Abstract
Ms1/STARS is a novel muscle-specific actin-binding protein that specifically modulates the myocardin-related transcription factor (MRTF)-serum response factor (SRF) regulatory axis within striated muscle. This ms1/STARS-dependent regulatory axis is of central importance within the cardiac gene regulatory network and has been implicated in cardiac development and postnatal cardiac function/homeostasis. The dysregulation of ms1/STARS is associated with and causative of pathological cardiac phenotypes, including cardiac hypertrophy and cardiomyopathy. In order to gain an understanding of the mechanisms governing ms1/STARS expression in the heart, we have coupled a comparative genomic in silico analysis with reporter, gain-of-function, and loss-of-function approaches. Through this integrated analysis, we have identified three evolutionarily conserved regions (ECRs), α, SINA, and DINA, that act as cis-regulatory modules and confer differential cardiac cell-specific activity. Two of these ECRs, α and DINA, displayed distinct regulatory sensitivity to the core cardiac transcription factor GATA4. Overall, our results demonstrate that within embryonic, neonatal, and adult hearts, GATA4 represses ms1/STARS expression with the pathologically associated depletion of GATA4 (type 1/type 2 diabetic models), resulting in ms1/STARS upregulation. This GATA4-dependent repression of ms1/STARS expression has major implications for MRTF-SRF signaling in the context of cardiac development and disease.
INTRODUCTION
The heart is the first organ to develop during mammalian embryogenesis, with proper embryonic formation and postnatal function being essential for the physiological well-being of the organism. Inherited and acquired defects in the structure and function of the embryonic and adult heart are the major causes of mortality in the developed world. At the molecular level, normal cardiac development and postnatal function are dependent on the correct temporal-spatial execution of cardiac gene regulatory networks (GRNs) underpinned by a core set of evolutionarily conserved cardiac enriched transcription factors (TFs) and cofactors (9, 40).
A pivotal family of TFs in the cardiac GRN is the GATA family of proteins. One particular family member, GATA-binding protein 4 (GATA4), has been extensively characterized as an essential modulator of cardiac gene expression (32). Numerous in vitro and in vivo gain- and loss-of-function studies have implicated GATA4 in modulating developmental and differentiated gene expression in the heart (45). Within the adult heart, GATA4 also mediates the hypertrophic response to stimuli including pressure overload, isoproterenol, phenylephrine, and endothelin-1 (5, 33, 39). Coupled to the hypertrophic response, GATA4 is also a regulator of cell survival that is able to specifically modulate cardiac apoptosis and autophagy in response to diverse cardiotoxic stimuli (3, 21).
Another important TF in the cardiac GRN is the MADS box domain-containing protein, serum response factor (SRF). In the cardiovascular system, SRF plays a critical role in modulating vascular smooth muscle cell (SMC) and cardiac myocyte differentiation in addition to the morphogenetic program regulating the development of the heart (6, 30, 35, 36). Studies utilizing conditional-gene-targeted mice demonstrated the obligatory role for SRF during cardiac development, with mutant mice succumbing to cardiac insufficiency and contractile dysfunction (37). Postnatal SRF is critical for maintaining the basal “trophic” state of the heart and is necessary for inducing myocardial growth in response to stress stimuli (31, 43, 56, 57).
SRF alone is a poor transcriptional activator, with its activity being dependent on its interaction with a range of cell-type-specific and signal-responsive cofactors (49). Myocardin and the myocardin-related transcription factors (MRTFs) MRTF-A (MAL or MKL-1) and MRTF-B (MKL-2) constitute a family of extraordinarily powerful SRF coactivators (7, 46, 50, 51). Myocardin, which is constitutively localized to the nucleus, is expressed specifically within cardiac and smooth muscle cells and is both necessary and sufficient for the normal expression of SRF-dependent smooth muscle genes (25, 50). In contrast, the MRTFs are expressed broadly in many different cell types (51) and act to couple actin dynamics and Rho signaling to SRF by shuttling from the cytoplasm to the nucleus in a tightly modulated manner.
Molecular pathways that promote muscle-specific MRTF nuclear translocation represent important regulatory modulators of the MRTF-SRF signaling axis. We, and others, have previously identified a novel striated muscle-specific actin-binding protein designated myocyte stress 1 (MS1) (also known as striated muscle-specific activator of Rho signaling [STARS] and actin-binding Rho activator [ABRA]) (2, 29). MS1 synergistically activates SRF-dependent transcription by inducing the nuclear accumulation of MRTF-A and -B through a RhoA-dependent mechanism, establishing a mechanism for MS1-dependent SRF activation. ms1 knockdown via RNA interference (RNAi) resulted in a significant attenuation of muscle-specific SRF activity, confirming that MS1 is an obligatory component of the muscle-specific MRTF-SRF regulatory circuit (2, 23).
Consequently, ms1 has been implicated in cardiac development, postnatal homeostasis, and cardiac dysfunction. During the initial characterization of ms1, we demonstrated that expression is transiently induced in a rat model of left ventricular hypertrophy (29). We subsequently showed that forced MS1 expression in vitro was sufficient to promote hypertrophy, which was associated with increased levels of prohypertrophic MRTF-SRF target genes, including interleukin-6 (IL-6) and brain natriuretic peptide (BNP) (22). MS1 has also been explicitly implicated in cardiac dysfunction in vivo. ms1 expression was upregulated in two independent mouse models of cardiac hypertrophy and in myopathic human hearts (23). By use of conditional transgenesis, the forced overexpression of MS1 sensitized the heart to pressure overload and calcineurin signaling, resulting in an exaggerated deterioration of cardiac function and the increased expression of SRF-dependent fetal cardiac genes (23). These findings demonstrate that ms1 is an important modulator of the cardiac stress response in the adult myocardium. Recent work in our laboratory has also implicated the MS1-MRTF-SRF axis in cardiac development. The knockdown of zebra fish ms1/STARS during development resulted in an SRF-dependent decrease in cardiac contractility (measured as a decrease in the ejection fraction), which was associated with the severe dilation of the myocardial chambers (N. W. Chong et al., submitted for publication).
Despite its importance, little is known of the molecular pathways governing ms1 expression. The characterization of such pathways will give exquisite insight into how the MS1-MRTF-SRF signaling axis is regulated in the heart and identify potential mechanisms for the pathological dysregulation of this important cardiac regulatory subcircuit. In the present study, we report a comprehensive comparative genomic analysis of a large genomic region flanking the rat ms1 gene. In addition to ECRs α and β (41), we have further identified two novel evolutionarily conserved regions (ECRs) (SINA [stress intergenic activator] and DINA [distal intergenic activator]) that contribute to the cardiac cell-specific modulation of ms1 expression. Additionally, we identify GATA4 as a novel cardiac cell-specific regulator of ms1 expression, acting positively and negatively on ECRs DINA and α, respectively. Overall, GATA4 negatively regulates ms1 expression in vivo, with the dysregulation of this negatively acting pathway having implications for the MS1-SRF regulatory axis. Perturbing GATA4 expression either in vitro (knockdown) or pathophysiologically (diabetic cardiomyopathy) has a direct effect on ms1 expression and the activity of the MRTF-SRF signaling pathway. This will have wide-ranging implications for this GRN subcircuit in both cardiac development and disease.
MATERIALS AND METHODS
Cell cultures.
The H9c2 rat myoblast, NIH 3T3 mouse fibroblast, and COS-7 monkey kidney cell lines were grown in a humidified atmosphere containing 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, and streptomycin and penicillin (each at 10 g/liter). Neonatal rat ventricular myocytes (NRVMs) were isolated from 0- to 2-day-old Harlan Sprague-Dawley rat neonates by using a kit from Worthington Biochemical Corporation (Lakewood, NJ). After the digestion of tissue overnight, cells were preplated for 1 h to remove nonmyocytes and then plated onto gelatinized cell culture dishes and cultured overnight in DMEM with 15% bovine serum and penicillin-streptomycin (100 units/ml). On the following day, the cells were washed in phosphate-buffered saline and cultured in serum-free DMEM containing 100 units/ml penicillin-streptomycin and 100 μM 5-bromo-2′-deoxyuridine (BrdU; Sigma, St. Louis, MO). BrdU was used to inhibit the growth of the contaminating nonmyocytes, including fibroblasts. Adult feline cardiomyocytes were isolated using enzymatic digestion and were cultured according to protocols described previously (18a).
Animals.
Transgenic (TG) mice expressing GATA4 in their hearts were described previously (26). Mice were housed in a fully accredited animal facility at the Cardiovascular Research Institute and were handled according to established guidelines. Both GATA4 TG and FVB wild-type (WT) mice were killed by cervical dislocation, and the heart tissues were snap-frozen until they were used for protein and total RNA isolation. GATA4flox/flox mice were generated by gene targeting followed by the Flp-mediated removal of the Kan-Neo resistance cassette, as described previously (44). In these mice, exon 2, containing the start codon and 46% of the coding sequence, is deleted upon the expression of Cre recombinase. The Nkx2.5Cre line was described previously (34). To obtain G4NK embryos, timed matings were set up between GATA4wt/flox Cre-positive and GATA4flox/flox Cre-null mice. To obtain G4S/S::troponin T (TNT)-Cre embryos, timed matings were set up between G4S/S mice (GATA4flox/flox, originally generated by Steve Duncan [54]) and TNT-Cre mice (originally generated by Scott Baldwin [17]). Embryonic day 0.5 (E0.5) was defined as noon of the day when the vaginal plug was detected. All procedures were carried out with the approval of the Institutional Animal Care and Use Committees of Beth Israel Deaconess Medical Centre and Boston Children's Hospital. Hearts from E9.5 (G4NK) and E12.5 (G4S/S::TNT-Cre) embryos were collected by dissection and snap-frozen. Cats were housed and maintained as previously described (18a).
Western blot analysis.
Protein extracts from cultured cardiomyocytes or heart tissue were prepared as described previously (26), except that 1% protease inhibitor cocktail (Sigma) was added to the extraction buffer. Protein samples were subjected to SDS-PAGE, transferred onto a polyvinylidene difluoride membrane (Amersham Pharmacia), and blocked in 5% nonfat dry milk in a 0.1% Tween 20–Tris-buffered saline (TBST) solution for 1 h. Primary antibodies against GATA4 (Santa Cruz Biotechnology, Santa Cruz, CA) and glyeraldehyde-3-phosphate dehydrogenase (GAPDH; Research Diagnostics, Flanders, NJ) were incubated overnight at room temperature in 3% milk in TBST. A horseradish peroxidase-conjugated secondary antibody was incubated for 1 h at room temperature in 3% milk in TBST and processed for chemiluminescent detection by using an ECL Plus Western blotting kit (Amersham Biosciences, Buckinghamshire, United Kingdom).
RNA isolation and reverse transcription-PCR.
Total RNA was extracted from H9c2 cells at 48 h posttransfection (WT/Δ-GATA4 overexpression plasmid) by using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. For whole embryonic hearts (G4NK), hearts from each genotype were pooled (n = 6), and RNA was prepared by using the RNeasy kit with on-column DNase digestion (Qiagen). Three pooled samples were used per genotype. RNA integrity was verified and concentrations were determined by using an Agilent Bioanalyser (model 2100; Agilent Technologies). The RNA (1 μg) was then reverse transcribed to cDNA using oligo(dT) and SuperScript II reverse transcriptase (Invitrogen). ms1 mRNA expression was analyzed by using quantitative PCR with fluorescent-labeled TaqMan probes (ms1 primers and probe, catalog no. Rn00598518_m1; Applied Biosystems). TATA-binding protein (TBP) was used as the internal control (catalog no. Mm00446973_m1; Applied Biosystems). PCR amplifications were performed in duplicate with 25-μl mixtures containing 2 μl cDNA template in 2× PCR Mastermix (Applied Biosystems). Amplification conditions were as follows: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Reactions were performed and products were detected by use of an ABI-Prism HT7900 sequence detector (Applied Biosystems). The level of expression of ms1 mRNA was normalized to the level of TBP expression. For semiquantitative reverse transcription (RT)-PCR, the reverse transcriptase PCR was carried out by using the SuperScript III first-strand synthesis system (Invitrogen, Carlsbad, CA). Briefly, 1 μg of total RNA isolated with TRIzol reagent (Invitrogen) from NRVMs or mouse hearts was used for first-strand cDNA synthesis in a 20-μl reaction mixture volume containing 500 ng of oligo(dT)12–18 as the primer. Two microliters of the reverse transcription reaction mixture was then used for PCR amplification with a volume of 50 μl containing primers specific for the ms1, Bcl2, or GAPDH gene. Aliquots of PCR mixtures were taken during different cycles for agarose gel analysis to determine the linear range of amplification. All reactions were run on a 1.5% agarose gel. The primers used were derived from mouse mRNA sequences, but they are 100% homologous to rat sequences. Primers designed specifically for mouse (Mus musculus) or rat (Rattus norvegicus) are designated. The sequences of the primers were as follows: forward primer 5′-GTGCCAACTGCCAGACTACC-3′ and reverse primer 5′-AGCCTTGTGGGGACAGCTTC-3′for GATA4 (GenBank accession no. NM008092), forward primer 5′-AGTTCGGTGGGGTCATGTGTG-3′ and reverse primer 5′-CCAGGTATGCACCCAGAGTG-3′ for Bcl2 (accession no. NM009741), forward primer 5′-GTGACAGCATAGACACAGAGGAC-3′ and reverse primer 5′-CACTGCTGCCCACCTGCCTT-3′ for ms1 (accession no. NM175844.2), forward primer 5′-AAGGTCATCCCAGAGCTGAAC-3′ and reverse primer 5′-TCATTGAGAGCAATGCCAGCC-3′ for GAPDH (accession no. NM001001978), forward primer 5′-TCACGACGACTCTTACGCAG-3′ and reverse primer 5′-CCTTGAGACCCCGATAGGGA-3′ for Mus musculus JunB, forward primer 5′-AGCCCTGGCAGTCTTTCTCT-3′ and reverse primer 5′-ACCCTTGAGACCCCGATAAG-3′ for Rattus norvegicus JunB, forward primer 5′-AGTCCTTCGGTCTCAAGGCA-3′ and reverse primer 5′-CCGATCCGGTCTATCTTGTGC-3′ for Mus musculus NPPB, and forward primer 5′-CAGCTCTCAAAGGACCAAGG-3′ and reverse primer 5′-CGGTCTATCTTCTGCCCAAA-3′ for Rattus norvegicus NPPB.
Plasmid constructs.
The rat ms1 gene sequence was obtained from GenBank (accession no. NC005106) and was used to design primers that would amplify truncations of the ms1 5′-flanking regions and the ECRs SINA and DINA. Oligonucleotides (see Table S2A in the supplemental material) were designed to amplify a portion of the DNA sequence starting 1,585 bp upstream of the transcription start site (position +1) and the ECRs SINA and DINA with primers tailed with restriction sites for SacI and HindIII restriction enzymes, respectively. In addition to the template (rat genomic DNA, WKY strain) and primers, the reaction mixture contained 0.2 mM deoxynucleoside triphosphates (dNTPs), Expand polymerase buffer, and 5 units of Taq Expand high-fidelity polymerase (Roche). Reaction mixtures were subjected to 35 cycles of amplification (45 s at 94°C, 45 s at 59°C, and 90 s at 72°C). The PCR products were cloned into the vector pGEMT-Easy (Promega) and sequenced to ensure the fidelity of amplification. The verified plasmid was then cut with SacI/HindIII, and the released promoter/ECR fragments were purified and cloned into the pGL3-Basic/Promoter reporter vector, itself SacI/HindIII digested. The WT GATA4 and Δ GATA4 expression plasmids were a gift from Mona Nemer (IRCM, Canada). The −638-atrial natriuretic factor (ANF) luciferase reporter was a gift from Junichi Sadoshima (New Jersey Medical School).
Chromatin immunoprecipitation assay.
Isolated adult feline cardiomyocytes and neonatal rat ventricular myocytes were treated with 1% (vol/vol) formaldehyde for 20 min at room temperature with slow rocking. A chromatin immunoprecipitation (ChIP) assay was performed as described in the manufacturer's manual (Upstate), with some modifications. Cells were washed two times with ice-cold phosphate-buffered saline and collected by centrifugation at 10,000 × g for 2 min. The cell pellet was suspended in lysis buffer and incubated on ice for 20 min. The cell lysate was sonicated 10 times for 10 s each, and the cell debris was spun down. The sample was precleared, and the immunoprecipitation antibody was added to the supernatant and incubated overnight at 4°C. After immunoprecipitation, the eluted protein-DNA complexes were de-cross-linked by heating at 65°C for 4 h. The DNA was ethanol precipitated, and the DNA was suspended in 50 μl of 10 mM Tris buffer. The feline Ncx1 proximal promoter, the murine Smarcd3 cardiac enhancer, and specific ms1 ECR/nonspecific sequences were PCR amplified from immunoprecipitated and nonimmunoprecipitated chromatin samples (primer sequences are shown in Table S2B in supplemental material). Antibody ChIP was performed on isolated E12.5 murine hearts as described previously (15).
Transient transfection and luciferase assay.
H9c2, NIH 3T3, and COS-7 cells were transfected by using the JetPei cationic transfection reagent (QBiogene) according to the manufacturer's protocols. Cells were seeded into six-well plates. At 24 h postplating, the cells were cotransfected with 0.5 μg of the promoter-luciferase construct and equimolar amounts for the other plasmids used (total of 0.6 μg). The total amount of DNA was kept constant by use of an empty vector (pcDNA3.1). To normalize for the transfection efficiency, the pRL-TK (Promega) expression plasmid containing Renilla luciferase (20 ng per well) was cotransfected. Firefly and Renilla luciferase activities were measured at 48 h posttransfection by using the Dual Luciferase assay system (Promega) and a Lumat LB9507 luminometer (Berthold Technologies). All plasmids were purified by using Qiagen columns, and at least two preparations per plasmid were tested. For NRVMs, 24 h after plating, myocytes were transfected with 500 ng of the appropriate reporter plasmid for 12 h by use of Lipofectamine 2000 (Invitrogen). After transfection, serum was removed from the growth medium. Cells were maintained for an additional 48 h before being lysed and processed for the luciferase reporter assay. The transfection efficiency was normalized by using Renilla luciferase activity levels, and each transfection was performed in triplicate and repeated in a minimum of three independent experiments.
Replication-deficient adenoviruses.
Adenoviruses expressing β-galactosidase (β-Gal) (Adβgal), GATA4 (AdGATA4), control RNAi (AdConi), and GATA4-targeting RNAi (AdG4i) were described previously (19, 21, 55). NRVMs were infected with Adβgal, AdGATA4, AdConi, or AdG4i at the multiplicities of infection (MOIs) indicated in Results for 2 h and then cultured in serum-free DMEM for an additional 48 h before processing.
Diabetic mouse models.
Type 1 diabetes was induced in mice by injecting 2-month-old FVB mice with a single dose of streptozotocin (STZ) (intraperitoneally) (150 mg/kg body weight in 10 mmol/liter sodium citrate [pH 4.5]), a well-established agent that destroys pancreatic beta cells (20, 47). A fasting blood glucose content of 15 mmol/liter or higher was considered diabetic, whereas vehicle-treated mice were used as controls. The heart tissues from type 2 diabetic db/db mice and their control db/+ mice were kindly provided by Paul Epstein, University of Louisville. All protocols involving animal use were reviewed and approved by the Internal Animal Care and Use Committee at the University of South Dakota.
Comparative genomic DNA analysis.
Comparative sequence analysis was performed by using Web-based software available at the National Center for Biotechnology Information DCODE Server (ECR browser; http://www.dcode.org/) and the Lawrence Berkley Laboratory Server (VISTA [http://genome.lil.gov/vista/index.shtml]). Orthologous sequences from Homo sapiens, Mus musculus, Rattus norvegicus, Canis familiaris, Monodelphis domestica, Gallus gallus, Xenopus laevis, and Fugu rubripes were obtained from the ENSEMBL (http://www.ensemble.org/) genome database and aligned by using CLUSTAL W, available from the European Bioinformatics Institute (http://www.ebi.ac.uk). For pattern-matching transcription factor-binding site (TFBS) analysis, MatInspector (http://www.genomatix.de/products/MatInspector/) and rVISTA (http://genome.lil.gov/vista/index.shtml) were used, utilizing default settings. The futility theorem states that the majority of TFBSs predicted by using pattern-matching algorithms will represent false-positive predictions, and therefore, the majority of our MatInspector-derived binding sites would be biologically inert. In order to enrich for potentially biologically active TFBSs and filter out false-positive matches, we formulated and executed the following manual filtering protocol. The primary enrichment filter/parameter involves the exclusion of all MatInspector hits with a matrix similarity score (MSS) lower than 0.8. All test sequences with MSSs of >0.8 (85% of total hits) were considered to be biologically more significant and were then manually interrogated for phylogenetic conservation. CLUSTAL W was used for cross-species sequence comparisons. Single-nucleotide conservation mismatches were tolerated at this step if they did not reduce the MSS to <0.8 (at this point, 45% of the initial hits were filtered out). The final filtering parameter was based on the integration and utilization of biologically relevant information regarding the context-specific expression profile of ms1. Based on ms1 expression both in vitro and in vivo, we suspect that context-specific regulatory factors modulating ms1 are likely to include factors implicitly involved in striated muscle-specific gene regulation. In order to construct a library of such factors, we performed a meta-analysis of all the published literature to extract information on TFBSs and cognate factors with characterized roles in striated muscle gene expression. The terms “cardiac transcription factor” and “skeletal muscle and/or myogenic transcription factor” were inputted into the Medline search engine, with the search strings generating 5,507 and 5,655 citations, respectively (June 2008). We then manually sorted the papers and extracted motif information on any TFBS and cognate factor with a role in striated muscle determination, differentiation, growth, proliferation, stress signaling, and postnatal homeostatic function. The remaining candidate TFBSs (approximately 100 hits) were examined for motifs corresponding to those present in our striated muscle library. After this final filtration step, our original 200 MatInspector hits were reduced to a total of 31 hits (86% of the original hits were filtered out), which we consider to be strong regulatory candidates as context-specific TFBSs (data not shown).
Statistical analysis.
Data were expressed as means ± standard errors (SE). Differences between experimental groups were evaluated for statistical significance by using Student's t test for unpaired data or a one-way analysis of variance (ANOVA) followed by Bonferroni's post hoc test. P values of <0.05 were considered to be statistically significant.
RESULTS
Identification of evolutionarily conserved regions and transcription factor-binding sites in the ms1 5′-flanking sequence.
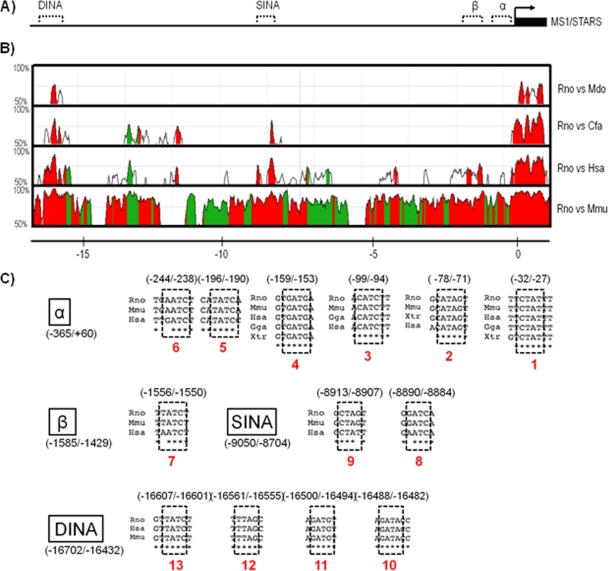
Comparative genomics provides a powerful means for the identification of putative cis-regulatory sequences and modules for experimental verification (28). We have previously utilized this strategy to analyze 5 kbp of the 5′-flanking sequence upstream of the rat ms1 transcriptional start site (TSS) and isolated two evolutionarily conserved regions (ECRs) (designated α and β) with functional myogenic regulatory activity (41). Here we have expanded this analysis to interrogate ∼20 kbp of the ms1 5′-flanking sequence. Using the MULAN alignment engine (available through the ECR browser [http://ecrbrowser.dcode.org/]) (42) with the rat ms1 locus as the reference sequence on the x axis (Fig. 1A), we plotted on the y axis the percent sequence conservation at the nucleotide level between orthologous sequences of mouse, human, dog, and opossum (Fig. 1B).
Fig 1.
Comparative genomic9 analysis of the rat ms1/STARS gene. (A) Schematic of the rat ms1/STARS gene (denoted by the small horizontal bar at the far right) within the ∼20-kbp 5′-flanking region of the rat ms1/STARS genomic locus. The bent arrow denotes the start site of transcription, while dashed lines denote identified evolutionarily conserved regions (ECRs). (B) Four-way nucleotide sequence homology (MULAN alignment engine) plot of the ∼20-kbp 5′-flanking region. The x axis represents the genomic distance (kbp) relative to the ms1/STARS annotated transcription start site (TSS). Green indicates repetitive DNA sequences, and red indicates the intergenic noncoding DNA sequence with significant evolutionary conservation (regions at least 100 bp long that show >70% nucleotide sequence conservation between orthologous sequences). (C) ECRs containing multiple evolutionarily conserved GATA-binding sites (dashed-line-boxed sequences, with locations relative to the TSS in rat) conserved in rat (Rattus norvegicus [Rno]), mouse (Mus musculus [Mmu]), chicken (Gallus gallus [Gga]), human (Homo sapiens [Has]), opossum (Monodelphis domestica [Mdo]), dog (Canis familiaris [Cfa]), and frog (Xenopus tropicalis [Xtr]).
In addition to ECRs α and β, we identified two additional noncoding regions that were evolutionarily well conserved: regions at least 100 bp long that show >70% nucleotide sequence conservation between orthologous sequences (Fig. 1B, red-filled peaks). The largest and most highly conserved ECR (>515 bp with 75% sequence identity versus human and >168 bp with 70% sequence identity versus opossum) was designated DINA (distal intergenic activator) and is located ∼16 kbp upstream of the rat ms1 TSS. The other major ECR was located ∼9 kbp upstream of the TSS (>340 bp with 75% sequence identify versus human) and was classified as SINA (stress intergenic activator). Consistent with our previous study (41), ECR β demonstrates species-specific conservation (conserved in rat, mouse, and human only), while ECR α is the most highly conserved ECR, present in opossum, chicken, and frog (data not shown). This evolutionary constraint is of no surprise considering the presence of the core promoter and TATA box required for basal promoter activity and transcriptional initiation (32, 41).
We then utilized rVISTA (http://rvista.dcode.org/) and MatInspector (http://www.genomatix.de/) pattern-matching-based algorithms to interrogate our four ECRs for highly conserved transcription factor-binding sites (TFBSs) using our manual filtering parameters as described in Materials and Methods. We identified 31 “striated muscle”-specific TFBSs (data not shown), including the previously characterized MEF2 (23) and E-box (41) elements. The most abundant motif within this set was the GATA elements, with 13 sites enriched within all four ECRs (Fig. 1C) (position weight matrix plots used to identify GATA motifs can be viewed in Fig. S1 in the supplemental material; Table S1 in the supplemental material also provides experimental evidence for the identified GATA motifs). Four of these GATA elements (sites 1 to 4) located in ECR α were conserved in frog and chicken and were therefore classified as ultraconserved TFBSs (Fig. 1C).
Functional characterization of GATA-enriched evolutionarily conserved regions.
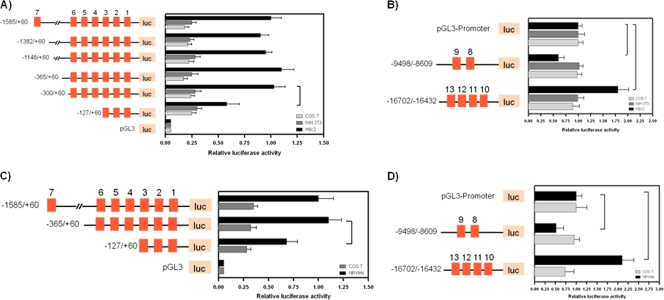
The enrichment of highly conserved GATA elements in all four ECRs suggests that GATA-binding proteins may impart an important regulatory control on ms1 gene expression. To test whether our GATA-enriched ECRs have cardiac cell-specific regulatory activity, we performed ECR-luciferase-reporter based assays. Specific combinations of forward and reverse primers were used to amplify 5′-flanking intervals of the proximal 1.6-kbp (relative to the ms1 TSS) sequence for cloning into the pGL3-Basic luciferase reporter plasmid. These combinations generated specific promoter truncations for the functional analysis of ECRs α and β in addition to GATA motifs (GATA elements 1 to 7) within these domains. Constructs containing serial 5′ deletions of the region from positions −1585 to −300 displayed a high and similar level of transcriptional activity in the cardiac tissue-derived H9c2 cells and isolated neonatal rat ventricular myocytes (NRVMs) (16) (Fig. 2A and C). This high level of activity was specific to cardiac cells, as the respective 5′-deleted reporters were approximately 75% less active in non-cardiac tissue-derived COS-7 and NIH 3T3 cells (P < 0.05 for all comparisons). Further truncation within the ultraconserved ECR α (positions −127 to +60) resulted in a significant 50% reduction (P < 0.05) in cardiac cell-specific promoter activity, with no effect being observed for COS-7 or NIH 3T3 cells.
Fig 2.
Characterization of the transcriptional activity of ECRs α, β, SINA, and DINA in a cardiogenic regulatory context. Shown are localizations of cardiac cell-specific regulatory regions of the rat ms1/STARS promoter identified by 5′ deletion analyses. Schematic representations of the ms1/STARS promoter 5′ deletion constructs used for transient transfection are shown on the left. Conserved GATA motifs (motifs 1 to 14) are denoted by filled red squares. (A and C) 5′ deletion constructs were cotransfected with pRL-TK into subconfluent H9c2, COS-7, NIH 3T3, and neonatal rat ventricular myocyte (NRVM) cells. Luciferase activity was normalized to Renilla luciferase activity and is shown relative to that of H9c2 (A) or NRMVs (C) transfected with the −1585/+60 reporter, the value of which was set to 1. (B and D) SINA and DINA SV40 enhancer reporters were cotransfected with pRL-TK into subconfluent H9c2, COS-7, and NIH 3T3 cells and NRVMs. Luciferase activity was normalized to Renilla luciferase activity and is presented relative to that of the appropriate cell line transfected with the pGL3-Promoter reporter, which was set to a value of 1. The results are expressed as means ± SE from at least three independent experiments, in triplicate for each reporter. Statistically significant differences are highlighted by a tailed line (P < 0.05).
The distal ECRs SINA (positions −9498 to −8609) and DINA (positions −16702 to −16432) were also isolated and cloned directly upstream of the simian virus 40 (SV40) basal promoter in the luciferase-based enhancer/repressor reporter vector pGL3-Promoter. These reporters were transiently transfected into H9c2, COS-7, and NIH 3T3 cells and NRVMs, with the relative activity of the SV40 basal promoter alone normalized to a value of 1 (Fig. 2B and D). The DINA and SINA domains conferred distinct cardiac cell-specific regulatory activity on the SV40 basal promoter. SINA repressed the activity of this basal promoter by 40% (H9c2 cells) and 50% (NRVMS) (P < 0.05), whereas DINA enhanced the basal SV40 promoter activity by 75% (H9c2 cells) and 110% (NRVMs) (P < 0.05). These distinct effects were confined to cardiac tissue-derived H9c2 cells and NRVMs, with no significant effects being observed for COS-7 or NIH 3T3 cells. These data demonstrate that the region at positions −300 to +60 is sufficient for basal cardiac cell-specific promoter activity, with elements located within the sequence at positions −300 to −127 being critical, suggesting that ECR α and its component TFBS (GATA elements 1 to 6) are of cardiac cell-specific regulatory importance. The distally located ECRs DINA and SINA also confer distinct regulatory activity specific to cardiac tissue-derived H9c2 cells and NRVMs. In addition, through analyses of publicly available ChIP sequencing data sets, we find that ECRs α, SINA, and DINA display cardiac cell-specific regulatory marks (epigenetic modifications including H3K4me1/me3 and p300 enrichment) in both the adult mouse heart (see Fig. S2A in the supplemental material) and fetal and adult human hearts (see Fig. S2B in the supplemental material). Thus, GATA-enriched ECRs α, SINA, and DINA represent potentially important cis-regulatory domains for the cardiac cell-specific cis hardwiring governing ms1 expression.
Modulation and targeting of ECRs α and DINA by GATA4.
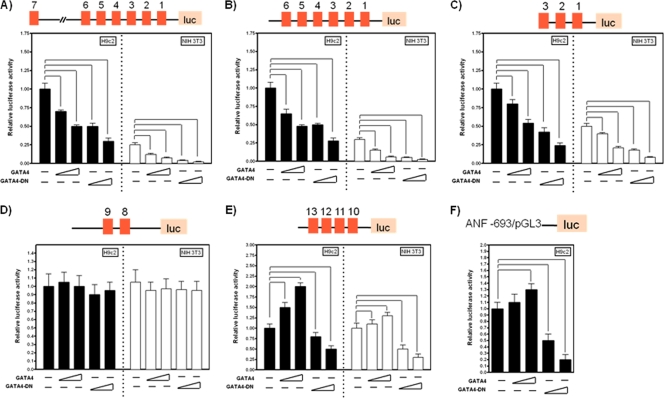
As our GATA-enriched ECRs α, SINA, and DINA confer cardiac cell-specific regulatory activity, we proceeded to determine whether GATA4, as the predominant GATA family member in the heart (32), modulated this effect and targeted these domains. Truncated reporters −1585/+60 (Fig. 3A), −365/+60 (Fig. 3B), and −125/+60 (Fig. 3C) were transiently cotransfected into H9c2 and NIH 3T3 cells in combination with wild-type and dominant negative (DN) GATA4 (ΔGATA4). ΔGATA4 contains the DNA-binding domain and was previously shown to inhibit basal and stress-inducible GATA4-dependent transcription (4, 8). ΔGATA4 repressed the activities of all three reporters in a dose-dependent manner in both H9c2 and NIH 3T3 cells (Fig. 3A to C). In H9c2 cells, the −127/+60 promoter was repressed by 50% (0.5 μg ΔGATA4; P < 0.05) and 70% (1.0 μg ΔGATA4; P < 0.05) (Fig. 3C), with a comparable level of repression being observed for NIH 3T3 cells. This finding is surprising considering the noncardiac nature of NIH 3T3 cells, suggesting that ΔGATA4-dependent repression may be mediated through the intrinsic DNA-binding ability of the protein rather than the canonical inhibition of endogenous GATA4 (or GATA6) binding. In support of this hypothesis, we found that wild-type GATA4 also repressed all three promoters in a dose-dependent fashion (Fig. 3A to C). In H9c2 cells, this repression was less potent than that with ΔGATA4, with the −127/+60 promoter being repressed by 30% (0.5 μg GATA4;P < 0.05) and 50% (1.0 μg GATA4; P < 0.05) (Fig. 3C) compared to the control (1.0 μg pcDNA3.1), with a correlated repression being observed for NIH 3T3 cells. Our GATA4 expression constructs were cotransfected with the GATA4-sensitive ANF-luciferase promoter reporter (−693-pGL3) in order to verify specificity. Consistent with previously reported findings (12), wild-type GATA4 activates the reporter, with concomitant repression by ΔGATA4 (Fig. 3F). These results demonstrate that GATA4 represses the ECR α promoter via GATA motifs within the proximal interval of positions −127 to +60. We repeated our GATA4 (WT GATA4 and ΔGATA4) cotransfection assays utilizing DINA and SINA SV40-luciferase reporters. DINA-SV40 was activated (in H9c2 cells) in a dose-dependent fashion by 60% (0.5 μg GATA4; P < 0.05) and 100% (1.0 μg GATA4; P < 0.05) (Fig. 3E). Activation was also observed for NIH 3T3 cells, although the level of activation was lower, with only a higher concentration of GATA4 being able to significantly activate the reporter by 40% (1.0 μg GATA4; P < 0.05). This finding suggests that cofactors specific to cardiac tissue-derived H9c2 cells likely collaborate with GATA4 to modulate the activity of DINA in H9c2 versus NIH 3T3 cells. ΔGATA4 repressed DINA by 20% and 50% (P < 0.05) in H9c2 cells, thereby supporting the importance of endogenous GATA4 targeting for the cardiac cell-specific activity of this module. Surprisingly, neither the wild type nor ΔGATA4 had any effect on the SINA-SV40-luciferase reporter in either H9c2 or NIH 3T3 cells (Fig. 3D). Therefore, the data presented here demonstrate that GATA4 is able to target both ECRs α and DINA, having distinct regulatory effects on both ECRs in vitro.
Fig 3.
Modulation of ECRs α and DINA by GATA4. (A to E) Wild-type (WT) GATA4 and dominant negative (DN) ΔGATA4 expression plasmids were cotransfected in combination with various 5′ deletion and ECR reporters into H9c2 (filled bars) and NIH 3T3 (open bars) cells. Luciferase activity was normalized to Renilla luciferase activity and is shown relative to that of H9c2 cells transfected with the appropriate reporter. (F) As a positive control, the GATA4-sensitive ANF promoter was cotransfected with WT GATA4 or DN ΔGATA4 into H9c2 cells only. pcDNA3.1 was used as a packing vector to maintain the concentration used for each individual transfection. Results are expressed as means ± SE from at least three independent experiments, in triplicate for each reporter. Statistically significant differences are highlighted by tailed lines (P < 0.05).
GATA4 binds ECRs α and DINA in embryonic, neonatal, and adult hearts.
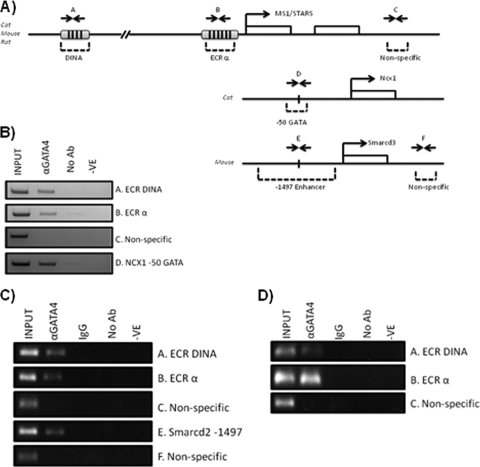
To determine whether GATA4 can target ECRs α and DINA in vivo, we performed chromatin immunoprecipitation (ChIP) assays on feline adult cardiomyocytes, neonatal rat ventricular myocytes, and murine hearts isolated on E12.5. ECRs α, SINA, and DINA are all fully conserved in the cat genome, with their syntenic organization also being preserved (see Fig. S3 in the supplemental material). Using primers specifically designed to generate amplicons spanning ECRs α and DINA in mouse, rat, and cat, semiquantitative PCR was performed on formaldehyde-cross-linked, sheared chromatin isolated from embryonic, neonatal, and adult cardiomyocytes, which was immunoprecipitated with anti-GATA4 antibody (Fig. 4). GATA4 antibody specifically pulled down DNA fragments containing ECRs α and DINA (Fig. 4B to D) in all developmental contexts. As a positive control, 5% of the input chromatin was used for PCR. The ChIP specificity was controlled by executing negative controls lacking antibody (Fig. 4B) and nonspecific IgG (Fig. 4C and D). To confirm the validity of the pulldown, we also demonstrated GATA4 enrichment at the NCX1 GATA motif, a bona fide GATA4-binding site in adult cardiomyocytes (Fig. 4B) (54), and the Smarcd3 enhancer in E12.5 murine heart (Fig. 4C) (15), while no GATA4 was enriched at the nonspecific intergenic sequences. These results demonstrate that GATA4 binds ECRs α and DINA in vivo, consistent with our transient reporter transfection assays in vitro. Therefore, a direct interaction of GATA4 with ECRs α and DINA is likely responsible for the cardiac cell-specific activation and repression of these domains.
Fig 4.
Binding of GATA4 to the endogenous ms1 locus in cardiomyocytes. Chromatin immunoprecipitation assays were performed on formaldehyde-cross-linked chromatin isolated from feline adult cardiomyocytes (B), neonatal rat ventricular myocytes (C), and isolated E12.5 murine hearts (D). The PCR results using primers spanning ms1 ECRs and nonspecific intergenic regions (A) are shown. As a positive control, GATA4 enrichment at the bona fide GATA4-targeted Ncx1 promoter and Smarcd3 enhancer was demonstrated (lanes D and E). Immunoprecipitations were performed with nonspecific IgG and without primary antibody (No Ab) as a negative control and with anti-GATA4 antibody. Input DNA is also shown as a positive control for PCR amplification. Similar results were observed for a total of four independent experiments. -VE, PCR negative control (water).
GATA4 modulates endogenous ms1 expression in neonatal rat ventricular myocytes.
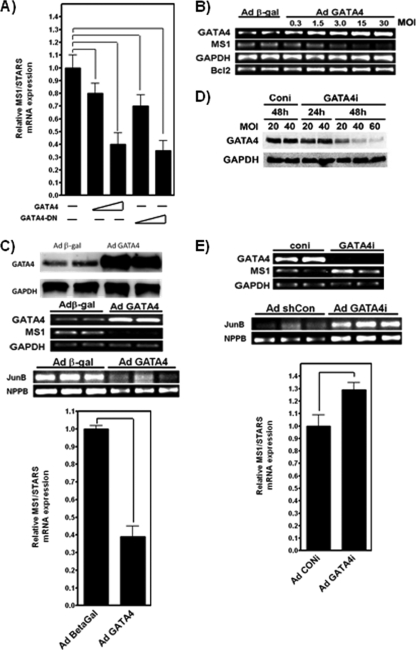
GATA4 targets ECRs α and DINA, and this is associated with both positive (DINA) and negative (ECR α) regulatory outputs. In order to reconcile the regulatory differences associated with GATA4 binding at ECRs α and DINA, the sensitivity of the endogenous ms1 cis-regulatory hardwiring to GATA4 expression was examined in H9c2 cells and neonatal rat ventricular myocytes (NRVMs). Wild-type GATA4 overexpression in H9c2 cells significantly decreased ms1 mRNA levels in a dose-dependent manner: 20% (0.5 μg GATA4; P < 0.05) and 70% (0.5 μg GATA4; P < 0.05) (Fig. 5A). The overexpression of ΔGATA4 also decreased ms1 mRNA levels in a dose-varying fashion: 20% (0.5 μg GATA4; P < 0.05) and 70% (0.5 μg GATA4; P < 0.05) (Fig. 5A). In order to confirm these findings with primary NRVMs, an adenoviral overexpression strategy was utilized. NRVMs were infected with adenovirus encoding β-Gal or GATA4 (AdGATA4). Similarly to the effect observed for H9c2 cells (Fig. 5A), GATA4 repressed endogenous ms1 expression in a dose-dependent manner (Fig. 5B). This repression was correlated with increased levels of Gata4 mRNA and Bcl2 expression, a proven GATA4 target in NRVMs (19). The greatest attenuation of ms1 expression was achieved at an AdGATA4 multiplicity of infection (MOI) of 30 PFU at 48 h after viral infection, resulting in a significant 50% downregulation of ms1 (n = 4; P < 0.05) (Fig. 5C). Immunoblots confirmed a significant increase in GATA4 protein levels in AdGATA4-infected NRVMs (Fig. 5C). In addition, the bona fide MRTF-SRF target genes JunB and NPPB were also significantly downregulated (Fig. 5C). To confirm the necessity of GATA4 for basal ms1 expression, an adenovirus encoding a short hairpin RNA targeted to GATA4 (AdGATA4i) was used to specifically knock down Gata4 mRNA and protein levels (Fig. 5D). The infection of NRVMs with AdGATA4i at an MOI of 60 PFU for 48 h resulted in a significant 1.3-fold increase in ms1 expression levels (n = 4; P < 0.05), compared to infection with adenovirus encoding a random-scrambled short hairpin RNA (AdConi) (Fig. 5E). As would be expected if MRTF-SRF signaling was increased, JunB and NPPB were also significantly upregulated (Fig. 5E). These data corroborate our ECR-luciferase reporter assay data and suggest that the net effect of GATA4 is to repress ms1 expression, presumably through the GATA4-sensitive ECR α. This raises interesting questions as to the role of the positively acting GATA4-sensitive ECR DINA.
Fig 5.
GATA4 modulates ms1 gene expression in H9c2 cells and neonatal rat ventricular myocytes (NRVMs). (A) Subconfluent H9c2 myoblasts were transiently transfected with increasing concentrations of the vector (0.5 μg and 1.0 μg, represented by the increasing gradient) expressing WT GATA4 or DN ΔGATA4. (B) At 48 h posttransfection, total RNA was isolated and reverse transcribed, and the expression levels of TATA-binding protein (TBP) and ms1 were determined by quantitative real-time PCR analysis. MS1 expression in each sample was normalized to that of TBP, with ms1 expression in empty-vector-transfected cells arbitrarily set at a value of 1. At 48 h after viral infection, AdGATA4 dose dependently overexpresses the GATA4 transcript, which correlates with decreased expression levels of MS1 mRNA and an increased abundance of the GATA4 target gene Bcl2. (C) NRVMs were infected with Adgal or AdGATA4 for 48 h at MOIs of 0.3, 1.5, 3.0, 15, and 30 PFU. GATA4 overexpression was associated with an attenuation of ms1 mRNA levels and the MRTF-SRF target genes JunB and NPPB (determined by RT-PCR and shown in an ethidium bromide-stained agarose gel). NRVMs were infected with Adgal or AdGATA4 for 48 h at an MOI of 30 PFU. Western blot analysis of GATA4 protein levels was performed by using 50 μg of whole-cell protein extract. ms1 expression in Adgal-infected cells is arbitrarily set at a value of 1. (D) GATA4 knockdown promotes ms1 expression and MRTF-SRF signaling in NRVMs. AdG4i knocks down GATA4 protein levels in a dose- and time-dependent manner. NRVMs were infected with AdConi or AdG4i at different MOIs for the indicated times, and Western blot analysis was performed by using goat anti-GATA4 antibody. (E) Transcript levels of ms1, Gapdh, JunB, and NPPB in NRVMs infected with AdConi or AdG4i at an MOI of 60 PFU and at 48 h after viral infection, as detected by semiquantitative RT-PCR (shown in an agarose gel image stained with ethidium bromide). Statistically significant differences are highlighted by tailed lines (P < 0.05). Results are expressed as means ± SE from at least three independent experiments.
GATA4 regulates ms1 expression in embryonic and adult murine hearts in vivo.
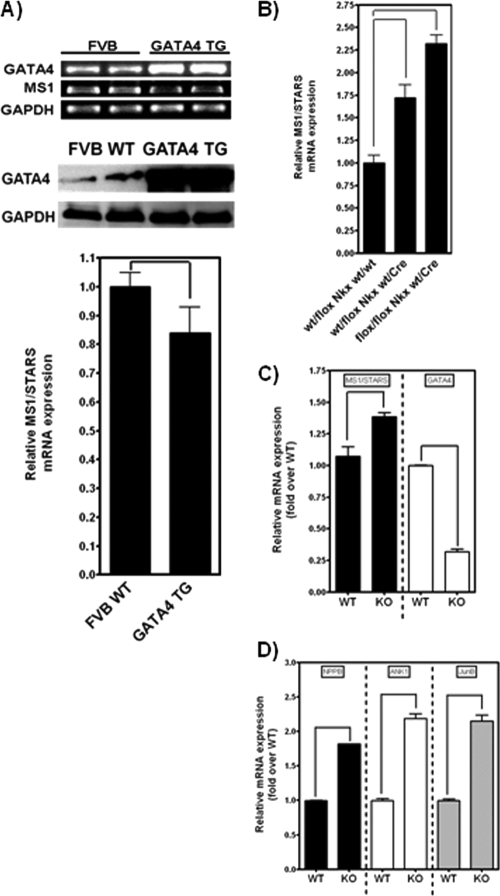
To examine the effect of GATA4 on ms1 expression in vivo, semiquantitative RT-PCR was used to determine mRNA levels of Gata4 and ms1 in hearts of FVB/N wild-type and GATA4 transgenic (TG) mice. These mice demonstrated a robust increase in Gata4 mRNA levels (Fig. 6A) that corresponded to a 2- to 4-fold increase in GATA4 protein levels relative to those of wild-type controls (Fig. 6A) (26, 27). ms1 expression was significantly downregulated in the TG hearts compared to the wild-type controls (Fig. 6A), therefore corroborating our in vitro findings and demonstrating the GATA4-dependent modulation of ms1 in vivo. Furthermore, we sought to determine whether basal GATA4 levels in the whole embryonic heart contribute to basal ms1 expression. To investigate this, we examined GATA4 gene-targeted knockout mice utilizing Cre/Lox technology to conditionally ablate GATA4 at embryonic day 9.5 (E9.5) and E12.5 specifically within cardiomyocytes of the developing myocardium. This early cardiac cell-specific GATA4 knockdown was achieved through the use of Nkx2-5-Cre and TNT-Cre (17), resulting in robust Cre-mediated recombination by E9.5 (34) and E12.5 (17). A combination of heterozygous and homozygous floxed GATA4 alleles in combination with Nkx2-5-Cre and TNT-Cre was used to dose-dependently ablate GATA4 (55). We executed quantitative RT-PCR on total RNA isolated from heterozygous and homozygous floxed GATA4/Nkx2-5-Cre and GATA4/TNT-Cre murine hearts in addition to wild-type hearts. In agreement with our data for adenovirus-based knockdown in vitro (Fig. 5E), we demonstrate a dose-dependent increase in ms1 expression levels in the heterozygous (1.75-fold; P < 0.05) and homozygous (2.25-fold; P < 0.05) floxed GATA4/Nkx2-5-Cre murine hearts compared to non-Cre Nkx2-5 wild-type hearts (Fig. 6B). We also demonstrate a significant increase in ms1 expression levels in homozygous (1.35-fold; P < 0.05) floxed GATA4/TNT-Cre murine hearts compared to non-Cre TNT wild-type hearts (Fig. 6C), which correlated with decreased GATA4 expression levels. We suspect that GATA4 levels are not as low as one might expect, due to the presence of noncardiomyocytes (e.g., epicardial and endocardial cells) and, therefore, non-TnT-expressing cells. Nonetheless, the observed increase in ms1 expression levels correlates with a significant upregulation of MRTF-SRF target genes (Fig. 6D). This finding supports our in vitro data and demonstrates that GATA4 is an important modulator of ms1 expression in both the developing embryonic myocardium and adult myocardium in vivo, which subsequently elicits a differential activity of the MRTF-SRF signaling axis.
Fig 6.
GATA4 modulates ms1 expression in vivo. (A) Protein extracts and RNA were prepared from the hearts of GATA4 TG and WT control mice. Western blot analysis demonstrated the overexpression of GATA4 in TG mice versus WT controls. ms1 mRNA was significantly downregulated in GATA4 TG mouse hearts (as determined by semiquantitative RT-PCR) (a representative PCR is shown with an ethidium bromide-stained agarose gel). Data are means ± SE (n = 4). P < 0.05 versus WT mice. (B) RNA isolated from the embryonic hearts (E9.5) of wt/flox Nkx wt/wt, wt/flox Nkx wt/Cre, and flox/flox Nkx wt/Cre mice was subjected to reverse transcription followed by quantitative PCR with mouse ms1 and TBP TaqMan probes. Data are means ± SE (n = 4). P < 0.05 versus wt/flox Nkx wt/wt mice. (C and D) RNA isolated from the embryonic ventricular apex (E12.5) of G4S/S::TNT-Cre and G4S/S mice was subjected to reverse transcription followed by quantitative PCR for ms1, Gapdh, Gata4, NPPB, ANK1, and JunB. Data are means ± SE (n = 3). Statistically significant differences are highlighted by tailed lines (P < 0.05).
ms1 expression levels are increased in type 1 and type 2 diabetic hearts.
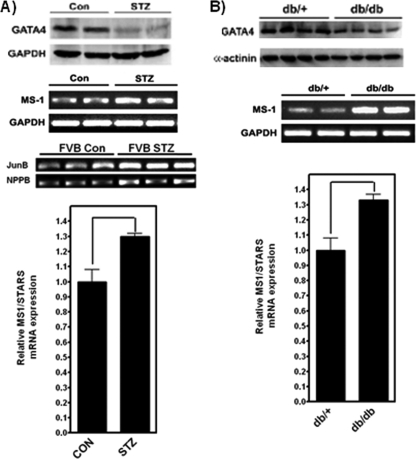
Our data demonstrate that ms1 transcription is exquisitely sensitive to GATA4 protein levels in vitro and in vivo. ms1 dysregulation has been implicated in both cardiac dysfunction and cardiomyopathy. We therefore wanted to determine whether ms1 expression is altered in a GATA4-dependent pathophysiological context. Diminished levels of GATA4 were shown previously to contribute to hyperglycemia-induced cardiomyocyte injury in both type 1 and type 2 diabetes, leading to the development of diabetic cardiomyopathy (20). To determine whether ms1 is dysregulated in this context, we measured expression levels in hearts from type 1 and type 2 diabetic mouse models. Type 1 diabetes was induced in 2-month-old FVB mice with streptozotocin (STZ), a well-established agent that destroys pancreatic β cells (47, 48). In this model, GATA4 protein levels were significantly reduced after 4 weeks of STZ dosing (Fig. 7A). Semiquantitative PCR showed that ms1 levels in the STZ heart were increased 1.3-fold versus those in untreated vehicle controls (P < 0.05) (Fig. 7A). Increased ms1 expression levels also correlated with a significant upregulation of the MRTF-SRF target genes JunB and NPPB. We also measured ms1 expression levels in 4-month-old db/db mice, a well-characterized type 2 insulin-resistant diabetic model with early-onset cardiomyopathy (1). The ms1 mRNA level was increased in db/db diabetic hearts compared with db/+ nondiabetic controls (1.3-fold increase; P < 0.05) (Fig. 7B), which correlated with a significant decrease in GATA4 protein levels (Fig. 7B). These data demonstrate that hyperglycemia-associated type 1 and type 2 diabetes diminishes cardiac GATA4 protein levels, which is associated with a significant increase in ms1 expression levels. This finding supports our in vitro and in vivo findings corroborating the hypothesis that endogenous GATA4 represses ms1 expression in the myocardium. These data have implications for the role of ms1 dysregulation and MRTF-SRF signaling in GATA4-dependent pathological phenotypes such as the development of diabetic cardiomyopathy.
Fig 7.
ms1 expression is modulated in GATA4-dependent pathophysiological models. Total RNA was isolated from the hearts of control and STZ-treated (A) as well as db/+ and db/db (B) mice and subjected to reverse transcription followed by semiquantitative PCR with mouse ms1-, JunB-, NPPB-, and GAPDH-specific primers. Amplified products were visualized on an ethidium bromide-stained agarose gel and quantified by densitometry. Values are presented as means ± SE (n = 4). P < 0.05 versus Con (A) and db/+ (B) mice. Statistically significant differences are highlighted by tailed lines.
DISCUSSION
MS1 has been implicated as an important regulator of cardiac development, postnatal myocardial homeostasis, and adaptation to stress. Despite its important role, little is known about the regulatory mechanisms governing its context-specific expression (developmentally and pathologically) within the myocardium.
Through the coupling of a comparative genomic sequence analysis with reporter assays, we have identified two novel distal ECRs with cardiac cell-restricted regulatory activity, designated DINA and SINA. Furthermore, we have dissected the previously identified ECRs (α and β) (41), demonstrating the necessity of the proximal 400 bp upstream of the MS1 transcriptional start site (TSS) for cardiac cell-specific activity. This proximal sequence is ultraconserved and highly enriched with cardiomyogenic transcription factor-binding sites, including six GATA motifs, in addition to the core promoter elements, including the TATA box. Kuwahara et al. (23) previously characterized the 5′-flanking proximal 1.5 kbp of the mouse ms1 promoter in cardiomyocytes using luciferase reporter assays and β-Gal transgenic reporter mice. Those authors demonstrated that the proximal 400 bp upstream of the TSS, encompassed within our designated ECR α, was critical for the cardiac cell-specific and stress-inducible capacity of the ms1 promoter.
In concordance with the data reported previously by Kuwahara et al. (23), ECR β does not appear to modulate cardiac cell-specific activity. Based on previous data (41), we suggest that ECR β acts exclusively as a myogenic enhancer dependent on myogenic regulatory factors for cis-dependent activity. The ECRs SINA and DINA directed distinct cardiac cell-specific regulatory activity, with DINA stimulating activity and SINA, at least in our regulatory model, repressing activity. In silico analyses identified GATA-binding proteins as putative regulators based on the enrichment of evolutionarily conserved GATA motifs within our cardiogenic ECRs (13 conserved GATA motifs). Utilizing cotransfection gene reporter and ChIP assays, we demonstrate that GATA4, the major cardiac GATA-binding protein and core regulator within the cardiac GRN (40), is able to target ECRs α and DINA. Somewhat unexpectedly, GATA4 potently repressed ECR α activity while enhancing the activity of DINA. We utilized gain-of-function and loss-of-function approaches to confirm an obligatory role for GATA4 in modulating cardiac ms1 expression in vitro and in vivo while also serving to reconcile the dichotomous response of ECRs α and DINA to GATA4. These approaches demonstrated that ms1 expression is negatively regulated by GATA4 in embryonic, neonatal, and adult cardiomyocytes, with the overexpression/transgenesis and knockdown/knockout of GATA4 resulting in the significant downregulation and upregulation of ms1, respectively. The overall “net” regulatory effect imparted on ms1 expression by GATA4 in our experimental models is negative. Gene expression profiling of MRTF-SRF tars and integrates the GATA4- and SRF-dependent regulatory “subcircuits,” having potentially major implications for the cardiac GRN. Perturbed GATA4 expression and/or function in the myocardium has been implicated in many developmental and postnatal pathological phenotypes, although the downstream effectors that modulate these phenotypes are poorly characterized. Considering the exquisite sensitivity of ms1 expression to GATA4, we suspect that GATA4-associated pathophysiological phenotypes (deveget genes confirmed the dependence of this signaling axis on MS1 expression levels in the GATA4-manipulated systems utilized in this study.
Our in vitro promoter characterization, ChIP assays, and gain-of-function/loss-of-function data suggest that this repression is mediated by GATA4 binding at ultraconserved GATA motifs 1, 2, and 3 within ECR α (located within the proximal 100 bp upstream of the TSS). ms1 is herein a novel bona fide regulatory target of GATA4. This direct control linklopmental and adult) are likely to be caused partly by the dysregulation of the MS1-SRF regulatory axis.
To elucidate the potential consequences of this regulatory connection and determine whether ms1, and, subsequently, the MRTF-SRF axis, is affected in GATA4-dependent pathophysiological contexts, we utilized type 1 and 2 diabetes-induced cardiomyopathy models. The hyperglycemia-induced depletion of GATA4 is an important mechanism that contributes to type 1/type 2 hyperglycemic cardiotoxicity and subsequent myopathy (20). Here we have shown that ms1 expression is upregulated in both type 1 and type 2 diabetic models, correlating with GATA4 protein depletion and increased MRTF-SRF signaling. Considering the established role for MS1 in the development of human dilated cardiomyopathy, it would be of interest to functionally determine whether the GATA4-dependent increase in the ms1 expression level observed for the diabetic mouse heart contributes to the development of diabetic myopathy.
Although not classically regarded as a cardiac cell-specific repressor, we believe that the GATA4-dependent repression of target genes will emerge as a common regulatory theme. During cardiac development as well as within the postnatal myocardium, GATA4 is a critical regulator of opposing cellular processes, including myocyte proliferation versus differentiation and cell survival versus apoptosis. We suspect that in addition to upregulating target genes involved in promoting such processes, GATA4 simultaneously represses positive regulators of the opposing process. For example, GATA4 was recently shown to be cardioprotective by repressing the doxorubicin-dependent induction of autophagy-promoting genes (e.g., Beclin-1 and ATG5) while simultaneously promoting the expression of antiapoptotic/autophagic genes (Bcl2) (21). During early cardiac development, GATA4 promotes cardiomyocyte proliferation, while during myocardial maturation, GATA4 in collaboration with SRF promotes cardiomyocyte differentiation. Therefore, in our model proposed here, GATA4 represses/blocks ms1 expression and, thus, the MRTF-SRF differentiation-promoting activity. Through this mechanism, GATA4 is able to simultaneously promote developmental proliferation (via the upregulation of targets, including Cyclin D1) and suppress differentiation-promoting regulatory subcircuits (MRTF-SRF) and “gene batteries” (10, 11, 24).
Postnatally, MRTF-SRF signaling needs to be finely balanced in order to maintain cardiac homeostasis through preserving sarcomerogenesis-associated gene expression. However, attenuated or precocious SRF activity is ultimately detrimental, leading to the development of cardiac hypertrophy, contractile dysfunction, myopathy, and cardiac failure (30, 35, 36, 56, 57). Therefore, postnatally, GATA4, via its robust repression of ms1, is able to prevent inappropriate MS1 expression (and, thus, MRTF-SRF activity), thereby maintaining the trophic/homeostatic state of the myocardium, a role in which GATA4 has already been implicated (3, 21, 39, 45). In all of our models, we observed a significant dysregulation of the MRTF-SRF signaling axis (demonstrated by the differential expressions of bona fide target genes) in contexts where GATA4 and ms1 are differentially expressed, thereby supporting this hypothesis.
The exact nature of ECR α-dependent GATA4 repression is unknown; however, there are a number of potential mechanisms for the transcriptional “blocking” observed. One potential mechanism is via ECR α-bound GATA4 sterically inhibiting transcription initiation complex formation at the TATA box. Steric hindrance has been observed for other GATA4-repressed targets, including the tryptophan oxygenase and COL1A2 promoters (18, 52). This mechanism has also been suggested for the GATA4-dependent repression of autophagy-inducing gene expression in response to doxorubicin (21).
We have demonstrated that via distinct modes of regulation at the proximal and distal ECRs (α and DINA), GATA4 is able to modulate ms1 expression. Although Kuwahara and colleagues (23) previously executed an initial analysis of the proximal −1.5 kbp, we have expanded upon this and identified two distal ECRs (DINA and SINA) with important cardiac cell-specific activities. We suspect that the activities of these distal ECRs will modify the context-dependent regulatory activity of this proximal −1.5 kbp. Through an understanding of the pathway through which ms1 is regulated, we gain a functional insight into general muscle-specific mechanisms directing MRTF-SRF activity. This provides a deeper understanding of the nature of this core regulatory subcircuit within the context of the global cardiac GRN, subsequently generating important insights into how, via MS1, the MRTF-SRF subcircuit becomes dysregulated during development and disease. We have identified single-nucleotide polymorphisms (SNPs) within human ECRs α, SINA, and DINA (N. W. Chong et al., unpublished observations). These variants may modify ms1 expression and therefore have profound effects on MRTF-SRF activity. Such variation could account for predispositions to SRF-associated pathophenotypes (cardiomyopathy). Interestingly, SNPs found in the noncoding sequence proximal to the porcine ms1 gene have been directly associated with lean muscle mass in Chinese porcine varieties, supporting the potential regulatory importance of such SNPs in ms1 control sequences (13).
From a regulatory network perspective, we have been able to functionally integrate the GATA4 and SRF regulatory subcircuits. Such integration and coordination of subcircuits are an important property of the cardiac GRN. Cross talk between these two subcircuits allows the establishment of positive and negative reinforcing regulatory loops, which underpin the activity of the cardiac GRN both temporally and spatially (10, 11, 40). Based on our proposed regulatory interaction between GATA4 and SRF subcircuits, we predict that many of the pathological processes associated with GATA4 (congenital heart defects and diabetic cardiomyopathy) are likely to be mediated partly via the MS1-MRTF-SRF axis. Identifying ms1 as a bona fide target of GATA4 and, hence, integrating the SRF and GATA4 subcircuits greatly increase our mechanistic understanding of GATA4-associated pathological phenotypes. Such knowledge will also allow us to better understand how GATA4-modulated processes, such as cardiac specification, proliferation, and differentiation, are directed during cardiac development.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Heart Research UK grant (RG2474/04/07) and a Coulson Trust UK grant to N.W.C., a research grant (1-2007-741) from the Juvenile Diabetes Research Foundation and a career development grant (1-09-CD-09) from the American Diabetes Association to Q.L., an NIH grant (R01 HL066223) to D.R.M., and an NHLBI grant (HL095712) to W.T.P. N.J.S. holds a British Heart Foundation Chair of Cardiology.
Footnotes
Published ahead of print 19 March 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Aasum E, Hafstad AD, Severson DL, Larsen TS. 2003. Age-dependent changes in metabolism, contractile function, and ischemic sensitivity in hearts from db/db mice. Diabetes 52:434–441 [DOI] [PubMed] [Google Scholar]
- 2. Arai A, Spencer JA, Olson EN. 2002. STARS, a striated muscle activator of Rho signaling and serum response factor-dependent transcription. J. Biol. Chem. 277:24453–24459 [DOI] [PubMed] [Google Scholar]
- 3. Aries A, Paradis P, Lefebvre C, Schwartz RJ, Nemer M. 2004. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc. Natl. Acad. Sci. U. S. A. 101:6975–6980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhalla SS, Robitaille L, Nemer M. 2001. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J. Biol. Chem. 276:11439–11445 [DOI] [PubMed] [Google Scholar]
- 5. Bisping E, et al. 2006. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc. Natl. Acad. Sci. U. S. A. 103:14471–14476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Camoretti-Mercado B, Dulin NO, Solway J. 2003. Serum response factor function and dysfunction in smooth muscle. Respir. Physiol. Neurobiol. 137:223–235 [DOI] [PubMed] [Google Scholar]
- 7. Cen B, et al. 2003. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol. Cell. Biol. 23:6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Charron F, et al. 2001. Tissue-specific GATA factors are transcriptional effectors of the small GTPase RhoA. Genes Dev. 15:2702–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cripps RM, Olson EN. 2002. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev. Biol. 246:14–28 [DOI] [PubMed] [Google Scholar]
- 10. Davidson EH. 2010. Emerging properties of animal gene regulatory networks. Nature 468:911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davidson EH, Levine MS. 2008. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 105:20063–20066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 16:5687–5696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durocher D, Nemer M. 1998. Combinatorial interactions regulating cardiac transcription. Dev. Genet. 22:250–262 [DOI] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15. He A, Kong SW, Ma Q, Pu WT. 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc. Natl. Acad. Sci. U. S. A. 108:5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hescheler J, et al. 1991. Morphological, biochemical, and electrophysiological characterization of a clonal cell (H9c2) line from rat heart. Circ. Res. 69:1476–1486 [DOI] [PubMed] [Google Scholar]
- 17. Jiao K, et al. 2003. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 17:2362–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaneoka H, Miyake K, Iijima S. 2008. GATA4 inhibits expression of the tryptophan oxygenase gene by binding to the TATA box in fetal hepatocytes. Cytotechnology 57:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a. Kent RL, Mann DL, Urabe Y, Hisano R, Hewett KW, Loughnane M, Cooper GT. 1989. Contractile function of isolated feline cardiocytes in response to viscous loading. Am. J. Physiol. Heart Circ. Physiol. 257:H1717–H1727 [DOI] [PubMed] [Google Scholar]
- 19. Kobayashi S, et al. 2006. Transcription factor gata4 regulates cardiac BCL2 gene expression in vitro and in vivo. FASEB J. 20:800–802 [DOI] [PubMed] [Google Scholar]
- 20. Kobayashi S, et al. 2007. Diminished GATA4 protein levels contribute to hyperglycemia-induced cardiomyocyte injury. J. Biol. Chem. 282:21945–21952 [DOI] [PubMed] [Google Scholar]
- 21. Kobayashi S, et al. 2010. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J. Biol. Chem. 285:793–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koekemoer AL, Chong NW, Goodall AH, Samani NJ. 2009. Myocyte stress 1 plays an important role in cellular hypertrophy and protection against apoptosis. FEBS Lett. 583:2964–2967 [DOI] [PubMed] [Google Scholar]
- 23. Kuwahara K, et al. 2007. Modulation of adverse cardiac remodeling by STARS, a mediator of MEF2 signaling and SRF activity. J. Clin. Invest. 117:1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li E, Davidson EH. 2009. Building developmental gene regulatory networks. Birth Defects Res. C Embryo Today 87:123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. 2003. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. U. S. A. 100:9366–9370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang Q, et al. 2001. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 276:30245–30253 [DOI] [PubMed] [Google Scholar]
- 27. Liang Q, et al. 2001. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 21:7460–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loots GG, et al. 2000. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science 288:136–140 [DOI] [PubMed] [Google Scholar]
- 29. Mahadeva H, Brooks G, Lodwick D, Chong NW, Samani NJ. 2002. ms1, a novel stress-responsive, muscle-specific gene that is up-regulated in the early stages of pressure overload-induced left ventricular hypertrophy. FEBS Lett. 521:100–104 [DOI] [PubMed] [Google Scholar]
- 30. Miano JM. 2010. Role of serum response factor in the pathogenesis of disease. Lab. Invest. 90:1274–1284 [DOI] [PubMed] [Google Scholar]
- 31. Miano JM, et al. 2004. Restricted inactivation of serum response factor to the cardiovascular system. Proc. Natl. Acad. Sci. U. S. A. 101:17132–17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molkentin JD. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949–38952 [DOI] [PubMed] [Google Scholar]
- 33. Morimoto T, et al. 2001. Calcineurin-GATA4 pathway is involved in beta-adrenergic agonist-responsive endothelin-1 transcription in cardiac myocytes. J. Biol. Chem. 276:34983–34989 [DOI] [PubMed] [Google Scholar]
- 34. Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. 2001. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 31:176–180 [DOI] [PubMed] [Google Scholar]
- 35. Niu Z, et al. 2008. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl. Acad. Sci. U. S. A. 105:17824–17829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niu Z, Li A, Zhang SX, Schwartz RJ. 2007. Serum response factor micromanaging cardiogenesis. Curr. Opin. Cell Biol. 19:618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Niu Z, et al. 2005. Conditional mutagenesis of the murine serum response factor gene blocks cardiogenesis and the transcription of downstream gene targets. J. Biol. Chem. 280:32531–32538 [DOI] [PubMed] [Google Scholar]
- 38.Reference deleted.
- 39. Oka T, et al. 2006. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ. Res. 98:837–845 [DOI] [PubMed] [Google Scholar]
- 40. Olson EN. 2006. Gene regulatory networks in the evolution and development of the heart. Science 313:1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ounzain S, Dacwag CS, Samani NJ, Imbalzano AN, Chong NW. 2008. Comparative in silico analysis identifies bona fide MyoD binding sites within the myocyte stress 1 gene promoter. BMC Mol. Biol. 9:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. 2004. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 32:W280–W286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Parlakian A, et al. 2004. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 24:5281–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peng YB, et al. 2008. Molecular characterization and expression pattern of the porcine STARS, a striated muscle-specific expressed gene. Biochem. Genet. 46:644–651 [DOI] [PubMed] [Google Scholar]
- 45. Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. 2004. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 63:196–207 [DOI] [PubMed] [Google Scholar]
- 46. Pipes GC, Creemers EE, Olson EN. 2006. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 20:1545–1556 [DOI] [PubMed] [Google Scholar]
- 47. Rossini AA, et al. 1977. Studies of streptozotocin-induced insulitis and diabetes. Proc. Natl. Acad. Sci. U. S. A. 74:2485–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rossini AA, Like AA, Dulin WE, Cahill GF., Jr 1977. Pancreatic beta cell toxicity by streptozotocin anomers. Diabetes 26:1120–1124 [DOI] [PubMed] [Google Scholar]
- 49. Shore P, Sharrocks AD. 1995. The MADS-box family of transcription factors. Eur. J. Biochem. 229:1–13 [DOI] [PubMed] [Google Scholar]
- 50. Wang D, et al. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851–862 [DOI] [PubMed] [Google Scholar]
- 51. Wang DZ, et al. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. U. S. A. 99:14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang L, Tanaka S, Ramirez F. 2005. GATA-4 binds to an upstream element of the human alpha2(I) collagen gene (COL1A2) and inhibits transcription in fibroblasts. Matrix Biol. 24:333–340 [DOI] [PubMed] [Google Scholar]
- 53.Reference deleted.
- 54. Xu L, et al. 2006. Regulation of Ncx1 expression. Identification of regulatory elements mediating cardiac-specific expression and up-regulation. J. Biol. Chem. 281:34430–34440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zeisberg EM, et al. 2005. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J. Clin. Invest. 115:1522–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X, et al. 2001. Cardiomyopathy in transgenic mice with cardiac-specific overexpression of serum response factor. Am. J. Physiol. Heart Circ. Physiol. 280:H1782–H1792 [DOI] [PubMed] [Google Scholar]
- 57. Zhang X, et al. 2001. Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J. Biol. Chem. 276:40033–40040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.