the Caldicellulosiruptor owensensis genome. Analysis of the gp36 sequence predicts that it has several alpha-helical regions, some with high propensity to form coiled coils, joined by regions of unstructured sequence, and little or no beta structure: these are all features of known scaffolding proteins, and this analysis strengthens the identification of gp36 as the scaffolding protein. It is somewhat unexpected to find peptides from the protease and the scaffolding protein in mature virions, as these proteins are thought to be lost from the structure during capsid maturation in most phages. However, there is evidence for residual amounts of both protease and scaffolding proteins being retained in virions of coliphage T4 (8, 50, 51), and the protease of coliphage P2 is retained in mature virions (9). Our results suggest that some of both protease and scaffolding proteins, or fragments of them, are similarly retained in the Marvin virions. An alternative explanation—that these proteins came from contaminating procapsids that had not packaged DNA—seems unlikely for these virions that were purified in a CsCl density gradient.
Marvin gp37 has weak matches to nonmycobacteriophage proteins, including gp37 of Streptomyces phage VWB, but its specific role has not been established. However, it is present in virions with greater than 66% coverage in the mass spectrometry analysis (Table 2). Many phages have abundant “decoration” proteins on the surface of the capsid which typically stabilize the capsid structure, and in some of those phages (e.g., coliphage lambda [26] and Bacillus phage G [GenBank accession no. JN638751.1]), the gene encoding the decoration protein is known to lie between the scaffolding protein and major capsid protein genes. We accordingly speculate that Marvin gp37 may be such a decoration protein.
Marvin gp38 contains a pfam03864 domain associated with major capsid subunits, and gp38 is presumably the capsid protein. It is the most abundant protein represented in the mass spectrometry analysis and a major band of the predicted size is seen by SDS-PAGE; we note that the Marvin capsid does not engage in wholesale covalent cross-linking, as seen in some other mycobacteriophages (14, 15, 23). Although it has no identifiable relatives among other mycobacteriophages, it has sequence similarity (35% identity) to the gp38 putative capsid subunit of Streptomyces phage VWB (1, 55). We suggest that genes 39 to 42 encode the head and tail completion proteins, and all of the products except gp41 are present in virions (Table 2), albeit in low abundance. Gene 43 encodes the major tail subunit, with weak sequence similarity (35% identity) to the putative major tail subunit (gp13) of mycobacteriophage LeBron (47). The gp43 product is the second most abundant protein seen by mass spectrometry, but it separates indistinctly by SDS-PAGE and migrates slower than anticipated by its predicted molecular mass (Fig. 6). However, this aberrant migration is not unusual among major tail subunit proteins, including those of mycobacteriophages (15, 23). Immediately downstream of gene 43 are two genes that are likely expressed by a −1 programmed translational frameshift, a highly conserved feature of phage genomes (57), with the protein products acting as tail assembly chaperones (Fig. 3); the predicted position of the frameshift is 15 to 16 codons prior to the termination codon of gene 44. These are not expected to be components of intact virions, and corresponding peptides are not observed (Table 2).
We identify Marvin gene 46 as encoding the tapemeasure protein (Tmp) based initially on its position in the gene order and its very large size (5,550 bp). Analysis of the predicted amino acid sequence shows a high propensity for alpha-helical and coiled-coil structure; these properties are characteristic of Tmp's. There is typically a correlation between the length of a phage tail and the length of the Tmp in the alpha-helical form that it is thought to assume during tail length determination (33, 34, 43). In the case of Marvin, the measured length of the tail (Fig. 1) is 250 nm, and the predicted length of the Tmp as an alpha-helix is 277.5 nm (1,850 amino acids × 0.15-nm rise per amino acid in an alpha helix), and it is plausible that some processing occurs prior to tail assembly. Although the product corresponding to gp46 cannot be unambiguously assigned by SDS-PAGE, there is a possible candidate at approximately 130 kDa (Fig. 6). This protein is too big to be encoded by any of the Marvin genes, except 46 (Tmp) and 87 (putative tail fiber), and there is a different band at the expected position for gp87. We therefore propose that the 130-kDa protein is derived from the gp46 Tmp. It is considerably smaller than the predicted 196 kDa of full-length gp46 and would therefore necessarily be a posttranslationally processed form of the Tmp; we note that such processing of Tmp's is seen quite commonly (27, 43, 58). Interestingly, the Marvin Tmp also contains two small motifs implicated in peptidoglycan hydrolysis. One of these is motif 3, described previously (43, 45), but the other is a putative lytic transglycosylase domain (cd00254), the first such motif to be identified in mycobacteriophage Tmp's. The roles of such domains in Tmp's have not been not fully resolved, but the motif 3 domain in the Tmp of phage TM4 enhances the ability of the phage to productively infect cells in the late stages of growth (45). The motifs in the Marvin Tmp may provide similar or related functions.
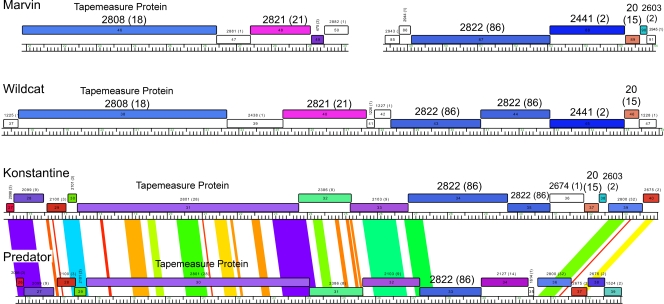
The arrangement of the minor tail protein genes—encoding the structure at the very tip of the tail and therefore important for host recognition and triggering the DNA injection process—in Marvin is one of its more unusual features. In all other mycobacteriophage genomes analyzed to date, the minor tail proteins are encoded by a group of 4 to 10 genes immediately downstream of the tapemeasure protein gene (19). However, in Marvin, this group of genes is split such that genes 47 to 50 likely encode four tail proteins, and the remaining proteins are encoded by genes 87 to 90 (Fig. 3 and 7), located among nonstructural genes and displaced by about 20 kbp from their normal location. Marvin gp47 and gp48 have sequence similarity to LeBron gp17 and gp18 (36% and 54% identity, respectively) and, more distantly, to Wildcat gp39 and gp40 (Fig. 7). Marvin gp49 shares 43% identity with Corndog gp72. Marvin gp50 has no database matches but is proline rich (12%), a feature sometimes found in minor tail proteins. Virion analysis confirms that gp87 and gp88, as well as gp47, gp48, and gp50, are structural components (Table 2 and Fig. 6); in a separate mass spectrometry experiment, gp89 was also identified as a virion protein (data not shown).
Fig 7.
Noncanonical arrangement of tail protein genes. The genomes of Marvin, Wildcat (singleton), Konstantine (subcluster H1), and Predator (subcluster H1) are represented with gene annotations as described for Fig. 3. Pairwise nucleotide sequence similarities are displayed using Phamerator and are colored as described for Fig. 5. Note that there are segments of DNA sequence similarity between Konstantine and Predator, but none between Marvin and Wildcat or between Wildcat and Konstantine. Two relevant segments of the Marvin genome are shown, encompassing genes 46 to 50 and 85 to 91, whereas contiguous regions of the other phages are displayed. The Pham designations of the minor tail protein genes shared between the phages are shown in large bold type.
One of the displaced genes, gene 87, encodes a large protein (1,318 residues) corresponding to gp43 and gp44 of Wildcat (Fig. 7), which are combined into a single open reading frame (Fig. 7). Wildcat gp44 contains a putative β-lactamase domain, and related proteins are widespread throughout mycobacteriophage genomes, although in each instance, the genes coding for these proteins are located among the minor tail protein genes (39). This is observed in Wildcat, as well as in the cluster H1 genome, Konstantine (Fig. 7), where they are positioned just downstream of tapemeasure protein genes; another H1 phage, Predator, lacks this function (Fig. 7). Marvin gp88 is a member of Pham2441, along with Wildcat gp45, although BlastP searches suggest that Konstantine gp36 is a more distantly related homologue (Fig. 7). Marvin gp89 and gp90 are homologues of Konstantine gp37 and gp38, respectively, and there is also a relative of Marvin gp89 in Wildcat (gp46). These relationships suggest that all four Marvin genes (87 to 90) code for minor tail proteins, although gp90 was not found by mass spectrometry.
Surprisingly, an additional protein, gp57, is found associated with virions (Table 2) although gene 57 lies to the right of the lysis cassette and outside the regions described above (Fig. 3). Marvin gp57 has weak sequence similarity to LeBron gp24 (27% identity), which is encoded at the extreme right end of the LeBron tail gene cluster and which therefore is also a candidate for a virion protein. None of the protein products of any of the surrounding genes were identified as virion-associated proteins.
Marvin lysis cassette.
The lysis cassette of Marvin is coded near the middle of the genome, a common location for mycobacteriophage genomes, and includes lysin A (gp51), the holin protein (gp54), and a putative lysin B (gp53) (16, 42) (Fig. 3). The lysin A is most closely related to the cluster B phages Pacc40 and Cooper (55% and 51% identity, respectively) and contains a PGRP domain associated with N-acetylmuramoyl-l-alanine amidase activity. Marvin gp53 is only a distant relative of other mycobacteriophage lysin B proteins, with the central portion having weak sequence similarity to Giles gp32, extending the considerable sequence diversity of this phamily of proteins (42). The 97-residue Marvin gp54 is a good candidate for the holin protein, containing two strongly predicted transmembrane domains at residues 8 to 30 and 51 to 73. The only other mycobacteriophage protein with significant sequence similarity is Barnyard gp41 (47% identity). The small protein encoded between lysins A and B (gp52) has no close relatives and is of unknown function. Although it is not related to the gp1 protein of mycobacteriophage Ms6 (7), it is plausible that it plays a similar chaperone-like role in the functioning of the lysis system.
Nonstructural genes 55 and 56, 58 to 86, and 92 to 107.
The 32-kbp right half of the Marvin genome (from 33.4 kbp to the right end) encodes mostly nonstructural proteins gp55 to gp86 and gp92 to gp107 (Fig. 3), the only exception being gp57, which is virion associated. These genes form three distinct groups: 55 to 91 in a rightwards-transcribed group that also includes the putative minor tail protein genes 87 to 90, the leftwards-transcribed genes 92 to 103, and four rightwards-transcribed genes at the right end, 104 to 107. Genes 55 to 92 may constitute a single operon, and most genes are closely linked, although there are intergenic gaps of >100 bp between genes 65 and 66, 70 and 71, and 75 and 76. Because of the difficulty in accurately predicting mycobacteriophage promoter sequences (other than canonical σ70-like candidates) it is unclear if these genes are transcribed from a single upstream promoter (presumably upstream of gene 55) or if there are additional promoters in the intergenic gaps. Twenty-seven of the 37 genes are orphams and have no close mycobacteriophage relatives (Fig. 3). However, several of these have either a weak match to other mycobacteriophage proteins or to other database matches. These include gp62, gp63, and gp72, which have weak sequence similarities to LeBron gp58 (34% identity), Tweety gp64 (46% identity), and Pacc40 gp68 (36% identity), respectively, all of which also match bacterial proteins of unknown function. Marvin gp64, gp71, and gp75 also have similarities to bacterial proteins of unknown function. Marvin gp65 and gp86 have similarity to HNH homing endonucleases (Fig. 3).
Perhaps the most informative of the database matches of genes in this region is to the gene coding for Marvin gp58, which has significant similarity to phage-encoded Xis proteins, including the putative Xis of the M. tuberculosis prophage-like element ϕRv2 (Rv2657c; 43% amino acid identity) (30). This is surprising because there is no evidence of an integrase gene in the Marvin genome. However, this is reminiscent of the genome structure observed in mycobacteriophage TM4. Until recently, TM4 was also a singleton phage, but it is now a member of cluster K, for which there are four other relatives (46). Although TM4 is not temperate and does not form stable lysogens, all of the other cluster K phages are temperate and contain easily recognizable integrase genes. The simple explanation is that TM4 is a derivative of a temperate parent in which the integrase and presumably the repressor genes have been lost (46); a similar event has been proposed for mycobacteriophage D29 (14). It is plausible that Marvin is also a derivative of a temperate parent that has lost its integrase gene but retained the Xis gene, 58.
Of the 10 putative gene products with mycobacteriophage relatives (Fig. 3), several have informative database matches to nonmycobacteriophage proteins or to conserved domains. For example, gp69 is predicted to have a domain of the family cl00695 that is associated with the SMF family of proteins, including Helicobacter pylori DprA, which binds to single-stranded DNA (ssDNA) to facilitate transformation. The gene segment from 79 to 83 is of particular note in that HHPred predicts that four of these genes' products (all except the small orpham gp82) are transferases, with gp79 being a methyltransferase and gp80, gp81, and gp83 being glycosyltransferases. Marvin gp80 and gp81 are predicted specifically to be polypeptide N-acetylgalactosaminyltransferases, and gp83 is predicted specifically to be an α-1,3-mannosyl-glycoprotein β-1,2-N-acetylglucosaminyltransferase. The four genes coding for these proteins perhaps contribute to a common biochemical pathway because they are conserved with a common synteny in phage Corndog (gp35 to gp38), although other mycobacteriophages have just a subset of the genes in mosaic relationships (see below). It is unclear whether the presumed protein targets of modification are phage or bacterial in nature.
The 11 leftwards-transcribed genes 93 to 103 have no close relatives in other mycobacteriophages, and only one's product, gp94, has weak matches to other mycobacteriophages as well as nonmycobacteriophage proteins. The closest mycobacteriophage relative is Konstantine gp57 (30% identity), and there are numerous related proteins of unknown functions. The role of this segment of the Marvin genome is therefore unclear. We note, however, that there are three putative σ70-like promoters positioned between genes 94 and 95, between genes 101 and 102, and upstream of 103 (Fig. 3).
The four rightwards-transcribed genes at the right end of the genome, 104 to 107, have no mycobacteriophage homologues, but gp105 and gp106 are closely related to families of host-encoded proteins. Marvin gp105 contains an amidoligase-2 (COOH-NH2 ligase superfamily) domain similar to that found in RflaF proteins of Ruminococcus flavefaciens (32% identity), and gp106 is related to glutamine amidotransferases of the type II class. The specific role of these genes is not known but could be involved in the synthesis of novel metabolites or peptide-tagging systems (32). We note that a pair of genes encoding related functions but only very distantly related are also present in phage phiEco32 (49), and these have been postulated to modify the bacterial cell wall to prevent infection by other bacteriophages (32). Curiously, located between Marvin genes 105 and 106 is a 26-bp palindrome composed of identical 13-bp inverted repeats (Fig. 3). The role of this is unclear, but it is a candidate for a binding site of a regulatory protein.
Mosaicism of the Marvin genome.
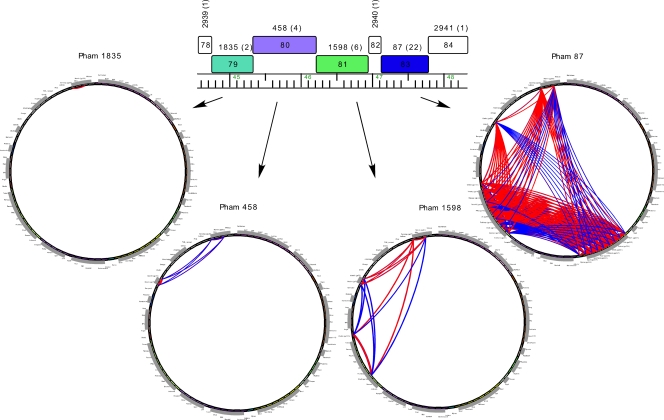
The prominent architectural feature of mycobacteriophage genomes is that they are mosaic, with different segments having distinctly different evolutionary origins (30, 43). Marvin is likely to be no exception to this, although the small number of genes with relatives in other mycobacteriophages makes this less obvious (Fig. 3). However, a particularly good example of genome mosaicism is seen in genes 78 to 84 (Fig. 8). Genes 78, 82, and 84 have no relatives, although 79, 80, 81, and 83 are related to other mycobacteriophage genes (Fig. 8). Phamily circle representations of the latter four genes show which mycobacteriophage genomes have the related genes and which do not (Fig. 8). For example, although all four have a related gene in Corndog, the presence in other genomes varies greatly. Pham 87 (containing Marvin gp83) has the largest number of members, none of which are in the subcluster I1 genomes Brujita or Island3. In contrast, Pham 458 and Pham 1598 have fewer members, but both Brujita and Island3 are included in both of them. These genes therefore have distinct phylogenies and have arrived in the Marvin genome through different evolutionary journeys.
Fig 8.
Marvin genome mosaicism. Mosaicism of the Marvin genome is illustrated by a segment of genes 78 to 84 and their relatives. Genes 78, 82, and 84 are orphams and have no relatives in other mycobacteriophages. In contrast, genes 79, 80, 81, and 83 have various numbers of relatives that are present in a wide variety of other mycobacteriophage genomes, as illustrated by the phamily circles. Each phamily circle has all 84 genomes in the “Marvin” phamerator database around the circumference, and arcs are drawn between genomes that contain members of that particular phamily. Blue arcs correspond to relationships revealed by BlastP comparison and red arcs to those by Clustal comparison.
DISCUSSION
We have described here a new singleton mycobacteriophage, Marvin, that reveals a number of new insights into the diversity and evolution of bacteriophages. Although the number of sequenced mycobacteriophage genomes has increased sharply over the past 10 years (19, 47), the continued discovery of new singleton phages such as Marvin demonstrates that our current collection is far from being a representative sample of the population at large. As the mycobacteriophage collection expands further, we anticipate that relatives of Marvin will be discovered, although we note that phages such as Giles, Corndog, and Wildcat (40, 43) persist as singleton phages many years after their initial isolation.
Marvin is the first mycobacteriophage in which we have observed an obvious interruption in the group of minor tail protein genes that are typically positioned immediately downstream of the tapemeasure protein gene. There are several examples of gene insertions within the structural gene operon, such as in Wildcat or Corndog (43), and the integration cassette appears to be “misplaced” within the Giles genome, such that it is flanked by tail genes (40). There are additional examples of interruptions within the head genes of siphoviral phages, including a large insertion between the head accessory protein and capsid protease genes in Vibrio phage SIO-2 (2). The Marvin genomic architecture is unusual, however, with a contiguous segment of the minor tail protein genes positioned about 20 kbp away from the other tail genes and within nonstructural genes. There is also a lone virion gene, 57, situated among nonstructural genes. In the absence of any close relatives of Marvin, it is not clear whether the evolutionary events giving rise to this are relatively recent or ones that are older and well established. The organization raises substantial questions as to how the structural genes are expressed, and if there are promoters for late gene expression upstream of genes 57 and 87.
Marvin is not a temperate phage, and we have been unable to recover stable lysogens. It does not contain an identifiable integrase gene, and although there are at least two candidate DNA-binding proteins (gp22 and gp58), we doubt that either acts as a phage repressor; Marvin gp22 may for example be an antitoxin component of a toxin-antitoxin system. Marvin gp58 is strongly predicted to contain a helix-turn-helix DNA binding motif and shows strong sequence similarity to Xis family proteins, including the RDF of the M. tuberculosis prophage-like element ϕRv2. This is a curious gene to find in a lytic phage and we therefore predict that Marvin is a derivative of a temperate parent and has lost—perhaps recently—its immunity and integration functions. This is not unprecedented, and similar conclusions can be drawn about the origins of mycobacteriophage D29 (14) as well as TM4 (15, 46). We thus predict that future phage discovery efforts will identify close relatives of Marvin but which are temperate, just as occurred with the finding of relatives of TM4 (46).
The Marvin genome contains several groups of genes that are not found in other mycobacteriophages. Although many of these have no known function, the presence of amidoligase and glutamine amidotransferase genes (105 and 106) suggests the possibility of intriguing new functions. Genes with these putative functions have been observed in the unrelated phage phiEco32, and it has been suggested that they could play a role in modifying the cell wall and thus preventing superinfection by other phages (32). This is certainly a plausible role in Marvin too, although they could also play roles in synthesis of secondary metabolites or in modulating expression of either phage or host genes. If Marvin is indeed derived from a temperate parent, then these genes could be expressed during lysogeny so as to influence the physiological state of the bacterial host.
Finally, although Marvin has no close relatives, comparisons with other mycobacteriophages clearly show its mosaic nature. This is observed with genes 79 to 83 encoding predicted transferases (Fig. 8) but also with the curious array of genes 7 to 11. This is the one segment of the Marvin genome that appears to have been acquired relatively recently and must be in rather rapid exchange among the mycobacteriophage genomes. We note that although Marvin gene 8 has 95% or greater nucleotide sequence similarity to at least 10 other mycobacteriophages, there do not appear to be any closely related host genes. Acquisition presumably therefore came from other mycobacteriophages, rather than from the host chromosome, and it is possible that these are new types of self-mobile elements.
ACKNOWLEDGMENTS
This work was supported in part by a grant to the University of Pittsburgh by the Howard Hughes Medical Institute in support of G.F.H. under HHMI's Professorship program, and by National Institutes of Health grant GM093901 to G.F.H. Cabrini College was supported by HHMI as a member of the Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science (HHMI's SEA-PHAGES) program.
We thank the Joint Genome Institute for DNA sequencing support and Daniel A. Russell and Michelle M. Boyle for assistance in sequence analysis and sample preparation. We also thank the Genomics and Proteomics Core Laboratories at the University of Pittsburgh and Lewis Brown at Columbia University for assistance with mass spectrometry. We are grateful to Steve Cresawn for help with Phamerator and for generating the Phamerator databases.
Footnotes
Published ahead of print 22 February 2012
REFERENCES
- 1. Anne J, et al. 1995. Analysis of the open reading frames of the main capsid proteins of actinophage VWB. Arch. Virol. 140:1033–1047 [DOI] [PubMed] [Google Scholar]
- 2. Baudoux AC, et al. 9 January 2012. Genomic and functional analysis of Vibrio phage SIO-2 reveals novel insights into ecology and evolution of marine siphoviruses. Environ. Microbiol. doi:10.1111/j.1462–2920.2011.02685.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belfort M, Roberts RJ. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borodovsky M, McIninch J. 1993. Recognition of genes in DNA sequence with ambiguities. Biosystems 30:161–171 [DOI] [PubMed] [Google Scholar]
- 5. Brussow H, Hendrix RW. 2002. Phage genomics: small is beautiful. Cell 108:13–16 [DOI] [PubMed] [Google Scholar]
- 6. Casjens SR. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8:451–458 [DOI] [PubMed] [Google Scholar]
- 7. Catalao MJ, Gil F, Moniz-Pereira J, Pimentel M. 2010. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 77:672–686 [DOI] [PubMed] [Google Scholar]
- 8. Champe SP, Eddleman HL. 1967. Poypeptides associated with morphogenetic defects in bacteriophage T4, p 55–70 In Colter JS, Paranchych W. (ed), The molecular biology of viruses. Academic Press, New York, NY [Google Scholar]
- 9. Chang JR, Poliakov A, Prevelige PE, Mobley JA, Dokland T. 2008. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology 370:352–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cresawn SG, et al. 2011. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics 12:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derbyshire V, Belfort M. 1998. Lightning strikes twice: intron-intein coincidence. Proc. Natl. Acad. Sci. U. S. A. 95:1356–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fineran PC, et al. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106:894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ford ME, Sarkis GJ, Belanger AE, Hendrix RW, Hatfull GF. 1998. Genome structure of mycobacteriophage D29: implications for phage evolution. J. Mol. Biol. 279:143–164 [DOI] [PubMed] [Google Scholar]
- 15. Ford ME, Stenstrom C, Hendrix RW, Hatfull GF. 1998. Mycobacteriophage TM4: genome structure and gene expression. Tuber. Lung Dis. 79:63–73 [DOI] [PubMed] [Google Scholar]
- 16. Gil F, et al. 2008. The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology 154:1364–1371 [DOI] [PubMed] [Google Scholar]
- 17. Gomathi NS, et al. 2007. In silico analysis of mycobacteriophage Che12 genome: characterization of genes required to lysogenise Mycobacterium tuberculosis. Comput. Biol. Chem. 31:82–91 [DOI] [PubMed] [Google Scholar]
- 18. Hatfull GF. 2006. Mycobacteriophages, p 602–620 In Calendar R. (ed), The bacteriophages. Oxford University Press, New York, NY [Google Scholar]
- 18a. Hatfull GF, et al. 2010. Comparative genome analysis of 60 mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hatfull GF. 2010. Mycobacteriophages: genes and genomes. Annu. Rev. Microbiol. 64:331–356 [DOI] [PubMed] [Google Scholar]
- 20. Hatfull GF, Hendrix RW. 2011. Bacteriophages and their genomes. Curr. Opin. Virol. 1:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hatfull GF, et al. 2006. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hatfull GF, Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science Program, KwaZulu-Natal Research Institute for Tuberculosis and HIV Mycobacterial Genetics Course Students, and Phage Hunters Integrating Research and Education Program 2012. The complete genome sequences of 138 mycobacteriophages. J. Virol. 86:2382–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hatfull GF, Sarkis GJ. 1993. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol. Microbiol. 7:395–405 [DOI] [PubMed] [Google Scholar]
- 24. Hendrix RW. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506–511 [DOI] [PubMed] [Google Scholar]
- 25. Hendrix RW. 2009. Jumbo bacteriophages. Curr. Top. Microbiol. Immunol. 328:229–240 [DOI] [PubMed] [Google Scholar]
- 26. Hendrix RW, Casjens S. 2006. Bacteriophage lambda and its genetic neighborhood, p 409–447 In Calendar R. (ed), The bacteriophages. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 27. Hendrix RW, Casjens SR. 1974. Protein cleavage in bacteriophage lambda tail assembly. Virology 61:156–159 [DOI] [PubMed] [Google Scholar]
- 28. Hendrix RW, Hatfull GF, Smith MC. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154:253–257 [DOI] [PubMed] [Google Scholar]
- 29. Hendrix RW, Lawrence JG, Hatfull GF, Casjens S. 2000. The origins and ongoing evolution of viruses. Trends Microbiol. 8:504–508 [DOI] [PubMed] [Google Scholar]
- 30. Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henry M, et al. 2010. In silico analysis of Ardmore, a novel mycobacteriophage isolated from soil. Gene 453:9–23 [DOI] [PubMed] [Google Scholar]
- 32. Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. 2009. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol. Biosyst. 5:1636–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Katsura I. 1987. Determination of bacteriophage lambda tail length by a protein ruler. Nature 327:73–75 [DOI] [PubMed] [Google Scholar]
- 34. Katsura I, Hendrix RW. 1984. Length determination in bacteriophage lambda tails. Cell 39:691–698 [DOI] [PubMed] [Google Scholar]
- 35. Krumsiek J, Arnold R, Rattei T. 2007. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23:1026–1028 [DOI] [PubMed] [Google Scholar]
- 36. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee E, Harris N, Gibson M, Chetty R, Lewis S. 2009. Apollo: a community resource for genome annotation editing. Bioinformatics 25:1836–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mediavilla J, et al. 2000. Genome organization and characterization of mycobacteriophage Bxb1. Mol. Microbiol. 38:955–970 [DOI] [PubMed] [Google Scholar]
- 40. Morris P, Marinelli LJ, Jacobs-Sera D, Hendrix RW, Hatfull GF. 2008. Genomic characterization of mycobacteriophage Giles: evidence for phage acquisition of host DNA by illegitimate recombination. J. Bacteriol. 190:2172–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nesbit CE, Levin ME, Donnelly-Wu MK, Hatfull GF. 1995. Transcriptional regulation of repressor synthesis in mycobacteriophage L5. Mol. Microbiol. 17:1045–1056 [DOI] [PubMed] [Google Scholar]
- 42. Payne K, Sun Q, Sacchettini J, Hatfull GF. 2009. Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 73:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pedulla ML, et al. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182 [DOI] [PubMed] [Google Scholar]
- 44. Petrovski S, Seviour RJ, Tillett D. 2011. Genome sequence and characterization of the Tsukamurella bacteriophage TPA2. Appl. Environ. Microbiol. 77:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piuri M, Hatfull GF. 2006. A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 62:1569–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pope WH, et al. 2011. Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4. PLoS One 6:e26750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pope WH, et al. 2011. Expanding the diversity of mycobacteriophages: insights into genome architecture and evolution. PLoS One 6:e16329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sampson T, et al. 2009. Mycobacteriophages BPs, Angel and Halo: comparative genomics reveals a novel class of ultra-small mobile genetic elements. Microbiology 155:2962–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Savalia D, et al. 2008. Genomic and proteomic analysis of phiEco32, a novel Escherichia coli bacteriophage. J. Mol. Biol. 377:774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Showe MK, Isobe E, Onorato L. 1976. Bacteriophage T4 prehead proteinase. I. Purification and properties of a bacteriophage enzyme which cleaves the capsid precursor proteins. J. Mol. Biol. 107:35–54 [DOI] [PubMed] [Google Scholar]
- 51. Showe MK, Isobe E, Onorato L. 1976. Bacteriophage T4 prehead proteinase. II. Its cleavage from the product of gene 21 and regulation in phage-infected cells. J. Mol. Biol. 107:55–69 [DOI] [PubMed] [Google Scholar]
- 52. Stein LD, et al. 2002. The generic genome browser: a building block for a model organism system database. Genome Res. 12:1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 54. Tori K, et al. 2009. Splicing of the mycobacteriophage Bethlehem DnaB intein: identification of a new mechanistic class of inteins that contain an obligate block F nucleophile. J. Biol. Chem. 285:2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Van Dessel W, et al. 2005. Complete genomic nucleotide sequence and analysis of the temperate bacteriophage VWB. Virology 331:325–337 [DOI] [PubMed] [Google Scholar]
- 56. Wommack KE, Colwell RR. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu J, Hendrix RW, Duda RL. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11–21 [DOI] [PubMed] [Google Scholar]
- 58. Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. 2003. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol. Microbiol. 50:303–317 [DOI] [PubMed] [Google Scholar]