Abstract
Although the rate at which proteins change is a key parameter in molecular evolution, its determinants are poorly understood in viruses. A variety of factors, including gene length, codon usage bias, protein abundance, protein function, and gene expression level, have been shown to affect the rate of protein evolution in a diverse array of organisms. However, the role of these factors in viral evolution has yet to be addressed. The polar 3′-5′ stepwise attenuation of transcription in the Mononegavirales, a group of single-strand negative-sense RNA viruses, provides a unique system to explore the determinants of protein evolution in viruses. We analyzed the relative importance of a variety of factors in shaping patterns of sequence variation in full-length genomes from 13 Mononegavirales species. Our analysis suggests that the level of gene expression, and by extension the relative genomic position of each gene, is a key determinant of the protein evolution in these viruses. This appears to be the consequence of selection for translational robustness, but not for translational accuracy, in highly expressed genes. The small genome size and number of proteins encoded by these viruses allowed us to identify other protein-specific factors that may also play a role in virus evolution, such as host-virus interactions and functional constraints. Finally, we explored the evolutionary pressures acting on noncoding regions in Mononegavirales genomes and observed that, despite being less constrained than coding regions, their evolutionary rates are also associated with genomic position.
INTRODUCTION
Understanding the determinants of protein evolution is one of the central tasks of molecular evolution. The rates of amino acid substitution vary substantially within and between species (72), and much of this variation reflects the differing types and intensities of natural selection acting on proteins. However, the factors that drive these selective differences are still the source of debate. The most common explanation is that the rate of protein evolution is largely set by the fraction of sites that are involved in protein function (i.e., “functional density”) (6, 71). Unfortunately, experimental tests of this theory are labor-intensive, since the evaluation of functional density involves extensive mutagenesis analysis. As a consequence, most work in this area has focused on measuring other, more accessible, variables, which are correlated in various degrees to functional density. Thus, substitution rates in proteins (often measured as the numbers of synonymous and nonsynonymous substitutions per site[dS and dN], respectively) and selection pressures (the ratio dN/dS) have been associated with a number of protein features, including length (39), interactions with other proteins (16), contribution to overall fitness (dispensability) (25), role in interaction networks (centrality) (22), codon usage bias (62), abundance (11), structure (3), and expression level (11, 12, 46).
Comparative analyses of the relative importance of a number of these factors in determining the rate of protein evolution in organisms ranging from bacteria to mammals have demonstrated that higher levels of protein expression are strongly correlated with lower dN, dS, and dN/dS values (11–13, 54). A possible explanation for this correlation is that the fitness effect of a mutation in a given protein is proportional to the contribution of that protein to the overall fitness of the organism (24, 54), with proteins expressed at higher levels contributing most. However, this explanation might not be of general applicability. For instance, under this theory, proteins that are expressed less despite being more abundant (for example, due to a slower turnover) are expected to evolve more slowly, which does not appear to be the case in yeast (11). It has also been hypothesized that increased expression might lead to selection for codons that are translated faster or more accurately (i.e., translational efficiency) (1) and/or selection for amino acid sequences that are able to fold properly despite mistranslation (i.e., translational robustness) (11). These factors would reduce the rate of protein evolution in highly expressed proteins due to the higher cost of slower/less accurate translation and/or protein misfolding compared to proteins expressed at lower levels. This idea is supported by mutagenesis experiments, in which proteins with higher expression levels evolve to greater stability despite accumulating more mutations, and sequence analyses, which show strong protein-level constraints in nonpreferred codons and asymmetry in the use of synonymous codons depending on expression level (3, 4, 11, 18).
Remarkably little is known about the factors that shape rates of protein evolution in viruses. As intracellular parasites, viruses need to overcome host resistance systems and use host cellular machinery to complete their life cycle. Accordingly, mutations that facilitate the evasion of host immune responses or genetic resistance, result in antiviral resistance, or improve interaction with cell proteins are clearly subject to strong selection pressure (17, 53). As a consequence, the selection pressures exerted by the host on viral proteins might be expected to have a stronger effect on protein evolution than the level of gene expression. In addition, while many of the mutations that increase translational efficiency occur at synonymous sites, optimizing codon usage, RNA viruses often utilize synonymous codons that tend to match the nucleotide biases across the viral genome as a whole (31). This suggests that selection on codon choice may often be set by background mutational pressure or selection for overall nucleotide composition, rather than optimizing the match between viral codon and host tRNA anticodon to increase the accuracy of protein translation (20, 26, 31), although exceptions have been reported (8, 34, 69).
To better understand the determinants of the rate of protein evolution in RNA viruses, we analyzed the role of several key factors—gene length, mRNA and protein abundance, gene relative position in the genome, and codon usage bias—in the evolution of viral species from the taxonomic order Mononegavirales which comprise an important set of human, animal, and plant pathogens. The Mononegavirales belong to four different viral families but share a number of important features. All possess an unsegmented negative-sense RNA genome that varies between 8.9 and 19 kb in length, encodes 5 to 10 proteins, and is encapsidated in virions with enveloped structures and a fringe of spike glycoproteins. The Mononegavirales also share a distinctive genome organization. The 3′-proximal genes encode the viral nucleoprotein (N), the phosphoprotein (P), and the matrix protein (M). Close to the 5′ are the two largest genes, which encode an attachment protein (either a glycoprotein [G], a hemagglutinin [H], or a hemagglutinin-neuraminidase [HN]) and the virus RNA polymerase (L protein) (37). The Paramyxoviridae have extra proteins encoded by different genes, including the fusion protein (F), or transcribed through RNA editing from the P gene (C and V proteins). Other unique proteins observed in some species include transcription factors (M2), other nonstructural proteins (NS1 and NS2), or membrane proteins (SH) (15, 38). In all Mononegavirales, genes are transcribed in a sequential interrupted synthesis from 3′ to 5′, which results in discrete mRNAs for each gene. This transcription is polar with stepwise attenuation, even if it can be further regulated (65). Critically, this generally results in a strong transcription gradient, such that the 3′ gene (N) is the most abundant RNA and the 5′ gene (L) is the least abundant. Since the products of the genes at the 3′ region of the genome are usually those required in the highest numbers, it is likely that this genome organization was selectively optimized as a way of controlling gene expression (27). Importantly, this also means that the relative position of each gene within the genome is a good predictor of the overall level of transcription. The exception are the filoviruses in which mRNA levels do not appear to strictly decrease 3′-5′, which could be the result of a strong effect of regulatory regions flanking certain genes or differences in mRNA turnover (43, 56, 57). In addition, the Mononegavirales contain noncoding regions of variable lengths (15 to 695 nucleotides, depending on the species). Although specific sequences within these regions are likely to possess a regulatory function (37) and so may be subject to selection pressures similar to those in coding regions, they provide a useful data set for the comparative analysis of substitution dynamics. This combination of factors makes the Mononegavirales a valuable system for analyzing the factors that shape the rate of protein evolution in viruses.
MATERIALS AND METHODS
Sequence data.
All available full-length genome sequences from species assigned to the order Mononegavirales were collated from GenBank. Species with more than 10 complete genome sequences were retained for analysis, so that we were able to utilize 13 taxa, and a total of 345 sequences from the four families of this order. For measles virus, 18 vaccine strains and seven isolates passaged in non-natural hosts were also included in the analysis, although their removal did not change our results in any meaningful way. A list of the isolates and GenBank accession numbers of the sequences used is provided in Table S1 in the supplemental material.
Sequence alignments were constructed for all genes from each species. By the convention used in this order, the term “gene” refers to the genomic RNA sequence encoding a single mRNA, even if that mRNA contains more than one ORF and encodes for more than one protein, as is the case for the P gene in some species. For the P gene, alignments of nonoverlapping regions were also obtained to analyze the evolutionary effect of additional ORFs. In all cases, alignments were constructed using MUSCLE 3.7 (14) and adjusted manually according to the amino acid sequences using Se-Al (50). Sequence alignments for the noncoding regions in each species were obtained using the same protocol. To study the differences in genetic distance between coding and noncoding regions, alignments of concatenated genes and noncoding regions were also generated.
Genomic factors.
Gene length (in nucleotides) and the relative genomic position (3′ to 5′) were extracted from GenBank and from Fauquet et al. (15). The values of gene expression level and protein abundance were obtained from several sources (see Table S2 in the supplemental material). To compile these data, several approaches were taken. (i) Measurements were directly extracted from relevant publications. (ii) Images of electrophoresis gels of total mRNA or protein extracts from infected cells (and likely to represent levels in natural infections) were obtained from the corresponding publications, and the bands of interest were evaluated densitometrically by using ImageJ v1.45 (51). The densitometry values of each band were automatically corrected by subtracting the background value from the densitometry value of the area delimited by each band. Where necessary, we also corrected the densitometry values for the length of each gene, or the number of labeled amino acids in the protein sequence. The densitometry values and subsequent calculations are presented in Table S3 in the supplemental material. (iii) When mRNA or protein amounts were represented as a densitometry graphic in a publication, the abundance was estimated as the area under the corresponding peak with ImageJ v1.45. No background correction was applied in these cases, but a length or number of labeled amino acids correction was performed as for densitometry values. These calculations are shown in Table S4 in the supplemental material. Measures of the relative mRNA abundance in mumps virus and of both mRNA and protein abundance in parainfluenza virus 3 were gathered from in vitro transcription assays. Using this information, the level of gene expression was compiled as the relative abundance of each gene mRNA compared to that of the N gene (the gene with the highest expression level in most Mononegavirales). Similarly, protein abundance was measured as the relative abundance of each protein compared to that of the N protein. Detailed information on the manuscripts and relevant figures used to extract these data are presented in Table S2 in the supplemental material. Codon usage bias in each gene was estimated as the effective number of codons (Nc′), which measures the departure of codon usage from that expected given the nucleotide composition of the data set. Nc′ takes the value of 61 when there is no deviation from the expected codon usage, and this value declines as codon usage bias increases (45).
Evolutionary factors.
The selection pressure for each gene was measured as the mean number of nonsynonymous (dN) to synonymous (dS) nucleotide substitutions per site (dN/dS ratio) and estimated using the single-likelihood ancestor counting (SLAC), the random effects likelihood (REL), and the fixed-effect likelihood (FEL) methods implemented in the HYPHY package (36). Since the three methods yielded similar results, only the SLAC results are shown here. In all cases, dN/dS ratio estimates were based on input neighbor-joining trees inferred using the general-time-reversible (GTR) nucleotide substitution model, with 95% confidence intervals (CI) calculated assuming a χ2 distribution. Estimates were considered to be significantly different if the mean value of the estimate from one data set fell outside of the 95% CI values of another (indicating that these ratios have been drawn from different distributions). Individual values of dN and dS were also obtained.
Genetic distances (d) were estimated for each coding and noncoding region and also for the concatenated data sets of the Mononegavirales species analyzed. For this analysis, the best-fit model of nucleotide substitution in each data set was determined using Modeltest 3.7 (49), and this was used to estimate pairwise distances with PAUP*4.0 (60).
Statistical analysis.
The contribution of each genomic factor to the variation in selection pressures and genetic distances was estimated using principal component analysis (PCA). The gene length, relative position, mRNA and protein abundance, and Nc′ were scaled to zero mean and unit variance, inserted into a regression matrix, and rotated to obtain the principal components (PCs). Significance thresholds for the load of each genomic factor on a PC were determined using a broken-stick model (47). A subsequent linear regression of d, dN/dS, dN, and dS on the PCs yielded the proportion of the variance in each of these variables explained by each component (R2), the significance of R2, and the fractional contribution of each original genomic factor to the component. The data on all of the factors considered were only available for 10 of the 13 species; consequently, Borna disease virus, Nipah virus, and the metapneumoviruses were excluded from this analysis. Ebola virus was excluded from the PCA that used the relative gene position as a variable, since the 3′-5′ gradient of mRNA levels is not observed in this species (57) (see Table S2 in the supplemental material), and therefore the relative gene position cannot be used as a proxy of the mRNA expression level. Indeed, the inclusion of Ebola virus in this analysis slightly reduced the R2 values, although it did not significantly change the results. Exclusion of species for which mRNA and/or protein relative abundance was obtained from in vitro assays did not alter the results.
All of the evolutionary parameters as well as measures of gene expression level were homoscedastic, i.e., these variables followed a normal distribution and presented homogeneous variances (55). Consequently, these variables were analyzed using an analysis of variance (ANOVA), and correlations between d, dN/dS, dN, and dS and the relative position of the coding and noncoding regions in the viral genome and gene expression level, as well as correlations between genetic distances in coding and noncoding regions, were assessed using Pearson coefficients. Correlation analyses were performed considering all 10 Mononegavirales species together. The presence of outliers, which potentially prevent significant linear correlation, was detected by calculating the studentized residual for each data point, dividing the residual by its standard deviation. Values outside the 95% CI of the Student t test distribution drawn with all of the studentized residuals were considered outliers (58). This widely used method of outlier detection allowed us to more accurately explore the relationships between the factors studied and to identify genes with unusual behavior. Similar correlation analyses were carried out for each species individually, excluding the outliers determined previously. In this case, 10 Mononegavirales species were used for the analysis involving mRNA abundance (see Table 2 in the supplemental material), and 13 species were used for the correlation test of the relative gene position and evolutionary parameters (see Table S3 in the supplemental material). Outlier detection in individual species data sets yielded similar results (data not shown). All statistical analyses were performed using SPSS 13.0 (SPSS, Inc., Chicago, IL).
Table 2.
Analyses of association between relative mRNA abundance and d, dN/dS, dN, and dS values of the coding regions in Mononegavirales speciesa
| Virus | No. of sequences |
d |
dN/dS |
dN |
dS |
Modificationb | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | rmod | rnov | r | rmod | rnov | r | rmod | rnov | r | rmod | rnov | |||
| Bornaviridae | ||||||||||||||
| Borna disease virus | 12 | |||||||||||||
| Rhabdoviridae | ||||||||||||||
| Vesicular stomatitis virus | 7 | –0.10 | –0.99* | –0.76* | 0.02 | –0.82* | –0.29 | 0.08 | –0.91* | –0.79* | –0.09 | –0.45 | –0.30 | (P) L |
| Rabies virus | 60 | –0.42 | –0.94* | –0.84 | –0.97* | –0.29 | –0.85* | –0.07 | 0.78 | P or L | ||||
| Filoviridae | ||||||||||||||
| Ebola virus | 18 | –0.48 | –0.83* | –0.32 | –0.67* | –0.65 | –0.90* | –0.27 | –0.05 | VP24 | ||||
| Pneumoviridae | ||||||||||||||
| Avian/human metapneumovirus | 19 | |||||||||||||
| Respiratory syncytial virus | 11 | –0.34 | –0.72* | –0.32 | –0.29 | –0.31 | –0.24 | - | –0.02 | 0.20 | G, L | |||
| Paramyxoviridae | ||||||||||||||
| Newcastle disease virus | 110 | 0.20 | –0.97* | –0.82* | 0.36 | –0.92* | –0.83* | 0.30 | –0.94* | –0.99* | –0.60 | –0.76 | –0.76 | (P) L |
| Sendai virus | 6 | –0.05 | –0.93* | –0.73* | –0.06 | –0.99* | –0.79* | –0.16 | –0.98* | –0.97* | –0.43 | –0.93* | –0.66 | (P) L |
| Mumps virus | 23 | –0.22 | –0.69 | –0.22 | –0.08 | –0.53 | –0.45 | –0.22 | –0.42 | –0.58 | –0.43 | –0.28 | –0.53 | L |
| Parainfluenza virus 3 | 12 | –0.43 | –0.87* | –0.83* | –0.04 | –0.75* | –0.93* | –0.19 | –0.71* | –0.90* | –0.40 | –0.92* | –0.54 | (P) L |
| Canine distemper virus | 18 | –0.48 | –0.93* | –0.92* | 0.12 | –0.25 | –0.72* | –0.25 | –0.98* | –0.80* | –0.59 | –0.73 | –0.60 | L |
| Measles virus | 37 | –0.62 | –0.96* | –0.94* | –0.78* | –0.84* | –0.97* | –0.06 | –0.82* | –0.82* | –0.38 | –0.54 | –0.32 | (P) L |
| Nipah virus | 12 | |||||||||||||
r, Pearson's correlation coefficient; rmod, Pearson's correlation coefficient, excluding P, G, and/or L; rnov, Pearson's correlation coefficient using nonoverlapping regions of the P gene and excluding L. *, P < 0.05.
Excluded gene(s). Cases in which values obtained using the full-length P gene were substituted for those obtained considering only nonoverlapping regions of this gene are shown in parentheses.
RESULTS
Relative contribution of different genomic factors to protein evolution in the Mononegavirales.
We used a PCA to determine the relative importance of five genomic factors on the evolution of the Mononegavirales. This analysis creates new uncorrelated variables (PCs), which group correlated factors, thereby avoiding potentially artificial correlations due to redundancy of the genomic factors. This analysis revealed that PC1 reflected the relative abundance of mRNA and relative position of the gene within the genome, as shown by squared loadings higher than the significance threshold. This is indicative of a high degree of association between the PC, and these genomic factors. Squared loading was higher for mRNA level than for relative genomic position (0.96 versus 0.93, respectively). PC1 explained around two-thirds of the total variance in the five genomic factors (60.9%); hence, of the genomic factors analyzed here, the relative position of the gene and mRNA abundance are the most important. In addition, PC1 was the only one significantly correlated with dN, dN/dS, and d (R2 = 0.174–0.226) (Table 1, “All” columns).
Table 1.
PCA of five genomic factors and four evolutionary traits in 10 Mononegavirales species
| Parameter | PCA resultsa |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All |
mRNA abundance |
Relative position |
||||||||||||||
| 1 | 2 | 3 | 4 | 5 | Sum | 1 | 2 | 3 | 4 | Sum | 1 | 2 | 3 | 4 | Sum | |
| % Variance explained byb: | ||||||||||||||||
| dN/dS | 17.4* | 8 | 5.4 | 2.5 | 2.1 | 35.4 | 14.5* | 8.4 | 4.6 | 3.9 | 31.4 | 19.3* | 12.4 | 1.9 | 1.6 | 35.2 |
| dS | 4.4 | 1 | 0 | 7.6 | 5.3 | 18.3 | 7.6 | 4.6 | 2.1 | 0 | 14.3 | 5.9 | 4.9 | 2.6 | 1.7 | 15.1 |
| dN | 22.6* | 3.9 | 13.4 | 1.7 | 0.4 | 42 | 21.6* | 4.3 | 3.9 | 1.4 | 31.2 | 20.5* | 0 | 1.2 | 2.3 | 24 |
| d | 20.1* | 2.7 | 1.7 | 5.1 | 2.6 | 32.2 | 15.6* | 3.4 | 12.8 | 0 | 31.8 | 20.2* | 12.6 | 0 | 0.6 | 33.4 |
| % Association (PC variable)c | ||||||||||||||||
| Relative mRNA abundance | 96.3 | 0.6 | 1.3 | 0.0 | 1.9 | 100 | 96.1 | 0.0 | 0.0 | 3.9 | 100 | |||||
| Relative position | 93.6 | 0.0 | 3.0 | 3.4 | 0.0 | 100 | 95.5 | 0.6 | 2.2 | 1.6 | 100 | |||||
| Protein abundance | 0.0 | 97.9 | 1.0 | 0.6 | 0.5 | 100 | 0.0 | 97.8 | 0.5 | 1.7 | 100 | 0.6 | 97.8 | 0.5 | 1.1 | 100 |
| Effective no. of codons | 3.7 | 1.5 | 86.1 | 1.7 | 7.0 | 100 | 0.0 | 0.5 | 98.7 | 0.9 | 100 | 2.7 | 0.7 | 87.6 | 8.9 | 100 |
| Length | 4.4 | 0.9 | 7.4 | 84.6 | 2.6 | 100 | 4.4 | 2.0 | 1.0 | 92.5 | 100 | 2.0 | 1.6 | 9.0 | 87.4 | 100 |
| Expected valuesd | 39.6 | 20.2 | 19.7 | 18.1 | 2.4 | 100 | 25.1 | 25.1 | 25.0 | 24.8 | 100 | 25.2 | 25.2 | 24.8 | 24.8 | 100 |
| Total variancee | 60.9 | 18.8 | 8.4 | 7.8 | 4.1 | 100 | 62.4 | 20.3 | 11.4 | 5.9 | 100 | 58.3 | 19 | 14.2 | 8.5 | 100 |
“All” refers to PCA for the five genomic factors used in this study. “mRNA abundance” refers to PCA, excluding the relative position as a genomic factor. “Relative position” refers to PCA, excluding relative mRNA abundance as a genomic factor. Individual PCs or collective PCs (Sum) are specified in the column subheadings. Boldfacing indicates a significant association based on broken-stick model thresholds. *, Significant linear correlation (P < 0.05).
That is, the percentage of the variance in each evolutionary factor is explained by the variance of each PC.
That is, the squared loadings of each genomic factor in each PC, representing the degree of association between both variables.
Squared loading thresholds were obtained by using the broken-stick model, determining significant association between PCs and genomic factors.
That is, the percentage of the total variance explained by each PC.
Due to the stepwise attenuation of gene transcription common to the Mononegavirales, the relative gene position and mRNA expression level largely reflect the same biological process. Indeed, both variables were not only associated with the same PC but also highly correlated in our data set (r = −0.75; P < 10−3). To assess the individual impact of these two variables on protein evolution, we also performed an independent PCA for each. When the relative gene position was excluded from the analysis, PC1, this time only reflecting the relative abundance of mRNA (squared loading = 0.96), explained ca. 62% of the total variance in the four genomic factors. Minor PCs 2, 3 and 4, comprising the protein abundance, the effective number of codons, and gene length, respectively (squared loadings > 0.92), each explained less than 20.3% of the variance. Interestingly, regression of these PCs against selection pressure and genetic distance revealed that only PC1 explained a significant fraction of the variance in dN, dN/dS, and d (R2 = 0.145 to 0.216). None of the four PCs was found to affect dS (Table 1, “mRNA abundance” columns). Similar results were obtained, excluding the mRNA abundance level from the analysis. In this case, PC1, reflecting the relative position of the gene within the genome, explained close to 58% of the total variance of the four variables and was the only one that significantly explained a fraction of the variation in dN, dN/dS, and d (R2 = 0.193 to 0.205) (Table 1, “Relative position” columns).
To examine the possible effects of assuming a linear model in the regression, we repeated our analyses using data ranks for mRNA level. The results of this analysis did not significantly differ from the parametric case (data not shown). However, the loadings of mRNA abundance onto PC1 were much lower, and this variable moved from being highly associated with PC1 to become a worse predictor than relative gene position (a rank variable by definition) (data not shown). Hence, information is contained in the relative magnitude of the mRNA expression level. Consequently, we focused on mRNA expression level for further analysis, considering gene relative position as an associated variable.
Overall, these results suggest that the level of mRNA expression, and its associated variable the relative position of the gene in the genome, is a major determinant of protein evolution in the Mononegavirales. Interestingly, the minor role of gene length in the PCA suggests that longer sequences do not contain higher variability.
Association between level of mRNA expression and genetic distance.
Genetic distances (d) were estimated for each coding region in each species and for the concatenated coding regions within each species (Fig. 1). In coding regions, d ranged from 0.016 to 0.477, with 80% of these values being <0.2.
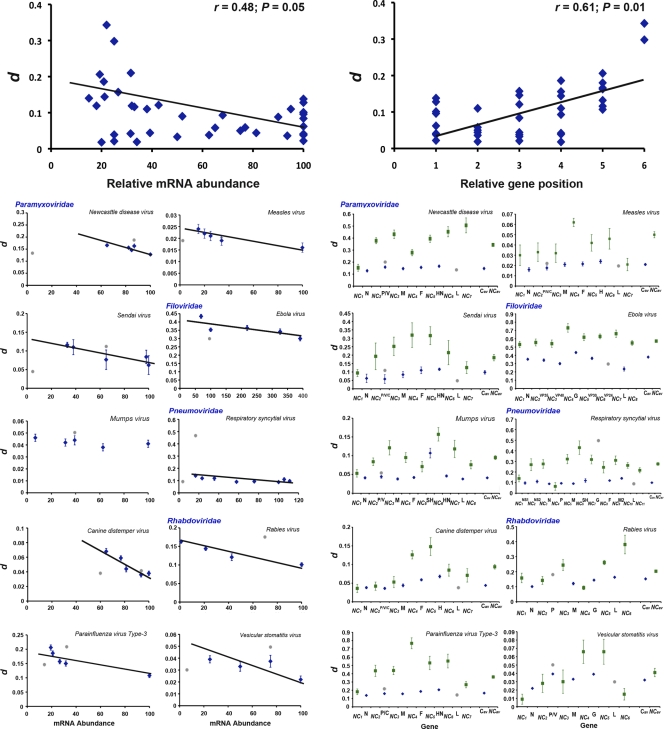
Fig 1.
Correlation between genetic distance (d) and relative mRNA abundance (left) or relative gene position (right), considering all of the Mononegavirales species together and each species individually. Coding regions are represented by blue diamonds, and noncoding regions are represented by green squares. Gray dots indicate outlier values. Values are means ± the standard deviations for each data set. Note the different scale in each panel.
The relationship between d and mRNA abundance was analyzed by Pearson correlation test considering all of the Mononegavirales species for which this information was available (10 species) as a whole. No significant association was found (r = 0.39; P = 0.17). This lack of correlation was due to a small number of outlier values (10/82), and their removal revealed a significant positive correlation between these two variables (r = 0.48; P = 0.05) (Fig. 1). A similar analysis using the relative gene position instead of mRNA abundance resulted in a significant correlation between this variable and the genetic distance after the removal of outliers (r = 0.61; P = 0.01) (Fig. 1).
Individual analyses for each species revealed that in most cases the outliers previously detected were the consequence of (i) lower levels of intraspecific genetic diversity in the L gene and (ii) higher values of d in the P and/or G genes than in the remaining coding regions (gray dots in Fig. 1). The exception was Ebola virus, for which the outlier value corresponded to the VP24 gene. In most species, the P gene encodes additional small overlapping proteins that may have affected our estimates of genetic distance. To address this possibility, we estimated d in nonoverlapping regions of the P gene and observed smaller values than in overlapping regions for all of the species studied (Fig. 1). To account for this effect, we considered both the complete P gene and the nonoverlapping region in further analyses.
Notably, we found no significant negative correlation between gene expression level and genetic diversity in any viral species (Table 2). However, this absence of correlation was again due to the levels of genetic diversity in the outliers described above, and their exclusion from the regression analyses resulted in significant positive correlation coefficients in 9 of 10 virus species (r ≥ −0.69; P ≤ 0.048). Mumps virus was the only species for which no significant correlation was found (r = −0.69; P = 0.201) (Fig. 1 and Table 2). Importantly, when the overall d values of the P genes were substituted by those in their nonoverlapping regions, these were not detected as outliers and significant correlations were obtained in 6 of 7 species. Equivalent analyses using the relative position in the genome gave similar results (Fig. 1 and Table 3), except for Ebola virus, for which no significant correlation was obtained (r = 0.47; P = 0.293; Table 3). This was expected given that mRNA levels do not strictly decrease 3′-5′ in this virus (see the introduction and Table S2 in the supplemental material). In addition, a significant correlation was observed in the three species for which gene expression level was not available (see Fig. S1 and Table S3 in the supplemental material).
Table 3.
Analysis of association between relative position in the genome and the genetic diversity of coding and noncoding regions in Mononegavirales speciesa
| Virus | No. of sequences | Coding regions |
Noncoding regions |
||||
|---|---|---|---|---|---|---|---|
| r | rmod | Modification | r | rmod | Modification | ||
| Bornaviridae | |||||||
| Borna disease virus | 12 | 0.87 (0.046) | 0.99 (0.012) | P | 0.74 (0.049) | 0.82 (0.042) | P |
| Rhabdoviridae | |||||||
| Vesicular stomatitis virus | 7 | 0.08 (0.904) | 0.99 (0.017) | L | 0.39 (0.447) | 0.95 (0.050) | 5′UTR |
| Rabies virus | 60 | 0.43 (0.469) | 0.99 (0.001) | P | 0.68 (0.137) | 0.93 (0.020) | G |
| Filoviridae | |||||||
| Ebola virus | 18 | –0.47 (0.293) | 0.65 (0.113) | 0.94 (0.005) | 5′UTR | ||
| Pneumoviridae | |||||||
| Avian/human metapneumovirus | 19 | 0.53 (0.176) | 0.77 (0.043) | L | 0.81 (0.008) | 0.81 (0.014) | F, 5′UTR |
| Respiratory syncytial virus | 11 | 0.24 (0.511) | 0.70 (0.041) | G, L | 0.38 (0.277) | - | |
| Paramyxoviridae | |||||||
| Newcastle disease virus | 110 | 0.01 (0.996) | 0.99 (0.012) | (P), L | 0.73 (0.048) | 0.80 (0.048) | P |
| Sendai virus | 6 | 0.24 (0.641) | 0.95 (0.014) | L | 0.66 (0.156) | 0.96 (0.011) | L, 5′UTR |
| Mumps virus | 23 | 0.20 (0.670) | - | - | 0.36 (0.379) | - | |
| Parainfluenza virus 3 | 12 | 0.09 (0.867) | 0.99 (0.003) | (P), L | 0.23 (0.613) | 0.90 (0.038) | F, 5′UTR |
| Canine distemper virus | 18 | 0.45 (0.273) | 0.94 (0.017) | L | 0.52 (0.235) | 0.94 (0.020) | L, 5′UTR |
| Measles virus | 37 | 0.53 (0.283) | 0.78 (0.022) | L | 0.52 (0.912) | 0.93 (0.023) | F, 5′UTR |
| Nipah virus | 12 | 0.33 (0.522) | 0.82 (0.009) | (P) | 0.35 (0.444) | 0.83 (0.053) | P, 5′UTR |
r, Pearson correlation coefficient (the P value is indicated in parentheses, and the correlation was calculated using all genes); rmod, Pearson correlation coefficient (the P value is indicated in parentheses, and the genetic distance was calculated excluding the P, G, and/or L genes). “Modification” columns refer to excluded gene(s) or noncoding region(s). Internal noncoding regions are named by the gene located downstream. Cases in which values obtained using the full-length P gene were substituted for those obtained considering only nonoverlapping regions of this gene are shown in parentheses.
In summary, these results suggest that level of gene expression is an important factor in determining the extent of genetic diversity in the Mononegavirales, in agreement with the PCA. However, other factors might account for the variability of P, G, and L, since the genetic diversity of these genes does not correlate with their expression level.
Association between mRNA expression level and protein evolution.
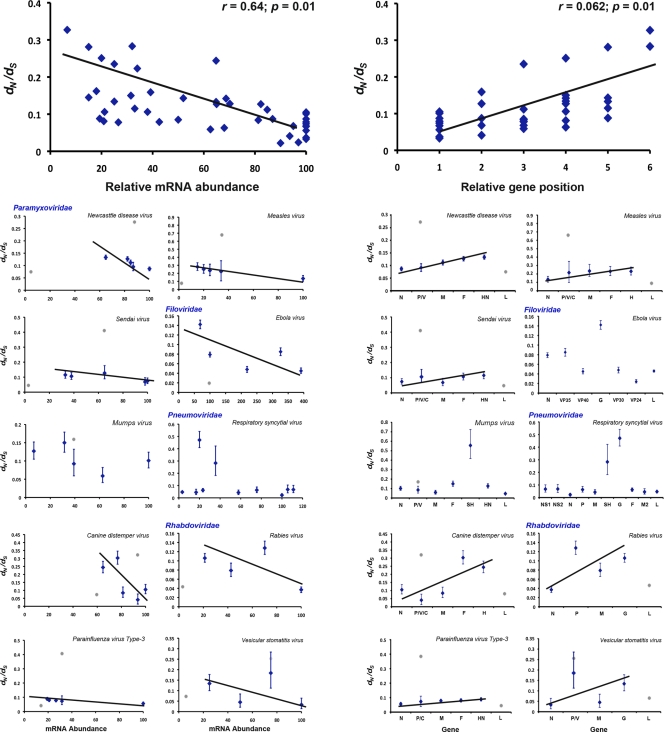
The selection pressures acting on each gene in each species were measured as the dN/dS ratio and individual dN values. Overall, the dN/dS ranged from 0.008 to 0.726, with 80% of the values being <0.2. Hence, the Mononegavirales are generally subject to relatively strong purifying selection. When all of the species were considered together, we observed a significant correlation between mRNA expression level and dN/dS upon removal of outlier values (r = 0.64; P = 0.01), with similar results obtained in an analysis using relative gene position (r = 0.62; P = 0.01) (Fig. 2). We accounted for the effect of gene overlap in P by estimating dN/dS values for both the complete P gene and the nonoverlapping region. Again, estimates in the former were significantly higher than in the latter (Fig. 2).
Fig 2.
Correlation between gene dN/dS ratio and relative mRNA abundance (left) or relative gene position (right), considering all of the Mononegavirales species together and each species individually. Values are means ± confidence intervals, assuming a χ2 distribution. Gray dots indicate outlier values. Note the different scale in each panel.
Analyses for each species indicated that excluding outlier values resulted in a significant positive correlation in 7 of 10 species (Table 2). Interestingly, the P and G genes had consistently higher dN/dS values than other genes, while the lowest values were observed for the L gene, such that they again represent outliers (gray dots in Fig. 2). When values in nonoverlapping regions of P were considered, a significant correlation between selection pressure and gene expression level was obtained in 6 of 7 cases (Fig. 2 and Table 2). Hence, selection pressure in most Mononegavirales species is associated with relative mRNA abundance, again in accordance with our PCA. Similar results were obtained when relative position of each gene in the genome was used rather than mRNA abundance (Fig. 2 and see Table S5 in the supplemental material), with the three species not considered in the analysis of gene expression level showing the same trend as the remaining Mononegavirales (see Fig. S1 in the supplemental material).
Analysis of the correlation between gene expression level and dN generally mirrored the results described above. No significant correlation was found considering all genes for any of the species, but when the same genes as with the dN/dS analysis were excluded, or only nonoverlapping regions of P were considered, both variables were correlated in the same species (Table 2), this correlation being significant in 8 of 10 species. Similar results were obtained using relative gene position, with 10 of 13 species showing a significant correlation (see Table S5 in the supplemental material).
Finally, correlation tests between dS and the level of gene expression or the relative position of the gene within the genome indicated that, with the exception of Sendai virus and parainfluenza virus type 3, these traits were not associated in any species regardless of the number of genes considered (Table 2). A significant positive correlation between dS and relative position of the gene within the genome was observed in parainfluenza virus type 3 and measles virus (see Table S5 in the supplemental material).
Evolution of noncoding regions.
To analyze the pace of evolution in noncoding regions, genetic distances were estimated for these regions in each species individually and for the concatenated noncoding regions within each species. Values ranged over a much larger interval than in coding regions (from 0.009 to 0.767, with only 50% of the values under 0.2) (Fig. 1), suggesting that noncoding regions are subject to relatively relaxed selective constraints. To address this point further, we compared the average genetic distance in coding and noncoding regions, concatenating all of the fragments of each type in each species. The average d values in noncoding regions were significantly higher than those in coding regions in all species (Fig. 1). Furthermore, one-way analysis of variance using the type of region (coding or noncoding) as a factor and considering each species individually showed significant differences in all viruses (F ≥ 6.11; P ≤ 0.023). Noncoding regions are therefore less constrained than those that encode proteins.
Mononegavirales possess conserved regulatory elements at the beginning and at the end of each noncoding region that are important in controlling transcription (65). Due to the key role of these elements, it can be hypothesized that evolutionary constraints in noncoding regions will be proportional to those acting on the genes that they control. To address this question, we first analyzed the correlation between the relative genomic positions of the noncoding regions and their associated d values considering all of the species together. A significant positive correlation between these two factors was found when outliers were excluded (r ≥ 0.47; P ≤ 0.021) (see Fig. S1 in the supplemental material). When each species was analyzed individually, a significant correlation was observed in Borna disease virus, metapneumoviruses, and Newcastle disease virus (r ≥ 0.73; P ≤ 0.049) and, overall, the correlation coefficients were higher than in the coding regions (Table 3). Moreover, a significant correlation was found in 10 of 13 species when one or two noncoding regions were excluded as outliers, mostly the 5′ noncoding region and those preceding the P and L genes (Table 3). These results suggest that genetic diversity in noncoding regions is influenced by the position in the genome and, by extension, might be correlated with the expression level or relative position of the gene they are regulating.
It is possible that this correlation is in fact the consequence of an association between d and region length rather than genomic position; that is, short noncoding regions might contain only essential regulatory elements, while longer regions may contain nonessential elements and could therefore accommodate more genetic variation. To explore this possibility, we analyzed the correlation between noncoding fragment length and genetic distance and between length and relative position, either considering all of the species together or each one individually. No significant correlation was found with the exception of the two species of the Pneumoviridae (see Table S6 in the supplemental material). Hence, fragment length does not appear to influence the level of genetic diversity in noncoding regions.
Finally, we analyzed the extent of the correlation between the genetic diversity of a given gene and that of the noncoding region immediately upstream of it and which contains the starting sequence for its transcription. A significant correlation was observed in 4 of 13 species (Table 4). Interestingly, three of these species are closely related paramyxoviruses (subfamily Paramyxovirinae) with very similar genomic structures (23), although the biological correlates of this pattern are unclear. Therefore, even if gene position does have an influence on genetic distance, this effect varies between coding and noncoding regions.
Table 4.
Analysis of association between genetic diversity in coding and noncoding regions in Mononegavirales species
| Virus | No. of sequences |
da |
|
|---|---|---|---|
| r | P | ||
| Bornaviridae | |||
| Borna disease virus | 12 | 0.74 | 0.042 |
| Rhabdoviridae | |||
| Vesicular stomatitis virus | 7 | 0.20 | 0.741 |
| Rabies virus | 60 | –0.04 | 0.944 |
| Filoviridae | |||
| Ebola virus | 18 | 0.26 | 0.577 |
| Pneumoviridae | |||
| Avian/human metapneumovirus | 19 | 0.47 | 0.236 |
| Respiratory syncytial virus | 11 | 0.24 | 0.495 |
| Paramyxoviridae | |||
| Newcastle disease virus | 110 | 0.34 | 0.506 |
| Sendai virus | 6 | 0.80 | 0.048 |
| Mumps virus | 23 | 0.18 | 0.738 |
| Parainfluenza virus type 3 | 12 | 0.43 | 0.399 |
| Canine distemper virus | 18 | 0.92 | 0.010 |
| Measles virus | 37 | 0.56 | 0.244 |
| Nipah virus | 12 | 0.79 | 0.043 |
d, genetic distance; r, Pearson's correlation coefficient; P, significance of the correlation coefficient (values < 0.05 are indicated in boldface).
DISCUSSION
Using a methodology that avoids artificial correlations (12), we provide evidence here that the level of gene expression is an important determinant of the rate of protein evolution in the Mononegavirales. Among the factors considered in our PCA, mRNA expression level—or its proxy, the relative gene position within the genome—had the greatest impact on the variance in dN, dN/dS, and d. In contrast, protein length, protein abundance and Nc′ do not appear to contribute meaningfully to the variance of these evolutionary parameters. Although this is the first analysis of its kind for RNA viruses, our results are largely in agreement with those obtained in a variety of other organisms (13, 46, 54, 67).
Studies on protein evolution in yeast and bacteria (11, 12) have led to the idea that high levels of gene expression result in lower rates of protein evolution through translational selection, rather than through relative protein importance. Two complementary mechanisms have been proposed to explain this phenomenon. First, highly expressed proteins undergo selection for combinations of mutations that allow proper folding despite mistranslation, thereby shaping dN. Second, these proteins also experience selection for preferred codons that increase both the efficiency and the accuracy of translation, in turn influencing dS (1, 11). However, the situation appears to be different in the Mononegavirales, in which codon usage bias (measured here as Nc′) and protein abundance are poorly associated with dN, dN/dS, and d. Moreover, dS did not correlate with any of the genomic variables studied here, and these variables explained a percentage of variance in dS ∼2-fold lower than in the other traits examined (see Table 1). This suggests that translational selection in the Mononegavirales acts only through translational robustness. Although this is at odds with previous analyses of cellular organisms (12), in these cases proteins are translated and folded in a constant environment. This is evidently not the case for subcellular parasites such as viruses, which are heavily dependent on their ability to infect and replicate in the host cellular environment, and hence are subject to strong host selection. Since many viruses are able to infect different cell types, as well as different hosts (15), this exposure to different cellular environments may lead to trade-offs between optimal translational conditions, likely mitigating the impact of selection for translational efficiency or accuracy. Indeed, in many RNA viruses the selection on codon choice appears to be relatively weak and is not driven by the requirement to match viral codons with host tRNA anticodons (31). Conversely, the strong correlation between gene expression level and dN is compatible with selection for translational robustness, which may be a particularly potent evolutionary force for proteins expressed in various environments. Moreover, viral proteins are produced in very large numbers during virus infection, representing the majority of the host cellular proteome (37). Therefore, the cost of protein misfolding on virus fitness would be expected to be comparatively higher than in cellular organisms, and consequently selection for translational robustness is likely to be stronger in RNA viruses. Interestingly, we also observed a strong correlation between gene expression and genetic distance, seemingly in parallel with variation in the dN. This suggests that variation in dN is a major component of the differences in variability between genes with different degrees of expression, since dS values are not correlated with this trait and generally exhibit only minor fluctuations within each species. Interestingly, comparison of dN in a subset of 10 protein pairs indicated that those with higher relative abundance and lower relative mRNA level evolved more rapidly (data not shown). This is in agreement with a major role for translational robustness rather than a functional importance in protein evolution (11).
Despite the importance of expression level in determining the rate of protein evolution, it is clear that other forces are at play. These can be difficult to disentangle when the organism in question has many genes or protein-protein interactions and redundant functions. However, because of their limited gene number, RNA viruses provide a unique opportunity to understand the role of a variety of factors in shaping protein evolution. Accordingly, while we found a strong positive correlation between gene expression level (and its proxy of gene relative position) and dN, dN/dS, and d, some genes had to be excluded from the analysis to achieve significant correlations. Importantly, similar trends were observed when the Mononegavirales were analyzed as a whole, suggesting that these results are robust. Moreover, they are in agreement with our PCA, in which all genes were considered. However, the presence of outlier genes does indicate that protein-specific factors other than expression level have at least some impact on protein evolution. Indeed, we have not considered several protein characteristics, such as centrality or dispensability, that have been proposed to partially determine the rate of protein evolution (9, 21, 25, 33, 68), albeit with a relatively weak effect (12, 63). Although centrality has been analyzed in several viruses (10, 30, 42), its effect on evolutionary parameters has been only addressed in hepatitis C virus (7). Dispensability has understandably not been addressed, since all of the proteins encoded in RNA virus genomes can be considered essential, and most sequence alterations result in deleterious effects (27).
The outlier genes that prevented a significant correlation between mRNA abundance/gene relative position and d, dN, and dN/dS were generally P, G, and L. The P protein, involved in RNA synthesis and encapsidation, is the least conserved protein among the Mononegavirales (41), as reflected in its capacity to undergo multiple amino acid changes or partial deletions without reducing its functionality (28, 55). This observation is perhaps surprising, considering that in many Mononegavirales the P gene encodes more than one protein. These proteins, such as the C and V proteins of the paramyxoviruses, are encoded in overlapping reading frames and expressed through RNA editing or the pseudotemplated addition of nucleotides (37). As a consequence, strong negative selection might be expected, since mutations in overlapping reading frames affect more than one gene and are likely to be deleterious (40). However, in our analysis the high variability of the P protein is maintained despite the presence of overlapping reading frames, perhaps because negative selection is strongest on the additional proteins encoded by this gene (32). The reduced dN/dS ratios observed in nonoverlapping regions of the P gene in most species analyzed supports this idea. Indeed, the rate of protein evolution in nonoverlapping regions exhibited values largely in agreement with what would be expected relative to their expression level.
While the trend for the P gene was general to all virus species, the G gene was detected as an outlier in only two species (Ebola virus and respiratory syncytial virus). The G protein acts as the attachment protein and is the major antigenic determinant of the Mononegavirales (37). Due to this interaction with the host immune system and to the presence of several hypervariable regions, the G protein is thought to be either under weaker negative selection or stronger positive selection than other proteins (70). That our analysis revealed higher d, dN, and dN/dS values for the G gene than expected from relative mRNA abundance suggests that in some species selection pressures related to its function, rather than level of gene expression, are the major determinant of its evolution.
Less genetic diversity than expected based on gene position was observed in the L gene, which encodes an RNA-dependent RNA polymerase (RdRp). The obvious functional importance of the RdRp likely explains its constrained amino acid sequence, with a number of functional motifs highly conserved across all RNA viruses (35, 48). Our study indicates that, despite being the least expressed, the L gene is the most conserved in the Mononegavirales. Similarly, the VP24 gene of Ebola virus was subject to stronger evolutionary constraints than expected based on its expression level. This protein is involved in many aspects of viral infection, such as nucleocapsid formation (29), viral budding or assembly (22), host range determination (59), and genome replication and transcription, as well as in blocking interferon signaling (52), which might explain its lower evolutionary rate. Hence, functional constraints may be the main factors shaping levels of genetic diversity in these two proteins. However, we cannot exclude that the differences observed for these proteins (P, G, L, and VP24) could also reflect, in part, variations in evolvability. Tolerance to amino acid substitutions has indeed been shown to vary between proteins (5) and be linked to protein structure and stability (4, 61). Interestingly, selection for translational robustness could also act on these traits and contribute to differences in mutational robustness (19).
There is a marked lack of experiment-based literature on the effect of gene expression on aspects of viral fitness and evolution, including for the Mononegavirales. To date, most research has focused on the fitness consequences of recombinant variants of vesicular stomatitis virus and rabies virus with rearranged genes (2, 44, 64, 66). Although these studies did not consider the role of gene expression in the pace of protein evolution, they did reveal that fitness is not correlated with the level of gene expression of P and G (2, 44, 66), two of our outlier genes, and which in turn suggests that the expression level is not a major evolutionary determinant in these genes. Conversely, virus fitness was associated with the expression level of the N and M genes (2, 44, 64), for which we propose that gene expression has an importantly evolutionary effect. As such, these experiments provide indirect support for the conclusions we draw here. However, it is also clear that a direct experimental test of the results obtained from our in silico analysis is an important avenue for future research.
Finally, our results indicate that not only are noncoding regions less conserved than those that encode proteins, but their evolutionary rates are associated with their relative genomic position (but not their length). Although the variation in d is not proportional between genes and the corresponding upstream noncoding region, the gradient in evolutionary rates with relative position follows the same trend in both coding and noncoding regions, perhaps because the latter contain key regulatory signals for gene expression. Interestingly, this correlation was also observed in Ebola virus for which there is no apparent 3′-5′ gradient in mRNA levels. This suggests that, at least for this virus, the relative position in the genome, rather than the expression level of the adjacent coding regions, contributes to the observed constraints on the evolution of the noncoding regions.
Our comparative study has taken advantage of the unique characteristics of the Mononegavirales. Although the processes described here contribute to our understanding of the evolution of this viral order, determining the generality of our observations will clearly require estimates of both genomic and evolutionary parameters in a wide array of virus species.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a Marie Curie Fellowship PIOF-GA-2009-236470 to I.P. and National Institutes of Health grant R01 GM080533-5 to E.C.H.
We thank Anthony Schmitt for assistance with an earlier draft of the manuscript and two anonymous reviewers for useful comments.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Akashi H. 2003. Translational selection and yeast proteome evolution. Genetics 164:1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ball LA, Pringle CR, Flanagan B, Perepelitsa VP, Wertz GW. 1999. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J. Virol. 73:4705–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloom JD, Drummond DA, Arnold FH, Wilke CO. 2006. Structural determinants of the rate of protein evolution in yeast. Mol. Biol. Evol. 23:1751–1761 [DOI] [PubMed] [Google Scholar]
- 4. Bloom JD, et al. 2007. Evolution favors protein mutational robustness in sufficiently large populations. BMC Biol. 5:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bloom JD, et al. 2005. Thermodynamic prediction of protein neutrality. Proc. Natl. Acad. Sci. U. S. A. 102:606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brookfield JFY. 2000. Evolution: what determines the rate of sequence evolution? Curr. Biol. 10:R410–R411 [DOI] [PubMed] [Google Scholar]
- 7. Campo DS, Dimitrova Z, Mitchell RJ, Lara J, Khudyakov Y. 2008. Coordinated evolution of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:9685–9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chantawannakul P, Cutler RW. 2008. Convergent host-parasite codon usage between honeybee and bee associated viral genomes. J. Invertebr. Pathol. 98:206–210 [DOI] [PubMed] [Google Scholar]
- 9. Chen Y, Xu D. 2005. Understanding protein dispensability through machine-learning analysis of high-throughput data. Bioinformatics 21:575–581 [DOI] [PubMed] [Google Scholar]
- 10. Doolittle JM, Gomez SM. 2011. Mapping protein interactions between dengue virus and its human and insect hosts. PLoS Negl. Trop. Dis. 5:e954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc. Natl. Acad. Sci. U. S. A. 102:14338–14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Drummond DA, Raval A, Wilke CO. 2006. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 23:327–337 [DOI] [PubMed] [Google Scholar]
- 13. Duret L, Mouchiroud D. 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17:68–74 [DOI] [PubMed] [Google Scholar]
- 14. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. 2005. Virus taxonomy: classification and nomenclature of viruses. 8th Report of the International Committee, 2nd ed Academic Press, Inc, San Diego, CA [Google Scholar]
- 16. Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. 2002. Evolutionary rate in the protein interaction network. Science 296:750–752 [DOI] [PubMed] [Google Scholar]
- 17. Garcia-Arenal F, McDonald BA. 2003. An analysis of the durability of resistance to plant viruses. Phytopathology 93:941–952 [DOI] [PubMed] [Google Scholar]
- 18. Geiler-Samerotte KA, et al. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc. Natl. Acad. Sci. U. S. A. 108:680–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldsmith M, Tawfik DS. 2009. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc. Natl. Acad. Sci. U. S. A. 106:6197–6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greenbaum BD, Levine AJ, Bhanot G, Rabadan R. 2008. Patterns of evolution and host gene mimicry in influenza and other RNA viruses. PLoS Pathog. 4:e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn MW, Kern AD. 2005. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol. Biol. Evol. 22:803–806 [DOI] [PubMed] [Google Scholar]
- 22. Han Z, et al. 2003. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77:1793–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harcourt BH, et al. 2001. Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287:192–201 [DOI] [PubMed] [Google Scholar]
- 24. Hartl DL, Dykhuizen DE, Dean AM. 1985. Limits of adaptation: the evolution of selective neutrality. Genetics 111:655–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hirsh AE, Fraser HB. 2001. Protein dispensability and rate of evolution. Nature 411:1046–1049 [DOI] [PubMed] [Google Scholar]
- 26. Hoelzer K, Shackelton LA, Parrish CR, Holmes EC. 2008. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J. Gen. Virol. 89:2280–2289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holmes EC. 2009. The evolution and emergence of RNA viruses. Oxford University Press, New York, NY [Google Scholar]
- 28. Hu C, Gupta KC. 2000. Functional significance of alternate phosphorylation in Sendai virus P protein. Virology 268:517–532 [DOI] [PubMed] [Google Scholar]
- 29. Huang Y, Xu L, Sun Y, Nabel GJ. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307–316 [DOI] [PubMed] [Google Scholar]
- 30. Jaeger S, Ertaylan G, van Dijk D, Leser U, Sloot P. 2010. Inference of surface membrane factors of HIV-1 infection through functional interaction networks. PLoS One 5:e13139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jenkins GM, Holmes EC. 2003. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 92:1–7 [DOI] [PubMed] [Google Scholar]
- 32. Jordan IK, Sutter BAT, McClure MA. 2000. Molecular evolution of the Paramyxoviridae and Rhabdoviridae multiple-protein-encoding P gene. Mol. Biol. Evol. 17:75–86 [DOI] [PubMed] [Google Scholar]
- 33. Jovelin R, Phillips PC. 2009. Evolutionary rates and centrality in the yeast gene regulatory network. Genome Biol. 10:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karlin S, Blaisdell BE, Schachtel GA. 1990. Contrasts in codon usage of latent versus productive genes of Epstein-Barr virus: data and hypotheses. J. Virol. 64:4264–4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koonin EV. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197–2206 [DOI] [PubMed] [Google Scholar]
- 36. Kosakovsky Pond SL, Frost SD. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533 [DOI] [PubMed] [Google Scholar]
- 37. Lamb R. 2007. Mononegavirales, p 1357–1361 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott/Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 38. Lamb R, Parks G. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott/Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 39. Marais G, Duret L. 2001. Synonymous codon usage, accuracy of translation, and gene length in Caenorhabditis elegans. J. Mol. Evol. 52:275–280 [DOI] [PubMed] [Google Scholar]
- 40. Miyata T, Yasunaga T. 1978. Evolution of overlapping genes. Nature 272:532–535 [DOI] [PubMed] [Google Scholar]
- 41. Morgan E. 1991. Evolutionary relationships of paramyxovirus nucleocapsid-associated proteins, p 163–176 In Kingsbury DW. (ed), The paramyxoviruses. Plenum Press, Inc, New York, NY [Google Scholar]
- 42. Munday DC, et al. 2010. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell Proteomics 9:2438–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neumann G, Watanabe S, Kawaoka Y. 2009. Characterization of Ebola virus regulatory genomic regions. Virus Res. 144:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Novella IS, Ball LA, Wertz GW. 2004. Fitness analyses of vesicular stomatitis strains with rearranged genomes reveal replicative disadvantages. J. Virol. 78:9837–9841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Novembre JA. 2002. Accounting for background nucleotide composition when measuring codon usage bias. Mol. Biol. Evol. 19:1390–1394 [DOI] [PubMed] [Google Scholar]
- 46. Pal C, Papp B, Hurst LD. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158:927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peres-Neto PR, Pedro R, Jackson DA, Somers KM. 2003. Giving meaningful interpretation to ordination axes: assessing loading significance in principal component analysis. Ecology 84:2347–2363 [Google Scholar]
- 48. Poch O, Sauvaget I, Delarue M, Tordo N. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 50. Rambaut A. 1996. Se-Al: sequence alignment editor. Department of Zoology, University of Oxford, Oxford, United Kingdom: http://evolve.zoo.ox.ac.uk/ [Google Scholar]
- 51. Rasband WS. 1997–2011. ImageJ. National Institutes of Health, Bethesda, MD [Google Scholar]
- 52. Reid SP, et al. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Restif O. 2009. Evolutionary epidemiology 20 years on: challenges and prospects. Infect. Genet. Evol. 9:108–123 [DOI] [PubMed] [Google Scholar]
- 54. Rocha EP, Danchin A. 2004. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol. Biol. Evol. 21:108–116 [DOI] [PubMed] [Google Scholar]
- 55. Ryan KW, Portner A. 1990. Separate domains of Sendai virus P protein are required for binding to viral nucleocapsids. Virology 174:515–521 [DOI] [PubMed] [Google Scholar]
- 56. Sanchez A, Geisbert T, Feldman H. 2007. Filoviridae: Marburg and Ebola viruses, p 1409–1448 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed Lippincott/Williams & Wilkins Co, Philadelphia, PA [Google Scholar]
- 57. Sanchez A, Kiley MP. 1987. Identification and analysis of Ebola virus messenger RNA. Virology 157:414–420 [DOI] [PubMed] [Google Scholar]
- 58. Sokal RR, Rohlf FJ. 1995. Biometry: the principles and practices of statistics in biological research. WH Freeman, New York, NY [Google Scholar]
- 59. Sullivan NJ, Sanchez A, Rollin PE, Yang ZY, Nabel GJ. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605–609 [DOI] [PubMed] [Google Scholar]
- 60. Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 61. Tokuriki N, Tawfik DS. 2009. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19:596–604 [DOI] [PubMed] [Google Scholar]
- 62. Wall DP, et al. 2005. Functional genomic analysis of the rates of protein evolution. Proc. Natl. Acad. Sci. U. S. A. 102:5483–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Z, Zhang J. 2009. Why is the correlation between gene importance and gene evolutionary rate so weak? PLoS Genet. 5:e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wertz GW, Perepelitsa VP, Ball LA. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 95:3501–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61–119 [DOI] [PubMed] [Google Scholar]
- 66. Wirblich C, Schnell MJ. 2011. Rabies virus (RV) glycoprotein expression levels are not critical for pathogenicity of RV. J. Virol. 85:697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wright SI, Yau CB, Looseley M, Meyers BC. 2004. Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol. Biol. Evol. 21:1719–1726 [DOI] [PubMed] [Google Scholar]
- 68. Zhang J, He X. 2005. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol. Biol. Evol. 22:1147–1155 [DOI] [PubMed] [Google Scholar]
- 69. Zhou J, Liu WJ, Peng SW, Sun XY, Frazer I. 1999. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J. Virol. 73:4972–4982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zlateva KT, Lemey P, Moes E, Vandamme AM, Van Ranst M. 2005. Genetic variability and molecular evolution of the human respiratory syncytial virus subgroup B attachment G protein. J. Virol. 79:9157–9167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zuckerkandl E. 1976. Evolutionary processes and evolutionary noise at the molecular level. I. Functional density in proteins. J. Mol. Evol. 7:167–183 [DOI] [PubMed] [Google Scholar]
- 72. Zuckerkandl E, Pauling L. 1965. Evolutionary divergence and convergence in proteins, p 97–166 In Bryson V, Vogel HJ. (ed), Evolving genes and proteins. Academic Press, Inc, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.