Abstract
Brucella ovis is a rough bacterium—lacking O-polysaccharide chains in the lipopolysaccharide—that is virulent in its natural host and whose virulence mechanisms remain almost unexplored. In a search for additional traits that distinguish B. ovis from smooth Brucella, which require O-polysaccharide chains for virulence, we have analyzed the significance in B. ovis of the main virulence factors described for smooth Brucella. Attempts to obtain strains of virulent B. ovis strain PA that are mutated in the BvrR/BvrS two-component regulatory system were unsuccessful, suggesting the requirement of that system for in vitro survival, while the inactivation of bacA—in contrast to the results seen with smooth Brucella—did not affect splenic colonization in mice or behavior in J774.A1 murine macrophages. Defects in the synthesis of cyclic ß-1,2 glucans reduced the uptake of B. ovis PA in macrophages and, although the intracellular multiplication rate was unaffected, led to attenuation in mice. Growth of strains with mutations in the type IV secretion system (encoded by the virB operon) and the quorum-sensing-related regulator VjbR was severely attenuated in the mouse model, and although the mutant strains internalized like the parental strain in J774.A1 murine macrophages, they were impaired for intracellular replication. As described for B. melitensis, VjbR regulates the transcription of the virB operon positively, and the N-dodecanoyl-dl-homoserine lactone (C12-HSL) autoinducer abrogates this effect. In contrast, no apparent VjbR-mediated regulation of the fliF flagellar gene was observed in B. ovis, probably due to the two deletions detected upstream of fliF. These results, together with others reported in the text, point to similarities between rough virulent B. ovis and smooth Brucella species as regards virulence but also reveal distinctive traits that could be related to the particular pathogenicity and host tropism characteristics of B. ovis.
INTRODUCTION
Brucella ovis is the etiological agent of contagious ram epididymitis. The typical traits of B. ovis infections are epididymitis, orchitis, and sterility in rams, but the microorganism has also been associated with occasional abortions in ewes and to an increase in perinatal mortality and low weight in lambs (8).
The virulence of Brucella is related to its resistance to bactericidal compounds released by the host and to its ability to survive and replicate inside phagocytes (38). Additionally, Brucella uses a stealth strategy to escape recognition by the innate immune response of the host, thus enabling the bacteria to reach their replication niche before the development of specific adaptive immunity (38). The particular characteristics of the Brucella outer membrane (OM), especially those associated with lipopolysaccharide (LPS), are thought to be crucial for this behavior (29, 38). Strains of the genus Brucella may be smooth (S) or rough (R), depending on the presence or absence, respectively, of O-polysaccharide (O-PS) chains in the LPS. O-PS masks other components of the bacterial surface (6), and since R mutant (M) strains derived from S strains are impaired in the interaction with phagocytes and are attenuated in animal models of infection (22, 40, 44), it is considered a key virulence factor in S Brucella species (such as B. melitensis, B. abortus, and B. suis).
Nevertheless, B. ovis is a naturally R Brucella species that is virulent in its natural host and in laboratory animal models (9, 33, 51). This ability of B. ovis to infect the host despite the absence of O-PS in the OM, together with its tropism for the male genital tract, is indicative of virulence mechanisms differing, at least in part, from those employed by S Brucella species, which are able to colonize the placenta and typically induce abortions. The differences that distinguish B. ovis from S B. melitensis, which also has sheep as a preferred host, are particularly remarkable. However, while B. melitensis induces abortions in ewes and is the Brucella species with the highest zoonotic potential—causing the most severe cases of human brucellosis—B. ovis has never been reported as a human pathogen and mainly infects rams. Little is known about the virulence of B. ovis, although some clues have been obtained from a study that deciphered the whole genome sequence (53) and from a small number of studies performed with individual genes (9, 33, 51). Additionally, it is known that—similarly to S Brucella and in contrast to R derivatives of S strains (45)—B. ovis enters the macrophage through cholesterol-rich lipid rafts (36). However, the mechanism is independent of the activity of phosphatidylinositol 3-kinase (PI3K) (36), an activity that has been reported to be necessary for S Brucella internalization (23).
Although Brucella cells lack many classical structures commonly involved in the establishment of bacterial infections—such as capsules, virulence plasmids, pili, and fimbriae—several virulence factors other than O-PS have been identified in S Brucella species. Among them, the type IV secretion system (T4SS), encoded by the virB operon, the quorum-sensing (QS)-related transcriptional regulator VjbR, the BvrR/BvrS two-component regulatory system, the periplasmic cyclic ß-1,2 glucans (CßGs), and BacA protein have been shown to be involved in survival inside phagocytes and virulence in animal models (7, 14, 15, 31, 42, 52). However, to date their significance along the course of infections caused by R Brucella species has not been assessed.
In the present report, we describe the characterization of a set of mutant strains obtained by inactivation of the virB operon or of the virB2, vjbR, cgs (coding for the CßG synthase), and bacA genes in virulent R B. ovis strain PA. Their OM-related properties, their behavior in murine macrophages, and their virulence in mice are analyzed in comparison to those previously reported for the corresponding mutants of S virulent strains. A preliminary characterization of the VjbR regulatory network in B. ovis PA and its connection with QS are also reported.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and cloning vectors.
The bacterial strains and plasmids used in this work are listed in Table S1 in the supplemental material. B. ovis PA—obtained from the Institut National de la Recherche Agronomique (INRA), Nouzilly, France—was used as parental strain. It is a virulent strain—isolated from a naturally infected ram—that has been extensively used as a challenge strain for the evaluation of vaccines in rams and mice (26, 40). B. ovis strains were cultured on tryptic soy agar (TSA; Conda, Madrid, Spain) or in tryptic soy broth (TSB; Conda) supplemented with 0.3% yeast extract (YE; Conda) and 5% horse serum (HS; Gibco-Invitrogen, Grand Island, NY) (TSA-YE-HS or TSB-YE-HS, respectively). Incubations were performed at 37°C in a 5% CO2 atmosphere, and liquid cultures were shaken at 115 rpm. When required, the medium was supplemented with 5% sucrose (Sigma-Aldrich, St. Louis, MO) and/or kanamycin (Kan) or ampicillin (Amp) (50 μg/ml), 5 μM N-dodecanoyl-dl-homoserine lactone (C12-HSL; Sigma) in acetonitrile (ACN; Sigma) or with the same volume of ACN alone. The number of CFU per milliliter in bacterial suspensions was estimated by optical density (OD) readings at 600 nm (OD600) and confirmed by retrospective quantification.
Plasmids pGEM-T Easy, pGEM-T, and pGEM-7Zf (Promega, Madison, WI), pUC19 (New England BioLabs [NEB], Ipswich, MA), pCVD442 and pCVD-KanD (this work), and pBBR1MCS-2 and pBBR1MCS-4 were used as cloning vectors. Plasmid pCVD-KanD was obtained at our laboratory by replacing the Amp resistance (Ampr) gene of pCVD442 by the Kanr gene in the same orientation. Briefly, the Ampr gene of pCVD442 was removed by enzymatic digestion with AspI and NdeI (Roche Diagnostics GmbH, Mannheim, Germany) and replaced—after treatment of the plasmid with Klenow (Roche) to obtain blunt ends—by the Kanr gene of pUC4K (GE Healthcare Europe GmbH, Barcelona, Spain) previously extracted by digestion with HindII (Roche). Plasmids were propagated either in Escherichia coli JM109 (Promega) or, in the case of pCVD442-derived plasmids, in E. coli CC118(λpir) as previously described (9).
Primers and nucleic acid techniques.
Primers (IDT, Leuven, Belgium) are listed in Table S2 in the supplemental material. PCR amplification with an Expand Long Template PCR system (Roche), Southern blot hybridization with digoxigenin (DIG)-labeled probes, and DNA sequencing were performed as described previously (9). ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) was used for nucleotide sequence alignments.
Endpoint reverse transcription-PCR (RT-PCR) and real-time quantitative RT-PCR (qRT-PCR) were performed with RNA extracted with an RNeasy minikit (Qiagen, Hilden, Germany) from bacteria cultured in TSB-YE-HS, TSB-YE-HS-ACN, or TSB-YE-HS-C12HSL (starting OD600 = 0.01). A culture volume equivalent to 1 ml at OD600 = 1.5 was used to obtain RNA at the exponential-growth phase (20 to 24 h of incubation; OD600 ∼ 0.7) and stationary-growth phase (48 h of incubation; OD600 ∼ 2.3). RNA was treated with RQ1 DNase (Promega) prior to the synthesis of cDNA with a Transcriptor first-strand cDNA synthesis kit (Roche), using the random hexamers provided as primers for reverse transcriptase. Control reactions omitting the reverse transcriptase were performed.
Specific primers for the bacA (BOV_0388), cgs (BOV_0108), virB2 (BOV_A0062), virB8 (BOV_A0057), vjbR (BOV_A0110), fliF (BOV_A1051), babR (or blxR; BOV_0183), bvrR (BOV_2010), and omp25c (BOV_0116) target genes were used for subsequent PCRs. For endpoint RT-PCR, PCRs were performed using the synthesized cDNA and Go Taq Green master mix (Promega). A control PCR performed with the IF-1 housekeeping gene (BOV_0267) (18) was also carried out. The products amplified by RT-PCR were visualized after electrophoresis in 2% agarose gels. The absence of DNA contamination in RNA samples was confirmed by a PCR using the IF-1 gene in the cDNA synthesis reactions performed without reverse transcriptase.
A qRT-PCR using cDNA samples of selected strains was performed with FastStart Universal SYBR green Master (ROX) (Roche) and an Applied Biosystems 7000 real-time PCR system (Life Technologies). Two cDNA samples per strain were used, and triplicate reactions were done for each gene. Data were normalized by the 2−ΔΔCt method (32), using the IF-1 gene as the endogenous control. The cDNA obtained in the exponential- or stationary-growth phase from parental B. ovis PA cultured on TSB-YE-HS-ACN was used as the calibrator sample for comparisons among strains and culture conditions.
Immunological techniques.
Polyclonal antibodies (Abs) reacting with the OM proteins (OMPs) of the Brucella Omp25/Omp31 family were obtained previously by immunization of rabbits with recombinant OMPs purified from E. coli (34). The monoclonal antibodies (MAbs) A68/03F03/D05 (D05), A59/05F01/C09 (C09), and A01/08H06/G02 (G02)—specific for R-LPS, Omp25, and Omp31, respectively (11, 60)—were also used.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analyses performed with whole-cell lysates from B. ovis strains and rabbit sera or MAbs were carried out as described previously (34, 60). Dot blot analyses were performed with 2-fold serial dilutions of bacterial suspensions in phosphate-buffered saline (PBS) (starting at OD600 = 20) spotted onto nitrocellulose membranes (2 μl/spot). Before incubation with the D05 and G02 MAbs, the membranes were soaked in SDS, as described previously for a colony-blotting technique (59). This step was omitted with the C09 MAb, because better reactivity was obtained under those conditions. Antigen-antibody binding was detected as previously described for Western blotting (34, 60).
Generation of mutant strains.
A polar mutation in the virB operon of B. ovis PA was induced by replacement of an MluI-EcoNI DNA fragment—containing part of the genes virB2 and virB3 (BOV_A0062 and BOV_A0061, respectively)—by the Kanr cassette of pUC4K. Briefly, the DNA region to be deleted—together with upstream and downstream DNA—was PCR amplified with primers virBMUT-F and virBMUT-R and cloned into pGEM-T Easy. The EcoRI insert of the resulting recombinant plasmid was cloned, in the direction opposite that of lacZ, in pGEM-7Zf, and then the SacI-XbaI insert was cloned in pUC19 to obtain plasmid pNVvirPA03. The Kanr gene was PCR amplified from pUC4K with primers KanF-MluI and KanR-EcoNI, cloned into pGEM-T, extracted by digestion with MluI (NEB) and partial digestion with EcoNI (NEB), and ligated to pNVvirPA03 cut with the same restriction enzymes. The SacI-XbaI insert DNA of the resulting plasmid, bearing inactivated virB2 and virB3, was ligated to SacI-XbaI-digested pCVD442 to afford pNVvirPA05. Plasmid pNVvirPA05 was introduced into B. ovis PA by electroporation with a Micropulser (Bio-Rad, Hercules, CA), and the recombinant bacteria were selected by plating on TSA-YE-HS-Kan plates. All the Kanr colonies were also Ampr, which is consistent with single homologous recombination events resulting in the integration of the entire plasmid in the bacterial chromosome (intermediate [I] strain). The second crossover event—leading either to the replacement of the wild-type DNA fragment by inactivated virB2-virB3 or to a revertant (Rv) bacterium recovering the wild-type genotype—was selected by plating the intermediate strain onto TSA-YE-HS with 5% sucrose. Mutant and revertant bacteria were detected by their resistance and susceptibility to Kan, respectively, and their genotypes were confirmed by PCR amplification, Southern blot hybridization, and sequencing of the target locus in the genome.
A ΔvirB2 nonpolar mutant was also obtained by in-frame deletion of virB2 by overlapping PCR (13). Briefly, the 5′ end of virB2 and upstream DNA were amplified by PCR using primers virBMUT-F and virB2-R Ovl, while the 3′ end and downstream DNA were amplified with primers virB2-F Ovl and virB-R3. Both fragments were fused—through the overlapping regions of primers virB2-R Ovl and virB2-F Ovl—by PCR with primers virBMUT-F and virB-R3. The amplified fragment was cloned in pGEM-T Easy, then in pGEM-7Zf, and finally in XbaI-SacI-digested pCVD-KanD to afford pNVvirB204. B. ovis PA was transformed with pNVvirB204 by electroporation, and the intermediary, revertant, and mutant strains were obtained as described for the polar mutant. Revertant and mutant strains were identified by PCR amplification and confirmed as described above.
In-frame deletion by overlapping PCR was also used to obtain Δcgs, ΔvjbR, and ΔbacA nonpolar mutants of B. ovis PA. For cgs inactivation, the 5′ end of the gene was amplified with primers cgsMUT-F and cgs-R Ovl whereas the 3′ end was amplified with primers cgs-F Ovl and cgsMUT-R. The overlapping PCR was performed with both fragments and primers cgsMUT-F and cgsMUT-R. For inactivation of vjbR, the first two PCRs—performed with primers vjbRMUT-F and vjbR-R Ovl (5′ end) and primers vjbR-F Ovl and vjbRMUT-R (3′ end)—were fused by PCR amplification with primers vjbRMUT-F and vjbRMUT-R. The ΔbacA mutant was obtained by overlapping PCR amplification with primers bacAMUT-F and bacAMUT-R of the 5′ and 3′ ends previously amplified with primers bacAMUT-F and bacA-R Ovl (5′) and bacA-F Ovl and bacAMUT-R (3′). For inactivation of bvrR, the 5′ end (amplified with primers bvrRS-F3 and bvrRS-R2 Ovl) and the 3′ end (amplified with primers bvrR-F2 Ovl and bvrR-R2) were fused by PCR with primers bvrRS-F3 and bvrR-R2. Overlapping PCR with primers bvrSMUT-F and bvrS-R of the 5′ and 3′ ends previously amplified with primers bvrSMUT-F and bvrS-SecR (5′) and primers bvrS-F Ovl and bvrS-R (3′) was used to inactivate bvrS (BOV_2011). The products of the final overlapping PCRs were cloned into pGEM-T or pGEM-T Easy and subcloned in pCVD-KanD to give recombinant pNVbacA03, pNVcgs03, pNVvjbR03, pPSbvrROVL02, and pPSbvrSOVL02. Electroporation of B. ovis PA with recombinant pCVD-KanD and the selection of intermediate, mutant, and revertant strains were performed as described for the ΔvirB2 mutant.
For each target gene, two independent mutant strains, from two intermediate colonies selected after the first crossover event, were obtained and analyzed. A strain reverting to the wild-type genotype was also selected as a control. Since the results obtained with the two mutants were consistently reproducible, data from only one mutant strain for each gene are shown in Table S1 in the supplemental material and reported here. The revertants behaved like the parental strain and are not described in Table S1 in the supplemental material.
Complementation of the mutant strains.
The virB2, cgs, and vjbR genes were amplified by PCR with primer pair virB2-F com and virB2-R com, primer pair cgs-F com and cgs-R com, and primer pair vjbR-F com and vjbR-R com, respectively. The virB2 and vjbR amplicons were cloned into pGEM-T Easy in the orientation opposite that of lacZ, sequenced to verify the open reading frame, extracted by digestion with ApaI and SacI, and cloned in pBBR1MCS2. The resulting plasmids pNVvirB202-com and pNVvjbR03-com were introduced by electroporation into B. ovis pNVvirB204M (ΔvirB2) and B. ovis pNVvjbR03M (ΔvjbR), respectively. The complemented strains were selected on the basis of their resistance to Kan by plating on TSA-YE-HS-Kan. Surprisingly, after several electroporation assays, no bacterial colonies were obtained following transformation with pNVvjbR03-com. Therefore, electroporation was also attempted with pNVvjbR04-com, a recombinant pBBR1MCS4 plasmid containing the KpnI-SacI insert of pNVvjbR03-com. The complemented mutant B. ovis pNVvjbR03M-com was selected based on its resistance to Amp. The cgs amplicon cloned into pGEM-T Easy in the same orientation as lacZ was extracted by partial digestion with NotI and cloned into the NotI site of pBBR1MCS2. The resulting plasmid, pNVcgs02-com, was electroporated into B. ovis pNVcgs03M (Δcgs), and the complemented mutant (B. ovis pNVcgs03M-com2) was selected by its resistance to Kan. Complementation of the mutant strains was checked by PCR amplification and detection of the corresponding transcripts by RT-PCR.
Susceptibility assays and autoagglutination.
Susceptibility to nonimmune human serum (4 h of exposure), acid stress (5 h of exposure to 25 mM citrate and 120 mM NaCl, pH 4), polymyxin B (Sigma) (1 mg/ml), and sodium deoxycholate (Sigma) (0.1 mg/ml) (1 h exposure) was expressed as the percentage of survival after exposure. It was determined by comparison of levels of CFU in untreated (100% survival) and treated bacterial suspensions, as described previously (9, 35). For human serum assays, 100% survival was attributed to serum heated at 56°C for 30 min to remove complement. Susceptibility to hydrogen peroxide was determined by measuring the diameter of the halo around a Whatman 3MM Chr disk (9-mm diameter) soaked in 10 μl of 30% H2O2 (Sigma) (9). Autoagglutination capacity was evaluated by measuring the evolution, over 48 h of static incubation, of the OD600 of bacterial suspensions with initial OD600 readings of 0.8 (100% OD600) in TSB-YE-HS (9).
Infection and intracellular survival of B. ovis strains in murine macrophages.
Experiments with murine macrophage-like J744.A1 cells (DSMZ ACC170) were performed as described previously (33). Briefly, macrophages were infected with bacterial suspensions (multiplicity of infection of 200 CFU/cell). Extracellular bacteria were killed by incubation with gentamicin, and at several intervals postinfection (p.i.), the numbers of viable intracellular bacteria were determined in three wells per experimental condition (33). The results were expressed as means ± standard deviations (SD) (n = 3) of the log CFU/well at each selected p.i. time point (0, 24, 48, or 72 h).
The cytotoxicity of B. ovis strains for macrophages was evaluated as described previously (36), measuring lactate dehydrogenase (LDH) activity with the CytoTox 96 nonradioactive cytotoxicity assay (Promega).
Virulence studies in mice.
Female 7-week-old BALB/c mice (Charles River Laboratories, Barcelona, Spain) were used. They were randomly distributed into experimental groups and kept with water and food made available ad libitum at the animal experimentation facilities of the University of Salamanca (registration number PAE SA-001). The procedures performed with mice were designed according to Spanish and European legislation regarding the use of animals in research (RD 1201/05 and directive 2010/63/UE).
The mice were inoculated by intraperitoneal injection of bacterial suspensions into PBS (106 CFU/mouse), and bacterial splenic CFU numbers were determined at 3 and 8 weeks p.i. in five mice per group, as previously described (9). The results are expressed as means ± SD (n = 5) of the log CFU/spleen.
Statistical analyses.
Statistical comparisons were performed using analysis of variance. The levels of significance of the differences between the experimental groups were determined with the post hoc Fisher's protected least significant difference (PLSD) test.
RESULTS
Inactivation of target genes in virulent B. ovis PA and complementation of the mutant strains.
Mutant (M) and revertant (Rv) strains were obtained for each target gene from an intermediate (I) B. ovis strain bearing one copy of the wild-type gene and one copy of the inactivated gene. M strains were obtained for all target genes, with the exception of bvrR and bvrS, because their corresponding I strains always reverted to the parental genotype when incubated with sucrose. All I, M, and Rv strains afforded the expected HindIII band pattern corresponding to the published genome sequence when analyzed by Southern blot hybridization with probes constituted by the respective recombinant plasmids used for electroporation. PCR amplification and sequencing of the target loci in the genome of the five mutant strains confirmed the proper inactivation of the respective genes. Lack of transcription of the inactivated gene in each mutant strain, in comparison with the parental strain, was confirmed by endpoint RT-PCR with the primers listed in Table S2 in the supplemental material. Transcription of virB8 was detected in B. ovis pNVvirB0204M (ΔvirB2), whereas virB2 transcription was not, which revealed that deletion of virB2 in this strain was a nonpolar mutation for downstream genes of the virB operon. In contrast, the polar effect on downstream genes caused by the replacement of virB2-virB3 by the Kanr cassette was revealed by the absence of both virB2 and virB8 transcripts in the B. ovis pNVvirPA05M (ΔvirBp) strain (data not shown).
All mutants behaved like the parental strain in classic bacteriological typing for the genus Brucella and showed no relevant differences in growth patterns in either liquid or solid medium compared to B. ovis PA (data not shown).
The isolation of the complemented ΔvjbR and Δcgs mutants was problematic, probably due to overexpression of the encoded proteins in the complemented strains. Several attempts to obtain colonies of the B. ovis ΔvjbR mutant complemented in trans with plasmid pNVvjbR03-com (Kanr; see Table S1 in the supplemental material) were unsuccessful. Using pNVvjbR04-com (Ampr; see Table S1 in the supplemental material) for complementation, a small number of normal-sized colonies was recovered from among hundreds of tiny colonies whose growth had ceased. These results, which were obtained with both the parental strain and the ΔvjbR mutant, suggest that VjbR levels must be finely tuned in B. ovis, which would explain why the wild-type phenotype was not recovered in the selected ΔvjbR-com strain (see below), a strain that was able to transcribe vjbR but with 7-fold-increased levels compared to the levels seen with the parental strain, as shown by qRT-PCR (data not shown). After several assays, only one Kanr colony of the B. ovis pNVcgs03M mutant was obtained after electroporation with pNVcgs02-com. Although the large size of cgs (8,601 bp) might account for this result, both the parental strain and the Δcgs mutant were readily transformed with pNV998-10, another pBBR1MCS2 recombinant plasmid, bearing a DNA insert of about 14 kb (57), but not with pNVcgs02-com. Since the transcription of cgs was not detected in this single complemented strain, it is likely that overexpression of the encoded protein is not tolerated in B. ovis. This interpretation is supported by the apparent weak transcription of cgs in B. ovis PA (data not shown).
OM-related properties of the B. ovis mutant strains.
Considering the relevance of the OM in the interaction of pathogens with the host, and since the inactivated genes are related—in one way or another—to the OM, several tests were performed to evaluate the OM characteristics of the mutant strains.
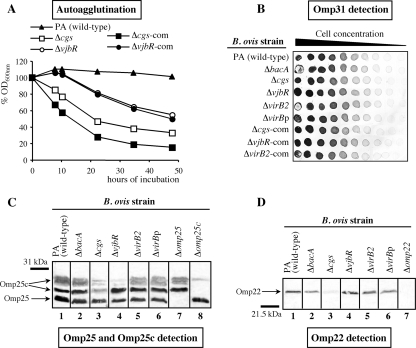
In agreement with previous results (9, 35), parental B. ovis PA remained in suspension after 48 h of static incubation (Fig. 1A). Similarly, the ΔbacA, ΔvirB2, and ΔvirBp mutants and all the revertant strains did not agglutinate (data not shown). The OD600 values determined for the ΔvjbR mutant and its complemented strain decreased progressively after the first 10 h of incubation, reaching about 55% of the initial value after 48 h (Fig. 1A). The Δcgs mutant and its complemented strain showed the most significant autoagglutinating ability, with OD600 readings below 40% of the initial value at the end of the experiment (Fig. 1A).
Fig 1.
Autoagglutination properties (A) and inmmunological detection of Omp31 (B), Omp25c (C), and Omp22 (D) in B. ovis strains. (A) For autoagglutination assays, the OD600s of bacterial suspensions with initial readings of 0.8 (100% OD600) were determined over 48 h of static incubation (9). The ΔbacA, ΔvirB2, and ΔvirBp mutants behaved like the parental strain and are not shown. (B) Detection of Omp31 was performed by semiquantitative dot blotting with MAb G02. Detection of Omp25 and R-LPS with the MAbs C09 and D05 did not evidence differences among strains and it is not shown in the figure. (C and D) Detection of Omp25c (C) and Omp22 (D) was performed with polyclonal antibodies obtained by immunizing rabbits with the recombinant proteins (34). Omp25 is also revealed with the Omp25c antisera (C) because of the antigenic similarities of the two proteins. All revertant strains behaved like the parental strain in all the tests and are not shown.
Although all B. ovis mutants showed at least one statistically significant difference compared to the parental strain, none of the induced mutations caused dramatic effects regarding susceptibility to polymyxin B, sodium deoxycholate, hydrogen peroxide, nonimmune human serum, or acid pH (Table 1). Strikingly, several mutant strains were more resistant than the parental strain in some assays. In particular, the ΔbacA mutant did not show any detrimental difference from the parental strain and was more resistant to polymyxin B and acid pH (Table 1).
Table 1.
Susceptibility to polymyxin B, sodium deoxycholate, hydrogen peroxide, acid pH, and nonimmune human serum of B. ovis PA and its derived mutant strainsa
| B. ovis strain | % survival after exposure to: |
Inhibition zone diam (cm) with H2O2 | |||
|---|---|---|---|---|---|
| Polymyxin B (1 mg/ml) (1 h) | Na deoxycholate (0.1 mg/ml) (1 h) | Nonimmune human serum (4 h) | Citrate buffer pH 4 (5 h) | ||
| B. ovis PA | 63.40 ± 1.51 | 95.62 ± 8.82 | 74.95 ± 4.32 | 61.15 ± 1.67 | 5.48 ± 0.15 |
| B. ovis ΔbacA | 82.03 ± 4.18* | 91.94 ± 2.76 | 83.05 ± 2.67 | 70.66 ± 7.69* | 5.28 ± 0.16 |
| B. ovis Δcgs | 65.94 ± 15.89 | 91.36 ± 12.95 | 54.48 ± 7.42** | 53.59 ± 5.01 | 5.69 ± 0.06 |
| B. ovis ΔvjbR | 47.77 ± 3.74* | 93.26 ± 4.80 | 85.97 ± 5.59* | 57.98 ± 4.31 | 5.80 ± 0.25* |
| B. ovis ΔvirB2 | 49.30 ± 2.81* | 91.06 ± 1.37 | 66.70 ± 6,40 | 73.52 ± 1.65* | 5.34 ± 0.13 |
| B. ovis ΔvirBp | 64.75 ± 1.99 | 91.64 ± 18.69 | 84.51 ± 6.05 | 72.50 ± 3.62* | 5.43 ± 0.18 |
Values represent means ± SD (n = 3). Statistical comparisons were performed with the PLSD test. Significant differences between the mutant strains and parental B. ovis PA are marked as follows: **, P ≤ 0.005; *, P ≤ 0.05.
No significant differences were found between the set of B. ovis mutants and the parental strain as regards their reactivity with MAbs specific for Omp25, Omp31, and R-LPS in assays using an indirect enzyme-linked immunosorbent assay (iELISA) with whole cells (data not shown), Western blotting (data not shown), or semiquantitative dot blotting (Fig. 1B for Omp31 and data not shown). In agreement with previous results (34), Western blotting with the anti-Omp25c serum gave the expected band pattern with B. ovis PA, revealing two protein bands of 27.5 and 29.5 to 30.5 kDa corresponding to Omp25c and, because of its antigenic similarity, the Omp25 protein band of 25.9 kDa (Fig. 1C, lane 1). This same profile of bands was found with the ΔbacA, ΔvirB2, and ΔvirBp mutants (Fig. 1C, lanes 2, 5, and 6), while the upper Omp25c band was less evident in the ΔvjbR mutant (Fig. 1C, lane 4), and both Omp25c bands were weaker in the Δcgs mutant (Fig. 1C, lane 3). With the predictable exception of the B. ovis Δomp22 mutant obtained previously (9, 34) (Fig. 1D, lane 7), Omp22 was detected as a band of the expected size in all the B. ovis strains (Fig. 1D), but it was almost unnoticeable in the Δcgs mutant (Fig. 1D, lane 3).
Contribution of the transcriptional regulator VjbR and the C12-HSL QS autoinducer to the expression of selected genes in B. ovis PA.
As a first step in elucidating the quorum-sensing (QS)-related regulation in B. ovis, we analyzed in B. ovis PA and its derived ΔvjbR mutant the transcription levels of several genes involved in the virulence of S Brucella and/or whose expression has previously been found to be regulated transcriptionally by VjbR and/or C12-HSL in B. melitensis 16M (14, 46, 55, 62).
Since VjbR is a transcriptional regulator of the virB operon in B. melitensis 16M (14, 54, 62), we initially performed endpoint RT-PCR with specific primers for virB2 (Fig. 2). Transcription of virB2 in the parental B. ovis PA strain during the exponential-growth phase was higher than that observed in the stationary-growth phase (Fig. 2A and B, lanes 3) and also higher than that detected for the ΔvjbR mutant in both growth phases (Fig. 2A and B, lanes 8). These results agree with the idea of a role for VjbR as a transcriptional activator of virB expression also in B. ovis PA.
Fig 2.
Endpoint RT-PCR with RNAs extracted from B. ovis PA and the ΔvjbR mutant during the exponential-growth phase (OD600 ∼ 0.7) (A) or stationary-growth phase (OD600 ∼ 2.3) (B) to evaluate the expression of virB. Each target gene was amplified with the primers described in Table S2 in the supplemental material and visualized after electrophoresis through the use of a 2% agarose gel. The constitutively expressed IF-1 gene (18) was used as an endogenous reference. Control RT-PCRs performed without reverse transcriptase and the IF-1 target gene (lanes 4 and 9) gave no amplification product. The results are representative of two independent bacterial cultures. An inverted image of the original picture is shown.
Additional information was obtained by qRT-PCR analysis of selected genes in the parental strain and the ΔvjbR mutant cultured in the presence or absence of C12-HSL to the exponential- and stationary-growth phases (Table 2). With the exception of the expected lack of expression of vjbR in the ΔvjbR mutant, no remarkable differences were found between B. ovis PA and the ΔvjbR mutant as regards the transcription of bvrR, cgs, fliF—coding for a flagellar MS-ring protein (20)—or vjbR (Table 2). Since the VjbR-mediated regulation of fliF transcription in B. melitensis 16M has been described previously, we performed an alignment of DNA sequences upstream of fliF in the published genomes of B. melitensis 16M and B. ovis 63/290 (Fig. 3). This analysis revealed two deletions in B. ovis that were flanked by direct repeats of 7 bp in the corresponding DNA region of B. melitensis (Fig. 3). These direct repeats were probably involved in the genesis of the B. ovis deletions through a slipped mispairing mechanism, leaving only one of the repeats.
Table 2.
Analysis by qRT-PCR of the relative transcriptional levels of selected genes in parental B. ovis PA and the ΔvjbR mutant cultured in the presence or absence of C12-HSL to the exponential- or stationary-growth phasea
| B. ovis strain | Culture medium supplement | Growth phase of RNA extraction | Relative transcription level of indicated target gene (2−ΔΔCT) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| babR (blxR) | bvrR | cgs | fliF | omp25c | virB2 | vjbR | |||
| PA | ACN | Exponential | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| PA | C12-HSL | Exponential | 2.570 | ND | 1.170 | 1.330 | 1.006 | 0.086 | 0.893 |
| ΔvjbR | ACN | Exponential | 2.093 | 1.078 | 0.963 | 1.219 | 1.601 | 0.009 | 0.000 |
| ΔvjbR | C12-HSL | Exponential | 2.013 | ND | ND | 1.086 | 1.583 | 0.008 | 0.000 |
| PA | ACN | Stationary | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| PA | C12-HSL | Stationary | 1.049 | ND | 1.215 | 0.721 | 0.482 | 0.328 | 0.931 |
| ΔvjbR | ACN | Stationary | 0.906 | 0.871 | 1.188 | 0.889 | 0.482 | 0.220 | ND |
| ΔvjbR | C12-HSL | Stationary | 0.925 | ND | ND | 0.842 | 0.619 | 0.138 | ND |
Relative transcription quantification was performed with the 2−ΔΔCT method (32), using the IF-1 gene as an endogenous reference gene and B. ovis PA cultured in the presence of ACN (solvent for C12-HSL) up to the exponential-growth phase (DO600 ∼ 0.7) or stationary-growth phase (DO600 ∼ 2.3) as the benchmark. Two cDNA samples—and three PCRs per gene and cDNA—were used to calculate relative transcription levels under each set of strain and culture conditions. Increased or reduced transcription of each gene in comparison with that seen with B. ovis PA cultured with ACN (2−ΔΔCT = 1) is indicated by values > 1 or < 1, respectively. Gene expression levels were considered different when the value was >1.5 or <0.5. ND, not determined.
Fig 3.
Alignment of DNA sequences upstream of fliF in B. melitensis 16M (ME) and B. ovis 63/290 (OV). The published genome sequences for chromosome II were used for alignment (NC_003318 and NC_009504) with ClustalW2. Only the 5′ end of fliF is shown (in italics), and its ATG start codon, determined according to the B. melitensis 16M genome annotation, is shown with an arrow. The two pairs of direct repeats flanking the deletions detected in B. ovis are framed in the B. melitensis 16M sequence. The predicted DNA-binding region for FtcR (30), a regulator of the flagellar system, is underlined.
In agreement with the results obtained by RT-PCR (Fig. 2), the most important differences in gene transcription were observed with virB2 (Table 2). In the exponential-growth phase, the transcription of virB2 in the parental strain was reduced to a considerable extent upon incubation in the presence of C12-HSL, and a more pronounced decrease was observed in the ΔvjbR mutant incubated with or without the autoinducer. During the stationary-growth phase, the transcription of virB2 was downregulated to similar extents in the parental strain incubated with C12-HSL and in the ΔvjbR mutant under both culture conditions. However, the levels of differences seen with the parental strain incubated without the autoinducer were lower than those detected in the exponential-growth phase (Table 2).
No differences regarding the expression of babR—also designated blxR and coding for another QS-related regulator (46, 55)—were found between strains and culture conditions in the stationary-growth phase. During the exponential-growth phase, however, the levels of babR transcripts increased in the ΔvjbR mutant, regardless of the presence or absence of the autoinducer in the culture medium, and also in the parental strain when it was incubated with C12-HSL (Table 2). Although the differences between strains and culture conditions regarding the relative levels of expression of omp25c—coding for a major OMP in B. ovis PA (34)—were not high, the transcription of omp25c seemed to be upregulated in the ΔvjbR mutant in the exponential-growth phase but downregulated in the stationary-growth phase (Table 2).
Dot blot analysis of R-LPS, Omp31, and Omp25 and Western blot analysis of Omp25c and Omp22 were performed during the stationary-growth phase with B. ovis PA and the ΔvjbR mutant cultured in the presence or absence of C12-HSL. No differences in antibody reactivity were observed between strains or culture medium compositions under the assay conditions tested (data not shown).
Internalization and intracellular multiplication of the B. ovis mutant strains in murine macrophages.
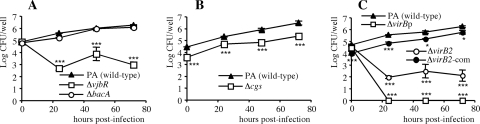
The intracellular CFU levels were determined at 0, 24, 48, and 72 h after the internalization period in J774.A1 murine macrophages. As expected from previous results (33, 36), the log values of CFU/well of the parental strain increased progressively from t = 0 (close to 5 log CFU/well) to the end of the experiment (about 6 log CFU/well at 72 h p.i.) (Fig. 4). Similar results were obtained with the B. ovis ΔbacA strain, pointing to the lack of attenuation of this mutant in the murine macrophage model (Fig. 4A).
Fig 4.
Uptake and intracellular survival in J774.A1 murine macrophages of strains ΔbacA and ΔvjbR (A), Δcgs (B), and ΔvirB2, ΔvirBp, and ΔvirB2-com (C) in comparison with parental B. ovis strain PA. Macrophages were infected with each B. ovis strain at a multiplicity of infection of 200 CFU/cell, extracellular bacteria were killed by incubation with gentamicin, and levels of intracellular bacteria were then determined at several time points (33). All the revertant strains behaved like the parental strain, and the ΔvjbR-com and Δcgs-com strains behaved like the respective mutants and are not shown in the figure. The results are expressed as the means ± SD (n = 3) of the log CFU/well at each time point. Significant differences (PLSD test) between the mutant strains and parental B. ovis PA at each time point are marked as follows: ***, P ≤ 0.0005; *, P ≤ 0.05.
In contrast, the remaining B. ovis mutants were impaired in their interactions with macrophages to different extents. Thus, internalization defaults were observed for the B. ovis Δcgs strain, which gave intracellular counts about 1 log lower than those obtained with the parental strain at t = 0. However, although the counts remained 1 log below those obtained with B. ovis PA, the increases in the CFU counts observed for the B. ovis Δcgs mutant throughout the experiment were similar to those obtained with the parental strain (Fig. 4B). The B. ovis ΔvjbR mutant internalized in macrophages in a manner similar to that observed with the parental strain, but 24 h after infection its CFU/well number had decreased by about 2 log (Fig. 4A). Thereafter, the ΔvjbR mutant survived inside macrophages but was impaired with respect to its multiplication (Fig. 4A).
The B. ovis ΔvirB2 nonpolar mutant showed behavior similar to that observed with the ΔvjbR mutant even though the bacterial counts after 24 h were about 0.5 log lower in the B. ovis ΔvirB2 mutant (Fig. 4C). The strongest attenuation in the macrophage model was observed with the strain that harbored a polar mutation with respect to the entire virB operon (ΔvirBp), which behaved like the parental strain regarding internalization but was cleared from the macrophages at 24 h p.i. (Fig. 4C).
The two independent mutant strains obtained for each inactivated gene gave the same results regarding their interactions with macrophages, and all the revertant strains behaved like the parental strain (data not shown). However, complementation with the wild-type gene cloned into a multicopy plasmid replicating in Brucella spp. restored the phenotype of the parental strain—and that not completely—only in the case of the nonpolar ΔvirB2 mutant (Fig. 4C). This is in agreement with the difficulties encountered with respect to reintroduction of wild-type vjbR and cgs cloned in the multicopy plasmid. Failure of efforts aimed at restoring the wild-type phenotype by complementation in trans with multicopy plasmids has often been reported previously for Brucella mutants (9, 16, 22); such failure has usually been attributed to overexpression of the protein, leading to negative effects on the cell.
The level of cytotoxicity for macrophages, measured by LDH release to the culture medium (33), was negligible for all mutant strains and parental B. ovis PA (data not shown), the latter strain behaving similarly to smooth B. abortus 2308 in this test (33).
Virulence of the B. ovis mutant strains in mice.
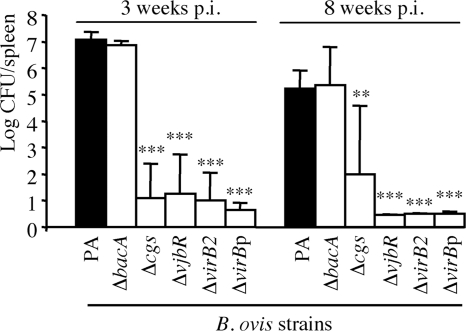
The spleen colonization by each mutant strain was evaluated at 3 and 8 weeks after intraperitoneal injection of an estimated inoculum of 106 CFU. The exact doses administered (CFU/mouse) for each B. ovis strain were determined retrospectively and were as follows: 1.1 × 106 for strain PA (parental), 1.15 × 106 for strain ΔbacA, 0.97 × 106 for strain Δcgs, 1.07 × 106 for strain ΔvjbR, 1.26 × 106 for strain ΔvirB2, and 1.26 × 106 for strain ΔvirBp.
Consistent with the results obtained for the interaction with murine macrophages in vitro, no statistically significant differences between the parental strain and the ΔbacA mutant in the numbers of CFU/spleen were found, with bacterial counts of about 7 and 5 log CFU/spleen at weeks 3 and 8 p.i., respectively (Fig. 5). In contrast, the Δcgs, ΔvjbR, ΔvirB2, and polar ΔvirB mutants showed a strong attenuation, 4 out 5 mice having cleared the bacterium from the spleen at week 3 p.i. At week 8 p.i., no bacteria were detected in the spleens of mice inoculated with the ΔvjbR, ΔvirB2, or ΔvirBp mutant and only one mouse of the group inoculated with the Δcgs mutant remained infected (Fig. 5).
Fig 5.
Colonization of B. ovis PA and the mutant strains in mouse spleen. Mice were inoculated intraperitoneally with an estimated dose of 1 × 106 CFU/mouse, and bacterial CFU in spleen were determined at 3 and 8 weeks p.i. The results are expressed as the means ± SD (n = 5) of the log CFU/spleen at each time point. The ΔvjbR, ΔvirB2, and ΔvirBp mutants were not detected at 8 weeks p.i. (the means and SD shown in the figure were obtained with the individual detection limit of each spleen). Significant differences (PLSD test) between the mutant strains and parental B. ovis PA are marked as follows: ***, P ≤ 0.0005; **, P ≤ 0.005.
DISCUSSION
Strains mutated for bacA, cgs, vjbR, virB2, and the virB operon were obtained from virulent B. ovis strain PA. Attempts to construct ΔbvrR and ΔbvrS mutants by in-frame deletion of the genes were unsuccessful, because the intermediate strains of the mutation procedure always reverted to the wild-type genotype during growth on sucrose-supplemented medium (data not shown). These results suggest that the two-component BvrR/BvrS regulatory system is required for the in vitro survival of B. ovis PA and are consistent with the multiple functions assigned to this regulatory system in B. abortus 2308 (control of metabolism, OM homeostasis, expression of key virulence genes such as the virB operon and vjbR, etc.) (37, 56). They are also in accord with the findings that bvrR and bvrS mutants have been obtained only by transposon mutagenesis in B. abortus 2308 and B. suis 1330 (28, 52) and that other attempts of deletion by homologous recombination have failed (49).
T4SSs are multiprotein complexes that span the cell envelope of diverse prokaryotes and translocate nucleoprotein complexes or proteins to or from the surrounding environment. The T4SS is recognized as a major virulence determinant in S Brucella, participating in the bacterial intracellular trafficking that leads to the biogenesis of the replicative niche in the endoplasmic reticulum of host cells (1, 10, 12). It is constituted of 12 proteins encoded by the virB genes (42) that are cotranscribed as an operon in S virulent Brucella strains (16, 17) and, according to our results, also in B. ovis PA (data not shown). Although the significance for the host-pathogen interaction is unknown, the optimal conditions for in vitro virB expression are dependent on the Brucella strain (48), and a new expression pattern was seen for B. ovis PA (Fig. 2). Unlike B. suis 1330, which requires minimal medium for expression (48), and in concordance with the results reported for B. abortus 2308, B. melitensis 16M, and B. ovis Reo198 (48), B. ovis PA is able to transcribe virB in rich medium (Fig. 2). However, maximal expression in B. ovis PA occurs in the exponential-growth phase (Fig. 2) whereas in B. abortus 2308 it occurs after entry into the stationary phase (17, 50).
In agreement with the observations reported for S Brucella strains (16, 17, 42, 44), the T4SS is also essential for the multiplication and survival of R virulent B. ovis strain PA in mice and in J774.A1 macrophages (Fig. 4 and 5). However, these results differ from those published for an R mutant (ΔmanBA) of S B. melitensis 16M. The intracellular counts of that mutant decreased over time in J774.A1 murine macrophages, whereas the double mutant 16M ΔmanBA ΔvirB2 showed an unexpected resistance to killing by macrophages in the absence of T4SS that was coupled to a loss of the cytotoxic effect of the single ΔmanBA mutant (44). In contrast, the cytotoxic effect for macrophages—which has also been observed in R mutants derived from S B. abortus (43)—was not evidenced with B. ovis PA or the mutant strains obtained in this work (data not shown). The cytotoxicity of R Brucella mutants has been suggested to be due to an increased secretion of unknown cytotoxin molecules into the host cell, which could be favored by the exposure of the T4SS on the bacterial surface due to the absence of O-PS in the LPS (43). Since important OM differences between B. ovis, S strains, and R derivatives of S strains have been previously reported (35, 58), the possibility that the T4SS is also shielded in B. ovis, in spite of the absence of O-PS, cannot be discounted.
Consistent with previous observations ruling out the participation of the T4SS in the uptake of Brucella spp. into phagocytes (16, 17, 50), neither the ΔvirB2 nor the ΔvirBp mutant of B. ovis PA exhibited internalization defects in murine macrophages (Fig. 4C). However, the ΔvirB2 nonpolar mutant showed a reduction in numbers of about 3 log at 24 h p.i. but persisted intracellularly thereafter (Fig. 4C), while the virBp mutant was cleared from murine macrophages between 0 and 24 h p.i. (Fig. 4C). These results suggest either that the loss of only virB2 still allows some T4SS function or that the removal of the whole T4SS structure, which connects the cytoplasm with the surrounding environment, produces alterations in the bacterial architecture that increase susceptibility to host defenses. In this respect, with the exception of the higher susceptibility of the ΔvirB2 nonpolar mutant to the cationic peptide polymyxin B (Table 1), no significant differences were detected between the two B. ovis mutants with respect to their OM-related properties, although hypotheses concerning other distinctive traits cannot be discarded. In contrast to the results reported by Wang et al. for a virB mutant derived from B. melitensis 55009 (61), we did not detect significant differences between B. ovis PA and the isogenic ΔvirB mutants regarding the Omp25/Omp31 family or autoagglutination ability (Fig. 1 and data not shown). Although the differences in the methodological approaches should be kept in mind, these discrepancies could be related to the OM differences that are known to exist between B. melitensis and B. ovis (34, 35, 58). However, since the characteristics of B. melitensis 55009 (smooth or rough, virulent or attenuated, etc.) were not detailed by Wang et al. (61), objective comparisons with rough virulent B. ovis PA are difficult to establish.
In B. melitensis 16M, the QS-related regulator VjbR, together with C12-HSL—an acyl homoserine lactone (AHL) signaling molecule—regulates the production of components of the bacterial surface, such as the T4SS, the flagellum, OMPs, and exopolysaccharide (14, 54, 55, 62), and the expression of a variety of genes involved in diverse cellular processes (55, 62). In similarity to our results obtained with B. ovis PA (Fig. 4 and 5), the ΔvjbR mutant of B. melitensis 16M has been reported to be severely impaired with regard to survival in macrophages and was almost completely cleared from the spleens of infected mice at 4 weeks p.i. (3, 14). In contrast, survival of the ΔvjbR mutant of the B. abortus S19 vaccine was only slightly attenuated in J774.A1 macrophages (2) and it showed levels of spleen infection and persistence in the mouse model similar to those of the parental strain and remarkably higher than those reported for the B. melitensis 16M mutant (2, 3, 14). These results suggest variability among the brucellae regarding the impact of VjbR on virulence, but other vjbR mutants should be obtained in the genus Brucella to clarify this point.
The overproduction of an exopolysaccharide is thought to be involved in the clumping phenotype detected in the ΔvjbR mutant of B. melitensis 16M (21) and could also be responsible for the autoagglutination observed in the B. ovis PA mutant in static culture (Fig. 1A). Although this aspect remain to be elucidated, the agglutinating phenotype has also been observed with B. ovis mutants in several OMPs (9) and is probably more suggestive of surface modifications than of the genesis of a biofilm-like matrix. Additionally, the exopolysaccharide has not been detected in B. abortus 2308 but the R strains of that species rapidly autoagglutinated under our experimental conditions (9, 35). Modifications in the cell surface could probably contribute to the attenuation of B. ovis ΔvjbR. However, considering that VjbR positively regulates the transcription of known virulence genes (such as virB and flagellar genes) in B. melitensis 16M and seems to control many other cellular characteristics (55, 62), the attenuation would most likely be due to the sum of several defaults caused by the lack of a global regulator. In this respect, the ΔvjbR mutant behaved like the parental strain regarding the expression of the flagellar fliF gene (Table 2) but exhibited an important downregulation of virB expression (Fig. 2 and Table 2). In light of previous reports of studies performed with B. melitensis 16M (14, 62), the absence of VjbR-mediated control of fliF transcription was surprising but seems justified by the two independent DNA deletions detected in B. ovis upstream of fliF (Fig. 3). These deletions account for 283 bp, affect the fliF promoter region, and could include DNA-binding sites for direct or indirect VjbR-mediated regulation. In fact, one of these deletions covers the DNA-binding region described for FtcR (Fig. 3), a master regulator of the B. melitensis 16M flagellar system (30), a system that is required for the establishment of persistent infections in mice (20). However, whether B. ovis is able to produce the appendage under specific in vitro or in vivo conditions and its potential role in virulence remain to be elucidated.
In both B. melitensis 16M (14, 55, 62) and B. ovis PA (Fig. 2 and Table 2), the addition of the QS signaling molecule C12-HSL mimics the effect of the absence of VjbR and decreases virB transcription, probably by triggering the dissociation of VjbR from the virB promoter (4). In both strains, C12-HSL also mediates an increase in the transcription of babR (or blxR) (Table 2) (55, 62), the gene that encodes the second QS-related regulator described in B. melitensis 16M (46, 55) and that shares a common set of target genes regulated with VjbR in opposite directions (55). In contrast, neither the downregulation of vjbR transcription in the presence of C12-HSL reported for B. melitensis 16M (55, 62) nor the downregulation of babR transcription in its derived ΔvjbR mutant (55) was observed in B. ovis under the conditions tested (Table 2). Although it should be borne in mind that the differences between the two Brucella species with respect to these results could have been due to the particular experimental conditions used, the upregulation of babR transcription in the ΔvjbR mutant and in B. ovis PA cultured in the presence of C12-HSL is suggestive of a direct or indirect VjbR-mediated downregulation of babR transcription that is abrogated by C12-HSL. This VjbR-BabR-AHL model of interaction would be consistent with the opposing roles reported for VjbR and BabR in the regulation of virulence genes (55), the former activating the genes necessary for the establishment of the B. ovis intracellular niche, such as virB (and silencing other undesirable or unnecessary genes, such as babR, for this purpose), and the latter switching them off when they are no longer required. Further studies are necessary for a better understanding of QS regulation in the genus Brucella, and a more exhaustive analysis of the B. ovis ΔvjbR mutant in comparison with the parental strain and a ΔbabR mutant—together with studies about the production of an AHL in B. ovis—would contribute significantly to this end.
Brucella cyclic ß-1,2-glucan synthase (Cgs) catalyzes the four enzymatic reactions required for the synthesis of CßGs (24, 25), periplasmic polymers of several bacteria belonging to the α-2 subclass of proteobacteria living in close association with eukaryotic cells, either as symbionts or pathogens (1, 5, 24). In B. abortus 2308, CßGs are required to avoid the fusion of the Brucella-containing vacuole with the lysosome, which has been attributed to the ability of CßGs to interact with the lipid rafts of eukaryotic membranes (1). In agreement with the notion that CßGs act in B. abortus once the bacterium has entered the host cell (1, 7, 24), the B. abortus 2308 and S19 cgs mutants did not show defects in internalization either in HeLa cells or in murine macrophages but were impaired in intracellular replication (7).
In contrast, our results suggest a role for CßGs in the penetration of B. ovis PA into macrophages (Fig. 4B). This could be related to the absence of O-PS chains in R B. ovis, which would leave periplasmic CßGs in closer proximity to the surrounding environment, thus facilitating their interaction with the eukaryotic cell. Alternatively, CßGs could be important molecules for maintaining the cellular architecture and surface topology in R strains, which could also affect the penetration of B. ovis into the eukaryotic cell. In this respect, it should be noted that the Δcgs mutant exhibits alterations in several properties related to the bacterial surface (Fig. 1 and Table 1) and that mutants of other proteobacteria unable to synthesize periplasmic glucans have a highly pleiotropic phenotype that affects their envelope properties (5). However, more studies are necessary to determine whether the alterations in B. ovis are due to the absence of periplasmic CßGs and/or to additional effects derived from the absence of CßG synthase. Considering that Cgs is one of the largest proteins of B. ovis (2,867 amino acids and 320 kDa) and is an integral membrane protein with six transmembrane-spanning segments (25), removal of the protein could also have important effects on the stability of the inner membrane and hence of the cell.
The results most divergent with respect to those published for S Brucella strains were obtained with the bacA gene, which encodes an integral inner membrane protein involved in cell envelope properties, peptide uptake, and lipid A acylation with very-long-chain fatty acids (19, 63), with the latter considered to be contributors to the particular OM properties and virulence of Brucella (41). The deletion of bacA did not affect the ability of B. ovis PA to penetrate and replicate in murine macrophages or its virulence in mice (Fig. 4A and 5); in contrast, the bacA mutant of S B. abortus 2308 was unable to establish chronic infections in mice and, in experiments performed with opsonized bacteria, did not survive inside murine macrophages (31). The characteristics distinguishing the two Brucella mutants can be extended to other phenotypic traits, and such characteristics could explain, at least in part, the divergences in their behavior in the murine model. Thus, instead of undergoing detrimental modifications in cell-envelope-related properties, the B. ovis ΔbacA mutant was significantly more resistant than the parental strain to the cationic peptide polymyxin B, acid pH, and hydrogen peroxide (Table 1), three conditions mimicking those found by the bacterium within the animal host. Contrariwise, although more resistant to the glycopeptide bleomycin, the B. abortus 2308 bacA mutant showed increased sensitivity to acid pH, SDS, and bovine serum (31, 47). These differences were not completely unexpected, considering the contrasting behavior previously reported for parental B. abortus 2308 and B. ovis PA in assays addressing susceptibility to polymyxin B, several detergents, and nonimmune human serum (35). Considering its relationship with the structure of the OM (19, 27), the variable role of bacA in host-microbe interactions—also described for rhizobia that establish symbiotic relationships with leguminous plants (39)—could be related to the particular OM composition of each Brucella species (58). However, a role for BacA in the outcome of B. ovis infections in its natural host cannot be ruled out.
This is the first report about the role of the main virulence factors described in S Brucella in B. ovis virulence, providing valuable information about the almost unexplored interaction of this rough bacterium with the host. Similarities to the better-studied S Brucella strains can be seen, but we also describe distinctive traits that could help to explain why B. ovis establishes persistent infections in its natural host whereas rough mutants derived from S Brucella do not or why its pathogenicity differs from that of B. melitensis in the same preferred host. A more exhaustive characterization of the B. ovis mutants, together with studies comparing those from other Brucella species, should provide a more precise picture of the significance of the inactivated pathways in the host-pathogen interactions in the genus Brucella, constituted by species that are highly homologous at the DNA level but with important differences in pathogenicity and host preferences.
Supplementary Material
ACKNOWLEDGMENTS
We thank Axel Cloeckaert for the gift of MAbs, Rosa Hermosa for her valuable help with real-time PCR, Nicholas Skinner for supervising the English version of the manuscript, and the personnel of the DNA sequencing and animal experimentation facilities of the University of Salamanca for their helpful collaboration.
This work and A.I.M.-M. were supported by projects AGL2008-04514-C03-02/GAN and AGL2011-30453-C04-02 of the Ministerio de Ciencia e Innovación, Spain.
Footnotes
Published ahead of print 5 March 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Arellano-Reynoso B, et al. 2005. Cyclic ß-1,2-glucan is a brucella virulence factor required for intracellular survival. Nat. Immunol. 6:618–625 [DOI] [PubMed] [Google Scholar]
- 2. Arenas-Gamboa AM, Ficht TA, Kahl-McDonagh MM, Gomez G, Rice-Ficht AC. 2009. The Brucella abortus S19 ΔvjbR live vaccine candidate is safer than S19 and confers protection against wild-type challenge in BALB/c mice when delivered in a sustained-release vehicle. Infect. Immun. 77:877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arenas-Gamboa AM, Ficht TA, Kahl-McDonagh MM, Rice-Ficht AC. 2008. Immunization with a single dose of a microencapsulated Brucella melitensis mutant enhances protection against wild-type challenge. Infect. Immun. 76:2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arocena GM, Sieira R, Comerci DJ, Ugalde RA. 2010. Identification of the quorum-sensing target DNA sequence and N-Acyl homoserine lactone responsiveness of the Brucella abortus virB promoter. J. Bacteriol. 192:3434–3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bohin JP. 2000. Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol. Lett. 186:11–19 [DOI] [PubMed] [Google Scholar]
- 6. Bowden RA, Cloeckaert A, Zygmunt MS, Bernard S, Dubray G. 1995. Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect. Immun. 63:3945–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Briones G, et al. 2001. Brucella abortus cyclic ß-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69:4528–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burgess GW. 1982. Ovine contagious epididymitis: a review. Vet. Microbiol. 7:551–575 [DOI] [PubMed] [Google Scholar]
- 9. Caro-Hernández P, et al. 2007. Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect. Immun. 75:4050–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Celli J, et al. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cloeckaert A, de Wergifosse P, Dubray G, Limet JN. 1990. Identification of seven surface-exposed Brucella outer membrane proteins by use of monoclonal antibodies: immunogold labeling for electron microscopy and enzyme-linked immunosorbent assay. Infect. Immun. 58:3980–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Comerci DJ, Martínez-Lorenzo MJ, Sieira R, Gorvel JP, Ugalde RA. 2001. Essential role of the VirB machinery in the maturation of the Brucella abortus-containing vacuole. Cell. Microbiol. 3:159–168 [DOI] [PubMed] [Google Scholar]
- 13. Conde-Alvarez R, et al. 2006. Synthesis of phosphatidylcholine, a typical eukaryotic phospholipid, is necessary for full virulence of the intracellular bacterial parasite Brucella abortus. Cell. Microbiol. 8:1322–1335 [DOI] [PubMed] [Google Scholar]
- 14. Delrue RM, et al. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell. Microbiol. 7:1151–1161 [DOI] [PubMed] [Google Scholar]
- 15. Delrue RM, et al. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell. Microbiol. 3:487–497 [DOI] [PubMed] [Google Scholar]
- 16. den Hartigh AB, Rolán HG, de Jong MF, Tsolis RM. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J. Bacteriol. 190:4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. den Hartigh AB, et al. 2004. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect. Immun. 72:5143–5149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eskra L, Canavessi A, Carey M, Splitter G. 2001. Brucella abortus genes identified following constitutive growth and macrophage infection. Infect. Immun. 69:7736–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ferguson GP, Datta A, Baumgartner J, Roop RM, II, Carlson RW, Walker GC. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. U. S. A. 101:5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fretin D, et al. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell. Microbiol. 7:687–698 [DOI] [PubMed] [Google Scholar]
- 21. Godefroid M, et al. 2010. Brucella melitensis 16M produces a mannan and other extracellular matrix components typical of a biofilm. FEMS Immunol. Med. Microbiol. 59:364–377 [DOI] [PubMed] [Google Scholar]
- 22. González D, et al. 2008. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS One 3:e2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guzmán-Verri C, et al. 2001. GTPases of the Rho subfamily are required for Brucella abortus internalization in nonprofessional phagocytes: direct activation of Cdc42. J. Biol. Chem. 276:44435–44443 [DOI] [PubMed] [Google Scholar]
- 24. Haag AF, Myka KK, Arnold MF, Caro-Hernández P, Ferguson GP. 2010. Importance of lipopolysaccharide and cyclic ß-1,2-glucans in Brucella-mammalian infections. Int. J. Microbiol. 2010:article ID 124509. doi:10.1155/2010/124509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Iñón de Iannino N, Briones G, Tolmasky M, Ugalde RA. 1998. Molecular cloning and characterization of cgs, the Brucella abortus cyclic ß(1–2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J. Bacteriol. 180:4392–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiménez de Bagüés MP, Barberán M, Marín CM, Blasco JM. 1995. The Brucella abortus RB51 vaccine does not confer protection against Brucella ovis in rams. Vaccine 13:301–304 [DOI] [PubMed] [Google Scholar]
- 27. Karunakaran R, et al. 2010. BacA is essential for bacteroid development in nodules of galegoid, but not phaseoloid, legumes. J. Bacteriol. 192:2920–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kohler S, et al. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. U. S. A. 99:15711–15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lapaque N, Moriyón I, Moreno E, Gorvel JP. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8:60–66 [DOI] [PubMed] [Google Scholar]
- 30. Léonard S, et al. 2007. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J. Bacteriol. 189:131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. LeVier K, Phillips RW, Grippe VK, Roop RM, II, Walker GC. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287:2492–2493 [DOI] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 33. Martín-Martín AI, Caro-Hernández P, Orduña A, Vizcaíno N, Fernández-Lago L. 2008. Importance of the Omp25/Omp31 family in the internalization and intracellular replication of virulent B. ovis in murine macrophages and HeLa cells. Microbes Infect. 10:706–710 [DOI] [PubMed] [Google Scholar]
- 34. Martín-Martín AI, et al. 2009. Analysis of the occurrence and distribution of the Omp25/Omp31 family of surface proteins in the six classical Brucella species. Vet. Microbiol. 137:74–82 [DOI] [PubMed] [Google Scholar]
- 35. Martín-Martín AI, Sancho P, Tejedor C, Fernández-Lago L, Vizcaíno N. 2011. Differences in the outer membrane-related properties of the six classical Brucella species. Vet. J. 189:103–105 [DOI] [PubMed] [Google Scholar]
- 36. Martín-Martín AI, Vizcaíno N, Fernández-Lago L. 2010. Cholesterol, ganglioside GM1 and class A scavenger receptor contribute to infection by Brucella ovis and Brucella canis in murine macrophages. Microbes Infect. 12:246–251 [DOI] [PubMed] [Google Scholar]
- 37. Martínez-Núñez C, et al. 2010. The two-component system BvrR/BvrS regulates the expression of the type IV secretion system VirB in Brucella abortus. J. Bacteriol. 192:5603–5608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martirosyan A, Moreno E, Gorvel JP. 2011. An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol. Rev. 240:211–234 [DOI] [PubMed] [Google Scholar]
- 39. Maruya J, Saeki K. 2010. The bacA gene homolog, mlr7400, in Mesorhizobium loti MAFF303099 is dispensable for symbiosis with Lotus japonicus but partially capable of supporting the symbiotic function of bacA in Sinorhizobium meliloti. Plant Cell Physiol. 51:1443–1452 [DOI] [PubMed] [Google Scholar]
- 40. Monreal D, et al. 2003. Characterization of Brucella abortus O-polysaccharide and core lipopolysaccharide mutants and demonstration that a complete core is required for rough vaccines to be efficient against Brucella abortus and Brucella ovis in the mouse model. Infect. Immun. 71:3261–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moriyón I, López-Goñi I. 1998. Structure and properties of the outer membranes of Brucella abortus and Brucella melitensis. Int. Microbiol. 1:19–26 [PubMed] [Google Scholar]
- 42. O'Callaghan D, et al. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210–1220 [DOI] [PubMed] [Google Scholar]
- 43. Pei J, Ficht TA. 2004. Brucella abortus rough mutants are cytopathic for macrophages in culture. Infect. Immun. 72:440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pei J, Wu Q, Kahl-McDonagh M, Ficht TA. 2008. Cytotoxicity in macrophages infected with rough Brucella mutants is type IV secretion system dependent. Infect. Immun. 76:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porte F, Naroeni A, Ouahrani-Bettache S, Liautard JP. 2003. Role of the Brucella suis lipopolysaccharide O antigen in phagosomal genesis and in inhibition of phagosome-lysosome fusion in murine macrophages. Infect. Immun. 71:1481–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rambow-Larsen AA, Rajashekara G, Petersen E, Splitter G. 2008. Putative quorum-sensing regulator BlxR of Brucella melitensis regulates virulence factors including the type IV secretion system and flagella. J. Bacteriol. 190:3274–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roop RM, II, et al. 2002. Seeking a niche: putative contributions of the hfq and bacA gene products to the successful adaptation of the brucellae to their intracellular home. Vet. Microbiol. 90:349–363 [DOI] [PubMed] [Google Scholar]
- 48. Rouot B, et al. 2003. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 71:1075–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Salhi I, et al. 2003. Characterization of new members of the group 3 outer membrane protein family of Brucella spp. Infect. Immun. 71:4326–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sieira R, Comerci DJ, Sánchez DO, Ugalde RA. 2000. A homologue of an operon required for DNA transfer in Agrobacterium is required in Brucella abortus for virulence and intracellular multiplication. J. Bacteriol. 182:4849–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Silva TM, et al. 2011. Putative ATP-binding cassette transporter is essential for Brucella ovis pathogenesis in mice. Infect. Immun. 79:1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sola-Landa A, et al. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125–138 [DOI] [PubMed] [Google Scholar]
- 53. Tsolis RM, et al. 2009. Genome degradation in Brucella ovis corresponds with narrowing of its host range and tissue tropism. PLoS One 4:e5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Uzureau S, et al. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 189:6035–6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uzureau S, et al. 2010. Global analysis of quorum sensing targets in the intracellular pathogen Brucella melitensis 16 M. J. Proteome Res. 9:3200–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Viadas C, et al. 2010. Transcriptome analysis of the Brucella abortus BvrR/BvrS two-component regulatory system. PLoS One 5:e10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vizcaíno N, Caro-Hernández P, Cloeckaert A, Fernández-Lago L. 2004. DNA polymorphism in the omp25/omp31 family of Brucella spp.: identification of a 1.7-kb inversion in Brucella cetaceae and of a 15.1-kb genomic island, absent from Brucella ovis, related to the synthesis of smooth lipopolysaccharide. Microbes Infect. 6:821–834 [DOI] [PubMed] [Google Scholar]
- 58. Vizcaíno N, Cloeckaert A. 2012. Biology and genetics of the Brucella outer membrane, p 133–161 In López-Goñi I, O'Callaghan D. (ed), Brucella molecular microbiology and genomics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 59. Vizcaíno N, Cloeckaert A, Zygmunt MS, Dubray G. 1996. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect. Immun. 64:3744–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vizcaíno N, Kittelberger R, Cloeckaert A, Marín CM, Fernández-Lago L. 2001. Minor nucleotide substitutions in the omp31 gene of Brucella ovis result in antigenic differences in the major outer membrane protein that it encodes compared to those of the other Brucella species. Infect. Immun. 69:7020–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang Y, et al. 2010. The type IV secretion system affects the expression of Omp25/Omp31 and the outer membrane properties of Brucella melitensis. FEMS Microbiol. Lett. 303:92–100 [DOI] [PubMed] [Google Scholar]
- 62. Weeks JN, et al. 2010. Brucella melitensis VjbR and C12-HSL regulons: contributions of the N-dodecanoyl homoserine lactone signaling molecule and LuxR homologue VjbR to gene expression. BMC Microbiol. 10:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wehmeier S, et al. 2010. Internalization of a thiazole-modified peptide in Sinorhizobium meliloti occurs by BacA-dependent and -independent mechanisms. Microbiology 156:2702–2713 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.