Abstract
Epstein-Barr virus (EBV) establishes a persistent latent infection in B lymphocytes and is associated with the development of numerous human tumors. Epstein-Barr nuclear antigen 3C (EBNA 3C) is essential for B-cell immortalization, has potent cell cycle deregulation capabilities, and functions as a regulator of both viral- and cellular-gene expression. We performed transcription profiling on EBNA 3C-expressing B cells and identified several chemokines and members of integrin receptor-signaling pathways, including CCL3, CCL4, CXCL10, CXCL11, ITGA4, ITGB1, ADAM28, and ADAMDEC1, as cellular target genes that could be repressed by the action of EBNA 3C alone. Chemotaxis assays demonstrated that downregulation of CXCL10 and -11 by EBNA 3C is sufficient to reduce the migration of cells expressing the CXCL10 and -11 receptor CXCR3. Gene repression by EBNA 3C was accompanied by decreased histone H3 lysine 9/14 acetylation and increased histone H3 lysine 27 trimethylation. In an EBV-positive cell line expressing all latent genes, we identified binding sites for EBNA 3C at ITGB1 and ITGA4 and in a distal regulatory region between ADAMDEC1 and ADAM28, providing the first demonstration of EBNA 3C association with cellular-gene control regions. Our data implicate indirect mechanisms in CXCL10 and CXCL11 repression by EBNA 3C. In summary, we have unveiled key cellular pathways repressed by EBNA 3C that are likely to contribute to the ability of EBV-immortalized cells to modulate immune responses, adhesion, and B-lymphocyte migration to facilitate persistence in the host.
INTRODUCTION
Epstein-Barr virus (EBV) is a potent transforming agent of resting B lymphocytes, promoting cell cycle entry and subsequent continuous proliferation. EBV is associated with the pathogenesis of numerous lymphoid tumors, including Burkitt's lymphoma (BL), Hodgkin's disease, posttransplant lymphomas, and certain T-cell and natural killer cell lymphomas, in addition to the epithelial cell tumor nasopharyngeal carcinoma (reviewed in reference 54). Like other members of the herpesvirus family, EBV has a biphasic life cycle involving a latent and a lytic phase. In infected B cells, EBV establishes a latent infection where the 172-kb double-stranded DNA viral genome is maintained as a closed circular episome and expresses a limited set of latent genes. These include the Epstein-Barr nuclear antigens (EBNAs) 1, 2, 3A, 3B, 3C, and -LP and latent membrane proteins (LMPs) 1, 2A, and 2B, the untranslated Epstein-Barr-encoded RNAs EBER 1 and EBER 2, and numerous microRNAs. Many of the EBV latent proteins are highly immunogenic, and effective immune control, combined with restricted expression of only subsets of latent proteins during viral persistence, enables over 90% of the world's population to carry EBV as a lifelong asymptomatic infection.
EBNA 3C is one of only six latent gene products crucial for B-cell transformation and is required for the continuous proliferation of EBV-immortalized lymphoblastoid cell lines (LCL) (28, 47). The first evidence for the role of EBNA 3C as a regulator of gene expression came from studies that detected upregulation of the B-cell activation antigen CD21 (CR2) on the surfaces of EBV-negative BL cells stably transfected with EBNA 3C-expressing plasmids (50). Further studies reported upregulation of LMP1 and the cellular proteins vimentin and CD23 on expression of EBNA 3C in the Raji BL cell line, which carries an EBNA 3C deletion virus (1). Subsequent reports mapped regions of EBNA 3C that possess transcriptional activation or repression activity when targeted to DNA as fusions with the DNA binding domain of the yeast transactivator Gal4 (4, 27). EBNA 3C does not appear to bind DNA directly and may be targeted to promoters through the cellular DNA binding proteins PU.1 and RBP-Jκ (40, 58). The association of EBNA 3C with RBP-Jκ, also the DNA-targeting partner of the EBNA 2 transcriptional activator, was shown to antagonize the activation of genes by EBNA 2 in reporter assays and to inhibit the association of RBP-Jκ with DNA in vitro (21, 42, 49). However, more recent work using a conditionally active form of EBNA 3C demonstrated that in the context of latently infected LCLs, loss of EBNA 3C function did not lead to increased expression of EBNA 2-regulated viral and cellular genes (28). The antagonistic effects of EBNA 3C on EBNA 2 targeting to gene promoters may therefore be less evident in EBV-infected cells.
Consistent with a role in the regulation of transcription, EBNA 3C has been reported to interact with both transcriptional coactivators and corepressors, e.g., p300, HDAC1, HDAC2, NcoR, mSin3A, and CtBP-1 (10, 18, 41, 48). Recent studies have provided important insights into the mechanism of transcriptional repression by EBNA 3C and have highlighted the role of cooperation between EBNA 3 family members in the control of cellular-gene expression. EBNA 3C and EBNA 3A are required for transcriptional repression of the gene encoding the proapoptotic protein Bim, thus providing a survival advantage to EBV-infected BL cell lines (3, 20). At the Bim locus, EBNA 3C and EBNA 3A establish a repressed chromatin state characterized by high levels of lysine 27 trimethylation on histone H3 (H3K27me3) that leads to subsequent DNA methylation at a CpG island (39).
EBNA 3C has also emerged as a key deregulator of the G1, G2, and mitotic cell cycle checkpoints, and expression of EBNA 3C alone in numerous cell types can lead to inappropriate cell cycle progression (19, 37, 38). The most compelling evidence for regulation of a cell cycle control gene by EBNA 3C comes from the observation that loss of EBNA 3C function in LCLs expressing conditionally active EBNA 3C leads to upregulation of the cyclin-dependent kinase inhibitor p16INK4a and growth arrest (28). Subsequent studies showed that EBNA 3C and EBNA 3A epigenetically silence p16INK4a transcription by promoting H3K27me3 in a manner dependent on binding to the corepressor CtBP-1 (44).
To fully elucidate the cellular-gene networks controlled by EBNA 3C in B cells and to dissect the specific role played by EBNA 3C in their regulation, we performed gene expression profiling using B cells expressing EBNA 3C and identified significant downregulation of integrin receptor signaling and chemokine genes. Follow-up studies based on data obtained from chromatin immunoprecipitation (ChiP) coupled with next-generation sequencing (ChIP-Seq) in latently infected cells expressing all latent proteins identified EBNA 3C binding sites at promoter and enhancer regions of many of these highly repressed gene loci. EBNA 3C-mediated repression correlated with reduced histone H3 9/14 lysine acetylation (H3K9/14ac) and increased H3K27me3 in EBNA 3C-expressing cells. Our results therefore provide the first demonstration of EBNA 3C binding to both promoter and long-range cellular-gene-enhancer elements in vivo and identify key target genes whose regulation is likely to play an important role in EBV-mediated pathogenesis.
MATERIALS AND METHODS
Cell lines.
Stable transfectants of the EBV-negative B-cell lymphoma cell line BJAB (pz1, pz2, pz3, E1-2, E1-5, U2-12, U2-15, LP-4, LP-12, E3A-1, E3B-2, E3C-3, E3C-4, E3C-7) (a gift from Alan Rickinson) were described previously (50). The Mutu III cell line is derived from an EBV-positive Burkitt's lymphoma (12). All cell lines were routinely passaged twice weekly and cultured using previously described conditions (51). The human CXCR3-transfected murine 300-19 cell line (a gift from Bernhard Moser) was cultured in normal RPMI growth medium supplemented with 5 ×10−5 M β-mercaptoethanol and 1 mg/ml of G418 (22).
RNA extraction and microarray analysis.
For microarray and real-time PCR analyses of cell line panels, cells were diluted to 2 × 105/ml and harvested after 3 days, and total RNA was extracted using Tri Reagent (Sigma). RNA samples were then purified using an RNeasy kit (Qiagen). Microarray analysis was carried out by UCL Genomics using two independent preparations of total RNA from BJAB pz2 and BJAB E3C-4 cells (biological duplicate) and U133A Affymetrix GeneChip arrays. Normalized data were analyzed using four different pairwise comparisons of the signals obtained from E3C-4 versus those from pz2.
Chemotaxis assays.
BJAB stable cell lines were resuspended in fresh media at a concentration of 5 × 105/ml, and cell culture supernatants were harvested after 3 days. Six hundred microliters of cell culture supernatant or 30 nM recombinant human CXCL10 (R and D systems) in growth media were added to the lower chamber of 24-well Transwell permeable supports with 5-μm pores (Corning). CXCR3-expressing cells were resuspended at a concentration of 5 × 105/ml, and 100 μl of cell suspension was added to the upper Transwell chamber. Migration was allowed to proceed for 2 h, and the number of cells in the lower chamber was determined by cell counting.
Flow cytometry.
Cells were resuspended at a concentration of 2 × 107/ml in phosphate-buffered saline (PBS) containing 0.1% (wt/vol) sodium azide (PBS-azide). Cells (106) were incubated either with 20 μl of IgG control antibody or 20 μl of a 1/3 dilution of anti-β1 integrin (anti-CD29; Cymbus Biotechnology) or anti-α4 integrin (anti-CD49d; Cymbus Biotechnology) antibody. Following a 30-min incubation, cells were washed three times and then resuspended in 50 μl of a 1/20 dilution of fluorescein isothiocyanate (FITC)–rabbit anti-mouse antibodies (F 0313; DakoCytomation). After a further 30-min incubation on ice, cells were washed three times and resuspended in 50 μl of a 1/20 dilution of FITC–goat anti-rabbit antibodies (F1262; Sigma). After a further 30-min incubation and final washing, cells were resuspended in 50 μl of PBS containing 1% fetal bovine serum (FBS) and 50 μl of 1% paraformaldehyde in PBS added as a fixative. Cells were analyzed using a FACSCanto flow cytometer (Becton Dickinson).
Immunoblotting.
SDS-PAGE and immunoblotting were carried out as described previously (5, 51). The following antibodies were used for immunoblotting: anti-EBNA 3C (mouse monoclonal; E3CA10 [29]), anti-EBNA 3A (mouse monoclonal; T2.78; a gift from Martin Rowe), anti-EBNA 3B (sheep polyclonal; F120P; Exalpha Biologicals), anti-EBNA 1 (human serum; a gift from Martin Rowe), anti-EBNA 2 (mouse monoclonal; PE2), anti-LP (mouse monoclonal; JF186), and anti-actin (A-2066; Sigma).
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP)-quantitative PCR (ChIP-QPCR) assays for EBNA 2 and acetylated K9 and K14 on histone H3 were carried out as described previously, using chromatin from BJAB stable cell lines (5, 36). ChIP for EBNA 3C and H3K27me3 was carried out as for acetylated histone H3, using 10 μl of sheep polyclonal anti-EBNA 3C antibodies (ab16128; Abcam) or 4 μg of anti-H3K27me3 antibodies (17-622; Millipore), respectively.
For ChIP sequencing, ChIP was carried out using a 6-fold scale-up of previously described ChIP methods, replacing salmon sperm DNA with a final concentration of 0.5% (wt/vol) bovine serum albumin (BSA) to preblock protein A Sepharose beads (Sigma) (5, 36). Briefly, 600 μl of chromatin from 30 × 106 cross-linked Mutu III cells was diluted 10-fold in IP dilution buffer (5) and precleared with protein A Sepharose beads. An input control sample was removed, and EBNA 3C was then precipitated by overnight rotation at 4°C with 60 μl of sheep polyclonal anti-EBNA 3C antibodies (ab16128; Abcam). BSA-coated protein A Sepharose beads (135 μl) were then added, and immune complexes were collected by rotation at 4°C for 3 h. Beads were washed as previously described (5). Cross-links were reversed, DNA was eluted, and an additional treatment with 0.2 μg/ml of RNase A was carried out for 1 h at 37°C to remove RNA. Following proteinase K digestion, DNA was purified using QIAquick gel extraction (Qiagen). Input samples were processed alongside ChIP samples.
Library preparation and sequencing.
ChIP and input DNA (10 ng) were used to generate sequencing libraries with a ChIP-Seq sample preparation kit (Illumina). DNA fragments were end repaired and phosphorylated, 3′-dA overhangs were added, and adapters were ligated according to the manufacturer's instructions. PCR-amplified samples (16 cycles) were separated on a 2% agarose gel in Tris-acetate–EDTA (TAE) buffer and visualized using SYBR-safe stain and a Dark Reader transilluminator instrument (Clare Chemical Research). The region of the gel containing 150- to 350-bp DNA fragments was excised, and DNA was purified using a gel extraction kit (Qiagen). The library was quantified using an Agilent bioanalzer instrument and subjected to 35-bp single-end read sequencing with an Illumina IIx genome analyzer.
ChIP-Seq data analysis.
Initial processing of sequencing images generated by the Illumina IIx genome analyzer was performed using the CASAVA pipeline. Polony identification, base calling, and quality control statistics were performed using GOAT and Bustard modules. Thirty-six-bp short reads were aligned to the hg19 build of the human genome using ELAND software. Only reads with zero to two mismatches were aligned, and only uniquely aligned reads passing the quality threshold were retained. When multiple reads matched to the same position, only a single read was considered for further analysis. Sequence reads were extended to 150 bp. WIG files were generated by calculating tag density in 10-bp windows and were normalized to reads per million total reads using in-house R scripts. The data for the EBNA 3 sequencing run were then background corrected using data from input DNA. Significant peaks of EBNA 3 binding were identified with MACS (P < 10−7) (55). Encyclopedia of DNA elements (ENCODE) ChIP-Seq data for PU.1 binding sites in EBV-immortalized LCLs were accessed through the human genome browser at http://genome.ucsc.edu/cgi-bin/hgGateway.
Real-time PCR.
For transcript analysis, cDNA was synthesized from total RNA using the ImProm II reverse transcription system and random oligonucleotides (Promega). QPCR was performed using the standard curve absolute quantification method on an Applied Biosystems 7500 real-time PCR instrument as described previously using either SensiMix SYBR green (Bioline) or GoTaq qPCR master mix (Promega) (5). Reaction mixtures contained 150 nM each primer (Table 1). Serial dilutions of cDNA or input DNA (for ChIP) were used to generate standard curves. For ChIP analysis, percentages of input signals, after subtraction of signals for controls with no antibody, were expressed relative to the highest signal obtained in all or the majority of experiments carried out with a series of primer sets.
Table 1.
Primer sets used for real-time PCR analysis of ChIP DNA or cDNA samples
| Primer set | Location/genome coordinatesa (bp) | Sequence (5′ to 3′) |
|---|---|---|
| PPIA | ||
| Forward, MW200 | −5 to +13 | GGGCCGAACGTGGTATAA |
| Reverse, MW201 | +65 to +87 | CCATGGCTAATAGTACACGGTTT |
| CXCL10 | ||
| Forward, MW476 | −893 to −873 | ACAGTGTCTTGGAGCTGAACC |
| Reverse, MW477 | −826 to −806 | GCACGCATAGAGACAGACCTT |
| Forward, MW474 | −177 to −157 | TCAAAGCAGGCCAGTCCTAT |
| Reverse, MW475 | −111 to −93 | AACAGCAAGTTGTGCCTGATT |
| Forward, MW488 | +130 to +156 | AAGGAACATCAAAGGATACTTAATTTG |
| Reverse, MW489 | +223 to +247 | TCCCTCTTATTTATAAGCATGCAGT |
| Forward, MW490 | +541 to +561 | TGATGTGATCCATTCCTCCTC |
| Reverse, MW491 | +635 to +659 | AGAGAAGGTTAGCACAGTGTTCATT |
| Forward, MW472 | +1426 to +1445 | ATGAATGCATAGCAGCAGGA |
| Reverse, MW473 | +1492 to +1512 | CTGACTCTGGCTTCAGATTGG |
| CXCL11 | ||
| Forward, MW482 | −822 to −803 | CGTGGCATGGAGTTACTGAA |
| Reverse, MW483 | −738 to −719 | AGCTCCTCCTGCCTTCTCTT |
| Forward, MW480 | −295 to −273 | CAGTCTTCCTGAATGAATGACAA |
| Reverse, MW481 | −209 to −190 | CCAGATGGTAACCAGCCTTC |
| Forward, MW492 | +46 to +65 | AAGCTGAAGTAGCAGCAGCA |
| Reverse, MW493 | +120 to +140 | GCACACAATATCACAGCCAAG |
| Forward, MW494 | +594 to +620 | CCAGTAACAGTTGTGTTAAGTGCTATT |
| Reverse, MW495 | +658 to +677 | TAGCTGGTGGTGAACGTGAG |
| Forward, MW478 | +1660 to +1685 | TTGATGCTTACAACTATTCTGTTGTG |
| Reverse, MW479 | +1754 to +1773 | CAATGTCTCCACCGTAACCA |
| ITGA4 | ||
| Forward, MW548 | −445 to −426 | GCTGTCTCTCTGGTTGCTGA |
| Reverse, MW549 | −366 to −347 | AACGCAACACACCTGAACTG |
| Forward, MW550 | −77 to −60 | GGGCTGCAGAGGAAGTGT |
| Reverse, MW551 | −8 to +15 | TGACAAAGACGTTATGGCTATTC |
| Forward, MW552 | +658 to +675 | CTGCGCCTCATCTCTTGG |
| Reverse, MW553 | +721 to +741 | AACACTAAACGGCCACTACCC |
| Forward, MW554 | +1145 to +1163 | GTGCGTTATGGTGCAAGGT |
| Reverse, MW555 | +1225 to +1242 | CGCCACAGATGTGCAGTC |
| ITGB1 | ||
| Forward, MW627 | −13574 to −13556 | TGCACCTTGGCAGATTGA |
| Reverse, MW628 | −13474 to −13456 | GGTTGAGAGCTGGTTGCTG |
| Forward, MW558 | −885 to −866 | CAGGATAGCAGCTTGCCAGT |
| Reverse, MW559 | −814 to −795 | GCATGTCGATTCGATGATTG |
| Forward, MW560 | −220 to −201 | CTTCGCAGAGGAGGAAACTG |
| Reverse, MW561 | −126 to −109 | GACCGGGACAAAGGAACC |
| Forward, MW562 | +1019 to +1037 | CGCGTGTCATTTCTTAGGC |
| Reverse, MW563 | +1116 to +1137 | TGATCATACCAATGAGTGCAAA |
| ADAM28 | ||
| Forward, MW506 | −587 to −567 | TCCTCATTGCCAGTTATGACA |
| Reverse, MW507 | −495 to −471 | CTCACACAAGTGTACTCTCAGGAAG |
| Forward, MW508 | −87 to −69 | AGGACCACAGCTTCGAGGT |
| Reverse, MW509 | −6 to +15 | CCTCCTCTCCAGTGAGACAGA |
| Forward, MW615 | +6412 to +6431 | GTGATCTTGGCTCACTGCAA |
| Reverse, MW616 | +6501 to +6518 | AATTAGCCGAGCGTGGTG |
| Forward, MW613 | +13274 to +13294 | CAACCAGAGAACTGAGGCACT |
| Reverse, MW614 | +13345 to +13364 | AGCAATGTGGCATGTGTGAT |
| Forward, MW510 | +19388 to +19407 | ACAGGAGCATGCACTCTTCA |
| Reverse, MW511 | +19476 to +19493 | GGCAATGTTCTGCTGCAA |
| ADAMDEC1 | ||
| Forward, MW514 | −1281 to −1260 | AGAGTTCACCATTCCAGGATTC |
| Reverse, MW515 | −1185 to −1163 | TCCAGAGATAGGCATCCTTCTTA |
| Forward, MW516 | −79 to −60 | GTAGATATGCACGCGACCAC |
| Reverse, MW517 | −5 to +16 | TTGAGGACTCACATCTGGACA |
| Forward, MW518 | +228 to +246 | GTGGGATCTCCCAGCTACC |
| Reverse, MW519 | +312 to +331 | AATGCTGGTGTGATGGTACG |
| ADAM (peak) | ||
| Forward, MW544 | 24224516 to 24224535 | ACCATAAAGCCATGGGTCAG |
| Reverse, MW545 | 24224580 to 24224604 | GGAGTACAGTTTGGAGCTCATATTT |
| Forward, MW540 | 24224967 to 24224987 | CTTCATGGCTACAGACTCTTGG |
| Reverse, MW541 | 24225040 to 24225059 | CCTATGTCTCGCTTCCTGCT |
| Forward, MW542 | 24225437 to 24225461 | TCCGGTGACTAAAGTATATGTAGGG |
| Reverse, MW543 | 24225505 to 24225526 | CTCCATGCTGGATTGATTACAA |
| cDNA | ||
| CCL3 | ||
| Forward, MW165 | CCAGTTCTCTGCATCACTTGCT | |
| Reverse, MW166 | GAATCTGCCGGGAGGTGTAG | |
| CCL4 | ||
| Forward, MW167 | CTGTCCTGTCTCTCCTCATGCTAGT | |
| Reverse, MW168 | GAGGGTCTGAGCCCATTGG | |
| CXCL10 | ||
| Forward, MW169 | GTCCACGTGTTGAGATCATTGC | |
| Reverse, MW170 | TCTTGATGGCCTTCGATTCTG | |
| CXCL11 | ||
| Forward, MW171 | GCTGTGATATTGTGTGCTACAGTTGTT | |
| Reverse, MW172 | CCAGGGCCTATGCAAAGACA | |
| ADAM28 | ||
| Forward, MW484 | GTACTGTCGCAGAGTGGATGAC | |
| Reverse, MW485 | GTCACTATCCGTCCTTTCCAGG | |
| ADAMDEC1 | ||
| Forward, MW486 | AGATCCACGACCATGCTCAGCT | |
| Reverse, MW487 | GTGACATCACTCCTACAAGAGCC |
Coordinates indicate distances from the transcription start site indicated for the gene entry or chromosome 8 coordinates where distances were long range (ADAM locus).
Microarray data accession number.
Duplicate raw microarray data files for the BJAB pz2 and BJAB E3C-4 cell lines have been deposited in the ArrayExpress database, available at http://www.ebi.ac.uk/arrayexpress/ under accession number E-MEXP-3585.
RESULTS
EBNA 3C independently downregulates genes that regulate chemotaxis, adhesion, and migration.
To identify cellular genes that were subject to regulation by EBNA 3C, we performed Affymetrix gene expression profiling using EBV-negative BJAB B-cell lymphoma cells stably expressing EBNA 3C (BJAB E3C-4) versus control cells containing empty vector (BJAB pz-2) (50). Conversion of Affymetrix probe set identities into Ensembl gene codes identified 47 genes that were downregulated (see Table S1 in the supplemental material) and 122 genes that were upregulated by EBNA 3C by 2-fold or more (see Table S2 in the supplemental material). EBNA 3C was previously reported to upregulate expression of the B-cell activation marker complement receptor 2 (CR2, CD21) in BJAB E3C-4 cells and in an EBNA 3C transcomplemented LCL (50, 56). Our analysis confirmed that this gene was upregulated 16-fold at the mRNA level, providing a good validation control for our study (see Table S2 in the supplemental material). Our results also confirmed a previous study demonstrating upregulation of ITGAV transcription by EBNA 3C in reporter and transient endogenous-B-cell assays (9).
In total, only 35 genes identified in our study have been previously described as genes differentially regulated by EBNA 3C in microarray analyses of either transcomplemented LCLs or EBV-negative BL cells infected with EBNA 3C knockout viruses (52, 56) (see Table S3 in the supplemental material). Our study has therefore identified an additional 134 EBNA 3C-regulated cellular genes. Significantly, our analysis revealed that 10 out of the 20 genes most repressed by EBNA 3C expression were either chemokines (CXCL10 [IP-10], CXCL11 [I-TAC], CXCL13 [BLC], CCL3 [MIP1-α], CCL3L1 and CCL4 [MIP1-β]) or members of integrin receptor-signaling pathways, including two integrin genes (ITGA4 and ITGB1) and two members of the a disintegrin and metalloprotease family (ADAM28 and ADAMDEC1) involved in integrin binding and/or adhesion (see Table S1 in the supplemental material). We therefore focused on validating and investigating the mechanisms involved in the repression of this subset of genes by EBNA 3C.
EBNA 3C downregulates integrin receptor-signaling genes.
The α4 integrin and β1 integrin subunits encoded by the ITGA4 and ITGB1 genes heterodimerize to form the major integrin receptor (α4β1) expressed in B-lymphoid cells. α4β1 integrin mediates migration through association with ligands, including fibronectin and vascular cell adhesion molecule-1 (VCAM-1) (2, 11, 17, 35). Interestingly, ADAM28 is highly expressed in B lymphocytes and also functions as a ligand for α4β1 integrin, with binding of soluble ADAM28 enhancing α4β1 integrin adhesion to VCAM-1 (7, 31). Less functional information is available for ADAMDEC1, but this gene is located in an ADAM gene cluster on chromosome 8p12 that includes ADAM28 and ADAM7, is expressed by immune cells, and is downregulated during tumorigenesis (6, 25).
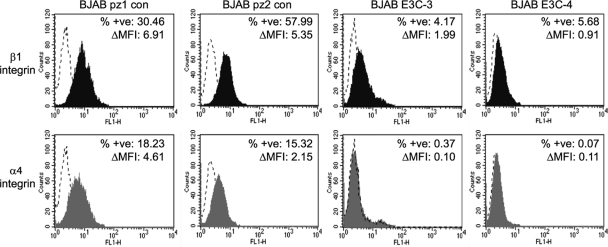
Since our microarray data were derived from comparisons between a single control (pz2) and an EBNA 3C-expressing cell line (E3C-4), we sought to validate our results by determining the expression of our gene subset in additional control BJAB and EBNA 3C-expressing clonal cell lines (Fig. 1). We confirmed that α4 and β1 integrin protein expression was downregulated in the EBNA 3C-expressing BJAB cell lines E3C-3 and E3C-4 compared to that in the control BJAB cell lines pz1 and pz2. The percentages of cells stained positive for β1 integrin were reduced from 30% and 58% in pz1 and pz2, respectively, to 4% and 6% in E3C-3 and E3C-4, respectively (Fig. 2). For α4 integrin, we found that the percentages of cells stained positive were reduced from 18% and 15% in pz1 and pz2, respectively, to 0.4% and 0.07% in E3C-3 and E3C-4, respectively (Fig. 2).
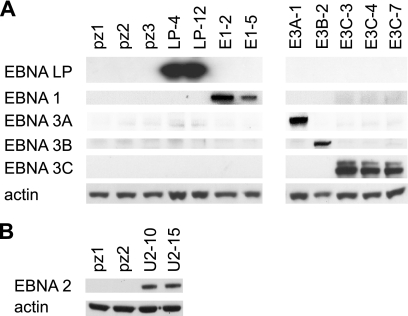
Fig 1.
Western blots of stable BJAB cell lines expressing EBV nuclear antigens. EBV nuclear antigen expression was detected by Western blotting using whole-cell lysates from independently derived control clonal cell lines (pz1, pz2, and pz3) and cell lines expressing each EBV nuclear antigen: EBNA-LP (LP-4 and LP-12), EBNA-1 (E1-2 and E1-5), EBNA 3A (E3A-1), EBNA 3B (E3B-2), EBNA 3C (E3C-3, E3C-4, E3C-7) (A), and EBNA 2 (U2-10 and U2-11) (B). Actin was used as a loading control.
Fig 2.
Flow cytometry analysis of β1 integrin and α4 integrin expression in control and EBNA 3C-expressing cell lines. Results of staining for control IgG (dotted lines), anti-β1 integrin (black fill), or anti-α4 integrin (gray fill) are shown. Increases in the percentages of positively stained cells between control and specific-antibody-stained cells (% +ve) and changes in mean fluorescence intensity (ΔMFI) are indicated.
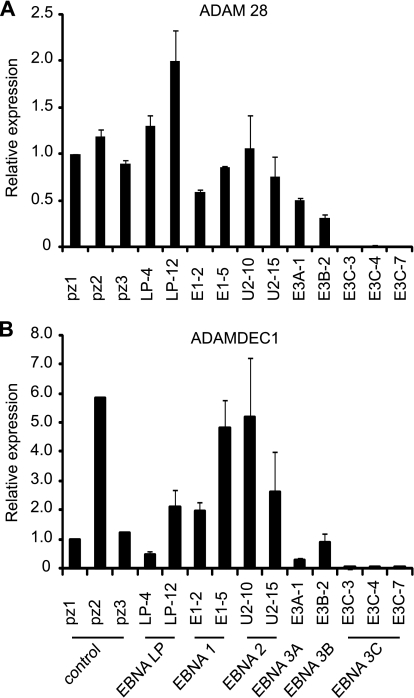
In further experiments, we validated ADAM28 and ADAMDEC1 downregulation by EBNA 3C using a panel of stable BJAB cell lines expressing all other EBV nuclear antigens (EBNA 1, EBNA-LP, EBNA 2, EBNA 3A, and EBNA 3B) (Fig. 1). Real-time PCR analysis confirmed that ADAM28 transcript levels were downregulated to barely detectable levels in all EBNA 3C-expressing cell lines examined (Fig. 3A). Previous reports have detected elevated ADAM 28 expression in EBV-negative BL cells (BL31) infected with a virus from which EBNA 3C has been deleted compared to wild-type virus-infected cells, implicating EBNA 3C as a repressor of ADAM28 (52). Our data further demonstrate that EBNA 3C can repress ADAM28 expression independently of other EBV latent proteins. Although individual expression of EBNA-LP, EBNA 1, and EBNA 2 had no consistent effect on ADAM28 expression, our analysis detected a lower level of ADAM28 expression in EBNA 3A- and EBNA 3B-expressing cell lines than in control lines (Fig. 3A). This observation is in agreement with previous studies where LCLs derived from viruses with EBNA 3A deleted were found to express higher levels of ADAM28 than control LCLs (14). However, since only one EBNA 3A- and one EBNA 3B-expressing cell line were available for analysis, the individual effects of these proteins on ADAM28 expression requires further verification.
Fig 3.
Real-time PCR analysis of ADAM28 and ADAMDEC1 mRNA expression in EBNA-expressing cells. Data are normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression levels and expressed relative to the signal obtained in the control cell line pz1 for ADAM28 (A) and ADAMDEC1 (B). Results are shown as means ± standard deviations of two independent experiments using different cDNA preparations.
Real-time PCR analysis also confirmed that ADAMDEC1 transcript levels were ablated in all EBNA 3C-expressing cell lines examined, confirming the identification of ADAMDEC1 as a new EBNA 3C target gene (Fig. 3B). Our analysis also indicated that EBNA 3A may have the capacity to independently repress ADAMDEC1 expression, since ADAMDEC1 levels were low in an EBNA 3A-expressing BJAB cell line, although variable ADAMDEC1 expression in control and EBNA-LP- and EBNA 1-expressing cell lines highlights the need for further verification of the independent effects of EBNA 3A (Fig. 3B). These observations are, however, in agreement with previous reports demonstrating that loss of EBNA 3A expression leads to increased ADAMDEC1 expression in both LCLs and a BL cell line (14, 52). Our analysis also detected increased expression of ADAMDEC1 in EBNA 2-expressing BJAB cells, although one control cell line (pz2) expressed similarly high levels of ADAMDEC1. A previous study also detected upregulation of ADAMDEC1 by EBNA 2 in EBV-negative Akata cells (23).
In summary, our results clearly demonstrate that expression of EBNA 3C alone is sufficient to significantly repress ITGA4, ITGB1, ADAM28, and ADAMDEC1 expression.
EBNA 3C associates with ITGA4, ITGB1, ADAM28, and ADAMDEC1 regulatory regions.
We hypothesized that EBNA 3C regulates the expression of chemokine and integrin receptor-signaling genes by directly targeting regulatory regions in the genome. To explore this possibility, we carried out chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-Seq) in a BL cell line expressing all EBV latent proteins (Mutu III).
ChIP-Seq using a sheep polyclonal antibody against EBNA 3C that also independently immunoprecipitates EBNA 3A and 3B (data not shown) revealed that EBNA 3 proteins directly associate with the human genome. ChIP-Seq data analysis detected 7,044 significant EBNA 3 binding sites, of which 752 (11%) were within 2 kb of a gene transcription start site, 1,417 (20%) were intragenic, and 4,875 (69%) were intergenic. Identification of the cellular genes closest to EBNA 3 binding peaks revealed 3,887 potential target genes. Forty percent of the EBNA 3C-regulated genes we identified in our microarray analysis were associated with EBNA 3 binding peaks (P = 7.8 × 10−8; hypergeometric), implicating targeted binding in their regulation. Seven hundred eighty-six (34%) out of a total number of 2,338 unique genes regulated by EBNA 3 proteins in our study or previous studies (8, 14, 52, 56) were associated with a significant EBNA 3 binding peak.
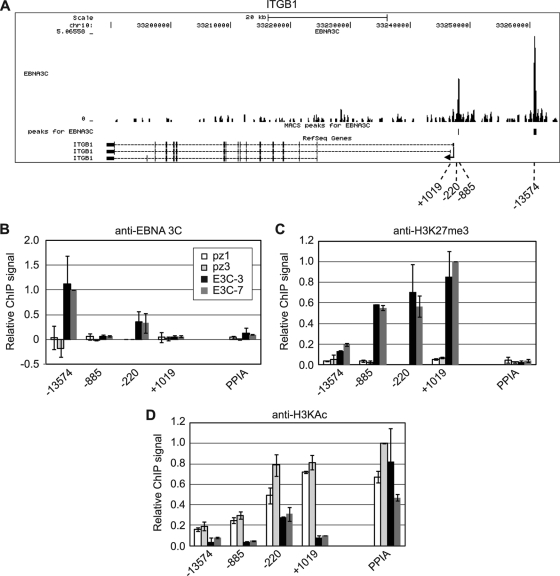
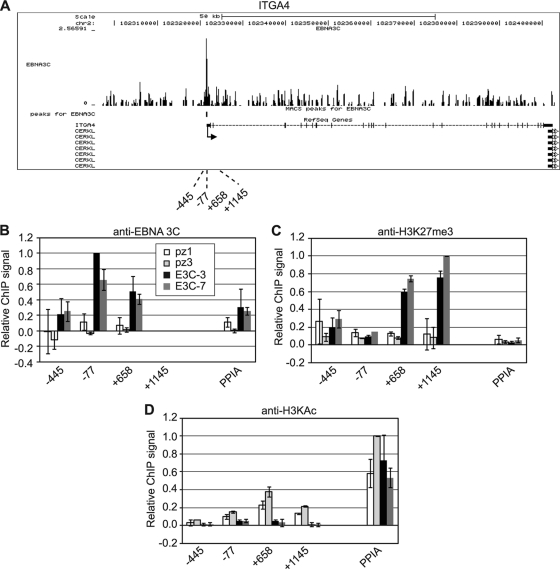
Significant peaks of EBNA 3 binding were detected upstream and close to the transcription start site of ITGB1 (Fig. 4 and Table 2). To verify that EBNA 3C was able to independently associate with these binding sites to regulate ITGB1 transcription, ChIP-QPCR was performed in two EBNA 3C-only-expressing cell lines and two control BJAB cell lines (Fig. 4A). This analysis confirmed that EBNA 3C was able to bind to these regions when expressed in the absence of other latent proteins, implicating EBNA 3C binding in the repression of ITGB1 expression. Since EBNA 3C-mediated gene silencing has been associated with increased levels of H3K27me3 (39, 44), we examined H3K27me3 at the ITGB1 locus in EBNA 3C-expressing cells. We detected large increases in H3K27me3 that were spread across a region upstream and within ITGB1, with no significant change in H3K27me3 detected at the cellular control gene, PPIA (peptidylprolyl isomerase A) (Fig. 4C). We also detected significant decreases in H3K9/14ac at ITGB1 consistent with EBNA 3C-mediated gene silencing (Fig. 4D).
Fig 4.
EBNA 3C binding and histone modification at ITGB1. (A) EBNA 3 binding at the ITGB1 locus. The numbers of sequencing reads from EBNA 3-enriched DNA are plotted per million background-subtracted total reads and aligned with the human genome. The positions of significant MACS peaks (P < 10−7) are indicated (black boxes). Gene structures are shown at the bottom of the panel. Numbers indicate the 5′ ends of the forward primers used for ChIP-PCR analysis relative to the transcription start site. Control primers spanned the transcription start site of the cellular gene encoding peptidylprolyl isomerase A (PPIA). (B) ChIP using anti-EBNA 3C antibodies in control (pz1 and pz3) and EBNA 3C-expressing (E3C-3 and E3C-7) BJAB stable cell lines. Percentage input signals, after subtraction of signals of controls with no antibody, are expressed relative to the highest signal obtained in all or the majority of experiments. Results are shown as means ± standard deviations of two independent experiments. (C and D) ChIP using anti-H3K27me3 antibodies (C) and anti- H3KAc antibodies (D).
Table 2.
Human genome coordinates for significant binding peaksa
| Gene or locus | EBNA 3 binding peak locationb |
||
|---|---|---|---|
| Chromosome | Coordinates (bp) |
||
| ADAM | 8 | 24224592 | 24225466 |
| ITGA4 (promoter) | 2 | 182321369 | 182321637 |
| ITGB1 (promoter) | 10 | 33247985 | 33248176 |
| ITGB1 (upstream) | 10 | 33260616 | 33261095 |
| CXCL | 4 | 76976240 | 76976555 |
Significance defined as a P value of <10−7.
Human genome assembly, February 2009 (GRCh37/hg19).
A promoter-proximal EBNA 3 binding site was also detected at the ITGA4 locus in Mutu III cells (Fig. 5A and Table 2), and ChIP-QPCR assays with EBNA 3C-expressing BJAB cells confirmed independent EBNA 3C association with this site (Fig. 5B). Substantial increases in H3K27me3 levels and reduced levels of H3K9/14ac were detected within ITGA4 in EBNA 3C-expressing BJAB cells, consistent with transcriptional repression mediated by EBNA 3C binding (Fig. 5C and D).
Fig 5.
EBNA 3C binding and histone modification at ITGA4. (A) EBNA 3 binding at the ITGA4 locus. (B) ChIP using anti-EBNA 3C antibodies in control (pz1 and pz3) and EBNA 3C-expressing (E3C-3 and E3C-7) BJAB stable cell lines. (C and D) ChIP using anti-H3K27me3 antibodies (C) and anti-H3KAc antibodies (D). Analysis and annotation are as for Fig. 4.
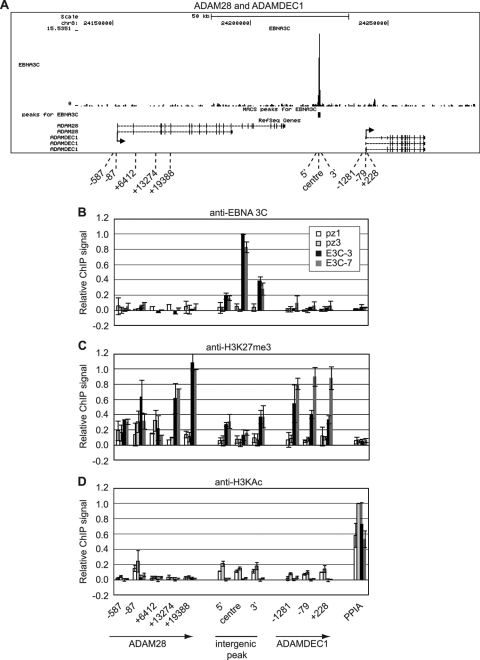
Although no significant binding sites were located around the transcription start site of ADAM28 or ADAMDEC1, EBNA 3 binding was detected in an intergenic region between these two genes in the ADAM gene cluster on chromosome 8 (Fig. 6A and Table 2). We confirmed high-level binding of EBNA 3C to this region in two BJAB cell lines expressing EBNA 3C alone (Fig. 6B). Again we found that these downregulated genes displayed large increases in H3K27me3, most evident upstream and within the genes rather than at the EBNA 3C binding site (Fig. 6C). H3K9/14ac levels were low across the ADAM gene locus in control cells, but some further reduction was observed in EBNA 3C-expressing cells at the intergenic EBNA 3C binding site, immediately upstream of ADAM28, and at the 5′ end of ADAMDEC1 (Fig. 6D). EBNA 3C binding to an intergenic element in the ADAM locus therefore appears to direct changes in histone modification within the adjacent ADAMDEC1 and ADAM28 genes.
Fig 6.
EBNA 3C binding and histone modification at the ADAM gene cluster. (A) EBNA 3 binding at the ADAM gene locus. (B) ChIP using anti-EBNA 3C antibodies in control (pz1 and pz3) and EBNA 3C-expressing (E3C-3 and E3C-7) BJAB stable cell lines. (C and D) ChIP using anti-H3K27me3 antibodies (C) and anti-H3KAc antibodies (D). Analysis and annotation are as for Fig. 4.
EBNA 3C downregulates chemokine genes and reduces CXCR3-mediated chemotaxis.
The CC family proinflammatory chemokines CCL3 and CCL4 play key roles in the regulation of immune responses, mediating the attraction of lymphocytes to sites of inflammation and infection. CCL3 and CCL4 function through interactions with the CCR1 and/or CCR5 receptors expressed on immune cells, including monocytes and T cells (30). The CXCR3-targeting CXC family chemokines CXCL10 and -11 also play important roles in modulating immune responses through the attraction of CXCR3-expressing cytotoxic T lymphocytes and exhibit potent antitumor activity in vivo (13, 24, 43).
Our initial microarray analysis detected downregulation of the chemokines CCL3, CCL3L1, CCL4, CXCL10, CXCL11, and CXCL13 by EBNA 3C in BJAB E3C-4 cells compared to that in the control cell line pz2. The Affymetrix probe set that detects CCL3 does not distinguish between CCL3 and the related chemokine CCL3L1 (see Table S1 in the supplemental material), indicating potential downregulation of one or both genes. Since CCL3L1 is commonly present in multiple copies, for simplicity we focused on CCL3 for further validation.
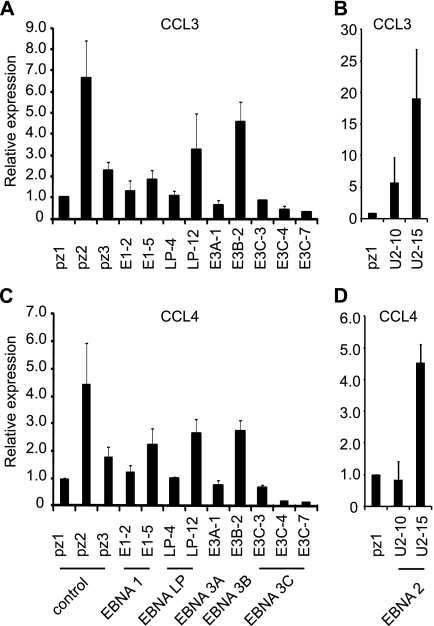
Real-time PCR analysis detected variable expression of CCL3 and CCL4 in control, EBNA 1-, and EBNA-LP-expressing BJAB cell lines, but nonetheless confirmed that expression of both chemokines was lowest in all three EBNA 3C-expressing BJAB cell lines (Fig. 7). The control cell line pz2 expressed much higher levels of CCL3 and CCL4, as was observed for ADAMDEC1. Some potential repression of CCL3 and -4 expression in the EBNA 3A-expressing cell line was evident, consistent with a previous microarray study that detected downregulation of CCL3(L1) and CCL4 by EBNA 3A in BL cells (52). Previously reported positive effects of EBNA 2 on CCL3 and CCL4 expression were also evident in at least one EBNA 2-expressing BJAB cell line (Fig. 7) (23, 26, 46).
Fig 7.
Real-time PCR analysis of CCL3 and CCL4 mRNA expression in EBV nuclear antigen-expressing cells. (A and B) Analysis of CCL3 expression in BJAB cells expressing EBV latent antigens. (C and D) Analysis of CCL4 expression in BJAB cells expressing EBV latent antigens. Results are shown as means ± standard deviations of three independent experiments using different cDNA preparations.
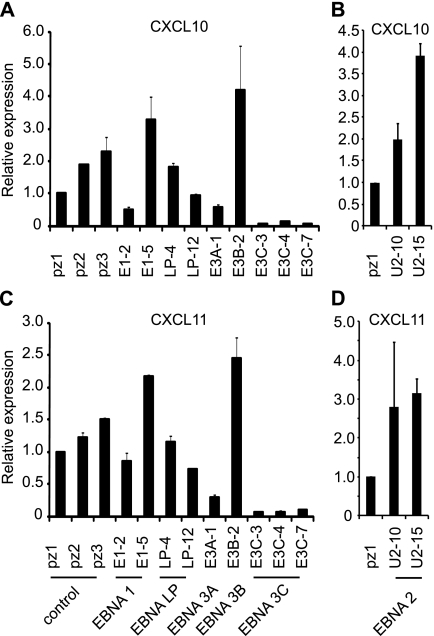
Real-time PCR analysis also confirmed significant downregulation of both CXCL10 and CXCL11 in EBNA 3C-expressing cell lines, with no consistent change in expression in EBNA 1- and EBNA-LP-expressing cells (Fig. 8). Analysis of cell lines expressing EBNA 3A or EBNA 3B also confirmed previous observations of negative and positive effects of EBNA 3A and 3B, respectively, on CXCL10 expression (14, 52). Positive effects of EBNA 2 on CXCL10 expression were also apparent in our study and in a study using a BL cell line (26). Our data also provide the first indication that CXCL11 expression may also be regulated in the same manner as CXCL10 by EBNA 2, 3A, and 3B (Fig. 8). Since only single EBNA 3A- and 3B-expressing cell lines were available for analysis, the effects of EBNA 3A and 3B will, however, require further corroboration. Although we confirmed that CXCL13 was downregulated by EBNA 3C in the cell lines used for microarray analysis (E3C-4 cells compared to pz2 cells), CXCL13 was not consistently downregulated in other EBNA 3C-expressing cell lines examined compared to control cell lines (data not shown). Taken together, our data demonstrate that expression of EBNA 3C alone is sufficient to downregulate expression of four key chemokines: CCL3, CCL4, CXCL10, and CXCL11.
Fig 8.
CXCL10 and CXCL11 are downregulated by EBNA 3C. Real-time PCR analysis of CXCL10 (A and B) and CXCL11 (C and D) mRNA expression in EBV nuclear antigen-expressing cells. Results are shown as means ± standard deviations of three independent experiments using different cDNA preparations.
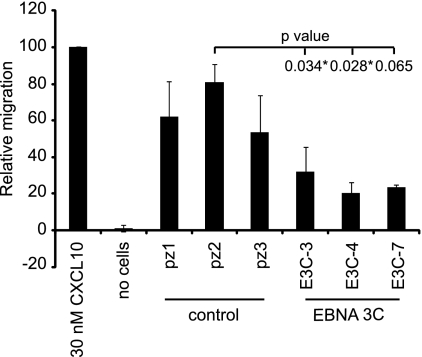
Since the CXCL10 and CXCL11 chemokines appeared to be most robustly downregulated by EBNA 3C, we investigated whether this reduction in expression suppressed the attraction of target cells. We therefore carried out chemotaxis assays to measure the level of attraction of CXCR3-positive cells by chemokines present in the culture media of control and EBNA 3C-expressing cells. Our results demonstrated that migration of CXCR3-expressing cells toward culture media obtained from EBNA 3C-expressing cell lines was reduced compared to that for control cells (Fig. 9). Migration of CXCR3-positive cells on exposure to chemokines secreted into culture media from E3C-3, E3C-4, and E3C-7 cell lines was reduced to 40%, 25%, and 29% of that of the control cell line pz2 (Fig. 9). These data therefore demonstrate that CXCL10 and CXCL11 downregulation by EBNA 3C produces a physiologically relevant reduction in CXCR3-mediated chemotaxis.
Fig 9.
EBNA 3C expression reduces chemotaxis of CXCR3-expressing cells. Chemotaxis assays using cell culture supernatant from control or EBNA 3C-expressing cells were performed. Shown is the migration of CXCR3-transfected cells expressed relative to the number of cells migrated in the presence of 30 nM recombinant CXCL10. Results are shown as means ± standard deviations of two independent experiments. Student's t test P values are indicated.
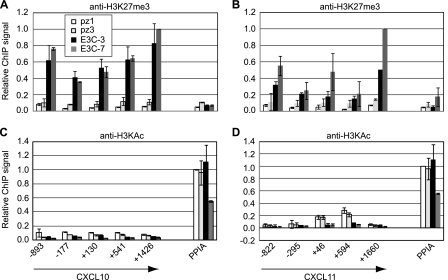
Our data also confirmed that increased H3K27me3 levels were detectable across the CXCL10 and CXCL11 locus in EBNA 3C-expressing cells, with some reduction in H3K9/14ac consistent with gene repression (Fig. 10). ChIP-Seq analysis in Mutu III cells identified one weakly enriched EBNA 3 binding peak approximately 20 kb to the right of the CXCL11, CXCL10, and CXCL9 gene cluster (Table 2), but binding at this site could not be verified by ChIP-QPCR in either Mutu III cells or EBNA 3C-expressing BJAB cells (data not shown). It is therefore possible that EBNA 3C binding to this region is below detectable limits in ChIP-QPCR assays or that this site is bound by EBNA 3A or EBNA 3B and not EBNA 3C in Mutu III cells. Given the large increase in H3K27me3 observed across CXCL10 and CXCL11 as a result of the expression of EBNA 3C alone in BJAB cells, it is, however, likely that CXCL10 and -11 represent gene targets that are downregulated indirectly by EBNA 3C.
Fig 10.
Histone modification at the CXCL gene cluster in EBNA 3C-expressing cells. (A and B) ChIP using anti-histone H3K27me3 antibodies in control (pz1 and pz3) and EBNA 3C-expressing (E3C-3 and E3C-7) BJAB stable cell lines analyzed with primers across the CXCL10 gene (A) and CXCL11 gene (B). (C and D) ChIP using anti-H3KAc antibodies analyzed with primers across the CXCL10 gene (C) and CXCL11 gene (D). Analysis and annotation are as for Fig. 4.
DISCUSSION
Taking an approach of examining the effects of expression of EBNA 3C alone in EBV-negative B cells, we aimed to dissect the role of EBNA 3C in the regulation of cellular-gene expression. Using gene expression profiling, we identified 169 cellular genes that were regulated by EBNA 3C. Half of the most highly downregulated genes either were chemokines or were involved in integrin receptor-signaling pathways, indicating that a role of EBNA 3C is to alter chemotaxis and/or migration of infected cells. Focusing our attention on these genes, we used additional stable cell lines expressing EBNA 3C to confirm that CCL3, CCL4, CXCL10, CXCL11, ITGB1, ITGB4, ADAM28, and ADAMDEC1 were significantly downregulated as a result of EBNA 3C expression. EBNA 3C may therefore promote immune evasion strategies involving reduced attraction of immune cells and the modulation of adhesion and migration of infected cells.
The CCL3 and CCL4 chemokines bind receptors on the surfaces of many immune cells, including monocytes and T lymphocytes, to promote host inflammatory responses at sites of infection and injury, largely by stimulating the recruitment of target cells. CCL3 and the related chemokine CCL3L1 bind to CCR1 and CCR5 receptors; CCL4, on the other hand, has specificity for CCR5 alone. CCL3 and CCL4 promote T-cell chemotaxis from the bloodstream to infected/inflamed areas and regulate endothelial migration of monocytes (for a review, see reference 30). Our data indicate that EBNA 3C expression alone is sufficient to downregulate CCL3 and CCL4 mRNA expression. Our analysis also identified and confirmed the CXC family chemokine genes CXCL10 and CXCL11 as targets repressed by the action of EBNA 3C alone. CXCL10 and CXCL11 also play key roles in the attraction of leukocytes to sites of infection and inflammation and possess antitumor activity in vivo through the recruitment of cytotoxic T lymphocytes to malignant tissues (13, 24, 43). Modulation of CCL3, CCL4, CXCL10, and CXCL11 expression in EBV-infected B cells by EBNA 3C may therefore represent a key mechanism to control antiviral responses and promote EBV persistence in the host. Our data indicate that the downregulation of CXCL10 and -11 expression by EBNA 3C, in particular, can be functionally significant, since it reduced the chemotaxis of cells expressing the CXCL10 and -11 receptor CXCR3.
It appears that regulation of CCL3 and CCL4 expression by EBNA 3 family proteins is cell type and context specific, since EBNA 3C and EBNA 3B positively regulate CCL3 in the EBV-negative BL cell line BL31 (52) (Table 3). Moreover, although our analysis of one stable BJAB cell line indicated that EBNA 3A may repress CCL3 and CCL4 expression (Fig. 7), positive effects of EBNA 3A have been reported in LCLs (14). CCL3 and CCL4 have also been shown in numerous microarray studies to be consistently upregulated by EBNA 2 (23, 26, 46) (Table 3). In our analysis, expression of EBNA 2 in BJAB cells increased CCL3 transcript levels, but not in excess of the level detected in the highly expressing control cell line pz2 (Fig. 7A and B). Upregulation of CCL4 was detected in only one EBNA 2-expressing cell line and was not appreciably higher than that of the pz2 control (Fig. 7C and D). Earlier studies demonstrated that CCL3 (MIP1-α), unlike CCL5 (RANTES) and CCL2 (MCP-1), is not induced by EBV infection of peripheral blood mononuclear cells (PBMCs) (15). EBV infection also prevented induction of CCL3 protein and mRNA, but not CCL2 production, on stimulation with lipopolysaccharide (15). Conflicting reports, however, have detected increases in CCL3 and CCL4 mRNA expression upon EBV B-cell infection and dramatic increases in the secretion of CCL3 and CCL4 in LCLs compared to that in peripheral blood B cells (33). Modulation of CCL3 and CCL4 expression in EBV-infected cells may therefore be influenced by the relative expression levels and activities of EBNA 2 and EBNA 3 family members and the cellular background. Nonetheless, our data demonstrate that CCL3 and CCL4 expression can be reduced by the actions of EBNA 3C alone.
Table 3.
Summary of expression data obtained from previous EBNA 2 and EBNA 3 microarrays for the chemokine and integrin receptor-signaling genes downregulated by EBNA 3C in BJAB cells
| Gene | EBNA 3 or EBNA 2 array and cell backgrounda |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E3A (LCL) | E3A (BL31) | E3B (LCL) | 3B (BL31) | 3B/3C (LCL) | 3C (LCL) | 3C (BL31) | 3A/3B/3C (BL31) | ER/E2 (BJAB) | ER/E2 (BL41) | ER/E2 (LCL) | ER/E2 type 1 (AK31) | ER/E2 type 2 (AK31) | |
| ADAM28 | − | − | − | + | |||||||||
| CXCL11 | |||||||||||||
| ITGB1 | + | ||||||||||||
| ITGA4 | + | − | + | + | + | + | |||||||
| ADAMDEC1 | − | − | + | + | |||||||||
| CXCL10 | − | + | + | + | + | + | |||||||
| CCL3 | + | + | + | + | + | + | + | ||||||
| CCL3L1 | + | − | + | + | + | + | + | + | + | + | |||
| CCL4 | + | − | + | + | + | + | + | ||||||
To facilitate comparison between knockout and conditional expression studies, we used a plus sign (+) to indicate that the EBV gene product has a positive effect on the expression of the cellular gene and a minus sign (−) to indicate that the EBV gene product has a negative effect on the expression of the cellular gene (for full gene descriptions and Ensembl IDs, see Table S1 in the supplemental material; data from references 45 and 57 were also compared, but no matches were found). Cell types are indicated in parentheses. ER/E2 indicates use of an estrogen receptor-EBNA 2 fusion protein. References: E3A (LCL), 14; E3A (BL31), E3B (LCL), 3B (BL31), 3C (BL31), and 3A/3B/3C (BL31), 52; 3B/3C (LCL), 8; 3C (LCL), 56; ER/E2 (BJAB) and ER/E2 (BL41), 26; ER/E2 (LCL), 46; ER/E2 type 1 (AK31) and ER/E2 type 2 (AK31), 23.
In contrast to the repressive effects of EBNA 3C we demonstrated here, positive regulation of CXCL10 in the EBV-negative BL cell line BL31 has previously been attributed to EBNA 3C, indicating that there may be cell-type-specific differences in the regulation of CXCL10 by EBNA 3C (52). Our analysis supports previous observations of negative and positive effects of EBNA 3A and 3B, respectively, on CXCL10 expression and demonstrates that these proteins can independently regulate CXCL10 (Fig. 8A and Table 3).
Previous studies on CXCL11 expression in EBV-infected cells have reported downregulation of CXCL11 in a newly established BL cell line on expansion of the EBV latency gene expression profile from a latency I-restricted to a full-latency III EBV gene expression profile (33). This reduction in CXCL11 gene expression correlated with increased expression of an EBV microRNA, miR-BHRF1-3, that was shown to specifically target and downregulate CXCL11 expression in vivo (53). Our study is the first to implicate EBNA 3 family members in the regulation of CXCL11 expression. In addition to the repression of CXCL11 expression by EBNA 3C, our data implicate EBNA 3A and 3B as repressors and activators, respectively, of CXCL11 (Fig. 5B). EBV therefore appears to have evolved multiple strategies to negatively regulate CXCL11.
The integrin receptor-signaling genes we confirmed as targets downregulated by EBNA 3C form part of a common pathway. They comprise the genes encoding the heterodimeric B-lymphocyte α4β1 integrin receptor (ITGB1 and ITGA4) and the novel α4β1 integrin ligand, ADAM28 (7, 31), along with a related gene, ADAMDEC1, located in the same gene cluster, that is downregulated during tumorigenesis (6, 25). In support of our results, while this work was in progress, EBNA 3C was reported to repress ITGA4 expression in transcomplemented LCLs, although in another BL cell background EBNA 3C is required to maintain ITGA4 expression (52, 56). Loss of EBNA 3B expression in LCLs results in downregulation of ITGB1, implicating EBNA 3B as a positive regulator of ITGB1 (52) (Table 3). These observations, taken together with the positive effects of the expression of EBNA 2 alone on ITGA4 expression previously observed (26), indicate that ITGA4 and ITGB1 expression, like that of CCL3, CCL4, and CXCL10, is also likely to be fine-tuned through interplay between other EBV latent proteins during the establishment and maintenance of viral persistence.
ADAM28 is highly expressed on B lymphocytes, and the soluble form of the protein binds to α4β1 integrin receptors, increasing their adhesion to VCAM-1. ADAM28 is therefore likely to be a key regulator of B-cell adhesion and migration through the endothelium. Interestingly, ADAM28 is overexpressed in non-small-cell lung cancers (NSCLC) and breast carcinomas, and its overexpression in NSCLC correlates with proliferation, tumor size, and lymph node metastasis (32, 34). EBNA 3C downregulation of these components of integrin receptor pathways may therefore be important in regulating the migration and adhesion of EBV-infected B cells. A recent report detected elevated ADAM 28 expression in EBV-negative BL cells (BL31) infected with a virus with lacking EBNA 3C compared to wild-type virus-infected cells, implicating EBNA 3C as a repressor of ADAM28 (52). Our data now clearly demonstrate that EBNA 3C has the capacity to repress ADAM28 expression independently from other EBV latent proteins. EBNA 3A also appears to repress ADAM28 expression, since LCLs derived from viruses lacking EBNA 3A express higher levels of ADAM28 than control LCLs (14). Our analysis detected some repression of ADAM28 expression in the E3A-1 cell line and may also implicate EBNA 3B as an ADAM28 repressor, although the variability in ADAM28 expression across the BJAB cell panel examined means that the effects of EBNA 3A and 3B alone require further verification (Fig. 3).
EBNA 3C has not previously been implicated as a repressor of ADAMDEC1, but loss of EBNA 3A expression has also been shown to result in increased ADAMDEC1 expression in both LCLs and a BL cell line, implicating EBNA 3A as a repressor of ADAMDEC1 (14, 52) (Table 3). Our analysis detected downregulation of ADAMDEC1 in the one available EBNA 3A-expressing BJAB cell line, indicating that EBNA 3A can downregulate ADAMDEC1 expression when expressed alone (Fig. 3B). EBNA 2 expression in EBV-negative Akata cells has been previously shown to upregulate ADAMDEC1 expression (23), and our results detected increased expression of ADAMDEC1 in EBNA 2-expressing BJAB cells, although one control cell line (pz2) expressed similarly high levels of ADAMDEC1. It is therefore likely that the overall level of ADAMDEC1 transcripts in EBV-infected cells is determined by the opposing effects of EBNA 2 and EBNA 3 family proteins and that EBNA 3A and 3C may act in concert to repress ADAMDEC1. Interestingly, recent studies have implicated reduced activation of ADAMDEC1 by EBNA 2 in the impaired transforming ability of type 2 EBV isolates. EBV strains are classified as type 1 or type 2 on the basis of variation in the sequences of the EBNA 2, EBNA-LP, and EBNA 3A, B, and C genes. Type 2 EBNA 2 fails to upregulate ADAMDEC1 and three other cellular genes (MARCKS, IL-1β, and CXCR7) to the same extent as type 1 EBNA 2 (23). It is therefore possible that the reduced effects of type 2 EBNA 2 on ADAMDEC1 may shift the balance between EBNA 2-mediated activation and EBNA 3A- and 3C-mediated repression and impede the outgrowth of immortalized cells. Control of ADAMDEC1 expression by the concerted actions of EBV latent proteins may therefore play a crucial role in the early stages of EBV-driven B-cell transformation.
Importantly, although previous reports have detected EBNA 3C association with the EBNA 2- and 3C-regulated viral LMP1 promoter (16), our study provides the first demonstration of EBNA 3C association with cellular DNA elements. We mapped EBNA 3C binding sites at the ITGB1 and ITGA4 promoters and confirmed an additional binding site upstream of ITGB1. We also detected a large EBNA 3C binding signal in an intergenic region between ADAM28 and ADAMDEC1. At the CXCL10 and -11 gene locus, however, the weakly enriched peak in the vicinity of these genes could not be confirmed by ChIP-QPCR assays in Mutu III cells or EBNA 3C-expressing BJAB cells, indicating that it may either be below the detection limits of ChIP-QPCR assays or be bound by EBNA 3A or EBNA 3B, since these proteins were also precipitated by the EBNA 3C antibody used for ChIP-Seq analysis. Further investigations are required to address this possibility, but the dramatic increase in H3K27me3 across the CXCL10 and -11 gene locus observed on expression of EBNA 3C alone indicates that EBNA 3C can act in isolation as a repressor of CXCL10 and -11 transcription, perhaps through indirect mechanisms.
EBNA 3C does not associate with regulatory elements by binding to DNA directly but has been shown to interact with the cellular DNA binding proteins, RBP-Jκ, and members of the PU.1 family that are also binding partners of EBNA 2 (40, 58). It is therefore possible that EBNA 3C is recruited to the binding sites we have identified through association with these cellular factors. Recent studies have used ChIP-Seq to map human genome binding sites for RBP-Jκ in an EBV-immortalized LCL (IB4) (59). Analysis of these published data sets revealed weakly enriched RBP-Jκ binding peaks at the ITGB1 and ITGA4 transcription start site (TSS), close to or encompassing the EBNA 3C binding sites. Examination of ENCODE ChIP-Seq data for PU.1 obtained from EBV-immortalized LCLs also identified binding sites for PU.1 at the ITGA4 TSS and at the intergenic ADAM peak. It is therefore possible that RBP-Jκ and/or PU.1 plays a role in EBNA 3C recruitment to the ITGA4, ITGB1, and ADAM gene loci. The lack of RBP-Jκ or PU.1 association with the EBNA 3C binding site upstream from ITGB1 may implicate additional, as-yet-unidentified, cellular transcription factors in the recruitment of EBNA 3C to this site.
Although our analysis of the effects of EBNA 3C on cellular-gene expression was carried out using stable transfectants of a B-cell line that express higher levels of EBNA 3C than are present in latently infected cells (50; also data not shown), 40% of the differentially regulated genes we detected were associated with EBNA 3 binding peaks in a latently infected BL cell line, indicating that our data are physiologically relevant. A large proportion of the genes we identified are therefore likely to represent bona fide direct targets of EBNA 3 proteins, with the remaining genes representing possible indirect targets. Our follow-up studies confirmed ITGA4, ITGB1, ADAM28, and ADAMDEC1 as direct EBNA 3C target genes and implicated CXCL10 and -11 as indirect targets. It is interesting that there are conflicting data from different studies on the direction of regulation of some of the chemokine and integrin receptor-signaling genes we have studied as EBNA 3C-repressed genes that cannot simply be explained by differences in expression levels of EBNA 3C (Table 3). For example, we report repression of ITGA4 by EBNA 3C, consistent with reports using a conditional EBNA 3C-expressing LCL system, where EBNA 3C function was switched off and the cells were transcomplemented by transfection of plasmids that express EBNA 3C at wild-type levels (56) (Table 3). However ITGA4 expression was reduced in a BL cell line (BL31) infected with EBNA 3C knockout EBV compared to that of wild-type EBV-infected cells expressing EBNA 3C at physiological levels, implicating EBNA 3C as an activator of ITGA4 in this context (52). However, the repressive effects of EBNA 3C on ADAM28 expression we have observed in BJAB cells were recapitulated in the context of BL31 cells (52). Conversely, the chemokine genes CXCL10, CCL3, and CCL3L1 were detected as targets for activation rather than repression by EBNA 3C in BL31 cells but were not detected as differentially regulated genes in the transcomplemented LCL system. Our study therefore confirms these genes as EBNA 3C targets and further highlights the ability of EBV latent genes to use different modes of cellular-gene regulation in a context-specific manner. The direction of regulation of cellular-gene expression by EBV-encoded factors is therefore likely to be determined by both the cellular background and the relative expression levels and/or activities of the individual latent proteins. An ability to switch between different modes of regulation may be crucial for infected cells to persist in the host.
In summary, our data provide the first demonstration of EBNA 3C binding to human genome elements and functionally link these binding sites to the downregulation of ITGB1, ITGA4, ADAM28, and ADAMDEC1 via increases in the polycomb repressor complex-associated repressive H3K27me3 mark. Significantly, our data reveal that EBNA 3C expression alone is sufficient to promote H3K27me3 at target genes. To further dissect the mechanism of EBNA 3C-mediated gene silencing, it will be important to determine whether EBNA 3C is able to associate with and/or promote the recruitment of polycomb repressor complexes at target gene loci. The mechanism of EBNA 3C recruitment to these repressed gene loci also remains to be fully elucidated, and it is possible that B-cell-specific factors in addition to PU.1 and RBP-Jκ may play a role in EBNA 3C targeting.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Wellcome Trust (064014 and 078291) and the Royal Society to M.J.W. M.J.M. is funded by a Ph.D. studentship from the Biotechnology and Biological Sciences Research Council, and S.N.S. and R.D.P. were funded by Ph.D. studentships from the University of Sussex. R.G.J. is supported by a Medical Research Council Career Development Award and A.S.K. by an MRC Centre grant to the MRC Centre for Medical Molecular Virology.
We thank Martin Rowe, Alan Rickinson, and Bernhard Moser for antibodies and/or cell lines.
Footnotes
Published ahead of print 22 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Allday MJ, Crawford DH, Thomas JA. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361–369 [DOI] [PubMed] [Google Scholar]
- 2. Alon R, et al. 1995. The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. J. Cell Biol. 128:1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderton E, et al. 2008. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene 27:421–433 [DOI] [PubMed] [Google Scholar]
- 4. Bain M, Watson RJ, Farrell PJ, Allday MJ. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bark-Jones SJ, Webb HM, West MJ. 2006. EBV EBNA 2 stimulates CDK9-dependent transcription and RNA polymerase II phosphorylation on serine 5. Oncogene 25:1775–1785 [DOI] [PubMed] [Google Scholar]
- 6. Bates EE, Fridman WH, Mueller CG. 2002. The ADAMDEC1 (decysin) gene structure: evolution by duplication in a metalloprotease gene cluster on chromosome 8p12. Immunogenetics 54:96–105 [DOI] [PubMed] [Google Scholar]
- 7. Bridges LC, et al. 2002. The lymphocyte metalloprotease MDC-L (ADAM 28) is a ligand for the integrin alpha4beta1. J. Biol. Chem. 277:3784–3792 [DOI] [PubMed] [Google Scholar]
- 8. Chen A, Zhao B, Kieff E, Aster JC, Wang F. 2006. EBNA-3B- and EBNA-3C-regulated cellular genes in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 80:10139–10150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choudhuri T, Verma SC, Lan K, Robertson ES. 2006. Expression of alpha V integrin is modulated by Epstein-Barr virus nuclear antigen 3C and the metastasis suppressor Nm23-H1 through interaction with the GATA-1 and Sp1 transcription factors. Virology 351:58–72 [DOI] [PubMed] [Google Scholar]
- 10. Cotter MA, II, Robertson ES. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin alpha. Mol. Cell. Biol. 20:5722–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dominguez-Jiménez C, Sánchez-Aparicio P, Albar JP, Garcia-Pardo A. 1996. The alpha 4 beta 1 fibronectin ligands CS-1, Hep II, and RGD induce different intracellular events in B lymphoid cells. Comparison with the effects of the endothelial ligand VCAM-1. Cell Adhes. Commun. 4:251–267 [DOI] [PubMed] [Google Scholar]
- 12. Gregory CD, Rowe M, Rickinson AB. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt's lymphoma cell line. J. Gen. Virol. 71:1481–1495 [DOI] [PubMed] [Google Scholar]
- 13. Hensbergen PJ, et al. 2005. The CXCR3 targeting chemokine CXCL11 has potent antitumor activity in vivo involving attraction of CD8+ T lymphocytes but not inhibition of angiogenesis. J. Immunother. 28:343–351 [DOI] [PubMed] [Google Scholar]
- 14. Hertle ML, et al. 2009. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathog. 5:e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jabs WJ, Wagner HJ, Maurmann S, Hennig H, Kreft B. 2002. Inhibition of macrophage inflammatory protein-1 alpha production by Epstein-Barr virus. Blood 99:1512–1516 [DOI] [PubMed] [Google Scholar]
- 16. Jiménez-Ramírez C, et al. 2006. Epstein-Barr virus EBNA-3C is targeted to and regulates expression from the bidirectional LMP-1/2B promoter. J. Virol. 80:11200–11208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim I, Uchiyama H, Chauhan D, Anderson KC. 1994. Cell surface expression and functional significance of adhesion molecules on human myeloma-derived cell lines. Br. J. Haematol. 87:483–493 [DOI] [PubMed] [Google Scholar]
- 18. Knight JS, Lan K, Subramanian C, Robertson ES. 2003. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J. Virol. 77:4261–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krauer KG, et al. 2004. The EBNA-3 gene family proteins disrupt the G2/M checkpoint. Oncogene 23:1342–1353 [DOI] [PubMed] [Google Scholar]
- 20. Leao M, Anderton E, Wade M, Meekings K, Allday MJ. 2007. Epstein-Barr virus-induced resistance to drugs that activate the mitotic spindle assembly checkpoint in Burkitt's lymphoma cells. J. Virol. 81:248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Roux A, Kerdiles B, Walls D, Dedieu JF, Perricaudet M. 1994. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology 205:596–602 [DOI] [PubMed] [Google Scholar]
- 22. Loetscher M, et al. 1996. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J. Exp. Med. 184:963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lucchesi W, et al. 2008. Differential gene regulation by Epstein-Barr virus type 1 and type 2 EBNA2. J. Virol. 82:7456–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luster AD, Leder P. 1993. IP-10, a -C-X-C- chemokine, elicits a potent thymus-dependent antitumor response in vivo. J. Exp. Med. 178:1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Macartney-Coxson DP, et al. 2008. Metastatic susceptibility locus, an 8p hot-spot for tumour progression disrupted in colorectal liver metastases: 13 candidate genes examined at the DNA, mRNA and protein level. BMC Cancer 8:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maier S, et al. 2006. Cellular target genes of Epstein-Barr virus nuclear antigen 2. J. Virol. 80:9761–9771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marshall D, Sample C. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maruo S, et al. 2006. Epstein-Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:19500–19505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maunders MJ, Petti L, Rowe M. 1994. Precipitation of the Epstein-Barr virus protein EBNA 2 by an EBNA 3c-specific monoclonal antibody. J. Gen. Virol. 75:769–778 [DOI] [PubMed] [Google Scholar]
- 30. Maurer M, von Stebut E. 2004. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 36:1882–1886 [DOI] [PubMed] [Google Scholar]
- 31. McGinn OJ, et al. 2011. Modulation of integrin alpha4beta1 by ADAM28 promotes lymphocyte adhesion and transendothelial migration. Cell Biol. Int. 35:1043–1053 [DOI] [PubMed] [Google Scholar]
- 32. Mitsui Y, et al. 2006. ADAM28 is overexpressed in human breast carcinomas: implications for carcinoma cell proliferation through cleavage of insulin-like growth factor binding protein-3. Cancer Res. 66:9913–9920 [DOI] [PubMed] [Google Scholar]
- 33. Nakayama T, et al. 2004. Selective induction of Th2-attracting chemokines CCL17 and CCL22 in human B cells by latent membrane protein 1 of Epstein-Barr virus. J. Virol. 78:1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohtsuka T, et al. 2006. ADAM28 is overexpressed in human non-small cell lung carcinomas and correlates with cell proliferation and lymph node metastasis. Int. J. Cancer 118:263–273 [DOI] [PubMed] [Google Scholar]
- 35. Oostendorp RA, et al. 1995. VLA-4 and VCAM-1 are the principal adhesion molecules involved in the interaction between blast colony-forming cells and bone marrow stromal cells. Br. J. Haematol. 91:275–284 [DOI] [PubMed] [Google Scholar]
- 36. Palermo RD, Webb HM, West MJ. 2011. RNA polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein-Barr virus. PLoS Pathog. 7:e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Parker GA, et al. 1996. Epstein-Barr virus nuclear antigen (EBNA)3C is an immortalizing oncoprotein with similar properties to adenovirus E1A and papillomavirus E7. Oncogene 13:2541–2549 [PubMed] [Google Scholar]
- 38. Parker GA, Touitou R, Allday MJ. 2000. Epstein-Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene 19:700–709 [DOI] [PubMed] [Google Scholar]
- 39. Paschos K, et al. 2009. Epstein-Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog. 5:e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Radkov SA, et al. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552–8562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Radkov SA, et al. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robertson ES, et al. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sgadari C, et al. 1996. Interferon-inducible protein-10 identified as a mediator of tumor necrosis in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:13791–13796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. 2010. Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathog. 6:e1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spender LC, Cornish GH, Sullivan A, Farrell PJ. 2002. Expression of transcription factor AML-2 (RUNX3, CBFα-3) is induced by Epstein-Barr virus EBNA-2 and correlates with the B-cell activation phenotype. J. Virol. 76:4919–4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Spender LC, et al. 2006. Cell target genes of Epstein-Barr virus transcription factor EBNA-2: induction of the p55alpha regulatory subunit of PI3-kinase and its role in survival of EREB2.5 cells. J. Gen. Virol. 87:2859–2867 [DOI] [PubMed] [Google Scholar]
- 47. Tomkinson B, Robertson E, Kieff E. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Touitou R, Hickabottom M, Parker G, Crook T, Allday MJ. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 75:7749–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waltzer L, Perricaudet M, Sergeant A, Manet E. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J. Virol. 70:5909–5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang F, et al. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. West MJ, Webb HM, Sinclair AJ, Woolfson DN. 2004. Biophysical and mutational analysis of the putative bZIP domain of Epstein-Barr virus EBNA 3C. J. Virol. 78:9431–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. White RE, et al. 2010. Extensive co-operation between the Epstein-Barr virus EBNA3 proteins in the manipulation of host gene expression and epigenetic chromatin modification. PLoS One 5:e13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xia T, et al. 2008. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 68:1436–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 55. Zhang Y, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao B, et al. 2011. Epstein-Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. U. S. A. 108:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhao B, et al. 2006. RNAs induced by Epstein-Barr virus nuclear antigen 2 in lymphoblastoid cell lines. Proc. Natl. Acad. Sci. U. S. A. 103:1900–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhao B, Sample CE. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 74:5151–5160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhao B, et al. 2011. Epstein-Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc. Natl. Acad. Sci. U. S. A. 108:14902–14907 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.