Abstract
The promiscuous presentation of epitopes by similar HLA class I alleles holds promise for a universal T-cell-based HIV-1 vaccine. However, in some instances, cytotoxic T lymphocytes (CTL) restricted by HLA alleles with similar or identical binding motifs are known to target epitopes at different frequencies, with different functional avidities and with different apparent clinical outcomes. Such differences may be illuminated by the association of similar HLA alleles with distinctive escape pathways. Using a novel computational method featuring phylogenetically corrected odds ratios, we systematically analyzed differential patterns of immune escape across all optimally defined epitopes in Gag, Pol, and Nef in 2,126 HIV-1 clade C-infected adults. Overall, we identified 301 polymorphisms in 90 epitopes associated with HLA alleles belonging to shared supertypes. We detected differential escape in 37 of 38 epitopes restricted by more than one allele, which included 278 instances of differential escape at the polymorphism level. The majority (66 to 97%) of these resulted from the selection of unique HLA-specific polymorphisms rather than differential epitope targeting rates, as confirmed by gamma interferon (IFN-γ) enzyme-linked immunosorbent spot assay (ELISPOT) data. Discordant associations between HLA alleles and viral load were frequently observed between allele pairs that selected for differential escape. Furthermore, the total number of associated polymorphisms strongly correlated with average viral load. These studies confirm that differential escape is a widespread phenomenon and may be the norm when two alleles present the same epitope. Given the clinical correlates of immune escape, such heterogeneity suggests that certain epitopes will lead to discordant outcomes if applied universally in a vaccine.
INTRODUCTION
Variation within the highly polymorphic major histocompatibility complex (MHC) region is the primary genetic component linked to immune control of HIV-1 (27, 39). This effect is due almost entirely to specific HLA-I alleles, many of which have been previously linked with rates of HIV disease progression in molecular epidemiology studies (22, 24, 33, 42, 44). HLA-I associated immune control of HIV is mediated by CD8+ T cells, which recognize viral epitopes presented by HLA-I proteins on the surfaces of infected cells. HIV-1, however, is able to evade recognition by HLA-restricted CD8+ T cells through the selection of immune escape mutations (32, 63).
Recently, HLA-restricted immune escape pathways were systematically identified through population-level analyses of linked HLA class I and HIV sequence data sets, yielding detailed “immune escape maps” of the HIV-1 proteome (14–16, 19, 60, 65). The discovery that immune escape pathways are generally predictable based on the host HLA repertoire represents a major step forward in HIV vaccine research (1, 18); however, substantial differences in the probability of escape have been observed between populations (4, 19, 41), between individuals (58, 67, 80), and even between members of the same HLA allelic family (41, 50). Achieving a deeper understanding of the host correlates of immune escape is therefore of utmost importance to T-cell-based HIV-1 vaccine design.
HLA class I peptide binding specificities are largely defined by polymorphisms in the peptide-binding groove of the HLA molecule (6, 7, 70). HLA alleles with similar sequences in the binding groove therefore tend to bind similar or even identical peptides, which allows HLA alleles to be grouped into families, or “supertypes,” based on shared peptide presentation (17, 71, 72). A large number of HLA-restricted CD8+ T-cell epitopes have been optimally defined in HIV-1 (52), and vaccine strategies based on the design of universal “supertope” immunogens have been proposed as a method to elicit broad immune responses using a limited number of epitopes (70, 72). However, despite these common patterns, substantial caveats remain. Although some epitopes display promiscuity of HLA binding (29), meaning that they can be presented by a variety of HLA-I alleles, both within (17, 29, 66, 76) and between (29, 54, 66) HLA supertypes, the frequency of epitope targeting and/or mutational escape may vary depending on which HLA allele presents the epitope. For example, members of the B7 supertype exhibit vastly different escape and functional characteristics despite similar epitope targeting frequencies (50), while members of the B58 supertype exhibit very different targeting frequencies (2, 46, 53, 57). Perhaps as a result of restricting different epitopes (25, 45, 46), or differential escape within commonly restricted epitopes (40), members of both the B7 and B58 supertypes have discordant associations with viral control (44, 49). More broadly, comparative studies of immune escape across cohorts, ethnicities, and geographic regions have revealed that alleles of the same supertype or type (formerly referred to as a two-digit allele group) are not always associated with the same immune escape patterns (41), and identical alleles may select different escape patterns in different ethnic groups (4). Taken together, these studies suggest that CD8+ targeting frequency and risk of immune escape are highly dependent on the genetic context in which the epitope is presented, a result that may have profound consequences for subsequent viral control. In this study, we explored in detail the relationship between HLA allele carriage (at the subtype level) and the risk of immune escape in HIV-1 and the ability to control viral replication.
Systematic analysis of context-dependent immune escape has been limited by a lack of appropriate statistical tools. Studies to date have relied on comparative analyses of HLA-associated polymorphisms identified in different HIV-1 cohorts worldwide (4, 41), an approach that is error prone due to high false-negative rates and statistical power that varies based on HLA allele frequency and cohort sample size. We therefore developed a statistical approach to compare the magnitude of immune selection pressure (and thus by extension the risk of immune escape) on a given HIV codon, in different host genetic contexts. We then applied this method to a population-based data set of linked CD8+ T-cell responses, HLA class I subtypes, and HIV sequences from southern Africa, to investigate the patterns and genetic correlates of immune escape within all optimally defined CD8+ T-cell epitopes in HIV-1 Gag, Pol, and Nef. Using this method, we identified members of the same HLA supertype that restrict the same optimally defined epitopes, as evidenced by the presence of HLA-associated polymorphisms at the population level (5, 9). We then systematically tested for differential selection among members of the same HLA supertype that restrict the same epitope. Finally, we explored the potential effects of differential selection on plasma viral load.
MATERIALS AND METHODS
Study subjects.
We studied 2,126 chronically HIV-1 subtype C-infected, antiretroviral-naïve adults from five established African cohorts, including subjects in (i) Durban, South Africa (n = 1,218) (49, 56), (ii) Bloemfontein, South Africa (n = 261) (38), (iii) Kimberley, South Africa (n = 31) (55), and (iv) Gaborone, Botswana (n = 514) (69), and (v) southern African subjects attending outpatient HIV clinics in the Thames Valley area of the United Kingdom (n = 102), originating from Botswana, Malawi, South Africa, and Zimbabwe (55). Ethics approval was granted by the University of KwaZulu-Natal Biomedical Research Ethics Committee and the Massachusetts General Hospital Review Board (Durban cohort); the University of the Free State Ethics Committee (Kimberley and Bloemfontein cohorts); the Office of Human Research Administration, Harvard School of Public Health and the Health Research Development Committee, Botswana Ministry of Health (Gaborone cohort); and the Oxford Research Ethics Committee (Durban, Kimberley, and Thames Valley cohorts). Study subjects from all cohorts gave written informed consent for their participation.
High-resolution sequence-based HLA typing was performed as previously described (55). For the present study, all HLA alleles that could not be resolved to the subtype level were considered missing (2,919 of 14,486; 20.2%). HLA supertype, type, and subtype frequencies are shown in Table S1 in the supplemental material. Population sequences of HIV-1 proviral DNA-derived gag (p17+p24, n = 1,327), pol (protease, n = 865; reverse transcriptase, n = 905; integrase, n = 344), and nef (n = 738) were obtained (see Table S2 in the supplemental material), as previously described (55).
Viral load in chronic infection was measured using the Roche Amplicor version 1.5 assay, and CD4+ T cell counts were measured by flow cytometry, as previously described (55). Individuals with <2,000 viral copies/ml plasma and >250 CD4+ T cells/mm3 were defined as viremic controllers. Due to the geographic heterogeneity of the Thames Valley cohort, this cohort was excluded from viral load analyses. Viral load and high-resolution HLA typing were available for 1,870 individuals from the remaining cohorts.
Phylogenetically corrected odds ratio.
To allow us to quantify and compare the strength of selection pressure exerted by a particular HLA allele on a given HIV-1 codon, we adapted standard logistic regression techniques to take into consideration underlying evolutionary relationships between the HIV-1 sequences in the data set, yielding a statistic we call the “phylogenetically corrected odds ratio” of escape, which measures the strength of selection exerted by an HLA allele on a given polymorphism.
Logistic regression is a model used for predicting the probability of occurrence of a binary event, making it useful for modeling the probability of observing particular viral amino acids as a function of various predictors (such as HLA alleles or viral load). For this reason, logistic regression was used in the first population-level immune escape study (60). The model can be described as follows. Suppose we are interested in the probability of seeing a particular amino acid at a particular position, for example, 242N in HIV-1 Gag. If P is the probability of observing 242N, then the odds of observing 242N are P/(1 − P). Logistic regression models the log of the odds (log-odds) as a linear function of predefined predictors. For example, if we assume that the odds of seeing 242N depend on whether an individual expresses HLA allele X or Y, then ln(P/(1 − P)) = aX + bY + c, where X and Y are taken to be 0/1 binary variables and a, b, and c are scalar parameters whose values are chosen so as to maximize the likelihood of the data. Conveniently, the maximum likelihood parameters have intuitive interpretations: c represents the log-odds of observing 242N among individuals who express neither X nor Y, and a is the log-odds ratio of 242N among individuals who express HLA X compared to individuals who do not express X (and similarly for b and Y). A positive log-odds ratio (a > 0) indicates that 242N is more likely to be observed among individuals expressing the allele than among those not expressing the allele, while a negative log-odds ratio (a < 0) indicates the opposite. Thus, if a typical escape is T242N mediated by X being B*57:03, then we would expect to see a negative weight when computing the odds of T and a positive weight when computing the odds of N.
Although logistic regression is broadly applied in biomedical research, it can yield surprisingly high false-positive and false-negative rates when applied to viral sequences, which share an evolutionary relationship (11, 21). This problem can be circumvented in the special case where the transmitted virus sequence is known; however, in the vast majority of cases, the transmitted viral sequence is unknown. To get around this issue, we perform maximum-likelihood phylogenetic reconstructions of the HIV-1 sequences observed in the data set (one maximum likelihood tree for gag-pol and another for nef, estimated using PhyML 3.0 [35]) in order to estimate the transmitted viral sequence for each subject. A statistical model can then be “phylogenetically corrected” by designing that model to make use of both the estimated transmitted and the observed current viral sequences and then averaging the likelihood over all possible phylogenetic histories, as previously described (19, 21).
To create a phylogenetically corrected logistic regression test, we therefore first need to define a logistic regression model for cases in which both the transmitted and current viral sequences are known for each individual. To this end we modified the above definition to be ln(P/(1 − P)) = aX + bY + cT, where T represents a binary variable indicating whether the transmitted sequence contained 242N. We model T as a −1/1 binary variable, whereas the HLA variables X and Y are modeled as 0/1 binary variables. Thus, if an individual expresses neither X nor Y, then the log-odds of observing 242N will be c if the transmitted sequence contained 242N and −c if it did not. After picking maximum-likelihood values for a, b, and c, we can then interpret a as the log-odds ratio comparing the odds of observing 242N among individuals expressing HLA-X compared to those not expressing HLA-X, conditioned on the transmitting sequence.
The distinction between an odds ratio conditioned on the transmitted sequence and a more traditional odds ratio is important. In the traditional case, we would model the odds of carriage of a specific polymorphism (regardless of whether it was acquired at transmission or subsequently selected in vivo) in individuals expressing the relevant HLA allele compared to those not expressing it. The magnitude of the traditional odds ratio is therefore influenced by the frequency of the polymorphism in persons expressing the relevant HLA allele, as well as its prevalence in the overall population. Thus, a high odds ratio may result from either a high probability of escape in individuals expressing the HLA allele or a high level of conservation among individuals not expressing the allele. In contrast, when we condition on the transmitting sequence, we effectively model the odds of observing the selection of this mutation in vivo (because both the observed and transmitted variants are included in the model). In the context of HLA-mediated escape, the magnitude of an odds ratio that is conditioned on the transmitted virus can therefore be viewed as a measure of the strength of selection in vivo.
In practice, we cannot observe the infecting sequence in large cross-sectional cohorts. Therefore, we perform a weighted average over all possible infecting sequences, where the weights are informed by the phylogeny (19).
Hypothesis testing with the phylogenetically corrected logistic regression model.
Various hypotheses can be tested using the likelihood ratio test, which compares the likelihood of the null model against the likelihood of a richer model. To test if an HLA allele is associated with a given polymorphism, we compare the null model ln(P/(1 − P)) = cT (meaning that the odds of observing a polymorphism are completely determined by the inferred transmitted sequence) to the alternative model ln(P/(1 − P)) = aX + cT (meaning that the odds of observing the polymorphism are mediated by selection pressure imposed by HLA allele X). We can also test whether HLA alleles X and Y exert differential selection pressure on 242N. First, we construct a new variable, max(X,Y), which is 1 only if an individual expresses either X or Y. We then compare the null model ln(P/(1 − P)) = a max(X,Y) + cT to the alternative model ln(P/(1 − P)) = a max(X,Y) + bX + cT to test if there is sufficient evidence that X and Y should be treated as separate variables. To test the hypothesis that HLA allele X exerts differential selection pressure on 242N when coexpressed with HLA allele Y, we construct an interaction term, XY, which is 1 only if an individual expresses both X and Y. We then compare the null model ln(P/(1 − P)) = aX + cT to the alternative interaction model ln(P/(1 − P)) = aX + bXY + cT. The parameter b can then be interpreted as the log-odds ratio of escape in individuals coexpressing both X and Y to escape in individuals expressing only X. This interaction model is also used when Y is a continuous variable (e.g., log viral load).
Multiple hypothesis testing.
In the present study, we performed thousands of statistical tests. In such scenarios, the standard interpretation of the P value has relatively little meaning. We therefore primarily report false-discovery rates, which addresses multiple hypothesis testing (8). The false-discovery rate (FDR) is a property of a P value (P0) in the context of a specific set of tests and is defined as the expected proportion of tests for which P ≤ P0 that are false positive. The false discovery rate can be estimated using FDR(P0) = π0P0N/R, where N is the total number of tests performed, R is the number of tests where P is ≤P0, and π0 is the (unknown) proportion of all tests that are truly null (74). A straightforward, robust estimate of π0 is π̂0 = 2 · avg(P), where avg(P) is the average P value of all the tests (64). To ensure monotonicity with respect to P values, the FDR is reported as a q value, which is the minimum false discovery rate for all P values that are ≥P0 (73).
The appropriate choice of q-value threshold is context specific and depends on how the results will be interpreted. In the present study, we typically report all tests where q is <0.2 (implying that we expect 20% of reported tests to be false positives) but will sometimes report lower q values when more conservative interpretations are appropriate.
Definition of expanded optimal epitopes.
Optimally defined (52), HLA-restricted cytotoxic-T-lymphocyte (CTL) epitopes in HIV-1 Gag, Pol and Nef proteins were retrieved from the Los Alamos Database (http://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html; last updated 31 August 2009) and hand edited to reflect recent published corrections. These optimal epitope definitions are derived from in vitro epitope fine mapping and HLA restriction experiments reported in the literature. Therefore, published epitopes have not necessarily been tested in the context of all possible HLA alleles that could present them, nor have the restricting HLA alleles been defined at the same level of resolution throughout. Indeed, many epitopes have been restricted to one or two alleles, whereas others have been attributed to broad serotypes. In recognition of the fact that alleles with shared similar binding grooves are likely to present similar peptides, we expanded the optimal epitope list to include all HLA subtypes belonging to the published HLA type, supertype, or serotype, as follows. For each optimal epitope, we expanded the list of restricting HLA alleles to include all members of the HLA supertype to which the original restricting allele belonged (71). For optimal epitopes restricted by HLA alleles defined by their serotype only, we expanded the list to include all HLA alleles belonging to that serotype (36). For HLA-C alleles, which do not have supertype definitions, we expanded the list to include all HLA subtypes belonging to the HLA type of the published restricting allele.
We next sought to identify putative HLA escape mutations for each optimal epitope by identifying polymorphisms at sites within or flanking each epitope that were positively or negatively associated with particular HLA alleles. Specifically, for each observed amino acid at each position within 3 amino acids of the optimal epitope boundary, we ran a forward selection procedure to identify all HLA alleles that were independently associated with the amino acid. Only HLA alleles that were expressed by at least three individuals in the present study were analyzed; likewise, only polymorphisms that were observed in at least three individuals, and were not observed in at least three individuals, were considered. For each round of forward selection, we tested each HLA allele using a likelihood ratio test that compared an alternative phylogenetically corrected logistic regression model that included the new allele to a null model that included all alleles that had been added in previous iterations. After each iteration, the most significant HLA allele was added to the model. The P value reported for each HLA allele was that computed when the allele was added to the model. As a postprocessing step, we filtered the final output to include only those HLA alleles that are in the expanded list of potential restricting HLA alleles and computed q values based on the resulting subset. In some cases, one escape association could be ascribed to multiple overlapping optimal epitopes, each of which is putatively restricted by the same HLA allele or HLA alleles in the same supertype (e.g., the overlapping Gag epitopes KIRLRPGGK, RLRPGGKKK, and RLRPGGKKKY are all published as A*03:01 optimal epitopes, while the overlapping B7-restricted epitopes VPLRPMTY and RPMTYKAAL are published as B*35:01 and B*07:02 restricted, respectively). In these cases, overlapping optimal epitopes were grouped by published restricting supertype so that each such polymorphism was analyzed only once. We tested for differential escape only between HLA alleles that restricted the same optimal epitope (as determined by the supertype/serotype expansion described above).
IFN-γ ELISPOT assays.
In vitro HIV-specific CD8+ T-cell responses were determined in a cohort of 1010 subtype-C infected individuals using gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISPOT) assays using a set of 410 overlapping 18-mer peptides (OLPs) spanning the whole HIV-1 subtype C proteome (2001 consensus sequence). Overlapping peptides were arranged in a matrix system with 11 to 12 peptides in each pool. Responses to matrix pools were deconvoluted by subsequent testing with the individual 18-mer peptides within each pool, and the identity of the individual 18-mers recognized were thus confirmed, as previously described (44). Each optimal epitope was mapped to the OLP(s) that completely contained the optimal epitope. The CTL targeting frequency of each optimal epitope was defined as the targeting frequency of the OLP containing it (or, in the case where it was contained in two OLPs, the maximum targeting frequency between them). Associations between HLA alleles and OLP responses were assessed using a stepwise Fisher's exact procedure. For each OLP, we identified the most significantly associated HLA allele using Fisher's exact test. We then removed all individuals who expressed that allele and repeated these steps until all HLA alleles had been added to the model. We then computed false discovery rates for each HLA allele–OLP pair using the method described in reference 20.
RESULTS
Systematic identification of escape mutations in optimally defined epitopes.
This study focuses primarily on differential escape within epitopes presented by similar HLA alleles. To this end, we developed a phylogenetically corrected logistic regression model, which estimates the relative odds of escape among individuals who express a given HLA allele compared to those who do not. As described in Materials and Methods, our model conditions on the transmitted sequence (as estimated from the phylogeny), thereby removing any confounding that may arise from evolutionary relatedness among the HIV sequences (11, 19, 21). By building on the logistic regression model, our model allows us to estimate the relative odds of escape, as well as to explicitly test for differential escape (difference of odds of escape between two alleles) or escape that is dependent on external factors (interaction effects).
We first applied this phylogenetically corrected model to a large population-based data set to identify associations between individual HLA alleles and HIV-1 polymorphisms occurring within 3 amino acids of all optimal epitopes potentially restricted by those alleles. Potential HLA-optimal epitope restriction was defined by expanding the published list of optimally defined epitopes (52) to include all HLA alleles in the same supertype family as the published restricting alleles (see Materials and Methods). A forward selection algorithm was used to reduce the risk of false positives arising from linkage disequilibrium among HLA alleles (19). We identified 301 significant (q < 0.2, P < 0.004) HLA-HIV associations in Gag (n = 147), Pol (n = 110), and Nef (n = 44), covering 90 of 157 (57%) optimal epitopes (see Table S3 in the supplemental material). In what follows, we say that an HLA allele “restricts” an epitope if that allele is in the expanded optimal list and is associated with at least one escape polymorphism. There was an average of 1.9 HLA alleles that restricted each of those 90 optimal epitopes. Thirty-eight epitopes were restricted by more than one HLA allele (Table 1), and 67 epitopes were restricted by an allele other than its published restricting allele. Thus, in addition to identifying putative HLA-specific escape mutations, this analysis expands the number of closely related HLA alleles capable of presenting each optimal epitope by using escape mutations as indicators of active immune selection pressure in vivo.
Table 1.
Associations between supertype members and polymorphisms in optimal epitopes at a q value of <0.2
| Protein | Epitope | Sequence |
Location | HLAc | Associationsd | Supertype(s)e | |
|---|---|---|---|---|---|---|---|
| Optimala | Consensusb | ||||||
| Gag | KK9 | KIRLRPGGK | KIRLRPGGK | 18–26 | A*03:01 | A*03:01(R18K, K28X), A*30:01(X20S, C/K28Q), A*31:01(X18Q), A*33:01(X18Q, X28Q), A*74:01(R20K, R28X, Q30X) | A3 |
| RLRPGGKKK | RLRPGGKKH | 20–28 | |||||
| RLRPGGKKKY | RLRPGGKKHY | 20–29 | |||||
| KW9 | KYKLKHIVW | HYMLKHLVW | 28–36 | A*24:02 | A*23:01(R28X, L34I), A*24:02(R28X) | A24 | |
| RY11 | RSLYNTVATLY | RSLYNTVATLY | 76–86 | A*30:02, B58, B63 | A*01:01(E73K, Y79F/H), A*29:02(L85I), A*29:11(C87X), A*30:02(T81A), A*36:01(Y79H), B*58:02(C87X) | A1, A24 | |
| LY9 | LYNTVATLY | LYNTVATLY | 78–86 | A*29:02, B*44:03 | A*01:01(Y79F/H), A*29:02(L85I), A*29:11(C87X), A*30:02(T81A), A*36:01(Y79H) | A1, A24 | |
| TK8 | TLYCVHQK | TLYCVHEK | 84–91 | A*11:01 | A*03:01(X91S), A*34:02(K90R), A*68:01(K90X), A*66:03(T81A, A83V), A*74:01(R91K, V94I) | A3 | |
| IL10 | IEIKDTKEAL | IEVRDTKEAL | 92–101 | B*40:01 | B*41:01(E93V), B*44:03(X91Q), B*45:01(E93X) | B44 | |
| VL10 | VHQAISPRTL | VHQAISPRTL | 143–152 | B*15:10 | B*14:02(V143I, L147I), B*15:10(A146S) | B27 | |
| IW9 | ISPRTLNAW | ISPRTLNAW | 147–155 | B*57:01, B63 | B*57:02(A146P, I147L), B*57:03(A146P, I147L/M), B*58:01(X146P), B*58:02(S146X) | B58 | |
| KF11 | KAFSPEVI | KAFSPEVI | 162–169 | B*57:03 | B*57:03(X161D, A163G/S, S165K/N), B*58:01(V168X) | B58 | |
| KAFSPEVIPMF | KAFSPEVIPMF | 162–172 | |||||
| EL9 | EVIPMFSAL | EVIPMFTAL | 167–175 | A*26:01 | A*26:01(V168X, T173 M), A*29:02(X173 M) | A1 | |
| TL9 | TPQDLNTML | TPQDLNTML | 180–188 | B*07:02, B*39:10, B*42:01, B*81:01, C*08:02 | B*39:10(E177D, T186X), B*42:01(X182T), B*67:01(T190A), C*08:02(X182H), B*81:01(E177D, Q/T182E/G/S, T186S, L188F, T190X, V191I) | B7 | |
| TPYDINQML | TPQDLNTML | ||||||
| TW10 | TSTLQEQIGW | TSTLQEQIAW | 240–249 | B*57:01, B*58:01 | B*57:02(T242N, X247 M), B*57:03(T242N, I247V), B*58:01(T242N, L243X, X248A) | B58 | |
| NY10 | NPPIPVGDIY | NPPIPVGDIY | 253–262 | B*35:01 | B*35:01(D260E), B*39:10(I250 M), B*53:01(X256T, R264X), B*81:01(X252A) | B7 | |
| QW9 | QASQEVKNW | QATQDVKNW | 308–316 | B*53:01, B*57:01 | B*53:01(A309X, N315G), B*58:01(S309A, T310S) | ||
| Pr | DL9 | DTVLEEWNL | DTVLEEINL | 30–38 | A*68:02 | A*02:02(X39S), A*02:05(X36V) | A2 |
| RT | AM9 | ALVEICTEM | ALTAICEEM | 33–41 | A*02:01 | A*02:01(X35I, X36V, X40D), A*02:02(E36A, X41I) | A2 |
| TI8 | TAFTIPSI | TAFTIPSI | 128–135 | B*51:01 | B*51:01(I135T), B*81:01(X134G) | B7 | |
| KY9 | KQNPDIVIY | AQNPEIVIY | 173–181 | A*30:02 | A*29:02(178X), A*30:01(E173X), A*36:01(X178V) | A1 | |
| IL9 | IEELRQHLL | IEELREHLL | 202–210 | B*40:01 | B*18:01(E207X), B*44:03(E204X, E207K/N) | B44 | |
| QR9 | QIYPGIKVR | QIYPGIKVR | 269–277 | A*03:01 | A*03:01(K277R), A*30:01(Q278X), A*33:03(X273R), A*34:02(R275X) | A3 | |
| IW9 | IAMESIVIW | IAMESIVIW | 375–383 | B*58:01 | B*15:16(X386V), B*58:01(I375V, X377Q/T, X379A) | B58 | |
| EY9 | ETKLGKAGY | ETKIGKAGY | 449–457 | A*26:01 | A*26:01(K451R, M452X), A*29:01(T450N) | A1 | |
| Nef | RM9 | RPQVPLRPM | RPQVPLRPM | 71–79 | B*42:01, B*07:02 | B*35:01(Y81F), B*81:01(X71T, L76T/V) | B7 |
| TPQVPLRPM | RPQVPLRPM | ||||||
| QK10 | QVPLRPMTYK | QVPLRPMTYK | 73–82 | A*03:01, A*11:01 | A*03:01(A83G, X85L), A*33:01(X71K), A*34:02(A83G), A*66:03(V70I), A*68:01(X82Q) | A3 | |
| RL9 | VPLRPMTY | VPLRPMTY | 74–81 | B*35:01, B*07:02 | B*35:01(Y81F), B*81:01(X71T, L76T/V) | B7 | |
| RPMTYKAAL | RPMTYKAAF | 77–85 | |||||
| PK8 | PLRPMTYK | PLRPMTYK | 75–82 | A*11:01 | A*03:01(A83G, X85L), A*34:02(A83G), A*68:01(X82Q) | A3 | |
| KL10 | KAAFDLSFFL | KAAFDLSFFL | 82–91 | B*57:03 | B*57:02(A83X), B*57:03(A83X), B*58:01(A83G) | B58 | |
| AK9 | AVDLSHFLK | AFDLSFFLK | 84–92 | A*03:01, A*11:01 | A*03:01(A83G, X85L), A*34:02(A83G), A*68:01(X82Q) | A3 | |
| HW9 | HTQGYFPDW | HTQGFFPDW | 116–124 | B*57:01 | B*57:03(H116N), B*58:01(X116N) | B58 | |
| TL10 | TPGPGVRYPL | TPGPGVRYPL | 128–137 | B*07:02, B*42:01 | B*35:01(V133T), B*53:01(V133I) | B7 | |
| RW8 | RYPLTFGW | RYPLTFGW | 134–141 | A*24:02 | A*23:01(F143Y), A*24:02(Y135F/L) | A24 | |
| YY9 | YPLTFGWCY | YPLTFGWCF | 135–143 | B*18:01, B*53:01 | B*35:01(V133T), B*53:01(V133I) | B7 | |
Optimally defined epitopes as defined at http://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html. Only optimal epitopes that were restricted by least two HLA alleles are shown. Overlapping optimals with published alleles in the same supertype are grouped together.
Consensus in the present cohort. Underlining marks differences from published optimal epitope.
HLA alleles associated with the epitope as defined in the optimal epitope definitions.
HLA alleles that are related to the published alleles via supertype were tested for associations with polymorphisms within or flanking the optimal epitope. Associations are of the form (YposZ), where Y is a residue that is negatively associated with the HLA allele and Z is a residue that is positively associated with the allele. X indicates no association in the positive/negative direction. All associations with a q value of <0.2 are reported.
Supertypes that are represented by HLA alleles that are associated with escape.
Widespread differential escape among HLA alleles restricting the same epitope.
Examination of HLA-associated polymorphisms in Table 1 gives the impression that different HLA alleles restricting the same epitope will select for the same escape mutation only rarely. However, it would be premature to draw this conclusion from the association lists alone, without undertaking rigorous statistical tests. For example, the absence of any particular association may be due to lack of statistical power. Furthermore, two apparently identical associations may actually occur at substantially different frequencies among individuals expressing two different HLA alleles despite achieving statistical significance in both cases. We therefore created a statistical test for differential escape based on the phylogenetically corrected logistic regression that allows us to explicitly test whether the odds of escape mediated by two different HLA alleles are different.
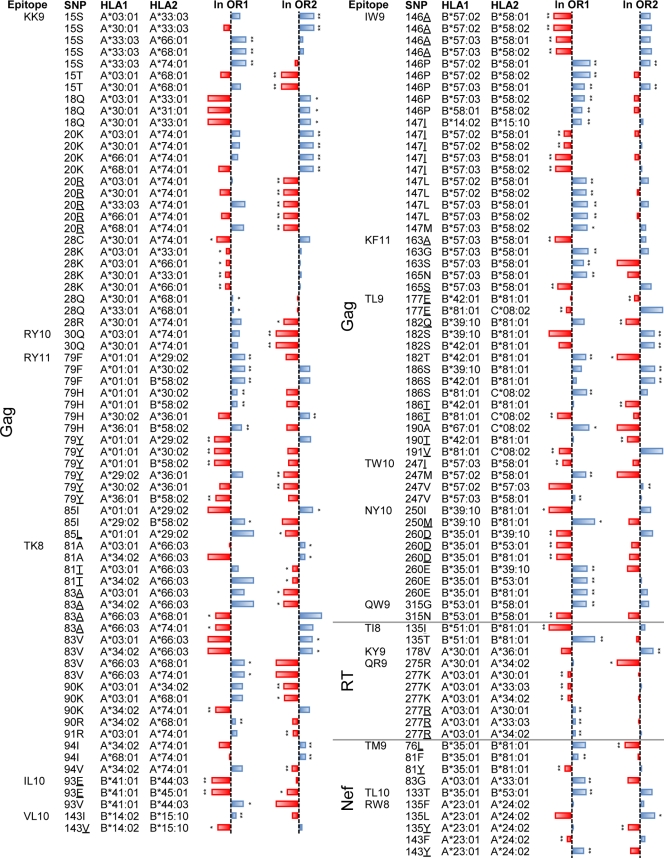
For each HLA-associated polymorphism in Table 1, we tested for differential selection between the reported allele and every other HLA allele that restricted the same epitope. In so doing, we confirm that HLA alleles restricting the same epitope exhibit differential escape in the vast majority of cases. Using the estimation method of Pounds and Cheng (64), which compares the observed distribution of P values for a large number of statistical tests against the expected distribution of P values under the null hypothesis, we estimate that roughly 70% of the 499 comparisons represent truly differential selection. Thus, differential selection appears to be the norm among HLA alleles that restrict the same epitope. Indeed, of the 38 epitopes that are restricted by multiple members of the same supertype, 37 (97%) exhibited differential escape in at least one position within or flanking the epitope. The only exception was RT-IL-9 (IEELRQHLL), which was restricted by B44 supertype members B*18:01 and B*44:03. Tests for differential escape did not achieve statistical significance despite the observation that the two alleles were associated with different polymorphisms (Table 1). Overall, a total of 278 instances of differential escape within the same epitope were observed at a P value of <0.05 (q < 0.025); these are listed in Table S4 in the supplemental material. Figure 1 displays the subset of these instances for which P was <0.005 (q < 0.006).
Fig 1.
Per-site differential escape between HLA alleles that restrict the same epitope. Bars represent the natural logarithm of the phylogenetically corrected odds ratio. Values between −20 (red, extending to the left) and +20 (blue, extending to the right) are shown; infinite log-odds ratios are set to values of ±20. Associations that are individually significant are labeled with single asterisks (q < 0.2) or double asterisks (q < 0.05). Polymorphisms are given in the SNP (single-nucleotide polymorphism) column; underlined amino acids signify cohort consensus. A phylogenetically corrected logistic model was used to derive a P value that tests the null hypothesis that both alleles select for escape with the same odds ratio. Comparisons with a P value of <0.005 (q < 0.006) are reported. The complete list of all comparisons with P values of <0.05 is available in Table S4 in the supplemental material.
Three broad categories of differential immune escape.
Differential escape (see Table S4 in the supplemental material) can be classified into three patterns. First, we observed cases where two alleles select for the same escape mutation, but to differing degrees. Second, we observed cases where one allele selects for escape whereas the other allele shows no association whatsoever. Finally, we observed cases where one allele is significantly positively associated with a polymorphism and the other allele is significantly negatively associated with the same polymorphism, a phenomenon termed “push-pull” escape (14).
The B7 supertype alleles B*42:01, B*81:01, B*39:10, and B*67:01, all of which are associated with escape in Gag-TL9 (TPQDLNTML), illustrate all three categories of differential escape. The first type (identical escape patterns that differ in statistical strength) is illustrated by the selection of T186X by both B*81:01 and B*39:10, but with a significantly higher absolute odds ratio for B*81:01 than for B*39:10 at this residue (ln odds ratios of −12 versus −10, q = 0.016; negative ln odds ratios indicate selection against a polymorphism, in this case the T variant). The second type (selection of escape by one but not the other of two related alleles) is illustrated by the lack of significant association between T186 and B*42:01. The third type, push-pull escape, is illustrated by the selection of X182T (wild type is Q) by B*42:01, but the specific selection against 182T by B*81:01 (which instead selects for Q182E/G/S). In this epitope, we also observed examples in which two alleles selected for the same escape patterns with the same frequencies: both B*39:10 and B*81:01 were associated with selection of E177D 3 amino acids upstream of TL9 with a ln odds ratio of 4 (P = 0.5 for differential escape between the two alleles).
Remarkably, there were only nine clear cases of differential escape in which two HLA alleles selected for the same polymorphisms but to a varying degree. These included B*57:03/B*58:01-mediated selection of T242N in Gag-TW10, A146P in Gag-IW9, and X116N in Nef-HW9 (where B*57:03 exhibited a higher odds of escape than B*58:01 in all three cases), B*81:01/B*39:10-mediated selection of T186X in Gag-TL9 (where B*81:01 exhibited higher odds of escape than B*39:10), B*35:01/B*53:01-mediated selection of V133X in Nef-TL10 (where B*35:01 exhibited higher odds of escape than B*53:01), and finally A*24:02/A*23:01-mediated selection of R28X (where A*24:02 exhibited higher odds of escape than A*23:01). Similarly, there were only two cases of significant push-pull: in addition to the B*81:01/B*42:01 example cited above, B*58:01 selected for S309A in Gag-QW9 (QASQEVKNW), while B*53:01 selected for A309X.
The remaining 267 (96%) examples of differential HLA-associated escape within the same epitope represented cases where one allele was significantly associated with a polymorphism at a given position and the other was not. Although some of these could represent cases of escape varying by degree where statistical power was insufficient to detect it, the observation that 182 (65% of total) of these instances represent cases where the log-odds ratios of the two alleles are in opposite directions argues against this interpretation in most cases. Similarly, although some of these could represent cases of push-pull escape where statistical power was insufficient to detect it, this is also not likely to be the explanation in most cases. Specifically, because odds ratios simply reflect the odds of selection among individuals who express the allele versus individuals who do not, observation of a statistically insignificant negative odds ratio by one allele alongside a significant positive odds ratio by another does not necessarily imply active selection against the polymorphism by the former allele. More likely, these insignificant negative odds ratios indicate a complete lack of selection on the part of the former restricting allele. What can thus be clearly concluded from the data is that at least 184 of 278 (66%) cases of observed differential selection represent instances in which the two HLA alleles drive distinct escape pathways within the epitope, as evidenced by opposing odds ratios.
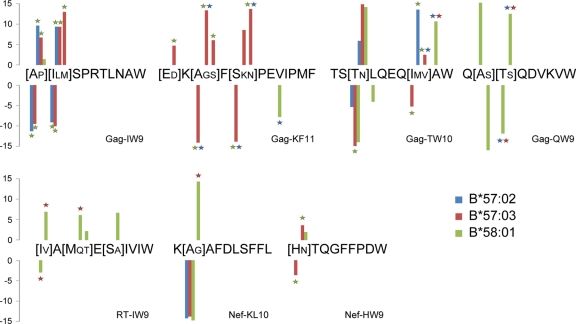
Differential escape among protective B58 supertype alleles.
We next used this approach to study in detail the escape pathways selected by the clinically important B58 supertype alleles B*57:02, B*57:03 and B*58:01 (note that B*57:01 frequency is negligible in African populations). We systematically compared the odds ratio of escape among the three alleles for every significant association reported in Table S3 (Fig. 2; q values computed separately for this analysis). The results highlight widespread variation in the selection patterns of these alleles, with an estimated 49% of comparisons representing true differences. For example, B*58:01, but not B*57:02 or B*57:03, selects for escape in Gag-QW9, with escape occurring most strongly at positions 309 (S309A) and 310 (T310S). These differences are statistically significant for T310S (q < 0.05) but not for S309X, for which B*58:01-mediated escape is comparably weaker. Gag-KF11 represents another striking example, with B*57:03 (but not B*57:02 or B*58:01) selecting for escape in positions −1, 2, and 4, and relatively weak B*58:01-mediated selection at position 5 of the epitope. Gag-TW10 is the only epitope for which all three alleles select for escape at the same position (T242N). At this position, we find that the odds of escape are significantly higher for B*57:03 than for B*58:01 (q = 0.05) and possibly B*57:02 (q = 0.2); no differences were observed between B*57:02 and B*58:01 (q > 0.4). B*57:03 selects for I247V, whereas B*57:02 selects for I247M, and B*58:01 does not appear to select for escape at this position. Rather, B*58:01 selects for 248A (which is the HIV-1 subtype C consensus residue), whereas there is no selection mediated at this position by B*57:02 or B*57:03. In the Gag-IW9 epitope, B*57:02 and B*57:03 both exhibit stronger selection pressure than B*58:01 at both positions 146 and 147 (q < 0.001). No significant differences between B*57:02 and B*57:03 were detected in this epitope, likely due to the relatively small number of individuals expressing B*57:02 (q > 0.2 for all comparisons).
Fig 2.
Differential escape among protective B58-supertype alleles. The phylogenetically corrected log-odds ratio of each B*57:02-, B*57:03-, or B*58:01-associated polymorphism from Table 1 was tested for differential selection against the other two alleles. Bars represent the log-odds of observing the indicated polymorphism. Stars indicate that the magnitude of the odds ratio is significantly (P < 0.05, q < 0.06) greater than that for the allele indicated by the color of the star. Large amino acid letters indicate cohort consensus; small letters indicate alternative polymorphisms associated with at least one allele.
Differential targeting frequency does not explain differential escape.
Selection of escape indicates that at least some individuals expressing the restricting allele have CTL that target the epitope in question. However, the absence of escape patterns at the population level does not necessarily imply a lack of targeting, nor do differential odds of escape necessarily imply differential odds of targeting. These observations are particularly evident for the B58-supertype epitopes, for which targeting was recently studied in detail (46). Comparing published B58 supertype-associated epitope targeting frequencies (46) with corresponding log-odds ratios of escape (Fig. 2) reveals several notable observations. First, the observation that Gag-KF11 is under strong B*57:03-mediated selection at multiple positions, whereas it is under only weak B*58:01-mediated selection and no B*57:02-mediated selection, is consistent with the observation that CTL frequently target KF11 when the epitope is presented by B*57:03 but rarely target KF11 when presented by B*58:01 and never target KF11 when presented by B*57:02 (46). In contrast, despite frequent targeting of RT-IW9 by both B*58:01-restricted and B*57:03-restricted (but not B*57:02-restricted) CTL (46), B*58:01 exhibits significantly higher odds of escape than either of the B*57 alleles at multiple positions within the epitope. Moreover, odds of B*57:03-mediated T242N escape within Gag-TW10 are significantly higher than those for B*58:01, despite the observation that CTL of B*58:01+ individuals target this epitope more frequently than do those of B*57+ individuals (46) (although decline of CTL responses following rapid escape in acute/early infection could provide an alternative explanation [1], as could the selection of the alternative 248A escape polymorphism in B*58:01+ individuals).
To test if odds of escape are correlated with odds of epitope targeting across all alleles in our study, we analyzed a data set of 1,010 adults with chronic C clade infection screened for responses to a panel of 18-mer peptides overlapping by 10 amino acids using IFN-γ ELISPOT assays. Defining odds of escape for a given HLA allele in a given epitope as the maximum absolute log-odds ratio over all significant HLA-associated polymorphisms in the epitope, we observed no correlation between odds of escape and odds of ELISPOT response (r2 < 0.01). When we compared the odds of observing an ELISPOT response between two alleles exerting selection pressure on the same codon but to potentially varying degrees (all allele pairs from Fig. 1 for which the sign of the log-odds ratios is the same for both alleles), we observed a weak negative trend between ELISPOT response frequency and odds of escape (P = 0.02, binomial test; data not shown). Although OLP data are inherently noisy, owing to the presence of multiple optimal epitopes per 18-mer, these data support the observation that differential escape is primarily the result of the selection of different escape pathways rather than differential frequencies of epitope targeting during chronic infection.
Risk of escape is not affected by HLA coexpression.
We hypothesized that the risk of escape could be modulated by the coexpression of other alleles. For example, a subdominant epitope may be less likely to be targeted (and thus escape) if the individual coexpresses an HLA allele that restricts one or more strongly immunodominant epitopes. Alternatively, the risk of escape may change if two overlapping epitopes are targeted at the same time. To test this hypothesis, we devised a statistical test that utilized a multiplicative interaction term between two alleles. Although several tests had P < 0.001, these were not significant after correction for multiple tests (q > 0.9 over 13,545 tests; data not shown). We next hypothesized that escape is more likely in individuals who are homozygous for a restricting allele. Once again, we observed no clear trends in the data (7 associations with 0.2 < q < 0.6, the rest with q > 0.9; data not shown). Overall, these results indicate that modulation of immune escape by HLA allele homozygosity or coexpression is not a general phenomenon; however, the observation of a number of results with low P values indicates that such interactions could occur in specific cases, though the present study is underpowered to identify such rare effects (note that the relationship between P and q values is a function of the number of tests exceeding the significance of a given P value relative to the total number of tests).
Risk of escape is independent of cohort.
One possible cause of differential escape is within-host T-cell receptor diversity, a factor that could also vary by population studied. Such variations could arise due to population-specific genetic characteristics or variations in antigenic exposure arising from region-specific vaccinations or diseases. Although we cannot explore the impact of T-cell-receptor (TCR) diversity on escape at the individual level, it is possible to investigate whether population level differences could confound the present analyses. To test this, we recomputed differential escape P and q values while conditioning on the cohort for which each individual was recruited. The resulting q values were nearly identical to the original analysis (r2 = 0.99; data not shown), indicating that differential escape could not be explained by region-specific variations (as approximated by cohort). We next tested if the odds of escape mediated by a specific allele were dependent on either cohort or country of origin (excluding the heterogeneous Thames Valley cohort). Once again, no significant cohort effects were observed (minimum q of 1 for both tests). Taken together, we found no evidence for odds of escape being a function of cohort or country of origin, suggesting that the dominant causal mechanism underlying the differential escape observed in the present study is more closely linked to specific HLA alleles than any unmeasured attributes that would be expected to correlate with ethnicity or region.
Population escape patterns predict the majority of intraepitopic variation.
The statistical evaluation of escape across individuals, such as the analyses described here, are inherently biased toward identification of common pathways of escape. Although the large size of our combined cohorts allows us to identify some uncommon escape pathways (over all associations, frequency of escape in individuals with the associated HLA allele ranged from 1.6% to 100%; interquartile range [IQR], 11% to 73%), very rare escapes, or rare escapes to uncommon HLA alleles, will go undetected (the statistical power falls precipitously for HLA alleles occurring in less than 1% of the population; data not shown).
To investigate the ability of population-based approaches to detect evidence of rare escape, we sought to identify whether optimal epitopes inherently display more sequence variation in individuals expressing the restricting allele than those who do not. For each optimal epitope, we tested for association between expression of any of the restricting HLA alleles and the presence of at least one nonconsensus residue within the epitope, excluding defined escape sites. This analysis will therefore identify epitopes in which variation commonly or occasionally occurs at any epitope position not identified in our previous analyses. Only 32 of 90 (36%) epitopes exhibited signs of increased general variation among individuals expressing the relevant HLA allele (q < 0.2). The majority of these (n = 24) were in Pol, for which the present study had the least statistical power due to low sequence coverage (e.g., integrase sequences were only available for 344 individuals). Overall, the median proportion of HLA-matched individuals with a nonconsensus residue at ≥1 non-HLA-associated site was 18%, compared to 13% in HLA mismatched individuals. To provide context, the median proportion of HLA-matched individuals with a nonconsensus residue at ≥1 HLA-associated site was 40%. This analysis suggests that the majority of escape mutations within HLA-optimal epitope pairs analyzed in this study is captured by the list of HLA-associated polymorphisms in Table S3 in the supplemental material but also supports the selection of unidentified rare escape pathways in some cases. This conclusion is broadly in line with a previous report on longitudinal clade B sequences from acutely infected individuals, in which 32 to 58% of observed substitutions (those achieving >25% frequency in a given quasispecies, as limited by “bulk” RT-PCR and sequencing protocols [47, 48]) in the first 2 years of infection exactly matched predicted HLA-associated polymorphisms identified in a chronically infected clade B cohort (13). Restricting that analysis to substitutions occurring inside optimally defined, HLA-matched epitopes shows that 80%, 52%, and 43% of intraepitopic substitutions in Nef, Gag, and Pol, respectively, are attributable to HLA associations used in that study (13; also our unpublished data).
Taken together, these data suggest that population studies with statistical power comparable to that of the present study are able to identify the majority of common escape mutations occurring in optimally defined epitopes, as well as some rarer mutations that smaller studies have missed. There is also, however, evidence of intraepitopic variation that is not captured by the present study and which may confer immune escape. It is unknown to what extent such rare escape pathways play a role in immune evasion. Furthermore, the current study focused exclusively on well-characterized epitopes, which may be more conserved than uncharacterized epitopes and may therefore display less variability in escape patterns.
Alleles exhibiting differential escape exhibit discordant associations with viral load.
The B58 supertype alleles B*57:03 and B*58:02 exhibit opposing correlations with plasma viral load (VL) in clade C infection, with B*57:03 being strongly correlated with low VL and B*58:02 strongly correlated with high VL (44, 49). These two alleles restrict completely different epitopes in HIV-1, which may account for these differences. Likewise, the B7 epitopes B*81:01, B*42:01, and B*07:01, which select for differential escape patterns within shared epitopes, also exhibit discordant associations with VL (44, 49, 50). We thus hypothesized that similar HLA alleles that select differential escape mutations within the same epitope commonly exhibit discordant associations with VL.
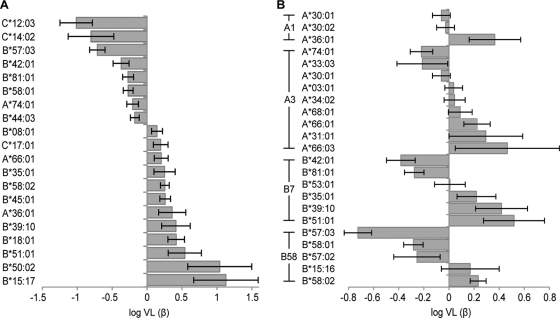
We therefore analyzed a data set of 1,870 chronically clade C-infected, antiretroviral-naïve adult Africans to test for associations between HLA alleles and VL. We first sought to identify which HLA alleles are independently and significantly associated with viral load. To this end, we tested all HLA subtypes using forward selection on a linear regression model, conditioned on the cohorts from which each sample was derived, with log10 VL as the dependent variable. From the distribution of P values, we estimate that 20% of the 98 HLA alleles tested are truly associated with VL. Using a P value of <0.05 (q < 0.13) as a threshold, we identified 20 HLA alleles that contribute to VL. These alleles were jointly added to a linear regression model to determine their independent contributions to VL (Fig. 3A). Eight of these alleles were associated with reduced VL (“protective” alleles), while 12 were associated with increased VL (“hazardous” alleles). Of note, 6 of the 12 (50%) hazardous alleles selected for escape in an epitope that was also restricted by at least one protective allele, and 5 of those cases were classified as differential escape.
Fig 3.
Relative contributions (β parameters) of HLA alleles to log VL. (A) All HLA alleles identified at a P value of <0.05 (q < 0.13) in a forward selection procedure; (B) HLA-A and HLA-B alleles, grouped by supertype, that were discordantly association with VL compared to an allele that was associated with differential escape in the same epitope, conditioned on the alleles from panel A. Relative bar heights depict the maximum likelihood β estimates from a joint linear model (estimated corrected average VL among individuals expressing that allele), conditioned on cohort labels. Error bars represent standard error estimates for β.
Simply identifying HLA alleles independently and significantly associated with VL, however, may be overly conservative. Indeed, two alleles that are not individually significantly associated with VL may have significantly discordant associations with VL if, for example, one allele tends to increase while the other tends to decrease VL. We therefore tested for discordant associations between HLA alleles and VL using the linear analogue of the differential selection model (with no correction for phylogeny, as none was needed). To reduce the possibility of confounding due to linkage disequilibrium, we conditioned all tests on the set of HLA alleles individually associated with VL (those in Fig. 3A). Using this model, an estimated 35% of HLA alleles that restrict the same epitope but select for differential escape also have discordant associations with VL. Twenty-seven pairs were significant at a q value of <0.2 (P < 0.1; see Table S5 in the supplemental material), and 11 were significant at a q value of <0.05 (P < 0.011; Fig. 3B). These differences were dominated by members of the A1, A3, B7, and B58 supertypes. Thus, these results indicate that similar HLA alleles that restrict the same epitope yet select for different escape pathways often have discordant associations with viral load.
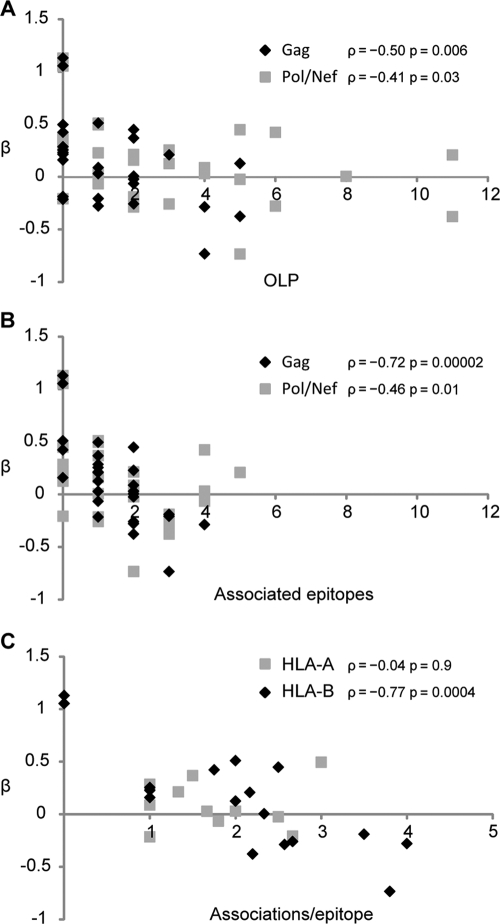
We next looked at whether various features of escape or targeting differentiated protective HLA alleles from hazardous ones. For this analysis, we built a single linear model that included all HLA alleles from Fig. 3A and B except the HLA-C alleles (for which there are few published epitopes) and interpreted the β estimates as the relative contribution of each allele to VL. We then correlated various HLA allele features against these β estimates. Over all 32 HLA alleles there was a strong correlation between the total number of Gag-OLPs associated with the allele and VL contribution (Spearman ρ = −0.50, P = 0.006), and a weak correlation between Pol/Nef-OLPs with VL contribution (ρ = −0.41, P = 0.03; Fig. 4A). An even stronger correlation was observed between VL contribution and the total number of optimal epitopes with associated escape polymorphisms in both Gag (ρ = −0.72, P = 1.7 × 10−5) and Pol/Nef (ρ = −0.46, P = 0.01; Fig. 4B). Furthermore, the total number of escape polymorphisms observed per epitope across Gag/Pol/Nef was strongly correlated with VL contribution in HLA-B alleles (ρ = −0.77, P = 3.5 × 10−4) but not HLA-A alleles (ρ = −0.04, P = 0.9; Fig. 4C), and the overall strength of escape associations was more statistically significant in protective alleles (median q = 0.001) than in hazardous alleles (median q = 0.03; P = 0.003, Mann-Whitney test). Of note, there was no difference in the entropy of epitopes restricted by protective versus hazardous alleles (P = 0.38), nor was there any difference in the entropy at the sites of associated escape (P = 0.96). Taken together, these results indicate that the presence of HLA-associated polymorphisms at the population level is a marker of effective epitope targeting, especially among CTL that target HLA-B-restricted Gag epitopes.
Fig 4.
HLA alleles that select for escape are associated with reduced viral load. Log viral load was modeled as a linear function of the HLA-A and -B alleles from Fig. 3, and the resulting β estimates were correlated against the number of OLP responses that associated with that allele (A), the number of optimal epitopes that were targeted by that allele as determined by the presence of associated escape polymorphisms (B), and the total number of escape polymorphisms per targeted optimal epitope (C). Spearman rank correlation coefficients (ρ) are reported for each plotted data set.
Although escape at the population level may indicate that CTL restricted by an HLA allele can be quite effective, escape in an individual may indicate that the epitope can no longer be effectively targeted in that individual. We therefore tested each HLA-associated polymorphism for an association with viremic controller status (VL of <2,000 copies/ml and CD4 counts of >250), using the interaction model described in Materials and Methods. Although only four associations were significant at a q value of <0.2 (data not shown), the overall trends were striking. Consistent with observations of reduced escape in clade B-infected elite controllers (59), 201 of 300 (67%) tests indicated that viremic controllers were less likely to have selected for a given escape than were noncontrollers (P = 3.9 × 10−9); 13 of 15 (88%; P = 0.0002) associations with a q value of <0.5 indicated that viremic controllers were less likely to have selected for escape. This effect was largely driven by conserved regions: when a site is relatively conserved, viremic controllers were much less likely to escape than were noncontrollers, whereas the odds of escape were similar between the two groups in nonconserved regions (Spearman correlation between entropy and relative log-odds of escape between controllers and progressors was ρ = −0.31, P = 0.0002; data not shown). Of note, protective alleles were not more likely than other alleles to exhibit differential odds of escape between viremic controllers and progressors (P = 0.77, Fisher's exact test).
DISCUSSION
The present study represents the first large-scale, systematic analysis of differential immune escape in HIV-1. Starting with optimally defined, published epitopes (52), we identified all related HLA alleles driving immune escape mutations in Gag (p17+p24), Pol and Nef. This list included 38 epitopes restricted by more than one HLA allele, which underscores the promiscuous nature of many CTL epitopes (17, 29, 54, 66, 76). Remarkably, distinct mutational patterns and risk of escape were observed in 37 of 38 of those epitopes, indicating that differential escape within promiscuous epitopes is typical. These numbers are almost certainly underestimates resulting from restricting the study to known, optimally defined epitopes.
There are several reasons why the odds of selecting a given escape polymorphism may differ based on the specific HLA allele restricting the epitope. One possibility is that epitope targeting frequency differs based on the restricting HLA allele. If this were the case, then differential selection pressure would tend to be simply a matter of degree, with the more frequently targeted HLA restricted-epitopes exhibiting higher odds of escape. Although we do observe a small number of distinct escape patterns that can be explained in this straightforward way (e.g., B*57:03, B*57:02, and B*58:01 all select for T242N escape in Gag-TW10 but to differing degrees), the vast majority cannot. Furthermore, in the relatively uncommon cases where two alleles select for the same amino acid polymorphism, no correlation between odds of escape and odds of OLP targeting in chronic infection was observed (although the elimination of CTL responses following escape in vivo must be acknowledged as a potential limitation of this analysis). Instead, between 66% and 97% of observed cases of differential escape reflect instances where two alleles select for different polymorphisms at the same site or at different sites within the epitope. Taken together, our observations indicate that differential immune selection by closely related alleles is a widespread phenomenon, and one that typically manifests itself via distinct escape pathways selected by the restricting HLA alleles, rather than common escape patterns that differ in their relative risk of occurrence. This observation is in line with previous studies, which have reported variations in functional avidity, TCR usage, and selection pressure, even in the absence of differential targeting frequency, for several B7- and B58-restricted epitopes (50, 53, 81).
Differential selection and epitope targeting between related HLA alleles suggests that such alleles will have discordant associations with viral load: indeed, this turns out to be true in approximately 35% of cases in which HLA alleles exhibit distinct escape patterns within the same restricted epitope. As such, our results complement previously described discordant associations with VL among alleles of the B58 supertype (2, 42, 44, 49), at least some of which appear to be due to the specific epitopes restricted by each allele (25). Differential escape mutations within A2-restricted (40), B58-restricted (51, 53, 57, 81), and B7-restricted (50, 81) epitopes have also been previously reported, while case studies of individual epitopes have linked differential escape pathways with discordant clinical outcomes (40) and recruitment of distinct TCR repertoires exhibiting differential functional avidities (50). The present study extends these observations by revealing that discordant associations with viral load are common among closely related HLA alleles restricting different epitopes and/or selecting for different escape mutations.
Historically, the relationship between immune escape and disease progression has been difficult to elucidate. The complexities of these relationships are illustrated by case studies describing loss of viral control following escape within the immunodominant B*27-restricted Gag-KK10 epitope (26, 34, 43), followed by a dramatic broadening of the CTL response (26) (though breadth of targeting appears to wane as many individuals progress to AIDS [37]). Thus, in these instances, KK10 escape appears to be a direct cause of viral breakthrough, whereas any escape in epitopes targeted by the subsequent broadened response would occur only after the VL increase. The complexities are compounded by the observation that escape is typically a marker of an (at least previously) effective in vivo CTL response (40). Indeed, expression of HLA class I alleles associated with a large number of population-level Gag escape associations (16, 30, 65), a large number of reverting associations (56), and/or a large number of associations in conserved regions (79), is predictive of relative viral control. Although escape inherently implies a net improvement of in vivo viral fitness, a number of escape polymorphisms have been linked to decreased in vitro (12, 23, 53, 62, 68, 77) and in vivo (30, 56) fitness in the absence of CTL pressure, suggesting an incomplete recovery of viral replicative capacity upon escape. Epidemiologically, the presence of costly escape positions could thus be a marker for immune control, as they identify cases of partial immune-mediated attenuation of HIV-1 (58, 65). Over all associations in the present study, escape was strongly linked to higher VL, an effect that was primarily driven by escape in conserved regions. However, HLA alleles that were associated with many escape polymorphisms, especially in Gag, were themselves associated with low viral load, a correlation that was much stronger than that observed with OLP-measured targeting of Gag. Taken together, these data suggest that, although the presence of population-level escape associations is a marker of the capacity of CTL restricted by that allele to effectively target the virus, loss of viral control is closely linked to actual immune escape in individuals, as was suggested in a chronically infected clade B cohort (16) and in elite controllers (59). Thus, the study of immune escape in general, and differential escape in particular, may shed light on which epitopes are most effective to target in vivo. From a vaccine design perspective, it is equally important to determine if it is possible to block escape from occurring, either through a polyvalent vaccine that primes the immune system to recognize escape variants (28) or by constraining escape pathways by blocking compensatory mutations through the targeting of other epitopes (Y. E. Wang et al., submitted for publication). The prospects of the latter approach may appear dim given that we found no instances in which the odds of escape were reduced in the context of the coexpression of another HLA allele; however, the present study was underpowered to identify such associations due to the large number (>13,000) of required statistical tests and the low frequency of any given pair of HLA alleles. Some of these associations may represent true interactions, and the analytical tool developed here may prove useful for future studies that consider a more restricted set of hypotheses.
One key assumption of the present study is that similar HLA alleles that restrict an epitope in a given region are likely to restrict the same optimal epitope. Violations of this assumption could lead to spurious identification of differential escape. Although this assumption remains largely untested, there are several lines of evidence supporting its validity in the majority of cases. First, HLA supertype definitions derive from shared binding profiles and epitope repertoires (17, 29, 71, 72, 76). The observation that supertypes tend to restrict the same epitopes has been demonstrated in a number of studies (3, 10, 31, 46, 50, 66, 76) and detailed studies of B7 (50) and B58 (46) supertypes consistently yielded identical optimal epitope definitions when multiple alleles were associated with the same OLP. Furthermore, many of the optimal epitopes used in the present study were previously tested in a cohort of 103 HIV-infected individuals (29). In addition to observing widespread promiscuity, titration experiments using truncated and extended peptides demonstrated that the same optimal epitope was presented in the majority of cases, though several exceptions were noted. Moreover, the same epitope was frequently optimal for alleles even of different loci, an effect that may be due to HLA-independent mechanisms such as proteasomal processing, epitope transport or trimming (78, 61, 75), suggesting that our present approach of limiting epitope expansion to supertype members is conservative. Taken together, the identification of an HLA-associated polymorphism within an optimal epitope known to be restricted by a similar HLA allele suggests that the associated HLA restricts the same optimal epitope. Nevertheless, a few known counter examples exist in the published optimal list, indicating that some instances of differential escape may be due to related alleles restricting overlapping epitopes. Future work is therefore required to validate proposed novel restrictions and to disentangle the causal mechanisms of apparent differential escape.
These studies were facilitated by a novel statistical model that enables quantifying and comparing the odds of immune escape while correcting for statistical confounding that may arise due to phylogenetic relatedness of HIV sequences. This model was first developed to compare the odds of escape between individuals who have progressed to AIDS and those who have not (37) and was here refined and extended to model differential escape. The resulting model is quite versatile, enabling direct tests for differential selection between two HLA alleles or differential selection mediated by one allele in various genetic or environmental contexts. The present studies demonstrate the widespread extent of differential escape in a relatively homogeneous population. Natural extensions will include studies of how escape varies among ethnic populations or viral clades, and studies of differential escape in the context of genetic variation outside the MHC-I locus or in the context of environmental factors, including antiretroviral treatment, which may alter immune function or the virus' ability to tolerate variation. A web server implementation of the differential escape methods described herein is available at http://research.microsoft.com/en-us/um/redmond/projects/MSCompBio/phyloDOddsRatio/.
Widespread differential immune selection pressure mediated by the specific HLA allele restricting the epitope raises additional challenges for an epitope-based CTL vaccine. Differential escape has been linked to differential CTL functional avidity (50) and in vivo efficacy (40), and the present study indicates that differential escape may be broadly related to differential viral control. These observations raise the possibility that an epitope-based vaccine will have varying results in different individuals, potentially reducing the efficacy of the vaccine or even representing a hazard to certain individuals by focusing their immune system on an ineffective response (50). In cases where differential escape has no direct in vivo consequence, understanding the specifics may help in the design of a polyvalent vaccine, as the escape routes of all common and rare alleles could be included in the vaccine (28). Although the present study confirms and extends our understanding of the nature and impact of differential immune selection by closely related HLA alleles, a number of limitations merit mention. The present study focused only on known optimal epitopes in Gag, Pol, and Nef and was restricted to a cohort of clade C-infected individuals. Furthermore, working with high-resolution HLA data reduces statistical power for most rare alleles, a problem that is quadratically compounded when coexpression of high resolution types is considered. Finally, although the large number of associations identified in this and other studies suggests that many escape polymorphisms are repeatedly selected in individuals expressing the same allele, the present study also identified a number of novel, rare escapes and suggested the presence of even rarer undetected escapes. It is unknown to what extent such rare escapes occur in vivo, to what extent they contribute to immune evasion, or whether their selection is attributable to specific environmental or genetic contexts. Large data sets that include thousands of ethnically diverse individuals, coupled with expanded high-fidelity epitope data, will be necessary to fully appreciate the extent and specifics of differential immune escape and the implication of alternative escape pathways on vaccine design.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chanson Brumme and Henrik Kloverpris for helpful discussions.
J.M.C., J.L., N.P., V.T., C.K., and D.H. are employed by Microsoft Corp. Z.L.B. is supported by a New Investigator Award from the Canadian Institutes of Health Research (CIHR). The Durban cohort was supported by the NIH (contract NO1-A1-15422, 2RO1AI46995-06, R01AI067073), the South African AIDS Vaccine Initiative, and the Mark and Lisa Schwartz Foundation. T.N. holds the South African DST/NRF Chair in Systems Biology of HIV/AIDS.
Footnotes
Published ahead of print 29 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Allen TM, et al. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 79:13239–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altfeld M, et al. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581–2591 [DOI] [PubMed] [Google Scholar]
- 3. Altfeld M, et al. 2000. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J. Virol. 74:8541–8549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avila-Rios S, et al. 2009. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology 6:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bansal A, et al. 2010. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J. Exp. Med. 207:51–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barber LD, et al. 1995. Overlap in the repertoires of peptides bound in vivo by a group of related class I HLA-B allotypes. Curr. Biol. 5:179–190 [DOI] [PubMed] [Google Scholar]
- 7. Barber LD, et al. 1997. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J. Immunol. 158:1660–1669 [PubMed] [Google Scholar]
- 8. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300 [Google Scholar]
- 9. Berger CT, et al. 2010. Viral adaptation to immune selection pressure by HLA class I-restricted CTL responses targeting epitopes in HIV frameshift sequences. J. Exp. Med. 207:61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bertoni R, et al. 1997. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Invest. 100:503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhattacharya T, et al. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583–1586 [DOI] [PubMed] [Google Scholar]
- 12. Brockman MA, et al. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brumme ZL, et al. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 82:9216–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brumme ZL, et al. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brumme ZL, et al. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One 4:e6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brumme ZL, et al. 2008. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS 22:1277–1286 [DOI] [PubMed] [Google Scholar]
- 17. Burrows SR, et al. 2003. Promiscuous CTL recognition of viral epitopes on multiple human leukocyte antigens: biological validation of the proposed HLA A24 supertype. J. Immunol. 171:1407–1412 [DOI] [PubMed] [Google Scholar]
- 18. Carlson JM, Brumme ZL. 2008. HIV evolution in response to HLA-restricted CTL selection pressures: a population-based perspective. Microbes Infect. 10:455–461 [DOI] [PubMed] [Google Scholar]
- 19. Carlson JM, et al. 2008. Phylogenetic dependency networks: inferring patterns of CTL escape and codon covariation in HIV-1 Gag. PLoS Comput. Biol. 4:e1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carlson JM, Heckerman D, Shani G. 2009. Estimating false discovery rates for contingency tables. Microsoft research technical report MSR-TR-2009-53. http://research.microsoft.com/en-us/um/redmond/projects/MSCompBio/FalseDiscoveryRate/ [Google Scholar]
- 21. Carlson JM, Kadie C, Mallal S, Heckerman D. 2007. Leveraging hierarchical population structure in discrete association studies. PLoS One 2:e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carrington M, et al. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748–1752 [DOI] [PubMed] [Google Scholar]
- 23. Christie NM, et al. 2009. Viral fitness implications of variation within an immunodominant CD8+ T-cell epitope of HIV-1. Virology 388:137–146 [DOI] [PubMed] [Google Scholar]
- 24. Deeks SG, Walker BD. 2007. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27:406–416 [DOI] [PubMed] [Google Scholar]
- 25. Dinges WL, et al. 2010. Virus-specific CD8+ T-cell responses better define HIV disease progression than HLA genotype. J. Virol. 84:4461–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feeney ME, et al. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fellay J, et al. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fischer W, et al. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13:100–106 [DOI] [PubMed] [Google Scholar]
- 29. Frahm N, et al. 2007. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur. J. Immunol. 37:2419–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goepfert PA, et al. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goulder PJ, et al. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses 12:1691–1698 [DOI] [PubMed] [Google Scholar]
- 32. Goulder PJR, Watkins DI. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630–640 [DOI] [PubMed] [Google Scholar]
- 33. Goulder PJR, Watkins DI. 2008. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8:619–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goulder PJR, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217 [DOI] [PubMed] [Google Scholar]
- 35. Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 36. Holdsworth R, et al. 2009. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens 73:95–170 [DOI] [PubMed] [Google Scholar]
- 37. Huang K-HG, et al. 2011. Progression to AIDS in South Africa is associated with both reverting and compensatory viral mutations. PLoS One 6:e19018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang K-HG, et al. 2009. Prevalence of HIV type-1 drug-associated mutations in pre-therapy patients in the Free State, South Africa. Antiviral Therapy 14:975–984 [DOI] [PubMed] [Google Scholar]
- 39. International HIV Controllers Study 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iversen AKN, et al. 2006. Conflicting selective forces affect T cell receptor contacts in an immunodominant human immunodeficiency virus epitope. Nat. Immunol. 7:179–189 [DOI] [PubMed] [Google Scholar]
- 41. John M, et al. 2010. Adaptive interactions between HLA and HIV-1: highly divergent selection imposed by HLA class I molecules with common supertype motifs. J. Immunol. 184:4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaslow RA, et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411 [DOI] [PubMed] [Google Scholar]
- 43. Kelleher AD, et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kiepiela P, et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769–775 [DOI] [PubMed] [Google Scholar]
- 45. Kiepiela P, et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 46. Kloverpris HN, et al. 2012. HLA-B*57 micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. J. Virol. 86:919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Larder BA, et al. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671–673 [DOI] [PubMed] [Google Scholar]
- 48. Leitner T, et al. 1993. Analysis of heterogeneous viral populations by direct DNA sequencing. Biotechniques 15:120–127 [PubMed] [Google Scholar]
- 49. Leslie A, et al. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J. Virol. 84:9879–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leslie A, et al. 2006. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 177:4699–4708 [DOI] [PubMed] [Google Scholar]
- 51. Leslie AJ, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 52. Llano A, Frahm N, Brander C. 2009. How to optimally define optimal cytotoxic T lymphocyte epitopes in HIV infection, p 3–24 In Yusim K, Korber BT, Brander C, Haynes BF, Koup RA, Moore JP, Walker BD, Watkins DI. (ed), HIV molecular immunology. Los Alamos National Laboratory, Theoretical Biology and Biophysics, Los Alamos, NM [Google Scholar]
- 53. Martinez-Picado J, et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Masemola AM, et al. 2004. Novel and promiscuous CTL epitopes in conserved regions of Gag targeted by individuals with early subtype C HIV type 1 infection from southern Africa. J. Immunol. 173:4607–4617 [DOI] [PubMed] [Google Scholar]
- 55. Matthews PC, et al. 2011. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J. Immunol. 186:5675–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matthews PC, et al. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Migueles SA, et al. 2003. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J. Virol. 77:6889–6898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miura T, et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte recognition. J. Virol. 83:2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Miura T, et al. 2009. HLA-associated viral mutations are common in human immunodeficiency virus type 1 elite controllers. J. Virol. 83:3407–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moore CB, et al. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439–1443 [DOI] [PubMed] [Google Scholar]
- 61. Neefjes J, Jongsma MLM, Paul P, Bakke O. 2011. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11:823–836 [DOI] [PubMed] [Google Scholar]
- 62. Peyerl FW, et al. 2004. Fitness costs limit viral escape from cytotoxic T lymphocytes at a structurally constrained epitope. J. Virol. 78:13901–13910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Phillips RE, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453–459 [DOI] [PubMed] [Google Scholar]
- 64. Pounds S, Cheng C. 2006. Robust estimation of the false discovery rate. Bioinformatics 22:1979–1987 [DOI] [PubMed] [Google Scholar]
- 65. Rousseau CM, et al. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J. Virol. 82:6434–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sabbaj S, et al. 2003. Cross-reactive CD8+ T cell epitopes identified in US adolescent minorities. J. Acquir. Immune Defic. Syndr. 33:426–438 [DOI] [PubMed] [Google Scholar]
- 67. Schneidewind A, et al. 2008. Structural and functional constraints limit options for cytotoxic T-lymphocyte escape in the immunodominant HLA-B27-restricted epitope in human immunodeficiency virus type 1 capsid. J. Virol. 82:5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Schneidewind A, et al. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shapiro RL, et al. 2010. Antiretroviral regimens in pregnancy and breast-feeding in Botswana. N. Engl. J. Med. 362:2282–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sidney J, et al. 1995. Several HLA alleles share overlapping peptide specificities. J. Immunol. 154:247–259 [PubMed] [Google Scholar]
- 71. Sidney J, Peters B, Frahm N, Brander C, Sette A. 2008. HLA class I supertypes: a revised and updated classification. BMC Immunol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sidney J, Southwood S, Sette A. 2005. Classification of A1- and A24-supertype molecules by analysis of their MHC-peptide binding repertoires. Immunogenetics 57:393–408 [DOI] [PubMed] [Google Scholar]
- 73. Storey JD. 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 64:479–498 [Google Scholar]
- 74. Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 100:9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tenzer S, et al. 2009. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 10:636–646 [DOI] [PubMed] [Google Scholar]
- 76. Threlkeld S, et al. 1997. Degenerate and promiscuous recognition by CTL of peptides presented by the MHC class I A3-like superfamily: implications for vaccine development. J. Immunol. 159:1648–1657 [PubMed] [Google Scholar]
- 77. Troyer RM, et al. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Van Endert P. 2011. Post-proteasomal and proteasome-independent generation of MHC class I ligands. Cell. Mol. Life Sci. 68:1553–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang YE, et al. 2009. Protective HLA class I alleles that restrict acute-phase CD8+ T-cell responses are associated with viral escape mutations located in highly conserved regions of human immunodeficiency virus type 1. J. Virol. 83:1845–1855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yang OO, et al. 2005. Genetic and stochastic influences on the interaction of human immunodeficiency virus type 1 and cytotoxic T lymphocytes in identical twins. J. Virol. 79:15368–15375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yu XG, et al. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J. Virol. 81:1619–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.