Abstract
Chronic myeloid leukemia (CML) is derived from a stem cell, and it is widely accepted that the existence of leukemia stem cells (LSCs) is one of the major reasons for the relapse of CML treated with kinase inhibitors. Key to eradicating LSCs is to identify genes that play a critical role in survival regulation of these stem cells. Using BCR-ABL-induced CML mouse model, here we show that expression of the stearoyl-CoA desaturase 1 (Scd1) gene is downregulated in LSCs and that Scd1 plays a tumor-suppressive role in LSCs with no effect on the function of normal hematopoietic stem cells. Deletion of Scd1 causes acceleration of CML development and conversely overexpression of Scd1 delays CML development. In addition, using genetic approaches, we show that Pten, p53, and Bcl2 are regulated by Scd1 in LSCs. Furthermore, we find that induction of Scd1 expression by a PPARγ agonist suppresses LSCs and delays CML development. Our results demonstrate a critical role for Scd1 in functional regulation of LSCs, providing a new anti-LSC strategy through enhancing Scd1 activity.
INTRODUCTION
Leukemia stem cells (LSCs) are defined as transformed hematopoietic stem cell (HSC) or progenitors that have acquired unlimited self-renewal ability. It is generally accepted that eradication of LSCs is required for effectively treating and eventually curing the diseases. Chronic myeloid leukemia (CML) is believed to originate from a HSC harboring Philadelphia chromosome formed through a reciprocal translocation between chromosomes 9 and 22. Human CML is induced by the BCR-ABL oncogene that produces the chimeric BCR-ABL protein functioning as a constitutively activated tyrosine kinase in leukemogenesis (19, 27, 31). Although BCR-ABL kinase inhibitors are highly effective in treating chronic-phase CML patients (5), they do not completely eradicate LSCs (5, 7, 10, 12). Therefore, there is an urgent need for developing anti-LSC strategies, and this approach relies on the identification of critical target genes in LSCs.
Accumulating evidence shows a close relationship between cancer and metabolism (15). Cancer cells display changes in fatty acid metabolism (2), which is associated with modulations of expression and activity of lipogenic enzymes. One such enzyme is stearoyl-CoA desaturase (Scd). Scd is an endoplasmic reticulum enzyme, belonging to a family of Δ9-fatty acid desaturase isoforms. Scd1 catalyzes the biosynthesis of monounsaturated fatty acids from saturated fatty acids, which are the most abundant fatty acids present in mammalian organisms (29). Scd1 expression is detected in almost all tissues, with a predominant expression in liver, and is involved in regulating metabolic pathways related to preadipocyte differentiation, insulin sensitivity, metabolism, and tumorigenesis (6, 8, 13, 28, 29). A recent study indicates that inhibition of Scd1 impairs lung cancer cell proliferation, survival, and invasiveness (8), suggesting that Scd1 plays a supporting role in lung cancer cells. Another study shows that the level of Scd1 expression correlates with predisposition to liver carcinogenesis (6). However, the role of Scd1 in leukemogenesis and hematopoiesis remains unknown, although it has been suggested that fatty acid metabolism plays a role in leukemogenesis and hematopoiesis (14, 20). In the present study, using the BCR-ABL induced CML mouse model, we found that the expression of the Scd1 gene was downregulated in LSCs and the deletion of Scd1 accelerated CML development through affecting the function of LSCs but not normal HSCs, suggesting that Scd1 plays a tumor-suppressive role in BCR-ABL leukemogenesis. Our study provides a new strategy for targeting LSCs by enhancing the Scd1 function.
MATERIALS AND METHODS
Mice.
Scd1−/−, C57BL/6J-CD45.1, and C57BL/6J-CD45.2 mice were purchased from the Jackson Laboratory. All mice had a C57BL/6J background, were bred and maintained in a temperature- and humidity-controlled environment, and given unrestricted access to 6% chow diet and acidified water.
Generation of retrovirus stocks.
The retroviral constructs MSCV-IRES-GFP, MSCV-BCR-ABL-IRES-GFP, MSCV-BCR-ABL-IRES-Scd1, and MSCV-Scd1-IRES-humanCD4 MSCV-BCR-ABL-IRES-Pten-IRES-GFP were used to generate a high-titer, helper-free, replication-defective ecotropic virus stock by transient transfection of 293T cells as previously described (16).
Lentiviral transduction and shRNA-mediated knockdown of p53 and Bcl2.
Lentiviral short hairpin RNA (shRNA) vector pLKO.1 was purchased from Open Biosystems. The sequences of p53 and Bcl2 shRNA were as follows: shRNA-p53 sense, 5′-CCAGTCTACTTCCCGCCATAA-3′; shRNA-p53 antisense, 5′-TTATGGCGGGAAGTAGACTGG-3′; shRNA-Bcl2 sense, 5′-CGTGATGAAGTACATACATTA-3′; and shRNA-Bcl2 antisense, 5′-TAATGTATGTACTTCATCACG-3′. For the transduction of shRNAs, bone marrow cells from CML mice were transfected with lentiviral shRNA and, 24 h later, were selected under puromycin (2.5 μg/ml) for 48 h to enrich shRNA-expressing cells before the next steps.
Bone marrow transduction/transplantation.
Eight- to twelve-week-old C57BL/6 mice were used for bone marrow transduction and transplantation. Retroviral transduction and transplantation of mouse bone marrow cells for inducing CML and B-cell acute lymphoblastic leukemia (B-ALL) by BCR-ABL had been described previously (8, 9, 14, 16, 20, 22). Briefly, in order to induce CML, donor mice were primed by intravenous injection with 5-fluorouracil (5-FU; 200 mg/kg) 4 days before collecting bone marrow cells. The cells were cultured at 2 × 107 cells per 10-cm plate in Dulbecco modified Eagle medium containing 6 ng of recombinant murine interleukin-3 (IL-3; PeproTech)/ml, 10 ng of recombinant murine IL-6 (PeproTech)/ml, and 50 to 100 ng of recombinant murine stem cell factor (SCF; PeproTech)/ml. After prestimulation for 24 h at 37°C, the cells were transduced with retroviral stocks in the same medium containing 50% retroviral supernatant, 10 mM HEPES (pH 7.4), and 2 μg of Polybrene/ml. To increase the transduction efficiency, the virus and cells were cosedimented at 1,000 × g for 90 min. A second round of transduction was performed at 48 h. For inducing B-ALL, bone marrow cells from non-5-FU-treated donor mice were transduced only once with retroviral stocks. Recipient mice were prepared by two doses of 550 cGy of gamma irradiation separated by 3 h, and bone marrow cells were transplanted into these recipient mice by intravenous injection. Diseased mice were evaluated by flow cytometric, histopathological, and molecular analyses. For secondary transplantation, equal numbers of bone marrow cells from primary CML mice were transplanted into lethally irradiated recipient mice. For drug treatment, rosiglitazone (GlaxoSmithKline) and imatinib (Novartis) were given orally in a volume of <0.3 ml by gavage (100 mg/kg, twice a day for rosiglitazone; 100 mg/kg, twice a day for imatinib) beginning 8 days after bone marrow transplantation (BMT) and continuing until the morbidity or death of the mice with leukemia.
Flow cytometry analysis.
Hematopoietic cells were collected from the bone marrow and peripheral blood (PB) of CML mice for fluorescence-activated cell sorting (FACS) analysis. For stem cell analysis, five mice from each experimental group were sacrificed at day 14 after BMT to collect bone marrow cells. Red blood cells (RBCs) were depleted using RBC lysis buffer (containing NH4Cl, KHCO3, and EDTA). To stain the cells with antibodies, bone marrow cells were suspended in staining medium (Hanks balanced salt solution with 2% heat-inactivated calf serum), followed by incubation with biotin-labeled lineage antibody cocktail containing a mixture of antibodies against CD3, CD4, CD8, B220, Gr-1, Mac-1, and Ter119 at 4°C for 15 min. After washing, the fluorochrome-labeled secondary antibody (allophycocyanin-cyanin 7 [APC-Cy7]-conjugated streptavidin) for recognizing biotin and phycoerythrin (PE)-conjugated c-Kit and APC-conjugated Sca-1 antibodies were added to the cells for 15 min at 4°C in the dark. Long-term and short-term LSCs were distinguished by the CD34 antibody. The CML stem cell population (GFP+ Lin− c-Kit+ Sca-1+) was analyzed by FACS. All of these antibodies were purchased from eBioscience.
Cell cycle analysis of LSCs was performed by staining cells with antibodies in combination with Hoechst 33342 for 90 min at 37°C, followed by flow cytometry analysis. To analyze apoptosis of bone marrow cells, the cells were stained with annexin V (eBioscience) for 15 min at room temperature. 7-Amino-actinomycin D was added before flow cytometry analysis.
Leukemia stem cell culture.
For mouse leukemia stem cell culture, bone marrow cells isolated from CML mice were cultured in vitro in the presence of stemspan SFEM, SCF, insulin-like growth factor 2, thrombopoietin, heparin, and anti-fibroblast growth factor as reported previously for the culture of HSCs. For in vitro treatment, rosiglitazone (Cayman Chemical) was dissolved in dimethyl sulfoxide (DMSO).
In vitro methylcellulose colony formation assay.
LSCs (GFP+ Lin− Sca-1+ c-Kit+) from the bone marrow of CML mice receiving BCR-ABL-transduced wild-type (WT) or Scd1−/− bone marrow cells were sorted by FACS and cultured in methylcellulose medium (Methocult GF M3434; Stem Cell Technologies) at 37°C in humidified air for 7 days. The colonies were counted under a microscope.
ChIP of RNA Pol II.
Chromatin immunoprecipitation (ChIP) assays were performed. Briefly, 3 × 107 leukemia cells from WT and Scd1−/− CML mice were incubated with 1% formaldehyde for 10 min at room temperature before cross-linking was quenched by the addition of 0.125 M glycine. The cells were collected by centrifugation and lysed in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 0.5% sodium dodecyl sulfate, proteinase inhibitors, and phosphatase inhibitors. The cell suspension was sonicated seven times for 10 s each time with 2-min intervals on ice using a Misonix Sonicator 3000 at output 8. Sonicated chromatin was then incubated at 4°C overnight with 5 μg of polymerase II (Pol II) antibody (Avalon). Immunoprecipitated DNA was amplified by real-time PCR using the following primers: Pten promoter-sense (5′-CGGGACTCTTTGTGCACTG-3′), Pten promoter-antisense (5′-GCGGCTCAACTCTCAAACTT-3′), p53 promoter-sense (5′-AACAGTGGCGGTCCACTTAC-3′), and p53 promoter-antisense (5′-GGGACTTGCAGAGTCAGGAT-3′), Bcl2 promoter-sense (5′-CCGGCGGAGAATGAAGTAA-3′), and Bcl2 promoter-antisense (5′-CGCACCCTTTCTCCTCCT-3′).
Real time reverse transcription-PCR (RT-PCR).
Total RNA was isolated from GFP+ Lin− Sca-1+ c-Kit+ bone marrow cells from mice using an RNeasy minikit (Qiagen, CA). cDNA was synthesized using the Ovation-Pico cDNA synthesis method. All real-time PCRs were done using the Applied Biosystems 7500. The 25-μl reaction system was composed of 12.5 μl of SYBR green, 2.5 μl of a 20 μM primer mixture, 10 ng of cDNA, and nuclease-free water. All experiments were performed in triplicate. β-Actin was used as the internal control. The primer sequences were as follows: Scd1 sense, 5′-TTCTTACACGACCACCACCA-3′; Scd1 antisense, 5′-GCAGGAGGGAACCAGTATGA-3′; Pten sense, 5′-ACACCGCCAAATTTAACTGC-3′; Pten antisense, 5′-TACACCAGTCCGTCCCTTTC-3′; p53 sense, 5′-AGAGACCGCCGTACAGAAGA-3′; p53 antisense, 5′-CTGGTAGCATGGGCATCCTTT-3′; Bcl2 sense, 5′-CTGGCTTCTTCTCCTTCCAG-3′; Bcl2 antisense, 5′-GACGGTAGCGACGAGAGGAAG-3′; β-actin sense, 5′-ATGGAATCCTGTGGCATCCA-3′; and β-actin antisense, 5′-CGCTCAGGAGGAGCAATGAT-3′.
Statistical analysis.
Results are given as means ± the standard error of the mean (SEM). Statistical analysis was performed by using the Student t test. For survival curves, P values were obtained by using a log-rank test.
RESULTS
Scd1 plays a tumor-suppressive role in CML development.
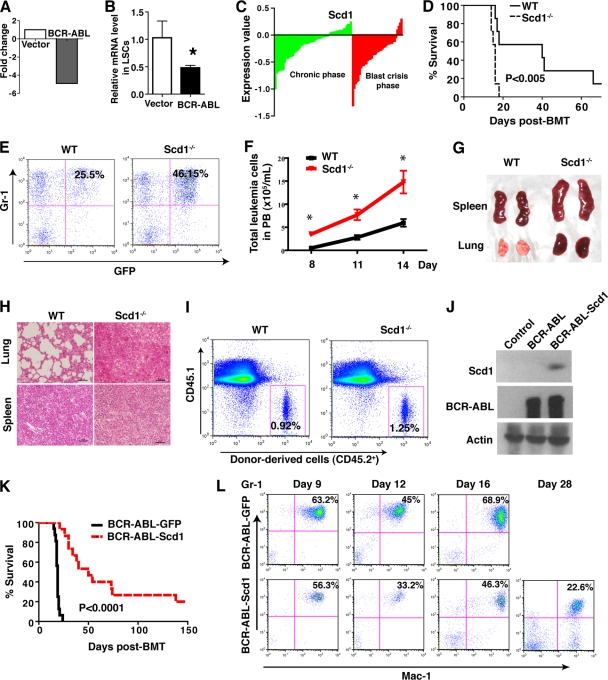
BCR-ABL-expressing HSCs (GFP+ Lin− Sca-1+ c-Kit+ cells) in mice function as LSCs (10). To examine whether BCR-ABL regulates Scd1 expression in LSCs, using our unique SDL global optimization method (17), we reanalyzed the results from our previously published DNA microarray study (GEO submission GSE10912), in which we compared the gene expression profiles between LSCs and normal HSCs. We found that expression of the Scd1 gene was dramatically lower in BCR-ABL-expressing LSCs compared to normal HSCs (Fig. 1A), and this downregulation of Scd1 in LSCs was confirmed by real-time RT-PCR (Fig. 1B). Furthermore, we analyzed a publicly available gene expression profiling database of human CML stem cells (25) and found that Scd1 expression was markedly downregulated in the majority of CML patients in chronic phase and blast crisis (Fig. 1C). These results suggest that Scd1 plays a tumor-suppressive role in CML development. To test this idea, we first investigated whether Scd1 deficiency causes acceleration of CML development using Scd1 homozygous knockout (Scd1−/−) mice. We transduced bone marrow cells from 5-FU-treated wild-type (WT) or Scd1−/− mice with the BCR-ABL-GFP retrovirus, followed by transplanting the transduced cells into lethally irradiated recipient mice to induce CML. We found that recipients of BCR-ABL-transduced Scd1−/− bone marrow cells developed and died of CML significantly faster than did recipients of BCR-ABL-transduced WT bone marrow cells (Fig. 1D). The accelerated CML development in the absence of Scd1 correlated with a higher percentage and the total numbers of GFP+ Gr-1+ myeloid leukemia cells in PB (Fig. 1E and F) and with more severe splenomegaly and lung hemorrhage (Fig. 1G). Histological examination showed more extensive infiltration of leukemia cells in the lung and spleen of CML mice receiving BCR-ABL-transduced Scd1−/− bone marrow cells (Fig. 1H). Because increased homing of donor bone marrow cells could lead to accelerated CML development, we compared the homing ability of Scd1−/− bone marrow cells to that of WT bone marrow cells 3 h after the transplantation of the cells into recipient mice. We found that the percentages of donor-derived WT and Scd1−/− bone marrow cells in recipient mice were similar (Fig. 1I), indicating that Scd1 deficiency does not significantly alter the homing ability of bone marrow cells. Reversely, we overexpressed Scd1 in donor bone marrow cells by transducing the cells with retrovirus expressing both BCR-ABL and Scd1 (Fig. 1J) to examine whether the enforced expression of Scd1 causes a delay of CML development. We did observe that Scd1 overexpression caused a significant delay of CML development in secondary recipients of bone marrow cells from the primary CML mice (Fig. 1K). This delayed CML development correlated with lower percentages of myeloid cells in the PB of the mice (Fig. 1L). Together, these results indicate that Scd1 plays an inhibitory role in CML development.
Fig 1.
Scd1 plays a tumor-suppressive role in CML development. (A) Microarray analysis showed that Scd1 expression was downregulated in BCR-ABL-expressing LSCs compared to normal stem cells that did not express BCR-ABL. (B) Real-time RT-PCR analysis showed that Scd1 expression was downregulated in LSCs. The expression data were normalized to β-actin expression and are shown as means ± the SEM (n = 3; *, P < 0.05). (C) Scd expression in human CML samples. Microarray analysis showed expression of Scd1 in CD34+ the bone marrow and peripheral blood (PB) cells from 42 chronic-phase (green) and 31 blast-crisis-phase (red) CML patients compared to normal human CD34+ cells. (D) Kaplan-Meier survival curve for recipients of BCR-ABL-transduced WT or Scd1−/− bone marrow cells (n = 7 to 8 mice per group; P < 0.005). (E) Percentages of GFP+ Gr-1+ cells in the PB of recipients of BCR-ABL-transduced bone marrow cells from WT or Scd1−/− donor mice at day 14 after BMT. (F) Total number of GFP+ Gr-1+ leukemia cells in PB of recipients of BCR-ABL-transduced WT or Scd1−/− bone marrow cells. (G) Gross appearance of lungs and spleens from mice receiving BCR-ABL-transduced WT or Scd1−/− bone marrow cells at day 14 after BMT. (H) Hematoxylin-eosin staining of tissue sections from the lungs and spleens of mice receiving BCR-ABL-transduced WT or Scd1−/− bone marrow cells at day 14 after BMT (scale bar, 50 μm). (I) The loss of Scd1 does not affect the homing ability of bone marrow cells. A total of 6 × 106 bone marrow cells from WT or Scd1−/− mice (CD45.2) were transplanted into lethally irradiated recipient mice (CD45.1). The donor-derived bone marrow cells (CD45.2) were detected by FACS in 3 h after BMT. (J) Western blot analysis showed the expression of Scd1 and BCR-ABL in 293T cells transfected with BCR-ABL-Scd1. (K) Kaplan-Meier survival curve for secondary recipients of equal number of bone marrow cells from primary CML mice receiving BCR-ABL-transduced or BCR-ABL-Scd1-transduced WT bone marrow cells (n = 14 to 16 mice per group; P < 0.0001). (L) The percentages of myeloid cells (Gr-1+ Mac-1+) in the PB of secondary recipients of bone marrow cells from primary CML mice receiving BCR-ABL-transduced or BCR-ABL-Scd1-transduced WT bone marrow cells at days 9, 12, 16, and 28 after BMT.
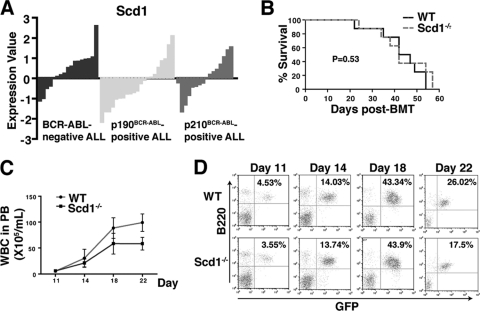
We also tested whether Scd1 is required for BCR-ABL induced B-ALL. We first assessed Scd expression in human B-ALL using a publicly available microarray data set (GSE5314) of 37 BCR-ABL-positive (21 of p190BCR-ABL and 16 of p210BCR-ABL) and 17 BCR-ABL-negative adult ALL (11). We did not observe significant difference in Scd expression between BCR-ABL-positive ALL and BCR-ABL-negative ALL and between p190BCR-ABL-positive and p210BCR-ABL-positive ALL (Fig. 2A). These data suggest that Scd expression may not be relevant to BCR-ABL in ALL. Consistent with this idea, recipients of WT or Scd1−/− bone marrow cells transduced with BCR-ABL retrovirus developed and died of ALL with similar disease latency and survival (Fig. 2B), correlating with similar numbers and percentages of GFP+ B220+ leukemia cells in the PB of the two groups of B-ALL mice (Fig. 2C and D). These results indicate that Scd1 is dispensable for BCR-ABL induced B-ALL.
Fig 2.
Scd1 does not suppress B-ALL induced by BCR-ABL. (A) Microarray analysis showing Scd expression in human B-ALL samples from 37 BCR-ABL-positive (21 of p190BCR-ABL and 16 of p210BCR-ABL) and 17 BCR-ABL-negative adult patients. (B) Kaplan-Meier survival curve for recipients receiving BCR-ABL-transduced WT or Scd1−/− bone marrow cells. Both groups of mice developed and died of ALL with similar disease latency (n = 8 mice per group). (C) Total number of GFP+ B220+ cells in the PB of recipients of BCR-ABL-transduced WT or Scd1−/− bone marrow cells. (D) FACS analysis revealed no differences in the percentages of GFP+ B220+ cells in the PB of recipients of BCR-ABL-transduced WT or Scd1−/− bone marrow cells.
Scd1 suppresses LSCs.
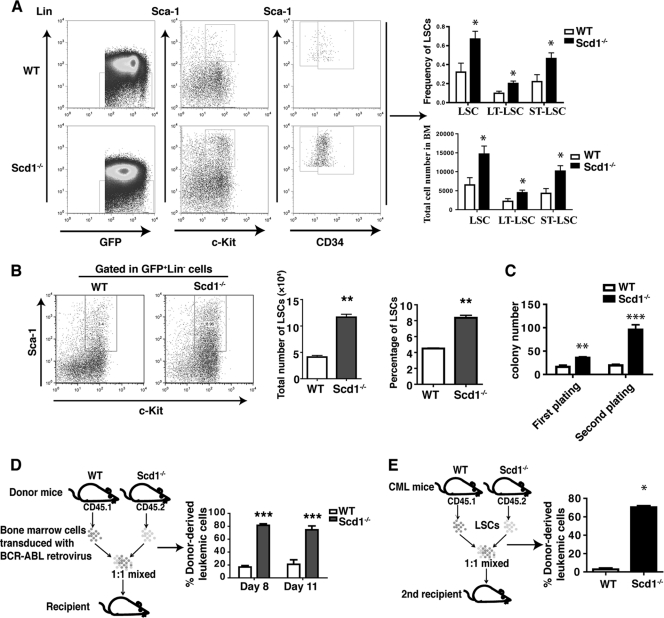
The tumor-suppressive effect of Scd1 on CML development could be caused by affecting LSCs. To test this possibility, we first examined whether deletion of Scd1 causes an increase of LSCs in bone marrow of CML mice. At 2 weeks after CML induction using BCR-ABL-transduced WT or Scd1−/− bone marrow cells, we analyzed bone marrow cells from CML mice by FACS (Fig. 3A). We found that the percentages and numbers of total LSCs and long-term (LT) (GFP+ Lin− c-Kit+ Sca-1+ CD34−) or short-term (ST) (GFP+ Lin− c-Kit+ Sca-1+ CD34+) LSCs were significantly higher in recipients of BCR-ABL-transduced Scd1−/− bone marrow cells than in recipients of BCR-ABL-transduced WT bone marrow cells (Fig. 3A). We also cultured bone marrow cells from CML mice in vitro for 7 days under the stem cell culture conditions (22) and found that the percentages and numbers of Scd1−/− LSCs were significantly higher than those of WT LSCs (Fig. 3B).
Fig 3.
Scd1 Suppresses LSCs. (A) Bone marrow cells from recipients of BCR-ABL-transduced WT or Scd1−/− bone marrow cells were analyzed by FACS at day 14 after BMT. The percentages and numbers of total LSCs, LT-LSCs, and ST-LSCs in the bone marrow of recipients of BCR-ABL-transduced Scd1−/− donor bone marrow cells were dramatically higher compared to those in the bone marrow of recipients of BCR-ABL-transduced WT donor bone marrow cells (n = 4 to 7 mice per group). The data are representative of one of four independent experiments, and mean values (± the SEM) are shown (*, P < 0.05). (B) Loss of Scd1 promotes the survival of LSCs in vitro. Bone marrow cells from recipients of BCR-ABL transduced WT or Scd1−/− bone marrow cells were cultured for 6 days under the in vitro stem cell culture conditions. The total numbers and percentages of LSCs were analyzed by FACS. (C) Loss of Scd1 increased the colony-forming ability of LSCs. The sorted GFP+ Lin− Sca-1+ c-Kit+ cells from CML mice were plated into methylcellulose medium, and the colonies were counted. The cells from colonies were serially replated. The data are representative of one of two independent experiments. Mean values (± the SEM) are shown (**, P < 0.01; ***, P < 0.001). (D) Equal numbers of BCR-ABL-transduced bone marrow cells from WT (CD45.1) or Scd1−/− (CD45.2) mice were mixed well and transplanted into lethally irradiated WT mice (CD45.2). At day 8 and 11 after BMT, FACS analysis gated in GFP+ cell population showed that the percentages of leukemia cells from Scd1−/− mice (CD45.2) were dramatically higher than those from WT mice (CD45.1). Mean values (± SEM) are shown (**, P < 0.01; **, P < 0.001). (E) A competitive repopulation assay showed that the loss of Scd1 increased the engraftment of LSCs. A total of 103 sorted LSCs from the bone marrow of primary CML mice induced by transplanting BCR-ABL-transduced Scd1−/− bone marrow cells (CD45.2) and BCR-ABL-transduced WT bone marrow cells (CD45.1) were mixed at 1:1 ratio, followed by transplantation into lethally irradiated recipients mice. The percentages of CD45.2 cells in GFP+ cell population from PB were compared to those of CD45.2 cells at 8 weeks after BMT (n = 3; ***, P < 0.0001).
To investigate whether Scd1 affects self-renewal of LSCs, we carried out an in vitro serial replating assay to examine the ability of LSCs to form progenitor colonies. We sorted LSCs from recipients of BCR-ABL-transduced WT or Scd1−/− bone marrow cells and plated the cells in vitro for colony formation. We observed that Scd1−/− LSCs formed significantly higher numbers of colonies compared to WT LSCs (Fig. 3C). To determine whether deletion of Scd1 causes an increase in LSC function in vivo, we mixed BCR-ABL-transduced WT (CD45.1) and Scd1−/− (CD45.2) bone marrow cells in a 1:1 ratio and transplanted into lethally irradiated mice. At days 8 and 11 after CML induction, FACS analysis showed much higher percentages of CD45.2+ leukemia cells in PB compared to the percentages of CD45.1+ leukemia cells (Fig. 3D). To further confirm this result, LSCs were sorted by FACS from the bone marrow of primary CML mice receiving BCR-ABL-transduced WT (CD45.1) or Scd1−/− (CD45.2) bone marrow cells, mixed in a 1:1 ratio, and then transplanted into secondary recipient mice. Eight weeks later, more than 75% of the GFP+ Gr-1+ leukemia cells in the PB were CD45.2+ (Fig. 3E), indicating that Scd1−/− LSCs grew predominantly over WT LSCs to induce CML in recipient mice. Together, these results demonstrate that Scd1 inhibits proliferation of LSCs.
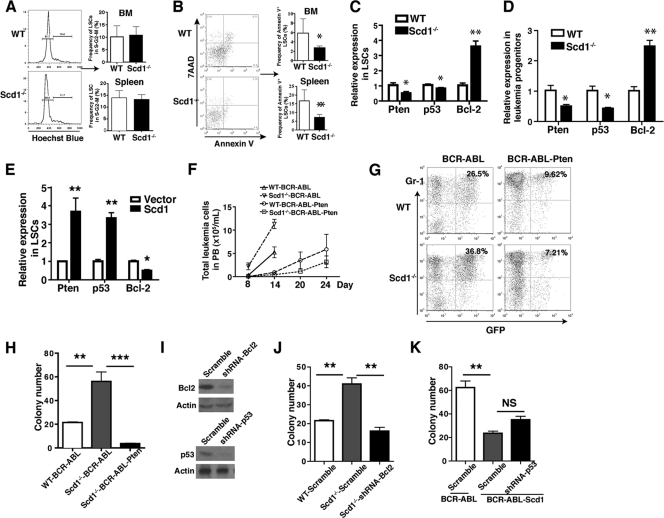
Scd1 deletion inhibits apoptosis but not the cell cycle of LSCs.
To explore cellular mechanisms by which Scd1 regulates LSC function, we investigated whether Scd1 suppresses LSCs through affecting cell cycle progression or apoptosis. We analyzed LSCs from the bone marrow and spleen of CML mice, and we did not observe a significant difference in the percentages of LSCs in the S-G2-M phase of the cell cycle in the presence or absence of Scd1 (Fig. 4A). However, the percentages of apoptotic LSCs (annexin V+) were significantly lower in the absence than in the presence of Scd1 (Fig. 4B), indicating Scd1 suppresses LSCs through reducing their survival. To study the molecular mechanisms by which Scd1 suppresses LSCs and attenuates CML development, we compared the expression of Pten, p53, and Bcl-2 in WT and Scd1−/− leukemia progenitor cells and LSCs. We found that a Scd1 deficiency caused a decreased expression of Pten and p53 and an increased expression of Bcl-2 in LSCs (Fig. 4C) and in GFP+ Lin− leukemia progenitor cells (Fig. 4D). Consistently, when we overexpressed Scd1 in LSCs, we found that Scd1 overexpression induced the expression of Pten and p53 and reduced the expression of Bcl-2 in LSCs (Fig. 4E). To examine functional significance of these genes in LSCs, we constructed the BCR-ABL-Pten-GFP retroviral construct to allow coexpression of BCR-ABL, Pten, and green fluorescent protein (GFP) in the same cells (23). We transduced bone marrow cells obtained from WT or Scd1−/− mice with BCR-ABL-Pten-GFP retrovirus and transplanted into lethally irradiated recipient mice. Recipients of BCR-ABL-transduced Scd1−/− bone marrow cells displayed higher percentages and numbers of leukemia cells in the PB compared to recipients of BCR-ABL-transduced WT bone marrow cells, whereas recipients of BCR-ABL-Pten-transduced WT and Scd1−/− bone marrow cells showed a lower number of leukemia cells (Fig. 4F and G). These results indicate that Pten functions downstream of Scd1 to suppress CML development. To further confirm the effect of Pten on Scd1−/− LSCs, we conducted a colony-forming assay and found that Pten suppressed the colony formation of Scd1−/− LSCs (Fig. 4H).
Fig 4.
Scd1 deletion promotes the survival but not the cell cycle progression of LSCs. (A) Cell cycle analysis showed comparable percentages of cycling LSCs from the bone marrow and spleens of CML mice receiving BCR-ABL-transduced WT and Scd1−/− bone marrow cells. The data shown are from a representative experiment of three independent experiments, with four to seven mice per group in each experiment. Mean values (± the SEM) are shown. (B) Apoptotic analysis of LSCs from the bone marrow and spleens of CML mice receiving BCR-ABL-transduced WT or Scd1−/− bone marrow cells. The data shown are from a representative experiment of three independent experiments, with four to seven mice per group in each experiment. Mean values (± the SEM) are shown (*, P < 0.05; **, P < 0.01). (C and D) Real-time RT-PCR analysis of expression of Pten, p53, and Bcl-2 in the sorted WT or Scd1−/− LSCs and progenitor cells from CML mice. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (*, P < 0.05; **, P < 0.01). (E) Real-time RT-PCR analysis demonstrated that Scd1 upregulated the expression of Pten and p53 and downregulated the expression of Bcl-2 in LSCs. Bone marrow cells from 5-FU-pretreated mice were cotransduced with MSCV-BCR-ABL-GFP and MSCV-Scd1-hCD4 or MSCV-hCD4. At 14 days after BMT, Scd1-overexpressing LSCs (hCD4+ GFP+ Lin− Sca-1+ c-Kit+) or control LSCs were sorted for extracting total RNA for real-time PCR analysis. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (*, P < 0.05; **, P < 0.01). (F) FACS analysis showing the total numbers of leukemia cells in PB of recipients of BCR-ABL or BCR-ABL-Pten transduced WT and Scd1−/− bone marrow cells at day 14 after BMT. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown. (G) FACS analysis showing the percentages of leukemia cells in the PB of recipients of BCR-ABL-transduced or BCR-ABL-Pten-transduced WT and Scd1−/− bone marrow cells at day 14 after BMT. (H) A colony-forming assay showed that Pten overexpression reduced the colony-forming ability of Scd1−/− LSCs. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (**, P < 0.01; ***, P < 0.001). (I) Western blotting of Bcl2 and p53 expression after lentiviral shRNA-mediated knockdown. (J) Colony-forming assay. CML bone marrow cells from recipients of BCR-ABL-transduced WT and Scd1−/− bone marrow cells were infected with shRNA-Bcl2 or control lentivirus and cultured in the presence of puromycin for 48 h. Equal numbers of infected cells were plated into methylcellulose medium. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (**, P < 0.01). (K) Colony-forming assay. CML bone marrow cells from recipients of BCR-ABL-transduced or BCR-ABL-Scd1-transduced WT bone marrow cells were infected with shRNA-p53 or control lentivirus and cultured in the presence of puromycin for 48 h. Equal numbers of infected cells were plated into methylcellulose medium. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (**, P < 0.01; NS, no significance).
To study whether Scd1 functions through Bcl2 and p53, we knocked down expression of these two genes with lentiviral shRNAs (Fig. 4I). Because Scd1 deficiency caused a higher level of Bcl2 expression in Scd1−/− LSCs (Fig. 4C), we assumed that Bcl2 knockdown would reverse the phenotype of Scd1−/− LSCs. As expected, we found that Scd1−/− LSCs with Bcl2 knockdown gave rise to fewer colonies compared to control Scd1−/− LSCs without Bcl2 knockdown (Fig. 4J), indicating that Bcl2 also mediates the function of Scd1 in LSCs. Furthermore, we knocked down p53 in Scd1-expressing LSCs and observed an increased number of colonies (Fig. 4K), although the increase was not statistically significant, suggesting that p53 plays a minor role downstream of Scd1 in LSCs.
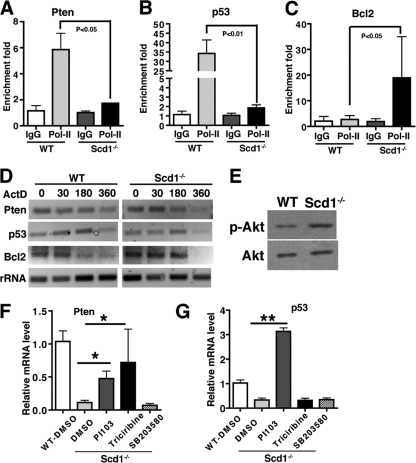
To examine whether Scd1 regulates expression of Pten, p53, and Bcl2 at a transcriptional or posttranscriptional level, we first performed ChIP of RNA Pol II using WT and Scd1−/− leukemia cells and found that the enrichments of Pol II in the promoters of Pten and p53 were significantly higher in the WT leukemia cells than in the Scd1−/− leukemia cells (Fig. 5A and B). We also found that the enrichment of Pol II in the promoter of Bcl2 was lower in the WT leukemia cells than in the Scd1−/− leukemia cells (Fig. 5C). These data suggest that Scd1 deletion reduces the transcription of Pten and p53 and induces Bcl2 transcription. We tested whether Scd1 affects mRNA stability of Pten, p53, and Bcl2. Leukemia cells from WT and Scd1−/− CML mice were treated with actinomycin D (5 μg/ml) to block the transcription, the total RNA was subsequently harvested at various time points, and the mRNA half-life for Pten, p53, or Bcl2 was examined by RT-PCR. We found that the mRNA half-lives for Pten and p53 were similar in WT and Scd1−/− leukemia cells, but the mRNA half-life for Bcl2 was markedly increased in Scd1−/− leukemia cells (Fig. 5D). These results indicate that Scd1 regulates the expression of Pten and p53 only at the transcriptional level but regulates the expression of Bcl2 through affecting both transcription and mRNA stability.
Fig 5.
Scd1 regulates Pten, p53, and Bcl2 transcription and mRNA stability. (A and B) Pol II ChIP analysis shows the accumulation of Pol II in the promoters of Pten and p53 of WT cells but not in Scd1−/− leukemia cells. (C) Pol II ChIP analysis showed the accumulation of Pol II in the promoter of Bcl2 of Scd1−/− cells but not in WT leukemia cells. (D) Decay time course of the mRNA levels for Pten, p53, and Bcl2 in WT and Scd1−/− leukemia cells. (E) Western blot analysis showed Akt phosphorylation in WT and Scd1−/− leukemia progenitor cells. (F) Real-time RT-PCR assay for Pten expression in Scd1−/− leukemia cells treated with PI103, triciribine, and SB203580. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (*, P < 0.05; **, P < 0.01). (G) Real-time RT-PCR assay for p53 expression in Scd1−/− leukemia cells treated with PI103, triciribine, and SB203580. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (*, P < 0.05; **, P < 0.01).
To further study the functional relationship between Scd1 and Pten/p53, we examined Akt phosphorylation in Scd1−/− leukemia cells, because a published study showed an increased Akt activity in Scd1−/− cells (26). We found that the level of Akt phosphorylation level was elevated in Scd1−/− leukemia cells (Fig. 5E). Therefore, we sought to determine whether the Akt pathway is involved in the regulation of Pten and p53 by Scd1. We treated Scd1−/− CML leukemia cells with the Akt inhibitor triciribine and found that inhibition of Akt activity partially reversed expression of Pten but not p53 (Fig. 5F and G). Inhibition of the PI3K/mTOR pathway by the dual inhibitor PI103 reversed the expression of both Pten and p53 (Fig. 5F and G). However, inhibition of mitogen-activated protein kinase (MAPK) activity by the inhibitor SB203580 did not have an effect on the expression of either Pten or p53 (Fig. 5F and G). These results suggest that the PI3K/Akt pathway but not the MAPK pathway mediates the regulation of Pten and p53 by Scd1.
Induction of Scd1 expression inhibits LSCs and attenuates CML development.
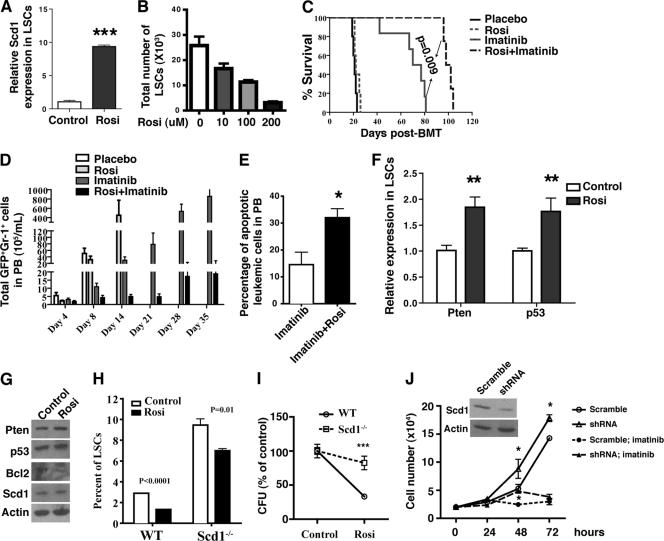
The tumor-suppressive role of Scd1 in CML development prompted us to test whether the upregulation of Scd1 expression delays CML development. It has been shown that the PPARγ agonists rosiglitazone and pioglitazone induce Scd1 expression, and this Scd1 upregulation is blocked by PPARγ antagonists in vitro and in vivo (24, 33). Therefore, we treated bone marrow cells from CML mice with rosiglitazone for 7 days in vitro and found that Scd1 expression was significantly induced in LSCs (Fig. 6A), accompanied by a dramatic decrease in the numbers of LSCs (Fig. 6B). To test the effect of rosiglitazone on CML development, we treated CML with a placebo, with imatinib alone, with rosiglitazone alone, and with the two agents in combination. Although treatment with rosiglitazone alone did not prolong the survival of CML mice, the combined treatment with rosiglitazone and imatinib synergized and more significantly prolonged survival of CML mice than did the use of imatinib alone (Fig. 6C). This therapeutic effect by rosiglitazone and imatinib correlated with much lower numbers of GFP+ Gr-1+ myeloid leukemia cells (Fig. 6D) and increased the apoptosis (Fig. 6E) of leukemia cells in the PB. Because Scd1 overexpression induced the expression of Pten and p53 in LSCs (Fig. 4E), we tested whether rosiglitazone treatment affects Pten and p53 expression. We observed higher levels of Pten and p53 expression in LSCs after rosiglitazone treatment (Fig. 6F). We also treated the human CML cell line K562 with rosiglitazone for 24 h and found that rosiglitazone treatment induced the expression of Pten, p53, and Scd1 but decreased the expression of Bcl2 (Fig. 6G).
Fig 6.
Induction of Scd1 expression suppresses LSCs and attenuates CML development. (A) Real-time RT-PCR showed the induction of Scd1 expression by rosiglitazone in LSCs. Bone marrow cells from primary CML mice were treated with rosiglitazone in vitro for 7 days, the LSCs were sorted by FACS, and the total RNA was isolated for RT-PCR analysis. Representative data from two independent experiments are shown. Mean values (± the SEM) are shown (***, P < 0.001). (B) Bone marrow cells from primary CML mice were treated with rosiglitazone in vitro for 7 days, and the number of LSCs was decreased upon rosiglitazone treatment. (C) Kaplan-Meier survival curve for CML mice treated with a placebo, with rosiglitazone alone, with imatinib alone, or with both rosiglitazone and imatinib (n = 6 to 12 mice per group; P = 0.009). (D) The total numbers of leukemia cells in the PB of CML mice treated with a placebo, with rosiglitazone alone, with imatinib alone, or with both rosiglitazone and imatinib were analyzed at days 4, 8, 14, 21, 28, and 35 after initiation of the treatments. The results are representative of two independent experiments. (E) FACS analysis of the percentages of apoptotic leukemia cells in PB of CML mice treated with imatinib alone (n = 5) or with both rosiglitazone and imatinib (n = 5) for 8 weeks. Mean values (± the SEM) are shown (*, P < 0.05). (F) Rosiglitazone treatment induced the expression of Pten and p53 in LSCs in vitro. Mean values (± the SEM) are shown (**, P < 0.01; ***, P < 0.001). (G) Western blot analysis showed the regulation of expression of Pten, p53, Bcl2, and Scd1 by rosiglitazone in human K562 cells. (H) Percentages of WT and Scd1−/− LSCs cultured in vitro in the presence of rosiglitazone. (I) Scd1 deletion reduced the inhibitory effect of rosiglitazone on colony formation of LSCs. Bone marrow cells from recipients of BCR-ABL-transduced WT and Scd1−/− bone marrow cells were plated in methylcellulose medium in the presence of rosiglitazone. Note that the CFU for cells treated with DMSO was defined as 100%. (J) Scd1 knockdown induced the proliferation of human K562 leukemia cells. The inset panel shows the decreased Scd1 expression mediated by lentiviral shRNA. Mean values (± the SEM) are shown (*, P < 0.05).
To confirm the requirement of Scd1 for the effect of rosiglitazone in suppressing leukemia development, we cultured bone marrow cells from recipients of BCR-ABL transduced WT and Scd1−/− bone marrow cells in vitro under the stem cell culture conditions in the presence of rosiglitazone. We found that rosiglitazone treatment caused a marked decrease (up to 55%) of WT LSCs (Fig. 6H). In contrast, rosiglitazone treatment only caused a modest decrease in the percentage of Scd1−/− LSCs (25% decrease) (Fig. 6H), indicating that rosiglitazone regulates LSCs through Scd1. In addition, we compared the impact of rosiglitazone on the colony-forming ability between WT and Scd1−/− LSCs. Since Scd1−/− LSCs had an increased colony-forming ability (Fig. 3C), we normalized the colony number in the control group (DMSO) to 100% and showed that the deletion of Scd1 caused a slight reduction in colony formation after rosiglitazone treatment (Fig. 6I), further demonstrating that the inhibitory effect of rosiglitazone on LSCs is mediated through the induction of Scd1 expression. To examine whether Scd1 plays a similar role in human leukemia cells, we silenced Scd1 expression in K562 cells using lentiviral shRNA and found that Scd1 knockdown markedly induced the proliferation of K562 cells and also significantly reduced imatinib sensitivity (Fig. 6J), indicating that Scd1 also plays a tumor-suppressive role in human leukemia cells.
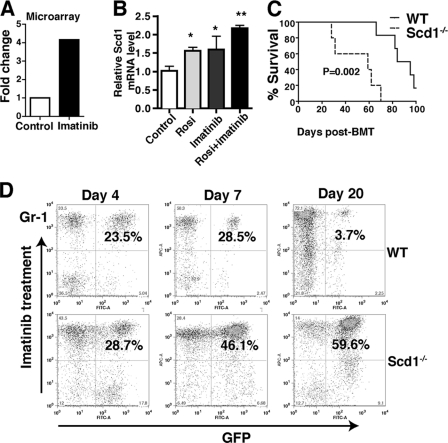
To investigate whether Scd1 synergizes with imatinib in delaying CML development, we first analyzed our LSC microarray data and found that imatinib treatment induced Scd1 expression in LSCs (Fig. 7A). We next treated leukemia cells with imatinib in vitro under the stem cell culture conditions and found that treatment with imatinib or rosiglitazone alone caused an increase in Scd1 expression and that treatment with both drugs further induced Scd1 expression (Fig. 7B). Furthermore, we treated recipients of BCR-ABL-transduced WT and Scd1−/− bone marrow cells with imatinib and found that, upon imatinib treatment, the survival of recipients of BCR-ABL-transduced WT bone marrow cells was significantly longer than recipients of BCR-ABL-transduced Scd1−/− bone marrow cells (Fig. 7C), a finding consistent with a lower percentage of leukemia cells in the presence of Scd1 (Fig. 7D).
Fig 7.
Scd1 mediates the inhibitory effect of imatinib on CML development. (A) Microarray analysis showed that imatinib treatment induced Scd1 expression in LSCs. (B) Real-time RT-PCR showed that imatinib treatment induced Scd1 expression in leukemia cells (*, P < 0.05; **, P < 0.01). (C) Kaplan-Meier survival curve for imatinib treated recipients of BCR-ABL-transduced WT and Scd1−/− bone marrow cells (n = 5 to 8 mice per group; P = 0.002). (D) FACS analysis showed the percentages of leukemia cells in PB of recipients of BCR-ABL-transduced WT and Scd1−/− bone marrow cells at days 4, 7, and 20 after imatinib treatment.
Scd1 does not affect normal HSCs.
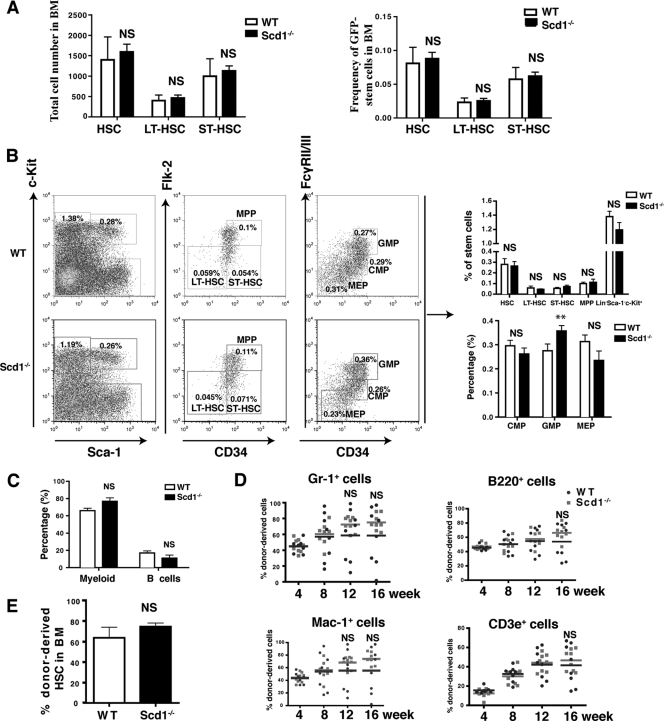
Since Scd1 is a suppressor of LSCs in CML mice (Fig. 3), we wondered whether this inhibitory effect is specific to LSCs with no effect on normal HSCs. We analyzed non-BCR-ABL-expressing (GFP−) HSCs (GFP− Lin− c-Kit+ Sca-1+) and LT (GFP− Lin− c-Kit+ Sca-1+ CD34−) and ST (GFP− Lin− c-Kit+ Sca-1+ CD34+) HSCs in CML mice receiving BCR-ABL-transduced WT or Scd1−/− bone marrow cells. We did not observe any significant differences in the percentages and numbers of these cell populations between the two groups (Fig. 8A). This result suggests that Scd1 does not suppress normal HSCs. To confirm this observation, we directly analyzed HSCs and progenitors in bone marrow of WT and Scd1−/− mice. We found that the percentages of total HSCs (Lin− Sca-1+ c-Kit+), LT-HSCs (Lin− Sca-1+ c-Kit+ CD34− Flk-2−), ST-HSCs (Lin− Sca-1+ c-Kit+ CD34+ Flk-2−), and MPP (Lin− Sca-1+ c-Kit+ CD34+ Flk-2+) in the bone marrow of Scd1−/− mice were similar to those in WT mice (Fig. 8B). Although the percentage of GMP (Lin− Sca-1+ c-Kit+ CD34+ FcyRII/IIIhi) was modestly higher in Scd1−/− mice than in WT mice, the percentages of CMP (Lin− Sca-1+ c-Kit+ CD34+ FcyRII/IIIlo) and MEP (Lin− Sca-1+ c-Kit+ CD34− FcyRII/IIIlo) in bone marrow were similar between Scd1−/− and WT mice (Fig. 8B). In addition, Scd1−/− mice had a normal distribution of mature myeloid cells (Gr-1+ Mac-1+) and B lymphoid cells (B220+) (Fig. 8C).
Fig 8.
Scd1 deficiency does not affect the function of normal HSCs. (A) The total numbers and percentages of HSCs (GFP− Lin− c-Kit+ Sca-1+), LT-HSCs (GFP− Lin− c-Kit+ Sca-1+ CD34−), and ST-HSCs (GFP− Lin− c-Kit+ Sca-1+ CD34+) in the GFP− cell population of recipients of BCR-ABL-transduced WT and Scd1−/− bone marrow cells were compared (NS, no significance). (B) The percentages of normal stem cells and progenitors from WT and Scd1−/− bone marrow (n = 4) were compared. HSC, Lin− c-Kit+ Sca-1+; LT-HSC, Lin− c-Kit+ Sca-1+ CD34− Flk-2−; ST-HSC, Lin− c-Kit+ Sca-1+ CD34+ Flk-2−; MPP, Lin− c-Kit+ Sca-1+ CD34+ Flk-2+; progenitor, Lin− c-Kit+ Sca-1−; CMP, Lin− c-Kit+ Sca-1− CD34+ FcγRII/IIIlo; GMP, Lin− c-Kit+ Sca-1− CD34+ FcγRII/IIIhi; MEP, Lin− c-Kit+ Sca-1− CD34− FcγRII/IIIlo. (C) The percentages of myeloid cells (Gr-1+) and B cells (B220+) in the bone marrow of WT and Scd1−/− mice were compared. Mean values (± the SEM) are shown. (D) Competitive repopulation assay for the effect of Scd1 on normal HSCs. FACS analysis of the percentages of donor-derived (CD45.2) myeloid cells (Gr-1+, Mac-1+), B cells (B220+), and T cells (CD3e+) in the PB of recipients of normal bone marrow cells from WT and Scd1−/− mice was performed. (E) FACS analysis of the donor-derived (CD45.2+) HSCs in the bone marrow of recipients of WT and Scd1−/− mice (n = 8 mice per group). Mean values (± the SEM) are shown.
To determine whether Scd1 affects the self-renewal capacity of HSCs, we performed a competitive repopulation assay. Bone marrow cells (2 × 105) from WT (CD45.2) or Scd1−/− (CD45.2) mice were transplanted into each lethally irradiated CD45.1 mouse, along with equal numbers of WT CD45.1 competitor bone marrow cells. Analysis of these chimeric mice was performed by FACS over 1 to 4 months after the BMT. We found that the percentages of WT and Scd1−/− donor-derived myeloid cells (Gr-1+, Mac-1+), B cells (B220+), and T cells (CD3e+) in PB were similar at 4 and 8 weeks after BMT; however, the percentages of Scd1−/− donor-derived myeloid cells at 12 and 16 weeks and the percentages of Scd1−/− donor-derived B and T cells at 16 weeks were slightly higher than those of WT donor-derived cells (Fig. 8D), although the difference was not statistically significant. At 16 weeks, we directly compared the percentages of donor-derived WT and Scd1−/− HSCs in bone marrow, and we did not observe a significant difference (Fig. 8E). Together, these results indicate that Scd1 does not affect the function of normal HSCs.
DISCUSSION
Our study supports a critical role for Scd1 in CML development through specifically regulating LSCs but not normal HSCs. In our previously published study, we identified another lipid-metabolic gene, arachidonate 5-lipoxygenase (Alox5), required for functional regulation of LSCs but not normal HSCs (1). Our identification of Scd1 in the present study strengthens the feasibility of specifically targeting LSCs in the treatment of CML.
Although dysregulated expression of Scd1 has been observed in several human cancers (32), to our knowledge the present study provides the first evidence for the role of Scd1 in hematopoiesis and leukemogenesis. Previous data indicated that Scd1 regulated cancer cell proliferation and survival. Knockdown of Scd1 by small interfering RNA significantly reduced the survival of multiple human tumor cell lines (21). In contrast, in the present study, we found that Scd1 plays a tumor-suppressive role in LSCs through regulating apoptosis of LSCs. This indicates that Scd1 plays different roles in different cancers. In addition, while CML development was accelerated in the loss of Scd1, the development of Ph+ B-ALL could be unimpaired. It is known that CML is a stem cell disease, while the cell of origin for ALL is a committed B-cell progenitor (16). Therefore, the distinct roles of Scd1 in different leukemia subtypes suggest that Scd1 acts in a cell-type-specific manner.
With respect to the molecular basis for the functional regulation of LSCs by Scd1, we show that Scd1 deficiency results in dysregulated expression of apoptosis-related genes, including decreased expression of Pten and p53 and increased expression of Bcl-2, which is consistent with the observation from other groups showing that Scd1 deficiency reduces cardiac apoptosis in ob/ob mice due to the increased expression of antiapoptotic factor Bcl-2 (4). In addition, we also show that treatment with PPARγ agonist induces the apoptosis of leukemia cells, which is associated with a higher expression of Scd1, Pten, and p53. A recent study showed that unsaturated fatty acid affected Pten expression in hepatoma cells through regulating microRNA-mediated Pten mRNA stability, which depends on the mTOR and NF-κB pathways (30). Our results show that Scd1 regulates Pten, p53, and Bcl2 at the transcriptional and/or posttranscriptional level. However, detailed mechanisms by which Scd1 regulates expression of Pten, p53, and Bcl2 need to be further investigated in the future.
The mechanisms by which a PPARγ agonist inhibits LSCs is unclear. PPARs are ligand-binding transcription factors belonging to the nuclear receptor family. PPARs are involved in regulating lipid metabolism, cell growth, and survival. It has been shown that activation of the PPAR pathway with the dual PPARα and PPARγ ligand TZD18 induces apoptosis and inhibits the proliferation of human CML cells (18, 34). Although rosiglitazone synergized with imatinib to delay CML development, rosiglitazone alone failed to do so. One possibility is that rosiglitazone alone is not effective enough to prevent lung hemorrhage, a major cause of death of CML mice. Another possibility is that the level of Scd1 induced by rosiglitazone is not high enough to delay CML development. PPAR agonists are anti-inflammatory agents since they inhibit the expression of several proinflammatory cytokines and most chemokines (3). A role of an anti-inflammatory agent on LSCs is supported by our previous study showing that the anti-inflammatory drug Zileuton, which inhibits the function of the Alox5 gene, prolongs the survival of CML mice (1). We treated LSCs from CML mice in vitro with rosiglitazone alone or both rosiglitazone and Zileuton and found that the dual drug treatment resulted in a significantly lower percentage of LSCs compared to treatment with rosiglitazone alone (data not shown), indicating that an Alox5 inhibitor synergizes with PPAR agonist to inhibit LSCs. Together, enhancement of Scd1 activity provides an attractive strategy for eliminating LSCs in CML.
ACKNOWLEDGMENTS
This study was supported by the grants from the Leukemia and Lymphoma Society and the National Institutes of Health (R01-CA122142 and R01-CA114199) to S.L. S.L. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Published ahead of print 19 March 2012
REFERENCES
- 1. Chen Y, Hu Y, Zhang H, Peng C, Li S. 2009. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat. Genet. 41:783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. 2008. Brick by brick: metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 18:54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dinarello CA. 2010. Anti-inflammatory agents: present and future. Cell 140:935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dobrzyn P, Dobrzyn A, Miyazaki M, Ntambi JM. 2010. Loss of stearoyl-CoA desaturase 1 rescues cardiac function in obese leptin-deficient mice. J. Lipid Res. 51:2202–2210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Druker BJ, et al. 2001. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N. Engl. J. Med. 344:1038–1042 [DOI] [PubMed] [Google Scholar]
- 6. Falvella FS, et al. 2002. Stearoyl-CoA desaturase 1 (Scd1) gene overexpression is associated with genetic predisposition to hepatocarcinogenesis in mice and rats. Carcinogenesis 23:1933–1936 [DOI] [PubMed] [Google Scholar]
- 7. Graham SM, et al. 2002. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood 99:319–325 [DOI] [PubMed] [Google Scholar]
- 8. Hess D, Chisholm JW, Igal RA. 2010. Inhibition of stearoyl-CoA desaturase activity blocks cell cycle progression and induces programmed cell death in lung cancer cells. PLoS One 5:e11394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Y, et al. 2004. Requirement of Src kinases Lyn, Hck, and Fgr for BCR-ABL1-induced B-lymphoblastic leukemia but not chronic myeloid leukemia. Nat. Genet. 36:453–461 [DOI] [PubMed] [Google Scholar]
- 10. Hu Y, et al. 2006. Targeting multiple kinase pathways in leukemic progenitors and stem cells is essential for improved treatment of Ph+ leukemia in mice. Proc. Natl. Acad. Sci. U. S. A. 103:16870–16875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Juric D, et al. 2007. Differential gene expression patterns and interaction networks in BCR-ABL-positive and -negative adult acute lymphoblastic leukemias. J. Clin. Oncol. 25:1341–1349 [DOI] [PubMed] [Google Scholar]
- 12. Kantarjian H, et al. 2002. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N. Engl. J. Med. 346:645–652 [DOI] [PubMed] [Google Scholar]
- 13. Kim YC, Ntambi JM. 1999. Regulation of stearoyl-CoA desaturase genes: role in cellular metabolism and preadipocyte differentiation. Biochem. Biophys. Res. Commun. 266:1–4 [DOI] [PubMed] [Google Scholar]
- 14. Kinder M, et al. 2010. Hematopoietic stem cell function requires 12/15-lipoxygenase-dependent fatty acid metabolism. Blood 115:5012–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kroemer G, Pouyssegur J. 2008. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell 13:472–482 [DOI] [PubMed] [Google Scholar]
- 16. Li S, Ilaria RL, Jr, Million RP, Daley GQ, Van Etten RA. 1999. The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189:1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li S, Li D. 2008. DNA microarray technology and data analysis in cancer research. World Scientific 4:37–54 [Google Scholar]
- 18. Liu H, et al. 2006. Growth inhibition and apoptosis in human Philadelphia chromosome-positive lymphoblastic leukemia cell lines by treatment with the dual PPARα/γ ligand TZD18. Blood 107:3683–3692 [DOI] [PubMed] [Google Scholar]
- 19. Melo JV, Barnes DJ. 2007. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat. Rev. Cancer 7:441–453 [DOI] [PubMed] [Google Scholar]
- 20. Middleton MK, et al. 2006. Identification of 12/15-lipoxygenase as a suppressor of myeloproliferative disease. J. Exp. Med. 203:2529–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan-Lappe SE, et al. 2007. Identification of Ras-related nuclear protein, targeting protein for xenopus kinesin-like protein 2, and stearoyl-CoA desaturase 1 as promising cancer targets from an RNAi-based screen. Cancer Res. 67:4390–4398 [DOI] [PubMed] [Google Scholar]
- 22. Peng C, et al. 2007. Inhibition of heat shock protein 90 prolongs survival of mice with BCR-ABL-T315I-induced leukemia and suppresses leukemic stem cells. Blood 110:678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peng C, et al. 2010. PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice. Blood 115:626–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin X, et al. 2007. Laminar shear stress upregulates the expression of stearoyl-CoA desaturase-1 in vascular endothelial cells. Cardiovasc. Res. 74:506–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Radich JP, et al. 2006. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 103:2794–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rahman SM, et al. 2003. Stearoyl-CoA desaturase 1 deficiency elevates insulin-signaling components and downregulates protein-tyrosine phosphatase 1B in muscle. Proc. Natl. Acad. Sci. U. S. A. 100:11110–11115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ren R. 2005. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat. Rev. Cancer 5:172–183 [DOI] [PubMed] [Google Scholar]
- 28. Scaglia N, Chisholm JW, Igal RA. 2009. Inhibition of stearoylCoA desaturase-1 inactivates acetyl-CoA carboxylase and impairs proliferation in cancer cells: role of AMPK. PLoS One 4:e6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scaglia N, Igal RA. 2005. Stearoyl-CoA desaturase is involved in the control of proliferation, anchorage-independent growth, and survival in human transformed cells. J. Biol. Chem. 280:25339–25349 [DOI] [PubMed] [Google Scholar]
- 30. Vinciguerra M, et al. 2009. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 upregulation in hepatocytes. Hepatology 49:1176–1184 [DOI] [PubMed] [Google Scholar]
- 31. Wong S, Witte ON. 2004. The BCR-ABL story: bench to bedside and back. Annu. Rev. Immunol. 22:247–306 [DOI] [PubMed] [Google Scholar]
- 32. Yahagi N, et al. 2005. Co-ordinate activation of lipogenic enzymes in hepatocellular carcinoma. Eur. J. Cancer 41:1316–1322 [DOI] [PubMed] [Google Scholar]
- 33. Yao-Borengasser A, et al. 2008. Stearoyl-coenzyme A desaturase 1 gene expression increases after pioglitazone treatment and is associated with peroxisomal proliferator-activated receptor-gamma responsiveness. J. Clin. Endocrinol. Metab. 93:4431–4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zang C, et al. 2006. Dual PPARα/γ ligand TZD18 either alone or in combination with imatinib inhibits proliferation and induces apoptosis of human CML cell lines. Cell Cycle 5:2237–2243 [DOI] [PubMed] [Google Scholar]