Abstract
Vibrio (Aliivibrio) salmonicida is the causal agent of cold-water vibriosis, a fatal bacterial septicemia primarily of farmed salmonid fish. The molecular mechanisms of invasion, colonization, and growth of V. salmonicida in the host are still largely unknown, and few virulence factors have been identified. Quorum sensing (QS) is a cell-to-cell communication system known to regulate virulence and other activities in several bacterial species. The genome of V. salmonicida LFI1238 encodes products presumably involved in several QS systems. In this study, the gene encoding LitR, a homolog of the master regulator of QS in V. fischeri, was deleted. Compared to the parental strain, the litR mutant showed increased motility, adhesion, cell-to-cell aggregation, and biofilm formation. Furthermore, the litR mutant produced less cryptic bioluminescence, whereas production of acylhomoserine lactones was unaffected. Our results also indicate a salinity-sensitive regulation of LitR. Finally, reduced mortality was observed in Atlantic salmon infected with the litR mutant, implying that the fish were more susceptible to infection with the wild type than with the mutant strain. We hypothesize that LitR inhibits biofilm formation and favors planktonic growth, with the latter being more adapted for pathogenesis in the fish host.
INTRODUCTION
The marine bacterium Vibrio (Aliivibrio) salmonicida is a motile Gram-negative curved rod and the etiological agent of cold-water vibriosis (CV) in farmed Atlantic salmon (Salmo salar L.), rainbow trout (Oncorhynchus mykiss), and Atlantic cod (Gadus morhua) (13, 14, 27, 30). The disease occurs mainly in late autumn to early spring and is a generalized septicemia characterized by anemia and extended hemorrhages, especially around the abdomen and in the integument surrounding the internal organs of the fish (14, 27, 52). V. salmonicida was recently proposed to be reclassified into the new genus Aliivibrio (65). However, A. salmonicida is the well-established abbreviation of Aeromonas salmonicida, the etiological agent of furunculosis in salmonids. To avoid possible nomenclature confusion, we use the name Vibrio salmonicida.
Current commercial vaccines give full protection against infection with V. salmonicida (12), and the bacterium is no longer an immediate threat to the salmonid aquaculture industry. However, the molecular mechanisms of host specificity, invasion, colonization and virulence are still largely unknown, making V. salmonicida an interesting bacterial species for studying host-pathogen interaction and pathogenesis. In addition, novel fish pathogens are frequently discovered and new variants or strains may arise, creating a constant need for knowledge about bacterial pathogenesis so that treatment and prophylactic strategies can be developed and improved. A few studies have identified some bacterial factors with possible roles in V. salmonicida virulence, such as the surface antigen VS-P1. This antigen is released by cells of V. salmonicida during growth in fish and is hypothesized to bind specific antibodies and thus protect the bacterium from complement-mediated killing and phagocytosis (25). Furthermore, temperature-sensitive iron sequestration is proposed as an important virulence mechanism for V. salmonicida (9). The V. salmonicida genome encodes three TonB systems and one heme uptake system probably involved in iron acquisition. Genomic analysis has also identified genes encoding multiple putative hemolysins, proteases, and several protein secretion systems (26). Finally, production of hydrogen peroxide has been suggested to be a possible virulence factor in addition to flagella and motility, which are linked to host colonization and virulence in several vibrios (23, 32, 36, 45, 47, 49, 50, 54).
By quorum sensing (QS), bacteria coordinate gene expression in a cell density-dependent manner, through the production of signal molecules called autoinducers (44, 46, 67). QS was first discovered in the two luminescent marine bacteria Vibrio harveyi and Vibrio (Aliivibrio) fischeri (21, 46). In V. fischeri, which is a symbiont of the Hawaiian bobtail squid, Euprymna scolopes, the LuxI-produced autoinducer N-(3-oxohexanoyl)-l-homoserine lactone (3-oxo-C6-HSL) forms a complex with the transcription activator LuxR. This complex enhances transcription of the lux operon, resulting in increased bioluminescence (21). V. fischeri encodes two additional QS systems that are involved in regulation of bioluminescence, namely, the LuxS/PQ and AinS/R systems. Both systems converge on the common phosphorelay signal transduction system LuxU-LuxO, which at high cell density activates the expression of the QS master regulator LitR (17, 39). Homologs of V. fischeri LitR are found in many vibrios. They control a number of activities, such as bioluminescence (17, 59), protease production (10, 29, 35), motility (17, 35), biofilm formation (10, 24, 35) and colony morphology (10, 17, 41, 42, 70).
V. salmonicida produces the signal molecules N-hexanoyl-l-homoserine lactone (C6-HSL), 3-oxo-C6-HSL, and, presumably, autoinducer 2 (5, 6, 26). In addition, several QS systems, including the LuxS/PQ, LuxI/R, and AinS/R systems, as well as a lux operon, have been identified in V. salmonicida (26), but no detailed or systematic functional studies of these systems have been performed. Although the lux operon is present, V. salmonicida is a cryptic bioluminescent bacterium that requires addition of exogenous aldehyde for production of light (18). An association between the lux operon and virulence has been demonstrated for V. salmonicida, and fish challenged with a luxA mutant showed delayed and reduced mortality compared to the wild-type strain NCMB2262 (47). In this report, we discuss the function of V. salmonicida LitR in the seawater environment and within the fish host, where the bacterium establishes a septicemic infection. This study demonstrates that a functional LitR protein is required for V. salmonicida virulence and that LitR downregulates adhesion, aggregation, and motility and upregulates cryptic bioluminescence. However, LitR is dispensable for the production of the autoinducers C6-HSL and 3-oxo-C6-HSL in V. salmonicida. Finally, our results indicate a salinity-sensitive regulation of LitR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. The wild-type V. salmonicida strain LFI1238 was originally isolated from the anterior kidney (head kidney) of a diseased cod (Gadus morhua) (26). V. salmonicida strains were grown on blood agar base no. 2 (BA) (Oxoid, Cambridge, United Kingdom) supplemented with 2.5% NaCl and 5% bovine blood (BA2.5+BB) or in Luria-Bertani broth (LB) supplemented with 1, 2.5, or 3% NaCl (LB1, LB2.5, or LB3) at 12°C for 4 days, unless otherwise stated. The different LB media were solidified with 1.2% Bacto agar (Difco, BD Diagnostics, Sparks, MD) (LA1, LA2.5, and LA3). The Escherichia coli strains β-2155 and S17-1 were cultivated in LB1 or on LA1 at 37°C (34, 60). Additionally, 0.3 mM diaminopimelic acid was added to the broths or plates for the β-2155 strain. The suicide plasmids pDM4 and pNQ701 were propagated in E. coli S17-1. For selection of E. coli transformants or V. salmonicida transconjugants, chloramphenicol at a final concentration of 25 μg/ml or 2 μg/ml, respectively, was added to the medium. For growth curve experiments, V. salmonicida strains were cultivated in LB1 or LB3 and incubated at 8°C and 12°C with agitation (200 rpm). The optical densities at 600 nm (OD600 values) were measured at 3- to 10-h intervals, using a Unicam 8625 UV-visible (UV-Vis) spectrometer. The experiments were performed with biological duplicates and repeated twice.
Table 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or sequence (5′–3′)a | Source or reference |
|---|---|---|
| Strains | ||
| V. salmonicida strains | ||
| LFI1238 | Wild type (wt); isolated from Atlantic cod | 26 |
| ΔlitR | LFI1238 containing an in-frame deletion in litR | This study |
| ΔlitRc | ΔlitR strain complemented with the LFI1238 litR gene; Cmr | This study |
| E. coli strains | ||
| S17-1 | Donor strain for conjugation; λ pir | 60 |
| β-2855 | Donor strain for conjugation; diaminopimelic acid auxotroph; λ pir | 34 |
| Plasmids | ||
| pDM4 | Cmr; suicide vector with an R6K origin (λ pir requiring) and sacBR | 45 |
| pNQ705 | Cmr; suicide vector with an R6K origin (λ pir requiring) | 45 |
| pDM4ΔlitR | pDM4 containing a fragment of litR harboring an internal deletion | This study |
| pNQ705litR+ | pNQ705 containing wt litR and flanking sequences | This study |
| Primers | ||
| Primers for construction of ΔlitR and ΔlitRc strains | ||
| LitR-A fwd | CCGCTCGAGTAGTTCCATAATCTTTTCTATG | This study |
| LitR-B rev | CTTACTCTACTTATATTTATTATATCCTTGCCAAC | This study |
| LitR-C fwd | TATAAGTAGAGTAAGCGTGC | This study |
| LitR-D rev | GGACTAGTGTTATTTTCGGGTTCAAC | This study |
| Primers for verification of plasmids, transconjugants, and mutants | ||
| LitR-G fwd | ACCAACGGCAGGACTTAGAC | This study |
| LitR-H rev | TTGATAACAATCGAGCAGAGC | This study |
| Pnq-fwd | TAACGGCAAAAGCACCGCCGGACATCA | Debra Miltonb |
| Pnq-rev | TGTACACCTTAACACTCGCCTATTGTT | Debra Milton |
| LitR-fwd | GTGTGGTTTGAATGGAGCAC | This study |
| LitR-rev | CTCAATCCCTTCAGCAAACA | This study |
| qRT-PCR primers | ||
| LitR-fwd | GTGTGGTTTGAATGGAGCAC | This study |
| LitR-rev | CTCAATCCCTTCAGCAAACA | This study |
| 16S-fwd | CTTGACGTTAGCGACAGAAGAA | This study |
| 16S-rev | CGCTTTACGCCCAGTAATTC | This study |
| AccD-fwd | TTGCTGGTCGTCGTGTTATT | This study |
| AccD-rev | TTTAGCCATCAAACCACCAA | This study |
| FtsZ-fwd | CGGATGTTCGTACGGTAATG | This study |
| FtsZ-rev | CAAGCAATGGGCTTGAGATA | This study |
| RpoD-fwd | AAGCCGAAGAAATTCGCTTA | This study |
| RpoD-rev | GCAAGATCTGATTCGCTCAA | This study |
Cmr, chloramphenicol resistance.
Umeå University, Sweden.
Phylogenetic analyses and software.
Assembly and ClustalW alignment of amino acid sequences were performed using BioEdit (version 5.0.9). A neighbor-joining (NJ) tree (55) was generated from the aligned sequences by using MEGA (version 4). Gaps in pairwise sequence comparisons were deleted. Bootstrap analyses with 500 replicates were conducted to provide confidence levels for the tree topology.
Construction of litR mutant and complemented strain.
DNA extraction, general recombinant DNA techniques, and transformations were performed using standard protocols (56). Restriction enzyme digestion, ligation, and plasmid purification were performed as recommended by the manufacturers (NEB Biolabs, Ipswich, MA, and Omega Bio-Tek, Norcross, GA). PCR (Phusion; FinnZyme, Espoo, Finland) and BigDye sequencing (Applied Biosystems, Carlsbad, CA) were performed with custom-made primers synthesized by Sigma (St. Louis, MO) and Operon (Leeds, United Kingdom). The primers used for sequencing, mutant construction, and real-time PCR (see below) are listed in Table 1.
The litR in-frame deletion mutant (ΔlitR) was made by allelic exchange as described by others (45). In brief, the ΔlitR allele was constructed by fusion of two PCR products amplified from genomic DNA flanking the region to be deleted. The upstream region (205 bp) was amplified using the primers LitR-A and LitR-B. The upstream PCR product included only the start codon of the litR open reading frame. The downstream region (210 bp) was amplified using the primers LitR-C and LitR-D and contained only the last three C-terminal codons of the litR open reading frame. The LitR-B and LitR-C primers contain complementary 3′ sequences which enable fusion of the two products by a second, overlap PCR. This overlap PCR was performed by mixing the two PCR products with deoxynucleoside triphosphates (dNTPs), DNA polymerase, and buffer and cycling them 7 times before the outermost primers (LitR-A and LitR-D) were added, followed by 25 more cycles. The resulting PCR product was digested with SpeI and XhoI (restriction sites are included in the LitR-A and LitR-D primers, respectively) and cloned into the corresponding restriction sites of pDM4, giving rise to pDM4ΔlitR. The complemented deletion mutant (ΔlitRc) was constructed by insertion of a full-length copy of the wild-type litR gene into the original locus of the ΔlitR mutant. For this purpose, the complete gene was PCR amplified using the primers LitR-A and LitR-D and cloned into the SpeI and XhoI restriction sites of pNQ705, giving rise to pNQ705litR+.
The pDM4ΔlitR construct was transferred to wild-type LFI1238 and the pNQ705litR+ construct transferred to the ΔlitR mutant by conjugation, mainly as described by others (45, 66). In brief, E. coli S17-1 or β-2155 was used as a donor in matings with V. salmonicida. S17-1 was found to be superior to β-2155 and significantly increased the number of transconjugants. The donor cells were grown to mid-exponential phase and the recipient cells to early stationary phase before they were harvested by centrifugation and washed twice in LB1 medium before being mixed and spotted onto LA1. The plates were incubated at 20°C for ∼6 h, followed by incubation overnight at 12°C. The spotted cells were resuspended in LB2.5 and incubated overnight at 12°C with agitation. Potential transconjugants were selected after 3 to 5 days on LA2.5 plates supplemented with chloramphenicol. To complete allelic exchange, transconjugants were spread onto LA2.5 plates containing 5% sucrose. After sucrose selection, chloramphenicol-sensitive colonies were analyzed for the deletion by PCR and verified by sequencing.
Adhesion and aggregation studies.
To study colony morphology and adhesion, V. salmonicida LFI1238 and the ΔlitRc and ΔlitR strains were grown on LA1, LA3, or BA containing 1% NaCl (BA1) or 3% NaCl (BA3) without supplements or supplemented with 5% bovine blood (BB), 5% washed bovine erythrocytes (ERY), 7% fetal bovine serum (FBS) (Sigma-Aldrich), or 1% bovine serum albumin (BSA) (A2153; Sigma-Aldrich). The plates were incubated at 4, 8, or 12°C, and the adherence of colonies to agar was determined at different time points by collecting single colonies from agar plates with a sterile plastic loop. The adherence was graded semiquantitatively as “none” (smooth and creamy colonies), “weak” (more viscous colonies that were slightly adherent in the periphery), “moderate” (adherent colonies that could partly be separated from the agar), or “strong” (adherent colonies that were nearly impossible to separate from the agar). Bacterial aggregation was analyzed in LB1, LB3, brain heart infusion (BHI) medium (Difco, BD Biosciences, Franklin Lakes, NJ) with 7% horse serum (HS), and Leibovitz-15 (L15) medium (Gibco, Invitrogen, San Diego, CA) supplemented with 200 mM l-glutamine, 50 mM β-mercaptoethanol, and 10% FBS. Precultures grown in LB3 were diluted to an OD600 of ∼0.05 in the above media and incubated in 24-well tissue culture trays at 8°C (200 rpm), and aggregation was monitored using an Olympus IX81 inverted phase-contrast microscope at regular intervals for up to 96 h. The amount of aggregation was graded as illustrated in Fig. S1 in the supplemental material.
Biofilm formation.
Precultures of V. salmonicida LFI1238 and the ΔlitRc and ΔlitR strains were diluted 1:20 in LB2.5 and incubated overnight with agitation. The cultures were then diluted to an OD600 of 1.3 before 1:10 dilutions of the cultures were made in L15 medium adjusted to 380 mosM by adding 29 mM NaCl. A total volume of 300 μl was added to each well of a 24-well tissue culture-treated tray (Falcon; BD Biosciences). The plates were incubated statically at 4°C, and biofilm formation and architecture were monitored by phase-contrast microscopy (Leica DM IRB; Leica Microsystems, Wetzlar, Germany). The biofilm at day 6 was photographed with a Canon D400 digital camera.
Motility and flagellation.
Motility was assayed using LB soft agar plates containing 0.25% agar supplemented with 1 or 3% NaCl. Overnight cultures of V. salmonicida LFI1238 and the ΔlitRc and ΔlitR strains were diluted 1:40 in LB1 and LB3 and grown to an OD600 of 0.4. Next, 3-μl culture samples were spotted onto the center of soft agar plates, incubated at 4, 8, and 12°C for 5 days, and monitored every 24 h. The experiment was performed in triplicate during each of three separate trials. Transmission electron microscopy (TEM) was used to study flagellation. Sample grids were prepared by touching hexagonal carbon-coated Formvar copper grids to bacterial colonies grown on LA2.5 or BA2.5+BB for 72 h at 12°C. Samples were also made by incubating grids for 10 min at room temperature on droplets of bacterial suspension collected either from fish implants (in vivo cultivation [see below]) or from colonies grown on BA2.5+BB and then resuspended in phosphate-buffered saline (PBS) or LB2.5. Samples were fixed for 4 min with 0.5% glutaraldehyde in PBS and washed 3 times on drops of PBS and once on distilled water (dH2O), followed by negative staining with 2% uranyl acetate (dissolved in dH2O) for 1 min. The stained preparations were viewed in a Philips CM100 transmission electron microscope.
Cryptic bioluminescence.
V. salmonicida LFI1238 and the ΔlitRc and ΔlitR strains were cultured in LB1 and LB3 at 8°C for 2 days with agitation (250 rpm), until an OD600 of 1.4 was reached. Next, 200 μl of each bacterial culture was transferred to a separate well of a nontransparent microtiter plate (OptiPlate-96; PerkinElmer, Waltham, MA), and immediately after addition of decyl aldehyde (Sigma-Aldrich) to a final concentration of 10 μM, luminescence was measured as counts per second (cps) with a VictorIII multilabel counter (PerkinElmer). The nonluminous strain Vibrio (Aliivibrio) wodanis FT5426 was included as a negative control (31). Each sample was assayed in triplicate, and the entire experiment was repeated three times. After subtracting background luminescence, luminescence per cell was calculated by dividing the cps of each sample by its OD600.
AHL production.
Production of N-acylhomoserine lactones (AHLs) in V. salmonicida LFI1238 and the ΔlitRc and ΔlitR strains was monitored in an agar well diffusion assay using the reporter strains Agrobacterium tumefaciens NT1 and Chromobacterium violaceum CV026 as described earlier (53). Briefly, V. salmonicida strains were grown in LB1 or LB3 at 8°C (250 rpm) for 4 days, to an OD600 of 1.8 ± 0.06. The bacterial cultures (30 ml) were extracted with an equivalent volume of ethyl acetate acidified by supplementation with 0.5% formic acid. After evaporation under nitrogen flow to dryness, the extracts were reconstituted in 1 ml acidified ethyl acetate and stored at −20°C until analysis. Sixty microliters of each extraction was added to punched wells of agar plates, on which one of the monitor strains was inoculated. C6-HSL (10 nM) and 3-oxo-C6-HSL (20 nM) (Sigma-Aldrich) were used as positive controls. The plates were incubated at 25°C for 2 days before the diameters of the AHL-induced zones were measured.
qRT-PCR.
Expression of litR was analyzed after in vitro or in vivo cultivation of V. salmonicida. For in vitro analysis, LFI1238 was grown to different cell densities in LB1 and LB3 at 8°C and 200 rpm before being harvested. Heart tissues from CV-diseased Atlantic salmon were used for in vivo analysis. Total RNA was stabilized using an RNAProtect bacterial reagent kit (Qiagen, Hilden, Germany) and isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) as described by the manufacturers. The RNA was precipitated and repurified using the Qiagen RNeasy minikit protocol (Qiagen). cDNA synthesis of 1 μg total RNA was performed using a QuantiTect reverse transcription kit (Qiagen) according to the manufacturer's instructions. Real-time PCR amplification was performed in a Stratagene Mx3000P thermal cycler (Agilent Technologies, Santa Clara, CA), using Express SYBR GreenER qPCR supermixes and two-step quantitative real-time PCR (qRT-PCR) kits (Invitrogen) as described by the manufacturer. Primarily, 16S rRNA was used as an endogenous control to adjust for different amounts of starting material. Additional experiments using accD (VSAL_I1076), ftsZ (VSAL_I2640), and rpoD (VSAL_I2697) as control genes for normalization were performed to confirm the obtained results. The experiments were performed with biological duplicates and technical triplicates and were repeated twice.
Procedures for fish handling during in vivo cultivation and challenge experiments.
Atlantic salmon were anesthetized in a water bath containing 0.0035% benzocaine (Benzoak VET; Euro-Pharma, Chemainus, Canada). Fish fins were clipped differentially to distinguish between groups. For challenge experiments, fish were exposed to V. salmonicida cells that were recently passaged through fish to verify and prepare virulence (16, 61). The challenge doses were established based on a prechallenge experiment as well as on previous studies (7, 12, 47, 48). Mortality was monitored daily for a period of 3 weeks. Samples from the head kidney were plated on BA2.5+BB and incubated at 12°C to verify the presence of V. salmonicida. The challenge experiments were approved by The Norwegian Animal Research Authority (approval no. ID1646, ID1728, and ID1913).
In vivo cultivation of V. salmonicida.
V. salmonicida cultures held in sealed dialysis tubing with a molecular mass cutoff of 12,000 to 14,000 Da (Spectra/Por, Los Angeles, CA) were surgically implanted into the peritoneal cavities of six Atlantic salmon (500 g) as described by Colquhoun and Sørum (8). Briefly, LFI1238, ΔlitRc, and ΔlitR cultures were grown at 8°C in LB2.5 with shaking (250 rpm) for 2 days and then centrifuged and resuspended in sterile PBS (pH 7.4) to an OD600 of 1.0 (representing 1 × 109 CFU/ml). Control implants were inoculated with sterile PBS. The implants were removed from fish after 5 days.
Intraperitoneal challenge experiment with Atlantic salmon.
One hundred Atlantic salmon (fry) with an average weight of 40 g were divided into three test groups and one control group. Each test group consisted of 30 fish, and the control group consisted of 10 fish. The four groups were kept together in the same 600-liter tank supplied with oxygenated and carbon-filtered fresh water at a temperature of 5 to 6°C. After sedation, the fish were injected intraperitoneally (i.p.) with 0.1-ml LB1 cultures (OD600 = 0.4) of LFI1238 and the ΔlitRc and ΔlitR strains, representing 3 × 107 CFU, 5 × 107 CFU, and 6.5 × 107 CFU, respectively. The control fish were injected i.p. with 0.1 ml LB1.
Immersion challenge experiment with Atlantic salmon.
Two hundred forty Atlantic salmon smolts with an average weight of 50 g were divided into four groups of 60 fish each. The groups were kept in separate tanks (1,400 liters) supplied with well-aerated, 8°C seawater taken directly from a depth of 60 m, with a salinity of 35 ppm. The fish were immersion challenged for 45 min in seawater with LB3-cultured bacteria. The final dose was determined by vital counting of the seawater and found to be 5 × 105, 1 × 106, and 2 × 106 CFU/ml for LFI1238 and the ΔlitRc and ΔlitR strains, respectively. The control group was immersed in seawater with 1% LB3 added.
RESULTS
LitR suppresses adhesion and cell-to-cell aggregation in V. salmonicida.
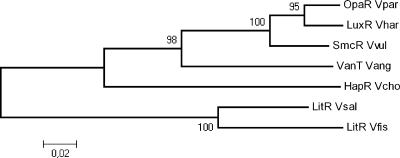
The litR gene (VSAL_I2619) was identified as a single gene in the genome of V. salmonicida strain LFI1238 (26) and is phylogenetically closely related to the V. fischeri homolog (Fig. 1), with which it shares 87% amino acid sequence identity. To analyze the functional role of LitR in LFI1238, we generated a ΔlitR deletion mutant and a ΔlitRc complementation mutant. The different strains were grown and analyzed in liquid or on solid media supplemented with either a low (1%) or high (3%) salt concentration to mimic a physiological “inside-the-host” or ocean environment. LFI1238 and the ΔlitRc and ΔlitR strains showed similar characteristics with regard to growth rate and colony morphology as previously described for V. salmonicida (14). They all grew faster with high salinity than with low salinity and at 12°C than at 8°C, but no differences in growth rates between strains were found (data not shown).
Fig 1.
Phylogenetic relationships of LitR homologs in vibrios. Accession numbers (NCBI) for the sequences are as follows: V. salmonicida LitR, gi:209696031; V. harveyi LuxR, gi:107933356; V. parahaemolyticus OpaR, gi:28899290; V. anguillarum VanT, gi:18104604; V. Vibrio cholerae HapR, gi:87133250; V. fischeri LitR, gi:59712784; and Vibrio vulnificus SmcR, gi:8101587. The bar indicates the number of substitutions/site. Bootstrap values are shown to show statistical support of branching.
In contrast to colonies of LFI1238 and the ΔlitRc strain, colonies of the ΔlitR strain grown at 12°C on blood agar transformed to highly adhesive colonies after storage at 4°C. This transformation was not observed on LB agar. Adhesion was further analyzed on agar media supplemented with different blood components and salt concentrations (Table 2). The adhesion of ΔlitR colonies was more pronounced at low temperatures (4°C) and on media containing a high salt concentration. The ΔlitR colonies were equally adherent on agar plates supplemented with washed erythrocytes, serum, or albumin, suggesting that albumin may be the blood component triggering adhesion. LFI1238 and the ΔlitRc strain produced nonadhesive, creamy colonies on the different media. To study cell-to-cell aggregation, the strains were cultivated in different broths. Compared to LFI1238 and the ΔlitRc strain, the ΔlitR strain was found to aggregate strongly in media containing serum (L15-FBS and BHI-HS). No difference in aggregation was observed between strains grown in LB medium, but the strains aggregated more in LB medium with a high salt concentration (Table 2).
Table 2.
Grading of adhesion and cell-to-cell aggregation propertiesa
| Parameter and medium | Result for: |
||
|---|---|---|---|
| LFI1238 wt | ΔlitR strain | ΔlitRc strain | |
| Adhesion | |||
| LA1 | − | − | − |
| LA3 | − | − | − |
| BA1 | − | + | − |
| BA3 | − | + | − |
| BA1 + BB | − | + | − |
| BA3 + BB | − | + + + | − |
| BA1 + FBS | − | + | − |
| BA3 + FBS | − | + + | − |
| BA1 + ERY | − | + | − |
| BA3 + ERY | − | + + | − |
| BA1 + BSA | − | + | − |
| BA3 + BSA | − | + + | − |
| Cell-to-cell aggregation | |||
| LB1 | (+) | (+) | (+) |
| LB3 | + | + | + |
| BHI-HS | ++ | +++ | ++ |
| L15-FBS | + | ++ | + |
Adhesion of bacterial colonies to different agar media was measured after 7 days of incubation at 4°C on LA1 or LA3, blood agar base with 1% or 3% NaCl without supplements (BA1 or BA3) or with 5% bovine blood (BB), 7% fetal bovine serum (FBS), 5% washed bovine erythrocytes (ERY), or 1% bovine serum albumin (BSA). The adherence was graded as follows: −, none (smooth and creamy colonies); +, weak (more viscous colonies that were slightly adherent in the periphery); ++, moderate (adherent colonies that were partly possible to scrape off); and +++, strong (adherent colonies that were almost impossible to scrape off). Cell-to-cell aggregation properties were measured after 4 days of incubation at 8°C (200 rpm) in LB1 or LB3, brain heart infusion medium with 7% horse serum (BHI-HS), or Leibovitz-15 medium supplemented with 200 mM l-glutamine, 50 mM β-mercaptoethanol, and 10% FBS (L15-FBS). Aggregation was graded as follows: −, none; +, weak; ++, moderate; and +++, strong.
LitR downregulates biofilm formation.
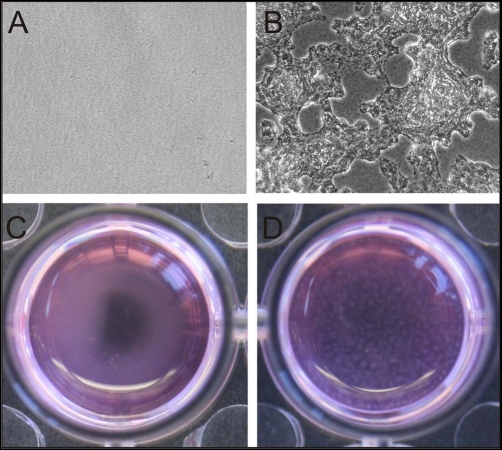
LitR homologs in different vibrios are known to regulate biofilm production (10, 24, 35, 72). The increased adhesion and cell-to-cell aggregation of the ΔlitR strain pointed to a similar role for LitR in V. salmonicida biofilm formation, as these activities are often coregulated (4). This led us to investigate biofilm formation at different temperatures. Using low temperatures and static conditions, we found that the ΔlitR strain formed a biofilm that could be visualized using phase-contrast microscopy after 3 days and macroscopically after 6 days, whereas LFI1238 and the ΔlitRc strain were found to be poor biofilm producers (if producing any) under the same conditions (Fig. 2). Biofilms start as microcolonies attached to the plastic surface and progress to a thick, very viscous, slimy, loosely attached biofilm, suggesting that there are large amounts of exopolymeric substances in the biofilm matrix (19). Due to the viscosity and loose attachment, quantification of the biofilm after traditional crystal violet staining was unsuccessful. Biofilm formation was not observed in LB1 or LB2.5 (data not shown).
Fig 2.
Static biofilms of V. salmonicida LFI1238 and the ΔlitR strain formed in L15 medium at 4°C. The biofilms were visualized by phase-contrast microscopy after 3 days (A and B) and macroscopically after 6 days (C and D). No biofilm was formed by LFI1238 (A and C), whereas the ΔlitR strain (B and D) formed a thick and viscous biofilm under the chosen conditions.
LitR suppresses motility but is not required for flagellum expression in V. salmonicida.
LitR homologs regulate motility in several vibrios (17, 42), and similarly, we analyzed the impact of litR deletion on the motility of V. salmonicida. Depending on the salt concentration and incubation temperature, the ΔlitR strain was 22 to 80% more motile than LFI1238 on soft agar plates. The differences in motility zones between the two strains were highest at 4°C and with 3% NaCl (LFI1238 x̄ = 16.0 ± 0.00 mm and ΔlitR x̄ = 28.7 ± 0.88 mm; P < 0.0001 by Student's t test) and lowest at 12°C and with 1% NaCl (LFI1238 x̄ = 39.3 ± 0.67 mm and ΔlitR x̄ = 48.0 ± 0.00 mm; P = 0.0001 by Student's t test). The ΔlitRc strain showed wild-type motility. As previously reported for the wild-type strain (32), all strains were more motile at a high salt concentration and high temperature (12°C) than at a low salt concentration and low temperature (4°C). Furthermore, we analyzed LFI1238 and the ΔlitRc and ΔlitR strains by TEM after in vitro (i.e., agar plates) and in vivo (i.e., bacteria grown in implants in the abdominal cavities of fish) cultivation (data not shown). No difference in flagellation was observed under the various growth conditions tested, nor could we detect any fimbriae or pili on the bacterial surface of either strain, which could have explained the nature of the increased adhesion of the ΔlitR strain. All strains were found to express both sheathed polar flagella and unsheathed lateral flagella.
Salinity-dependent regulation of cryptic bioluminescence by LitR.
To elucidate the role of LitR regulation of the lux operon of V. salmonicida, we analyzed bioluminescence production after addition of decyl aldehyde. All strains produced 7 to 10 times more bioluminescence in media containing a high salt concentration than in media with low salt (P < 0.001 by Student's t test). Interestingly, the ΔlitR strain produced 20-fold (LFI1238 x̄ = 3,069 ± 291 cps/cell and ΔlitR x̄ = 158 ± 22 cps/cell; P < 0.0001 by Student's t test) and 4-fold (LFI1238 x̄ = 161 ± 16 cps/cell and ΔlitR x̄ = 40 ± 4 cps/cell; P = 0.0002 by Student's t test) less bioluminescence than the wild type in media containing 3% and 1% NaCl, respectively. V. wodanis strains isolated from fish lack the lux operon (31), and hence the FT5426 strain used in this assay produced only background levels of bioluminescence.
AHL production is dependent on salinity but is LitR independent.
Because LitR influences cryptic bioluminescence in V. salmonicida, we wanted to determine whether LitR regulates the production of AHLs. For this purpose, an AHL well diffusion assay was performed using the reporter strains C. violaceum CV026 and A. tumefaciens NT1 (53). The V. salmonicida strains produced significantly more AHLs in LB3 than in LB1 with C. violaceum as the reporter (diffusion zone x̄ for wild type in LB1 = 50.0 ± 0.8 mm, and in LB3 = 61.3 ± 1.3 mm [P = 0.0003 by Student's t test]; diffusion zone x̄ for ΔlitR strain in LB1 = 51.3 ± 0.8 mm, and in LB3 = 63.8 ± 1.3 mm [P = 0.0002 by Student's t test]) but not with A. tumefaciens as the reporter. No differences in AHL production were observed between LFI1238 and the ΔlitRc and ΔlitR strains, using either reporter strain, indicating that deletion of litR does not affect the production of C6-HSL and 3-oxo-C6-HSL.
QS and salinity regulate litR expression.
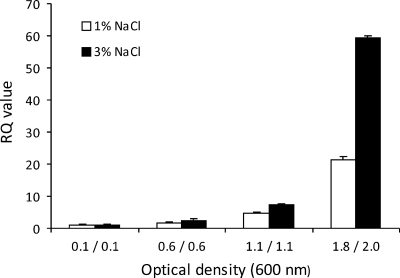
qRT-PCR was performed to quantify litR expression in V. salmonicida grown in vitro with a high or low salt concentration and to different cell densities. The PCR amplification signals were normalized to the 16S rRNA level. The results show that litR is expressed in a cell density-dependent manner, with approximately 21 and 59 times higher expression at high optical densities (1.8 and 2.0, respectively) than at an OD600 of 0.1 in LB1 and LB3, respectively (Fig. 3). This also suggests that a high salt concentration in the growth medium affects litR expression. Additional experiments using accD, ftsZ, and rpoD as endogenous controls gave similar results (see Fig. S2 in the supplemental material). To investigate if litR was transcribed in vivo, we isolated total RNA from the heart tissue of fish infected with the wild type and the ΔlitR mutant. The RT-PCR analysis identified litR mRNA in heart tissue from fish challenged with the wild type (threshold cycle [CT] value = 14.64) but not in that from the ΔlitR infected fish or control fish (CT value of >40).
Fig 3.
Expression of litR mRNA in V. salmonicida LFI1238 as determined by qRT-PCR. The wild type was cultivated in LB medium supplemented with 1% NaCl and 3% NaCl and harvested at different optical densities (at 600 nm). The relative expression (RQ) was calculated by the 2−ΔΔCT method to determined the level of expression relative to the transcription level in cultures harvested at an OD600 of 0.1 in LB1 and LB3, respectively (38). The CT values were normalized using the 16S rRNA level of V. salmonicida as a reference.
LitR is required for full virulence in V. salmonicida.
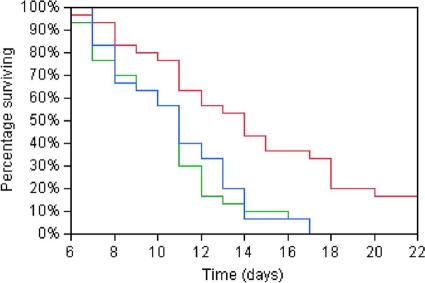
Having established that LitR regulates several disease-associated phenotypes and that its mRNA is expressed in fish suffering from CV, it was of interest to investigate the role of LitR in virulence. Atlantic salmon infected with LFI1238 and the ΔlitRc and ΔlitR strains expressed similar symptoms of CV to those described previously (13, 14, 52). No significant differences in symptoms or pathological signs were observed in fish infected with the different strains, except for a tendency for more tissue inflammation at the injection site in fish infected with the LFI1238 strain. In the i.p. challenge experiment, all salmon infected with LFI1238 and the ΔlitRc strain died during the trial. In contrast, 17% of the fish infected with the ΔlitR mutant survived. The difference between the LFI1238 and ΔlitR mortality rates was statistically significant (log rank test P < 0.0001; Wilcoxon test P < 0.0041) (Fig. 4). In the immersion challenge experiment, a growth attenuation of LFI1238 was observed before the start of challenge. However, because LFI1238 and the ΔlitRc strain expressed the same phenotype in each bioassay and in the i.p. challenge experiment, the immersion challenge experiment was continued without LFI1238. At the end of the trial, 13% of the ΔlitRc group survived, compared to 60% of the ΔlitR group. In both challenge experiments, V. salmonicida was grown from the head kidney for all diseased fish. No control fish showed symptoms of infection or mortality. Neither V. salmonicida nor any other bacterial species was identified after cultivation from the head kidneys of survivors or control fish.
Fig 4.
Survival plots after i.p. challenge of Atlantic salmon (Salmo salar) with V. salmonicida LFI1238 (blue line), the ΔlitRc strain (green line), and the ΔlitR strain (red line).
A comparison of the phenotypes expressed by the LFI1238 and ΔlitR strains is summarized in Table 3.
Table 3.
Phenotypes of LFI1238 and ΔlitR
| Parameter | Phenotypec |
|
|---|---|---|
| LFI1238 | ΔlitR | |
| Adhesiona | − | + |
| Aggregationb | + | ↑ |
| Biofilm formation | − | + |
| Motility | + | ↑ |
| Cryptic bioluminescence | + | ↓ |
| AHL production | + | + |
| Virulence | + | ↓ |
Adhesion on agar containing blood or blood components.
Aggregation in L15-FBS and BHI-HS media.
Upward- and downward-facing arrows indicate increased and reduced properties, respectively, compared to those of LFI1238.
DISCUSSION
For several bacterial pathogens, QS has been shown to regulate the production of virulence determinants (2, 3, 68, 69), and LitR homologs such as SmcR and HapR, from Vibrio vulnificus and Vibrio cholerae, respectively, regulate virulence genes and adaptive phenotypes (28, 35, 42, 57, 58, 74). A cell density-dependent system to ensure the optimal timing of virulence is a sophisticated strategy to overwhelm the host responses. Most vibrios, including V. fischeri, are known to regulate expression of litR or corresponding homologs in a cell density-dependent manner where a high cell density is necessary for stabilizing the mRNA of this master regulator (17, 44). However, one exception is the homolog vanT, in Vibrio anguillarum, which appears to be expressed equally regardless of cell density (11). Furthermore, VanT is not required for virulence but is believed to regulate the production of metalloprotease, pigment, and biofilm outside the fish host (10). In our study, we demonstrated that litR mRNA is expressed in a cell density-dependent manner in V. salmonicida and that LitR is involved in the pathogenesis of CV. Atlantic salmon infected with the ΔlitR mutant showed reduced mortality compared to fish infected with LFI1238 or the ΔlitRc strain, implying that the fish were more susceptible to infection with the wild type than with the mutant strain. No significant differences in symptoms or pathological signs were observed in diseased fish infected with the various strains. Symptoms of disease are usually established after the initial adherence, colonization, and invasion phase, suggesting that the factor(s) responsible for attenuation of the ΔlitR strain probably manifests early in infection.
Inactivation of litR changed several in vitro phenotypes of V. salmonicida, and the most prominent features of the ΔlitR strain were increased adhesion, aggregation, and ability to form a biofilm (Table 3). Adhesion is an important virulence factor in the early phase of infection but may also come at a cost, as bacterial attachment to immune cells can facilitate phagocytosis and clearing (33, 62). The litR homolog opaR in Vibrio parahaemolyticus is known to control the production of a thick capsular polysaccharide which promotes opacity, surface adhesion, and cell-cell adhesion (15, 41). Similarly, HapR in V. cholerae regulates the Vibrio polysaccharide synthesis (vps) genes, which are important for biofilm formation and colony morphology (20, 71, 73). Thus, increased adhesion and formation of a thick, slimy biofilm by the ΔlitR strain could be due to increased polysaccharide production and will be investigated further.
Little is known about the life cycle of V. salmonicida outside the fish. Our results make it tempting to suggest that adhesion, aggregation, and biofilm formation are different stages of a phenotype expressed in response to stress-related conditions frequently present in the marine environment. A biofilm protects bacteria from unfavorable environmental conditions (22). In addition, aggregation and biofilm formation could be a way to protect the bacterium from the immune system of the host and would explain the less severe tissue inflammation observed at the injection site for ΔlitR infected fish. During infection in a nutrient-rich environment such as fish tissue, increasing cell densities of V. salmonicida, and hence expression of LitR, should eventually downregulate aggregation and the biofilm mode. This downregulation may be necessary for adequate disease development, as reported for HapR of V. cholerae (37, 74). The ΔlitR strain mimics low-cell-density behaviors, and in the absence of LitR, the bacterium is captured in the biofilm mode, potentially resulting in a smaller number of planktonic cells that are able to disseminate within the host. Thus, biofilm formation in V. salmonicida is regulated by QS through LitR, where LitR is important for disassembly of the biofilm. In this way, aggregation and biofilm formation work as antivirulence factors in the ΔlitR strain.
Motility is linked to colonization, and attenuation of virulence due to loss of motility has been described thoroughly for many bacterial species (23, 36, 45, 49, 50, 54). However, motility demands considerable metabolic energy, and the flagella are also one of the major antigenic targets of the immune system (43). Therefore, the synthesis of the motility apparatus must be subjected to strict control (36, 40, 51). Differences in motility and adhesion could be explained by a differential flagellum, pilus, or fimbria expression by the ΔlitR strain compared to LFI1238 and the ΔlitRc strain. The V. salmonicida genome carries genes for two type IV pili, among which the pilC and pilQ analogs appear to be pseudogenes (26). Our study shows that the absence of LitR increases motility without affecting the number or morphology of the flagella. A similar phenotype has been reported for V. vulnificus, where SmcR downregulates motility (42). Thus, the cost of having an increased motility could attenuate the ΔlitR strain.
Our results strengthen the hypothesis that LitR positively regulates the lux operon, as described earlier for other bioluminous bacteria (18, 59). Any advantages that V. salmonicida gains from having a dysfunctional bioluminescence system are only speculative. Nelson et al. (47) hypothesized that the incomplete ability to produce bioluminescence in V. salmonicida leads to the formation of oxidative radicals (e.g., H2O2) potentially harmful to the host. This might explain the reduced mortality observed in fish infected with the ΔlitR strain. Various virulence mechanisms, including protease activity and cytotoxicity, have been described for different Vibrio species (1, 10, 29, 64). The tissue damage observed in fish suffering from CV may also be due to extracellular toxin activity (27, 63). However, we examined the cytotoxicity in cell cultures according to protocols by Tunsjø et al. (64), and we detected no cytopathogenic effects in any of six fish cell lines infected with LFI1238 or the ΔlitR mutant (unpublished results). These negative results could be explained by experimental conditions. However, it should be noted that Hjelmeland et al. (25) also failed to identify any extracellular cytotoxins or proteolytic enzymes in their study. Additionally, in the intestines of moribund salmon with CV, where V. salmonicida dominates the microbial flora, an absence of tissue damage of the epithelium has been reported (63). This suggests that V. salmonicida probably relies on mechanisms other than extracellular toxins to attack the host and escape its immune system.
From our challenge studies, we can also make some assumptions about the port of entrance of V. salmonicida into its natural host. i.p. injection is an artificial way of challenge compared to a bath challenge that simulates the natural route of infection. After an i.p. injection, bacteria must enter the bloodstream through the peritoneal serosa, whereas the port of entrance during a natural infection is unknown. In our challenge experiments, similar survival rates were found irrespective of infection route. This could point to the gills as the port of entrance during a natural infection, as the gills, in contrast to the intestinal mucosa and the skin epidermis, have an epithelial lining that is roughly similar to the peritoneal serosa.
The reporter strains we used for AHL detection were not selective for monitoring the concentrations of specific types of AHL compounds in the extracts, and this could have camouflaged a real difference in AHL production, as the diffusion zone induced by C6-HSL could have hidden the absence of a 3-oxo-C6-HSL diffusion zone and vice versa. To elucidate the qualitative nature of the produced AHLs, more sophisticated methods, such as high-performance liquid chromatography–high-resolution mass spectrometry, should be performed. However, our results suggest that LitR has little or no impact on the AHL-regulated part of the QS system in V. salmonicida. On the other hand, our results indicate a salinity-sensitive regulation of LitR. The production of AHLs, as well as adhesion, motility, and bioluminescence, was increased when V. salmonicida was grown at high salt concentrations. This indicates that the regulation of the different phenotypes is very complex and may be controlled not only by QS but also by other bacterially encoded factors, environmental conditions, and host-pathogen interactions.
We suggest that the QS master regulator LitR is required for cells of V. salmonicida to exhibit normal virulence in the fish host. The reduced virulence of the ΔlitR mutant appears to be due to the inability of the cells to transition from the biofilm mode to the planktonic mode, which is important for disease development. Future investigations may reveal other phenotypes regulated by LitR and generate more knowledge regarding the impact of the QS systems on V. salmonicida virulence and the mechanisms underlying the development of CV.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Debra Milton (Umeå University, Sweden) for kindly providing the suicide plasmids pDM4 and pNQ701. We also thank Stein Helge Skjelde (SørSmolt AS) for providing Atlantic salmon parr and smolts for the challenge experiments, Tove Hansen (Fellesakvariet, Norwegian School of Veterinary Science [NSVS]) and Oddbjørn Pettersen and coworkers (NIVA, Solbergstrand) for fish experiment facility management and technical assistance, and Ellen Dahl and coworkers (Hormonlab, NSVS), Ragnar Høie (The National Veterinary Institute), Norbert Roos (Electron Microscopy Laboratory, Department of Molecular Biosciences, UiO), and Aud Kari Fauske (Bacteriology Lab, NSVS) for technical support.
This work was supported by The Research Council of Norway (grant 174968/S10). H.C.W.-L. was supported by The Research Council of Norway (grant 158882/I10).
Footnotes
Published ahead of print 27 February 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Adin DM, Engle JT, Goldman WE, Fall-Ngai MJ, Stabb EV. 2008. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J. Bacteriol. 191:2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antunes LC, Ferreira RB. 2009. Intercellular communication in bacteria. Crit. Rev. Microbiol. 35:69–80 [DOI] [PubMed] [Google Scholar]
- 3. Antunes LC, Ferreira RB, Buckner MM, Finlay BB. 2010. Quorum sensing in bacterial virulence. Microbiology 156:2271–2282 [DOI] [PubMed] [Google Scholar]
- 4. Basson A, Flemming LA, Chenia HY. 2008. Evaluation of adherence, hydrophobicity, aggregation, and biofilm development of Flavobacterium johnsoniae-like isolates. Microb. Ecol. 55:1–14 [DOI] [PubMed] [Google Scholar]
- 5. Bruhn JB, et al. 2005. Quorum sensing signal molecules (acylated homoserine lactones) in gram-negative fish pathogenic bacteria. Dis. Aquat. Organ. 65:43–52 [DOI] [PubMed] [Google Scholar]
- 6. Chen X, et al. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549 [DOI] [PubMed] [Google Scholar]
- 7. Colquhoun DJ, Alvheim K, Dommarsnes K, Syvertsen C, Sørum H. 2002. Relevance of incubation temperature for Vibrio salmonicida vaccine production. J. Appl. Microbiol. 92:1087–1096 [DOI] [PubMed] [Google Scholar]
- 8. Colquhoun DJ, Sørum H. 1998. Outer membrane protein expression during in vivo cultivation of Vibrio salmonicida. Fish Shellfish Immunol. 8:367–377 [Google Scholar]
- 9. Colquhoun DJ, Sørum H. 2001. Temperature dependent siderophore production in Vibrio salmonicida. Microb. Pathog. 31:213–219 [DOI] [PubMed] [Google Scholar]
- 10. Croxatto A, et al. 2002. VanT, a homologue of Vibrio harveyi LuxR, regulates serine, metalloprotease, pigment, and biofilm production in Vibrio anguillarum. J. Bacteriol. 184:1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Croxatto A, et al. 2004. A distinctive dual-channel quorum-sensing system operates in Vibrio anguillarum. Mol. Microbiol. 52:1677–1689 [DOI] [PubMed] [Google Scholar]
- 12. Eggset G, Mikkelsen H, Killie JE. 1997. Immunocompetence and duration of immunity against Vibrio salmonicida and Aeromonas salmonicida after vaccination of Atlantic salmon (Salmo salar L.) at low and high temperatures. Fish Shellfish Immunol. 7:247–260 [Google Scholar]
- 13. Egidius E, Andersen K, Clausen E, Raa J. 1981. Cold-water vibriosis or “Hitra disease” in Norwegian salmonid farming. J. Fish Dis. 4:353–354 [Google Scholar]
- 14. Egidius E, Wiik R, Andersen K, Hoff KA, Hjeltnes B. 1986. Vibrio salmonicida sp. nov., a new fish pathogen. Int. J. Syst. Bacteriol. 36:518–520 [Google Scholar]
- 15. Enos-Berlage JL, McCarter LL. 2000. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J. Bacteriol. 182:5513–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernandez AI, Perez MJ, Rodriguez LA, Nieto TP. 1995. Surface phenotypic characteristics and virulence of Spanish isolates of Aeromonas salmonicida after passage through fish. Appl. Environ. Microbiol. 61:2010–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131–143 [DOI] [PubMed] [Google Scholar]
- 18. Fidopiastis PM, Sørum H, Ruby EG. 1999. Cryptic luminescence in the cold-water fish pathogen Vibrio salmonicida. Arch. Microbiol. 171:205–209 [DOI] [PubMed] [Google Scholar]
- 19. Flemming HC, Neu TR, Wozniak DJ. 2007. The EPS matrix: the “house of biofilm cells.” J. Bacteriol. 189:7945–7947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fong JC, Syed KA, Klose KE, Yildiz FH. 2010. Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology 156:2757–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goller CC, Romeo T. 2008. Environmental influences on biofilm development. Curr. Top. Microbiol. Immunol. 322:37–66 [DOI] [PubMed] [Google Scholar]
- 23. Graf J, Dunlap PV, Ruby EG. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J. Bacteriol. 176:6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104 [DOI] [PubMed] [Google Scholar]
- 25. Hjelmeland K, Stensvaag K, Jørgensen T, Espelid S. 1988. Isolation and characterization of a surface layer antigen from Vibrio salmonicida. J. Fish Dis. 11:197–205 [Google Scholar]
- 26. Hjerde E, et al. 2008. The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holm KO, Strøm E, Stensvaag K, Raa J, Jørgensen TØ. 1985. Characteristics of a Vibrio sp. associated with the “Hitra disease” of Atlantic salmon in Norwegian fish farms. Fish Pathol. 20:125–129 [Google Scholar]
- 28. Jeong HS, Lee MH, Lee KH, Park SJ, Choi SH. 2003. SmcR and cyclic AMP receptor protein coactivate Vibrio vulnificus vvpE encoding elastase through the RpoS-dependent promoter in a synergistic manner. J. Biol. Chem. 278:45072–45081 [DOI] [PubMed] [Google Scholar]
- 29. Jobling MG, Holmes RK. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023–1034 [DOI] [PubMed] [Google Scholar]
- 30. Jørgensen TØ. 1987. Microbiological and immunological aspects of “Hitra disease” of coldwater vibriosis (a summary), p 113–119 In Stenmark A, Malmberg G. (ed), Parasites and diseases in natural waters and aquaculture in Nordic countries. Naturhistoriska Riksmuseet, Stockholm, Sweden [Google Scholar]
- 31. Karlsen C. 2011. Marine cold water vibrio-like Atlantic salmon pathogens—adaptation, virulence and interactions. Ph.D. thesis University of Tromsø, Tromsø, Norway [Google Scholar]
- 32. Karlsen C, et al. 2008. Motility and flagellin gene expression in the fish pathogen Vibrio salmonicida: effects of salinity and temperature. Microb. Pathog. 45:258–264 [DOI] [PubMed] [Google Scholar]
- 33. Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592 [DOI] [PubMed] [Google Scholar]
- 34. Kolter R, Helinski DR. 1978. Construction of plasmid R6K derivatives in vitro: characterization of the R6K replication region. Plasmid 1:571–580 [DOI] [PubMed] [Google Scholar]
- 35. Lee JH, et al. 2007. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J. Microbiol. Biotechnol. 17:325–334 [PubMed] [Google Scholar]
- 36. Lee JH, et al. 2004. Role of flagellum and motility in pathogenesis of Vibrio vulnificus. Infect. Immun. 72:4905–4910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Z, et al. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 105:9769–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 39. Lupp C, Ruby EG. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lupp C, Ruby EG. 2005. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. J. Bacteriol. 187:3620–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCarter LL. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McDougald D, Rice SA, Kjelleberg S. 2001. SmcR-dependent regulation of adaptive phenotypes in Vibrio vulnificus. J. Bacteriol. 183:758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Millikan DS, Ruby EG. 2002. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl. Environ. Microbiol. 68:2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Milton DL. 2006. Quorum sensing in vibrios: complexity for diversification. Int. J. Med. Microbiol. 296:61–71 [DOI] [PubMed] [Google Scholar]
- 45. Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178:1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nealson KH. 1977. Autoinduction of bacterial luciferase. Occurrence, mechanism and significance. Arch. Microbiol. 112:73–79 [DOI] [PubMed] [Google Scholar]
- 47. Nelson EJ, Tunsjø HS, Fidopiastis PM, Sørum H, Ruby EG. 2007. A novel lux operon in the cryptically bioluminescent fish pathogen Vibrio salmonicida is associated with virulence. Appl. Environ. Microbiol. 73:1825–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nordmo R, Sevatdal S, Ramstad A. 1997. Experimental infection with Vibrio salmonicida in Atlantic salmon (Salmo salar L.): an evaluation of three different challenge methods. Aquaculture 158:23–32 [Google Scholar]
- 49. Ormonde P, Horstedt P, O'Toole R, Milton DL. 2000. Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J. Bacteriol. 182:2326–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. O'Toole R, Milton DL, Horstedt P, Wolf-Watz H. 1997. RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology 143:3849–3859 [DOI] [PubMed] [Google Scholar]
- 51. Ottemann KM, Miller JF. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109–1117 [DOI] [PubMed] [Google Scholar]
- 52. Poppe TT, Håstein T, Salte R. 1985. “Hitra disease” (haemorrhagic syndrome) in Norwegian salmon farming: present status, p 223–229 In Ellis AE. (ed), Fish and shellfish pathology. Academic Press, London, United Kingdom [Google Scholar]
- 53. Ravn L, Christensen AB, Molin S, Givskov M, Gram L. 2001. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 44:239–251 [DOI] [PubMed] [Google Scholar]
- 54. Richardson K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 56. Sambrook JE, Fritsch F, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 57. Shao CP, Hor LI. 2001. Regulation of metalloprotease gene expression in Vibrio vulnificus by a Vibrio harveyi LuxR homologue. J. Bacteriol. 183:1369–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shao CP, Lo HR, Lin JH, Hor LI. 2011. Regulation of cytotoxicity by quorum-sensing signaling in Vibrio vulnificus is mediated by SmcR, a repressor of HlyU. J. Bacteriol. 193:2557–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Showalter RE, Martin MO, Silverman MR. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 172:2946–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 61. Somerville GA, et al. 2002. In vitro serial passage of Staphylococcus aureus: changes in physiology, virulence factor production, and agr nucleotide sequence. J. Bacteriol. 184:1430–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taylor RK. 1991. Bacterial adhesion to mucosal surfaces. J. Chemother. 3:190–195 [PubMed] [Google Scholar]
- 63. Totland GK, Nylund A, Holm KO. 1987. An ultrastructural study of morphological changes in Atlantic salmon, Salmo salar L., during the development of cold water vibriosis. J. Fish Dis. 11:1–13 [Google Scholar]
- 64. Tunsjø HS, Paulsen SM, Berg K, Sørum H, L'Abée-Lund TM. 2009. The winter ulcer bacterium Moritella viscosa demonstrates adhesion and cytotoxicity in a fish cell model. Microb. Pathog. 47:134–142 [DOI] [PubMed] [Google Scholar]
- 65. Urbanczyk H, Ast JC, Higgins MJ, Carson J, Dunlap PV. 2007. Reclassification of Vibrio fischeri, Vibrio logei, Vibrio salmonicida and Vibrio wodanis as Aliivibrio fischeri gen. nov., comb. nov., Aliivibrio logei comb. nov., Aliivibrio salmonicida comb. nov. and Aliivibrio wodanis comb. nov. Int. J. Syst. Evol. Microbiol. 57:2823–2829 [DOI] [PubMed] [Google Scholar]
- 66. Valla S, et al. 1992. Development of a gene transfer system for curing of plasmids in the marine fish pathogen Vibrio salmonicida. Appl. Environ. Microbiol. 58:1980–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365–404 [DOI] [PubMed] [Google Scholar]
- 68. Williams P, et al. 2000. Quorum sensing and the population-dependent control of virulence. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:667–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Winzer K, Williams P. 2001. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int. J. Med. Microbiol. 291:131–143 [DOI] [PubMed] [Google Scholar]
- 70. Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. 2004. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol. Microbiol. 53:497–515 [DOI] [PubMed] [Google Scholar]
- 71. Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028–4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656 [DOI] [PubMed] [Google Scholar]
- 74. Zhu J, et al. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99:3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.