Abstract
The detection of noncoding transcription at multiple enhancers within the mammalian genome raises critical questions regarding whether and how this activity contributes to enhancer function. Here, using in vivo analysis of a human growth hormone (hGH) transgene locus, we report that activation of a domain of noncoding transcription adjacent to the long-range hGH-N enhancer, HSI, is established by the enhancer independent of any interactions with its target promoter. We further demonstrate that the appearance of this enhancer-linked noncoding transcription is temporally and spatially concordant with induction of hGH-N in the embryonic pituitary. Finally, we show that the level of transcriptional enhancement of hGH-N by HSI is directly related to the intensity of HSI-dependent noncoding transcription and is fully independent of the structure of the locally transcribed RNA. These data extend our understanding of the relationship of long-range enhancer activity to enhancer-dependent noncoding transcription and establish a model that may be of general relevance to additional mammalian loci.
INTRODUCTION
Alterations in the cellular transcriptome drive critical developmental pathways. These changes in mRNA representation are heavily dependent on selective controls of gene transcription. Whereas the accuracy of transcriptional initiation is established by promoter elements, the timing and levels of gene expression are often controlled by regulatory determinants that are remotely situated from their target promoters (10, 45). The activities of these remote determinants track with alterations in the structure and higher-order configuration of defined chromatin domains (22, 38, 48). What remains unclear is how these remote elements are themselves activated and organized and how they impact target promoters.
Mechanisms of enhancer function are intimately linked to the recruitment of macromolecular complexes that impart covalent and higher-order alterations in chromatin structure (4, 19, 20). These epigenetic modifications can be confined to trans-factor recruitment sites or they can be quite extensive. A number of models link specific subsets of structural alterations at enhancers to the transcriptional activation (13, 34). The way in which distal enhancers impact their target promoters is of current interest. One model invokes a linear progression (“tracking”) of factors and/or polymerase II (PolII) from the enhancer recruitment site to the target promoter (17, 51). In another model, the enhancer-promoter interactions are mediated via long-range “looping” that brings the corresponding sites into direct contiguity (6, 35, 47). The extensive use of technologies designed to capture native chromatin conformations has revealed that major alterations in higher-order chromatin structure may be a common occurrence in both the activation and the repression of target genes by remote regulatory elements (11, 26, 28). Finally, alterations in the composition and configuration of a locus can be linked to reorganization of the nuclear architecture, localizing a gene within regions conducive to either transcriptional enhancement or to solidification of gene silencing (36).
Recent studies indicate that the function of an enhancer may be accompanied by its own transcription (12, 16, 33). A role for this enhancer-linked “noncoding” transcription in gene regulation has been inferred from genome-wide surveys (30, 31). Such observations, while intriguing, are essentially descriptive. Mechanistic links between enhancer transcription and promoter activity need to be more fully explored. The structures of these noncoding transcription units are in most cases undefined, and whether their establishment is enhancer autonomous or dependent on cooperative interactions between the enhancer and its target promoter has been only minimally explored. Finally, it remains to be established whether noncoding transcriptional activity at enhancer elements plays an essential role in gene expression and whether such activity relates to transcription per se or is dependent on actions of the enhancer-encoded RNAs. It is clear from recent studies that maximally informative experimental approaches to these complex problems necessitate the exploration of transcriptional regulatory circuits in intact, physiologically relevant settings (43).
The mammalian growth hormone gene (GH) gene is selectively and robustly expressed in somatotrope cells of the anterior pituitary. In the case of the human GH locus (hGH), this expression is under the control of a set of remote regulatory elements that constitute the hGH locus control region (hGH LCR) (27). The hGH LCR encompasses four DNase I hypersensitive sites (HS) in pituitary chromatin located 14.5 to 32 kb 5′ to the hGH-N transcription start site (27) (Fig. 1A). These elements are collectively sufficient to establish an autonomous chromatin domain that supports robust, pituitary-specific, and developmentally appropriate expression of an hGH transgene irrespective of its site of integration in the mouse genome (2, 27, 44). A single, defined pituitary-specific component of this LCR, HSI, located 14.5 kb 5′ to the hGH gene promoter, serves an essential function in the hGH-N transcriptional enhancement (22). Site-specific inactivation of critical trans-acting factor binding sites within HSI results in a 20-fold decrease in hGH-N expression in vivo (22). Thus, HSI constitutes a model of a potent long-range enhancer of in vivo gene expression.
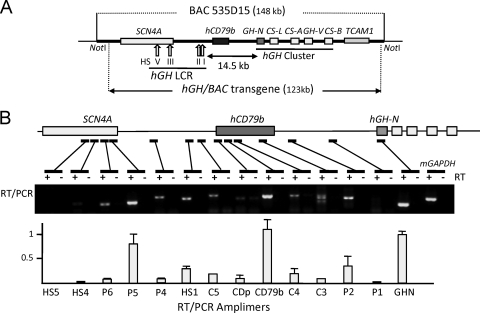
Fig 1.
Transcript mapping across the hGH/BAC transgene in the mouse pituitary revealed a peak of transcriptional activity across the hCD79b region. (A) Map of the hGH/BAC transgene. The 123-kb hGH/BAC transgene, released from the originating BAC clone by NotI digestion, was used to generate the hGH/BAC transgenic mouse lines (49). Each structural gene is indicated by a labeled box (exonic substructures are not shown). The vertical arrows labeled with roman numerals indicate the positions of DNase I hypersensitive sites (HS) that form in pituitary chromatin and constitute the hGH LCR. HSI,II are pituitary-specific DNase I HS of the hGH LCR. (B) Transcriptional profile across the hGH/BAC transgene. The presence of transcripts corresponding to each of 14 sites across the hGH/BAC transgene locus was determined by RT-PCR of transgenic mouse pituitary RNA. The position of each amplimer is indicated on the below the map by a bar and the sequences are listed on Table 1. Reactions were carried out in the presence or absence of reverse transcriptase (+ and −, respectively). Amplified cDNAs were analyzed on a 1% agarose gel. The concentration of each amplified RNA segment, shown in the histogram, was determined by PhosphorImager quantification of corresponding [α-32P]dCTP-labeled RT-PCR products generated by a separate set of reactions. Each value was normalized to a parallel amplification of genomic DNA to adjust for minor differences in amplification efficiency. The data are normalized to an arbitrary value of 1.0 for hGH-N mRNA. A robust transcriptional domain was detected over hCD79b, located between HSI and its target hGH-N.
The hGH LCR, extending 5′ from HSI to HSV, is itself bidirectionally transcribed by PolII in the pituitary, and this transcriptional activity is HSI dependent (23). Remarkably, a gene encoding a B-cell-specific, transmembrane receptor protein is situated immediately 3′ to HSI, between HSI and its target hGH-N promoter (3) (Fig. 1A). This hCD79b locus is robustly transcribed in the pituitary, as well as in B cells, although Igβ, the encoded protein, is only produced in B cells (5). Site-specific inactivation of HSI results in a loss of hCD79b transcription in the pituitary with a corresponding loss of hGH-N gene expression (5). This same mutation has no adverse effect on hCD79b in B cells (5). Remarkably, insertion of a PolII termination element between HSI and hCD79b represses hCD79b transcription in the pituitary with a comparable loss in hGH-N expression. This PolII terminator insertion has no effect on the formation of HSI itself, nor does it repress the bidirectional transcription between HSI and HSV (23). These data support a model in which noncoding transcription across the hCD79 region, immediately 3′ to HSI, plays an essential and specific role in HSI-mediated long-range enhancement of hGH-N gene transcription. In the present study, we explore the mechanistic basis for the activation and function of this domain of noncoding transcription. The data revealed a quantitative relationship between noncoding transcription 3′ to HSI and the enhancement of hGH-N transcription. These data further demonstrated that the HSI enhancer activity is a direct effect of noncoding transcription per se and is fully independent of the structure of this encoded RNA.
MATERIALS AND METHODS
BAC transgene modifications and generation of transgenic mouse lines.
Modifications were introduced in the hGH/BAC transgene according to a published protocol (15). The primer sets for constructing shuttle vectors are shown in Table 1. Modified BAC DNAs were linearized with NotI prior to microinjection. The released 123-kb DNA fragment was microinjected into fertilized mouse oocytes (C57BL/6×SJL) to generate the transgenic lines. The University of Pennsylvania Transgenic & Chimeric Mouse Core carried out all microinjection procedures under IACUC approved protocols. Transgenic founders were identified by PCR and Southern blot analysis of tail genomic DNA. The −8.0CD79b and −1.3CD79b transgenic lines had been previously described (5).
Table 1.
Oligonucleotides used in this study
| Method and oligonucleotide | Sequence (5′–3′) |
|---|---|
| hGH/BAC transgene modification | |
| (CDΔ1.6)hGH/BAC | |
| L-arm-5′ | CAGGGCGCGCCGACGATTTAGCATCTCTTCCTCTCCTGGG |
| L-arm-3′ | GTGGAATTCACTGGGAGAAGATTCAGTCCAGGTC |
| R-arm-5′ | GGGCGAATTCTGTGCGACTATGTCCTGTGTCC |
| R-arm-3′ | GCCATTAATTAATATGCCCATTACAACAGCCT |
| (ΔCDλ)hGH/BAC | |
| L-arm-5′ | Same as with (CDΔ1.6)hGH/BAC |
| L-arm-3′ | CCACCTCGAGAAGATTCAGTCCAGGTC |
| R-arm-5′ | AGGAGTCATCTCGAGGTCGCCCCATGACCTGGGTGCAG |
| R-arm-3′ | CCCACTTAATTAAGGGCTCACAGATGCCACATTC |
| λ-5′ | CAATCTCGAGCCGGATTCGGTATGGCTG |
| λ-3′ | GCTTCTCGAGACACCTTATGTTCTATAC |
| RT-PCR | |
| HSV-5′ | CTTGCCCAGTCCTCACACTT |
| HSV-3′ | CTGAGGCTTCTGTCCTCCTT |
| HSIV-5′ | TGCCTCTACGTGGACATCTC |
| HSIV-3′ | TATCAGCAGAGAGTGCACAA |
| P6-5′ | CTGGGTGGCGTAGAGATG |
| P6-3′ | GACCCACGTTGTCGTAGTTG |
| P5-5′ | GCCTCAAAACCTGATTGG |
| P5-3′ | GGAGATCTCTGAGGCTGG |
| P4-5′ | GCTGTATTCTTCCAGACAAG |
| P4-3′ | GAGCTAAGCTATGAGGATGC |
| HS1-5′ | CCAAGCCTTTCCCAGTTATAC |
| HS1-3′ | GATCTTGGCCTAGGCCTC |
| P3-5′ | CTTCCCCCAGAAGACTGA |
| P3-3′ | GGAGGAAAACGTGTAACTGC |
| C5-5′ | AGACCCTCTGCCTTCCAACCATGGCAT |
| C5-3′ | CACTGGGAGAGATTCAGTCCAGGTC |
| CDp-5′ | GGACAGGTGCCTATTTCGCTC |
| CDp-3′ | GACCCCAAACCCGTGACAAC |
| CD79b-5′′ | GACCATGGCCAGGCTGGCGTTGTCTCCTGT |
| CD79b-3′ | AGCGTCTGGATCATGATGATACCATCCTTC |
| C4-5′ | GGAGGAAGATCACACCT |
| C4-3′ | ATCCCCAGAGAACTCCC |
| C3-5′ | GTTCTCTGGGGATGGACGGGACCCAGCC |
| C3-3′ | TGGCATGCAGCCCCGTTCCAG |
| P2-5′ | TGCTCAGACCAGCCTATGCA |
| P2-3′ | TCAACAGGAAGTGGAGCACA |
| P1-5′ | GATTACAAGCGCCCACTACC |
| P1-3′ | GAGAGAATAAGCCAGGAGGTG |
| GHN-5′ | TTTGACAACGCTATGCTCCG |
| GHN-3′ | GCCAAAAGGGTCATCATCTC |
| mGAPDH-5′ | GCCAAAAGGGTCATCATCTC |
| mGAPDH-3′ | CTGCTTCACCACCTTCTTGA |
| λ5-5′ | CGCTTATGCGGATTATTGCCGTAG |
| λ5-3′ | ACCTCTCTGCCTGCGATGGTTGGAG |
| λm-5′ | TACCGCAAGCAGCTTGGCCTG |
| λm-3′ | AGCATCAGCTAACTCCTTCG |
| λ4-5′ | GTCAGTCAGTGCGTGAAGCC |
| λ4-3′ | CTACTCGCTACTGCGCTGGC |
| λ3-5′ | GGACGTTTCTATAAGATGCGTG |
| λ3-3′ | GTCGGTTGTATTTCCCTCCAG |
RNA isolation and Northern blot analysis.
Total RNA was extracted from tissue samples with RNA-Bee (Tel-Test) according to the manufacturer's procedure. A 10-μg portion of each RNA sample was separated on a 1.2% agarose gel containing 2.2 M formaldehyde in 1× morpholinepropanesulfonic acid and transferred to a Zeta-probe membrane (Bio-Rad). The membrane was incubated with 32P-labeled probes in hybridization buffer at 65°C overnight and washed (2× SSC–0.1% sodium dodecyl sulfate [SDS] and 1× SSC–0.1% SDS) at 65°C, and signals were detected by exposure to X-ray film (Kodak).
Reverse transcription-PCR (RT-PCR) and quantitative RT-PCR (qPCR).
Isolated total RNAs were treated with RQ DNase (Promega) and purified using an RNeasy minikit (Qiagen). In all cases, 1 μg of purified RNA was reverse transcribed by SuperScript III reverse transcriptase (Invitrogen) with random primers, amplified using a primer set (Table 1), and separated on 1% agarose gel. For qPCR analyses, 1 μg of purified RNA was reverse transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). The levels of hGH-N (Hs00236859_m1), hCD79b (Hs00236881_m1), λ, and mGAPDH (Mm99999915_g1) RNAs were assessed by using TaqMan Universal PCR Master mix (Applied Biosystems), and real-time PCRs were performed by a 7900HT machine and analyzed using SDS2.2 software.
DIG-labeled RNA probe synthesis, embryo sectioning, and in situ hybridization.
0.5 kb cDNA of each gene (hGH-N, hCD79b, Pit-1, Prop-1, Shh, and Pitx1) in the pGEM-T Easy vector (Promega) was transcribed in vitro to generate digoxigenin (DIG)-labeled antisense RNA probes. Embryos were collected at specified days postcoitus and fixed overnight at 4°C in 4% paraformaldehyde. The fixed embryos were then washed in phosphate-buffered saline (PBS), immersed in 30% sucrose in PBS overnight at 4°C, embedded in OCT (Sakura), quick frozen on dry ice, and cryosectioned at 20 μm. Sectioning and in situ hybridizations were performed as described previously (37). Briefly, sections were washed in PBS, fixed in 4% paraformaldehyde for 10 min, washed in PBT (1× PBS with 0.1% Tween 20), and permeabilized with 1.0 μg of proteinase K/ml for 10 min. Sections were then washed in PBT, fixed in 4% paraformaldehyde for 5 min, acetylated in 0.25% acetic anhydride in 0.1 M triethanolamine for 10 min, washed in PBT, and air dried. The sections were then incubated with RNA probes in 150 μl of hybridization buffer (10 mM Tris [pH 7.5], 600 mM NaCl, 1 mM EDTA, 0.25% SDS, 10% dextran sulfate, 1× Denhardt solution, 200 μg of yeast tRNA/ml, 50% formamide), covered with Hybri-slips (Sigma), and incubated overnight at 65°C in a humidified box. The next day, coverslips were removed and washed for 30 min in 1× SSC–50% formamide at 65°C. The slides were then transferred to TNE (10 mM Tris [pH 7.5], 500 mM NaCl, 1 mM EDTA) at 37°C for 10 min, incubated in RNase A in TNE for 30 min at 37°C, and washed in TNE for 10 min. Sections were washed in 2× SSC for 20 min at 65°C, washed twice in 0.2× SSC for 20 min at 65°C, and transferred into MABT (100 mM maleic acid, 150 mM NaCl, and 0.1% Tween 20 [pH 7.5] with NaOH). The slides were blocked in 2% blocking reagent (Roche) in MABT with 20% heat-inactivated goat serum (Sigma) for at least 1 h and then incubated overnight at 4°C with anti-DIG alkaline phosphatase-conjugated antibodies (Roche) in MABT blocking buffer with 5% serum. Slides were washed in MABT and equilibrated in NTM (100 mM Tris [pH 9.5], 100 mM NaCl, 50 mM MgCl2) for 10 min. Color detection was performed using BM purple (Roche) in a humidified box at room temperature for 4 to 12 h.
RESULTS
Transcription within the hGH transgene locus 3′ to HSI is robust and noncontiguous with the hGH-N promoter.
We previously mapped the location of the B-cell-specific gene, hCD79b, to a site immediately 3′ to HSI, between the hGH LCR and hGH-N (3, 5) (Fig. 1A). Although hCD79b is transcribed in the pituitary, its encoded product, the immunoglobulin receptor Igβ subunit, remains B cell restricted (5). This noncoding transcription of hCD79b in the pituitary is dependent on the activity of HSI, whereas the transcription of hCD79b in B cells is controlled by a distinct set of B-cell-specific promoter-proximal determinants (1, 39, 46). Our studies initially suggested a model in which hCD79b transcription represented a functionally neutral “bystander” event reflecting its fortuitous location in an “activated” chromatin environment between hGH and its LCR in pituitary (5). This proposed functional neutrality was subsequently brought into question by the observation that targeted repression of hCD79b transcription in the pituitary resulted in a dramatic loss of hGH-N expression (23). These data led us to conclude that noncoding transcription across the hCD79b region plays an essential role in the HSI-dependent enhancement of hGH-N expression.
Prior to exploring the basis for hCD79b transcription in the pituitary and its mechanistic relationship to HSI enhancer function, we mapped the transcriptional architecture across an intact hGH locus in the pituitary of an hGH/BAC transgenic mouse. The hGH/BAC 123-kb transgene encompasses the full hGH LCR, the five-gene hGH cluster, and the intervening hCD79b (Fig. 1A). Analysis of pituitary RNA from an hGH/BAC transgenic mouse revealed various levels of transcription throughout the LCR and a discrete robust peak of transcription coincident with hCD79b. The level of hCD79b transcription was equivalent to that at the hGH-N gene, as assessed by mRNA abundance (Fig. 1B). Of note, there was a gap in transcription between the 3′ end of hCD79b and the hGH-N promoter (“P1” amplimer). This gap is concordant with a corresponding gap of PolII occupancy in this region (23). These data led us to conclude that transcription across hCD79b is robust and circumscribed and that PolII does not linearly track between hCD79b and hGH-N.
The domain of noncoding transcription 3′ to HSI can be established in the pituitary independent of the hCD79b promoter.
hCD79b transcription in pituitary is HSI dependent (5). The basis for this transcriptional activation is unclear. We sought to determine whether the hCD79b promoter, as previously defined in B-cell-expression studies (1, 21, 39, 46), is necessary for this activity. The promoter and contiguous 5′ end of the hCD79b gene (extending through exon 2) was deleted from the hGH/BAC transgene (Fig. 2A). Six corresponding (CDΔ1.6)hGH/BAC transgenic mouse lines were established. The structure of the (CDΔ1.6)hGH/BAC transgene was validated by detailed mapping of the corresponding mouse genome (data not shown). Remarkably, the expression of hCD79b mRNA in the mouse pituitary was unaltered by this extensive 5′ deletion. In contrast, this same 5′-terminal deletion resulted in complete loss of hCD79b expression in the B cells of the (CDΔ1.6)hGH/BAC mice (Fig. 2B). It was of note that the amount and size of the hCD79b mRNA, as assessed by Northern analysis of each of the (CDΔ1.6)hGH/BAC mouse pituitaries, was essentially the same as in the pituitary of the mouse carrying the intact hGH/BAC transgene (Fig. 2B and data not shown). The lack of any apparent change in size of the hCD79b mRNA generated from the CDΔ1.6)hGH/BAC transgene most likely reflects the small sizes of exon 1 (76 bp) and exon 2 (51 bp). We further demonstrated that the mRNA generated from the 5′-deleted hCD79b gene [(CDΔ1.6)hGH/BAC] was initiated within intron 2 (5′ rapid amplification of cDNA ends assay; unpublished data). The position of this new intronic transcription start site was fully consistent with the size of the resultant hCD79b mRNA in the pituitary, as detected on Northern analysis. These data led us to conclude that hCD79b transcription in the pituitary is not dependent on the hCD79b promoter/enhancer elements that had been previously defined to operate in B cells.
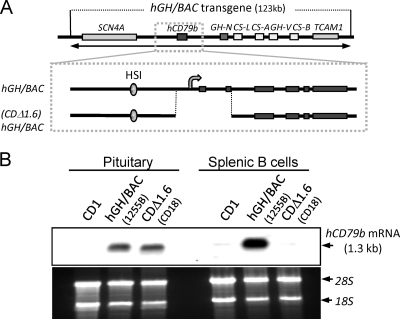
Fig 2.
hCD79b transcription can be activated in the pituitary in the absence of the hCD79b promoter. (A) Diagram of the hGH/BAC transgene and a derivative transgene lacking the 5′ terminus of hCD79b. A 1.6-kb deletion from the hGH/BAC encompassing the promoter and first two exons was created by homologous recombination in E. coli to generate (CDΔ1.6)hGH/BAC. The site of transcription initiation (promoter) encompassed by the deletion in (CDΔ1.6)hGH/BAC is indicated by a bent arrow. The structure of the transgene was confirmed by restriction mapping, targeted amplification, and sequencing across critical regions. The modified BAC DNA was released from the plasmid by NotI digestion, purified, and injected into fertilized mouse oocytes to generate transgenic mouse lines. (B) Deletion of the promoter and 5′ end of hCD79b [(CDΔ1.6)hGH/BAC transgene] selectively ablates hCD79b mRNA expression in B cells but not in the pituitary. Pituitary and B-cell RNAs from a mouse transgenic for the intact hGH/BAC (line 1255B) and the (CDΔ1.6)hGH/BAC line (CD18) were analyzed by Northern hybridization, along with a sample from a nontransgenic mouse (CD1) as a negative control. Membranes were probed with a segment of the hCD79b cDNA. The position of the hCD79b mRNA (∼1.3 kb) is indicated to the right of the autoradiograph. Visualization of 28S and 18S rRNA (lower panel) was used to control for gel loading and RNA quality. The data reveal that the promoterless transgene was expressed in pituitary, but not in splenic B cells of the same (CDΔ1.6)hGH/BAC mouse.
Enhancer action of the noncoding transcription domain 3′ to HSI is independent of RNA structure.
The key role played by hCD79b transcription in HSI enhancer function could reflect the structure of the encoded hCD79b RNA. Alternatively, it could reflect transcriptional activity through this region in a manner that is independent of hCD79b RNA sequence or structure. To distinguish between these two models, we replaced the hCD79b gene with an identically sized fragment of bacteriophage λ DNA [(ΔCDλ)hGH/BAC; Fig. 3A]. The replaced hCD79b segment extended from bp −500 (relative to the native hCD79b transcription start site) through the coding region, preserving only the 3′-processing signals. Six (ΔCDλ)hGH/BAC transgenic mouse lines were generated and studied. Remarkably, a prominent 3.3-kb λ RNA band was observed by Northern blotting of the pituitaries isolated from all but one of these lines (Fig. 3B and E). The expression of this “λ RNA” was pituitary specific (Fig. 3C). Based on the size of this RNA and the position of the retained hCD79b 3′-processing signal, the initiation of λ RNA transcription was mapped to a site located 0.7 kb within the λ DNA replacement cassette, ∼1.4 kb 3′ to HSI. These data led us to conclude that transcription can be initiated 3′ to HSI in a manner independent of native promoter elements. This conclusion was fully consistent with the analysis of the (CDΔ1.6)hGH/BAC transgene (Fig. 2).
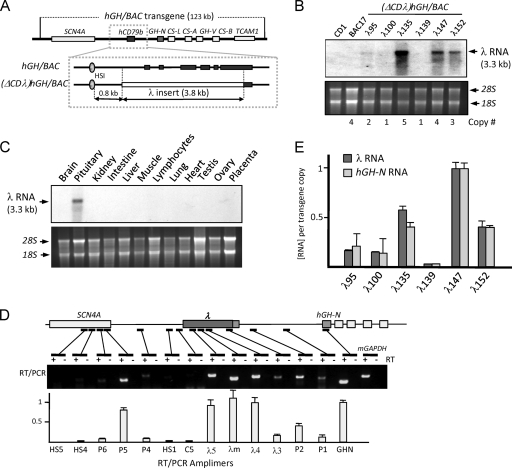
Fig 3.
The intensity of transcription 3′ to HSI is a direct, quantitative, and RNA-independent determinant hGH-N expression. (A) Replacement of the hCD79b gene in the hGH/BAC transgenes by a segment of λ phage DNA. An expanded view of the HSI/hCD79b region of the hGH/BAC is shown below the map of the hGH/BAC transgene. The hCD79b gene (exons; dark rectangles), extending from −0.5 kb to the middle of exon 6, was substituted with an identically sized fragment of bacteriophage λ DNA (3.8 kb, open rectangle). The segment of the hCD79b exon 6 containing its 3′ processing determinant was retained 3′ to the λ DNA insert. The resultant (ΔCDλ)hGH/BAC transgene was released from vector sequences and microinjected into fertilized mouse oocytes. Six transgenic lines were established and studied. (B) Northern blot analysis of pituitary RNA from (ΔCDλ)hGH/BAC transgenic mice reveals the expression of λ RNA. Total pituitary RNA from each of the six (ΔCDλ)hGH/BAC lines, from a nontransgenic control mouse (CD1), and from a mouse line carrying the intact hGH/BAC (line BAC17) were resolved on a 1.2% denaturing agarose gel, and Northern blot hybridization was performed using a λ DNA probe. Ethidium bromide-stained rRNA on the gel was used to assess equivalent RNA loading and quality control. The transgene copy number of each line is indicated below the gel. Five of the six (ΔCDλ)hGH/BAC lines expressed a 3.3-kb λ RNA. The levels of λ RNA varied among the lines (see also panel E). (C) Expression of the λ RNA from the (ΔCDλ)hGH/BAC transgene is restricted to the pituitary. A Northern blot of RNAs isolated from the indicated tissues of a (ΔCDλ)hGH/BAC transgenic mouse (λ135) was probed with a 32P-labeled λ DNA segment. rRNA controls are as described in panel B. The 3.3-kb λ transcript was expressed only in pituitary. (D) Transcription mapping across the (ΔCDλ)hGH/BAC transgene in the pituitary. Transgenic (ΔCDλ)hGH/BAC pituitary RNA was subjected to RT-PCR using the 14 primer sets (Table 1) shown below the transgene map. Amplification of mGAPDH mRNA served as a positive control. RT-PCR products were assayed on an ethidium bromide-stained agarose gel. The concentration of each amplified segment shown in the histogram was measured by PhosphorImager quantification of corresponding [α32P]dCTP-labeled RT-PCR. Each value was normalized to a parallel amplification of genomic DNA to adjust for minor differences in amplification efficiency. The peak of transcription 3′ to HSI (corresponding to the λ DNA insert) and the gap of transcription between this region and the hGH-N promoter from the (ΔCDλ)hGH/BAC transgene demonstrated a remarkable conservation with the pattern of transcription across the native locus (compare to Fig. 1). (E) Direct and quantitative relationship between λ RNA levels and hGH-N mRNA levels in the pituitaries of six (ΔCDλ)hGH/BAC transgenic lines. The histogram shows λ RNA levels and hGH-N mRNA levels as determined by real-time PCR. The results (± the standard deviation) were from averages of four independent experiments. Each value was normalized to the corresponding transgene copy number and was related to the level of the highest expressing line (λ147; defined as 1.0).
To further explore the relationship between the noncoding transcription 3′ to HSI and the activity of the hGH-N gene, we assessed the distribution of transcription in the pituitary across the (ΔCDλ)hGH/BAC transgene locus. The study revealed a peak of transcription coinciding with the λ DNA insert, followed by a gap in transcripts between the (λ) insert 3′ terminus containing the hCD79b 3′ processing signals and the hGH-N promoter (Fig. 3D). This transcriptional profile was remarkably consistent with that observed at the native locus (Fig. 1B); both studies demonstrated variable transcription at sites within the LCR region and robust transcription 3′ to HSI. Further, these studies revealed the same discontinuity between the HSI-dependent noncoding transcription domain (hCD79b or λ) and the hGH-N promoter. These two concordant sets of data led us to conclude that transcription does not track in a linear fashion from the HS1-dependent transcriptional domain to the hGH-N promoter.
The levels of λ RNA were next quantified in each of the six (ΔCDλ)hGH/BAC lines and compared to the corresponding levels of hGH-N mRNA. Each value, normalized to transgene copy, was plotted in arbitrary units (Fig. 3E). This analysis revealed a direct and quantitative relationship between the level of noncoding transcription 3′ to HSI and the level of hGH-N expression (Pearson r = 0.97). This quantitative relationship between hGH-N expression and λ RNA levels further allowed us to conclude that the enhancing activity of HSI is directly linked to the act of transcription 3′ to HSI rather than to the structure of the locally generated hCD79b transcripts.
The initiation of transcription across hCD79b parallels the activation of hGH-N during pituitary development.
The observation that transcription of hCD79b is tightly correlated with, and essential to, the activation of hGH-N led us to question whether hCD79b transcription was activated with the same developmental specificity as hGH-N. To test this, sagittal sections of the anterior region of hGH/BAC embryos 8.5 to 17.5 days postcoitus (dpc) were probed with DIG-labeled synthetic antisense RNAs. A series of well-established controls for pituitary developmental were used to validate embryo staging of each sample (Fig. 4A and B). Consistent with expectations, Pit-1, the POU-homeodomain protein essential to GH gene expression (52), was activated at 14.5 days within the nascent pituitary. This was followed 2 days later by the appearance of robust signals corresponding to hGH-N mRNA and hCD79b transcripts, as well as the endogenous mGH mRNA (Fig. 4C). Both hGH-N and hCD79b mRNAs appear within the same time window, between embryonic day 15.5 (e15.5) and e16.5. The coexpression of hGH-N and hCD79b RNAs was subsequently maintained through gestation (Fig. 4C) and into the postnatal period (data not shown). hGH-N activation was also observed to occur between e15.5 and e16.5 in an in situ hybridization analysis of an (ΔCDλ)hGH/BAC line (data not shown). These combined data support a functional linkage between the act of noncoding transcription 3′ to HSI (either hCD79b or λ) and hGH-N gene activation during pituitary development.
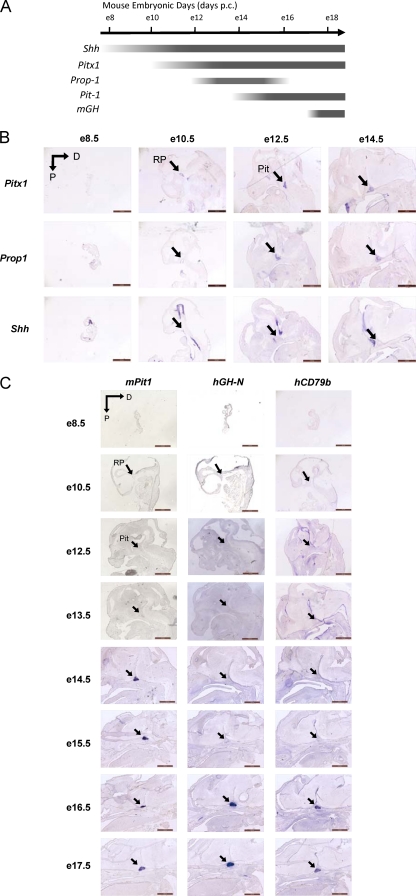
Fig 4.
Transcriptional activation of the hCD79b domain and activation of hGH-N expression are temporally concordant during pituitary development. (A) Timing of pituitary marker gene expression during mouse embryonic development. The approximate timing of mRNA expression of pituitary marker genes shown here is adapted and modified from Dattani and Preece (9). (B) Developmental time course of expression of Pitx1, Prop-1, and sonic hedgehog (Shh). In situ hybridization signals in the e8.5 through e14.5 mouse embryo heads of hGH/BAC (line BAC17) are shown. The transcription factor Pitx1 is an early pituitary-restricted maker initially expressed in Rathke's pouch at e9.5 (14, 52). This is followed by sequential activation of the genes encoding the transcription factors Prop-1 and Pit-1, at 12.5 and 13.5 dpc, respectively. Shh is expressed throughout the oral ectoderm except in the Rathke's pouch, creating a boundary between two ectodermal domains of Shh-expressing and -nonexpressing cells. Abbreviations: RP, Rathke's pouch; Pit, pituitary; D, dorsal; P, posterior. Scale bars, 1 mm. This study confirmed the embryonic dates of the samples as estimated from copulation timing. (C) Developmental time course of Pit1, hGH-N, and hCD79b. The transcriptional activation of Pit-1, hGH-N, and hCD79b was studied by in situ hybridization on hGH/BAC transgenic embryos (line BAC17) from e8.5 through e17.5, as indicated. Pit-1 mRNA signal was first detected at e14.5 (1). Its positioning is within the caudomedial region of the pituitary gland, a region that ultimately gives rise to somatotropes, lactotropes, and thyrotropes. Signals corresponding to hGH-N and hCD79b RNA first appear in the pituitary between e15.5 and e16.5. The timing of hGH-N activation was also assessed in (CDΔλ)hGH/BAC transgenic embryos; hGH-N mRNA was found to appear within the same time window and with the same pituitary-specific distribution as observed in the hGH/BAC embryos (data not shown).
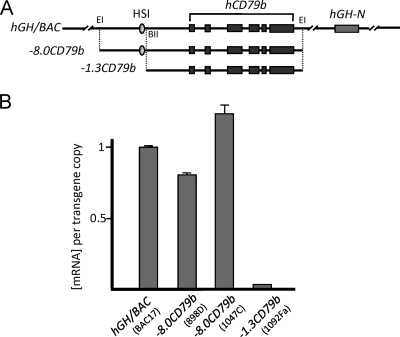
Transcription across hCD79b is established independent of interactions with the hGH-N promoter.
Recent studies have suggested that activation of enhancer-associated, noncoding transcription, generating “enhancer RNAs,” is dependent upon cooperative interactions between the enhancer and its target promoter (30). To test the relevance of this model to the hGH locus, we compared hCD79b transcription in the presence or absence of hGH-N (Fig. 5A). The −8.0CD79b transgene isolates the intact HSI-hCD79b region from the downstream hGH-N (5). Analysis of hCD79b mRNA expression in the −8.0CD79b transgenic pituitaries revealed that it was indistinguishable from the intact hGH/BAC transgene (Fig. 5B). In contrast, a more extensive deletion that also removes HSI (−1.3CD79b transgene; Fig. 5A) ablated hCD79b transcription in pituitary (Fig. 5B). These data confirmed that the domain of noncoding transcription across hCD79b is HSI-dependent and demonstrated that it can be fully activated in the pituitary in the absence of interactions with the target hGH-N promoter. This observation, along with the previous demonstration that transcription in this region is independent of the hCD79b promoter (Fig. 2), support a model in which HSI has a direct and possibly sufficient role in the activation of transcription across the hCD79b noncoding domain in the pituitary.
Fig 5.
Transcription of the hCD79b domain in the pituitary is HSI dependent and established independent of the target hGH-N promoter. (A) Map of the hGH/BAC transgene and two derived transgenes lacking hGH-N. The −8.0CD79b transgene is a 12-kb genomic fragment of the hGH/BAC that encompasses HSI of the hGH LCR and the full hCD79b gene. The −1.3CD79b transgene is a 5.6-kb genomic subfragment that contains the full hCD79b but excludes the HSI region. Abbreviations: EI, EcoRI; BII, BglII. (B) Quantitative determination of hCD79b mRNA in transgenic pituitaries. Pituitary RNAs from mice carrying the hGH/BAC, −0.8CD79b, and −1.3CD79b transgenes were assessed for levels of hCD79b mRNA in the pituitary by qPCR. Each result was normalized to the corresponding transgene copy number and is represented as a value relative to the expression of the intact hGH/BAC transgene (defined as 1.0). Each value and standard deviation on the histogram is derived from three separate studies. The transgene name and specific line designation are shown below the respective bars. Note that pituitary hCD79b expression requires HSI, but not the target hGH-N promoter.
DISCUSSION
Transcriptional regulation in higher organisms depends on an array of long-range regulatory elements. These elements can mediate locus insulation, transcriptional enhancement, and/or sublocalization of the region within the nucleus (4, 10). Communications between enhancers and their target promoters are of particular importance for control of tissue-specific gene expression as well as for establishing patterns of temporally restricted expression (20, 32, 50). Recent genome-wide studies have revealed that many transcriptional enhancers are transcribed and generate a variety of noncoding RNAs (30). The presence of transcription coincident with, or closely linked to, an enhancer determinant has been shown in some cases to be concordant with the activity of target genes (30). Of significant interest, these and related studies have yet to delineate critical relationships between enhancer-linked transcription and target gene expression. Outstanding questions include: how are these enhancer-linked transcriptional units established, are they temporally linked to and/or dependent upon cooperative interactions with their target promoters, and is their impact on target gene expression dependent on sequences and/or structures of the locally generated noncoding transcripts?
In the present study, we focused our analyses on the control of the human growth hormone gene (hGH-N) by its remote enhancer, HSI. Our previous observations revealed that a domain of noncoding transcription located immediately 3′ to HSI, encompassing hCD79b, plays an essential role in HSI-mediated enhancement of hGH-N expression in the pituitary (23). Selective repression of transcription through this region in transgenic mouse pituitary, by insertion of a PolII terminator immediately 3′ to HSI, results in a major loss of hGH-N expression. Importantly, the insertion of this transcription termination element does not alter the formation of HSI itself, nor does it impact on noncoding transcription within the regions of the LCR 5′ to HSI (23). These observations support a direct role for transcription through the hCD79b region in hGH-N gene activation.
Noncoding RNAs have been studied as key players in imprinting and gene dosage compensation (7, 18, 29). More recently, it has been shown that depletion of particular long noncoding RNAs can result in repression of neighbor coding genes (40). This suggests that at least in some situations noncoding RNAs may contribute to the enhancement of target gene expression (8, 25). This enhancement might reflect RNA-mediated recruitment of basal transcription factors, transcriptional activators, chromatin remodeling factors, and/or displacement of transcriptional repressors (40). Our present studies demonstrated that noncoding transcription may also work in an RNA-independent manner. Of particular note is the observation that the HSI-dependent enhancement of hGH-N expression is fully maintained when the structure of the noncoding transcription domain 3′ to HSI is switched from the native hCD79b sequence to a comparably sized segment of bacteriophage λ DNA [Fig. 3A, (ΔCDλ)hGH/BAC]. A quantitative analysis of six genetically distinct (ΔCDλ)hGH/BAC transgenic lines further revealed a positive concordance between the levels of λ DNA transcription and hGH-N expression (Fig. 3E). These results indicated that enhancement of hGH-N expression is directly and quantitatively related to the intensity of the noncoding transcriptional activity 3′ to HSI. The data further revealed that this enhancing activity is independent of the locally generated RNA transcripts. From these data, we conclude that the mechanistic linkage between the HSI-dependent domain of transcription and hGH-N expression reflects noncoding transcription per se and not some attribute of the locally generated RNA transcript sequence or structure.
The dependence of hGH-N expression on HSI activity, and the mechanistic linkage between HSI-enhancing activity and transcription across hCD79b, predicts that hCD79b transcripts should appear in the embryonic pituitary concordant with, or preceding, the appearance of hGH-N mRNA. This prediction was confirmed by in situ hybridization studies. The hCD79b and hGH-N transcripts were both initially detected between 15.5 and 16.5 dpc in the developing pituitary and were subsequently maintained into postnatal life (Fig. 4C). This coordinate spatial and temporal specificity of transcription support the mechanistic linkage between the HSI-dependent noncoding transcription and hGH-N gene activation.
The present study also addresses the questions of how transcription through the hCD79b region is activated in the pituitary and how this might relate to hGH-N activation. Kim et al. recently published a model of enhancer and target gene relations based on a tissue-specific enhancer in neuronal tissue (30). These researchers concluded that enhancer transcription is dependent on interaction of the enhancer with its target promoter. In the context of the hGH locus, this appears not to be the case. Analysis of a series of transgenic models revealed that transcription 3′ to HSI is dependent on HSI activity and independent of the target promoter. Several lines of evidence support this conclusion. The first was the observation that a transgene encompassing the HSI-hCD79b region could fully activate hCD79b transcription in the pituitary independent of the target hGH-N promoter (Fig. 5). Second, we found that activation of the hCD79b region was maintained in a transgene lacking the hCD79b promoter. This same deletion of the 5′ terminus of hCD79b effectively eliminated its transcription in B cells (5). Finally, transcription in this region was supported when the native sequences were replaced by a segment of λ DNA (Fig. 3). Based on these data we propose that this transcriptional domain is established in an HSI-dependent manner and does not require the presence or actions of the hGH-N target promoter. The findings lead us to conclude that the establishment of this domain of transcription serves as an initiating event in the pathway of hGH-N activation (Fig. 6).
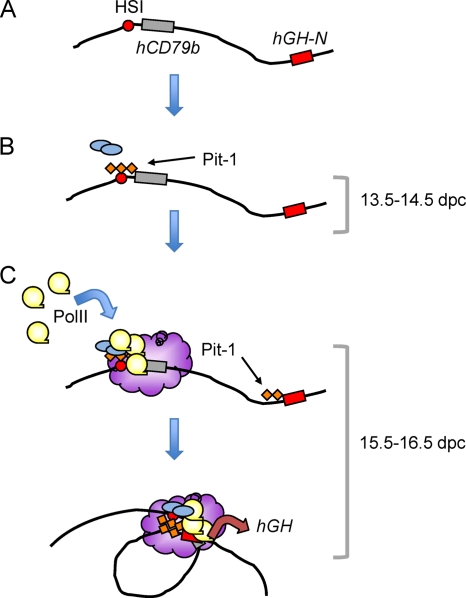
Fig 6.
Model of HSI-mediated long-range enhancer action. (A) The inactive hGH chromatin locus lacks a specific higher-order chromatin conformation (24). (B) The initial step in hGH-N LCR activation is triggered at e13.5 to e14.5 by binding of Pit-1 (orange diamonds) to an array of cognate binding sites at HSI. Pit-1 recruits additional transcriptional complexes (blue ovals), including HAT complexes that extend acetylation to, and increase the exposure of, the chromatin-embedded hGH-N promoter (53). (C) A domain of intense PolII activity is established by HSI over the hCD79b region and is brought into contiguity with the hGH-N promoter between e15.5 and e16.5 via a chromatin “looping” configuration. The formation of a localized region of intense transcriptional activity (purple cloud) occurs as an autonomous function of HSI, being independent of both hCD79b and hGH-N promoters (Fig. 2 and 3). Pit-1 is recruited to the array of cognate sites at the hGH-N promoter during this developmental window. Interactions between the HSI/hCD79b region and the hGH-N promoter are stabilized. This “looping” within the locus, possibly mediated by Pit-1 dimerization (24, 41), brings the HSI-generated “transcriptional factory” into close contact with the hGH-N promoter. This conformation supports stable and robust expression of hGH-N in the adult pituitary somatotrope.
In prior studies we had demonstrated that the chromatin configuration of the hGH locus in the adult pituitary places the hGH-N promoter in close proximity to HSI (24). This configuration is not present in the chromatin of nonexpressing tissues. In addition, we have previously shown that the pituitary-specific POU-homeodomain transcription factor Pit-1, a protein essential for hGH-N gene expression, occupies an array of cis-acting binding sites within HSI prior to binding its cognate sites within the hGH-N promoter (41, 42). The sequential assembly of the Pit-1 complexes at HSI and then the hGH-N promoter, separated as they are by 14.5 kb, may play a key role in establishing the “looping” structure between HSI/hCD79b region and hGH-N promoter in definitive somatotropes (24). This looping would position the intensely transcribed hCD79b domain, or the surrogate transcribed λ DNA domain, in close proximity to the hGH-N promoter (Fig. 6). Thus, the impact of the domain of noncoding transcription on hGH-N promoter activity may reflect the juxtaposing of a cis-acting “mini-PolII factory” established 3′ to the HSI enhancer element in close proximity to the hGH-N promoter. This model would be consistent with the direct impact of the transcription per se through this region, rather than a function of the encoded RNA transcripts, on enhancer function.
Altogether, our studies lead us to propose a model in which HSI-linked activation of the hCD79b domain of noncoding transcription may constitute an initial and critical step in the developmental pathway of the anterior pituitary. The present data suggest that the higher-order chromatin configuration of the hGH locus in the pituitary, in concert with the intense transcription immediately 3′ to the long-range enhancer, may activate high levels of hGH-N expression and sustain this robust activity throughout adult life. However, this model does not exclude roles for additional factors and interactions that solidify the connections between the enhancer-linked domain of noncoding transcription and the target hGH-N promoter. The identification and characterization of these additional determinants of long-range enhancer function present critical challenges for future studies.
ACKNOWLEDGMENTS
We thank the University of Pennsylvania Transgenic and Chimeric Mouse Facility (supported by National Institutes of Health [NIH] P30 grants DK019525, DK050306, and CA016520) for generation of the transgenic mice. We also thank Douglas Epstein, University of Pennsylvania, for critical review and comments, and Diane Dolson and Staci Rakowiecki for help with the in situ hybridization.
This study was supported by NIH grants R01 HD/DK25147 and R01 HD/DK046737 (to N.E.C. and S.A.L.).
Footnotes
Published ahead of print 19 March 2012
REFERENCES
- 1. Andersen B, Rosenfeld MG. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endo. Rev. 22:2–35 [DOI] [PubMed] [Google Scholar]
- 2. Bennani-Baiti IM, et al. 1998. DNase I-hypersensitive sites I and II of the human growth hormone locus control region are a major developmental activator of somatotrope gene expression. Proc. Natl. Acad. Sci. U. S. A. 95:10655–10660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennani-Baiti IM, Cooke NE, Liebhaber SA. 1998. Physical linkage of the human growth hormone gene cluster and the CD79b (Igβ/B29) gene. Genomics 48:258–264 [DOI] [PubMed] [Google Scholar]
- 4. Bulger M, Groudine M. 2010. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 339:250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA. 2004. Bystander gene activation by a locus control region. EMBO J. 23:3854–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Carter D, Chakalova L, Osborne CS, Dai Y-F, Fraser P. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623–626 [DOI] [PubMed] [Google Scholar]
- 7. Chan SW, et al. 2004. RNA silencing genes control de novo DNA methylation. Science 303:1336. [DOI] [PubMed] [Google Scholar]
- 8. Corcoran AE. 2010. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin. Immunol. 22:353–361 [DOI] [PubMed] [Google Scholar]
- 9. Dattani M, Preece M. 2004. Growth hormone deficiency and related disorders: insights into causation, diagnosis, and treatment. Lancet 363:1977–1986 [DOI] [PubMed] [Google Scholar]
- 10. Dean A. 2006. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 22:38–45 [DOI] [PubMed] [Google Scholar]
- 11. Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295:1306–1311 [DOI] [PubMed] [Google Scholar]
- 12. Drewell RA, Bae E, Burr J, Lewis EB. 2002. Transcription defines the embryonic domains of cis-regulatory activity at the Drosophila bithorax complex. Proc. Natl. Acad. Sci. U. S. A. 99:16853–16858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu Q, Manolagas SC, O'Brien CA. 2006. Parathyroid hormone controls receptor activator of NF-κB ligand gene expression via a distant transcriptional enhancer. Mol. Cell. Biol. 26:6453–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gage PJ, Suh H, Camper SA. 1999. The bicoid-related Pitx gene family in development. Mamm. Genome. 10:197–200 [DOI] [PubMed] [Google Scholar]
- 15. Gong S, et al. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425:917–925 [DOI] [PubMed] [Google Scholar]
- 16. Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5:377–386 [DOI] [PubMed] [Google Scholar]
- 17. Hatzis P, Talianidis I. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467–1477 [DOI] [PubMed] [Google Scholar]
- 18. Heard E. 2004. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16:247–255 [DOI] [PubMed] [Google Scholar]
- 19. Heintzman ND, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39:311–318 [DOI] [PubMed] [Google Scholar]
- 20. Heintzman ND, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hermanson GG, Eisenberg D, Kincade PW, Wall R. 1998. B29: a member of the immunoglobulin gene superfamily exclusively expressed on beta-lineage cells. Proc. Natl. Acad. Sci. U. S. A. 85:6890–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ho Y, Elefant F, Cooke NE, Liebhaber SA. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291–302 [DOI] [PubMed] [Google Scholar]
- 23. Ho Y, Elefant F, Liebhaber SA, Cooke NE. 2006. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23:365–375 [DOI] [PubMed] [Google Scholar]
- 24. Ho Y, Tadevosyan A, Liebhaber SA, Cooke NE. 2008. The juxtaposition of a promoter with a locus control region transcriptional domain activates gene expression. EMBO Rep. 9:891–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huarte M, et al. 2010. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142:409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jing H, et al. 2008. Exchange of GATA factors mediates transitions in looped chromatin organization at a developmentally regulated gene locus. Mol. Cell 29:232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones BK, Monks BR, Liebhaber SA, Cooke NE. 1995. The human growth hormone gene is regulated by a multicomponent locus control region. Mol. Cell. Biol. 15:7010–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kagey MH, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawasaki H, Taira K. 2005. Transcriptional gene silencing by short interfering RNAs. Curr. Opin. Mol. Ther. 7:125–131 [PubMed] [Google Scholar]
- 30. Kim T-K, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koch F, Jourquin F, Ferrier P, Andrau J-C. 2008. Genome-wide RNA polymerase II: not genes only! Trends Biochem. Sci. 33:265–273 [DOI] [PubMed] [Google Scholar]
- 32. Ling J, et al. 2004. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 279:51704–51713 [DOI] [PubMed] [Google Scholar]
- 33. Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. 2003. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4:132–137 [DOI] [PubMed] [Google Scholar]
- 34. Meyer MB, Goetsch PD, Pike JW. 2010. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1α,25-dihydroxyvitamin D3. J. Biol. Chem. 285:1599–15610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Miele A, Dekker J. 2008. Long-range chromosomal interactions and gene regulation. Mol. Biosyst. 4:1046–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitchell JA, Fraser P. 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22:20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nissim S, Allard P, Bandyopadhyay A, Harfe BD, Tabin CJ. 2007. Characterization of a novel ectodermal signaling center regulating Tbx2 and Shh in the vertebrate limb. Dev. Biol. 304:9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nolis IK, et al. 2009. Transcription factors mediate long-range enhancer-promoter interactions. Proc. Natl. Acad. Sci. U. S. A. 106:20222–20227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Omori SA, Wall R. 1993. Multiple motifs regulate the B cell-specific promoter of the B29 gene. Proc. Natl. Acad. Sci. U. S. A. 90:11723–11727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ørom UA, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shewchuk BM, Asa SL, Cooke NE, Liebhaber SA. 1999. Pit-1 binding sites at the somatotrope-specific DNase I hypersensitive sites I, II of the human growth hormone locus control region are essential for in vivo hGH-N gene activation. J. Biol. Chem. 274:35725–35733 [DOI] [PubMed] [Google Scholar]
- 42. Shewchuk BM, Liebhaber SA, Cooke NE. 2002. Specification of unique Pit-1 activity in the hGH locus control region. Proc. Natl. Acad. Sci. U. S. A. 99:11784–11789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. 2010. Functional analysis of CTCF during mammalian limb development. Cell Dev. 19:819–830 [DOI] [PubMed] [Google Scholar]
- 44. Su Y, Liebhaber SA, Cooke NE. 2000. Human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in the transgenic mouse. J. Biol. Chem. 275:7902–7909 [DOI] [PubMed] [Google Scholar]
- 45. Szutorisz H, Dillon N, Tora L. 2005. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem. Sci. 30:593–599 [DOI] [PubMed] [Google Scholar]
- 46. Thompson AA, et al. 1996. The promoter and 5′ flanking sequences controlling human B29 gene expression. Blood 87:666–673 [PubMed] [Google Scholar]
- 47. Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453–1465 [DOI] [PubMed] [Google Scholar]
- 48. Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26:2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoo EJ, et al. 2006. Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions. Mol. Cell. Biol. 26:5569–5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao H, Dean A. 2004. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between and enhancer and gene. Nucleic Acids Res. 32:4903–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu X, et al. 2007. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 35:5532–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu X, Cleiberman AS, Rosenfeld MG. 2007. Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol. Rev. 87:933–963 [DOI] [PubMed] [Google Scholar]
- 53. Xu L, et al. 1998. Signal-specific coactivator domain requirements for Pit-1 activation. Nature 395:301–306 [DOI] [PubMed] [Google Scholar]