Abstract
MicroRNAs (miRNAs) are a class of noncoding small RNAs that regulate multiple cellular processes, as well as the replication and pathogenesis of many DNA viruses and some RNA viruses. Examination of cellular miRNA profiles in West Nile virus (WNV)-infected HEK293 and SK-N-MC cells revealed increased expression of multiple miRNA species. One of these miRNAs, Hs_154, was significantly induced not only in WNV-infected neuronal cells in culture but also in the central nervous system tissues of infected mice and, upon transfection, caused a significant reduction in viral replication. Analysis of mRNA transcripts enriched through immunoprecipitation of the RNA-induced silencing complex identified several transcripts that contain seed sequence matches to Hs_154 in their 3′ untranslated regions (UTRs). Two of these targets, the CCCTC-binding factor (CTCF) and the epidermal growth factor receptor (EGFR)-coamplified and overexpressed protein (ECOP/VOPP1) proteins display reduced expression in WNV-infected cells, and the 3′ UTRs of these transcripts were sufficient to cause downregulation of expression in infected cells or in cells transfected with Hs_154, findings consistent with miRNA targeting of these transcripts. CTCF and ECOP have been shown to be associated with cell survival, implicating miRNA-directed repression of these targets in WNV-induced cell death. Consistent with this hypothesis, expression of these genes in WNV-infected cells results in a reduction in the number of cells undergoing apoptosis. These observations suggest that induction of Hs_154 expression after WNV infection modulates the apoptotic response to WNV and that cellular miRNA expression can be quickly altered during WNV infection to control aspects of the host response.
INTRODUCTION
West Nile virus (WNV) is a member of the flavivirus family that has emerged as a significant threat to the U.S. population since its introduction into North America in 1999 (3, 34). Although the majority of infections remain asymptomatic or cause mild febrile illness in healthy individuals, viral neuroinvasion can result in meningitis or encephalitis, in some cases leading to long-term neurologic sequelae with both physical and cognitive manifestations or death (27). Immunocompromised and aged populations have been shown to be at greatest risk for neuroinvasion and more severe forms of WNV-associated disease (41). To date, over 30,000 cases of WNV disease have been reported in the United States (9). Of these, ca. 40% represent cases of WNV neuroinvasive disease.
The cellular response to WNV infection involves induction of the interferon response through the triggering of various pathogen-associated molecular pattern recognition receptors, such as RIG-I/IPS-1, TLR3, and PKR. In addition, WNV infection initiates cellular apoptosis through a variety of caspase-dependent mechanisms, including induction of the unfolded protein response (UPR) (32), interaction of WNV proteins with apoptosis factors (42, 57, 58), and activation of inflammatory pathways (51). Cell death via these mechanisms contributes to the neuropathogenicity observed in WNV infection (44).
The recent identification and functional characterization of miRNAs has revealed a previously underappreciated mechanism of posttranscriptional gene regulation in mammalian cells. miRNAs are 21- to 23-nucleotide single-stranded RNAs. They are derived from larger transcripts containing stem-loop structures, which are processed by the RNase III enzymes Drosha and Dicer into 22-bp duplexes. A single strand of the duplex (the mature microRNA [miRNA]) associates with a multiprotein complex termed the RNA-induced silencing complex (RISC). In this context, the miRNA acts as guide strand, directing the RISC to targeted mRNAs, resulting in inhibition of translation initiation and destabilization of the mRNA through deadenylation (5). Targeting of an individual mRNA is dependent on Watson-Crick base pairing of nucleotides 2 to 8 of the miRNA (the “seed sequence”) with a complementary region in the 3′ untranslated region (3′UTR) of the mRNA.
Several of the DNA viruses, most notably members of the herpesvirus family, encode miRNAs that function to regulate both viral and cellular gene expression (reviewed in reference 50). In general, cytoplasmic RNA viruses such as flaviviruses are not believed to express their own miRNAs (although small RNAs derived from the viral genome can be identified in infected cells [38]). However, recent studies have demonstrated that viral infection can result in changes in expression of cellular miRNAs (9, 24, 33, 42, 45, 55, 58). Several reports have demonstrated that specific miRNAs can exert positive or negative influences on viral replication through direct interaction with viral nucleic acid sequences (25, 28, 37, 40), while other reports have demonstrated that individual miRNAs can affect viral replication via targeting cellular proteins (24, 50). Because individual miRNAs have the potential to regulate the expression of multiple mRNAs, the changes in miRNA expression following viral infection are predicted to have profound effects on the host response and may represent either an arm of the innate response to viral infection or may be induced by the virus in order to promote a cellular environment more conducive to viral replication.
We report here changes in cellular miRNA expression induced by WNV infection and the role that the most highly induced miRNA, Hs_154, plays in the virus-stimulated apoptosis via miRNA-mediated repression of two proteins, CCCTC binding factor (CTCF) and EGFR-coamplified and overexpressed protein (ECOP). We demonstrate that transfection of a Hs_154 mimic significantly reduces viral replication, suggesting that induction of this miRNA is a cellular antiviral response to infection. Since virus-induced apoptosis itself contributes to WNV-induced pathogenesis (44), Hs_154, and perhaps additional cellular miRNAs, may be key factors in directing the pathogenic outcome of WNV infection in the host.
MATERIALS AND METHODS
Virus strains and cell culture.
HEK293 and SK-N-MC cells were grown in modified Eagle medium (Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone), 2 mM l-glutamine (Invitrogen), 100 U of penicillin G sodium/ml, 100 μg of streptomycin sulfate (Invitrogen)/ml, and 1× nonessential amino acids (Gibco). HeLa, Huh7, and Huh7.5 cells were grown in Dulbecco modified Eagle medium supplemented with 10% FBS, 2 mM l-glutamine, 100 U of penicillin G sodium/ml, and 100 μg of streptomycin sulfate/ml. WNV 385-99 (55) and dengue virus 2 (New Guinea C strain; American Type Culture Collection) were passaged twice on C6/36 cells and purified by centrifugation as previously described (31). Virus titers were determined using a by focus formation assay. Serial dilutions of virus were plated on Vero cells and allowed to adsorb for 1 h, followed by overlay with 0.5% carboxymethyl cellulose (Sigma). At 48 h postinfection (p.i.), the cells were fixed with 4% paraformaldehyde, washed twice with phosphate-buffered saline (PBS), blocked, and permeabilized for 1 h in PBS supplemented with 2% normal goat serum (NGS; Sigma) and 0.4% Triton X-100. The cells were then incubated with 0.3 μg of anti-flavivirus monoclonal antibody 4G2 (22)/ml in PBS supplemented with 2% NGS for 1 h, washed twice more with PBS, incubated with anti-mouse IgG-horseradish peroxidase (Santa Cruz Biotech) for 1 h, and washed twice with PBS. Foci were visualized by incubation using a Vector VIP peroxidase substrate kit (Vector Labs) according to the manufacturer's specifications.
All infections were performed by incubation of the virus at the indicated multiplicities of infection (MOIs) in medium supplemented with 2% FBS at a low volume for 1 h with rocking. The cells were then washed with PBS and refed with complete growth medium.
miRNA microarray.
HEK293 cells and SK-N-MC cells were infected with WNV at a multiplicity of 3 PFU/cell. At 48 h p.i., the total RNA was isolated with TRIzol (Invitrogen) according to the manufacturer's protocol. RNA was prepared for hybridization using a miRNA expression profiling reagent kit (Illumina) according to the manufacturer's protocol. Briefly, 200 ng of total RNA was polyadenylated with poly(A) polymerase. The polyadenylated RNA was converted to cDNA using a biotinylated oligo(dT) primer with a universal PCR sequence at its 5′ end. The biotinylated cDNA was hybridized to chimeric oligonucleotides containing (i) a universal PCR priming site on the 5′ end, (ii) an address sequence that complements a specific capture sequence on the array, and (iii) a miRNA-specific sequence. This hybridization complex was recovered using streptavidin-conjugated paramagnetic beads. The chimeric oligonucleotide was extended with DNA polymerase. The complex was amplified by PCR using the incorporation of a primer with a fluorescent tag. The PCR products were hybridized to a miRNA bead array composed of oligonucleotides complementary to the address sequence. The array was scanned on a BeadArray scanner (Illumina), and the data were prepared using Genome Studio. The heat map was constructed using Multi-Experiment Viewer software (43).
Reagents.
Where indicated, universal type I interferon (PBL Biomedical Laboratories) was added to cells to a final concentration of 1,000 U/ml. QVD-OPh (RnD Systems, catalog no. OPH001-01M) was added to 20 μM. The antibodies used here were anti-dicer (Cell Signaling Technologies, catalog no. 3363), anti-Drosha (Cell Signaling Technologies, catalog no. 3364) anti-CTCF (Cell Signaling Technologies, catalog no. 2899S), anti-FLAG (Sigma, catalog no. F3165), anti-β-actin (Sigma, catalog no. A5441), anti-PARP (Cell Signaling Technologies, catalog no. 9532), and anti-firefly luciferase (Sigma, catalog no. L2164).
Plasmids.
The 3′UTRs of CTCF and ECOP were amplified by PCR (CTCF-A, 5′-ATCCTAGGCTTGTGCGTCGCCAGGACTTC-3′; CTCF-B, 5′-ATGAGCTCGGAGCACTTGTTAATCCGTTA-3′; ECOP-A, 5′-CTCGAGGTGCAAGAGGAGAGACAGGAG-3′; and ECOP-B, 5′-GCGGCCGCGCACACATAGGAAACAGGATC-3′), digested, and ligated directly into pMirKan (Ambion), with the ampicillin cassette replaced with a kanamycin cassette as previously described (20), or psiCheck2 (Promega). Full-length CTCF and ECOP clones were produced by PCR amplification (CTCF, 5′-AAGAATTCACCATGGAAGGTGATGCAGTCGAA-3′ and 5′-TTCTCGAGTCACCGGTCCATCATGCT-3′; and ECOP, 5′-GGAAGCTTATGAGGCGCCAGCCTGCG-3′ and 5′-TTGCGGCCGCCTACTTGTCGTCATCCTTCACTAC-3′). In the case of ECOP, a Flag tag was incorporated at the 3′ end during amplification to facilitate visualization. PCR products were digested and ligated into pcDNA3 (Invitrogen). Mutants were produced by site-directed mutagenesis of Hs_154 seed sites in CTCF and ECOP 3′UTRs (CTCFm1, positions 431 to 437 of the 3′UTR; CTCFm2, positions 578 to 584; CTCFm3, positions 264 to 269; ECOPm1, positions 2148 to 2154; ECOPm2, positions 1941 to 1946).
miRNA Northern blotting.
Total RNA was separated on a 15% polyacrylamide–7 M urea gel, transferred to a GeneScreen Plus membrane (Perkin-Elmer), and cross-linked to the membrane with UV light. Probes were prepared by end labeling antisense oligonucleotides (Hs_154, 5′-TCCTCCCCCTTCCTTTTCCC-3′; Hs_243.1, 5′-TCTCCGCCGGGCCTTCAC-3′; Hs_188, 5′-TCCGCCGCTCCGCCTTCCGC-3′; miR-16, 5′-CGCCAATATTTACGTGCTGCTA-3′) with [γ-32P]dATP (Perkin-Elmer) using T4 polynucleotide kinase (Fermentas). Probes were hybridized to membranes overnight at 38°C in PerfectHyb buffer (Sigma) and visualized by autoradiography.
qRT-PCR.
Portions (25 ng) of total RNA or RISC-associated RNA were subjected to quantitative reverse transcription-PCR (qRT-PCR) using CTCF (Hs00902008_m1)-, ECOP (Hs00697470_m1)-, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Hs99999905_m1)-specific TaqMan gene expression assays and One-Step RT-PCR reagents on a StepOne Plus real-time PCR machine (Applied Biosystems) according to the manufacturer's instructions. The relative expression in samples was calculated by the ΔΔCT method using GAPDH as the endogenous control.
RISC-IP.
RISC-immunoprecipitation (RISC-IP) analysis was carried as out previously described (26). HEK293 cells stably transfected with c-myc-tagged argonaute 2 (Ago2) protein were infected with WNV at a multiplicity of 3 PFU/cell. At 48 h postinfection, the cells were lysed, and Ago2 and associated proteins and RNAs were immunoprecipitated using anti-c-myc antibody beads (Sigma). Immunoprecipitations were performed in duplicate using biological replicates. RNA was isolated using TRIzol and analyzed for quality by using an Agilent Bioanalyzer, and transcript levels were determined on an Illumina HumanRef-6 platform. Microarray data were analyzed using Gene Sifter software (Geospiza). Enrichment of specific transcripts, through association with miRNA-protein complexes was determined by dividing the immunoprecipitated levels of transcripts by the total levels as previously described (21).
Infection and analysis of mice.
B6.129S7-Rag1tm1Mom/J mice (33) were purchased from The Jackson Laboratory (Bar Harbor, ME), bred at the VGTI Vivarium (Oregon Health and Science University [OHSU]), and used at 12 weeks of age. All animals were housed and bred under specific-pathogen-free conditions at OHSU. All WNV experiments were completed within a U.S. Department of Agriculture-approved biosafety level 3 facility and were approved by the Institutional Animal Care and Use Committee (permit 0724) and the Institutional Biosafety Committee in accordance with the applicable federal, state, and local regulations. This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals (National Institutes of Health). Mice were infected with 250 PFU of WNV subcutaneously. At days 3 and 11 p.i., they were sacrificed, the brains were collected and homogenized, and the total RNA was isolated using TRIzol. Immunofluorescent staining was performed as previously described (31).
siRNA, miRNA, and plasmid transfections.
Small interfering RNAs (siRNAs) against Dicer and Drosha proteins (Applied Biosystems) were transfected into HEK293 cells with RNAiMAX reagent (Invitrogen). miRNA duplexes were synthesized by Integrated DNA Technologies and transfected using RNAiMAX reagent. The antisense locked-nucleic-acid (LNA) Hs_154 inhibitor (Exiqon; listed as the hsa-miRPlus-F1050 miRCURY LNA microRNA power inhibitor) and negative control (Exiqon; miRCURY LNA microRNA power inhibitor control, negative control A) were transfected with RNAiMAX and 50 nM LNA inhibitor. Poly(I:C) (Sigma, catalog no. P9582) transfections were performed with 2.5 μg of poly(I:C)/ml and Lipofectamine LTX reagent (Invitrogen). Plasmids expressing CTCF and ECOP cDNAs were transfected using FuGene reagent (Roche). All transfections were carried out according to the manufacturer's recommendations. Unless otherwise stated, samples were processed or infected at 48 h posttransfection. Cotransfections with Hs_154 miRNA duplex and psiCheck reporter plasmids were performed with Lipofectamine 2000 and processed for luciferase detection at 24 h posttransfection.
Luciferase assays.
HEK293 cells transfected with psiCheck2 reporter plasmids and negative control/Hs_154 were lysed at 24 h posttransfection. Luciferase assays were performed using a dual luciferase reporter assay system (Promega) according to the manufacturer's recommendations.
TUNEL assay.
Control, CTCF, ECOP/FLAG, control LNA, or Hs_154 LNA-transfected cells were infected with WNV at 48 h posttransfection. The cells were fixed at 48 h p.i. and stained for apoptotic cells using the DeadEnd Fluorimetric TUNEL system (Promega). The number of TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling)-positive cells per frame was determined using ImageJ software.
miRNA annotation in miRBase.
The sequences for Hs_154, Hs_188, and Hs_243 have been submitted to the public miRNA database miRBase (www.mirbase.org) and assigned the following names: Hs_154, hsa-mir-6124; Hs_188, has-mir-6125; and Hs_243, hsa-mir-6126.
RESULTS
WNV infection alters the expression level of multiple cellular miRNAs.
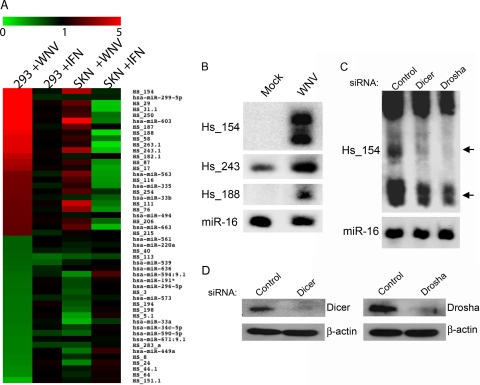
In order to examine the effect of WNV infection on miRNA expression, HEK293 and SK-N-MC cell lines were infected at a multiplicity of 3 PFU/cell, RNA was collected at 48 h p.i., and miRNA expression was examined by miRNA-specific microarray. This time postinfection has been reported to show maximal changes in mRNA expression relative to uninfected cells, including strong induction of multiple interferon-responsive genes (17). In parallel, we also treated both cell lines with type I interferon and analyzed RNA from these cells at 6 h posttreatment, at which time we would expect to see increased expression of multiple miRNAs (40). We observed differential expression of multiple miRNAs in WNV-infected cells compared to uninfected controls (Fig. 1A and Files S1 and S2 in the supplemental material). Surprisingly, the most highly induced miRNAs following WNV infection did not correspond to those upregulated after interferon treatment, suggesting that the induction observed is WNV specific or due to a cellular pathway distinct from the interferon response to viral infection. Induction of the interferon response was confirmed by qRT-PCR analysis of interferon-stimulated gene 54 (ISG54) expression (data not shown). Interestingly, several of the most highly induced miRNAs (e.g., Hs_154, Hs_243, and Hs_188; Fig. 1 and Table 1) in WNV-infected HEK293 and SK-N-MC cells were previously identified as low-abundance miRNAs present in RNA derived from fetal brain and other human tissues (6, 7). Upregulation of these miRNAs was confirmed by Northern blotting (Fig. 1B). Expression of miR-16 appeared to be unchanged following WNV infection and serves as a loading control.
Fig 1.
WNV alters expression of cellular miRNAs. (A) Heat map analysis showing cellular miRNA expression after the indicated treatment. Values indicate increased (red) or decreased (green) expression of miRNAs versus controls and are ranked in descending order based on the fold change in HEK293 cells infected with WNV. The 25 miRNAs showing the greatest induction of expression and the 25 miRNAs showing greatest repression of expression in WNV-infected HEK293 cells relative to uninfected controls are shown. miRNAs annotated “Hs” are described in references (6, 7). The annotation “0.1” appended to some indicates those described in reference 7. Cells were infected with WNV at a multiplicity of infection (MOI) of 3 PFU/cell or treated with type I interferon. At 48 h p.i., total RNA was extracted, and miRNA expression was analyzed by microarray. For interferon treatment, the cells were harvested at 6 h posttreatment. Expression values and the fold change are provided in Files S1 and S2 in the supplemental material. (B) HEK293 cells were mock treated or infected at an MOI of 3 PFU/cell. Total RNA was subjected to denaturing gel electrophoresis and probed for the indicated miRNAs using end-labeled antisense oligonucleotide probes. miR-16 was detected as a loading control. (C) The effects of siRNA knockdown of the RNases, Dicer and Drosha, on Hs_154 induction were detected by siRNA transfection of HEK293 followed by WNV infection (MOI = 3 PFU/cell) at 48 h posttransfection. The total RNA was extracted at 48 h p.i. and analyzed by Northern blotting. Arrows indicate pre-miRNA (top) and mature miRNA (bottom) species. (D) Western blots demonstrating knockdown of Dicer and Drosha proteins following siRNA transfection.
Table 1.
Sequences of miRNAs induced by WNV
To confirm that Hs_154 is produced through the canonical miRNA biogenesis pathway, the effect of siRNA knockdown of two essential factors of miRNA formation, Dicer and Drosha, on Hs_154 induction by WNV was examined. Western blot analysis indicated efficient knockdown of Dicer and Drosha following siRNA transfection (Fig. 1D). As shown (Fig. 1C), depletion of Dicer or Drosha diminished the ability of WNV to induce Hs_154 expression. It has previously been shown that the average miRNA half-life is approximately 5 days (18), so miRNAs present at high levels before siRNA transfection and subsequent WNV infection are not predicted to show dramatic changes in expression. Consistent with this, miR-16 levels appeared to be unaffected by Dicer and Drosha knockdown. These results confirm that Dicer and Drosha activity are required for Hs_154 expression following WNV infection. The signal for Hs_154 appears as a doublet, indicating a 21- to 22-nucleotide (nt) form, as well as slightly larger form. This may be due to heterogeneity at the miRNA 3′ ends, a phenomenon frequently observed in miRNA biogenesis (30, 47, 54).
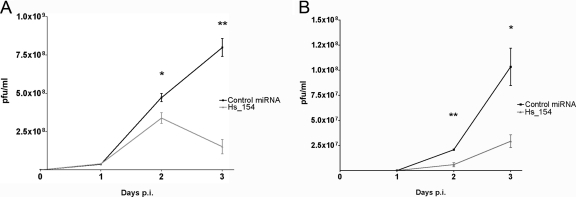
To assess the effect of Hs_154 on viral replication, a double-stranded RNA designed to specifically load the mature Hs_154 sequence into the RISC (46, 49) was transfected in to HeLa and SK-N-MC cells, followed by infection with WNV. Expression of Hs_154 significantly reduced virus titers at days 2 and 3 p.i. in HeLa (Fig. 2A) and SK-N-MC cells (Fig. 2B), demonstrating an antiviral role for this miRNA. Due to the particularly strong induction of this miRNA by WNV infection and its effect on viral replication, the requirements for induction of Hs_154 and its downstream targets were investigated further.
Fig 2.
Hs_154 expression inhibits WNV replication. HeLa (A) or SK-N-MC cells (B) were transfected with a negative control miRNA (black squares) or a double-stranded RNA mimic of Hs_154 (gray triangles). At 48 h posttransfection, the cells were infected with WNV at an MOI of 5 PFU/cell (HeLa) or 0.01 PFU/cell (SK-N-MC). Supernatants were collected at 1, 2, and 3 days p.i., and titers were determined on Vero cells. Titers are shown are averages of three biological replicates ± the SD. Significant reductions in virus titers were detected at day 2 (HeLa, P < 0.05; SK-N-MC, P < 0.005) and day 3 (HeLa, P ≤ 0.001; SK-N-MC, P < 0.05) postinfection in Hs_154-transfected cells. P values were determined by Student t test.
Hs_154 is induced by WNV in multiple cell types, with late kinetics, and occurs in vivo.
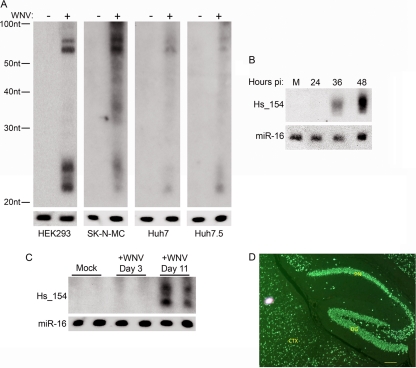
To determine whether Hs_154 upregulation by WNV is a cell-type-specific phenomenon, the effect of WNV infection on Hs_154 levels was examined in other cell types. The neuroblastoma-derived cell line SK-N-MC also showed Hs_154 induction in the microarray, and this induction was confirmed by Northern blot analysis (Fig. 3A). We also examined Hs_154 induction in Huh7 cells (a hepatocellular carcinoma-derived cell line) and its derivative Huh7.5, which carries a mutation inactivating the double-stranded RNA sensor RIG-I (48). Hs_154 induction by WNV infection was observed in these cell types as well (Fig. 3A), indicating that induction of Hs_154 expression does not require a RIG-I-dependent pathway. We next performed a time course experiment of WNV-infected cells to establish how early after infection this Hs_154 is expressed. As shown, Hs_154 upregulation was not observed at 24 h p.i. but was detected at 36 h p.i., with the strongest induction at 48 h p.i. (Fig. 3B).
Fig 3.
Hs_154 is induced with late kinetics in multiple cell types and in WNV-infected mouse brains. (A) HEK293, SK-N-MC, Huh7, and Huh7.5 cells were mock treated or infected with WNV at an MOI of 3 PFU/cell, and the total RNA was extracted at 48 h p.i. and subjected to Northern blot analysis for Hs_154. (B) HEK293 cells were infected with WNV (MOI = 3 PFU/cell). At the indicated times postinfection, RNA was isolated from infected cells and analyzed for Hs_154 levels. (C) RNA was isolated from the neural tissue of WNV-infected rag−/− mice at the indicated times postinfection, and Hs_154 and miR-16 expression was analyzed by Northern blotting. (D) Paraffin-embedded brain from a moribund WNV-infected adult rag−/− mouse was sectioned and immunostained for WNV. Strong green fluorescent labeling of all neuronal soma in the hippocampal cortex (dentate gyrus [DG] and pyramidal neurons [PN]) was detected. Adjacent neocortex (CTX) shows similar strong labeling of neuronal soma in all cortical layers. Bar, 50 μm.
In order to determine whether Hs_154 induction occurs in vivo, we examined Hs_154 expression in the brains of WNV-infected mice. A hairpin structure homologous to that encoding Hs_154 in humans has been identified in mice, suggesting that Hs_154 expression is conserved in mammals (6). We infected Rag−/− mice, which are exquisitely sensitive to WNV and show a high percentage of infection within the neurons of the brains by day 9 p.i. (Fig. 3D). Rag−/− mice were infected with WNV, and the total RNA was isolated from the brains of individual mice at days 3 and 11 p.i. Hs_154 was not detected in the brains of uninfected mice or in mice at 3 days p.i. In contrast, high levels of Hs_154 were observed at day 11 p.i., indicating that the induction of Hs_154 occurs in vivo and correlates temporally with WNV pathogenesis (Fig. 3C).
Hs_154 is not induced by interferon, double-stranded RNA, or other flaviviruses and is independent of caspase activity.
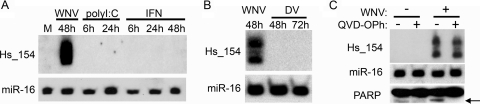
Since the induction of Hs_154 by WNV appears to be a cellular response to infection, one might expect that other treatments, such as antiviral cellular pathways or related viral infections, might also result in increased expression of this miRNA. Therefore, we examined Hs_154 levels after treatment with triggers of antiviral pathways that are activated by WNV infection. Neither poly(I:C) transfection to activate RIG-I nor treatment with recombinant interferon resulted in the induction of Hs_154 (Fig. 4A), a finding consistent with the observed induction of Hs_154 in Huh7.5 cells (Fig. 3A) and the previously discussed results of the microarray (Fig. 1A). In addition, Hs_154 was not upregulated after infection of HEK293 cells with dengue virus (DENV), another member of the flavivirus family (Fig. 4B). DENV replication proceeds more slowly than WNV in HEK293 cells, so cells were examined through 72 h p.i. (peak replication is achieved at between 48 and 72 h p.i.). Poly(I:C) transfection, interferon addition, and DENV infection all induced ISG54 expression, verifying that the cells responded to these stimuli as expected (data not shown). Since the strongest Hs_154 induction by WNV was at 48 h p.i., at which time highly cytopathic effect can be observed and the cells are undergoing apoptosis (32), we hypothesized that apoptosis activation by WNV infection could be related to the induction of Hs_154 expression. Thus, we determined whether increased expression of this miRNA was a downstream effect of WNV-induced apoptosis. The cells were infected with WNV in the presence or absence of QVD-OPh, a caspase inhibitor previously demonstrated to inhibit WNV-induced apoptosis without affecting infection (44). Treatment with QVD-OPh did not inhibit Hs_154 upregulation by WNV (Fig. 4C, top panel), suggesting that Hs_154 induction is either unrelated to or occurs upstream of caspase activation. Cleavage of the caspase substrate poly(ADP-ribose) polymerase (PARP) in WNV-infected cells and inhibition of PARP cleavage by QVD-OPh were confirmed by Western blotting (Fig. 4C, bottom panel).
Fig 4.
Hs_154 induction is independent of interferon, dsRNA, other flaviviruses, and caspase activation. (A) The effects of poly(I:C) (2.5 μg/ml) or interferon (3,000 U/ml) treatment on Hs_154 levels were determined by Northern blot analysis. (B) HEK293 cells were infected with WNV or DENV at an MOI of 3 PFU/cell. RNA was extracted at indicated times postinfection and analyzed for Hs_154 by Northern blotting. (C) HEK293 cells were treated with QVD-OPh and infected with WNV at a multiplicity of 3 PFU/cell. RNA was extracted at 48 h p.i. and analyzed by Northern blotting.
CTCF and ECOP are targets of Hs_154 in WNV-infected cells.
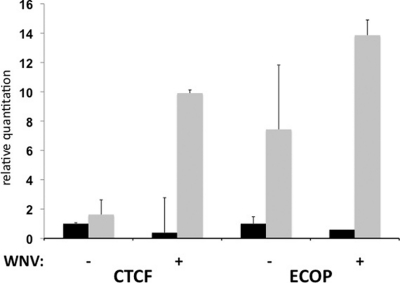
miRNA target identification is complicated by the fact that miRNA-mRNA interactions commonly occur through complementarity of as few as 7 bp. Thus, bioinformatic approaches often produce a high number of false-positive results. Therefore, we used RISC-IP to determine which mRNAs are specifically targeted by miRNAs during WNV infection. HEK293 cells expressing a myc-tagged Ago2 protein (26) were mock infected or infected with WNV, and RISC-IP was performed with an antibody specific for the myc epitope tag. Total and RISC-associated RNA were isolated and analyzed by microarray. The presence of an individual mRNA at a higher level in the IP fraction compared to the total RNA is indicative of the mRNA being associated with the RISC. Since targeting of mRNAs to the RISC is most commonly mediated by complementarity between the miRNA seed site (nt 2 to 8) and nucleotides within the 3′UTR of the targeted gene, we examined the 3′UTRs of highly enriched mRNAs following WNV infection for Hs_154 seed site matches. The presence of several seed site matches within a 3′UTR, coupled with a high degree of conservation of these sequences, suggests that these mRNAs are indeed Hs_154 targets. Table 2 displays the top 20 RISC-enriched mRNAs containing matches to the Hs_154 seed site, the level of enrichment within the RISC following WNV infection, the number of Hs_154 seed site matches found in the 3′UTR of these genes, and whether or not these matches are conserved across species. Two genes, CTCF and ECOP, were determined to have several highly conserved Hs_154 seed site matches within their 3′UTRs and were chosen for further investigation. To confirm the RISC association of these mRNAs, RNA was isolated using an independently performed RISC-IP, and the CTCF and ECOP mRNAs were quantified by qRT-PCR. Both CTCF and ECOP mRNAs showed enrichment in the immunoprecipitated fraction (Fig. 5). Interestingly, ECOP mRNA appears to be enriched (i.e., RISC associated) in uninfected cells as well, which is consistent with previous reports demonstrating targeting of the ECOP by miR-218 (19).
Table 2.
Genes enriched in the RISC in WNV-infected cells containing Hs_154 seed matchesa
| Gene | RISC-IP enrichment | No. of Hs_154 seed site matches in 3′UTR | Seed site match conservation |
|---|---|---|---|
| PCNA | 3.4787 | 1 | Yes |
| C20ORF95/ARHGAP40 | 2.3271 | 2 | |
| NPHP1 | 2.3211 | 1 | No |
| ECOP | 2.3001 | 2 | Yes |
| CTCF | 2.2306 | 3 | Yes |
| C6ORF78/FAM26D | 2.2302 | 1 | No |
| C10ORF63/ENKUR | 2.2096 | 3 | No |
| LOC440957 | 2.1951 | 1 | Yes |
| APP | 2.1775 | 2 | Yes |
| FLJ32679/GOLGA8F | 2.1653 | 1 | |
| SS18 | 2.1539 | 1 | Yes |
| MGC13017 | 2.1255 | 1 | No |
| ZMAT3 | 2.1066 | 1 | No |
| SORBS2 | 2.0874 | 2 | No |
| FOXD4 | 2.0836 | 1 | No |
| MTMR12 | 2.0829 | 1 | No |
| WDR61 | 2.0636 | 1 | No |
| IL-33 | 2.0080 | 4 | Yes |
| CCDC56 | 2.0026 | 1 | No |
Genes showing >2-fold enrichment in the RISC-associated fraction following RISC immunoprecipitation and microarray analysis were examined for seed matches within the 3′UTR. The number of potential seed matches is indicated, as well as conservation in other species. Seed matches were considered conserved if a corresponding sequence was identified in the 3′UTR of the gene from one or more of the following: chimpanzees, rhesus macaques, mice, rats, and chickens.
Fig 5.
CTCF and ECOP mRNA association with RISC is enhanced in WNV-infected cells. Total RNA (black bars) or RNA immunoprecipitated with the RISC from WNV or mock-infected cells was analyzed by qRT-PCR to determine the relative levels CTCF and ECOP mRNA. mRNA levels were normalized to β-actin mRNA and expressed relative to the expression in total RNA from uninfected cells. Values represent the averages of two biological replicates.
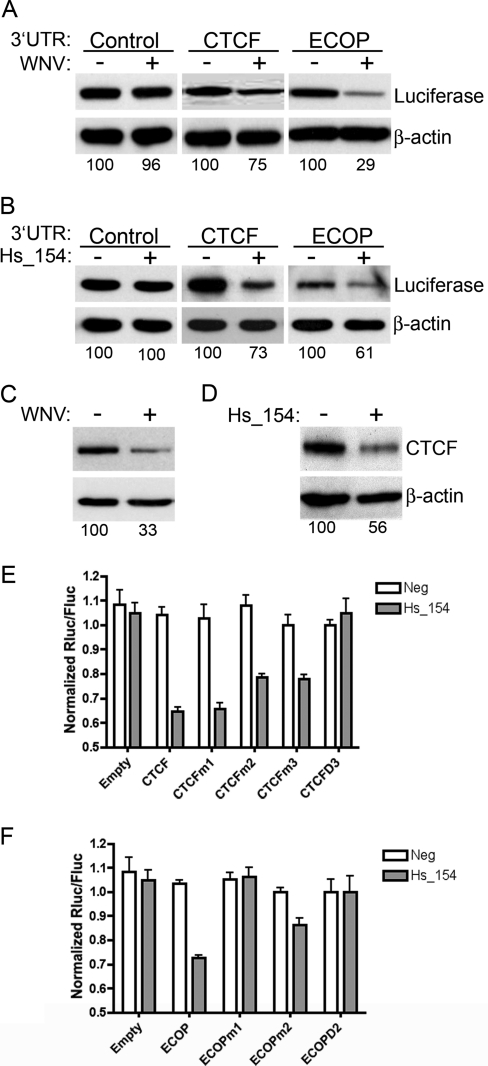
In order to determine whether CTCF and ECOP were targets of Hs_154, the mRNA 3′UTRs of CTCF and ECOP were inserted downstream of a luciferase reporter, and HEK293 cells were transfected with these or control (i.e., no exogenous 3′UTR) luciferase expression vectors. At 48 h posttransfection, the cells were infected with WNV, and the total protein was analyzed by Western blotting for luciferase expression at 48 h p.i. WNV infection resulted in a reduction in the expression of luciferase from vectors bearing CTCF or ECOP—but not control—UTRs (Fig. 6A). In addition, the three expression vectors were transfected together with an RNA duplex designed to mimic the mature Hs_154 sequence or a control miRNA, and total protein was analyzed at 48 h posttransfection. Inclusion of the 3′UTRs of both CTCF and ECOP downstream of luciferase caused downregulation following Hs_154 duplex transfection (Fig. 6B), demonstrating that both of these genes are downstream targets of the WNV-induced miRNA, Hs_154. Consistent with these observations, both WNV infection and Hs_154 duplex transfection resulted in the downregulation of endogenous levels of CTCF protein at 48 h posttreatment (Fig. 6C and D). Finally, CTCF has three potential seed sequence matches to Hs_154, while ECOP has two such matches (see Fig. S1 and S2 in the supplemental material). Although mutation of individual sites in the 3′UTR of CTCF did not significantly restore luciferase expression in cells transfected with Hs_154, mutation of all three sites returned luciferase expression to levels equivalent to cells transfected with the negative control miRNA (Fig. 6E). Interestingly, mutation of one of the Hs_154 seed matches in the ECOP 3′UTR fully restored luciferase expression, whereas mutation of the other resulted in a partial restoration of expression (Fig. 6F).
Fig 6.
CTCF and ECOP, targets of Hs_154, are repressed after WNV infection and Hs_154 transfection. HEK293 cells were transfected with luciferase reporter plasmids encoding control, CTCF, or ECOP 3′UTRs. At 48 h posttransfection, cells were infected with WNV at an MOI of 3 PFU/cell (A) or transfected with a double-stranded RNA corresponding to Hs_154 (B). Protein was isolated at 48 h p.i. and subjected to Western blot analysis with antibodies to firefly luciferase or β-actin. Endogenous levels of CTCF were analyzed by Western blotting after WNV infection (C) or Hs_154 transfection (D). Relative protein levels compared to untreated controls normalized to β-actin were determined by using ImageJ software and shown below individual bands. Luciferase reporter vectors encoding CTCF (E) or ECOP (F) 3′UTRs with seed sequence sites mutated individually or in combination to disrupt Hs_154 targeting were transfected into HEK293 cells. Renilla (test) and firefly (control) luciferase activity was measured after transfection with a negative control or Hs_154. Values represent the averages of three replicates ± the standard deviations.
Expression of CTCF and ECOP inhibit WNV-dependent induction of apoptosis.
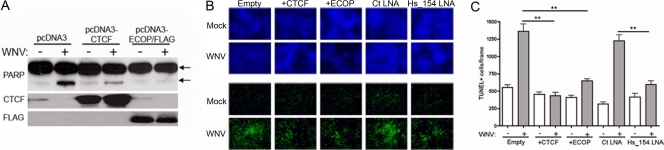
CTCF and ECOP have been shown to have antiapoptotic, prosurvival functions in which downregulation of these genes results in cellular apoptosis (15, 29, 39). Since the time course of induction of Hs_154 by WNV coincides with virus-mediated apoptosis, we hypothesized that Hs_154 upregulation in response to WNV infection leads to the downregulation of these two genes and contributes to subsequent cell death. This hypothesis predicts that expression of these proteins will protect cells from WNV-mediated apoptosis. In order to test this hypothesis, CTCF- and Flag-tagged ECOP-expressing vectors were transfected into HEK293 cells, followed by WNV infection. Total protein was collected at 48 h p.i. and analyzed for PARP cleavage by Western blotting. As expected, WNV infection induced PARP cleavage in cells transfected with the empty expression vector (pcDNA3). In contrast, expression of CTCF and ECOP significantly reduced PARP cleavage (Fig. 7A) after WNV infection. In parallel, the effects of CTCF and ECOP expression on WNV-induced apoptosis were also investigated by TUNEL assay (Fig. 6B and C). SK-N-MC cells transfected with CTCF and ECOP expression vectors were infected with WNV and, at 48 h p.i., were fixed and stained. As shown in Fig. 7, WNV infection significantly increases the number of apoptotic cells in cultures transfected with the control vector but not in cells transfected with CTCF or ECOP. Finally, an antisense LNA inhibitor (36) of Hs_154 was transfected into cells, followed by WNV infection and TUNEL staining. In contrast to the control LNA, the Hs_154 inhibitor significantly reduced the number of apoptotic cells in WNV-infected cultures (Fig. 7B and C). Taken together, these results indicate that CTCF and ECOP expression block WNV-induced apoptosis and that downregulation of these proteins by Hs_154 contributes to apoptosis of the infected cell.
Fig 7.
Overexpression of CTCF or ECOP or a Hs_154-targeting LNA protects cells from WNV-mediated apoptosis. (A) HEK293 cells were transfected with empty vector (control) or plasmids encoding CTCF or FLAG-tagged ECOP. At 48 h posttransfection, cells were WNV infected (MOI = 3 PFU/cell), and protein was isolated at 48 h p.i. for Western blot analysis of PARP cleavage (arrow), CTCF, FLAG-ECOP, and β-actin. (B) SK-N-MC cells were transfected as in panel A or transfected with a control LNA or Hs_154-targeting LNA and infected at an MOI of 3 PFU/cell. Cells were analyzed by TUNEL assay for apoptotic cells at 48 h p.i. The top panel represents total cells (blue). The bottom panel shows TUNEL-positive cells (green). (C) The number of TUNEL positive cells per frame was quantified by using ImageJ software (National Institutes of Health).
DISCUSSION
We have identified multiple miRNAs that display altered expression following WNV infection. One such miRNA, Hs_154, which is strongly induced by WNV and whose expression significantly inhibits viral replication, was shown to target two antiapoptotic proteins, CTCF and ECOP. Indeed, the expression of CTCF and ECOP blocked apoptosis in WNV-infected cells, demonstrating that the downregulation of expression of these proteins is critical to the induction of apoptosis. Our results therefore demonstrate that cellular miRNAs can be mobilized during viral infection to influence the response of the cell.
Several of the miRNAs displaying altered expression levels in WNV-infected cells have been previously identified as low-abundance miRNAs in various tissue types using massively parallel sequencing or bioinformatics, followed by RNA-primed array-based Klenow extension (RAKE) analysis (6, 7). Our results confirm that these miRNAs are expressed at relatively low levels in uninfected cells but are strongly induced during WNV infection. Hs_154 demonstrates similar induction patterns in multiple WNV-infected cell types, as well as in the neural tissues of WNV-infected mice. Interestingly, induction of this miRNA is not observed after treatment of cells with interferon, poly(I:C), or DENV, suggesting that the expression of Hs_154 is driven by a pathway unique to WNV. It is possible that Hs_154 induction is due, in part, to the particularly robust growth of WNV in the cell types examined. In general, WNV grows with faster kinetics and to higher titers than DENV in HEK293 cells, which may be required for detectable Hs-154 induction. In addition, in contrast to previously published results (40), we observed little miRNA induction following interferon treatment, although other reports have reported a similar lack of change in miRNA expression in the presence of interferon (30).
Examples of miRNAs that respond to viral infection and affect viral replication include those believed to target viral sequences directly (23, 28, 37, 40), as well as those that target host proteins to modulate the cellular environment and thereby influence viral replication. Several miRNAs with antiviral activity have been described. miR-100 and miR-101 have been demonstrated to inhibit human cytomegalovirus (HCMV) replication via inhibition of the mammalian target of rapamycin (mTOR) pathway (52), and miR-27 has been shown to block murine cytomegalovirus (MCMV) replication (8). Notably, these miRNAs are downregulated during HCMV and MCMV infection, respectively, suggesting that these viruses have evolved to manipulate cellular miRNA expression to promote their replication. Furthermore, a large screen of miRNA mimics and antagonists identified several miRNAs that affected the replication of multiple viruses, including miRNAs that demonstrate proviral and others that show antiviral activity (45). One of these, miR-199a-3p, was active against a range of viruses, inhibiting the replication of several members of the herpesvirus family, as well as of Semliki Forest virus, a member of the alphavirus family of positive-sense RNA viruses. This miRNA has been hypothesized to function through modulation of the mitogen-activated protein kinase pathway, although other studies have suggested that it can inhibit hepatitis B virus and hepatitis C virus replication through direct interactions with the viral genomes (35, 59).
We have identified mRNA transcripts enriched in the RISC (i.e., targeted by miRNAs) in WNV-infected cells, and we noted several with potential seed sequence matches to Hs_154. Two of these mRNAs encode the proteins CTCF and ECOP, both of which have been reported to have antiapoptotic activity (15, 29, 39). CTCF is a DNA-binding zinc-finger protein involved in the regulation and maintenance of chromatin domains. Its antiapoptotic function has been hypothesized to occur via the repression of transcription of proapoptotic genes, such as Bax (15). Loss of ECOP expression has been correlated with an increase in reactive oxygen species and mitochondrial dysfunction (4). Depending on the cell type and culture conditions, loss of ECOP may also contribute to decreased NF-κB activity and subsequent apoptosis (39). We have confirmed that the Hs_154 seed matches in the 3′UTRs of the CTCF and ECOP mRNAs do indeed mediate Hs_154-dependent repression of protein expression. Furthermore, endogenous CTCF protein expression was downregulated by WNV infection or Hs_154 transfection, which is consistent with a role for Hs_154 in downregulating the expression of these proteins during infection. Finally, we observed that expression of CTCF and ECOP blocks apoptosis in WNV-infected cells, supporting the hypothesis that the reduction of expression of these proteins is necessary for efficient completion of the apoptotic process.
Multiple miRNAs have been shown to impact apoptosis (reviewed in reference 53). Several miRNAs possessing antiapoptotic activity, such as miR-21, have been shown to be overexpressed in a variety of human cancers and to target numerous mRNAs encoding proapoptotic host proteins (10, 16, 24). Inhibition of apoptosis is also the function of several miRNAs encoded by herpesviruses, which establish long-term latent or persistent infections (1, 12, 56). In contrast, other miRNAs have been shown to promote apoptosis through targeting of prosurvival proteins, such as Bcl-2, and are often deleted or downregulated in human cancers. miR-15a, miR-16, and the miR-34 family of miRNAs have been shown to fall into this class (11, 13).
WNV has previously been shown to induce apoptosis in infected cells through multiple mechanisms. Expression of the viral nonstructural proteins results in activation of multiple arms of the UPR, induction of the cyclic AMP response element-binding transcription factor homologous protein (CHOP), and caspase activation (2, 32). Apoptosis has also been shown to be mediated in a paracrine fashion in infected brains via secretion of neurotoxic factors by infected astrocytes (51). In most cases of viral infection, the apoptotic response is considered an antiviral mechanism by which infected cells are deleted and targeted for phagocytosis by macrophages or other phagocytic cells (14). Consistent with this mechanism, we observed an inhibitory effect of Hs_154 on viral replication, implicating Hs_154 induction as an aspect of the host antiviral response. We detected Hs_154 in abundant amounts in infected brains from rag−/− mice, showing that neurons in vivo utilize this pathway in WNV infection. Although it is possible, and even likely, that the absence of the fully functional immune system in these mice makes this phenomenon more pronounced, we are confident that the upregulation of Hs_154 is a primary cellular response to WNV infection. We note that although Hs_154 (and apoptosis) may demonstrate and antiviral effect in cell culture, the results of cellular apoptosis following viral infection may, in fact, be detrimental to the host. Most importantly, within the brain of a WNV-infected host, apoptosis of neural cells is a key determinate of viral pathogenesis, and disruption of the apoptotic pathway via knockout of caspase-3 results in less neuronal death in several regions of the brain and greater survival of infected mice without significantly influencing viral loads (44). The association of Hs_154 with apoptosis may therefore make this miRNA a possible candidate for therapeutic intervention.
Supplementary Material
ACKNOWLEDGMENTS
Microarray assays (and data analysis) were performed in the OHSU Gene Microarray Shared Resource. We thank C. Wiley (University of Pittsburgh) for performing immunofluorescent staining on mouse tissue and M. Hancock and J. Nelson for critical readings of the manuscript and useful discussions.
This study was supported by the Pacific Northwest Regional Center of Excellence (National Institutes of Health grant U54 AI081680) and National Institutes of Health contracts HHSN266200500027C and HHSN272201100017C.
Footnotes
Published ahead of print 15 February 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Abend JR, Uldrick T, Ziegelbauer JM. 2010. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J. Virol. 84:12139–12151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ambrose RL, Mackenzie JM. 2011. West Nile virus differentially modulates the unfolded protein response to facilitate replication and immune evasion. J. Virol. 85:2723–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Artsob H, et al. 2009. West Nile virus in the New World: trends in the spread and proliferation of West Nile virus in the Western Hemisphere. Zoonoses Public Health 56:357–369 [DOI] [PubMed] [Google Scholar]
- 4. Baras AS, Solomon A, Davidson R, Moskaluk CA. 2011. Loss of VOPP1 overexpression in squamous carcinoma cells induces apoptosis through oxidative cellular injury. Lab. Invest. 6:1170–1180 [DOI] [PubMed] [Google Scholar]
- 5. Behm-Ansmant I, Rehwinkel J, Izaurralde E. 2006. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harbor Symp. Quant. Biol. 71:523–530 [DOI] [PubMed] [Google Scholar]
- 6. Berezikov E, et al. 2006. Diversity of microRNAs in human and chimpanzee brain. Nat. Genet. 38:1375–1377 [DOI] [PubMed] [Google Scholar]
- 7. Berezikov E, et al. 2006. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 16:1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buck AH, et al. 2010. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA (New York, NY). 16:307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention 2011, posting date West Nile virus. Centers for Disease Control and Prevention, Atlanta, GA: http://www.cdc.gov/ncidod/dvbid/westnile/index.htm [Google Scholar]
- 10. Chan JA, Krichevsky AM, Kosik KS. 2005. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 65:6029–6033 [DOI] [PubMed] [Google Scholar]
- 11. Chang TC, et al. 2007. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell 26:745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choy EY, et al. 2008. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 205:2551–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cimmino A, et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. U. S. A. 102:13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Devitt A, Marshall LJ. 2011. The innate immune system and the clearance of apoptotic cells. J. Leukoc. Biol. 90:447–457 [DOI] [PubMed] [Google Scholar]
- 15. Docquier F, et al. 2005. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 65:5112–5122 [DOI] [PubMed] [Google Scholar]
- 16. Esquela-Kerscher A, Slack FJ. 2006. Oncomirs: microRNAs with a role in cancer. Nat. Rev. Cancer 6:259–269 [DOI] [PubMed] [Google Scholar]
- 17. Fredericksen BL, Smith M, Katze MG, Shi PY, Gale M., Jr 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737–7747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gantier MP, et al. 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 39:5692–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao C, et al. Reduced microRNA-218 expression is associated with high nuclear factor κB activation in gastric cancer. Cancer 116:41–49 [DOI] [PubMed] [Google Scholar]
- 20. Grey F, Meyers H, White EA, Spector DH, Nelson J. 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grey F, et al. 2010. A viral microRNA downregulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathog. 6:e1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henchal EA, Gentry MK, McCown JM, Brandt WE. 1982. Dengue virus-specific and flavivirus group determinants identified with monoclonal antibodies by indirect immunofluorescence. Am. J. Trop. Med. Hyg. 31:830–836 [DOI] [PubMed] [Google Scholar]
- 23. Huang J, et al. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 24. Jazbutyte V, Thum T. 2010. MicroRNA-21: from cancer to cardiovascular disease. Curr. Drug Targets 11:926–935 [DOI] [PubMed] [Google Scholar]
- 25. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]
- 26. Karginov FV, et al. 2007. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. U. S. A. 104:19291–19296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klee AL, et al. 2004. Long-term prognosis for clinical West Nile virus infection. Emerg. Infect. Dis. 10:1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lecellier CH, et al. 2005. A cellular microRNA mediates antiviral defense in human cells. Science 308:557–560 [DOI] [PubMed] [Google Scholar]
- 29. Li T, Lu L. 2007. Functional role of CCCTC binding factor (CTCF) in stress-induced apoptosis. Exp. Cell Res. 313:3057–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marti E, et al. 2010. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 38:7219–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Medigeshi GR, et al. 2009. West nile virus capsid degradation of claudin proteins disrupts epithelial barrier function. J. Virol. 83:6125–6134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Medigeshi GR, et al. 2007. West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J. Virol. 81:10849–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mombaerts P, et al. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68:869–877 [DOI] [PubMed] [Google Scholar]
- 34. Mostashari F, et al. 2001. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet 358:261–264 [DOI] [PubMed] [Google Scholar]
- 35. Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. 2009. Regulation of the hepatitis C virus genome replication by miR-199a. J. Hepatol. 50:453–460 [DOI] [PubMed] [Google Scholar]
- 36. Orom UA, Kauppinen S, Lund AH. 2006. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 372:137–141 [DOI] [PubMed] [Google Scholar]
- 37. Otsuka M, et al. 2007. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity 27:123–134 [DOI] [PubMed] [Google Scholar]
- 38. Parameswaran P, et al. 2010. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog. 6:e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park S, James CD. 2005. ECop (EGFR-coamplified and overexpressed protein), a novel protein, regulates NF-κB transcriptional activity and associated apoptotic response in an IκBα-dependent manner. Oncogene 24:2495–2502 [DOI] [PubMed] [Google Scholar]
- 40. Pedersen IM, et al. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petersen LR, Marfin AA. 2002. West Nile virus: a primer for the clinician. Ann. Intern. Med. 137:173–179 [DOI] [PubMed] [Google Scholar]
- 42. Ramanathan MP, et al. 2006. Host cell killing by the West Nile Virus NS2B-NS3 proteolytic complex: NS3 alone is sufficient to recruit caspase-8-based apoptotic pathway. Virology 345:56–72 [DOI] [PubMed] [Google Scholar]
- 43. Saeed AI, et al. 2006. TM4 microarray software suite. Methods Enzymol. 411:134–193 [DOI] [PubMed] [Google Scholar]
- 44. Samuel MA, Morrey JD, Diamond MS. 2007. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J. Virol. 81:2614–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Santhakumar D, et al. 2010. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc. Natl. Acad. Sci. U. S. A. 107:13830–13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schwarz DS, et al. 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208 [DOI] [PubMed] [Google Scholar]
- 47. Starega-Roslan J, Koscianska E, Kozlowski P, Krzyzosiak WJ. 2011. The role of the precursor structure in the biogenesis of microRNA. Cell. Mol. Life Sci. 68:2859–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sumpter R, Jr, et al. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 79:2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. 2004. A protein sensor for siRNA asymmetry. Science 306:1377–1380 [DOI] [PubMed] [Google Scholar]
- 50. Umbach JL, Cullen BR. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Marle G, et al. 2007. West Nile virus-induced neuroinflammation: glial infection and capsid protein-mediated neurovirulence. J. Virol. 81:10933–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang F-Z, et al. 2008. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J. Virol. 82:9065–9074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang Z. 2010. MicroRNA: a matter of life or death. World J. Biol. Chem 1:41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu H, et al. 2007. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One 2:e1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiao SY, Guzman H, Zhang H, Travassos da Rosa AP, Tesh RB. 2001. West Nile virus infection in the golden hamster (Mesocricetus auratus): a model for West Nile encephalitis. Emerg. Infect. Dis. 7:714–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu S, Xue C, Li J, Bi Y, Cao Y. 2010. Marek's disease virus type 1 microRNA miR-M3 suppresses cisplatin-induced apoptosis by targeting Smad2 of the transforming growth factor beta signal pathway. J. Virol. 85:276–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang JS, et al. 2002. Induction of inflammation by West Nile virus capsid through the caspase-9 apoptotic pathway. Emerg. Infect. Dis. 8:1379–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang MR, et al. 2008. West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol. 10:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang G-L, et al. 2010. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 88:169–175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.