Abstract
Somatic hypermutation (SHM) of immunoglobulin (Ig) genes is initiated by the activation-induced cytidine deaminase (AID). However, the influence of chromatin on SHM remains enigmatic. Our previous cell-free studies indicated that AID cannot access nucleosomal DNA in the absence of transcription. We have now investigated the influence of nucleosome stability on mutability in vivo. We introduced two copies of a high-affinity nucleosome positioning sequence (MP2) into a variable Ig gene region to assess its impact on SHM in vivo. The MP2 sequence significantly reduces the mutation frequency throughout the nucleosome, and especially near its center, despite proportions of AID hot spots similar to those in Ig genes. A weak positioning sequence (M5) was designed based on rules deduced from published whole-genome analyses. Replacement of MP2 with M5 resulted in much higher mutation rates throughout the nucleosome. This indicates that both nucleosome stability and positioning significantly influence the SHM pattern. We postulate that, unlike RNA polymerase, AID has reduced access to stable nucleosomes. This study outlines the limits of nucleosome positioning for SHM of Ig genes and suggests that stable nucleosomes may need to be disassembled for access of AID. Possibly the variable regions of Ig genes have evolved for low nucleosome stability to enhance access to AID, DNA repair factors, and error-prone polymerases and, hence, to maximize variability.
INTRODUCTION
The somatic hypermutation (SHM) of antibody genes is initiated by the activation-induced cytidine deaminase (AID) that creates cytosine (C)-to-uracil (U) mutations, starting after ∼100 to 200 bp from the promoter and extending for about 2 kb. During SHM, these U's are repaired in error-prone fashion via translesion DNA polymerases, leading to mutations at and near the U (reviewed in reference 38). Absence of AID results in a variety of immunodeficiencies (6), but on the other hand, AID is a dangerous oncogenic mutator (reviewed in reference 24). Additionally, DNA demethylation via AID may be essential for normal early development and perhaps some aspects of DNA methylation in general (9, 12, 21, 30, 31). Thus, the study of the molecular mechanisms of AID action is essential for understanding the roles of AID in immunity and oncogenesis, as well as development. The process of SHM requires transcription without requiring a specific promoter (3, 4) but is linked to transcription initiation (25). We have postulated that AID is crucially associated with the transcription complex and may target negative supercoils, as they arise in the wake of the transcription complex during transcript elongation (34). Transcription occurs in the context of chromatin, which sterically occludes DNA binding complexes (15, 18, 28, 29). To begin to understand the role of chromatin in SHM, we previously investigated the effect of a strong nucleosome positioning sequence (NPS) (MP2) on the function of AID in a cell-free system (33). Nucleosomes positioned within a circular plasmid that was susceptible to AID-induced cytosine deamination when the DNA was naked inhibited AID access specifically to the sequences associated with histone octamers. However, when the nucleosomal region was transcribed by the phage RNA polymerase T7, it underwent efficient cytosine deamination, suggesting that AID, unlike RNA polymerase, cannot access tightly wrapped DNA. Since transcription in these cell-free assays was by the small T7 polymerase, it was possible that the eukaryotic polymerase pol II was less able to unwrap tight nucleosomes sufficiently. Indeed, the effect of tightly positioned nucleosomes and chromatin on the targeting of AID in vivo remained unknown. We report here a study in which the same MP2 sequence was introduced into the variable (V) region of an immunoglobulin (Ig) gene by homologous integration in cells that normally undergo SHM in culture. Surprisingly, the presence of the MP2 sequence affected the efficiency of SHM in vivo.
MATERIALS AND METHODS
Cell culture and transgenic clones.
DT40 ψV knockout (KO) cells derived from avian leukosis virus-induced chicken bursal B cells were a gift of H. Arakawa and J. M. Buerstedde (Institute of Molecular Radiology, Neuherberg, Germany) (2). The cells were cultured in RPMI 1640 with 1% penicillin/streptomycin, 1% l-glutamine (Invitrogen), 1% chicken serum, and β-mercaptoethanol (Sigma-Aldrich) at 39.5°C with 5% CO2. MP2-MP2 (M5-MP2, MP2-M5, and M5-M5) (M5 is a weak positioning sequence designed based on rules deduced from published whole-genome analyses [see Fig. 2]) knock-in constructs were linearized with SalI and transfected as previously described (2). After 12 h, transfected cells were treated with 20 μg/ml blasticidin for selection of blasticidin resistance, and single clones were isolated by subsequent limiting dilutions. Single clones from limiting dilutions were expanded to perform fluorescence-activated cell sorter (FACS) analysis for surface IgM-negative clones and to subsequently collect their genomic DNA for Southern blotting. The genomic DNA was digested with SbfI and MluI, respectively, and a radioactive probe for the MP2 region was used for Southern blot analysis.
Fig 2.
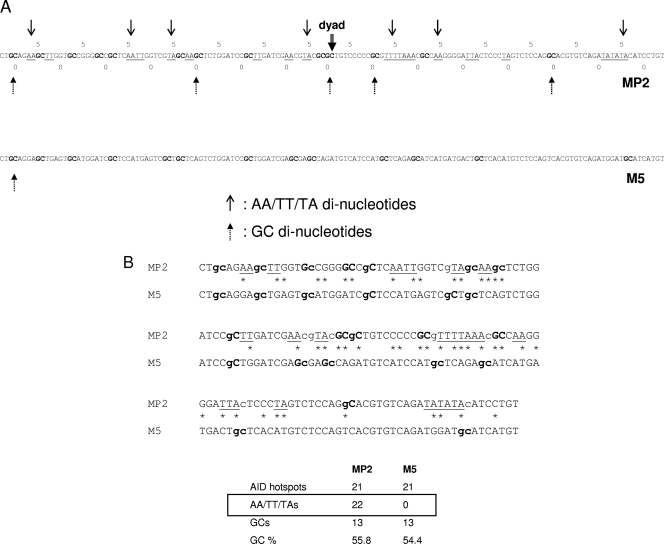
M5 control sequence. (A) Alignment of the MP2 and M5 sequences showing the periodicity of AA/TT/TA and GC dinucleotides. Stretches of AA, TT, and TA are underlined; GCs are in boldface. The numbers 0 and 5 relate to the dyad center. (B) Sequences of MP2 and M5. Stretches of AA, TT, and TA are underlined, GCs are in boldface, c and g (WRC and GYW) in the AID hotspots are in lowercase, and 47 changes in the MP2 sequence to create M5 are marked with asterisks.
Flow cytometric analysis and cell sorting.
DT40 knock-in clones for MP2/M5 sequences were stained with phycoerythrin (PE)-conjugated anti-chicken IgM antibody (Santa Cruz Biotechnology, Inc.) and were analyzed for loss of surface IgM and the presence of AID- internal ribosome entry site (IRES)-green fluorescent protein (GFP) expression of 50,000 live cells on an LSR II (BD) using DT40 CL18 cells and GFP+ ψV KO cells (a gift of H. Arakawa and J. M. Buerstedde [2]) as gating controls. DT40 knock-in clones treated with tamoxifen were sorted for AID-IRES-GFP+ single cells on a cell sorter (FACSAria; BD) at the University of Chicago Flow Cytometry Facility.
Quantitative PCR (Q-PCR) analysis.
Real-time (RT) PCRs were run and analyzed on a MYiQ system with SYBR green SuperMix (both from Bio-Rad Laboratories). The primers used were pk58 and pk59 (see Table S1 in the supplemental material) for the MP2 region and pk61 and pk62 (see Table S1) for the spacer between the MP2 regions. For the analysis of the M5 mononucleosomes, the primers used were pk71 and pk72 (see Table S1 in the supplemental material) for the M5 region and pk156 and pk157 (see Table S1 in the supplemental material) for the spacer between the M5 regions. The PCR conditions were 95°C for 30 s, 64°C for 45 s, and 72°C for 60 s for 40 cycles. The values were normalized for the copy number and primer efficiencies using the Pfaffl method (26).
RT-PCR analysis of transcripts in MP2/M5 knock-in clones.
Total RNA was made from DT40 cells with RNA STAT-60 (Tel-Test Inc.), recovered in 50 μl, and stored at −80°C. Equal amounts of RNA were used for making cDNA with the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Real-time PCRs were run and analyzed on a MYiQ system with SYBR green SuperMix (both from Bio-Rad Laboratories). The primers used were pk142 and pk24 (see Table S1 in the supplemental material) for the IgL V region and gg-actin1 and gg-actin2 (see Table S1) for the chicken actin region. The PCR conditions were 95°C for 30 s, 64°C for 45 s, and 72°C for 60 s for 40 cycles. The data from chicken actin were used as a reference for the relative quantification of IgL V region levels using the Pfaffl method (26).
Identification of somatic mutations.
Mutations in knock-in clones were detected by PCR cloning using Pfu polymerase (Agilent Technologies), and primers PK64 and PK65 (see Table S1 in the supplemental material) were used for PCR cloning with Pfu polymerase at 95°C for 30 s, 67°C for 30 s, and 72°C for 160 s for 25 cycles and cloning in a PCR cloning kit (Zero Blunt Topo; Invitrogen). DNA sequencing was performed by the University of Chicago Cancer Research Center DNA Sequencing Facility.
DNA synthesis.
The 147-bp MP2, M5, or Lytechinus variegatus 5S (35) nucleosome positioning sequences were cloned into pUC19 so that the nucleotide sequences flanking MP2, M5, or 5S were homologous. MP2-247, M5-247, and 5S-247 (5S rRNA sequence) were amplified by PCR to contain 50 bp of DNA flanking each side of the positioning sequence, Cy5 on the 5′ end of the forward strand, and Cy3 on the 5′ end of the reverse strand. 5′-Amine-labeled primers (Sigma) were conjugated with Cy3-N-hydroxysuccinimide (NHS) ester or Cy5-NHS (GE Healthcare) and purified by reverse-phase high-performance liquid chromatography (HPLC) (Vydac C18). The forward primer was mgp1, and the reverse primer was mgp2 (see Table S1 in the supplemental material). Amplified DNA was purified by HPLC on a Gen-Pak Fax ion-exchange column (Waters).
Nucleosome preparation.
Nucleosomes for exonuclease III (ExoIII) mapping were reconstituted by salt double dialysis as previously reported (19) with 1 μg of Cy3/Cy5-labeled MP2-247 or M5-247 DNA, 3 μg of lambda DNA (Invitrogen), and 1.5 μg of purified histone octamer (HO). The DNA and HO were mixed in 50 μl of 0.5× Tris-EDTA (TE) (pH 8.0) with 2 M NaCl and 1 mM benzamidine (BZA). The sample was loaded into an engineered 50-μl dialysis chamber, which was placed in a large dialysis tube with 80 ml of 0.5× TE (pH 8.0), 2 M NaCl, and 1 mM BZA. The large dialysis tube was extensively dialyzed against 0.5× TE with 1 mM BZA. The 50-μl sample was extracted from the dialysis button and purified by sucrose gradient centrifugation.
Competitive reconstitutions.
Competitive reconstitutions were performed as previously described (19). Reconstitutions were prepared in 2 M NaCl, 0.5× TE, and 1 mM BZA with 6 ng/μl labeled MP2-247, M5-247, or 5S-247 DNA (5S rRNA sequence); 50 ng/μl buffer DNA; and 10 ng/μl of HO in a volume of 50 μl. To minimize variation in DNA and HO concentrations, we first prepared an HO and buffer DNA master mixture that was split and combined with each DNA stock. Each DNA sample was then split into thirds and dialyzed separately. Each sample was dialyzed against the same reservoir containing 0.2 liter of 2 M NaCl, 0.5× TE, and 1 mM BZA. The concentration of salt in the dialysis reservoir was slowly reduced to 200 mM over 24 h; the samples were then dialyzed overnight against 0.5× TE and 1 mM BZA to reduce the final NaCl concentration to 1 mM NaCl. The reconstitution products were examined by PAGE, scanned with a Typhoon 8600 variable-mode imager (GE Healthcare), and analyzed with ImageQuant (GE Healthcare).
Electrophoresis mobility shift assay (EMSA).
The population of positioned and depositioned nucleosomes on the MP2 and M5 sequences was resolved by 5% native acrylamide gel with 0.3× TBE at 300 V for 1 h and imaged with Cy5 using a Typhoon 8600 variable-mode imager (GE Healthcare). The fraction of centrally positioned nucleosomes was calculated by measuring the image intensity within a box drawn around the positioned nucleosome band and dividing it by the intensity within a box drawn around all nucleosome bands using ImageQuant software (Invitrogen) with local median background subtraction enabled.
Exonuclease III mapping.
The nucleosome positions within the MP2-247 or M5-247 DNA molecules were determined by ExoIII mapping as previously reported (19). Reactions were carried out with 10 nM nucleosomes in 30 U/ml of ExoIII (New England BioLabs [NEB]) and buffer 1 (NEB) at 37°C. At each time point, 7 μl of the reaction mixture was quenched with a final concentration of 20 mM EDTA. A final concentration of 1 mg/ml of proteinase K and 0.02% SDS was added at each time point to remove the histone octamer from the DNA. Samples were separated by 8% denaturing PAGE in 7 M urea and 1× TBE. The sequence markers were prepared with a SequiTherm Excel II DNA-sequencing kit (Epicentre) using the Cy5- or Cy3-labeled primers, MP2-247 or M5-247 DNA template, and either ddATP or ddTTP. The results were imaged by a Typhoon 8600 variable-mode imager (GE Healthcare), which detects Cy3 and Cy5 separately in the same gel. The Cy3 and Cy5 ladders could be loaded in the same lanes to increase the accuracy of the mapping-gel readout.
RESULTS
Controlling nucleosome positioning and stability within the IgL locus.
To explore how SHM is influenced by nucleosomes within chromatin, we placed two copies of the strong NPS, MP2, that inhibited AID access in vitro (33) into the active lambda gene of mutating DT40 B cells by homologous recombination (Fig. 1). The cell line we used is a variant of DT40 cells that is an AID knockout and expresses AID as a transgene (AID-IRES-GFP) (2); all 25 ψV IgL genes are deleted (2) to make sure that these cells do not undergo IgL gene conversion. We chose a 147-bp MP2 sequence as an NPS for our study, since it is reported to be a strong nucleosome positioning sequence (28). It has a significant number of AID hot spots (14.3%), very similar to the IgL gene (11.6%). We integrated two MP2 sequences, with a spacer of 76 bp between them, into the IgL locus in such a way that the MP2 sequences would be at 235 bp and 458 bp from the transcription start site, which is the peak region of SHM in Ig genes.
Fig 1.
Map of the rearranged Ig light chain locus in the chicken B-cell line DT40. The locus contains the leader, V, J, and C regions of the IgL gene. The strategy of knocking in 2 MP2s (440 bp) by targeted integration is shown.
Nucleosome occupancy and positioning appear to be partially regulated by the underlying DNA sequence (16, 22, 32, 43). AA/TT and/or TA dinucleotides spaced every 10 bp and out of phase with GC dinucleotides that are also spaced at 10 bp appear to have the highest preference for forming nucleosomes (17). The MP2 sequence is a variant of the strong 601 positioning sequence, which was determined by Selex experiments (17). The MP2 sequence has a significant number of AA, TT, and TA dinucleotides at around 10-bp intervals that are out of phase with GC dinucleotides (Fig. 2A).
To validate our results with the stable nucleosome positioning sequence, MP2, we created a control sequence that is less favored to interact with histone proteins, lacking all the AA/TT/TA repeats that are a signature of a strong NPS, eventually reducing its affinity for the histone octamer, which is expected to make it a weaker positioning sequence, and to test the influence of nucleosome stability and positioning on AID accessibility. During the design of the 147-bp control sequence, we replaced all AA/TT/TA repeats; however, we kept the number of GCs and AID hot spots the same as that of the MP2 sequence (Fig. 2B). When this control sequence M5 is aligned with MP2, there is no periodicity of either of AA/TT/TA dinucleotides or GC dinucleotides (Fig. 2A and B).
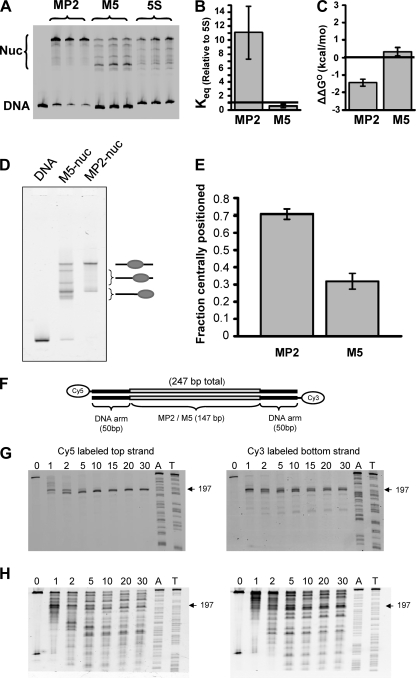
To assess the influence of the DNA base changes, which convert MP2 to M5, on the DNA-histone binding affinity, we carried out competitive nucleosome reconstitutions (40). Nucleosomes were reconstituted with histone octamer; an excess of low-affinity competitor DNA; and either the MP2-247, M5-247, or 5S-247 DNA molecules that were fluorophore labeled at the 5′ ends. The 5S positioning sequence was used to allow comparison to previous competitive reconstitution studies (40). A dynamic equilibrium between free DNA and DNA wrapped around a histone octamer is established, and EMSA (Fig. 3A) is used to determine the equilibrium constant, Keq, between these DNA states. We determined Keq relative to the 5S sequence for MP2 (11 ± 4) and M5 (0.6 ± 0.2) (Fig. 3B) from the intensities of the nucleosome bands relative to the DNA band (Fig. 3C). The ratio of the relative Keq for MP2 and M5 indicates that MP2 is 18 (11/0.6) times more likely to form a nucleosome than M5. We also determined the relative free energy of nucleosome formation with MP2-247 (−1.4 ± 0.2 kcal/mol) and M5-247 (0.4 ± 0.3 kcal/mol) relative to the 5S-247 DNA molecule from the following equation: ΔΔG = −kBT(ln(Keq/Keq-5S), where kBT = 0.6 kcal/mol (where kBT is the thermal energy, which is the Boltzmann constant times room temperature, 25°C) (Fig. 3C). This demonstrates that MP2 has a 1.8-kcal/mol lower free energy than M5 and that nucleosomes containing MP2 are significantly more stable than nucleosomes containing M5.
Fig 3.
Properties of MP2 and M5 NPSs. (A, B, and C) Competitive reconstitution. The probability and differences in the relative free energy of nucleosome formation were determined by competitive reconstitution. (A) An electromobility shift assay was done to quantify the fraction of nucleosomes that formed within MP2-247 (lanes 2 to 4), M5-247 (lanes 5 to 7), and 5S-247 (lanes 8 to 10) in the presence of low-affinity and unlabeled carrier DNA. Competitive reconstitution was done in triplicate for the MP2, M5, and 5S sequences, and each lane represents a separate reconstitution. (B) Equilibrium constants (Keq) for the formation of nucleosomes with MP2 (Keq = 11 ± 4) and M5 (Keq = 0.6 ± 0.2) relative to the 5S positioning sequence. (C) The difference in the free energy for nucleosome formation between 5S-247 and either MP2 (ΔΔG = −1.4 ± 0.2) or M5 (ΔΔG = 0.4 ± 0.3). (D and E) Electrophoretic mobility shift assay. (D) Five percent native PAGE gel of nucleosomes reconstituted on MP2-247 or M5-247 DNA. The mobility of a nucleosome through the gel is dependent upon its position on the DNA. The symbols on the right of the gel represent nucleosomes that are either centrally located or located towards/at the end of the DNA molecule. (E) Fraction of centrally positioned nucleosomes with respect to all nucleosome positions from MP2-247 and M5-247. (F, G, and H) Exonuclease III mapping of MP2 and M5 nucleosome positioning sequences. (F) Map of the nucleosome and flanking DNA. (G and H) Exonuclease assays (for details, see Materials and Methods) (33). The Cy5- or Cy3-labeled nucleosomes were treated with exonuclease III (G, lanes [from left] 1 to 8, MP2, and H, lanes 1 to 7, M5, show increasing incubation times from 0 to 30 min). The sequencing ladders (lanes A and T) were prepared with ddATP and ddTTP. The error bars in panels B, C, and E represent the standard deviations of three independent measurements.
To determine the influence of this reduction in DNA-histone binding on nucleosome positioning, we quantified nucleosome positions by EMSA of nucleosomes reconstituted with the MP2 sequence and the M5 sequence (Fig. 3D). Each sequence was centrally located within a 247-bp DNA test molecule (Fig. 3F). In the EMSA, the lowest-mobility band contained nucleosomes that are centrally located within the DNA molecule and the highest-mobility band contained nucleosomes that are located at either end of the 247-bp DNA molecule (19) (Fig. 3D). Figure 3D clearly shows that nucleosomes within the MP2 sequence are largely centrally located, with the only other position being at the ends of the DNA molecule. The M5 control sequence has a reduced fraction of nucleosomes at the central position and additional shifted positions and is largely positioned at the ends of the DNA molecule (Fig. 3D). Quantification of the band intensities found that 70% of nucleosomes are centrally positioned within the MP2 DNA molecule, whereas less than 35% of the nucleosomes within M5 are centrally positioned (Fig. 3E).
To confirm that the findings in the gel shift analysis are due to changes in nucleosome position, we performed exonuclease III mapping of both the MP2 positioning sequence and the M5 sequence (Fig. 3F to H). In this experiment, a 247-bp linear DNA was used, in which the central 147-bp DNA is either the MP2 or the M5 sequence (Fig. 3F). Purified nucleosomes were reconstituted with the same 247-bp DNA molecules that were labeled at the 5′ end with Cy5 and at the other end with Cy3. The nucleosomes were treated with exonuclease III (Fig. 3F) and subsequently analyzed by denaturing polyacrylamide gel electrophoresis. Nucleosomes containing the MP2 sequence had a single stall position about 50 bp into the DNA molecule for both strands (Fig. 3G). This confirms that the nucleosomes are centrally located, with a 147-bp footprint. However, exonuclease III mapping of nucleosomes containing the M5 positioning sequence showed a number of stall positions (Fig. 3H) that are consistent with nucleosome positions observed by EMSA (Fig. 3D and E), which confirms that positioning within DNA molecules containing M5 is significantly reduced relative to MP2.
After confirming that the M5 sequence has low affinity for histones, we replaced either the first, the second, or both copies of the MP2 positioning sequence with the less efficient M5 positioning sequence (Fig. 4A). Similar to the MP2-MP2 knock-in construct (Fig. 1), we created three control knock-in constructs, namely, M5-MP2, MP2-M5, and M5-M5.
Fig 4.
Replacing either the first, second, or both MP2s with M5. (A) Four combinations of MP2 and M5 inserted in the DT40 Ig lambda locus. (B) Transcription levels in the IgL V region in the four knock-in clones. The histograms show mRNA levels relative to those of the MP2-MP2 knock-in clone; the values are normalized to chicken β-actin levels. The data represent means and standard deviations (SD) of three independent experiments.
The knock-in plasmids were constructed by cloning genomic sequences that flank the integration site in the IgL gene. A surface IgM-positive clone of DT40 ψV knockout cells was used for transfection so that targeted integration into the IgL locus could be easily detected by the loss of surface IgM expression. The targeted integrations were confirmed by Southern blotting, with the MP2/M5 region and V region of the IgL gene as probes (data not shown). The blasticidin (Bsr) drug marker gene present between two LoxP sites (Fig. 1) was excised by treating the cells with tamoxifen. Incidentally, the AID-IRES-GFP transgene is also flanked by two LoxP sites and is likely to be excised after treatment with tamoxifen. We used a very low concentration of tamoxifen (25 nM) for 24 h so that a fraction of cells retained the AID-IRES-GFP transgene. Cell clones that were GFP positive and had the blasticidin drug marker gene excised were selected, and the blasticidin drug marker gene excision was confirmed by PCR and Southern blotting (data not shown).
To test whether the presence of a strong or weak NPS influenced transcription through the IgL gene, we performed RTQ-PCR in the V region of the IgL gene in MP2-MP2, M5-MP2, MP2-M5, and M5-M5 knock-in clones, respectively, and observed that knocking in a pair of strong positioning sequences (MP2) does not decrease transcription through the IgL gene (Fig. 4B).
To confirm whether nucleosomes are assembled at the MP2 sequence in the DT40 cells, we performed a micrococcal nuclease (MNase) assay for the MP2-MP2 knock-in clones. We treated nuclei from these cells with MNase, gel eluted mononucleosomes (i.e., a 150-bp band), and then performed PCR amplification for the positioning sequence, as well as the spacer region. In Fig. 5A, lanes 1 and 2 are PCR bands with MP2 sequence-specific primers (pk58 and pk59) (see Table S1 in the supplemental material), whereas lanes 3 and 4 are specific for the spacer region (pk61 and pk62) (see Table S1 in the supplemental material). Lanes 6 and 7 are control PCRs, with the genomic DNA from the MP2-MP2-containing cell clones as templates. We observed a very strong band for the nucleosome region compared with the spacer region, suggesting that indeed nucleosomes were assembled at the MP2 sequence in the DT40 cells. Quantitation by Q-PCR analysis showed that the MP2 region was ∼7-fold more abundant than the spacer region (Fig. 5B), confirming nucleosome assembly at the MP2 sequence. Similarly, to compare the relative stability of nucleosomes at the M5 sequence, Q-PCR analysis was performed with the gel-eluted mononucleosomes from the M5-M5 knock-in clones using primers specific for the M5 sequence (pk71 and pk72) (see Table S1 in the supplemental material), as well as the spacer region (pk156 and pk157) (see Table S1), and observed that the M5 region was only ∼2-fold more abundant than the spacer region (Fig. 5B). These results, combined with our observation that MP2 is 7 times more abundant than the spacer region, suggest that nucleosome occupancy at the M5 sequence is about four times less than at the MP2 sequence in DT40 cells. This is consistent with our in vitro results showing that MP2 is significantly more stable than M5 (Fig. 3).
Fig 5.
Nucleosomes are assembled at the MP2 and M5 sequences in vivo. (A) MP2-MP2 cells. Lanes 1 and 2, 147-bp amplification band with MP2-specific primers using the mononucleosomes as the template; lanes 3 and 4, 144-bp amplification band with spacer-specific primers using the mononucleosomes as the template; lane 5, 100-bp DNA ladder (100 to 1,000 bp); lane 6, 147-bp and 370-bp amplification bands with MP2-specific primers using the genomic DNA (gDNA) from the MP2-MP2 knock-in clones; lane 7, 144-bp amplification band with spacer-specific primers using the genomic DNA from the MP2-MP2 knock-in clones as the template. (B) Fold stability of the MP2 and the M5 positioning sequences in the MP2-MP2 and M5-M5 DT40 knock-in clones. The histograms represent the relative abundance of either the MP2 or the M5 positioning sequence compared to the respective spacer regions, as analyzed by Q-PCR. The values are normalized for the copy number and primer efficiencies. The data represent means and SD of two independent experiments.
Nucleosomes significantly influence the mutation pattern of the IgL locus.
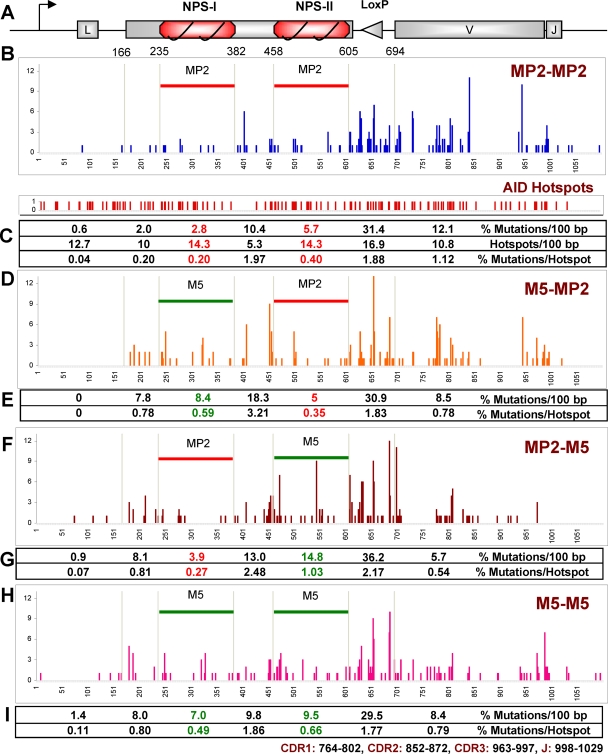
The MP2-MP2 knock-in clones were cultured for 5 weeks to acquire mutations in the IgL gene. A 1.2-kb PCR product of genomic DNA encompassing MP2, as well as the VJ region, was amplified and sequenced (Fig. 6A). Figure 6B shows the somatic hypermutation pattern of the MP2-MP2 knock-in clones. In the strong nucleosome positioning sequences (MP2), we observed a reduction in mutation frequencies compared with the flanks (Fig. 6C). The number of mutations was 2- to 4-fold reduced in both copies of the MP2 nucleosome positioning sequence compared with the spacer region and the neighboring IgL V region. Moreover, mutation frequencies per AID hot spot in the two nucleosome positioning sequences were also considerably lower, despite a similar proportion of AID hot spots (Fig. 6C). We observed a predominance of single-nucleotide substitutions with few insertions and deletions (see Fig. S1A in the supplemental material). The clones show very few mutations in A/T bases and a preference for transversion mutations (see Fig. S1B and S2 in the supplemental material), as was found by others in DT40 cells (2). Mutations at G bases were more numerous than at C bases in the regions surrounding MP2 (Fig. 7B); this relationship was, however, reversed in MP2 (Fig. 7A). Finally, we found mutations were most significantly suppressed in the central region of the MP2 nucleosome, while mutations near the entry/exit regions of the nucleosome were the least suppressed (Fig. 7C and E). These results indicate that the presence of stably positioned nucleosomes in the immunoglobulin gene significantly affects the accessibility of AID to and the mutation patterns within Ig genes.
Fig 6.
Ig light chain sequence analysis of the nucleosome positioning sequence knock-in clones. (A) Map of lg gene with 2 MP2s (not to scale); the triangle represents the two recombined loxP sites. (B, D, F, and H) Mutations in 1.1 kb from the start of transcription (= 1); the numbers on the y axis represent point mutations at the indicated positions in MP2-MP2 (B), M5-MP2 (D), MP2-M5 (F), and M5-M5 (H). 1 to 165, IgL gene containing the leader region; 235 to 382, first NPS (MP2/M5); 383 to 457, spacer between two NPSs; 458 to 605, second NPS; 606 to 695, loxP site generated from the Bsr marker excision; 695 to 1100, IgL gene containing V and J regions. (C, E, G, and I) Summary of mutations. CDR, complementarity determining region.
Fig 7.
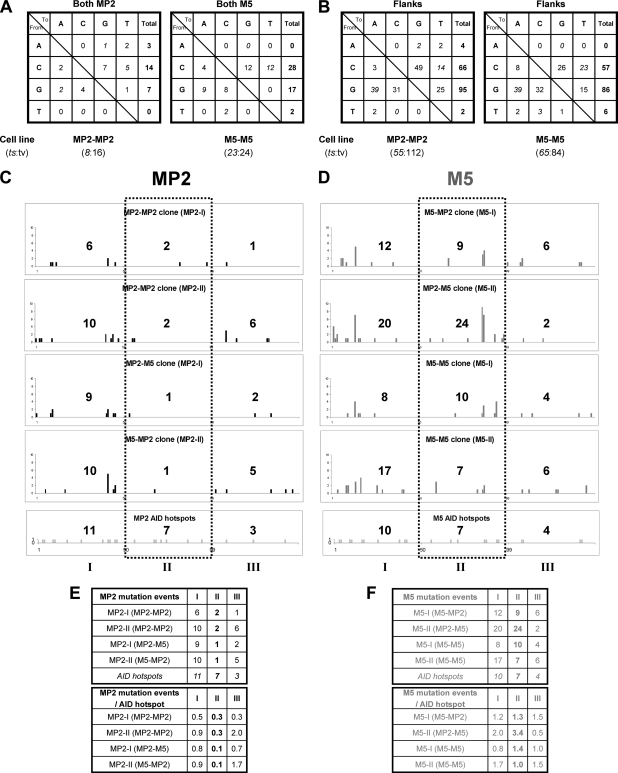
Mutation events in the MP2 and the M5 sequences. (A and B) Patterns of nucleotide substitutions within 2 NPSs (147 bp) of MP2-MP2 and M5-M5 nucleosome positioning knock-in clones (A) and 0.8 kb of flanking DNA that includes 5′ of the first NPS, the spacer between the two NPSs, and 3′ of the second NPS (B). The ratios of transitions (ts) to transversions (tv) are also shown. (C and D) Histograms showing distribution of mutations across the 147-bp region of either MP2 or M5 sequence. The 147-bp positioning sequence was divided into three 49-bp regions (I, II, and III), and the total number of mutation events is shown in each region. The bottom row shows the positions and the number of AID hot spots (WRC and GYW) in the three regions. (E and F) Summary of mutations in the 147-bp region of either the MP2 or M5 sequence. MP2-I (MP2-MP2) is the first of the two MP2 inserts, and so on. (Top) All mutation events. (Bottom) Mutations per AID hot spot in the three 49-bp regions of the MP2 and M5 sequence, respectively.
Reduction in nucleosome stability alters the mutation pattern within the IgL locus.
After 5 weeks of culturing, we analyzed the mutation profiles of the M5-MP2, MP2-M5, and M5-M5 knock-in clones and compared them with the SHM profile of MP2-MP2 knock-in clones. With the M5-MP2 construct, the percentage of mutations in the M5 region was around 3 times higher than in the corresponding MP2 region of the MP2-MP2 construct (P = 0.0026), whereas mutations in the second MP2 region were almost the same (Fig. 6D). We also observed significant increases in the percentage of mutations in the neighboring regions of M5 (Fig. 6E). Similarly, with MP2-M5, the percentage of mutations in the M5 region was around 3 times higher than in the corresponding MP2 region in the MP2-MP2 construct (P = 0.0001), whereas the mutation frequency in the first MP2 region was almost the same (Fig. 6F and G). Finally, when we replaced both copies of the MP2 sequence with M5, we observed a significant increase in the percentage of mutations in both copies of M5 (Fig. 6H). Very similar to the M5-MP2 and MP2-M5 constructs, the first M5 again showed around a 3-fold increase in the percentage of mutations compared to MP2 (P = 0.0089), and the second M5 showed around a 2-fold increase in the percentage of mutations (P = 0.0447), but not in the region 3′ of the second NPS (Fig. 6I). Replacing the MP2 sequence with the M5 sequence also changed the mutation pattern within the nucleosome. The mutations within the MP2 sequence are suppressed in the center of the nucleosome relative to the DNA entry/exit region of the nucleosome (Fig. 7C). In contrast, the distribution of mutations within the M5 sequence remained relatively constant (Fig. 7D). Furthermore, these patterns were not influenced by whether the adjacent NPS was M5 or MP2.
We observed a predominance of single-nucleotide substitutions with few insertions and deletions in MP2-MP2, M5-MP2, MP2-M5, and M5-M5 knock-in clones (see Fig. S1A and S2 in the supplemental material). All four types of knock-in clones showed very few mutations in A/T bases and a preference for transversion mutations (see Fig. S1B in the supplemental material); two independent cell clones for each of the four types of nucleosome combinations were very similar (data not shown). In the total 1.1 kb sequenced for each cell type, there were more mutations at G bases than at C bases (see Fig. S1B in the supplemental material); this reflects the SHM pattern outside the NPSs (Fig. 7B), but within the NPSs, the C and G frequencies were reversed (Fig. 7A). Considering the three 49-bp regions of the NPSs separately, the central region is less mutated in the MP2 sequence than the entry/exit points, i.e., sequences on the left and right (Fig. 7C and D). However, the M5 sequence has more mutation events in the central 49-bp region, although both the MP2 and the M5 sequences have the same number (seven) of AID hotspots in this region (Fig. 7E and F). Thus, we conclude that when we replaced a strong nucleosome positioning sequence (MP2) with a weak positioning sequence (M5), mutations in M5 were much higher than in MP2.
DISCUSSION
The rules for nucleosome assembly deduced from total-genome analyses (22, 32) enabled us to change the MP2 sequence with high affinity for histone cores to the low-affinity M5 sequence. While regulatory mechanisms also play a major role in chromatin structure (16, 43), the striking difference in the biophysical properties of MP2 and M5 nucleosomes validates previous conclusions that the primary DNA sequence can considerably affect the propensity for its assembly into nucleosomes (22, 32).
The findings show that both the presence and stability of the nucleosome strongly influence mutation patterns during SHM: the number of mutations was significantly reduced in both copies of the NPS MP2, and the numbers of mutations per AID hot spot in the MP2 sequence were considerably lower than those of the IgL gene, despite a similar proportion of hot spots. Moreover, replacement of a stable NPS (MP2) with a less stable sequence (M5) resulted in higher numbers of mutations. We conclude that the stability of nucleosomes in the IgL gene significantly affects the outcome of the somatic hypermutation process.
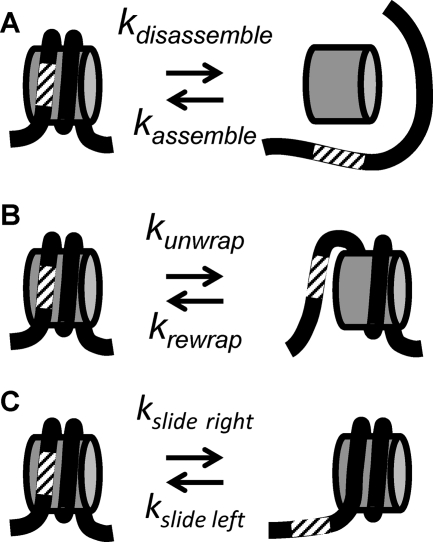
There are two mechanisms by which AID could gain access to nucleosomal DNA. One possible mechanism is that the DNA must be nucleosome free for AID to access DNA (Fig. 8A). Nucleosomes could be disassembled by RNA transcription, which is important for deamination by AID (33). Histone chaperones (7) and/or chromatin remodeling (5, 36) could further enhance nucleosome disassembly. In this model, the mutation rate should remain constant through the nucleosome positioning sequence, since mutations occur only when the DNA is nucleosome free. Alternatively, the nucleosomes could be retained during transcription, and AID could gain access to DNA through partial DNA unwrapping (Fig. 8B) and/or nucleosome repositioning (Fig. 8C). These nucleosome alterations expose DNA that is originally wrapped into a nucleosome. Transcription through a nucleosome (13) and nucleosome remodeling could induce nucleosome repositioning (5) and enhance nucleosomal-DNA unwrapping fluctuations, which occur rapidly, many times a second (14). In this model, the mutation rates are expected to be highest near the entry/exit regions and lowest near the nucleosome center. DNA site exposure by unwrapping is greatest near the entry/exit regions and exponentially reduced for DNA sites further into the nucleosome (1, 29). Also, since nucleosomes are spaced by 30 to 60 bp (42), nucleosome repositioning is restricted, so again, sites near the DNA entry/exit regions are the most accessible. The unwrapping/repositioning models are not mutually exclusive, so both could occur in vivo.
Fig 8.
Models of nucleosomal DNA exposure for SHM. (A) The nucleosome can be disassembled and reassembled, which requires all of the DNA-histone contacts to be broken. (B and C) Alternatively, the DNA can partially unwrap from (B) or reposition (C) with respect to the histone octamer. Both of these models maintain DNA-histone contacts.
Our studies of the mutation levels within both MP2 and M5 and our biophysical characterization of MP2 and M5 nucleosomes indicate that both the disassembly and unwrapping/repositioning mechanisms occur in vivo. M5 has both a reduced affinity for the histone octamer and reduced nucleosome positioning strength relative to MP2. Therefore, M5 has the ability to enhance mutations by both nucleosome repositioning and disassembly. Furthermore, the mutation patterns within MP2 and M5 indicate that both mechanisms occur. The mutation frequency within the MP2 sequence is greatest near the entry/exit regions, which is consistent with the unwrapping/repositioning models (Fig. 8B and C). This mutation pattern suggests that the dominant mechanism by which AID gains access to highly stable nucleosomes is by either the unwrapping (Fig. 8B) or the repositioning (Fig. 8C) model. Repositioning would likely be affected by the neighboring nucleosomes. Since there is no increase in the mutability of MP2 when the other NPS is M5 rather than MP2 (Fig. 6), the current study supports unwrapping (Fig. 8B). However, the mutation frequency within the M5 sequence is relatively constant, in relative concordance with the AID hot spot distributions across M5 (Fig. 7D and F); this finding supports the disassembly mode for M5 (Fig. 8A). Since the M5 sequence has a lower affinity for the histone octamer, this DNA sequence could reduce nucleosome occupancy by both enhancing the rate of nucleosome disassembly and reducing reassembly. The ∼4-fold reduction in nucleosome occupancy produced by replacing MP2 with M5 (Fig. 3 and 5) also resulted in a 3-fold increase in the mutation frequency (Fig. 6). The combination of these results strongly suggests that AID accesses less stable nucleosomes largely by the disassembly model.
These in vivo findings are interesting, given our previous results with in vitro assays of the mutability by AID of MP2 embedded in a supercoiled circular plasmid, pKMP2 (33). In contrast to naked pKMP2 DNA, MP2-containing nucleosomes in the plasmid were not mutated by AID alone. However, there were ample mutations in the MP2 nucleosome sequences when the plasmids were transcribed. The arrangement and sequences of the two MP2 and spacer elements were the same as used in this paper. The conclusion was that AID cannot access nucleosomes unless they are transcribed (33). Clearly the Ig lambda gene in the DT40 cells used here is continuously transcribed and MP2 is mutated, although at a reduced frequency, compared with the flanking DNAs. It is not simple to make a direct comparison between MP2 and flanking DNAs in the in vitro experiments; some regions without a defined nucleosome were slightly more mutable than MP2, while others had very few mutations (see Fig. 5K in reference 33). We do not know whether and where nucleosomes were randomly placed in the ∼3.9-kb plasmid outside the MP2 regions. Interestingly, in the in vitro experiments, the pKMP2 plasmid was transcribed by T7 RNA polymerase that is considerably smaller than the pol II operating in vertebrate cells. It has been shown that nucleosomes containing a T7 promoter are completely displaced by T7 pol (39). The results reported here show that RNA polymerase Pol II can deal with nucleosomes more efficiently than AID and suggest that subtle epigenetic events may be best investigated in vivo.
AID requires single-stranded DNA for access (8) and operates processively (27). Our previous data support the idea that negative supercoils behind the RNA polymerases (pol) extrude single-stranded C's as AID targets (34). The propagation of negative supercoils is probably inhibited at the next nucleosome. This is supported by comparing the processivity of AID in cell-free assays with that in vivo (37). In vitro, up to 16 consecutive C's are deaminated by AID in stretches of up to ∼60 total nucleotides (nt) (37). In vivo, a maximum of 4 or 5 consecutive C's, but usually only 2, are mutated in up to a total of 11 nt (37), but nevertheless, the AID processivity is also significantly greater than expected in vivo (P < 0.01). Thus, the average length of the spacers between nucleosomes apparently allows sufficiently large stretches of negative supercoils to develop and become accessible to AID.
We find that while nucleosome occupancy influences SHM within the Ig locus, the level of transcription is not significantly influenced by a strong NPS. Only a 2-fold change in transcription was also observed in budding yeast when the high-affinity NPS, 603, was inserted at the beginning of the CUP1 gene (10). Interestingly, Gaykalova et al. (10) found that 603 did not position nucleosomes well in vivo, while our studies found that the MP2 sequence significantly positions nucleosomes. In addition, a high-throughput sequencing study (11) found that a 601 NPS inserted in the EF1a promoter of the human factor IX gene contained well-positioned nucleosomes that then became depositioned as the gene was silenced.
An important difference between these previous studies and ours is the location of the NPS with respect to the transcription start site. We inserted the two NPSs into the transcribed region of the gene (235 and 458 bp, respectively, from the transcription start site). The 603 sequence in the studies by Gaykalova et al. was at the first nucleosome within the transcribed region of the gene (56 bp from the transcription start site), and the 601 sequence in the Gracey et al. studies was inserted in the promoter upstream from the transcription start site. The combinations of these results are consistent with the idea that chromatin remodeling may selectively influence the nucleosome position near the promoter regions of genes. Interestingly, in vitro measurements by Gaykolova et al. suggested that chromatin remodeling near the promoter could be responsible for their observed depositioning at the 603 NPS.
SHM experiments in mice showed a certain periodicity of the mutation patterns in a highly mutable Ig transgene, RS (20). The results were consistent with the conclusion that the Ig gene was organized into nucleosomes but that different cells had different nucleosome phasings and that the nucleosome pattern was relatively stable for a given cell for several generations throughout the hypermutation process.
Clearly, in the current study, the inserted MP2 must have caused rather stable nucleosome phasing. In MP2-MP2, on average, there are seven times more MP2 sequences than spacer sequences in nucleosomes (Fig. 5B). Even the M5 sequence is slightly more nucleosomal than the spacer (Fig. 5B), and about 30% of the M5 sequences are stably associated with histone cores (Fig. 3E). We found that the free energy of the MP2 sequence, which is a variant of the 601 sequence, is 1.4 kcal/mol lower than that of the 5S sequence, while that of the M5 sequence is 0.4 kcal/mol higher than that of the 5S sequence. The free energy of the MP2 sequence is about 1 kcal/mol (23, 40) higher than that of the original 601 sequence and is similar to that of one of the highest in vivo NPSs (40, 41). This indicates that the MP2 sequence is at the extreme of high-affinity nucleosome positioning sequences in vivo and that it is not representative of the typical nucleosomal DNA in vivo. The M5 sequence is lower affinity than the well studied 5S NPS but similar to mouse minor satellite DNA (40). These sequences are about 0.6 kcal/mol lower than the average affinity of mouse DNA to histone octamers. This indicates that the M5 sequence is representative of typical nucleosomal DNA in vivo and that our observations of the influence of nucleosomes on SHM at the M5 sequence may apply to nucleosomes in the native Ig locus. Thus, this study suggests that the limits of nucleosome positioning for Ig genes may be below MP2 stability and around or below that of M5. It will be interesting to investigate the propensity for nucleosome positioning of endogenous Ig genes in mice and humans. It seems possible that the variable regions of Ig genes have evolved for low nucleosome stability to enhance the chance for increased access to AID, DNA repair factors, and error-prone DNA polymerases and hence the creation of maximal variability by somatic hypermutation.
Supplementary Material
ACKNOWLEDGMENTS
We are greatly indebted to J. Widom (Northwestern University, Evanston, IL) for advice on the design of the M5 sequence. We thank H. Arakawa and J. M. Buerstedde (Institute of Molecular Radiology, Neuherberg, Germany) for the DT40 CL18 and ψV knockout cells, W. Buikema and C. Hall for DNA sequencing, and R. Duggan for flow cytometric cell sorting. We are grateful to B. Kee for technical advice and use of her instrument for real-time PCR. We thank T. E. Martin for constructive discussions of these experiments and critical reading of the paper.
P.K. thanks the Lady Tata Memorial Trust, United Kingdom, and the Cancer Research Institute for postdoctoral fellowships. J.A.N. acknowledges support from an Ohio State Comprehensive Cancer Center Pelotonia predoctoral fellowship. This work is supported by the NIH (AI047380 and AI053130 to U.S. and GM083055 to M.G.P.) and a Burroughs-Wellcome Career award to M.G.P.
We have no conflicting financial interests.
Footnotes
Published ahead of print 5 March 2012
Supplemental material for this article may be found at http://mcb.asm.org.
REFERENCES
- 1. Anderson JD, Widom J. 2000. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. Y 979–987 [DOI] [PubMed] [Google Scholar]
- 2. Arakawa H, Saribasak H, Buerstedde JM. 2004. Activation-induced cytidine deaminase initiates immunoglobulin gene conversion and hypermutation by a common intermediate. PLoS Biol. 2:E179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Betz AG, et al. 1994. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell 77:239–248 [DOI] [PubMed] [Google Scholar]
- 4. Chaudhuri J, et al. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature 422:726–730 [DOI] [PubMed] [Google Scholar]
- 5. Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304 [DOI] [PubMed] [Google Scholar]
- 6. Conley ME, et al. 2009. Primary B cell immunodeficiencies: comparisons and contrasts. Annu. Rev. Immunol. 27:199–227 [DOI] [PubMed] [Google Scholar]
- 7. Das C, Tyler JK, Churchill ME. 2010. The histone shuffle: histone chaperones in an energetic dance. Trends Biochem. Sci. 35:476–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dickerson SK, Market E, Besmer E, Papavasiliou FN. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fritz EL, Papavasiliou FN. 2010. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 24:2107–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaykalova DA, et al. 2011. A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res. 39:3520–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gracey LE, et al. 2010. An in vitro-identified high-affinity nucleosome-positioning signal is capable of transiently positioning a nucleosome in vivo. Epigenetics Chromatin 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hochedlinger K, Plath K. 2009. Epigenetic reprogramming and induced pluripotency. Development 136:509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jin J, et al. 2010. Synergistic action of RNA polymerases in overcoming the nucleosomal barrier. Nat. Struct. Mol. Biol. 17:745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li G, Levitus M, Bustamante C, Widom J. 2005. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12:46–53 [DOI] [PubMed] [Google Scholar]
- 15. Li G, Widom J. 2004. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11:763–769 [DOI] [PubMed] [Google Scholar]
- 16. Locke G, Tolkunov D, Moqtaderi Z, Struhl K, Morozov AV. 2010. High-throughput sequencing reveals a simple model of nucleosome energetics. Proc. Natl. Acad. Sci. U. S. A. 107:20998–21003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lowary PT, Widom J. 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276:19–42 [DOI] [PubMed] [Google Scholar]
- 18. Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 19. Manohar M, et al. 2009. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J. Biol. Chem. 284:23312–23321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michael N, et al. 2002. Effects of sequence and structure on the hypermutability of immunoglobulin genes. Immunity 16:123–134 [DOI] [PubMed] [Google Scholar]
- 21. Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. 2004. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 279:52353–52360 [DOI] [PubMed] [Google Scholar]
- 22. Morozov AV, et al. 2009. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Res. 37:4707–4722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Partensky PD, Narlikar GJ. 2009. Chromatin remodelers act globally, sequence positions nucleosomes locally. J. Mol. Biol. 391:12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perez-Duran P, de Yebenes VG, Ramiro AR. 2007. Oncogenic events triggered by AID, the adverse effect of antibody diversification. Carcinogenesis 28:2427–2433 [DOI] [PubMed] [Google Scholar]
- 25. Peters A, Storb U. 1996. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity 4:57–65 [DOI] [PubMed] [Google Scholar]
- 26. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pham P, Bransteitter R, Petruska J, Goodman MF. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature 424:103–107 [DOI] [PubMed] [Google Scholar]
- 28. Poirier MG, Bussiek M, Langowski J, Widom J. 2008. Spontaneous access to DNA target sites in folded chromatin fibers. J. Mol. Biol. 379:772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polach KJ, Widom J. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130–149 [DOI] [PubMed] [Google Scholar]
- 30. Popp C, et al. 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rai K, et al. 2008. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135:1201–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Segal E, et al. 2006. A genomic code for nucleosome positioning. Nature 442:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shen HM, et al. 2009. The activation-induced cytidine deaminase (AID) efficiently targets DNA in nucleosomes but only during transcription. J. Exp. Med. 206:1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen HM, Ratnam S, Storb U. 2005. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol. Cell. Biol. 25:10815–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simpson RT, Stafford DW. 1983. Structural features of a phased nucleosome core particle. Proc. Natl. Acad. Sci. U. S. A. 80:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith CL, Peterson CL. 2005. ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65:115–148 [DOI] [PubMed] [Google Scholar]
- 37. Storb U, Shen HM, Nicolae D. 2009. Somatic hypermutation: processivity of the cytosine deaminase AID and error-free repair of the resulting uracils. Cell Cycle 8:3097–3101 [DOI] [PubMed] [Google Scholar]
- 38. Storck S, Aoufouchi S, Weill JC, Reynaud CA. 2011. AID and partners: for better and (not) for worse. Curr. Opin. Immunol. 23:337–344 [DOI] [PubMed] [Google Scholar]
- 39. Studitsky VM, Clark DJ, Felsenfeld G. 1994. A histone octamer can step around a transcribing polymerase without leaving the template. Cell 76:371–382 [DOI] [PubMed] [Google Scholar]
- 40. Thastrom A, et al. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213–229 [DOI] [PubMed] [Google Scholar]
- 41. Widlund HR, et al. 1997. Identification and characterization of genomic nucleosome-positioning sequences. J. Mol. Biol. 267:807–817 [DOI] [PubMed] [Google Scholar]
- 42. Widom J. 1992. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl. Acad. Sci. U. S. A. 89:1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y, et al. 2009. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat. Struct. Mol. Biol. 16:847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.