Abstract
Collapsin response mediator protein 2 (CRMP-2) is known as a regulator of neuronal polarity and differentiation through microtubule assembly and trafficking. Here, we show that CRMP-2 is ubiquitously expressed and a splice variant (CRMP-2L), which is expressed mainly in epithelial cells among nonneuronal cells, regulates myosin II-mediated cellular functions, including cell migration. While the CRMP-2 short form (CRMP-2S) is recognized as a substrate of the Rho-GTP downstream kinase ROCK in neuronal cells, a CRMP-2 complex containing 2L not only bound the catalytic domain of ROCK II through two binding domains but also trapped and inhibited the kinase. CRMP-2L protein levels profoundly affected haptotactic migration and the actin-myosin cytoskeleton of carcinoma cells as well as nontransformed epithelial cell migration in a ROCK activity-dependent manner. Moreover, the ectopic expression of CRMP-2L but not -2S inhibited fibronectin matrix assembly in fibroblasts. Underlying these responses, CRMP-2L regulated the kinase activity of ROCK II but not ROCK I, independent of GTP-RhoA levels. This study provides a new insight into CRMP-2 as a controller of myosin II-mediated cellular functions through the inhibition of ROCK II in nonneuronal cells.
INTRODUCTION
The actin cytoskeleton is pivotal in many aspects of cellular behavior, including adhesion, migration, extracellular matrix assembly, and mitosis, key processes in development (40, 52). Therefore, it is spatiotemporally regulated at multiple levels, such as actin filament formation, severing, and bundling (9, 42). The small GTPase Rho is a major regulator of actin cytoskeleton organization, with downstream targets including the Rho kinases (ROCK I and II) and mDia (8). In many cell types, the two homologous Rho kinases modulate actin-myosin II-mediated cell contractility through the control of the myosin II regulatory light chain (MLC) phosphorylation state (3, 44). These 160-kDa serine/threonine kinases, encoded by separate genes, are essential in homeostatic and developmental processes (24, 49) but are also potential therapeutic targets for diverse diseases, including cancer, hypertension, fibrosis, and central nervous system scarring (18).
The activation states of many protein kinases can be readily detected or measured in cells by antibodies against specific phosphorylation sites in their catalytic domains (41). However, crystal structures of ROCK catalytic domains revealed that their phosphorylation is not required for kinase activity (23, 64). ROCKs can exist in an unfolded (active) or a folded (inactive) configuration, where the catalytic domain is silenced by interactions with an autoinhibitory domain (2). The activation of ROCKs is induced by Rho-GTP or acidic lipid binding, the phosphorylation of autoinhibitory domains, or protease cleavage, resulting in a released, constitutively active catalytic domain (3, 30). Although the two ROCKs share similar domain structures and substrate specificities, evidence for nonoverlapping cellular functions is accumulating. Our previous studies showed that ROCK I is required for focal adhesion and stress fiber formation in fibroblasts, whereas ROCK II regulates phagocytic activity (65) and fibronectin (FN) matrix assembly (66). Previous reports of ROCK isoform-specific knockout mice also revealed their specific functions in cardiovascular disease and diabetes (29, 49). Therefore, it is important to understand the specific mechanisms by which the cellular activity of each ROCK is regulated.
Collapsin response mediator protein 2 (CRMP-2), also named TOAD-64/DRP-2/Ulip2/TUC-2, belongs to the CRMP family, consisting of 5 members in mammals. These proteins can be highly phosphorylated by various protein kinases, including ROCK (4, 47). CRMP-1 to -4 exist in two isoforms (long and short), with a common core polypeptide but different N-terminal domains that are products of alternative mRNA splicing. This study denotes the long form as CRMP-L, which was previously called CRMP-A or TUC-b, and the short form as CRMP-S. Since a mutant of unc-33, the Caenorhabditis elegans homologue of CRMP, showed abnormal axon termination (20), most studies have focused on neurobiology, and functional analyses of CRMP-2 have been limited almost exclusively to the short form. Some CRMP-2S-binding molecules are known, including the cytoskeletal protein tubulin and the motor protein dynein (21). CRMP-2S has been linked to neuronal differentiation and polarity during nervous system development and regeneration and to neurological disorders such as Alzheimer's disease (21). In fact, CRMP-2S is expressed ubiquitously (19), although only rarely have its potential functions in nonneuronal cells been considered (54, 56). Moreover, the functional sequelae of CRMP splice variant expression are unclear.
Here, CRMP-2L is shown to be expressed in epithelial cells and to be endogenous inhibitor of ROCK II but not ROCK I rather than simply a substrate. CRMP-2 exercised a control of ROCK II activity through interactions with its catalytic domain, independent of RhoA-GTP levels. Cell migration and the actin cytoskeleton of carcinoma cells were strongly regulated by the CRMP-2–ROCK II interaction. Moreover, the ectopic expression of CRMP-2L or its ROCK II-binding domains inhibited carcinoma cell migration and matrix assembly in fibroblasts in a ROCK-dependent manner. Taken together, the interaction between these two proteins provides a novel mechanism by which CRMP-2 controls myosin II-mediated cellular functions through the inhibition of ROCK II in nonneuronal cells and a potential therapeutic strategy for cancer and fibrosis.
MATERIALS AND METHODS
Antibodies.
The following antibodies were used: antibodies against ROCK II (C-20), ROCK I (K-18), RhoA (119), and actin (I-19) from Santa Cruz Biotechnology; CRMP-2 clone C4G from IBL Co. Ltd. (Gunma, Japan); polyclonal antibodies against phosphothreonine (pThr) and the phosphorylated myosin light chain (pMLC) at Ser19 from Cell Signaling Technology; anti-V5 monoclonal antibody, Alexa Fluor (488 or 546)-labeled secondary antibodies, and Alexa Fluor 546-phalloidin from Invitrogen; anti-MLC monoclonal antibody from Sigma-Aldrich; rabbit polyclonal antibodies against phosphorylated CRMP-2 at Thr522 or at Thr555 from ECM Biosciences LLC; rabbit polyclonal and mouse monoclonal anti-green fluorescent protein (anti-GFP) antibodies from Clontech; monoclonal antibodies against paxillin and actin from Millipore; monoclonal antihemagglutinin (anti-HA) antibody from Covance; horseradish peroxidase-labeled secondary antibodies from Dako A/S; rabbit polyclonal antibodies against CRMP-1 and CRMP-4 from Abcam; and a β1 integrin-inhibitory rat monoclonal antibody (AIIB2) from the Developmental Studies Hybridoma Bank, maintained by the University of Iowa. Rabbit polyclonal antifibronectin antibody R2/7 was described previously (34).
Cell culture.
Rat embryo fibroblasts (REFs) were maintained in α minimal essential medium Eagle (Lonza Walkersville, Inc.) with 5% heat-inactivated fetal bovine serum (FBS; HyClone). Rat1 rat fibroblasts, A7r5 rat smooth muscle cells, MDA-MB-231 human breast carcinoma cells, and PANC1 human pancreatic carcinoma cells were purchased from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle medium (DMEM) with 10% FBS. MCF7 human breast carcinoma cells were obtained from the ATCC and were maintained in DMEM with 10% FBS on collagen I (Advanced BioMatrix)-coated dishes. SW480 and SW620 human colon carcinoma cells were obtained from the ATCC and maintained in DMEM-F12 medium with 10% FBS on collagen I-coated dishes. MDCK II cells were obtained from the ATCC and maintained in DMEM with 5% FBS. All mammalian cells were grown at 37°C in a 5% CO2 humidified atmosphere. Drosophila Schneider 2 (S2) cells were a gift from F. Werner (University College London, United Kingdom) and were maintained in Schneider's Drosophila medium (Lonza) with 10% FBS at room temperature (RT).
Immunoprecipitation and Western blotting.
Immunoprecipitations of ROCKs with antibodies against ROCK C-terminal regions but not N-terminal catalytic domains or epitope-tagged proteins were carried out as described previously (65). Immunoprecipitated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies as appropriate. Signals were analyzed with NIH Image, version 1.61. In some experiments, cells were pretreated with the ROCK inhibitor Y-27632 (30 μM; Calbiochem) in growth medium for 1 h at 37°C prior to lysis. Thin white vertical lines indicate that the blots were obtained from different parts of the same membranes.
Protein identification by tandem mass spectrometry.
Immunoprecipitates (IPs) from REF cell lysates (eight round 15-cm-diameter dishes) were prepared as described above for immunoprecipitation procedures. Proteins were separated by SDS-PAGE and detected with silver staining. Silver-stained gel bands were excised with a scalpel, and proteins were digested in gel with trypsin, using an Investigator Progest robot (Genomic Solutions) as previously described (57).
Samples were analyzed by high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry (MS/MS). High-performance liquid chromatography was carried out with a CapLC liquid chromatography system (Waters). Aliquots (6 μl) of peptide mixtures were injected onto a Pepmap C18 column (300 μm by 0.5 cm; LC Packings) and eluted with an acetonitrile–0.1% formic acid gradient to the nanoelectrospray source of a Q-Tof spectrometer (Micromass) at a flow rate of 1 μl/min. The spray voltage was set to 3,500 V, and data-dependent MS/MS acquisitions were performed on precursor peptides with charge state 2, 3, or 4 over a survey mass range of 440 to 1,400 kDa, using argon collision gas. Product ion spectra were recorded over the range of 100 to 1,800 kDa and transformed onto a singly charged m/z axis using a maximum entropy method (MaxEnt3; Waters), and centroided peaklist (pkl) files were extracted by using MassLynx Routine Peptide Auto (Waters).
Proteins were identified by the correlation of uninterpreted spectra to entries in Swiss-Prot (Release 2010_04; 516,081 entries), using a local installation of Mascot (version 2.2; Matrix Science). MS/MS ion searches specified up to two missed cleavages per peptide, a precursor mass tolerance of ±100 ppm, and a fragment ion mass tolerance of ±0.5 Da. Carbamidomethylation of cysteines and methionine oxidation were specified as fixed and variable modifications, respectively.
Criteria for protein identification.
MS/MS-based peptide and protein identifications were validated by using Scaffold (version 3.01; Proteome Software Inc.) Peptide identifications were accepted if they could be established with a greater than 95.0% probability, as specified by the Peptide Prophet algorithm (26). Protein identifications were accepted if they were established with a greater than 99.0% probability and contained at least 2 matched peptides. Protein probabilities were assigned by the Protein Prophet algorithm (38).
Reverse transcription-PCR.
Total RNA was prepared from REF and MDA-MB-231 cells by using RNA-Bee (Tel-Test Inc.) according to the manufacturer's protocols and was transcribed with Superscript II (Invitrogen) and oligo(dT)15 primer. To analyze mRNA levels of CRMP-4 in REFs, total RNA (1 μg) treated with RNase-free DNase I (Sigma) was transcribed by using high-capacity cDNA reverse transcription kits (Applied Biosystems) and random primers. PCR was carried out by using KOD Hot Start DNA polymerase (Novagen) and primers 5′-GGA GCA GGC ACG AAT GCT GG and 5′-TTA ACT CAG GGA TGT GAT GTT AGA ACG GCC for rat CRMP-4 and primers 5′-ATG GAT GAC GAT ATC GCT GCG and 5′-CTA GAA GCA TTT GCG GTG CAC for rat β-actin mRNAs.
Plasmid construction.
Rat CRMP-2S, rat CRMP-4L, or human CRMP-2L cDNAs were amplified by PCR from a REF cDNA library or an MDA-MB-231 cell cDNA library and subcloned into pET41b(+) and pET24a (Novagen). Rat CRMP-2S and human CRMP-2L cDNAs were also subcloned into pEGFP-N1 and pIRES2-EGFP (Clontech). To create HA-tagged CRMP-2S or -2L, an oligonucleotide sequence encoding HA was inserted at the 5′ end of the cDNA in frame by PCR, using the pIRES2-CRMP construct as a template. The cDNAs encoding fragments of CRMP-2S (fragment A [amino acids {aa} 1 to 275], fragment B [aa 276 to 440], or fragment C [aa 441 to 572]) or the cDNAs encoding fragments of CRMP-2L (fragment A′ [aa 1 to 380] or fragment A″ [aa 1 to 118]) were subcloned into pET41b(+). The cDNAs encoding fragment C and fragment A′ were also subcloned into pEGFP-N1. Phosphorylation-resistant (replacement of Thr with Ala) mutants of the CRMP-2 short form at Thr555 and the CRMP-2 long form at Thr660 were created by PCR using wild-type cDNA in pET41b(+) and pET24 as a template.
Human ROCK I cDNA (a gift from S. Narumiya, Kyoto University, Japan) was subcloned into pMT/V5-His A (Invitrogen), and bovine ROCK II cDNA (a gift from K. Kaibuchi, Nagoya University, Japan) was subcloned into pMT/V5-His C (Invitrogen). A fragment containing the rat ROCK II catalytic domain (aa 1 to 543) was subcloned into pMT/V5-His C. All constructs were verified by DNA sequencing.
Transfection.
S2 cells were cotransfected with pMT/V5-His plasmids and pCoHygro by use of a calcium phosphate transfection kit (Invitrogen) according to the manufacturer's protocol. Stable transfectants were selected in growth medium containing 300 μg/ml hygromycin B (Roche Applied Science). ROCK protein expression was induced with 0.5 mM cupric sulfate for 2 days. Recombinant protein expression was confirmed by Western blotting with monoclonal anti-V5 antibody. REF and MCF7 cells were transfected with plasmids by use of Lipofectamine LTX reagent (Invitrogen) and Plus reagent (Invitrogen) according to the manufacturer's protocols. SW480 cells were transfected with short hairpin RNA (shRNA) plasmids against human CRMP-2 (4 individual shRNAs targeting both isoforms of human CRMP-2, shRNA1 to shRNA4, in the pREFP-C-RS vector; Origene) individually or a plasmid carrying a scrambled control shRNA (pREFP-C-RS vector; Origene) by using Nucelofector with kit V (Amaxa) according to the manufacturer's protocol. Stable transfectants were established for each shRNA construct by puromycin resistance (1 μg/ml; Sigma). SW480 cells expressing shRNA were also transfected with small interfering RNA (siRNA) against human CRMP-2 (Hs_DPYSL2_3; Qiagen) or AllStars negative-control siRNA (Qiagen) using Oligofectamine (Invitrogen) according to the manufacturer's protocol. MDA-MB-231 cells were transfected with siRNA against human CRMP-2 (Hs_DPYSL2_3 and Hs_DPYSL2_4; Qiagen), one against human ROCK II (D-004610-05; Dharmacon), or AllStars negative-control siRNA by using Oligofectamine. Two different siRNAs against human CRMP-2L (target sequences are 5′-ACG AAG AGG TCC CTG CTT TTT and 5′-GAG AGA AAG CAA TCC GGG ATT) were designed and synthesized by Dharmacon. MDCK II cells were transfected with either the shRNA-3 plasmid against human CRMP-2 or control shRNA using Lipofectamine LTX reagent and Plus reagent according to the manufacturer's protocol. Transfectants were selected by puromycin resistance (5 μg/ml). SW620 cells were transfected with plasmids by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. REFs were transfected with siRNAs using Oligofectamine as previously described (65). An siRNA against rat CRMP-2 (target sequence of 5′-AAA CTC CTT CCT CGT GTA CAT) was designed and synthesized by Ambion, and an siRNA against rat CRMP-4 (48833) was purchased from Ambion (45).
Recombinant protein expression in E. coli and binding assay.
According to the manufacturers' protocols, recombinant glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli strain BL21 (Promega) transformed with pET41b(+) constructs, and the preparation of glutathione-agarose (Sigma) coated with recombinant proteins was carried out, except that recombinant proteins were extracted with 1% (vol/vol) Triton X-100 after sonication.
S2 cells expressing ROCK proteins as described above were harvested, washed with Tris-buffered saline (TBS) (pH 7.5) twice, and lysed with TBS (pH 7.5) containing 1% Triton X-100, protease inhibitor cocktail (Roche), 10 mM MgCl2, 1 mM EDTA, and phosphatase inhibitors (20 mM β-glycerophosphate, 25 mM NaF, and 1 mM orthovanadate). After centrifugation, cleared supernatants were incubated with GST fusion protein-coated glutathione-agarose beads at 4°C for 1 h. Beads were washed with ice-cold lysis buffer four times. The proteins pulled down were analyzed by Western blotting with a V5 antibody.
ROCK and GTP-RhoA activity assay.
ROCK isoform-specific activities from SW480 cells expressing shRNA were analyzed by using a GST-MLC substrate as described previously (65). Briefly, both isoforms were specifically immunoprecipitated (preclearing step for 15 min and immunoprecipitation step for 30 min at 4°C), and the phosphorylation activity for 10 min at 30°C in immunoprecipitates (IPs) was measured by using GST-MLC as the substrate and [γ-32P]ATP (3 μCi, 370 GBq/mmol, 6 μM final concentration; Perkin-Elmer) as a phosphate source. MLC was absorbed onto P81 paper, and the radioactivity was measured by scintillation counting. To measure the inhibition ability of CRMP-2, a similar protocol was used. The catalytic domain of ROCK II expressed in S2 cells was immunoprecipitated on anti-V5-agarose antibody (Sigma). Equal amounts of beads were individually incubated with 5 μM GST, CRMP-2L(T660A)–His6, CRMP-2S(T555A)–His6,or a mixture of GST–CRMP-2A′ and GST–CRMP-2C on ice for 15 min in phosphorylation buffer (65). Phosphorylation reactions were then carried out by using 4 μg (3.8 μM) of GST-MLC as a substrate and [γ-32P]ATP (3 μCi) for 10 min at 30°C. In control reactions, Y-27632 was added to 2 μM in the presence of GST and MLC. CRMP-2L(T660A)–His6 and CRMP-2S(T555A)–His6 were expressed in BL21 cells transformed with pET24a constructs and purified as described in a previous study (53), which reported that CRMP-2S forms tetramers.
Conventional GTP-RhoA and GTP-Rac1 pulldown assays were performed as described previously (65).
Cell migration assay.
SW480 cell migration assays were carried out as described previously (16), with minor modifications. Briefly, the lower sides of transwell inserts (polycarbonate filter, 8-μm pore size; Costar) were coated with 10 μg/ml of rat tail type I collagen (BD Biosciences) in phosphate-buffered saline (PBS) at 4°C overnight. Cells were serum starved overnight and then harvested by using EDTA-trypsin (Invitrogen). Trypsin was inactivated by a soy bean trypsin inhibitor (Sigma), and cells were suspended in migration medium for 30 min at 37°C. In some experiments, Y-27632 (10 μM) was added to cell suspensions, before 4 × 104 cells were seeded in 100 μl onto inserts. After 6 h, the cells that migrated through the filter were counted as reported previously (33). SW620 cell migration assays with serum-free DMEM-F12 medium were performed similarly but for 8 h. MDCK II cell migration assays with serum-free DMEM were performed similarly but with phorbol 12-myristate 13-acetate (PMA) (125 nM; Calbiochem) as a chemoattractant and for 18 h.
Quantification of ROCK and CRMP-2 proteins in cell lysates.
Antibodies against ROCK I (K-18) and ROCK II (C-20) recognize the C-terminal domains of ROCK I and II, respectively. GST-pleckstrin homology domains of ROCK I and ROCK II were purified as described previously (65) and used as standard proteins. As the epitope for the CRMP-2 monoclonal antibody, C4G, located within the CRMP-2 C fragment (17), GST-CRMP-2 C was purified as described above and used as a standard. Concentrations of standard proteins were calculated by comparison with bovine serum albumin (BSA) with Coomassie blue staining of SDS-PAGE gels. Total cell lysates lysed with Laemmli sample buffer and serial dilutions of standard proteins for each antibody were loaded into the same gel and analyzed for protein expression. Signals were analyzed as described above.
Blue native polyacrylamide gel electrophoresis.
Blue native polyacrylamide gel electrophoresis (BN-PAGE) was performed as described previously by Wittig et al. (61). Briefly, confluent cell cultures in 6-cm dishes were harvested in 50 mM imidazole-HCl (pH 7.0), 500 mM 6-aminocaproic acid, 1 mM EDTA, 4% digitonin (Sigma), 5% glycerol, EDTA-free protease cocktail (Roche), 20 mM β-glycerophosphate, and 25 mM NaF. After centrifugation (12,600 × g for 2.5 min at 4°C), 0.5% (wt/vol) Coomassie blue G-250 was added to the soluble fraction. Samples were divided into two fractions, separated by the same 3-to-10% gradient gel with a 3% stacking gel at 4°C. After the gels were treated with 1× SDS-PAGE running buffer for 10 min, proteins in gels were electroblotted onto a polyvinylidene difluoride (PVDF) membrane at 4°C. To determine the molecular sizes, a NativeMark unstained protein standard (Invitrogen) was resolved in the same gel and stained with Coomassie blue R-250. Membranes were cut into two pieces: one piece was incubated with monoclonal anti-ROKα antibody (BD), and the other was incubated with anti-CRMP-2 antibody C4G, followed by horseradish peroxidase-conjugated secondary antibodies. Detection was carried out by using Amersham ECL Prime Western blotting reagent. Molecular sizes were calculated by using Adobe Photoshop and Microsoft Excel 2010.
Immunofluorescence microscopy and confocal laser microscopy.
SW480 cells seeded in growth medium onto glass coverslips coated with collagen I (rat tail) and MDA-MB-231 cells seeded in growth medium onto coverslips were fixed with 4% paraformaldehyde for 10 min and permeabilized with 0.1% Triton X-100 in PBS for 15 min, and ROCK and CRMP-2 were detected as reported previously (65). Coverslips were mounted by using ProLong Gold antifade reagent (Invitrogen). Controls for nonspecific cross-reactions of secondary antibodies gave no staining above background levels. Fluorescence was viewed with a confocal laser scanning microscope (Axiovert 200 M LSM510; Carl Zeiss Inc.) using a 63× 1.2-numerical-aperture (NA) c-Apochromat objective in water at RT. Images were processed by using LSM510 META software plus LSM image examiner software (Carl Zeiss Inc.) and Photoshop (Adobe). MDA-MB-231 cells in growth medium were fixed at 48 h posttransfection with siRNA. In some experiments, cells were treated with 30 μM Y-27632 for 1 h prior to fixation. Cells were stained for pMLC or paxillin as reported previously (65). At 48 h posttransfection with siRNA, REF cells were serum starved for 4 h and then treated with lysophosphatidic acid (LPA) (100 ng/ml; Sigma) for 30 min. Cells were fixed with paraformaldehyde and then stained for pMLC together with paxillin as reported previously (65). At 24 h after transfection with plasmids, REF cells were stained for F-actin or FN antibodies as reported previously (66). Images were taken with a Zeiss Axioplan-2 microscope (Plan-Apochromat ×63/1.4-NA oil objective) equipped with a CoolSNAP cf camera (Photometrics) at RT and processed by using MetaMorph software (MDS Analytical Technologies) and Adobe Photoshop. For each transfection, at least 5 different fields for MDA-MB-231 cells and a minimum 50 cells for REFs were analyzed, and data from three separate experiments were analyzed.
DOC lysis.
REFs (4 × 104 cells) seeded into 12-well plates were transfected with plasmids. One day later, cells were analyzed for FN deoxycholate (DOC) insolubility as reported previously (48). Statistical analysis was performed by Kruskal-Wallis analysis with the post hoc Student-Newman-Keuls method.
Cell attachment assay.
Cell suspensions of SW480 cells expressing shRNA were prepared as described above for the cell migration assay. Cells were preincubated with β1 integrin-blocking antibody (AIIB2) (10 μg/ml) or normal rat IgG (control) (10 μg/ml) in serum-free medium on ice for 30 min and then replated onto collagen I (rat tail) in the presence of antibodies at 37°C for 1 h. Cells were fixed with paraformaldehyde, and phase-contrast images of cells were taken with an Olympus IX71 inverted microscope (objective, UPlanFl ×10/0.30) with a DP50 camera at RT and processed with Viewfinder Lite, version 1.0, and Studio Lite, version 1.0.
RESULTS
ROCK II is less sensitive to LPA-mediated activation of cells than ROCK I.
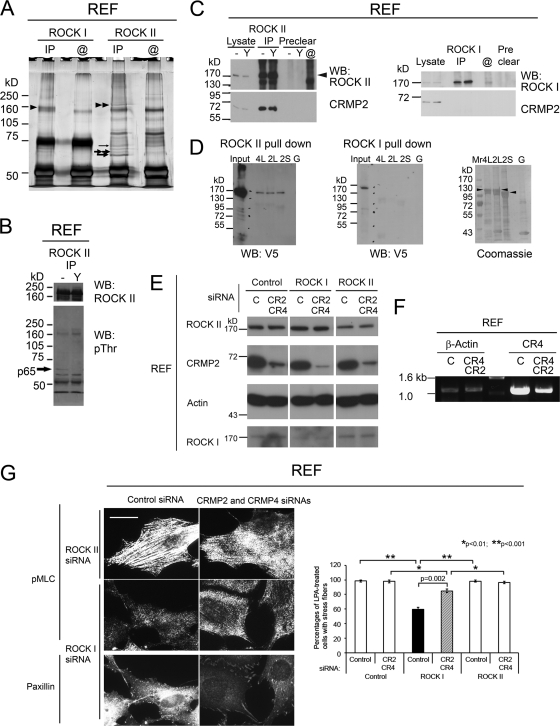
Our previous study demonstrated that the level of ROCK II kinase activity, compared to that of ROCK I, was low during REF adhesion and less sensitive to LPA treatment (65). LPA is a bioactive lipid that can stimulate tumor formation and metastasis (36) and induces the formation of stress fibers and focal adhesions in a Rho-GTP-ROCK-dependent manner (1, 43). Stress fibers are thick actin bundles that are decorated with pMLC and terminate at focal adhesions detected by marker proteins such as paxillin (65). To examine whether ROCK II, in the absence of ROCK I, mediates LPA-induced actin cytoskeleton reorganization, ROCK proteins in REFs were knocked down by siRNAs. After serum starvation, control siRNA-transfected REFs formed fine microfilaments rather than thick stress fibers, which could be restored by LPA treatment (Fig. 1). ROCK II-depleted REFs possessed thin filaments under starved conditions, and LPA treatment induced stress fibers decorated with pMLC. However, ROCK I-depleted REFs failed to form actin filament bundles and focal adhesions under both starved and LPA-treated conditions (Fig. 1). The depletion of one isoform of ROCK did not affect the expression of the other (65). ROCK II was therefore less sensitive to the elevation of cellular GTP-RhoA levels in fibroblasts, suggesting the presence of a ROCK II-specific regulatory molecule(s).
Fig 1.
ROCK II is less sensitive to LPA treatment than ROCK I. REFs transfected with the indicated siRNAs were serum starved and then stimulated with LPA. Cells were stained for pMLC and paxillin. While control and ROCK II-depleted REFs formed stress fibers terminating at focal adhesions (arrowheads) upon LPA stimulation, ROCK I-depleted REFs, which express ROCK II, did not. Insets show high-power images of the boxed areas. Scale bar, 25 μm. Images are representative of 3 separate experiments.
CRMP-2 selectively interacts with ROCK II but not ROCK I.
To search for ROCK isoform-specific substrates or binding proteins, immunoprecipitates (IPs) from REF lysates were analyzed by Western blotting with pThr antibodies and silver staining. Comparable amounts of ROCK I and ROCK II were immunoprecipitated, and several specific proteins were detected in ROCK II IPs (Fig. 2A). The reactivity of a 65-kDa protein (p65) with an anti-pThr antibody was sensitive to the ROCK inhibitor Y-27632 (22), suggesting that p65 could be an endogenous ROCK substrate (Fig. 2B). p65 was identified by tandem mass spectrometry as a short form of CRMP-2 (CRMP-2S) (Fig. 2A) (mass spectrometry data are available upon request). CRMP-2S is a 62-kDa protein that is highly phosphorylated in neuronal cells (21) and is phosphorylated on Thr555 by ROCK II, although it was not established whether ROCK I could phosphorylate CRMP-2S (4). Mass spectrometry also detected that some of the CRMP-2S was phosphorylated on Ser522, a Cdk5 phosphorylation site (7). A second polypeptide with a slightly higher apparent molecular mass was also identified as CRMP-2S (Fig. 2A, thick double arrows), suggesting multiple phosphorylation states of CRMP-2S. Moreover, the CRMP-4 long form (CRMP-4L) was detected at a molecular mass of 72 kDa (Fig. 2A, thin arrow), suggesting that CRMP-2S may form heterotetramers with CRMP-4L, as described previously (58).
Fig 2.
CRMP-2 selectively interacts with ROCK II but not ROCK I. (A) Silver staining of ROCK II and ROCK I IPs from REF lysates. Beads coated with ROCK antibodies but not incubated with lysates are shown as @. Comparable amounts of ROCK II (double arrowheads) and ROCK I (arrowhead) proteins were immunoprecipitated. Proteins coimmunoprecipitated with ROCK II specifically were identified as CRMP-2S, including a phosphorylated peptide (thick single arrow); CRMP-2S (thick double arrows); and CRMP-4L (thin arrow). (B) Western blotting (WB) of ROCK II IPs from REF lysates with antibodies against ROCK II (top) and pThr (bottom). A 65-kDa protein was sensitive to treatment with 30 μM Y-27632 (Y). (C) Western blotting of CRMP-2 (bottom) from ROCK II IPs of REF lysates (left) or ROCK I IPs (right). Arrowhead, ROCK II proteins. CRMP-2 was detected in IPs of ROCK II independent of its kinase activity but not in those of ROCK I. (D) Pulldown of recombinant full-length ROCK II (left) but not ROCK I (middle) on beads coated with full-length CRMP-4L, -2L, or -2S but not with GST (G). Bound proteins were detected by Western blotting with a V5 antibody. The right panel shows Coomassie staining of GST proteins eluted from beads. Black arrowheads indicate intact GST-CRMPs. Blots are representative of 3 separate experiments. (E) Western blot analysis of total cell lysates of REF cells transfected with siRNAs targeting CRMP-2 (CR2), CRMP-4 (CR4), ROCK I, ROCK II, or control siRNA (C). The reduction in CRMP-2 levels had no effect on ROCK protein levels, and vice versa. (F) Total RNA from siRNA-treated REF cells was isolated for reverse transcription-PCR analysis of CRMP-4 and β-actin mRNA levels. (G) Cotransfection of siRNAs against CRMP-2, CRMP-4, and ROCKs. REFs transfected with siRNAs were stimulated with LPA as described in the legend of Fig. 1 and stained for pMLC and paxillin. The knockdown of CRMP-2 partially restored LPA-induced stress fiber formation in ROCK I-depleted REFs and increased focal adhesion formation. Scale bar, 25 μm. Images are representative of the total population of cells. Shown are percentages of cells with stress fibers (means ± standard errors of the means [SEM]; n = 3). Comparisons among ROCK knockdown experiments were analyzed by a one-way analysis of variance (ANOVA) with the post hoc Holm-Sidak method, and the effect of the CRMP-2 knockdown on ROCK I siRNA-treated cells was analyzed by a t test.
The coimmunoprecipitation of CRMP-2S with ROCK II was demonstrated by Western blotting, whereas ROCK I IPs never contained CRMP-2S (Fig. 2C), suggesting that the coimmunoprecipitation of CRMP-2 with ROCK II is ROCK isoform specific. Notably, Y-27632 treatment to inhibit Rho kinases did not affect the CRMP-2–ROCK II association (Fig. 2C). Due to the inability of the CRMP-2 monoclonal antibody to immunoprecipitate from REF cell lysates, CRMP-2–ROCK II interactions were examined by alternative assays, where V5-tagged full-length ROCK proteins expressed in S2 cells were pulled down by GST-fused CRMP-coated beads. In addition to the CRMPs identified by mass spectrometry analysis, CRMP-2L was assessed. Smaller polypeptides other than intact CRMPs seen by Coomassie staining may be degradation products, as shown previously (13). While the CRMP-2S, -2L, and -4L proteins could pull down ROCK II, no interactions with ROCK I were detected (Fig. 2D).
Next, we examined whether the knockdown of CRMPs provided ROCK I siRNA-treated REFs with a capacity for LPA-induced stress fiber formation. The knockdown of CRMP-2 together with CRMP-4 partially rescued stress fiber formation in ROCK I knockdown cells but had no effect on cells treated with either control or ROCK II siRNA (Fig. 2E to G). These data suggest that CRMPs may regulate ROCK II activity.
CRMP-2L is expressed widely in carcinoma cells, and CRMP-2 complexes containing 2L preferentially associate with ROCK II.
CRMP-2S expression in nonneuronal cells was reported previously for fibroblasts (54), T cells (56), and colorectal tumor cells (63), although its functions are unknown. The screening of many normal and tumor cells confirmed its ubiquitous expression (Fig. 3A to C and Table 1). The coimmunoprecipitation of CRMP-2S with ROCK II was also observed for A7r5 rat aortic smooth muscle cells and Rat1 fibroblasts (Fig. 3A). Many human carcinoma cell lines (10 out of 16 carcinoma cell lines), including MDA-MB-231 breast carcinoma cells and PANC-1 pancreatic carcinoma cells, expressed doublet polypeptides (75 kDa and 65 kDa) cross-reacting with CRMP-2 antibody (Fig. 3B and C and Table 1). The 75-kDa polypeptide corresponded to CRMP-2L, and its cDNA was subcloned from MDA-MB-231 cells and sequenced. On the other hand, other CRMP family members, for example, CRMP-1 and CRMP-4, were detected in only a few cell lines by Western blotting (Table 1 and Fig. 3B). Both CRMP-2S and -2L were coimmunoprecipitated with ROCK II from MDA-MB-231 cells, and their phosphorylation was detected by Western blotting with both pSer522- and pThr555-specific antibodies (Fig. 3D). The specificities of the antibodies were confirmed by the Western blotting of cell lysates prepared without phosphatase inhibitors or treated with Y-27632 (data available upon request). This result indicated that CRMP-2L could be phosphorylated on Ser627 and on Thr660 (corresponding to Ser522 and Thr555 of CRMP-2S) by Cdk5 and ROCK, respectively, although phosphorylation by ROCK did not affect the CRMP-2–ROCK II interaction (Fig. 2C).
Fig 3.
ROCK II associates with CRMP-2 complexes but preferentially with those containing long isoforms. (A and C) Coimmunoprecipitation of CRMP-2 (bottom) with ROCK II (top) from normal (A) and cancer (C) cells. Arrows indicate the degradation product. (B) Expression of CRMPs in various human cancer cell lines. Total cell lysates were analyzed by Western blotting for CRMP-2, CRMP-1, CRMP-4, and actin. (C) Abundant CRMP-2 proteins were coimmunoprecipitated with ROCK II from MDA-MB-231 and PANC1 cells that express endogenous CRMP-2L, while nearly undetectable amounts of CRMP-2S were detected in ROCK II IPs from MCF7 cells, where CRMP-2S was dominant. (D) CRMP-2L coimmunoprecipitated with ROCK II was phosphorylated. ROCK II IPs from MDA-MB-231 cell lysates were divided into 3 parts and analyzed by Western blotting with the indicated antibodies. (E) Preferential association of CRMP-2L with ROCK II. ROCK II IPs from MCF7 cells transfected with pIRES2-EGFP vector cDNAs, as indicated, were probed with antibodies against ROCK II (top) and CRMP-2 (bottom). (F) Reciprocal coimmunoprecipitation of ROCK II and CRMP-2L. Lysates of MCF7 cells transfected with a cDNA encoding CRMP-2L–GFP were divided for immunoprecipitation with ROCK II antibodies (right) or with GFP antibody (left). IPs were analyzed with antibodies against ROCK II (top) and GFP (bottom). (G) CRMP-2L and -2S form a protein complex. MCF7 cells were cotransfected with the pIRES2-EGFP vector containing cDNAs encoding CRMP-2L and -2S, where one isoform was tagged with an HA epitope. IPs with HA antibody were divided for Western blotting with HA antibody (right) or CRMP-2 antibody (left). Both isoforms were coimmunoprecipitated. The asterisk indicates the IgG heavy chain derived from the HA antibody. Blots are representative of 3 separate experiments. (H) ROCK II and CRMP-2 were detected in the same complexes of ≥800 kDa. Freshly prepared total cell lysates were subjected to BN-PAGE and then Western blotting. ROCK II exists in multiple complexes with a major pool of ∼1,100 kDa (arrow). Similarly, CRMP-2 was present in multiple populations. Doubly ROCK II- and CRMP-2-positive pools were repeatedly observed at ∼800 to 900 kDa, 1,000 to 1,100 kDa, and 1,200 kDa (arrowheads). Blots are representative of separate experiments.
Table 1.
CRMP expression
| Cell type | Origin | Presence of CRMPa |
|||
|---|---|---|---|---|---|
| 2S | 2L | 1S | 4 | ||
| Human carcinoma cell lines | |||||
| PANC1 | Pancreas | + | + | − | − |
| MDA-MB-231 | Breast | + | + | − | − |
| HMT3909 S13 | Breast | + | + | − | − |
| MCF-7 | Breast | + | − | − | − |
| T-47D | Breast | + | − | − | − |
| A498 | Kidney | + | + | − | S |
| PC3 | Prostate | + | + | − | − |
| HepG2 | Liver | + | + | − | S |
| HeLa | Cervix | + | − | − | − |
| A431 | Epidermis | − | − | − | − |
| SW480 | Colon | + | + | − | − |
| SW620 | Colon | + | − | − | − |
| RKO | Colon | + | + | − | − |
| CX-1 | Colon | + | − | − | − |
| Clone A | Colon | + | + | − | − |
| CALU-1 | Lung | + | + | − | − |
| Human noncarcinoma cell lines | |||||
| SHSY5Y | Neuroblastoma | + | + | + | − |
| K562 | Erythroleukemia | + | − | − | − |
| G361 | Melanoma | + | − | − | − |
| A375 | Melanoma | + | − | − | − |
| SK-MEL-28 | Melanoma | + | − | − | − |
| Lisa-2 | Liposarcoma | + | − | − | − |
| A204 | Rhabdomyosarcoma | + | − | − | − |
| RD | Rhabdomyosarcoma | + | − | − | − |
| HT1080 | Fibrosarcoma | + | − | − | − |
| SAOS2 | Osteosarcoma | + | − | − | − |
| U2OS | Osteosarcoma | + | − | − | − |
| Normal cell lines | |||||
| REF | Rat embryo fibroblast | + | − | − | L |
| Rat1 | Rat embryo fibroblast | + | − | − | L |
| A7r5 | Rat smooth muscle | + | − | − | L |
| MDCK | Canine kidney epithelial | + | + | −b | −b |
+ denotes “detected” and − denotes “not detected” by Western blotting. S indicates short form, and L indicates long form.
It is not known to cross-react with canine CRMPs.
Both CRMP-2 isoforms were readily detected by Western blotting of ROCK II IPs from cells expressing CRMP-2L (Fig. 3C). In contrast, where CRMP-2S was dominant and CRMP-2L was expressed at levels below detection, such as G361 melanoma cells and MCF7 breast carcinoma cells (Fig. 3C) (data available upon request), the coimmunoprecipitation of CRMP-2 with ROCK II was less or, in MCF7 cells, almost negligible. Moreover, an enrichment of CRMP-2L in ROCK II IPs was often observed compared to the ratio of the two endogenous CRMP-2 isoforms in cell lysates (Fig. 3C). To test whether the ectopic expression of CRMP-2L increases the binding of CRMP-2 to ROCK II, cDNAs encoding CRMP-2L were transfected into MCF7 cells. The expression of CRMP-2L significantly increased the amounts of CRMP-2 in ROCK II IPs (Fig. 3E), whereas that of CRMP-2S had little effect, suggesting a preferential interaction of ROCK II with CRMP-2L. To further examine whether ROCK II could be coimmunoprecipitated with CRMP-2L, CRMP-2L–GFP in MCF7 cells was subjected to immunoprecipitation with either GFP antibodies or ROCK II antibodies. Endogenous ROCK II was coimmunoprecipitated with CRMP-2L–GFP in reciprocal immunoprecipitation experiments, confirming an association between the two molecules (Fig. 3F) (control data available upon request).
CRMP family members form tetramers (58). To investigate whether CRMP-2L and -2S combine in complexes, MCF7 cells were cotransfected with cDNA encoding CRMP-2S and -2L, one with an HA tag and the other without, and IPs with HA antibody were analyzed by Western blotting with both HA and CRMP-2 antibodies (Fig. 3G). These experiments confirmed that CRMP-2L and -2S endogenously formed hetero-oligomeric complexes. In total, the data together suggest a preferential association of ROCK II with CRMP-2 complexes containing 2L.
Expression levels of ROCKs and CRMP-2 proteins in some normal and transformed cell lines were analyzed (Table 2). In general, ROCK II was expressed at higher levels than ROCK I, while CRMP-2 was in excess over ROCK II, but a large proportion of this was CRMP-2S rather than CRMP-2L. To estimate the stoichiometry of ROCK II–CRMP-2 complexes, freshly prepared total cell lysates were subjected to BN-PAGE (61). As ROCK II was suggested previously to exist in high-molecular-mass complexes of 660 kDa or higher in rat brain extracts (10), BN-PAGE revealed that ROCK II in all cell lines tested existed as multiple complexes with molecular masses ranging from 700 to 1,200 kDa, with a major pool of ∼1,100 kDa (Fig. 3H). Similarly, CRMP-2 was detected as multiple populations, with a major pool of ∼500 kDa. Doubly ROCK II- and CRMP-2-positive pools were repeatedly observed at ∼800 to 900 kDa, 1,000 to 1,100 kDa, and 1,200 kDa. Although both proteins can form polymers on their own with various potential stoichiometries, we can nevertheless conclude that both proteins can be detected in the same complexes of ≥800 kDa.
Table 2.
Amounts of ROCK I, ROCK II, and CRMP-2 proteins in cells analyzed in the study
| Protein | Mean amt of protein (pmol/106 cells) ± SEM in cell line |
|||||
|---|---|---|---|---|---|---|
| REF | MDCK | MDA-MB-231 | MCF-7 | SW480 | SW620 | |
| ROCK I | 2.92 ± 1.26 | 0.27 ± 0.04 | 0.47 ± 0.19 | 0.16 ± 0.03 | 0.07 ± 0.01 | 0.08 ± 0.2 |
| ROCK II | 5.37 ± 0.75 | 1.14 ± 0.16 | 0.66 ± 0.21 | 0.23 ± 0.12 | 0.42 ± 0.04 | 0.27 ± 0.06 |
| CRMP-2S | 46.7 ± 24.5 | 2.15 ± 0.57 | 6.71 ± 1.68 | 1.22 ± 0.32 | 16.3 ± 4.0 | 7.22 ± 2.92 |
| CRMP-2L | 0.13 ± 0.01 | 1.19 ± 0.49 | 1.18 ± 0.19 | |||
| ROCK II/ROCK I | 1.84 | 4.24 | 1.41 | 1.42 | 6.19 | 3.17 |
| CRMP-2/ROCK II | 8.71 | 2.01 | 12.0 | 5.24 | 41.5 | 27.0 |
| CRMP-2L/ROCK II | 0.12 | 1.80 | 2.81 | |||
| CRMP-2S/ROCK II | 8.71 | 1.89 | 10.2 | 5.24 | 38.7 | 27.0 |
| CRMP-2L/2S | 0.06 | 0.18 | 0.07 | |||
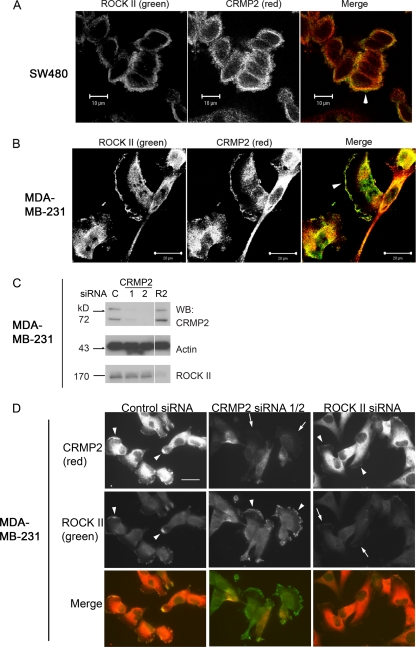
ROCK II and CRMP-2 colocalize at the motile periphery of human carcinoma cells.
Since CRMP-2 tetramers containing 2L preferentially associated with ROCK II, the cellular localizations of ROCK II and CRMP-2 in SW480 colon carcinoma and MDA-MB-231 cells, where CRMP-2L is expressed endogenously (Fig. 3B and C), were analyzed by indirect immunofluorescence staining and laser confocal microscopy. A prominent colocalization of ROCK II and CRMP-2 in SW480 cells was detected in cortical membrane areas and occasional cell-cell contacts in cell clusters (Fig. 4A). Colocalization in MDA-MB-231 cells was also detected in ruffling membrane areas (Fig. 4B). The cellular localization of each protein in response to siRNA treatment was analyzed. The efficient suppression of CRMP-2 or ROCK II proteins in MDA-MB-231 cells was elicited by the transfection of specific siRNAs compared to control siRNA-treated cells, while a reduction in levels of CRMP-2 had no effect on ROCK II protein levels, and vice versa (Fig. 4C). The specificities of staining in MDA-MB-231 cells were confirmed by the overall reduction of the fluorescence intensity after siRNA treatments by epifluorescence microscopy (Fig. 4D, arrows). The localization of the two proteins at the cell periphery was not dependent on their mutual interaction (Fig. 4D, arrowheads).
Fig 4.
CRMP-2 and ROCK II colocalize at the periphery of human carcinoma cells independently of their interaction with each other. (A and B) Colocalization of ROCK II and CRMP-2 at the periphery (arrowheads) of SW480 (A) or MDA-MB-231 (B) cells. Images were acquired by confocal laser scanning microscopy. (C) Western blot analysis of total cell lysates of MDA-MB-231 cells transfected with siRNAs targeting CRMP-2 (CR2) (siRNAs 1 and 2), ROCK II (R2), or control siRNA (C). An efficient suppression of CRMP-2 or ROCK II proteins was observed. (D) Double staining for CRMP-2 and ROCK II proteins of MDA-MB-231 cells transfected with siRNAs. Images were acquired by epifluorescence microscopy. Arrowheads indicate a pericellular protein localization. After the knockdown of target proteins, decreased fluorescence was observed, especially at cell margins (arrows). Exposure times were equal for each protein. Scale bar, 25 μm. Images are representative of 3 separate experiments.
CRMP-2 controls cell migration through the regulation of ROCK II kinase activity but independently of GTP-RhoA levels.
Some CRMP family members may be metastasis/invasion regulators, as suggested by previous proteomic/microarray analyses, although the underlying molecular mechanisms are unknown (15, 50). Moreover, ROCK activity has been implicated in the regulation of cell migration in various types of cancer cells (37, 46). To examine whether the CRMP-2–ROCK II interaction affects cancer cell behavior, endogenous CRMP-2 levels were reduced by stable shRNA expression in SW480 cells. The stable knockdown of CRMP-2 proteins (70 to 90% reduction) by 2 individual shRNAs was achieved in SW480 cells, while ROCK I and II protein levels were not altered by CRMP-2 shRNA expression (normalized to actin) (Fig. 5A). Since there are no tools to detect ROCK isoform-specific activity in cells, the average activity of each ROCK was measured by using immunoprecipitation from SW480 cells expressing shRNA, as reported previously (65). The knockdown of CRMP-2 with shRNA 1 increased mean ROCK II activity in SW480 cells over that of control shRNA transfectants (25% increment) (Fig. 5B), while the mean ROCK I activity was unaffected. Furthermore, CRMP-2 shRNA expression affected neither GTP-RhoA nor GTP-Rac1 levels (Fig. 5C and D). Taken together, our data strongly suggested that CRMP-2 bound ROCK II and inhibited its kinase activity despite similar levels of endogenous GTP-RhoA.
Fig 5.
CRMP-2 regulates cell migration in a ROCK II-dependent manner but independently of GTP-RhoA levels. (A) Western blot of total lysates from SW480 cells stably expressing shRNAs targeting CRMP-2 (shRNA 1 or 2) or control shRNA (control). Reductions of CRMP-2 protein levels (70 and 90%) were observed following shRNA 1 and 2 treatments, respectively. SW480 cells expressing CRMP-2 shRNA 2 or the control shRNA were further transfected with CRMP-2 siRNA (CR2) or the control siRNA. An average ∼90% reduction of CRMP-2 levels was observed with the combination of shRNA and siRNA, without a significant alteration in ROCK protein levels (normalized to β-actin). (B) Rho kinase activities from SW480 shRNA transfectants. Activities were normalized to ROCK protein levels (n = 4). ∗, P < 0.02 (Mann-Whitney test). (C and D) Pulldown assays of GTP-RhoA (C) and GTP-Rac1 (D) from SW480 shRNA/siRNA transfectants (n = 3 to 6). No significant differences were observed (Student's t test). (E) Haptotactic cell migration of SW480 shRNA/siRNA transfectants. In some cases, cells were treated with 10 μM Y-27632 (Y). (n = 3 [Y-27632 treated] or 6 [untreated]). Comparisons between different RNA interference (RNAi) transfectants were analyzed by a Mann-Whitney test, and the effect of inhibitor treatment was analyzed by Student's t test. (F) Western blots of total lysates of MDCK cells stably expressing shRNA targeting CRMP-2 (shRNA 3) or control shRNA (control). An average 80% reduction in CRMP-2 levels, compared to those of control cells, was observed with shRNA 3. (G) PMA-induced cell migration of MDCK shRNA transfectants. In some cases, cells were treated with 10 μM Y-27632 (Y) (n = 3 [Y-27632 treated] or 4 [untreated]). Comparisons between different shRNA transfectants were analyzed by a Mann-Whitney test, and the effect of inhibitor treatment was analyzed by Student's t test (CRMP2 shRNA) and by a Mann-Whitney test (control shRNA). Data in graphs show means ± SEM, and values for control shRNA/siRNA transfectants were set at 1.0 throughout. a.u., arbitrary units.
To reveal the roles of the CRMP-2–ROCK II interaction in cancer cell migration, the haptotactic cell migration of shRNA-expressing SW480 cells toward collagen I was analyzed in transwells. The level of cell migration after the CRMP-2 depletion was significantly higher than that with control shRNA transfectants (Fig. 5E). In a separate clone (shRNA 2), the residual CRMP-2 protein level was further decreased by siRNA transfection to <10% (Fig. 5A). CRMP-2 protein suppression by the combination of shRNA 2 and siRNA also resulted in increased cell migration compared to that of control-treated cells (Fig. 5E) without affecting GTP-RhoA and GTP-Rac1 levels (Fig. 5C and D). The increased cell migration resulting from the CRMP-2 knockdown by shRNA 1 was dependent on ROCK activity, since the addition of Y-27632 reduced cell migration to the same level as that of cells expressing control shRNA (Fig. 5E). The Y-27632 inhibitor did not affect that migration of control cells, suggesting that the baseline haptotactic migration of SW480 was not dependent on ROCK activity but confirming CRMP-2 as an important ROCK activity regulator. SW480 cells express α2β1 integrin as a collagen receptor (12), but an alteration of CRMP-2 levels had no influence on adhesion receptor characteristics. In each case, a β1 integrin-inhibitory antibody (AIIB2) almost completely blocked cell attachment to collagen I substrates (data available upon request). Therefore, the CRMP-2 protein regulated the haptotactic migration of SW480 cells in a ROCK II activity-dependent manner but independently of GTP-RhoA.
CRMP-2L was detected in various epithelial cells of chicken embryo and normal epithelial MDCK cells (67), although its cellular functions were not investigated. The treatment of MDCK cells with PMA stimulates ruffle membrane formation and cell scattering in a protein kinase C (PKC)-RhoGDI-RhoGTP-ROCK-dependent manner (14, 25). To examine the significance of CRMP-2 in PMA-induced MDCK cell migration, CRMP-2-deficient cells were established by expressing a human CRMP-2 shRNA 3, which completely and uniquely matched with an oligonucleotide sequence in canine CRMP-2 cDNA (Fig. 5F). The expression of CRMP-2 shRNA 3 did not alter ROCK protein levels. In comparison with cells expressing control shRNA, CRMP-2-deficient cells migrated more in response to PMA (Fig. 5G). Importantly, PMA responses depended on ROCK activity, since Y-27632 completely inhibited cell migration. The CRMP-2 depletion did not increase cell proliferation during PMA treatment (after PMA treatment, control shRNA-expressing cells were 91% of control cell numbers, and CRMP-2 shRNA-expressing cells were 70% of control cell numbers). These data suggested that CRMP-2 regulated the PMA-induced, ROCK-dependent migration of normal epithelial cells.
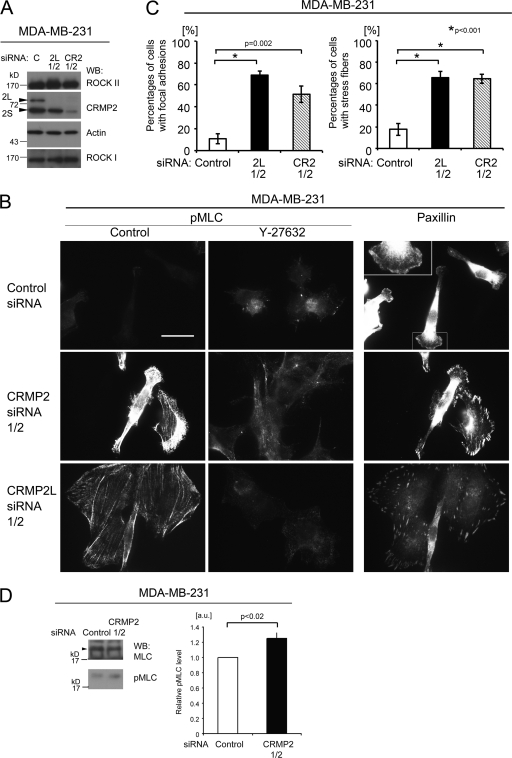
CRMP-2L regulates the actin cytoskeleton in human breast carcinoma cells.
To examine whether altered CRMP-2 levels, including 2L alone, in cancer cells affected ROCK-myosin II-mediated cell contractility, To examine whether altered CRMP-2 levels, including 2L alone, in cancer cells affected ROCK-myosin II-mediated cell contractility, the MLC phosphorylation status as an indicator of myosin II activity (55) was analyzed in MDA-MB-231 cells that have a more uniform morphology than SW480 cells. pMLC occasionally localized to protrusions of control siRNA-treated cells (Fig. 6B). On the other hand, pMLC staining in both CRMP-2 siRNA-treated and CRMP-2L siRNA-treated cells showed its presence in cortical filaments and microfilament bundles, resulting in increased percentages of cells with stress fibers, although there are normally few of these in this cell line (Fig. 6A to C). This staining pattern was sensitive to ROCK inhibitor treatment (Fig. 6B, middle). Upregulated MLC phosphorylation levels were confirmed by Western blotting. The knockdown of total CRMP-2 induced an average 25% increase in pMLC levels compared to those of control siRNA-treated cells (Fig. 6D). Moreover, differences in focal adhesion formation, which requires myosin II-mediated cellular contractility (62), were observed. (Fig. 6B, right). The localization of the focal adhesion marker protein paxillin in small adhesion structures at the cell periphery was evident in CRMP-2 or CRMP-2L siRNA-treated cells, whereas it was detected in ruffling membranes in control siRNA-treated cells. Therefore, CRMP-2L regulates actin cytoskeleton contractile activity through the ROCK II-pMLC pathway in transformed cells.
Fig 6.
CRMP-2 regulates the actin cytoskeleton of breast carcinoma cells. (A) Western blots of total lysates of MDA-MB-231 cells transfected with siRNAs targeting CRMP-2 (CR2) (siRNA mixture), CRMP-2L (2L) (a mixture of siRNAs 1 and 2), or control siRNA (C). Efficient suppression of CRMP-2 or CRMP-2L protein was observed. The reduction in CRMP-2 levels had no effect on ROCK II protein levels, and vice versa. (B) Staining for pMLC (left and middle) or paxillin (right) of MDA-MB-231 cells transfected with the siRNAs indicated. In some cases, cells were treated with 30 μM Y27632 (middle). The inset shows a high-power image of the boxed area. Images are representative of the total population of cells. Scale bar, 25 μm. (C) Percentages of cells with focal adhesions (left) or stress fibers (right) (means ± SEM; n = 3). A one-way ANOVA with the post hoc Holm-Sidak method was used. (D) Western blots for pMLC and MLC levels in total lysates of MDA-MB-231 cells transfected with the indicated siRNAs. The graph shows relative MLC phosphorylation levels. Values for control shRNA/siRNA transfectants were set at 1.0, and data show means ± SEM (n = 4) (Mann-Whitney test). Blots are representative of separate experiments.
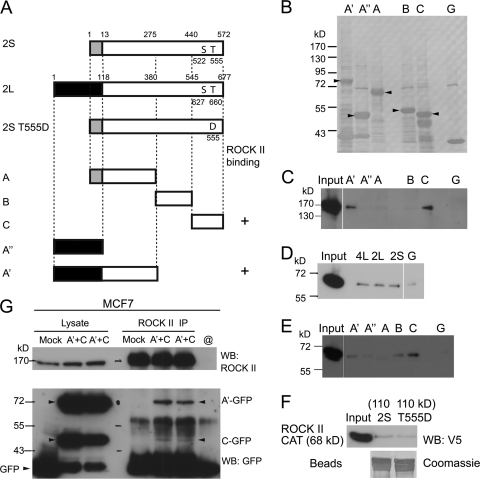
CRMP-2L has a binding site for ROCK II not shared with CRMP-2S.
To further understand the molecular basis and cellular functions of CRMP-2–ROCK II complexes, their interaction sites were mapped. Five different polypeptides of CRMP-2 proteins (polypeptides A′ [aa 1 to 380] and A″ [aa 1 to 118] from CRMP-2L and polypeptides A [aa 1 to 275], B [aa 276 to 440], and C [aa 441 to 572] from CRMP-2S) were produced as GST fusion proteins in E. coli (Fig. 7A and B), and pulldown assays were performed. Recombinant full-length ROCK II bound to the A′ polypeptide of CRMP-2L and the common C-terminal domain of CRMP-2L and -2S (Fig. 7C). The ability of the recombinant ROCK II catalytic domain (aa 1 to 543) to interact with full-length CRMP-2L, -2S, and -4L was also confirmed by pulldown assays (Fig. 7D), although no preferential binding to 2L or 4L over 2S was observed. Similar results were obtained with full-length ROCK II (Fig. 2D), but the results may have been influenced by GST at the N termini or because CRMP proteins on beads do not form tetramers. The catalytic domain of ROCK II bound to the A′ polypeptide and to the common C domain, although a weak binding of the B domain to the ROCK II catalytic domain was also observed (Fig. 7E). As A″ and A polypeptides did not bind to ROCK II, the linkage region of the long-form-specific N-terminal domain and the CRMP-2 core polypeptide may be important for ROCK II recognition. The phosphorylation of CRMP-2 by ROCK had no effect on the interaction, as confirmed by pulldown assays with a phosphorylation mimetic mutant [2S(T555D), with a replacement of Thr555 with Asp]. Both wild-type and mutant GST-tagged full-length CRMP-2S proteins could pull down the ROCK II catalytic domain to the same extent (Fig. 7F). In summary, these data showed that the ROCK II catalytic domain interacted with the C-terminal domain of CRMP-2, while the N-terminal domain of CRMP-2L provided an additional binding site. The ROCK II-binding properties of these two polypeptides derived from CRMP-2L were confirmed by coimmunoprecipitation assays with mammalian cells. Both GFP-A′ and -C polypeptides expressed in MCF7 cells were coimmunoprecipitated with endogenous ROCK II proteins (Fig. 7G).
Fig 7.
CRMP-2L has a binding site for ROCK II not shared with CRMP-2S. (A) Diagram of CRMP-2 constructs used for pulldown assays. Amino acid numbering from methionine 1 is shown above CRMP-2L and -2S. Shaded or filled boxes at the N termini of CRMP-2 proteins indicate regions unique to each isoform, whereas white boxes show common core polypeptides. +, ROCK II-binding polypeptide. (B) Coomassie staining of recombinant GST proteins eluted from agarose beads used for pulldown assays. G, GST protein alone. Arrowheads indicate the predicted masses of CRMP polypeptides. (C to F) Pulldown assay. Recombinant full-length ROCK II (C) or the catalytic domain of ROCK II (D and E) expressed in S2 cells was used for pulldown assays with GST fusions of CRMP-2 polypeptides (C and E) or full-length CRMPs (D), as described in the legend for Fig. 2D. (F) The catalytic domain of ROCK II was pulled down by wild-type and mutant [2S(T555D)] CRMP-2S proteins to the same extent. The Coomassie-stained blot shows recombinant GST proteins eluted from beads used in the pulldown assay. (G) Coimmunoprecipitation of CRMP-2 polypeptides with ROCK II. ROCK II IPs from MCF7 cells transfected with cDNAs encoding the GFP fusion proteins indicated were analyzed by Western blotting with antibodies against ROCK II (top) and GFP (bottom). Arrowheads indicate enhanced GFP (EGFP) fusion proteins. Blots are representative of 3 separate experiments.
CRMP-2L and its ROCK II-binding domains inhibit ROCK-dependent carcinoma cell migration and FN matrix assembly.
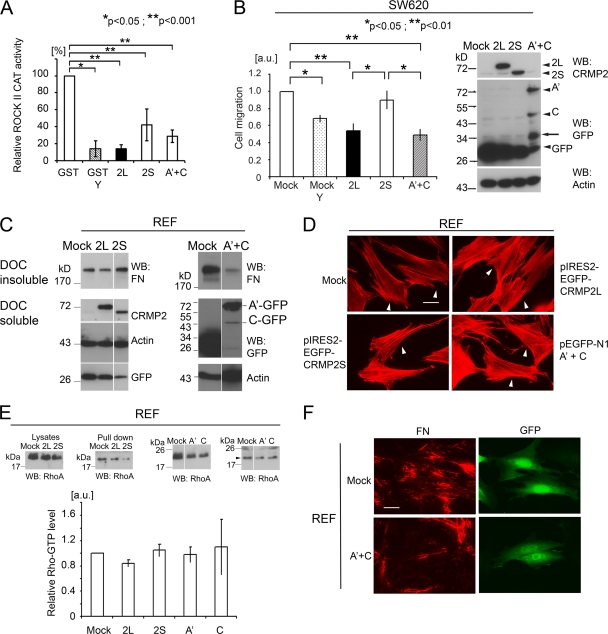
The data suggest that CRMP-2L may endogenously inhibit ROCK II. As further confirmation, the inhibitory activities of CRMP-2 proteins, and their ROCK II-binding domains, were tested in kinase assays with the ROCK II catalytic domain and GST-MLC as a substrate. Both CRMP-2L and -2S inhibited the ROCK II catalytic activity, although 2L was more efficient (Fig. 8A). The mixture of A′ and C domains also inhibited the activity.
Fig 8.
CRMP-2L and its ROCK II-binding domains inhibit ROCK-dependent carcinoma cell migration and FN matrix assembly. (A) CMRP-2L inhibits ROCK II catalytic activity. The kinase activity in the presence of GST was set at 100% (n = 3 to 5). Comparisons among different proteins were analyzed by Kruskal-Wallis analysis with a post hoc Sheffé test, and effects of Y-27632 treatment were analyzed by a Mann-Whitney test. (B) Haptotactic cell migration of SW620 cells transfected with plasmids encoding CRMP-2 cDNAs, as indicated. In some cases, cells were treated with 10 μM Y-27632 (Y). Data show means of data from at least 3 separate experiments, with the value for mock transfectants set at 1.0. Comparisons among different plasmid transfectants were analyzed by Kruskal-Wallis analysis with a post hoc Sheffé test, and the effect of inhibitor treatment was analyzed by Welch's t test. Western blots show the expressions of CRMP-2 proteins in SW620 transfectants. Arrowheads indicate the intact sizes of proteins. Arrows indicate degradation products of GFP-fused CRMP-2 proteins. Blots are representative of 3 separate experiments. (C) Reduced amounts of FN in DOC-insoluble fractions from cells expressing CRMP-2L or its ROCK II-binding domains. REF cells were transfected with pIRES2-EGFP vector cDNAs (left) or pEGFP-N1 vector cDNAs (right), as indicated. FN in the DOC-insoluble fraction (top) and CRMP-2, actin, and GFP in the DOC-soluble fraction (bottom) were detected by Western blotting. Blots are representative of 4 separate experiments. (D) CRMP-2L expression in REF cells does not affect stress fiber organization. Arrowheads indicate transfectants. No apparent alteration of the actin cytoskeleton by CRMP-2 expression was observed. Scale bar, 25 μm. (E) Pulldown assay of GTP-RhoA from REF transfectants. Images are representative of 3 separate experiments. The arrowhead indicates the RhoA protein. The graph shows mean GTP-RhoA levels, which were normalized to data from mock transfectants (n = 3). No statistically significant difference among transfectants was observed (Kruskal-Wallis test). (F) Reduced FN matrix assembly by expression of ROCK II-binding domains derived from CRMP-2L. REF cells transfected with the plasmids indicated were stained for FN. Transfected cells were detected as being GFP positive. Scale bar, 25 μm. Images are representative of 3 separate experiments. Error bars indicate standard errors of the means.
SW620 colon carcinoma cells were established from lymph nodes of a patient whose primary tumor was the source of SW480 cells (31). In contrast to SW480 cells, the CRMP-2L protein was undetectable in SW620 cells (Fig. 3B). Therefore, its effect on SW620 cell migration was tested by ectopic CRMP-2L expression, which led to significantly reduced haptotactic cell migration toward collagen I compared to that of a mock transfectant. Ectopic CRMP-2S expression had no effect (Fig. 8B). Importantly, Y-27632 treatment of mock transfectants reduced cell migration to a level similar to that of CRMP-2L transfectants. This verified the essential role for ROCK activity in SW620 cell migration, in contrast to SW480 cells. Together, the data suggest an inverse correlation between CRMP-2L protein levels and ROCK-mediated colon cancer cell migration. The abilities of ROCK II-binding polypeptides (polypeptides A′ and C) from CRMP-2 to affect cell migration were also tested in SW620 cells. Consistent with the results described above, the expression of these polypeptides significantly reduced SW620 cell migration to levels comparable to those of CRMP-2L transfectants (Fig. 8B), confirming that they can be functional inhibitors of ROCK II in tumor cells. Taken together with the data shown in Fig. 5, the CRMP-2L protein strongly influenced carcinoma cell migration, with its ROCK II-binding domains being capable of inhibiting ROCK II cellular functions.
The FN matrix is essential for development, and its uncontrolled assembly is linked to various pathological conditions, including fibrosis and tumor microenvironments (6, 52, 60). FN assembly is a myosin II-dependent cellular process and is reduced by ROCK II protein suppression by siRNA treatment or the inhibition of ROCK activity in fibroblasts (66). Therefore, we examined whether the expression of CRMP-2L affects FN matrix assembly. When FN assembles into a fibrillar matrix, it becomes deoxycholate (DOC) insoluble (35). The overexpression of CRMP-2L in REF cells reduced the amount of FN in DOC-insoluble fractions compared to that in the mock transfectant (45% reduction; P < 0.05) (Fig. 8C). Importantly, however, the overexpression of CRMP-2S did not. CRMP-2L expression did not affect stress fiber formation (97% [n = 116] of mock-transfected, 90% [n = 119] of CRMP-2L-transfected, and 96% [n = 114] of CRMP-2S-transfected cells retained stress fibers) (Fig. 8D). This was in good agreement with previously reported data showing that ROCK I, rather than ROCK II, is required for stress fiber formation in these cells (65). Moreover, CRMP-2L inhibited ROCK II-mediated matrix assembly independent of GTP-RhoA levels (Fig. 8E).
An effective inhibition of FN matrix assembly was achieved by the expression of ROCK II-binding polypeptides (polypeptides A′ and C) from CRMP-2. The coexpression of both polypeptides potently reduced the amount of DOC-insoluble FN compared to that in mock-transfected cells (70% reduction; P < 0.05) (Fig. 8C), in good agreement with data from immunofluorescence microscopy (Fig. 8F). Most fibroblasts expressing ROCK II-binding polypeptides of CRMP-2 retained stress fibers (83% [n = 130] for polypeptides A′ and C, 79% [n = 145] for polypeptide A′, and 93% [n = 124] for polypeptide C were positive for stress fibers) (Fig. 8D), and the expression of these polypeptides also did not affect GTP-RhoA levels (Fig. 8E). In summary, the expression of ROCK II-binding domains of CRMP-2L or the full-length molecule inhibited a ROCK II-dependent process in fibroblasts.
DISCUSSION
The current data revealed that the neuron polarity regulator CRMP-2 is a widespread protein and that its splice variant CRMP-2L, which is expressed mainly in epithelial cells among nonneuronal cells, is both a substrate and an inhibitor of ROCK II with no discernible activity against ROCK I (Fig. 9). The two homologous ROCKs are major regulators of myosin II activity and the actin cytoskeleton. In turn, cell adhesion, migration, and differentiation are strongly influenced by ROCKs, particularly the invasive migration of tumor cells (37). As with most kinases, protein expression levels reveal little concerning ROCK activity, since there are tight posttranslational regulatory mechanisms. For the ROCKs, activation can be elicited through interactions with GTP-Rho and acidic lipids, phosphorylation, and cleavage (3, 30). However, major regulation through the inhibition of active kinases is achieved by specific, endogenous binding partners that target a kinase domain. The knockdown of CRMP-2 increased the mean kinase activity of ROCK II in SW480 cells and, consistent with this, increased the mean phosphorylation state of MLC in MDA-MB-231 cells. However, these assays probably underestimated the CRMP-2 inhibitory activity: since total cytosolic pools were used, inhibition might be greater where ROCK II and CRMP-2 codistribute. To our knowledge, this is a novel mechanism by which an endogenous substrate traps the kinase by binding to the catalytic domain rather than the regulatory domain. The latter scenario is exemplified by the kinase PAK1, whose substrate, merlin, competes with GTP-Rac for the Rac-binding domain on PAK1 (27).
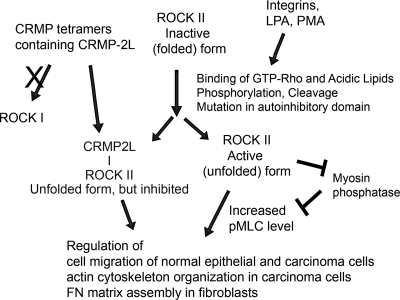
Fig 9.
Schematic summary of CRMP-2 regulation of ROCK-myosin II-mediated cellular behavior. Under quiescent conditions, ROCKs are in inactive (folded) forms. Stimuli increase cellular levels of Rho-GTP, which binds to ROCKs to activate them (unfolded). The active kinases or freed kinase domains phosphorylate downstream substrates, including MLC, resulting in actin-myosin II-mediated contraction. However, in CRMP-2L-expressing cells, the kinase domain of unfolded ROCK II is silenced by CRMP-2 tetramers containing 2L. CRMP-2–ROCK interactions are specific to ROCK II and not ROCK I. CRMP-2-inhibited ROCK II kinase impacts the migration of normal and transformed epithelial cells and FN matrix assembly.
Consistent with CRMP-2L being a specific inhibitor of ROCK II, a major binding site was identified in its N-terminal region, which is unique to the long isoform. Another site is more C terminal. CRMPs have been reported to form tetramers, both homotypic and heterotypic. The crystal structure of CRMP-1S and CRMP-2S revealed lung-lobe-shaped monomers, although these structures lacked their C-terminal regions, which contain a ROCK II-binding site (13, 53). Nothing is known regarding the regulation of CRMP alternate splicing. Our data show that the balance of long and short isoforms of CRMP-2 proteins in normal and tumor cells can vary, suggesting a regulatory mechanism worthy of future investigation. Furthermore, most studies have concerned the short isoform, while here, the long form, for which less structural data are available, has properties not shared with the short form. This provides the first clear evidence, at the molecular level, that CRMP-2 proteins have splice variants with distinct functions in vivo. It has been reported, however, that different microtubule organizations were achieved by the overexpression of CRMP-2 isoforms in chicken embryonic fibroblasts, although the molecular basis for this remains unknown (67).
A single report has suggested a significance of CRMP-2 in cancer. It was identified as a potential marker of human colorectal carcinoma from cancer cell secretomes (63). CRMP-2 positivity was significantly increased in early-stage tumors and lymph node metastasis. Splice form expression was not monitored, and it is apparent that the relative amounts of CRMP-2S and -2L can be divergent, even across two related colon carcinoma cell lines (Fig. 3B). However, as we demonstrate, CRMP-2L expression in a carcinoma cell line from lymph node metastasis can repress haptotactic cell migration (Fig. 8B), suggesting pathways by which CRMP-2 can regulate a key aspect of tumor progression.
Several studies have suggested a significance for ROCK activity in tumor cell migration, invasion, metastasis, and progression (11, 18, 37). This may relate partly to the cellular contractility mediated by ROCKs (46). Frequent overexpressions of ROCK isoforms in various tumors, including breast tumor, have been reported (18). Although ROCK II protein levels were consistent in tumor and normal breast tissues (28), we detected high levels of CRMP-2 associated with ROCK II in MDA-MB-231 cells, suggesting that ROCK II activity was suppressed. Supporting this hypothesis, the knockdown of CRMP-2 in these cells, while not affecting ROCK II localization, increased the ROCK-dependent phosphorylation of MLC, accompanied by the development of prominent cortical actin cytoskeleton and adhesion structures.
Furthermore, these findings provide a potential therapeutic strategy to target ROCK II in cancer and fibrosis. Specific somatic mutations found in some human cancers encode an unfolded, constitutively active ROCK II, due to the disruption of intramolecular interactions between catalytic and C-terminal domains (32). Essential roles of FN in lung fibrosis and the ability of a Rho kinase inhibitor to suppress bleomycin-induced lung fibrosis have been reported (51, 60), which correlate with a known role of ROCK II in matrix assembly (61). Thus, our data provide a paradigm to explore ROCK II function specifically.
A single previous report suggested that the small GTPase Ras family members Gem and Rad are ROCK I and II inhibitors, respectively (59). More recently, active Raf1 was proposed to inhibit ROCK II in the context of Ras/Rho cross talk in tumorigenesis and cell motility (39), but they have not been reported as substrate traps in the manner of CRMP-2.
In conclusion, we describe a novel substrate trap mechanism where the endogenous ROCK substrate CRMP-2L acts as a selective inhibitor of ROCK II but without an influence on ROCK I. Moreover, this regulation does not affect GTP-Rho levels in cells. Known CRMP-2-interacting protein partners bind to regions common between the long and short forms (5, 47), while this regulation of ROCK II is isoform specific. CRMP-2L–ROCK II interactions influence processes such as cell motility and extracellular matrix assembly, both required for development and implicated in disease, including tumor progression and fibrosis. It will be important to investigate how the alternate splicing of CRMP-2 mRNA is governed, notably in transformed cells, and whether this is an area for possible future interventions in tumor progression.
ACKNOWLEDGMENTS
We thank S. Narumiya and K. Kaibuchi for cDNA constructs and D. Stautz for some tumor cell lysates.
We gratefully acknowledge support from The Novo Nordisk Foundation and The Danish Cancer Research Foundation (A.Y.), The Danish Medical Research Council (A.Y. and J.R.C.), The Danish National Research Foundation and Lundbeck Fonden (J.R.C.), the Faculty of Health Sciences at the University of Copenhagen (J.R.C. and M.M.-F.), and The Danish Cancer Society and Arvid Nilsson's Fond (U.M.W.). R.W. thanks Arthritis Research UKand the Medical Research Council for their support.
Footnotes
Published ahead of print 19 March 2012.
REFERENCES
- 1. Amano M, et al. 1997. Formation of actin stress fibers and focal adhesions enhanced by Rho-kinase. Science 275:1308–1311 [DOI] [PubMed] [Google Scholar]
- 2. Amano M, et al. 1999. The COOH terminus of Rho-kinase negatively regulates rho-kinase activity. J. Biol. Chem. 274:32418–32424 [DOI] [PubMed] [Google Scholar]
- 3. Amano M, Nakayama M, Kaibuchi K. 2010. Rho-kinase/ROCK: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton (Hoboken) 67:545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arimura N, et al. 2000. Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J. Biol. Chem. 275:23973–23980 [DOI] [PubMed] [Google Scholar]
- 5. Arimura N, Menager C, Fukata Y, Kaibuchi K. 2004. Role of CRMP-2 in neuronal polarity. J. Neurobiol. 58:34–47 [DOI] [PubMed] [Google Scholar]
- 6. Barkan D, Green JE, Chambers AF. 2010. Extracellular matrix: a gatekeeper in the transition from dormancy to metastatic growth. Eur. J. Cancer 46:1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown M, et al. 2004. Alpha2-chimaerin, cyclin-dependent kinase 5/p35, and its target collapsin response mediator protein-2 are essential components in semaphorin 3A-induced growth-cone collapse. J. Neurosci. 24:8994–9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burridge K, Wennerberg K. 2004. Rho and Rac take center stage. Cell 116:167–179 [DOI] [PubMed] [Google Scholar]
- 9. Campellone KG, Welch MD. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11:237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen XQ, et al. 2002. Characterization of RhoA-binding kinase ROKalpha implication of the pleckstrin homology domain in ROKalpha function using region-specific antibodies. J. Biol. Chem. 277:12680–12688 [DOI] [PubMed] [Google Scholar]
- 11. Croft DR, et al. 2004. Conditional ROCK activation in vivo induces tumor cell dissemination and angiogenesis. Cancer Res. 64:8994–9001 [DOI] [PubMed] [Google Scholar]
- 12. Defilles C, et al. 2009. Alphavbeta5/beta6 integrin suppression leads to a stimulation of alpha2beta1 dependent cell migration resistant to PI3K/Akt inhibition. Exp. Cell Res. 315:1840–1849 [DOI] [PubMed] [Google Scholar]
- 13. Deo RC, et al. 2004. Structural bases for CRMP function in plexin-dependent semaphorin3A signaling. EMBO J. 23:9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dovas A, et al. 2010. Serine 34 phosphorylation of rho guanine dissociation inhibitor (RhoGDIalpha) links signaling from conventional protein kinase C to RhoGTPase in cell adhesion. J. Biol. Chem. 285:23296–23308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao X, et al. 2010. Expression profiling identifies new function of collapsin response mediator protein 4 as a metastasis-suppressor in prostate cancer. Oncogene 29:4555–4566 [DOI] [PubMed] [Google Scholar]
- 16. Gong W, et al. 2009. P21-activated kinase 5 is overexpressed during colorectal cancer progression and regulates colorectal carcinoma cell adhesion and migration. Int. J. Cancer 125:548–555 [DOI] [PubMed] [Google Scholar]
- 17. Gu Y, Hamajima N, Ihara Y. 2000. Neurofibrillary tangle-associated collapsin response mediator protein-2 (CRMP-2) is highly phosphorylated on Thr-509, Ser-518, and Ser-522. Biochemistry 39:4267–4275 [DOI] [PubMed] [Google Scholar]
- 18. Hahmann C, Schroeter T. 2010. Rho-kinase inhibitors as therapeutics: from pan inhibition to isoform selectivity. Cell. Mol. Life Sci. 67:171–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hamajima N, et al. 1996. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene 180:157–163 [DOI] [PubMed] [Google Scholar]
- 20. Hedgecock EM, Culotti JG, Thomson JN, Perkins LA. 1985. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111:158–170 [DOI] [PubMed] [Google Scholar]
- 21. Hensley K, Venkova K, Christov A, Gunning W, Park J. 2011. Collapsin response mediator protein-2: an emerging pathologic feature and therapeutic target for neurodisease indications. Mol. Neurobiol. 43:180–191 [DOI] [PubMed] [Google Scholar]
- 22. Ishizaki T, et al. 2000. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol. Pharmacol. 57:976–983 [PubMed] [Google Scholar]
- 23. Jacobs M, et al. 2006. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J. Biol. Chem. 281:260–268 [DOI] [PubMed] [Google Scholar]
- 24. Kamijo H, et al. 2011. Impaired vascular remodeling in the yolk sac of embryos deficient in ROCK-I and ROCK-II. Genes Cells 16:1012–1021 [DOI] [PubMed] [Google Scholar]
- 25. Kawano Y, et al. 1999. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147:1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 27. Kumar R, Gururaj AE, Barnes CJ. 2006. p21-activated kinases in cancer. Nat. Rev. Cancer 6:459–471 [DOI] [PubMed] [Google Scholar]
- 28. Lane J, Martin TA, Watkins G, Mansel RE, Jiang WG. 2008. The expression and prognostic value of ROCK I and ROCK II and their role in human breast cancer. Int. J. Oncol. 33:585–593 [PubMed] [Google Scholar]
- 29. Lee DH, et al. 2009. Targeted disruption of ROCK1 causes insulin resistance in vivo. J. Biol. Chem. 284:11776–11780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee HH, Chang ZF. 2008. Regulation of RhoA-dependent ROCKII activation by Shp2. J. Cell Biol. 181:999–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leibovitz A, et al. 1976. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 36:4562–4569 [PubMed] [Google Scholar]
- 32. Lochhead PA, Wickman G, Mezna M, Olson MF. 2010. Activating ROCK1 somatic mutations in human cancer. Oncogene 29:2591–2598 [DOI] [PubMed] [Google Scholar]
- 33. Lydolph MC, et al. 2009. Alpha9beta1 integrin in melanoma cells can signal different adhesion states for migration and anchorage. Exp. Cell Res. 315:3312–3324 [DOI] [PubMed] [Google Scholar]
- 34. McCarthy KJ, Bynum K, St John PL, Abrahamson DR, Couchman JR. 1993. Basement membrane proteoglycans in glomerular morphogenesis: chondroitin sulfate proteoglycan is temporally and spatially restricted during development. J. Histochem. Cytochem. 41:401–414 [DOI] [PubMed] [Google Scholar]
- 35. McKeown-Longo PJ, Mosher DF. 1983. Binding of plasma fibronectin to cell layers of human skin fibroblasts. J. Cell Biol. 97:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mills GB, Moolenaar WH. 2003. The emerging role of lysophosphatidic acid in cancer. Nat. Rev. Cancer 3:582–591 [DOI] [PubMed] [Google Scholar]
- 37. Narumiya S, Tanji M, Ishizaki T. 2009. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 28:65–76 [DOI] [PubMed] [Google Scholar]
- 38. Nesvizhskii AI, Keller A, Kolker E, Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 39. Niault T, et al. 2009. From autoinhibition to inhibition in trans: the Raf-1 regulatory domain inhibits Rok-alpha kinase activity. J. Cell Biol. 187:335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parsons JT, Horwitz AR, Schwartz MA. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pearce LR, Komander D, Alessi DR. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22 [DOI] [PubMed] [Google Scholar]
- 42. Pellegrin S, Mellor H. 2007. Actin stress fibres. J. Cell Sci. 120:3491–3499 [DOI] [PubMed] [Google Scholar]
- 43. Ridley AJ, Hall A. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389–399 [DOI] [PubMed] [Google Scholar]
- 44. Riento K, Ridley AJ. 2003. Rocks: multifunctional kinases in cell behaviour. Nat. Rev. Mol. Cell Biol. 4:446–456 [DOI] [PubMed] [Google Scholar]
- 45. Rosslenbroich V, et al. 2005. Collapsin response mediator protein-4 regulates F-actin bundling. Exp. Cell Res. 310:434–444 [DOI] [PubMed] [Google Scholar]
- 46. Sanz-Moreno V, et al. 2008. Rac activation and inactivation control plasticity of tumor cell movement. Cell 135:510–523 [DOI] [PubMed] [Google Scholar]
- 47. Schmidt EF, Strittmatter SM. 2007. The CRMP family of proteins and their role in Sema3A signaling. Adv. Exp. Med. Biol. 600:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sechler JL, Takada Y, Schwarzbauer JE. 1996. Altered rate of fibronectin matrix assembly by deletion of the first type III repeats. J. Cell Biol. 134:573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi J, Zhang L, Wei L. 2011. Rho-kinase in development and heart failure: insights from genetic models. Pediatr. Cardiol. 32:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shih JY, et al. 2001. Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. J. Natl. Cancer Inst. 93:1392–1400 [DOI] [PubMed] [Google Scholar]
- 51. Shimizu Y, et al. 2001. Contribution of small GTPase Rho and its target protein rock in a murine model of lung fibrosis. Am. J. Respir. Crit. Care Med. 163:210–217 [DOI] [PubMed] [Google Scholar]
- 52. Singh P, Carraher C, Schwarzbauer JE. 2010. Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 26:397–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stenmark P, et al. 2007. The structure of human collapsin response mediator protein 2, a regulator of axonal growth. J. Neurochem. 101:906–917 [DOI] [PubMed] [Google Scholar]
- 54. Tahimic CG, et al. 2006. Evidence for a role of collapsin response mediator protein-2 in signaling pathways that regulate the proliferation of non-neuronal cells. Biochem. Biophys. Res. Commun. 340:1244–1250 [DOI] [PubMed] [Google Scholar]
- 55. Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vincent P, et al. 2005. A role for the neuronal protein collapsin response mediator protein 2 in T lymphocyte polarization and migration. J. Immunol. 175:7650–7660 [DOI] [PubMed] [Google Scholar]
- 57. Wait R, et al. 2002. Strategies for proteomics with incompletely characterized genomes: the proteome of Bos taurus serum. Electrophoresis 23:3418–3427 [DOI] [PubMed] [Google Scholar]
- 58. Wang LH, Strittmatter SM. 1997. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J. Neurochem. 69:2261–2269 [DOI] [PubMed] [Google Scholar]
- 59. Ward Y, et al. 2002. The GTP binding proteins Gem and Rad are negative regulators of the Rho-Rho kinase pathway. J. Cell Biol. 157:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. White ES, Baralle FE, Muro AF. 2008. New insights into form and function of fibronectin splice variants. J. Pathol. 216:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wittig I, Braun HP, Schagger H. 2006. Blue native PAGE. Nat. Protoc. 1:418–428 [DOI] [PubMed] [Google Scholar]
- 62. Wolfenson H, Henis YI, Geiger B, Bershadsky AD. 2009. The heel and toe of the cell's foot: a multifaceted approach for understanding the structure and dynamics of focal adhesions. Cell Motil. Cytoskeleton 66:1017–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu CC, et al. 2008. Identification of collapsin response mediator protein-2 as a potential marker of colorectal carcinoma by comparative analysis of cancer cell secretomes. Proteomics 8:316–332 [DOI] [PubMed] [Google Scholar]
- 64. Yamaguchi H, Kasa M, Amano M, Kaibuchi K, Hakoshima T. 2006. Molecular mechanism for the regulation of rho-kinase by dimerization and its inhibition by fasudil. Structure 14:589–600 [DOI] [PubMed] [Google Scholar]
- 65. Yoneda A, Multhaupt HA, Couchman JR. 2005. The Rho kinases I and II regulate different aspects of myosin II activity. J. Cell Biol. 170:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yoneda A, Ushakov D, Multhaupt HA, Couchman JR. 2007. Fibronectin matrix assembly requires distinct contributions from Rho kinases I and -II. Mol. Biol. Cell 18:66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yuasa-Kawada J, et al. 2003. Axonal morphogenesis controlled by antagonistic roles of two CRMP subtypes in microtubule organization. Eur. J. Neurosci. 17:2329–2343 [DOI] [PubMed] [Google Scholar]