Abstract
Proliferating cell nuclear antigen (PCNA) is a ubiquitous protein that interacts with multiple partners and regulates nuclear activities, including chromatin assembly, histone modifications, replication, and DNA damage repair. The role of specific partners in regulating PCNA activities is not fully understood. Here we identify the nucleosome binding protein HMGN1 as a new PCNA-interacting protein that enhances the binding of PCNA to chromatin but not to purified DNA. Two tetrapeptides in the conservative domain of HMGN1 contain amino acids necessary for the binding of HMGN1 to PCNA. Deletion of both tetrapeptides abolishes the HMGN1-PCNA interaction. PCNA preferentially binds to the linker DNA adjacent to an HMGN-containing nucleosome. In living cells, loss of HMGN1 decreases the rate of PCNA recruitment to damaged DNA sites. Our study identifies a new factor that facilitates the interaction of PCNA with chromatin and provides insights into mechanisms whereby nucleosome binding architectural proteins affect the cellular phenotype.
INTRODUCTION
The interaction of regulatory factors with their cognate binding sites in chromatin plays a key role in the orderly progression of DNA-dependent processes such as transcription, repair, and replication. These interactions are facilitated by dynamic changes in chromatin structure resulting from the action of various chromatin modifiers, including that of the various chromatin-binding architectural proteins such as the linker histone H1 (9, 46) and members of the high-mobility-group (HMG) protein superfamily (3, 8). Numerous studies established that HMG proteins affect distinct chromatin functions and impact the cellular phenotype (3, 19); however, these proteins do not bind to specific DNA sequences, and the molecular mechanisms responsible for their biological effects are not fully understood. Conceivably, the biological effects of HMGs could in part be due to their ability to associate with specific nuclear factors. Here we demonstrate that HMGN1, a prominent member of the high-mobility-group N (HMGN) protein family (6, 33), interacts specifically with proliferating cell nuclear antigen (PCNA), an important nuclear regulatory factor, and that this interaction facilitates the binding of PCNA to chromatin.
HMGN is a family of ubiquitous vertebrate proteins that bind specifically to the 147-bp nucleosome core particle (CP), the building block of chromatin (13, 21). The interaction of the HMGN variants with chromatin is highly dynamic, the proteins continuously roam throughout the nucleus, and the residence time of any HMGN molecule on any CP is relatively short (10, 11). Although HMGN variants bind to all CPs, their global organization is not random: genome-wide studies indicate that the HMGN1 variant preferentially localizes to chromatin regulatory regions such as DNase I hypersensitive sites, promoters, and functional enhancers (14). Analysis of mice and cells that either overexpress or lack HMGN proteins revealed that changes in HMGN levels alter the cellular transcription profile (37, 48) and affect the rate of repair of DNA damaged by either UV or ionizing radiation (4, 5). All these effects are contingent on the ability of HMGN to bind to CPs; yet by itself, this binding does not fully explain some of the specific effects of HMGN variants on transcription (37) and histone modification (25).
HMGN proteins could affect the cellular phenotype by partnering with other nuclear proteins and affecting their chromatin binding. In this study, we used affinity chromatography and mass spectrometry (MS) to search for proteins that interact with HMGN1, a major member of the HMGN protein family which has been shown to affect both transcription and DNA repair, but only in the context of chromatin. We identify several partners and concentrate on PCNA, a ubiquitous nuclear protein known to play a major role in DNA replication (27). In addition, PCNA affects DNA repair, chromatin assembly, and chromatin modification (26, 40, 41), processes that do not always occur during the S phase of the cell cycle. While the factors affecting the binding of PCNA to replicating DNA have been studied in detail (27), less is known about the factors affecting the interaction of PCNA with interphase chromatin.
Here we show that the nucleosome binding protein HMGN1 affects the interaction of PCNA with chromatin. We demonstrate that the interaction between PCNA and HMGN1 is specific, identify the protein regions involved in this interaction, and demonstrate that HMGN1 enhances the interaction of PCNA with chromatin both in vitro and in vivo. These studies provide insights into the molecular mechanisms whereby HMGN proteins affect the cellular phenotype and into factors that affect the intranuclear organization of PCNA. We propose that HMGN1 facilitates the interaction of PCNA with chromatin and enhances its rate of recruitment to damaged DNA sites.
MATERIALS AND METHODS
Materials.
Affinity-purified rabbit anti-HMGN1 and anti-H3 were described previously (7). The following materials were used: anti-CREB (C-21) from Santa Cruz, antiactin (clone AC-74) from Sigma, horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Millipore), anti-PCNA from Santa Cruz Biotechnology (PC10; catalog no. sc-56), human PCNA (Cell Sciences; catalog no. CRP150), and TNT Quick coupled transcription/translation systems for 35S-PCNA labeling (Promega). MIN6 (43) and simian virus 40 (SV40)-transformed mouse embryonic fibroblasts (MEFs) (5), human HMGN1, and human HMGN1 S20,24E mutants were as described previously (42). Vectors for glutathione S-transferase (GST) fusion proteins were cloned into pGEX-4T (GE Healthcare; catalog no. 28-9545-49) and expressed in Escherichia coli BL21(DE3)-RIL or Rosetta-Gami (Stratagene). Mammalian expression vectors for HMGN1-Cherry and yellow fluorescent protein (YFP)-PCNA fluorescent fusion proteins were cloned in pEYFP vector (Clontech; GenBank accession no. U57608). Retroviral vectors for expressing hemagglutinin (HA) and FLAG-tagged HMGNs were described previously (37).
PCNA deletion mutants have been constructed with the QuikChange site-directed mutagenesis kit (Stratagene; catalog no. 200518) using custom-designed primers and the open reading frame of mouse PCNA cloned into pTnT vector (Promega).
Cross-linking and immunoprecipitation of HMGN1 protein partners.
MIN6 cells expressing HA-tagged HMGN1 were collected from 10 15-cm culture dishes and cross-linked by 1% formaldehyde (FA) (Sigma) in phosphate-buffered saline (PBS) for 15 min at room temperature (RT). Following quenching with 2.5 M glycine (final concentration, 125 mM) for 5 min, cells were pelleted, lysed in DNase I digestion buffer (15 mM Tris-HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 3 mM MgSO4, 0.5 mM dithiothreitol [DTT], 0.5% Triton X-100, and protease inhibitors), and briefly sonicated. The nuclei were collected by centrifugation at 700 × g for 10 min at 4°C and treated with 500 U/ml DNase I for 20 min at RT, and the digestion was stopped by adding 2 volumes of 2× radioimmunoprecipitation (RIPA) buffer containing 20 mM EDTA and protease inhibitors. The samples were sonicated briefly, clarified, and loaded on immobilized anti-HA agarose resin. After 3 washes with NETNM buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10% glycerol, 5 mM MgCl2, and 0.5 mM DTT), the resin was boiled for 20 min at 100°C to reverse formaldehyde cross-links. Following centrifugation, the eluted proteins were then fractionated on a 4 to 20% SDS-PAGE gradient gel and visualized with a SilverQuest silver staining kit (Life Technologies). The following modifications were used for SV40-transformed MEFs expressing HA-HMGN1. Cells were cross-linked with 2 mM dithiobis(succinimidyl propionate) (DSP) in PBS for 30 min at RT, and the reaction was terminated by adding 1 M Tris-HCl (pH 7.5) to a final concentration of 20 mM for 15 min at RT. After being washed with PBS, the cells were lysed in buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, and protease inhibitors) on ice for 10 min. Triton X-100 was added to a final concentration of 0.1%, and the pellet was homogenized, briefly sonicated, resuspended in buffer C (20 mM Tris-HCl [pH 7.5], 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, 0.5 mM DTT, and protease inhibitors) and sonicated briefly. The suspension was clarified by centrifugation and treated as described for MIN6 cells.
Protein identification by MS.
Silver-stained bands excised from SDS-PAGE gels were trypsin digested. MS analysis was performed as described previously (38).
GST-HMGN1 pulldown assay.
GST pulldown assays were performed as previously described (15), with minor modifications. For the in vitro binding assay, cells were lysed with buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5 mM EDTA, 0.5% Triton X-100, 1 mM DTT, and protease inhibitors) and sonicated. Extracts were incubated with 20 μl of glutathione-Sepharose beads (GE Healthcare) for 1 h. The beads were washed twice with binding buffer 150 (20 mM HEPES [pH 7.9], 0.1% IGEPAL CA630, 1 mM EDTA, 1 mM DTT, 150 mM NaCl, and protease inhibitor cocktail) and once with binding buffer 500 (same components except 500 mM NaCl) sequentially and then washed once more with binding buffer 150. Full-length and various deletion mutants of PCNA were synthesized with [35S]Met using an in vitro transcription/translation system according to the manufacturer's instructions (Promega). Then, 10 μl of reaction product was mixed with GST or GST fusion proteins in 1 ml of binding buffer 150. After being rocked at 4°C for 2 h, the resin was washed four times with binding buffer 150. Then, the bound proteins were released by reaction with SDS-sample buffer and boiling. The proteins were analyzed by SDS-PAGE followed by autoradiography.
Fusion proteins were expressed in Escherichia coli strain BL21(DE3)-RIL (Stratagene) and purified using glutathione-Sepharose beads (GE Healthcare). For the in vitro binding assay, PCNA was labeled using a plasmid and reticulocyte cell-free transcription/translation extract (Promega; TNT Quick coupled transcription/translation systems, catalog no. L1170). The recombinant wild-type and deletion mutant HMGN1-GST fusion proteins, along with control GST, were isolated, checked for purity, and divided into equal amounts. Purified GST fusion proteins (10 μg) were incubated with 20 μl of glutathione-Sepharose beads (Amersham Biosciences) and in vitro 35S-labeled translated PCNA for 1 h at 4°C. The bound fractions were resolved on an SDS-PAGE gel. Gels were dried and exposed to storage phosphorimage screens, scanned with a Storm 840 (Amersham), and visualized by autoradiography.
Chromatin preparations.
Isolation of nuclei, preparation of nucleosome core particles, fractionation of chromatin fragments on sucrose gradients, and H5/H1 removal by ion-exchange chromatography were as described previously (2, 34, 35).
PCNA-chromatin binding assay.
Mobility shift assays were done as described previously (34). Recombinant HMGN1 or PCNA or mixtures of both (20 to 200 nM) were incubated with 50 nM core particles in 5 μl of 2× Tris-borate-EDTA (TBE) with 2% Ficoll for 15 min on ice. Samples were then loaded directly onto a 5% polyacrylamide gel made in 2× TBE and electrophoresed at 4°C. For gel filtration, 1 μg of HMGN1 was incubated with either 0.2 μg of PCNA or 10 μl of 35S-PCNA for 5 min at RT in 10 μl of TEENT-20 [10 mM Tris-HCl (pH 8), 1 mM EDTA, 1 mM EGTA, 100 μM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) 20 mM NaCl, 0.05% Tween 20] and added to 10 μg of chromatin in 50 μl of TEENT-20. After 5 min, the mixture was loaded onto a Superose 12 HR 10/150 column equilibrated with TEENT-20. Fractions containing chromatin-bound and free PCNA were pooled and analyzed either by Western blotting with anti-PCNA antibodies or by a Storm phosphorimaging system. The analyzed volume of the free fraction was 5-fold higher than that of the bound fraction.
For replicating factor C (RFC)-assisted assays, the reaction mixture (slightly modified from reference 32) contained 20 mM Tris-HCl (pH 7.6), 20 mM NaCl, 5 mM MgCl2, 1 mM glutathione, 0.05% Tween 20, 1 pmol of various DNA preparations or polynucleosomal fraction from chicken, 10 μl of in vitro-labeled 35S-PCNA, and 1 pmol of yRFCΔN in a total volume of 50 μl. Also, 1 pmol of HMGN1, 1.5 mM ATP, or 2 units of potato apyrase (Sigma-Aldrich) was optionally added as indicated in Fig. 4F. After incubation at 37°C for 5 min, the reaction mixture was either spin dialyzed with an Amicon Ultra 100K or loaded onto a size exclusion column and processed as described above.
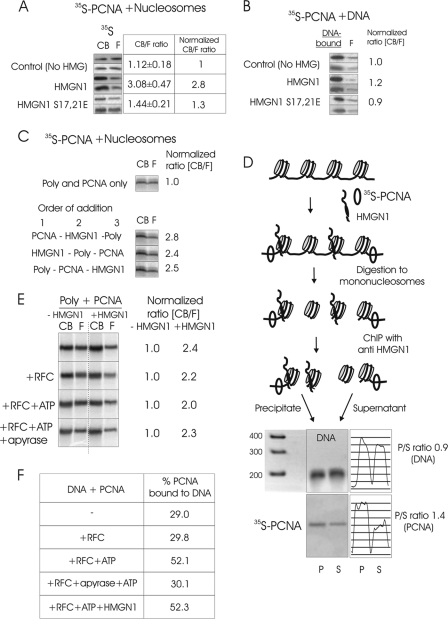
Fig 4.
(A) The interaction of HMGN1 with nucleosomes enhances the binding of PCNA to chromatin. 35S-PCNA was added to salt-stripped polynucleosomes in the presence of either native HMGN1 or mutant HMGN1 which does not bind to chromatin, and the mixture was fractionated on size exclusion columns. The relative amounts of chromatin-bound (CB) and free (F) PCNA were determined by ImageQuant analyses of autoradioagraphic images. Shown are images from 2 separate experiments. The CB/F values were obtained from triplicate experiments. (B) HMGN1 does not affect the binding of PCNA to purified DNA. (C) HMGN1 improves the binding of PCNA to polynucleosome (poly) independently of order of addition. (D) PCNA binds preferentially adjacent to HMGN-containing nucleosomes. Mixtures of polynucleosomes, HMGN1, and 35S-PCNA were formaldehyde cross-linked, digested to mononucleosomes, and immunoprecipitated with anti-HMGN1. The amount of 35S-PCNA in the precipitate (HMGN-containing nucleosomes) and supernatant was analyzed by polyacrylamide gels followed by autoradiography. Scans of the DNA gel and of the autoradiograph, and the ratio of DNA and PCNA in the precipitate (P) to that in the supernatant (S), are on the right. (E) HMGN1-mediated enhancement of PCNA to polynucleosomes is not dependent on RFC. Shown are autoradioagraphs of 35S-PCNA in the CB and F fractions in the presence and absence of HMGN1. The RFC, ATP, and apyrase present in the various experiments are indicated on the left. The ratio of CB/F for 35S-PCNA in the absence of HMGN1 was normalized to 1.0. (F) HMGN1 does not affect the RFC-mediated loading of PCNA onto purified DNA.
Confocal microscopy, FRAP, and microirradiation.
For fixed-cell imaging, cells were plated onto poly-d-lysine-coated coverslips. Fluorescence recovery after photobleaching (FRAP) analysis and microirradiation experiments were done with an LSM710 confocal microscope, with live cells plated onto MatTek glass bottom plasticware dishes (catalog no. P35GC-1.5-14-C). A constant temperature of 37°C and 5% CO2 were maintained during the experiment using an XL-LSM710S1 incubator (PeCon). FRAP experiments were done using 458-, 488-, and 514-nm lines of an argon laser and the 543-nm line of an HeNe laser as described previously (12). In a typical experiment, several spots, in 6 to 10 cells, were used for FRAP. Each experiment was repeated at least three times. UV A laser microirradiation assays were done with a 405-nm diode laser at 100% output as described previously (28). Cells were sensitized for microirradiation by incubation in a medium containing bromodeoxyuridine (BrdU) (10 mg/ml) for 24 to 48 h. The microirradiated spot was 0.58 μm in diameter. At each time point, the fluorescence signal of the irradiated region was corrected for background and normalized to the total fluorescence signal.
Determination of free and chromatin-bound PCNA after DNA damage.
MEFs grown to 90% confluence were exposed to 20 J/m2 UV light and harvested at different times following irradiation as described previously (20), with slight modifications. Briefly, the harvested cells were resuspended in buffer A [100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 1 mM EGTA, 0.2% Triton X-100, 100 μM Na3VO4, 50 mM NaF, and protease inhibitors (Roche Applied Science)], incubated for 5 min on ice with gentle shaking, and centrifuged at 14,000 × g for 3 min at 4°C. The supernatants contain the soluble, non-chromatin-bound PCNA. The pellet was washed once in buffer A and resuspended in buffer B (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 100 μM Na3VO4, 50 mM NaF, and protease inhibitors) for immunoblotting. The samples were sonicated and then incubated for another 10 min on ice before centrifugation to isolate the chromatin-bound fraction.
RESULTS
Specific interactions between HMGN1 and PCNA.
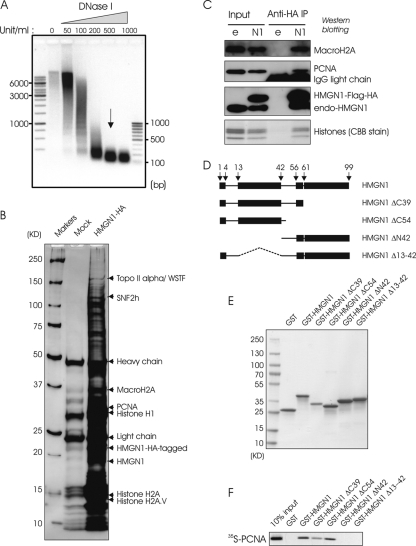
In the nucleus, part of the HMGN1 forms metastable complexes with various nuclear proteins, most of which have not yet been identified (24). Because these interactions are weak, we used formaldehyde-treated cells to stabilize putative complexes and identify HMGN1 interacting proteins. MIN6 cells stably expressing either HA-tagged HMGN1 protein or empty vector were cross-linked with formaldehyde, the nuclei were isolated and treated with DNase I to digest the chromosomal DNA to nucleosome size fragments (Fig. 1A), and the cross-linked material was fractionated by HA-affinity chromatography. Fractionation of the de-cross-linked proteins on SDS-PAGE gradient gels reveals that in the cells expressing HMGN1-HA, but not in the mock-transfected cells, a large number of proteins were retained on the anti-HA-conjugated resin (Fig. 1A).
Fig 1.
HMGN1 interacts with PCNA. (A) DNase I digestion kinetics of cross-linked chromatin used for affinity chromatography. Shown is 1% agarose gel electrophoresis of the digested chromatin. The arrow indicates the fraction taken for affinity chromatography. (B) Identification of nuclear proteins specifically bound to HA-tagged HMGN1 in MIN6 cells that stably express either HA-tagged HMGN1 or only HA tag (for mock affinity isolation). Shown is a silver-stained 4 to 20% SDS-PAGE gradient gel of the proteins recovered from anti-HA-conjugated resin. Arrows point to specific HMGN1 partners identified by mass spectrometry. WSTF, Williams syndrome transcription factor. (C) Immunoprecipitation of PCNA by HA-HMGN1 in mouse embryonic fibroblasts. Shown are Western blotting experiments of HA immunoprecipitates from cells stably expressing either HA-tagged HMGN1 or HA-empty vector (e), with the antibodies indicated on the right of each lane. CBB, Coomassie brilliant blue. (D) Schematic representation of HMGN1 deletion mutants designed to identify the PCNA interaction domain. (E) SDS-PAGE analysis of the deletion mutants shown in panel D. (F) Identification of HMGN1 regions that interact with PCNA by GST pulldown assays. GST-HMGN1 or GST-HMGN1 deletion mutants were incubated with in vitro-translated 35S-PCNA, purified on glutathione-Sepharose, fractionated on SDS-PAGE gels, and visualized by autoradiography.
Mass spectrometry analysis of the proteins fractionated on polyacrylamide gels identified several putative HMGN1-interacting proteins (Fig. 1A and supplemental material available upon request), including PCNA, a ubiquitous protein involved in several biologically important functions. Five peptides originating from PCNA were conclusively and unambiguously identified by MS/MS and SEQUEST program analysis (supplemental material available upon request). The coverage of the observed peptide mass map was 30% of the full possible coverage PCNA in the database. Western analysis verified the presence of PCNA in the cross-linked affinity-fractionated material (data not shown).
To further verify the specificity of the interaction between HMGN1 and PCNA in living cells, and to test whether the interaction is cell type or cross-linking agent specific, we stably transfected SV40-transformed mouse embryonic fibroblasts (SV-MEFs) with vectors expressing either HA-tagged HMGN1 or just HA, cross-linked the cells with dithio(bissuccinimidyl propionate) (DSP), and purified the complex by immunoaffinity chromatography on immobilized anti-HA. Western analysis of input and bound fractions revealed that PCNA was bound to the HA-HMGN1 (Fig. 1B). Thus, the experiments in SV-40-MEFs fully support the notion that HMGN1 forms specific complexes with PCNA.
Identification of the interaction sites in HMGN1 and PCNA.
To test whether HMGN1 and PCNA interact directly, and in an attempt to identify the major HMGN1 protein domains involved in this interaction, we used reticulocyte cell-free transcription/translation to synthesize 35S-labeled PCNA and bacterial expression vectors to prepare GST fusion proteins of HMGN1 and several HMGN1 deletion mutants (Fig. 1D). The deletion mutants were prepared to selectively lack the major functional domain of HMGN1 (33). The recombinant wild-type and deletion mutant GST-HMGN1 fusion proteins, as well as the control GST, were purified to homogeneity by affinity chromatography (Fig. 1F).
Equal amounts of the GST fusion proteins were incubated with 35S-labeled PCNA, the complexes were purified on glutathione-Sepharose, and the bound fractions were resolved on an SDS-PAGE gel and visualized by autoradiography. The results indicate that the full-length HMGN1 protein and the two C-terminally truncated mutants associate specifically with the PCNA, while the deletion mutants lacking either the 42 N-terminal amino acids or just the region spanning amino acids 13 to 42 did not (Fig. 1F).
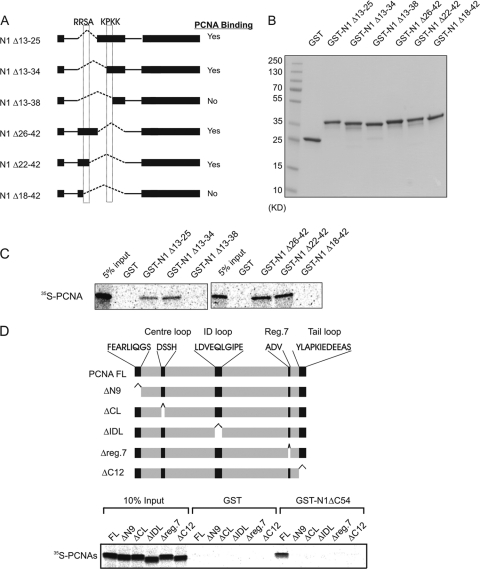
To further define the protein region through which HMGN1 interacts with PCNA, we created an additional set of GST-fused expression vectors coding for deletion mutants lacking various regions in the HMGN1 region spanning amino acids 13 to 42 (Fig. 2A and B). Binding assays of the purified GST-HMGN1 deletion mutants (Fig. 2B) revealed that only two deletion mutants, HMGN1Δ13–38 and HMGN1Δ18–42, failed to interact with 35S-labeled PCNA (Fig. 2A and C). The data demonstrate that either one of two tetrapeptides, 18RRSA21 and 34KPKK37, is necessary for the interaction of HMGN1 with PCNA. Deletion of either one of these, by itself, does not eliminate PCNA binding. Only deletion of both regions causes total ablation of the HMGN1-PCNA interaction.
Fig 2.
Identification of the PCNA-binding regions in HMGN1. (A) Schematic representation of HMGN1 deletion mutants for fine mapping of the region responsible for interaction with PCNA. The dotted boxes depict the locations of the critical HMGN1 tetrapeptides necessary for PCNA binding. (B) SDS-PAGE analysis of purified deletion mutants shown in panel A. (C) Autoradioagraphic identification of HMGN1 regions that interact with PCNA using cell-free 35S labeling and a GST pulldown assay. (D) Identification of PCNA deletions which abrogate the interaction between PCNA and HMGN1. 35S-labeled PCNA or PCNA deletion mutants were incubated with either purified ΔC54 HMGN1-GST or only GST proteins, and the complex was purified on glutathione-Sepharose, fractionated on an SDS-PAGE gel, and visualized by autoradiography.
PCNA is known to interact with numerous proteins through several overlapping sites, which have been mapped (25, 38). To further test the binding of HMGN1 to PCNA, we carried out pulldown experiments with a set of PCNA deletion mutants that span sites that were previously identified as protein binding sites on PCNA (Fig. 2D). Pulldown experiments with 35S-labeled full-length PCNA or with these mutants revealed that only full-length PCNA bound efficiently GST-HMGN1ΔC54. In contrast, none of the five PCNA deletion mutants tested was able to do so (Fig. 2D). Thus, the interaction between HMGN1 and PCNA involves distinct sites in both PCNA and HMGN1. It is dependent on two distinct tetrapeptides in HMGN1 and on PCNA regions known to serve as binding sites for protein partners and to play a role in PCNA trimerization.
HMGN1 enhances the binding of PCNA to chromatin.
In the eukaryotic nucleus, PCNA affects not only DNA replication but also DNA repair, chromatin assembly, and additional activities that occur in the context of chromatin. A fraction of the nuclear PCNA is associated with chromatin, an association that is affected by posttranslational modifications in PCNA (27, 29, 45). Given that HMGN1 interacts specifically with PCNA and also binds specifically to the 147-bp core particle (CP), we tested whether it affects the interaction of PCNA with the chromatin fiber.
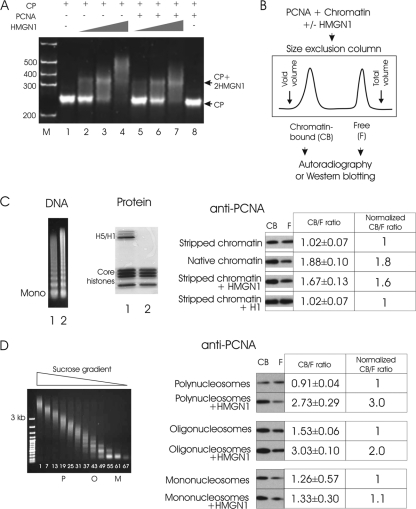
To test whether PCNA can bind to CPs and if HMGN1 affects this binding, we used gel mobility assays, a commonly used method to detect the interaction of chromatin-binding proteins with nucleosomes. As previously demonstrated, HMGN1 binds cooperatively to CPs to form complexes containing 2 molecules of HMGN1 per CP (Fig. 3A, lanes 1 to 4). In this assay, PCNA did not bind to the CPs (Fig. 3A, lane 8). Reaction mixtures containing HMGN1, PCNA, and CP did not contain “supershifts” (high-molecular-weight [MW] complexes) that run slowly on polyacrylamide gels, an indication that HMGN1 does not facilitate the binding of PCNA to CPs (Fig. 3A, lanes 5 to 7).
Fig 3.
HMGN1 facilitates PCNA binding to chromatin in vitro. (A) HMGN1 does not facilitate the binding of PCNA to the 147-bp CP. A mobility shift assay demonstrates that HMGN1 (lanes 2 to 4) but not PCNA (lane 8) binds to CPs. The lack of supershifts in lanes 5 to 7 indicates that HMGN1 does not promote the binding of PCNA to CPs. (B) Size exclusion chromatography assay for analysis of the effect of HMGN1 on the binding of PCNA to chromatin. PCNA is added to chromatin in either the presence or absence of HMGN1. Chromatin-bound PCNA elutes in the void volume, while free PCNA elutes in the total volume. (C) HMGN1 enhances the binding of PCNA to chromatin. Left side, DNA and protein gels of micrococcal nuclease-digested chromatin. Lane 1, not salt stripped; lane 2, stripped with 0.5 M NaCl. Right side, Western analysis of free (F) and chromatin-bound (CB) PCNA present in various chromatin preparations. The ratio of CB to F PCNA, determined by scanning the film, is indicated. An average of three replicates is presented, with standard deviations. Note that addition of HMGN1, but not of H1, to chromatin increased the amount of chromatin-bound PCNA. (D) HMGN1 preferentially enhances PCNA binding to longer chromatin fragments. Left, sucrose gradient fractionation of salt-stripped micrococcal nuclease-digested chromatin; right, Western analysis of the relative amounts of chromatin-bound and free PCNA. PCNA was added to either polynucleosomes (P, fragments ranging from 9 to 15 nucleosomes), oligonucleosomes (O, 2 to 6 nucleosomes), or mononucleosomes (M) in either the presence or absence of HMGN1 and fractionated on size exclusion columns (see panel B).
Although PCNA did not bind to the 147-bp CP, it is still possible that it could interact with chromatin or even with isolated mononucleosomes that contain linker DNA. To test this possibility, we developed an in vitro binding assay in which we used size exclusion chromatography to separate chromatin-bound PCNA from free PCNA. A typical experiment consisted of mixing chromatin or nucleosomes with either nonradioactive or 35S-labeled recombinant PCNA, with or without HMGN1, and fractionating the mixtures on Superose 12 HR 150/5 into high- and low-molecular-weight fractions. The high-molecular-weight fraction, which elutes near the void volume, contains chromatin fragments, while the unbound PCNA elutes near the total volume fraction (Fig. 3B). The ratio of chromatin-bound to free PCNA is determined either by Western blotting or by autoradiography.
We found that the amount of PCNA bound to native chromatin fragments was about 2-fold higher than that bound to chromatin fragments fully stripped of both H1 and nonhistone proteins (Fig. 3C). Significantly, efficient binding of PCNA to the stripped chromatin could be restored by adding purified HMGN1 but not by adding purified H1 (Fig. 3C). Thus, HMGN1 enhances the binding of PCNA to chromatin.
To determine whether the size of the chromatin fragment affects the ability of HMGN1 to enhance the binding of PCNA to chromatin, we fractionated the salt-extracted micrococcal nuclease-digested chromatin on sucrose gradients (Fig. 3D) and tested the effect of HMGN1 on the interaction of PCNA with three nucleosome preparations differing only by length. We found that HMGN1 enhances the binding of PCNA to polynucleosomes (fraction P, 12 nucleosomes on average) more efficiently than to oligonucleosomes (fraction O, 4 nucleosomes), and it did not affect the binding of PCNA to mononucleosomes (Fig. 4D). Thus, HMGN1 enhances the binding of PCNA to longer chromatin fragments more efficiently than to shorter fragments.
HMGN1 binds specifically to CPs, while PCNA binds preferentially to DNA. We therefore tested whether the ability of HMGN1 to enhance the chromatin binding of PCNA is indeed contingent on the ability of HMGN1 to bind to nucleosomes. We found that wild-type HMGN1 enhances the binding of PCNA to polynucleosomes, but the double mutant HMGN1 S17,21E, which does not bind to CPs (36, 42), does not (Fig. 4A). Furthermore, neither the wild type nor the mutant affects the interaction of PCNA with purified DNA (Fig. 4B). Thus, HMGN1 enhances the interaction of PCNA with chromatin by binding to nucleosomes.
HMGN1 may enhance the interaction of PCNA with chromatin either by first binding to PCNA and targeting it to chromatin or by first binding to chromatin and inducing structural changes that enhance the subsequent binding of PCNA. To gain insight into this question, we tested whether the binding of PCNA to polynucleosomes depends on the order of addition of the three components of reaction: polynucleosomes, HMGN1, and PCNA (Fig. 4C). The results did not show a preference for order of addition, most likely because the binding of both HMGN1 and PCNA to chromatin and the formation of the PCNA-HMGN1 complex are reversible processes leading to a fast equilibration of the system.
PCNA does not bind to purified CPs (Fig. 3A), but it does to mono- and oligonucleosomes (Fig. 3D), suggesting that it interacts mainly with the linker DNA between adjacent CPs. We therefore tested whether HMGN1 targets the PCNA to the linker region adjacent to the CPs containing HMGN1. In this experiment, polynucleosomes stripped of H1 and nonhistones were incubated with 35S-PCNA and limited amounts of HMGN1, sufficient to bind only 50% of the nucleosomes (Fig. 4D). The mixture was formaldehyde fixed, nuclease digested, and immunoprecipitated using antibodies to HMGN1, which have been shown to efficiently immunoprecipitate chromatin fragments containing HMGN1 (14). After reversal of the cross-links, equal amounts of mononucleosomes (normalized by measuring DNA [Fig. 4D]) from immunoprecipitated (HMGN1-containing) and nonbound (HMGN1-free) fractions were fractionated on polyacrylamide gels, and the relative amount of 35S-PCNA in each fraction was determined by autoradiography. Quantification of the ratios of 35S-PCNA indicates a 40% preference for PCNA binding in the immunoprecipitated fraction, an indication that PCNA binds preferentially to the linker region adjacent to the HMGN-containing CPs.
PCNA is uploaded to replicating DNA in ATP-dependent manner by replicating factor C (RFC) (27). We therefore tested whether the HMGN1-dependent binding of PCNA to chromatin is affected by RFC. To this end, we incubated polynucleosomes with PCNA in either the presence or absence of HMGN1 and then added to these reaction mixtures either RFC alone, RFC and ATP, or RFC, ATP, and apyrase. After a 5-min incubation period, each mixture was applied to a Superose 12 column and the ratio of chromatin-bound to free PCNA was quantified as described above. The results indicate that in the absence of HMGN1, RFC and ATP did not enhance the binding of PCNA to oligonucleosomes (Fig. 4E). Likewise, RFC and ATP did not affect the HMGN1-facilitated recruitment of PCNA to the oligonucleosomes (Fig. 4E). In contrast, RFC and ATP did facilitate the binding of PCNA to purified DNA (Fig. 4F). The effect was abolished by apyrase, attesting to the ATP dependence of the PCNA loading onto the DNA. Significantly, HMGN1 did not affect the binding of PCNA to DNA either in the presence of RFC and ATP (Fig. 4F) or in the absence of these components (Fig. 4B).
We conclude that HMGN1 enhances the interaction of PCNA to chromatin but not to purified DNA and that this effect does not involve the RFC-dependent uploading of the PCNA to chromatin.
HMGN1 enhances the rate of recruitment of PCNA to DNA damage sites.
The intranuclear organization and chromatin interactions of PCNA change during the cell cycle and in response to DNA damage. During G1/G2, PCNA is dispersed in multiple small foci and binds weakly to chromatin, while during S phase the PCNA is strongly bound at sites of replication (16, 39). To test whether HMGN1 affects the intranuclear organization and chromatin binding of PCNA in living cells, we transfected mouse embryonic fibroblasts (MEFs) derived from Hmgn1+/+ and their littermate Hmgn1−/− mice with vectors expressing YFP-PCNA and used fluorescence recovery after photobleaching (FRAP) to assess the mobility of PCNA. Since the FRAP analyses involve single-cell studies, the cell cultures were not synchronized. Cells in which the YFP-PCNA localized to multiple small foci were considered to be in G1/G2, while cells with large and distinct clusters of YFP-PCNA were considered to be in S phase (Fig. 5A, left).
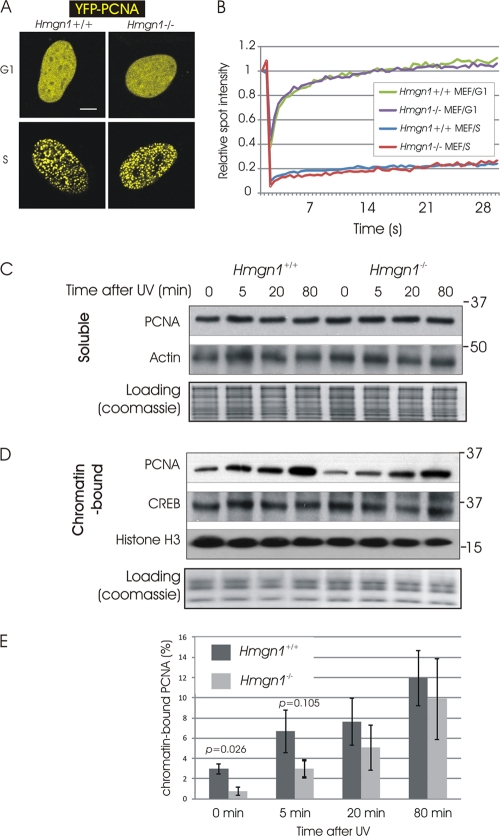
Fig 5.
(A) Organization of PCNA in mouse embryonic fibroblasts during G1/G2 and S. Shown are confocal images of cells expressing YFP-PCNA. (B) FRAP analysis of PCNA at G1/G2 and S in Hmgn1−/− and Hmgn1+/+ MEFs. (C) Western analysis of the effects of DNA damage on the levels of soluble (non-chromatin-bound) PCNA in Hmgn1−/− and Hmgn1+/+ MEFs. (D) Western analysis of the effects of DNA damage on the levels of chromatin-bound PCNA in Hmgn1−/− and Hmgn1+/+ MEFs. (E) Increased binding of PCNA to UV-damaged chromatin of Hmgn1−/− and Hmgn1+/+ MEFs. Western analyses were quantified using ImageQuant software. The graph shows the means of three biologically independent experiments, plus or minus standard deviations.
FRAP analysis revealed that in both Hmgn1+/+ and Hmgn1−/− MEFs, PCNA moved throughout the nuclei of G1/G2-phase cells much faster than in S-phase cells (Fig. 5A, right). In these FRAP assays, HMGN1 did not affect the nuclear dynamics of PCNA. Likely, HMGN1 did not affect the mobility of PCNA during G1/G2 because only a small fraction of the cellular PCNA is associated with chromatin. Indeed, cellular-fractionation experiments indicate that in these MEFs, less than 5% of the nuclear PCNA is chromatin bound (Fig. 5E). HMGN1 affects only the binding of proteins to chromatin, and therefore any putative effects would be masked by the vast excess of non-chromatin-bound PCNA. Likewise, during S phase, most of the PCNA is bound to the replicating DNA rather than to chromatin; therefore, HMGN1 is not expected to affect this interaction.
A key event in the nuclear response to DNA damage is the accumulation of PCNA at the damaged sites in chromatin (16, 39, 44). In light of our finding that HMGN1 affects the interaction of PCNA with chromatin, we UV irradiated Hmgn1−/− and Hmgn1+/+ MEFs, used Triton X-100 to fractionate the PCNA into soluble (non-chromatin-bound) and chromatin-bound fractions (20, 23), and measured the relative amount of free and chromatin-bound PCNA by Western blotting (Fig. 5C and D). In these studies, we used cells at high confluence (∼90%) to minimize cells in S phase, since PCNA accumulates in replication factories and is resistant to Triton X-100 extraction. We found that prior to irradiation, only 3% of the PCNA in Hmgn1+/+ MEFs and less than 1% of the PCNA in Hmgn1−/− MEFs is associated with chromatin. Thus, loss of HMGN1 does reduce the binding of PCNA to chromatin, a finding that is in agreement with the in vitro studies.
Following UV treatment, the fraction of PCNA associated with chromatin increases until 80 min postirradiation, when it is approximately 10% of the cellular PCNA. Loss of HMGN1 decreased significantly the amount of chromatin-bound PCNA only within first 5 min after the UV irradiation, most likely because HMGN1 affects the initial step of recruitment of PCNA to the damaged sites.
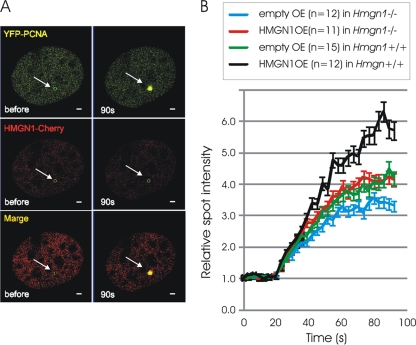
To test whether HMGN1 indeed affects the rate of PCNA accumulation at the damaged site, we performed microirradiation assays (28) in live Hmgn1+/+ MEFs coexpressing HMGN1-Cherry and YFP-PCNA fusion proteins. These assays reveal that PCNA, but not HMGN1, accumulates rapidly at the damaged site (Fig. 6A). Likely, the metastable HMGN1-PCNA complexes dissociate rapidly, and while PCNA remains associated with the DNA at the damaged site, HMGN1 does not. Analysis of the PCNA recruitment kinetics in Hmgn1−/− and Hmgn1+/+ MEFs reveals that loss of HMGN1 reduced the rate of PCNA accumulation to the damage sites (compare blue line to green line in Fig. 6B). Furthermore, expression of exogenous HMGN1 in either Hmgn1−/− or Hmgn1+/+ MEFs increased the amount of PCNA accumulated at the site of DNA damage (Fig. 6B). Control studies with empty plasmids verified that the effect is not due to the experimental manipulations involved in the transfection steps. Thus, HMGN1 enhances the rate of PCNA recruitment to the damaged sites in the chromatin of living cells.
Fig 6.
HMGN1 enhances the rate of recruitment of PCNA to UV-damaged sites in chromatin. (A) Fluorescence microscopy of a MEF coexpressing HMGN1-Cherry and YFP-PCNA before and 90 s after microirradiation. Arrows point to the irradiated site. (B) Recruitment kinetics of PCNA to DNA damage site in Hmgn1−/− and Hmgn1+/+ cells transfected either with control vectors (empty) or with vectors expressing HMGN1 (HMGN1 OE). The number of cells analyzed is indicated. Error bars indicate standard deviations.
DISCUSSION
The intranuclear organization and chromatin interaction of PCNA are highly dynamic and change during the cell cycle and in response to DNA damage. Photobleaching analyses revealed that during the S phase of the cell cycle, PCNA is tightly associated with replication foci, while during G1 and G2, the protein is evenly distributed throughout the nucleus and its residence time at any specific site is very short. Throughout all the phases of the cell cycle, PCNA accumulates rapidly at sites of damaged DNA, where it is tightly bound, as judged by its long residence time (16, 39). While the factors affecting the loading of PCNA to replication foci and to damaged DNA sites have been studied in some detail, little is known about factors that affect the steady-state organization of PCNA in nonreplicating nuclei.
In agreement with previous findings, our FRAP analyses also indicate that in living cells at G1/G2, the residence time of PCNA is very short. Nevertheless, the FRAP recovery curve is similar to those of some transcription factors which are known to interact with chromatin (31), and our chromatin immunoprecipitation (ChIP) analyses reveal that PCNA is associated with chromatin. Thus, like most nuclear proteins, PCNA binds transiently and continuously to nucleosomes, albeit with a very short chromatin residence time.
We found that HMGN1 interacts specifically with PCNA and enhances its steady-state interaction with the chromatin fiber. HMGN1 interacts with PCNA not only in living cells but also in vitro, in the absence of any additional cellular components, an indication that the proteins bind directly to each other. HMGN1 enhances the binding of PCNA to chromatin but not to naked DNA, and HMGN1 mutants that do not bind to nucleosomes fail to enhance the interaction of PCNA with chromatin. Furthermore, HMGN1 enhances the interaction of PCNA to an array containing 12 nucleosomes more efficiently than to an array containing only 4 nucleosomes and does not affect the binding of PCNA to either mononucleosomes or to the 147-bp CP. HMGN1 is known to induce both local and global changes in chromatin. Together with our finding that PCNA is preferentially bound adjacent to a nucleosome containing HMGN1 (Fig. 4D), the results suggest that HMGN1 facilitates the interaction of PCNA with the linker DNA by altering the local structure of the chromatin fiber. In addition, it is possible that HMGN1-mediated changes in PCNA conformation would also influence this process.
The interaction of HMGN proteins with nucleosomes is known to affect the structure and activity of chromatin and enhance the rate of DNA repair in the context of chromatin. Rescue experiments with Hmgn1−/− cells revealed that HMGN1 mutations that abolish either its nucleosome binding or chromatin “unfolding” activity also reduce its ability to enhance the repair of UV-damaged DNA (4, 5). Given the role of PCNA in DNA repair, it is likely that factors that affect its recruitment to damaged sites impact the efficiency of the repair process. Thus, our present findings that HMGN1 enhances the rate of recruitment of PCNA to damaged sites, a process that can be independent of RFC (30), suggest an additional mechanism whereby HMGN1 affects DNA repair. Conceivably, the delayed accumulation of PCNA at the DNA damage site in Hmgn1−/− cells (Fig. 6) might ultimately affect the rate of repair and genome stability.
By virtue of their highly disordered structure (33, 37), HMGN proteins can form complexes with multiple proteins (24). Increasing evidence links the interaction of HMGNs with other nuclear proteins to their biological function. Thus, the interaction of HMGN3 with PDX1 in beta cells affects insulin secretion (43), the interactions of HMGN2 with PITX2 (1) or with the phosphorylated prolactin receptor-Stat5a complex (17) modulate specific transcriptional events, HMGN1 modulates estrogen activity by interacting with estrogen receptor α (47), and Cockayne syndrome protein A interacts with HMGN1 at DNA sites damaged by UV (18). We now find that the interaction of HMGN1 with PCNA modulates the binding of PCNA to chromatin and its recruitment to damaged DNA sites. Taken together with previous analyses of genetically altered mice and Hmgn1−/− cells (4, 5, 18) and with the known role of PCNA in DNA repair processes (27), our studies strengthen the conclusion that HMGN1 affects the rate of DNA repair.
We note that HMGN1 is not a bona fide DNA repair factor and does not affect the RFC-dependent loading of PCNA onto damaged DNA. HMGN1 is a nucleosome binding protein that affects the structure of chromatin and the levels of histone modifications (6, 25, 33). Changes in chromatin are one of the earliest cellular responses to DNA damage. These changes affect the interaction of nuclear proteins such as histone H1 (10) or ATM (22) with chromatin. By interacting with PCNA and by modulating chromatin structure, HMGN1 may affect the intranuclear organization of PCNA, most likely its dynamic steady-state interaction with the chromatin fiber, which could be altered during the initial phases of the DNA damage response. Thus, our findings provide insights into the mechanisms whereby architectural proteins affect the interaction of regulatory factors with chromatin and impact the cellular phenotype.
ACKNOWLEDGMENTS
We thank T. Veenstra (Laboratory of Proteomics and Analytical Technologies, NCI—Frederick) for MS analysis, S. Garfield (Confocal Core Facility, LEC, NCI) for help with imaging experiments, and S. Khan and K. Kraemer (CCR, NCI) for help with irradiation experiments and discussions.
This project was supported by the Center for Cancer Research, intramural program of the NCI, NIH, by contract number N01-CO-12400 granted by NCI, NIH, and by a JSPS research fellowship from the Japanese Biomedical and Behavioral Research at NIH to T.K. The yeast RFC and human RFC preparations were generous gifts from Paul Modrich (Duke University).
Footnotes
Published ahead of print 5 March 2012
REFERENCES
- 1. Amen M, et al. 2008. Chromatin-associated HMG-17 is a major regulator of homeodomain transcription factor activity modulated by Wnt/beta-catenin signaling. Nucleic Acids Res. 36:462–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ausio J, Dong F, van Holde KE. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206:451–463 [DOI] [PubMed] [Google Scholar]
- 3. Bianchi ME, Agresti A. 2005. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 15:496–506 [DOI] [PubMed] [Google Scholar]
- 4. Birger Y, et al. 2005. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 65:6711–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birger Y, et al. 2003. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 22:1665–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bustin M. 2001. Chromatin unfolding and activation by HMGN* chromosomal proteins. Trends Biochem. Sci. 26:431–437 [DOI] [PubMed] [Google Scholar]
- 7. Bustin M. 1989. Preparation and application of immunological probes for nucleosomes. Methods Enzymol. 170:214–251 [DOI] [PubMed] [Google Scholar]
- 8. Bustin M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bustin M, Catez F, Lim JH. 2005. The dynamics of histone H1 function in chromatin. Mol. Cell 17:617–620 [DOI] [PubMed] [Google Scholar]
- 10. Catez F, Brown DT, Misteli T, Bustin M. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3:760–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Catez F, Hock R. 2010. Binding and interplay of HMG proteins on chromatin: lessons from live cell imaging. Biochim. Biophys. Acta 1799:15–27 [DOI] [PubMed] [Google Scholar]
- 12. Catez F, Ueda T, Bustin M. 2006. Determinants of histone H1 mobility and chromatin binding in living cells. Nat. Struct. Mol. Biol. 13:305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crippa MP, Alfonso PJ, Bustin M. 1992. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J. Mol. Biol. 228:442–449 [DOI] [PubMed] [Google Scholar]
- 14. Cuddapah S, et al. 2011. Genomic profiling of HMGN1 reveals an association with chromatin at regulatory regions. Mol. Cell. Biol. 31:700–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai P, et al. 1996. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 10:528–540 [DOI] [PubMed] [Google Scholar]
- 16. Essers J, et al. 2005. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell. Biol. 25:9350–9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fiorillo AA, et al. HMGN2 inducibly binds a novel transactivation domain in nuclear PRLr to coordinate Stat5a-mediated transcription. Mol. Endocrinol. 25:1550–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fousteri M, Vermeulen W, van Zeeland AA, Mullenders LH. 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23:471–482 [DOI] [PubMed] [Google Scholar]
- 19. Hock R, Furusawa T, Ueda T, Bustin M. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 17:72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kannouche PL, Wing J, Lehmann AR. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14:491–500 [DOI] [PubMed] [Google Scholar]
- 21. Kato H, et al. Architecture of the high mobility group nucleosomal protein 2-nucleosome complex as revealed by methyl-based NMR. Proc. Natl. Acad. Sci. U. S. A. 108:12283–12288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim YC, et al. 2009. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat. Cell Biol. 11:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee KY, et al. 2010. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through its interactions with PCNA and USP1. J. Biol. Chem. 285:10362–10369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim JH, Bustin M, Ogryzko VV, Postnikov YV. 2002. Metastable macromolecular complexes containing high mobility group nucleosome-binding chromosomal proteins in HeLa nuclei. J. Biol. Chem. 277:20774–20782 [DOI] [PubMed] [Google Scholar]
- 25. Lim JH, et al. 2004. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell 15:573–584 [DOI] [PubMed] [Google Scholar]
- 26. Mello JA, Almouzni G. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136–141 [DOI] [PubMed] [Google Scholar]
- 27. Moldovan GL, Pfander B, Jentsch S. 2007. PCNA, the maestro of the replication fork. Cell 129:665–679 [DOI] [PubMed] [Google Scholar]
- 28. Mortusewicz O, Rothbauer U, Cardoso MC, Leonhardt H. 2006. Differential recruitment of DNA ligase I and III to DNA repair sites. Nucleic Acids Res. 34:3523–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ogi T, et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell 37:714–727 [DOI] [PubMed] [Google Scholar]
- 30. Overmeer RM, et al. 2010. Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 30:4828–4839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phair RD, et al. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pluciennik A, et al. 2010. PCNA function in the activation and strand direction of MutLα endonuclease in mismatch repair. Proc. Natl. Acad. Sci. U. S. A. 107:16066–16071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Postnikov Y, Bustin M. 2010. Regulation of chromatin structure and function by HMGN proteins. Biochim. Biophys. Acta 1799:62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Postnikov YV, Bustin M. 1999. Analysis of HMG-14/-17-containing chromatin. Methods Mol. Biol. 119:303–310 [DOI] [PubMed] [Google Scholar]
- 35. Postnikov YV, Bustin M. 1999. Reconstitution of high mobility group 14/17 proteins into nucleosomes and chromatin. Methods Enzymol. 304:133–155 [DOI] [PubMed] [Google Scholar]
- 36. Prymakowska-Bosak M, et al. 2001. Mitotic phosphorylation prevents the binding of HMGN proteins to chromatin. Mol. Cell. Biol. 21:5169–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rochman M, et al. 2011. Effects of HMGN variants on the cellular transcription profile. Nucleic Acids Res. 39:4076–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simkus C, Bhattacharyya A, Zhou M, Veenstra TD, Jones JM. 2009. Correlation between recombinase activating gene 1 ubiquitin ligase activity and V(D)J. recombination. Immunology 128:206–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Solomon DA, Cardoso MC, Knudsen ES. 2004. Dynamic targeting of the replication machinery to sites of DNA damage. J. Cell Biol. 166:455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stoimenov I, Helleday T. 2009. PCNA on the crossroad of cancer. Biochem. Soc. Trans. 37:605–613 [DOI] [PubMed] [Google Scholar]
- 41. Strzalka W, Ziemienowicz A. 2011. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 107:1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ueda T, Catez F, Gerlitz G, Bustin M. 2008. Delineation of the protein module that anchors HMGN proteins to nucleosomes in the chromatin of living cells. Mol. Cell. Biol. 28:2872–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ueda T, Furusawa T, Kurahashi T, Tessarollo L, Bustin M. 2009. The nucleosome binding protein HMGN3 modulates the transcription profile of pancreatic beta cells and affects insulin secretion. Mol. Cell. Biol. 29:5264–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Umar A, et al. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65–73 [DOI] [PubMed] [Google Scholar]
- 45. Wang SC, et al. 2006. Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8:1359–1368 [DOI] [PubMed] [Google Scholar]
- 46. Woodcock CL, Skoultchi AI, Fan Y. 2006. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14:17–25 [DOI] [PubMed] [Google Scholar]
- 47. Zhu N, Hansen U. 2007. HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors. Mol. Cell. Biol. 27:8859–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu N, Hansen U. 2010. Transcriptional regulation by HMGN proteins. Biochim. Biophys. Acta 1799:74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]