Abstract
The Hsp70 chaperone plays a central role in multiple processes within cells, including protein translation, folding, intracellular trafficking, and degradation. This protein is implicated in the replication of numerous viruses. We have shown that rabies virus infection induced the cellular expression of Hsp70, which accumulated in Negri body-like structures, where viral transcription and replication take place. In addition, Hsp70 is present in both nucleocapsids purified from infected cells and in purified virions. Hsp70 has been shown to interact with the nucleoprotein N. The downregulation of Hsp70, using specific chaperone inhibitors, such as quercetin or RNA interference, resulted in a significant decrease of the amount of viral mRNAs, viral proteins, and virus particles. These results indicate that Hsp70 has a proviral function during rabies virus infection and suggest that Hsp70 is involved in at least one stage(s) of the viral life cycle, such as viral transcription, translation, and/or production. The mechanism by which Hsp70 controls viral infection will be discussed.
INTRODUCTION
In eukaryotic cells, the posttranslational folding of proteins and quality control are performed by molecular chaperones and heat shock proteins (HSPs). The ubiquitous chaperone family of the 70-kDa HSPs plays a central role in protein homeostasis and protection against proteotoxic stresses by preventing protein misfolding and aggregation by directing damaged protein to the ubiquitin-proteasome system for degradation (4, 15, 34, 40). Hsp70 chaperones not only survey the folding status of proteins as part of the quality control function, which is very important under stress conditions, but also are involved in the regulation of fundamental cellular processes, such as signal transduction, cell cycle regulation, apoptosis, and innate immunity (23, 25).
Viral infection depends on the successful recruitment of host cellular factors at different steps of the viral life cycle: genome replication, viral protein synthesis, viral assembly, and counterdefense against cell apoptosis and innate immunity. During viral infection, large amounts of viral proteins are synthesized and protein folding can become a limiting step. Therefore, on the one hand viruses need cellular chaperones for their own protein folding processes; on the other hand, as chaperones are involved in the regulation of fundamental cellular processes, viruses have to interact with them. Hsp70 chaperone, the major inducible heat shock protein, is frequently recruited by viruses. Research during the past 30 years has revealed the involvement of Hsp70 in all steps of viral life cycle replication and for viruses from numerous families of diverse orders (24, 28). Several DNA viruses, such as herpesvirus (HSV1) (10, 38), vaccinia virus (18), adenovirus type 5 (24), and simian virus 40 (SV40) (27, 31), induce the specific expression of Hsp70 (33). In most cases this induction has a proviral effect. Evidence is growing that the Hsp70 induction is also important for the replication of many positive-strand RNA and negative-strand RNA viruses (28). In the case of measles virus, Hsp70 interacts with nucleocapsids (42) and induces the increased expression of viral genes (5). Nevertheless, in some cases Hsp70 was found to inhibit viral infection. An increase of its expression confers to cells a protection against rotavirus (2), vesicular stomatitis virus (9), respiratory syncytial virus (41), and influenza virus (16) infections. In the latter case, it has been reported that Hsp70, which interacts with the ribonucleoprotein complex, interferes with the polymerase activity and negatively regulates viral transcription and replication (16, 22). Interestingly, Hsp70 was found to have both positive and negative regulatory effects on the RNA replication of flock house virus, a nodavirus (39), highlighting the complexity of this particular virus-chaperone interaction.
Rabies virus, the prototype of the Lyssavirus genus that belongs to the Rhabdoviridae family (Mononegavirales order), causes a fatal disease that is associated with intense viral replication in the central nervous system. Its single-stranded negative-sense RNA genome (∼12 kb), encoding five viral proteins, is encapsidated by the nucleoprotein (N; 40 kDa) to form the nucleocapsid that is associated with the RNA-dependent RNA polymerase L (220 kDa) and its cofactor, phosphoprotein (P; 33 kDa). Inside the viral particle, the nucleocapsid has a tightly coiled helical structure that is associated with the matrix protein (M; 22 kDa) and is surrounded by a membrane containing a unique glycoprotein (G; 62 kDa). The virus enters the host cell through the endosomal transport pathway via a low-pH-induced membrane fusion process catalyzed by G (14). The nucleocapsid released into the cytoplasm serves as a template for transcription and replication processes that are catalyzed by the L-P polymerase complex (for a review see reference 1). It has been shown recently that rabies virus transcription and replication take place within Negri body-like (NBL) structures, which are inclusion bodies formed during viral infection (20). During transcription, a positive-stranded leader RNA and five capped and polyadenylated mRNAs are synthesized. The replication process yields nucleocapsids containing full-length antigenome-sense RNA, which in turn serve as templates for the synthesis of genome-sense RNA. During their synthesis, both the nascent antigenome and the genome are encapsidated by N protein. This encapsidation is regulated by the P protein, which plays the role of chaperone, preventing the N protein from binding to cellular RNA and from aggregating (32). The neosynthesized genomic RNA either serves as a template for secondary transcription or is transported to the cell membrane and assembles with the M and G proteins for the budding of neosynthesized virions.
We have previously shown that Hsp70 protein accumulates in the NBL structures, which are sites of rabies virus transcription and replication (20). In addition, Hsp70 has been previously shown to be incorporated in purified rabies virus (37). Therefore, we investigated the role of this chaperone during viral infection. We report here that rabies virus infection induces the expression of Hsp70. We also show that Hsp70 interacts with the nucleoprotein N, the major component of NBL structures, and that Hsp70 downregulation results in the inhibition of different steps of the viral cycle. The inhibition of viral protein synthesis and of viral production can reflect the inhibition of viral RNA synthesis, but it also could be due to a specific role of Hsp70 in these further steps.
MATERIALS AND METHODS
Virus stock and cell infection.
BSR cells, cloned from BHK 21 (baby hamster kidney) cells, were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Human neuroblastoma SK-N-BE cell lines were grown in RPMI 1640 plus l-glutamine (Gibco) supplemented with 15% fetal calf serum (FCS).
The CVS strain of rabies virus was grown in BSR cells. Virus titers were determined by standard plaque assays of BSR cells.
Antibodies and drugs.
The mouse polyclonal anti-P antibody has been described previously, and the anti-P monoclonal antibodies (MAb) 25C2 and 30F2 have been previously described (36). The monoclonal anti-N MAb 62B5 was produced in mice immunized with the virus. Rabbit polyclonal anti-P and anti-M antibodies were obtained by the repeated injection of purified recombinant protein produced in Escherichia coli. The mouse anti-Hsp70 MAb (SPA-810) and rabbit polyclonal anti-Hsp70 (SPA-812) antibody were obtained from Stressgen, and the anti-α-tubulin MAb (N356) was from Amersham. Quercetin (Qct; Q4951) and the rabbit polyclonal anti-actin (20–33) antibody were obtained from Sigma. The mouse anti-green fluorescent protein (GFP) MAb (11814460001) was purchased from Roche.
Plasmids and siRNA.
The plasmids pCDM8-P, encoding the P protein (CVS strain), and pCDM8-N, encoding the N protein (CVS strain), have been described previously (6). The plasmid pRL-TK was described elsewhere (11, 13) and was a generous gift of D. Garcin (University of Geneva School of Medicine). The plasmids pTit-N, pTit-P, and pTit-L were described previously (7) and were obtained from K. K. Conzelmann (Ludwig-Maximilians University of Munich). The plasmid pDImut (21) was provided by N. Tordo (Institut Pasteur). The plasmid pCMV-Hsp70, encoding the Hsp70 protein, and pCMV-Neo were kindly provided by G. Trugnan (2). The siRNA-Hsp70 duplex and scrambled siRNA were purchased from Eurogentec.
Cell infection and transfection.
BSR cells were grown on glass coverslips in 6-well plates and were infected with rabies virus at various multiplicities of infection (MOI) and at various times postinfection (p.i.). BSR cells grown to 90% confluence in 60-mm-diameter dishes also were transfected with 2 μg of plasmid using Lipofectamine 2000 as described by the manufacturer (Invitrogen).
Purification of virus and nucleocapsids.
Purified virus was obtained as follows. Virions were pelleted through a cushion of 25% glycerol TNE (10 mM Tris, pH 7.5, 1 mM EDTA, 50 mM NaCl) in TD (0.8 mM Tris, pH 7.4, 150 mM NaCl, 5 mM KCl, 0.7 mm Na2HPO4, 10 mM EDTA) and further purified by centrifugation in a sucrose gradient (10 to 40%, wt/vol) in TD.
Rabies virus nucleocapsids were isolated from infected BSR cells as described previously (17).
Treatment of cells with drugs.
Cells were kept in DMEM containing 25 to 200 μM quercetin for 1 h before virus inoculation and during infection. As all drugs were reconstituted in dimethyl sulfoxide (DMSO), untreated cells were maintained in medium containing the same concentration of DMSO. The expression of Hsp70 was induced by a heat shock (HS) at 45°C for 20 min, and then cells were reincubated at 37°C.
siRNA experiments.
The siRNA-Hsp70 sequence of 21 nucleotides (nt) was chosen based on the rules proposed by Elbashir et al. (12) and was designed to interfere with the two mRNAs encoding Hsp70 (GenBank accession number NM_005345 for HSPA1A and BC057397 for HSPA1B) but not with the two Hsc70 mRNA variants. The sequences were the following: siRNA-Hsp70 sense, 5′-CACGGCAACCUGGAGAUCA-deoxythymidine (dTdT)-3′; siRNA-Hsp70 antisense, 5′-UGAUCUCCACCUUGCCGUG-dTdT-3′; siRNA-scrambled sense, 5′-ACAAUUCCGGGGGAGAGAG-dTdT-3′; and siRNA-scrambled antisense, 5′-CUCUCUCCCCCGGAAUUGA-dTdT-3′. Transfections were obtained using an Invitrogen Lipofectamine kit. BSR cells (in serum-free DMEM) were transfected with 400 pmol of scrambled siRNA or 200 to 400 pmol of siRNA-Hsp70. After 6 h at 37°C, the medium was completed with 5% FCS. After 48 h at 37°C, it was possible to induce a heat shock and to infect cells. After incubation, the culture supernatant was collected to quantify viral production. The cell layer was recovered to obtain total extract for Western blot analysis.
Detection of viral mRNA and Hsp70 mRNA by RT-PCR.
The presence of Hsp70 mRNA or rabies N and P mRNA was determined by RT-PCR. Total cellular RNA was isolated from cell culture using an RNA kit according to the manufacturer's instructions (RNA NOW). Oligo(dT) primer was used for the synthesis of the first strand of cDNA. The expression of Hsp70 and rabies genes was carried out by the PCR amplification of the Hsp70 or rabies N and P genes using the following primers: Hsp70 sense (Hsp70-A), 5′-GCCGGATCCATATGGCCAAAGCC-3′; Hsp70 antisense (XhoI-B), 5′-GCCCTCGAGCTAATCTACCTCCT-3′; CVS-N sense (N-A), 5′-GCCGGATCCATGGATGCCGACAAGA-3′; CVS-N antisense (N-B), 5′-GCCGGATCCTTATGAGTCACTCGA-3′; CVS-P sense (P-A), 5′-GCCGCTAGCATGAGCAAGATCTTTGTT-3′; and CVS-P antisense (P-B), 5′-CTCGAGTTAGCAGGATGTATAGCG-3′. The PCRs were performed using three different volumes of RT mixture (cDNA; 1, 3, and 5 μl). The PCR products were electrophoresed in a 1.2% agarose gel and visualized with ethidium bromide.
Northern blot analysis.
RNA was isolated from cells with the RNA NOW kit (Ozyme). Total RNA was separated on a 1.5% agarose gel under denaturing conditions and blotted onto nylon membranes (Roche Molecular Biochemicals). Hybridizations were performed with digoxigenin (DIG)-labeled oligonucleotides recognizing the rabies virus (RAV) N or P gene sequence and by incubation with anti-DIG antibody conjugated to alkaline phosphatase, followed by CDP Star analysis.
Minireplicon system assay and luciferase assay.
The minireplicon system assay was slightly modified from Le Mercier et al. (21). BSR cells were grown in a 12-well plate (3 × 105 cells per well) in DMEM supplemented with 5% FCS and incubated for 24 h at 37°C (in 5% CO2).
The cells were transfected with pTit-N (1.2 μg), pTit-P (1.2 μg), pTit-L (0.3 μg), pT7 (1 μg; to allow the cytoplasmic expression of the T7 RNA polymerase), pRL-TK (0.3 μg), and pDImut (0.75 μg) using the FuGENE 6 reagent as described by the manufacturer (number 11814443001 from Roche). The N, P, and L proteins and the RNA minigenome formed a functional RNP template resulting in luciferase gene transcription, thus the amount of luciferase was related to the transcriptional activity of the reconstituted RNP. The inhibition of Hsp70 expression was obtained by siRNA-Hsp70 transfection 48 h before minireplicon transfection, and the overexpression of Hsp70 was induced by transfection with pCMV-Hsp70 (1.2 μg). In the absence of pCMV-Hsp70, all cells were transfected with pCMV-Neo. Thirty-six hours after the transfection of the minireplicon system, the measurement of Firefly and Renilla luciferase activities were performed according to the manufacturer's protocol (dual-luciferase assay system from Promega). Relative expression levels were calculated by dividing the Firefly luciferase values by those of the Renilla luciferase.
Pulse-chase experiments and immunoprecipitation.
At 16 h postinfection, the culture medium was replaced with methionine- and cysteine-free medium, and the culture was incubated for 1 h. The cells then were labeled with 2 ml of prewarmed medium containing 20 μCi of [35S]methionine and [35S]cysteine (0.8 Mbq) (express protein labeling mix; Perkin Elmer) for 5 min. Prewarmed medium containing 5 mM cold methionine and 1 mM cold cysteine then was added for the chase period. The cells were washed in cold PBS and lysed on ice in 500 μl of buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and an antiprotease cocktail (2 μg of leupeptin per ml, 2 μg of antipain per ml, 2 μg pepstatin per ml, 2 μg of chymostatin per ml, and 16 μg of aprotinin per ml). Nuclei were eliminated from the lysate by centrifugation at 12,000 × g for 2 min at 4°C. The cytoplasmic fractions were incubated for 2 h at 4°C with specific antibodies (anti-P, anti-N, or anti-M). Immune complexes were precipitated by incubation with protein A-Sepharose for 1 h at 4°C, washed three times, denatured in Laemmli buffer, and analyzed by SDS-PAGE. The quantification of radioactivity was performed by a phosphorimager.
Coimmunoprecipitation.
Cells were harvested by being scraped into cold phosphate-buffered saline (PBS), and cell extracts were prepared as described above. The cytoplasmic fraction was incubated overnight at 4°C with specific antibodies (anti-Hsp70 or anti-N). Immune complexes were precipitated by incubation with protein A-Sepharose for 1 h at 4°C, washed three times, and denatured in Laemmli buffer. Immunoprecipitated proteins were analyzed by Western immunoblotting using different antibodies.
Western blot analysis.
Cells were washed and resuspended in PBS, lysed in hot Laemmli sample buffer, and boiled for 5 min. About 20 μg of protein was analyzed on a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane. The proteins were blocked on the membranes with 10% skimmed milk in PTBS for 2 h and incubated overnight at 4°C with rabbit polyclonal anti-Hsp70, anti-P, anti-N, anti-actin, or anti-tubulin antibody. The blots then were washed extensively in PBS-Tween and incubated for 1 h with the appropriate peroxidase-coupled secondary antibodies (Amersham). All of the blots then were subjected to chemiluminescence (ECL; Amersham). Protein spot levels were determined by using ImageJ quantification software.
Immunoaffinity columns and proteomic analysis. (i) Immunoaffinity columns.
Monoclonal anti-N antibodies (62B5 and 64B6), which were previously described (36), were first purified by binding to protein A Sepharose, eluted with 100 mM glycine (pH 3), and immediately readjusted to neutral pH by adding Tris (pH 8).
Second, the purified antibodies were applied on a protein A-Sepharose column equilibrated in Tris M, pH 8. Protein A was then washed and resuspended in 0.2 M borate buffer, pH 9.
The cross-linking reaction takes place in the presence of 20 mM dimethyl pimelimidate during 30 min at room temperature under gentle rotation. Protein A then is washed and resuspended with 0.2 M ethanolamine (pH 8) for 2 h. After the removal of ethanolamine, the free antibodies (not cross-linked to protein A) were eluted with 100 mM glycine buffer (pH 3).
(ii) Purification of viral and cellular proteins associated with N.
Cytoplasmic fractions of uninfected or infected SK-N-BE cells (1 × 108 cells at an MOI of 10) were loaded separately on the anti-N immunoaffinity columns. The columns then were washed extensively with PBS, and the proteins bound to the column were eluted using 100 mM glycine buffer (pH 3).
(iii) Analysis of proteins bound to columns of anti-N.
The eluates were resolved on SDS-PAGE, and the proteins were revealed using silver nitrate (silver stain plus kit; Bio-Rad) or colloidal Coomassie blue. Unique protein bands that had been compared to proper controls were trypsinized for proteomic analysis by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF; Platform of Proteomics in the Research Unit of Biochemistry and Structure Protein, INRA, Jouy en Josas, France).
RESULTS
Hsp70 interacts with the nucleoprotein.
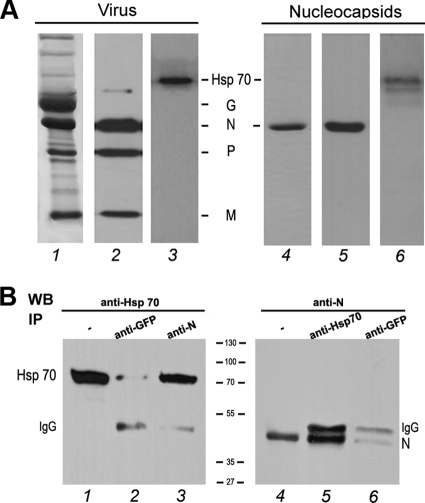
Hsp70 has been shown to be present in rabies virions (37) and within NBL structures containing the viral L, N, and P proteins and viral RNAs (20, 26). We confirmed here that Hsp70 was also incorporated in purified virions (Fig. 1A, lanes 1 to 3). NBL structures are sites of viral transcription and replication (20). To identify the viral partner(s) of Hsp70, we first investigated whether Hsp70 protein was present in the nucleocapsids composed of the viral RNA tightly associated with the N protein. Rabies nucleocapsids were purified from infected cells by cesium chloride gradient and analyzed by Western blotting with anti-N and anti-Hsp70 antibodies. Hsp70 was found to be associated with the nucleocapsids containing exclusively the N protein (Fig. 1A, lanes 4 to 6). This result suggests that Hsp70 binds to the N protein.
Fig 1.
Hsp70 interacts with the rabies virus N protein. (A) Hsp70 is detected in purified virus and in purified nucleocapsid. Purified virus (5 μg) and purified nucleocapsid (5 μg), prepared as described in Materials and Methods, were analyzed by SDS-PAGE followed by Coomassie blue staining (lanes 1 and 4) and Western blotting using a mixture of anti-N, anti-P, and anti-M antibodies (lanes 2 and 5) and a polyclonal anti-Hsp70 (lanes 3 and 6). (B) BSR cells were cotransfected with plasmids expressing Hsp70 and rabies N protein. Two days after transfection, cells were harvested and lysed. Cell lysate was incubated with the irrelevant polyclonal anti-GFP antibody (lanes 2 and 6), with the mouse MAb anti-N antibody (lane 3), or with the polyclonal anti-Hsp70 antibody (lane 5). Immune complexes were analyzed by Western immunoblotting using anti-Hsp 70 (lanes 2 and 3) and anti-N (lanes 5 and 6) antibodies. Direct cellular extracts of transfected cells (input) also were analyzed by WB with anti-Hsp70 (lane 1) and anti-N (lane 4). Bands corresponding to Hsp70, N, and heavy-chain IgG are indicated.
To examine whether Hsp70 interacts with the N protein in rabies-infected cells, two types of experiments were conducted. In the first one, an immunoaffinity column immobilized with anti-N MAbs was used to identify cellular factors interacting with the N protein. The cytoplasmic fraction of human neuroblastoma SK-N-BE-infected cells was loaded on an anti-N immunoaffinity column. In parallel, uninfected cell extracts or infected cell extracts incubated without antibodies were used as controls. The proteins retained on the column were eluted, analyzed by SDS-PAGE, and identified by MALDI-TOF proteomic analysis after excision from the gels. Among the cellular proteins, the chaperone Hsp70 was identified as a cellular partner of the N protein (data not shown). As expected, the viral phosphoprotein P was also detected as a viral partner of N. In the second experiment, BSR cells were cotransfected by two plasmids individually expressing Hsp70 and N protein. Cell extracts were immunoprecipitated with a mouse monoclonal anti-N antibody. The proteins present in the immune complexes then were analyzed by Western blotting with anti-Hsp70 antibodies. Hsp70 was detected in the immunoprecipitates obtained with the anti-N antibody but not with the unrelated anti-GFP antibody, indicating the specificity of the interaction (Fig. 1B, lanes 2 and 3). The reverse experiment showed that the N protein also could be coimmunoprecipitated with Hsp70 (Fig. 1B, lane 5). Thus, all of these data indicated that Hsp70 interacts with the viral N protein in the absence of other viral proteins, and this supports the idea that this interaction results in the localization of Hsp70 in NBL structures (20).
Rabies infection induces Hsp70 expression.
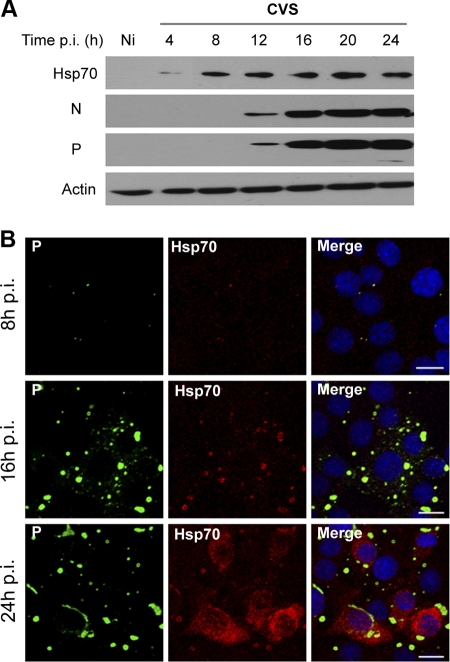
We then investigated the effect of viral infection on the expression of Hsp70. Infected cells were analyzed at different times postinfection (p.i.) by Western blotting. Hsp70 was detected at early times postinfection (4 h p.i.), whereas no detectable trace was observed in uninfected cells (Fig. 2A). The Hsp70 level was enhanced during viral infection (Fig. 2A). This was confirmed by the confocal immunofluorescence analysis of infected cells, which showed that Hsp70 levels increased in the NBL structures concomitantly with those of viral N and P proteins (Fig. 2B, 16 h p.i.) before spreading throughout the cytoplasm (Fig. 2B, 24 h p.i.). These results indicated that Hsp70 is induced by rabies infection.
Fig 2.
Expression of Hsp70 during rabies virus infection. (A) BSR cells were uninfected (Ni) or infected at an MOI of 1. At different times p.i. as indicated, cell extracts were analyzed by Western blotting using anti-Hsp70, anti-N, anti-P, and anti-actin antibodies. (B) BSR cells were infected as described for panel A. Double-immunofluorescence staining was performed with anti-Hsp70 and anti-P antibodies. The scale bars correspond to 20 μm.
Modulation of cellular Hsp70 and effect on rabies virus infection.
It is not known whether the induction of Hsp70 expression is the result of a cellular stress response due to viral infection or if the protein is induced specifically by some viral mechanism because it would play a role during the viral life cycle. To analyze the function of this Hsp70 induction during rabies infection, different experiments were performed to modulate the intracellular level of Hsp70.
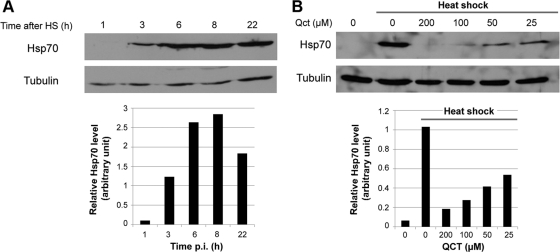
First, we determined the conditions to modulate the Hsp70 expression in noninfected cells. To induce the expression of Hsp70 protein, BSR cells were subjected to heat shock (HS) for 20 min at 45°C, and then the cells were reincubated at 37°C for different times. The Western blotting indicated that the induction of the expression of Hsp70 was maximal at 6 h postshock and was maintained thereafter (Fig. 3A). In contrast, treatment with an inhibitor of Hsp synthesis, quercetin (Qct), before HS inhibited the synthesis of Hsp70 proteins. The application of 100 μM Qct resulted in a drastic inhibition of Hsp70 expression (Fig. 3B) without causing visible cell damage to the cell (data not shown). Thus, the following experiments were performed at 6 h after HS or with 100 μM Qct to induce or inhibit Hsp70 expression, respectively.
Fig 3.
Modulation of Hsp70 expression in the absence of infection. (A) Uninfected BSR cells were incubated for 20 min at 45°C and then at 37°C for various times after heat shock (HS) as indicated. Cell extracts were analyzed by Western blotting using anti-Hsp70 and anti-tubulin antibodies (used as a loading control). The level of Hsp70 was quantified by immunoblot scanning and normalized with respect to the amount of tubulin (lower panel). (B) Uninfected BSR cells were untreated (0) or were treated with different concentrations of quercetin (Qct) (as indicated) for 1 h before HS at 45°C for 20 min in the presence of quercetin. After HS, the cells were incubated at 37°C for 6 h. Cell extracts were analyzed by Western blotting using anti-Hsp70 and anti-tubulin antibodies. The level of Hsp70 was quantified by immunoblot scanning and normalized with respect to the amount of tubulin (lower panel).
We first analyzed the effect of the modulation of Hsp70 on the synthesis of viral proteins during viral infection (Fig. 4A and B). When Hsp70 was overexpressed (HS), we observed an increase of P protein synthesis (factor of 4 at 12 h p.i.) compared to control cells (Fig. 4A), although this increase became less pronounced with the progression of infection (Fig. 4A, 16 to 20 h p.i.). In contrast, the inhibition of Hsp70 (Qct) resulted in a drastic reduction of the level of P protein at 12 and 16 h p.i. (factors of 5 and 20, respectively). The accumulation of the other viral proteins was also inhibited (data not shown). This inhibition was not due to drug toxicity, as the number of infected BSR cells obtained by counting the fluorescent cells was unchanged in the absence of Hsp70 (data not shown). This drastic effect on the viral protein level was attenuated at later stages of infection concomitantly with the induced Hsp70 expression (Fig. 4A). To investigate whether the inhibition of viral protein accumulation was due to the inhibition of viral protein translation or to an increase of viral protein degradation, we analyzed the effect of Qct on viral translation by pulse-chase experiments with [35S]methionine/cysteine. Infected cells treated with Qct were labeled for 5 min and chased for periods of up to 60 min. Cells then were lysed and immunoprecipitated with anti-N and anti-P or anti-M antibodies. The immunoprecipitates were analyzed by SDS-PAGE. Treatment with Qct dramatically affected viral protein synthesis (Fig. 4B). To analyze the effect of Qct on cellular protein synthesis, mock-infected cells left untreated or treated with Qct were labeled for 5 min and analyzed directly by SDS-PAGE. In contrast to viral proteins, cellular protein synthesis did not seem to be affected (Fig. 4B, lower, lanes 17 and 18). This indicates that the drug led to the inhibition of viral protein translation but did not affect protein stability.
Fig 4.
Effect of modulation of Hsp70 on viral protein synthesis. (A) BSR cells were left untreated (DMSO) or were treated with quercetin (Qct; 100 μM) for 1 h and infected (MOI of 1) in the presence of Qct, or cells were submitted to heat shock (HS) (after 1 h of DMSO treatment) and infected (MOI of 1) 6 h after HS. Cells were lysed at different times p.i. as indicated, and cell extracts were analyzed by Western blotting using anti-Hsp70, anti-P, and anti-tubulin antibodies. The levels of P and Hsp70 proteins present in different conditions (DMSO, Qct, and HS) were quantified by Western blot scanning and normalized with respect to the amount of tubulin (expressed in logarithmic scale) (lower panel). An arbitrary level of 100 is applied to P and Hsp70 levels (*) in a classical infection (DMSO) for each hour (p.i.) to allow comparisons. (B) BSR cells were left untreated (−) or were treated with 100 μM Qct (+) for 1 h and infected (MOI of 1) in the presence of Qct. After 16 h p.i., cell were pulse labeled with [35S]methionine and [35S]cysteine for 5 min and then chased in cold medium for the times indicated before lysis. The lysates were immunoprecipitated with anti-N and anti-P antibodies (upper panel) or anti-M antibodies (lower panel). The immunoprecipitates were analyzed by SDS-PAGE followed by autoradiography. Mock-infected cells were labeled for 5 min (without chase), and cell extracts (CE) were directly analyzed by SDS-PAGE followed by autoradiography (lower panel, lanes 17 and 18).
We next studied the effect of Hsp70 on viral production. Similar experiments were performed and supernatants were used for viral titration (Fig. 5A and B). When Hsp70 was overexpressed (HS), viral production at 16 h p.i. was slightly higher (factor 2) than that in normal conditions (Fig. 5A and B). In contrast, the inhibition of Hsp70 (with Qct) induced a dose-dependent inhibition of the viral production during the viral cycle (Fig. 5A). Viral yield was 10- to 20-fold and up to 100-fold lower than that in control cells in the presence of 100 and 200 mM Qct, respectively (Fig. 5A and B). These data indicated that the reduction of Hsp70 significantly impairs viral protein synthesis and virus production, suggesting that Hsp70 has a proviral effect during rabies infection. However, the overexpression of Hsp70 in infected cells has at most a small effect on viral life cycle. This could be explained by the fact that Hsp70 is efficiently induced by rabies infection, and this induction would be sufficient to support the viral life cycle.
Fig 5.
Effect of Qct treatment on viral production. BSR cells were infected at an MOI of 1 without any treatment (DMSO) or in the presence of Qct as indicated or were subjected to HS treatments as described in the text. Culture media were collected at 16 h p.i. (A) or at different times pi (B), and viral titers were determined as described in Materials and Methods. P values (**, P < 0.001; ***, P < 0.0001) were calculated using the Student's test.
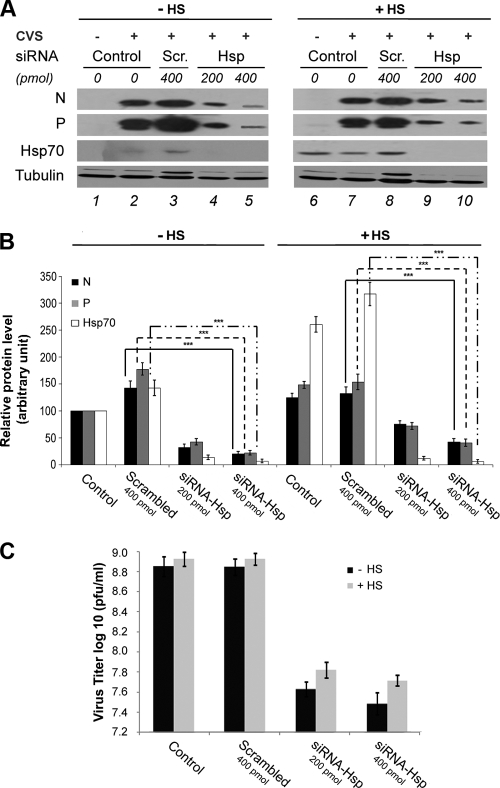
Specific inhibition of Hsp70 expression by siRNA and effect on viral infection.
As quercetin is not a specific inhibitor of Hsp70 but inhibits the synthesis of other heat shock proteins, we performed silencing experiments to investigate more specifically the function of Hsp70 during rabies viral infection. BSR cells were transfected with two different concentrations of Hsp70 siRNA (200 and 400 pmol) for 48 h, and then cells were infected under normal (control) or heat shock conditions. Hsp70 was efficiently downregulated, as shown in HS-treated cells (Fig. 6A, lanes 9 and 10) compared to control and scrambled siRNA (Fig. 6A, lanes 6 to 8). In the absence of HS, the induction of Hsp70 by viral infection (Fig. 6A, lanes 2 and 3) was also inhibited after Hsp70 silencing (Fig. 6A, lanes 4 and 5). These observations were confirmed by the quantification of the cellular Hsp70 protein level (Fig. 6B). Under these conditions, the knockdown of Hsp70 expression was associated with a significant reduction of the amounts of viral N and P proteins of 3- to 8-fold depending on the siRNA dose (Fig. 6A, lanes 4 and 5 versus lane 3, and B). The decrease of the amount of viral N and P proteins was maintained in HS conditions between a factor of 2 to 4 depending on siRNA-Hsp70 quantities (Fig. 6A, lanes 9 and 10 versus lane 8, and B). As expected, the N and P protein levels were similar in the presence of scrambled siRNA (Fig. 6A, lanes 2 and 3 versus 7 and 8, and B). These results confirmed that the specific inhibition of Hsp70 synthesis led to a reduction of viral protein levels and indicated that Hsp70 has a proviral role.
Fig 6.
Effect of specific inhibition of Hsp70 by siRNA interference on rabies replication. (A) BSR cells were not transfected (Control) or were transfected with scrambled siRNA (Scr) or siRNA-Hsp70 (Hsp) (two concentrations as indicated). After 48 h, cells were submitted to HS (+HS) or not (−HS), and 6 h later cells were infected at an MOI of 3 for 16 h. Cell extracts were analyzed by Western blotting using anti-Hsp70, anti-N, anti-P, and anti-tubulin antibodies. (B) Quantitative analysis of cellular P, N, and Hsp70 protein levels in cells treated as described for panel A. The resulting Western blots (three independent experiments) were quantified using immunoblot scanning and normalized with respect to the amount of tubulin. The amounts of N (black histograms), P (gray histograms), and Hsp70 (white histograms) were measured, and an arbitrary level of 100 is applied to N, P, and Hsp70 levels in a classical infection (lane 2 of panel A) for comparisons. P values (***, P < 0.0001) were calculated using the Student's test. (C). Culture media of infected cells treated as described for panel A were collected at 16 h p.i., and viral titers were determined as described in Materials and Methods. Viral titers represent averages from three independent experiments. Error bars indicate the standard deviations.
The effect of siRNA-Hsp70 on viral production was also analyzed in the same conditions as those described above. The siRNA-Hsp70 treatment resulted in an inhibition of the viral production of 10- to 15-fold compared to control and scrambled siRNA, whatever the dose of siRNA and conditions (Fig. 6C). These results confirmed that Hsp70 plays a positive role in rabies infection.
Effect of Hsp70 on viral RNA synthesis.
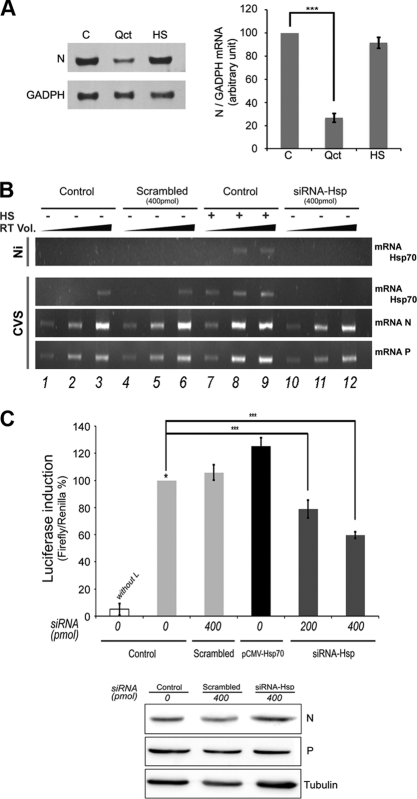
We next tested whether the inhibition of viral protein synthesis (observed in the presence of Qct or in siRNA experiments) could reflect an inhibition of viral transcription. First, we analyzed the effect of Qct on viral mRNA synthesis. Total RNAs were extracted from untreated infected cells, cells treated with Qct, or cells submitted to HS and were analyzed for the presence of the viral N mRNA by Northern blot analysis. In the presence of Qct, the amount of N mRNA was reduced by a factor 5 (Fig. 7A), indicating that Qct impaired viral transcription. However, the induction of Hsp70 expression (HS) did not result in an increase of the amount of rabies N mRNA (Fig. 7A).
Fig 7.
Effect of Hsp70 on viral RNA synthesis. (A) Effect of Quercetin on viral RNA by Northern blotting. BSR cells were left untreated (C) or were treated with quercetin (Qct; 100 μM) for 1 h and infected (MOI of 1) in the presence of Qct, or cells were submitted to heat shock (HS) (after 1 h of DMSO treatment) and infected (MOI of 1) 6 h after HS. Total RNA was extracted from cells, and samples (10 μg of RNA per lane) were analyzed for the presence of rabies N mRNA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA, as described in Materials and Methods. (B) Detection of viral RNA by RT-PCR. BSR cells were left untreated (Control) or were treated with scrambled siRNA or siRNA-Hsp70 (two concentrations as indicated). After 48 h, cells were submitted to HS (+) or not (−), and 6 h later cells were infected by CVS at an MOI of 3 for 16 h and total RNA was extracted. The presence of Hsp70 mRNA or rabies N and P mRNA was determined by RT-PCR as described in Materials and Methods. The PCRs were performed using three different volumes of RT mixture (cDNA; 1, 3, and 5 μl) and were analyzed on a 1.2% agarose gel with ethidium bromide. (C) Luciferase expression from a rabies minireplicon system. BSR cells were left untreated (control), were treated with scrambled siRNA or siRNA-Hsp70 (different concentrations), or were transfected by a plasmid encoding Hsp70 (pCMV-Hsp70). After 48 h, cells were transfected with plasmids encoding L, N, and P proteins, T7 polymerase, and the plasmid (pRL-TK) encoding Renilla luciferase in the presence of a rabies minireplicon system plasmid for the expression of Firefly luciferase. The L plasmid was missing from one experiment, and this served as a negative control (without L). After 36 h, cells extracts were analyzed for luciferase expression. Data represent the separate assays for Firefly luciferase normalized to the expression of Renilla luciferase and are expressed as percentages of the control (*). Error bars indicate standard deviations (from three independent experiments). In the bottom panel of B, the expression of N, P, and tubulin (Tub) present in the cell extracts of control (C), scramble (Sc), and siRNA Hsp was analyzed by Western blot analysis with anti-N, anti-P, and anti-tubulin antibodies.
We then tried to confirm these effects on viral RNA synthesis in the presence of a more specific Hsp70 depletion. As Northern blotting is not the most sensitive method, we first used RT-PCR to detect the viral mRNA of N and P proteins and the mRNA of Hsp70 in infected cells after Hsp70 silencing experiments and in cells infected under normal (control) or heat shock (HS) conditions (Fig. 7B). Increasing amounts of RT reactions (1, 3, and 5 μl) were used for PCR amplification. As expected, HS or/and rabies virus infection induced the expression of Hsp70 mRNA (Fig. 7A, lanes 1 to 3 and 7 to 9) and Hsp70 was efficiently downregulated in the presence of siRNA-Hsp70 compared to control and scrambled siRNA (Fig. 7B, lanes 4 to 6 and 10 to 12). Hsp70 induction resulted in a slight increase of the amount of viral mRNA of N and P (Fig. 7B, lanes 7, 8, and 9 versus lanes 1, 2, and 3); however, Hsp70 silencing did not result in a decrease of the amount of viral mRNA by this method (Fig. 7B, lanes 10, 11, and 12 versus lanes 4, 5, and 6).
We then used a more quantitative assay, the luciferase reporter system in a minireplicon system, that allows the reconstitution of a functional rabies virus RNP. The luciferase gene reporter is framed by 3′ leader and 5′ trailer sequences and is under the control of rabies transcription signals (21). The production of luciferase activity is the result of the encapsidation of the minigenome followed by its transcription by the L-P complex and by its replication in the presence of N protein. The siRNA-Hsp70-treated BSR cells were cotransfected with plasmids encoding L, N, and P proteins and the minigenome. As expected, luciferase activity background was obtained in the absence of the L protein (Fig. 7C). The overexpression of Hsp70 induced a slight stimulation of the luciferase activity (to about 20%) compared to that of the control (Fig. 7C). Similarly, the Hsp70 silencing weakly reduced the luciferase activity (to about 20 to 40%) in a dose-dependent manner. We ensured that the amounts of viral N and P proteins expressed from the plasmids were unchanged in the different conditions (Fig. 7C). Taken together, these data suggested that Hsp70 plays a minor role even in viral transcription. However, whether this role is sufficient to explain further effects on viral protein synthesis and viral replication should be further investigated.
DISCUSSION
In this study, we have investigated the role of Hsp70 during rabies virus infection. Coimmunoprecipitation and proteomic analyses indicated that Hsp70 interacts with N. However, we cannot exclude that this interaction is indirect and mediated by one or more other factors. Whatever the case, this interaction probably results in the presence of Hsp70 in purified nucleocapsids, within NBL structures, and in purified rabies particles. In addition, we have shown that rabies infection induces an increase of cellular Hsp70 levels as soon as 4 h p.i. in BSR cells and in neuroblastoma cells (data not shown). The increase in Hsp70 levels following viral infection has been widely observed. This can be explained by different mechanisms. The induction may occur indirectly through the production of a large number of unfolded proteins during viral infection. Second, the viruses could interfere with stress signal transduction pathways independently of the folding status of proteins. Indeed, Hsp70, which is involved in many cellular processes, such as cell cycle, apoptosis, and cellular innate immunity pathways, is frequently diverted by viruses for their benefit (23, 25). In the case of rabies virus, Hsp70 enrichment might simply be correlated with excess amounts of viral proteins (particularly N and P); nevertheless, the recruitment of Hsp70 inside the NBLs, which are sites of viral transcription and replication, in the purified nucleocapsids and in the purified virions suggests that Hsp70 plays an active role in the viral life cycle.
The downregulation of Hsp70, using specific chaperons inhibitors such as quercetin or RNA interference, significantly reduced the intracellular level of viral proteins and viral production. The reduction of viral protein accumulation appeared to be the result of an inhibition of viral translation more than protein instability, as shown by pulse-chase labeling (in the presence of Qct). Viral transcription was also decreased. Interestingly, the inhibition at different steps of the viral cycle (transcription, translation, and viral production) was more drastic in the presence of Qct than siRNA, suggesting that other chaperones are involved in the rabies virus infection cycle. Unexpectedly, the overexpression of Hsp70 following heat shock treatment or pCMV-Hsp70 transfection resulted in a slight increase of viral protein synthesis or viral production. This is likely because infected cells already expressed a sufficiently high level of Hsp70, and the overexpression before the infection did not have much effect. The siRNA-induced reduction of viral yield may be due to a decrease of the amount of viral particles released, but it could well be related to the reduced infectivity of the progeny virions lacking Hsp70.
All of these data indicate that Hsp70 has a proviral effect on rabies virus replication by acting during at least one step of the viral cycle. As these steps are connected, it is difficult to clearly determine the mechanism by which Hsp 70 controls viral infection. Indeed, the resulting inhibition in viral protein synthesis and in viral production could be a direct consequence of the viral transcription inhibition, but it could also be amplified by an Hsp70-specific role in these further steps of the viral infection.
Interestingly, the potential role of Hsp70 in viral RNA synthesis is consistent with the interaction of Hsp70 with N and its association with the NBLs, which are sites of viral transcription and replication, suggesting a possible interaction with the polymerase complex. Indeed, previous studies of three paramyxoviruses, measles virus, canine distemper virus, and respiratory syncytial virus, have provided evidence for an interaction between the N protein and Hsp70 (3, 29, 30). In these cases, Hsp70 has been shown to stimulate viral polymerase in vitro activity, but it is not clear whether Hsp70 acts directly as a cofactor of the enzymatic process or whether it assists the folding of the N protein in an active form. Interestingly, the positive effect of Hsp70 on viral infection is not a common property of negative-strand RNA viruses belonging to the same order, highlighting the complexity of the virus-Hsp connections.
Other chaperones could play a role in rabies viral RNA synthesis. For example, Hsp60 has been detected in the transcriptase complex of VSV, although its function has not been investigated so far (35), and Hsp40 has been shown to enhance the binding of Hsp70 with the N protein of measles virus (8). More recently, it has been reported that Hsp70 is associated with complexes formed between Hsp40 and the viral protein Nef in HIV-1-infected cells and that the differential expression of Hsp40 and Hsp70 reciprocally regulates viral gene expression and replication (19).
In conclusion, these data support a positive regulatory role for Hsp70 in the rabies virus infection cycle and suggest that Hsp70, consistently with its multifunctional role, participates at the different stages of the viral life cycle, such as the transcriptional and/or translational level and/or viral assembly and budding.
ACKNOWLEDGMENTS
We are grateful to Yves Gaudin for careful reading of the manuscript. We acknowledge Noël Tordo (Unit Antiviral Strategies, Institut Pasteur, France) for providing the rabies minireplicon system. We thank the Platform of Proteomics (Unités de Recherches de Biochimie et Structures des Protéines, Jouy en Josas, France). Confocal microscopy was performed at the Plate-forme Imagerie et Biologie Cellulaire of the CNRS campus, which is supported by the Institut Fédératif de Recherche 87 La Plante et Son Environnement and the ASTRE program of the Conseil Général de l'Essonne.
We acknowledge support from the CNRS.
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1. Albertini AA, Ruigrok RW, Blondel D. 2011. Rabies virus transcription and replication. Adv. Virus Res. 79:1–22 [DOI] [PubMed] [Google Scholar]
- 2. Broquet AH, et al. 2007. Hsp70 negatively controls rotavirus protein bioavailability in caco-2 cells infected by the rotavirus RF strain. J. Virol. 81:1297–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown G, et al. 2005. Evidence for an association between heat shock protein 70 and the respiratory syncytial virus polymerase complex within lipid-raft membranes during virus infection. Virology 338:69–80 [DOI] [PubMed] [Google Scholar]
- 4. Bukau B, Deuerling E, Pfund C, Craig EA. 2000. Getting newly synthesized proteins into shape. Cell 101:119–122 [DOI] [PubMed] [Google Scholar]
- 5. Carsillo T, Traylor Z, Choi C, Niewiesk S, Oglesbee M. 2006. hsp72, a host determinant of measles virus neurovirulence. J. Virol. 80:11031–11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chenik M, Chebli K, Gaudin Y, Blondel D. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75:2889–2896 [DOI] [PubMed] [Google Scholar]
- 7. Conzelmann KK. 1998. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu. Rev. Genet. 32:123–162 [DOI] [PubMed] [Google Scholar]
- 8. Couturier M, et al. 2010. High affinity binding between Hsp70 and the C-terminal domain of the measles virus nucleoprotein requires an Hsp40 co-chaperone. J. Mol. Recognit. 23:301–315 [DOI] [PubMed] [Google Scholar]
- 9. De Marco A, Santoro MG. 1993. Antiviral effect of short hyperthermic treatment at specific stages of vesicular stomatitis virus replication cycle. J. Gen. Virol. 74:1685–1690 [DOI] [PubMed] [Google Scholar]
- 10. Diaz-Latoud C, et al. 1997. Herpes simplex virus Us11 protein enhances recovery of protein synthesis and survival in heat shock treated HeLa cells. Cell Stress Chaperones 2:119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Didcock L, Young DF, Goodbourn S, Randall RE. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elbashir SM, Harborth J, Weber K, Tuschl T. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199–213 [DOI] [PubMed] [Google Scholar]
- 13. Garcin D, Curran J, Kolakofsky D. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823–8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaudin Y. 2000. Rabies virus-induced membrane fusion pathway. J. Cell Biol. 150:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hartl FU, Hayer-Hartl M. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852–1858 [DOI] [PubMed] [Google Scholar]
- 16. Hirayama E, Atagi H, Hiraki A, Kim J. 2004. Heat shock protein 70 is related to thermal inhibition of nuclear export of the influenza virus ribonucleoprotein complex. J. Virol. 78:1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iseni F, Barge A, Baudin F, Blondel D, Ruigrok RW. 1998. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J. Gen. Virol. 79:2909–2919 [DOI] [PubMed] [Google Scholar]
- 18. Jindal S, Young RA. 1992. Vaccinia virus infection induces a stress response that leads to association of Hsp70 with viral proteins. J. Virol. 66:5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumar M, et al. 2011. Reciprocal regulation of human immunodeficiency virus-1 gene expression and replication by heat shock proteins 40 and 70. J. Mol. Biol. 410:944–958 [DOI] [PubMed] [Google Scholar]
- 20. Lahaye X, et al. 2009. Functional characterization of Negri bodies (NBs) in rabies virus infected cells: evidence that NBs are sites of viral transcription and replication. J. Virol. 83:7948–7958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le Mercier P, Jacob Y, Tanner K, Tordo N. 2002. A novel expression cassette of lyssavirus shows that the distantly related Mokola virus can rescue a defective rabies virus genome. J. Virol. 76:2024–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li G, Zhang J, Tong X, Liu W, Ye X. 2011. Heat shock protein 70 inhibits the activity of influenza a virus ribonucleoprotein and blocks the replication of virus in vitro and in vivo. PLoS One 6:e16546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mayer MP. 2010. Gymnastics of molecular chaperones. Mol. Cell 39:321–331 [DOI] [PubMed] [Google Scholar]
- 24. Mayer MP. 2005. Recruitment of Hsp70 chaperones: a crucial part of viral survival strategies. Rev. Physiol. Biochem. Pharmacol. 153:1–46 [DOI] [PubMed] [Google Scholar]
- 25. Mayer MP, Bukau B. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62:670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menager P, et al. 2009. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri bodies. PLoS Pathog. 5:e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Michalovitz D, Fischer-Fantuzzi L, Vesco C, Pipas JM, Oren M. 1987. Activated Ha-ras can cooperate with defective simian virus 40 in the transformation of nonestablished rat embryo fibroblasts. J. Virol. 61:2648–2654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagy PD, Wang RY, Pogany J, Hafren A, Makinen K. 2011. Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411:374–382 [DOI] [PubMed] [Google Scholar]
- 29. Oglesbee M, Ringler S, Krakowka S. 1990. Interaction of canine distemper virus nucleocapsid variants with 70K heat-shock proteins. J. Gen. Virol. 71:1585–1590 [DOI] [PubMed] [Google Scholar]
- 30. Oglesbee MJ, Kenney H, Kenney T, Krakowka S. 1993. Enhanced production of morbillivirus gene-specific RNAs following induction of the cellular stress response in stable persistent infection. Virology 192:556–567 [DOI] [PubMed] [Google Scholar]
- 31. Omar RA, Lanks KW. 1984. Heat shock protein synthesis and cell survival in clones of normal and simian virus 40-transformed mouse embryo cells. Cancer Res. 44:3976–3982 [PubMed] [Google Scholar]
- 32. Peluso RW, Moyer SA. 1988. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162:369–376 [DOI] [PubMed] [Google Scholar]
- 33. Phillips B, Abravaya K, Morimoto RI. 1991. Analysis of the specificity and mechanism of transcriptional activation of the human hsp70 gene during infection by DNA viruses. J. Virol. 65:5680–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pratt WB, Toft DO. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228:111–133 [DOI] [PubMed] [Google Scholar]
- 35. Qanungo KR, Shaji D, Mathur M, Banerjee AK. 2004. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc. Natl. Acad. Sci. U. S. A. 101:5952–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raux H, Iseni F, Lafay F, Blondel D. 1997. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J. Gen. Virol. 78:119–124 [DOI] [PubMed] [Google Scholar]
- 37. Sagara J, Kawai A. 1992. Identification of heat shock protein 70 in the rabies virion. Virology 190:845–848 [DOI] [PubMed] [Google Scholar]
- 38. Santoro MG. 1994. Heat shock proteins and virus replication: hsp70s as mediators of the antiviral effects of prostaglandins. Experientia 50:1039–1047 [DOI] [PubMed] [Google Scholar]
- 39. Weeks SA, et al. 2010. A targeted analysis of cellular chaperones reveals contrasting roles for heat shock protein 70 in flock house virus RNA replication. J. Virol. 84:330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Young JC, Barral JM, Ulrich Hartl F. 2003. More than folding: localized functions of cytosolic chaperones. Trends Biochem. Sci. 28:541–547 [DOI] [PubMed] [Google Scholar]
- 41. Zeng R, Zhang Z, Mei X, Gong W, Wei L. 2008. Protective effect of a RSV subunit vaccine candidate G1F/M2 was enhanced by a HSP70-Like protein in mice. Biochem. Biophys. Res. Commun. 377:495–499 [DOI] [PubMed] [Google Scholar]
- 42. Zhang X, et al. 2005. Hsp72 recognizes a P binding motif in the measles virus N protein C-terminus. Virology 337:162–174 [DOI] [PubMed] [Google Scholar]