Abstract
Pseudomonas aeruginosa undergoes cell elongation and forms robust biofilms during anaerobic respiratory growth using nitrate (NO3−) as an alternative electron acceptor. Understanding the mechanism of cell shape change induced upon anaerobiosis is crucial to the development of effective treatments against P. aeruginosa biofilm infection. Here, we uncovered the molecular basis of anaerobiosis-triggered cell elongation and identified vitamin B12 to be a molecule that can reinstate defective anaerobic growth of P. aeruginosa. The ratio of total cellular DNA content to protein content was significantly decreased in the PAO1 strain grown under anaerobic conditions, indicating that DNA replication is impaired during anaerobic growth. Anaerobic growth of PAO1 reached a higher cell density in the presence of vitamin B12, an essential coenzyme of class II ribonucleotide reductase. In addition, cell morphology returned to a normal rod shape and transcription of stress-response genes was downregulated under the same anaerobic growth conditions. These results suggest that vitamin B12, the production of which was suppressed during anaerobic growth, can restore cellular machineries for DNA replication and therefore facilitate better anaerobic growth of P. aeruginosa with normal cell division. Importantly, biofilm formation was substantially decreased when grown with vitamin B12, further demonstrating that anaerobiosis-induced cell elongation is responsible for robust biofilm formation. Taken together, our data reveal mechanistic details of a morphological change that naturally occurs during anaerobic growth of P. aeruginosa and illustrates the ability of vitamin B12 to modulate the biofilm-forming capacity of P. aeruginosa under such condition.
INTRODUCTION
Pseudomonas aeruginosa, an opportunistic human pathogen, establishes persistent infections in the mucous airways of patients suffering from bronchiectasis, including cystic fibrosis (CF) (46). In the CF-affected lung, defective ion transport due to the lack of a functional cystic fibrosis transmembrane conductance regulator (CFTR) results in the formation of thickened mucous plaque on the airway epithelium, and such abnormal mucous layers are readily colonized by P. aeruginosa, which eventually proliferates into microbial communities known as biofilms (4, 9, 59). Of note, it was clearly demonstrated that the oxygen potential was decreased inside this thick mucous layer (56). Importantly, the results of previous studies by our group and others have revealed that P. aeruginosa, when grown by anaerobic respiration, forms robust biofilms (29, 57, 60). These data further implicate the clinical relevance of the biofilm mode of bacterial growth inside the mucous airway of CF patients.
As an obligate respirer, P. aeruginosa is equipped with highly sophisticated regulatory mechanisms that allow it to grow anaerobically using alternative electron acceptors, such as nitrate (NO3−) (44) or nitrite (NO2−) (58). These two compounds, presumed to be derived from nitric oxide (NO) produced by inflammatory responses, were present in relatively large quantities in the mucous airway of CF patients (16, 22, 26, 28). P. aeruginosa senses the lack of oxygen through an FNR (fumarate/nitrate regulator)-like transcriptional activator, ANR (anaerobic nitrate regulator) (1, 55), which becomes dimerized upon exposure to anaerobic environments through its oxygen-labile [4Fe-4S]2+ cluster (61). Active ANR recognizes a specific conserved promoter sequence (5′-TTGA-N6-TCAA-3′) called an ANR box and initiates the transcription of genes under its control. The genome of PAO1, a prototype strain of P. aeruginosa, contains a total of 170 ANR boxes in its genome-wide promoter regions (52). Included among these are promoters that direct the expression of genes encoding major anaerobic respiratory enzymes (61). Consistent with this function, a microarray analysis reported that 691 genes, or ∼12.4% of its total number of genes, were differentially expressed in response to anaerobiosis (14). Together, these results suggest that the genetic regulatory system of P. aeruginosa allows it to respond flexibly to changes in ambient oxygen potential.
Biofilm is a sessile microbial community, and its formation is often considered a complex developmental process (6). The general steps involved include (i) initial attachment of planktonic bacteria to a surface (30), (ii) microcolony formation (47), (iii) secretion of polymeric matrix and further proliferation into a macrocolony (40), (iv) maturation into a biofilm with a three-dimensional structure (33), and (v) liberation of planktonic bacteria from the biofilm (41). At each stage, bacterial cell-to-surface or bacterial cell-to-cell contact within a biofilm can be modulated by alterations in cell surface properties. In recent work, we demonstrated that unique changes in cellular morphology (i.e., cell elongation) intrinsically accompany anaerobic NO3− respiration and that such changes in cell shape positively influence the biofilm formation of PAO1, possibly accounting for the anaerobiosis-induced stimulation of biofilm formation in P. aeruginosa (57). Given that cell elongation normally occurs in bacterial cells under conditions when DNA replication is interrupted (10), these results suggested that P. aeruginosa, which has long been regarded to be a proficient anaerobic respirer, may indeed encounter stress associated with DNA replication during anaerobic growth. In this study, we uncovered the molecular mechanisms that underlie the cell elongation elicited only under anaerobic growth conditions and identified vitamin B12 to be a compound that can alleviate the stress associated with anaerobic growth. We also investigated the effect of supplementary vitamin B12 on the expression profiles of the whole transcriptome and on the biofilm formation. This report reveals previously undescribed molecular features associated with the anaerobic growth of P. aeruginosa and provides better insight into its pathogenic potential, leading us to formulate novel strategies to treat chronic P. aeruginosa airway infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains and plasmids used in this study are listed in Table 1. Bacterial cultures were grown at 37°C in Luria-Bertani medium (LB; 10 g tryptone, 5 g yeast extract, and 10 g NaCl per liter). Anaerobic growth of P. aeruginosa strains was achieved in a Coy anaerobic chamber (Coylab Inc., Grass Lake, MI). To support anaerobic growth, 1% KNO3 (Sigma-Aldrich) was added to the medium (termed LBN).
Table 1.
Bacterial strains and plasmid used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| P. aeruginosa strains | ||
| PAO1 | Wild type | Lab collection |
| ΔRNR_I | PAO1, PA1155 and PA1156 deleted | This study |
| ΔRNR_II | PAO1, PA5496 and PA5497 deleted | This study |
| ΔRNR_III | PAO1, PA1920 deleted | This study |
| ΔRNR_II&III | PAO1, PA5496, PA5497, and PA1920 deleted | This study |
| ΔnirS | PAO1, PA0519 deleted | 58 |
| ΔoprE | PAO1, PA0291 deleted | This study |
| E. coli SY327/λpir | F−ara del(lac-pro) argE(Am) recA56 rifR nalA λpir | Lab collection |
| Plasmid pCVD442 | sacB suicide vector from plasmid pUM24 | Lab collection |
Flow cytometry analysis.
The average protein or DNA content per cell was determined as average fluorescein isothiocyanate (FITC) or Hoechst 33258 fluorescence per cell, respectively. Bacterial cell staining and flow cytometry analysis were performed as described previously (27). In brief, PAO1 cells grown overnight either aerobically or anaerobically in LBN were harvested and resuspended in phosphate-buffered saline (PBS). The bacterial suspensions were fixed for 1 h by incubation with 1 ml of 75% ethanol. Fixed cells were spun down and washed with ice-cold PBS. Next, the fixed cells were sequentially stained with 500 μl of 2 μg/ml FITC in PBS, followed by 500 μl of 2 μg/ml Hoechst 33258. Flow cytometry analysis was performed using ∼104 cells with an LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ) equipped with an argon ion laser emitting 0.5 W at 488 nm (Spectra-Physics, Santa Clara, CA) and a krypton laser emitting 0.5 W in multiline UV mode (351 and 357 nm; Spectra-Physics). The flow data were analyzed using FACSDiva software (Becton Dickinson). The average DNA and protein contents per cell were determined as the average Hoechst 33258 and FITC fluorescence per cell, respectively.
DPA, vitamin B12, and CV biofilm assays.
The quantitative measurement of total DNA content was performed by diphenylamine (DPA) assay following the procedures described previously (3). In summary, the DPA reagent that consists of 2 g DPA (Sigma-Aldrich) dissolved in 100 ml of pure acetic acid and 2.75 ml of concentrated sulfuric acid was mixed with bacterial cell extract in the proportion of 2:1. The mixture was incubated at 37°C for 4 h, followed by measurement of absorbance at 595 nm. The amount of vitamin B12 present in culture supernatants was measured as described previously (31). Crystal violet (CV) biofilm staining assays were performed as described previously (57).
qRT-PCR analysis.
Transcript levels of ribonucleotide reductase (RNR)-coding genes (PA1155, PA1156, PA1920, PA5496, and PA5497) were measured by quantitative real-time PCR (qRT-PCR). To verify our microarray results, qRT-PCR analysis was also performed on a subset of genes whose expression levels were determined to be significantly altered. The detailed procedure used for the analysis has been described previously (57). Transcript levels of the rpoD gene were similar in cells grown under aerobic or anaerobic conditions, and transcript levels of rpoD were thus used to normalize the transcript levels of tested genes. The primers used for qRT-PCR are listed in Table 2.
Table 2.
Primers used for qRT-PCR
| Gene name | Orientationa | Primer sequence (5′-3′) |
|---|---|---|
| nrdA (PA1156) | F | GCAAGGCCATCGATCACGAG |
| R | TGCAGGCCGAGGTAGGTGAA | |
| nrdB (PA1155) | F | GTTCCTGCGCAACCTGATCG |
| R | GCCCATGGAGAGGATCTGGG | |
| nrdJa (PA5497) | F | CGGAACTGATCGAGGCGTTG |
| R | GCATTGTTGGCCAGGCTCAG | |
| nrdJb (PA5496) | F | TCAACGAAGCCGAGGAGCAG |
| R | TCGAACAGCGGCGACTTGAT | |
| nrdD (PA1920) | F | GCACCCTGCAGAACGAGTGG |
| R | GCGTAGTCGAGGCGGTCCTT | |
| PA4554 | F | CAATTGCTGAACGACTCGAA |
| R | AAGAAGTTCACCCGGTGTTG | |
| PA2009 | F | GATCTTCACCGTGCTGACCT |
| R | GGGAAGATCACGAAGTCGAT | |
| PA3152 | F | ATGGGCTGGAATCAAGTGTC |
| R | TAGAATCGGCTTTGCTCGTT | |
| PA2171 | F | GGTCAAGGTGCTCAAGGAAC |
| R | TAGTTCGCCCTCTTCCTCCT | |
| PA0620 | F | TATGCGAATCAGACGAGTGC |
| R | ACCTGTACTGGCATCCGTTC | |
| PA1150 | F | AGGACCAAGTCCGTATGTCG |
| R | CCCGCTTCACTCTTGAGTTC | |
| PA3875 | F | AGTACCTGCTCGGGGCGAAG |
| R | ATGCGGAAGTCCAGGGTGGT | |
| PA4761 | F | ACATCCGCCTGATCGACTAC |
| R | CAGCTCGATCTTGGCTTTCT | |
| PA5054 | F | CAACGAGGAAGAGCTCAAGG |
| R | TCTCGTCGATGAAGACGATG | |
| rpoD (internal control) | F | AAGGCCCTGAAGAAGCACGG |
| R | GATCGGCATGAACAGCTCGG |
F, forward; R, reverse.
Construction of nrdAB, nrdJab, and nrdD deletion mutants.
RNR deletion mutants were created by allele replacement as previously described (34). An allelic exchange reaction to construct a class I RNR mutant was performed under strict anaerobic growth conditions. Five hundred base pairs flanking sequences at both ends of the nrdAB, nrdJab, or nrdD locus was PCR amplified with primers harboring specific restriction enzyme sites. The nrdAB upstream sequence was digested with SalI and BamHI, while the downstream sequence was digested with BamHI and SacI. In this manner, the 3′ end of the upstream sequence and the 5′ end of the downstream sequence can be connected with no further treatment. Likewise, the nrdJab upstream and downstream sequences were digested with pairs of SphI/SacI and SacI/SmaI, respectively. Pairs of restriction enzymes (SalI/SphI and SphI/SacI) were used to process the nrdD PCR products. pCVD442-Gm, a suicide vector for gene replacement carrying a gentamicin resistance marker, was cleaved with each set of restriction enzymes (i.e., SalI/SacI for nrdAB and nrdD and SphI/SmaI for nrdJab) and ligated with the corresponding PCR products. The resultant suicide vectors carrying each gene deletion were electroporated into Escherichia coli SY327/λpir for subsequent conjugation into P. aeruginosa. Transconjugants were selected on LB agar plates containing 200 μg/ml gentamicin, and the second crossover of allele exchange was induced by 6% sucrose. The deletion of each gene locus was confirmed by PCR, and the nrdAB, nrdJab, and nrdD mutants were named ΔRNR_I, ΔRNR_II, and ΔRNR_III, respectively. Further mutation of the nrdD gene was induced by using the ΔRNR_II mutant as a recipient strain to create the ΔRNR_II&III double mutant.
Confocal microscopy image analysis.
Differential interference contrast (DIC) images to show bacterial cell shape and three-dimensional fluorescent biofilm images were acquired using a confocal microscope, as described in previous literature (57). For DIC images, bacterial cells grown for 16 h under specified conditions were mounted in eight-well Lab-Tek chambered cover glass (catalog no. 155411; Nalge Nunc International). PAO1 cells transformed with a plasmid expressing green fluorescent protein (GFP) were used for biofilm analysis. Two microliters of preculture grown aerobically was inoculated into 200 μl of LBN or LBN supplemented with 1 μM vitamin B12 (Sigma-Aldrich) placed in the same chambered cover glass. After biofilm growth for 24 h at 37°C inside the anaerobic chamber, each well was washed with PBS. A 488-nm laser excited the samples, and the emission was detected through a 520-nm filter. The green fluorescence images were collected at 2 μs/pixel speed. xy images of 57.232 μm by 57.232 μm were acquired, and 40 sliced images of 20.28 μm total depth (0.507 μm/slice) were scanned in the z direction. Images were saved as TIF files with embedded xyz scale lines. The average green fluorescence intensity of each sliced image was measured using ImageJ software (http://rsbweb.nih.gov/ij) and plotted against distance from the bottom of the biofilm.
Microarray analysis.
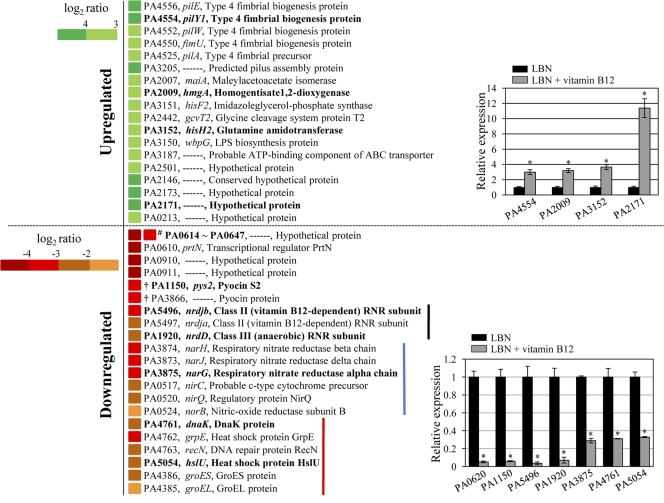
Microarray-based expression analysis of the whole genome of PAO1 was performed using GeneChip P. aeruginosa genome arrays (Affymetrix, Santa Clara, CA). The PAO1 strain was grown anaerobically in LBN or LBN with 1 μM vitamin B12 for 12 h. Total bacterial RNA was isolated from each of three independent cultures per growth condition. RNA was extracted using TRIzol reagent (Invitrogen, Burlington, ON, Canada) following the manufacturer's instructions, and extracted RNA was further purified by using an RNeasy kit (Qiagen). Purified RNA samples were then pooled together and submitted to DNA Link Inc. (Seoul, South Korea), where RNA quality was monitored using an Agilent 2100 bioanalyzer. Per RNA sample, 10 μg was used as input into the Affymetrix procedure as recommended by the manufacturer's protocol (Affymetrix). Briefly, 10 μg of total RNA was converted to double-stranded cDNA using random primers. Double-stranded cDNA was purified with a MinElute PCR purification kit (Qiagen, Hilden, Germany) and quantified by an ND-1000 spectrophotometer (NanoDrop Technologies, Inc., DE). The purified double-stranded cDNA was fragmented using 0.6 U/μl of DNase I and end labeled by terminal transferase reaction incorporating a biotinylated dideoxynucleotide. Fragmented end-labeled cDNA was hybridized to the GeneChip P. aeruginosa genome arrays for 16 h at 45°C and 60 rpm as described in the Affymetrix technical manual. After hybridization, the chips were stained and washed in a GeneChip Fluidics Station 450 apparatus (Affymetrix) and scanned by using a GeneChip Array Scanner 3000 7G (Affymetrix). The image data were extracted through Affymetrix Command Console software (version 1.1), and the raw CEL file was saved for subsequent data analysis. The Robust MultiAverage (RMA) algorithm implemented in Affymetrix Expression Console software (version 1.1) was used to normalize the raw data. Genes that showed significantly altered expression levels in response to the growth with vitamin B12 were selected and displayed as a heat map in Fig. 7.
Fig 7.
Microarray analysis of PAO1 grown by anaerobic NO3− respiration with added vitamin B12. Heat map of differentially expressed genes upon growth with versus without vitamin B12. The heat map shows the log2 ratio of transcripts that were upregulated (green) or downregulated (red) in response to the anaerobic growth with 1 μM vitamin B12. Genes encoding similar traits were clustered together and are denoted by colored vertical lines. A list of all the genes that were differentially regulated is also provided as a table in the supplemental material. #, expression of 34 genes (PA0614 to PA0647) originated from a phage genome was invariably downregulated and, thus, presented as one indication unit; among 34 genes, log2 ratios of 29 genes were lower than −4 and log2 ratios of the other 5 genes (PA0622, PA0645, PA0621, PA0634, and PA0616) were between −3.7 and −4.0; †, some of the strains of P. aeruginosa (i.e., C3719 and PACS2) do not possess these two genes in their genomes. Genes shown in boldface were selected, and their altered expression levels, shown in bar graphs to the right, were further confirmed by qRT-PCR analysis. Experimental conditions for qRT-PCR were identical to those described for Fig. 2B. *, P < 0.05 versus transcript levels of PAO1 cells grown in LBN.
Statistical analysis.
Data are expressed as mean ± standard deviation (SD). An unpaired Student's t test was used to analyze the data. A P value of <0.05 was considered statistically significant. All the experiments were repeated for reproducibility.
Microarray data accession number.
The entire microarray results are available in NCBI's GEO database under accession number GSE34836.
RESULTS
The average DNA content per cell was decreased in PAO1 cells grown by anaerobic versus aerobic respiration.
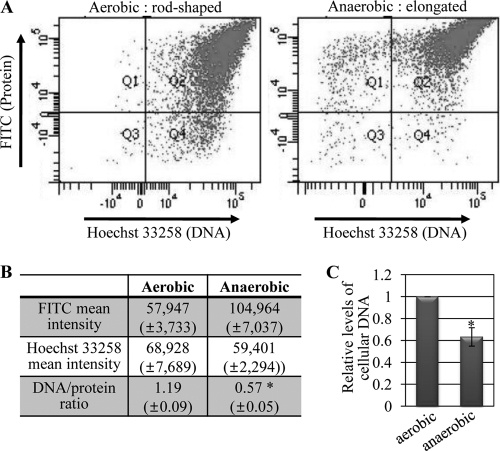
Since cell elongation is known to be caused by stimuli that induce DNA damage in bacteria (27, 42), we postulated that DNA replication may not occur optimally in PAO1 during anaerobic respiration. To address this issue, we measured the DNA/protein ratios of PAO1 cells grown by either aerobic or anaerobic respiration. The DNA/protein ratio was reported to be kept constant in Gram-negative bacteria (8, 20), and thus, this ratio has been used as a parameter to indicate whether DNA replication is orchestrated appropriately with the bacterial cell division cycle (27, 49). As described in Materials and Methods, the DNA content per cell was determined on the basis of Hoechst 33258 fluorescence intensity, while the protein content was presented as the fluorescence intensity of FITC that labels the amine groups of cellular proteins. Figure 1A shows double-fluorescent dot plots of the bacterial cells (∼10,000 cells) stained with both Hoechst 33258 and FITC. Clear and distinct differences in staining patterns were observed between the rod-shaped and elongated PAO1 cells. As summarized in Fig. 1B, the mean FITC intensity of the anaerobically grown (and, thus, elongated) PAO1 cells increased more than 2-fold compared to that of the rod-shaped cells. The total DNA content, represented as the mean intensity of Hoechst 33258 staining, however, was somewhat decreased in anaerobically grown PAO1 cells, yielding a DNA/protein ratio of 0.57, a value significantly lower than that found in aerobically grown PAO1 cells. This suggests that DNA synthesis was not accordingly increased in PAO1 during the anaerobiosis-induced cell elongation process. Next, to corroborate our flow cytometry results, we compared the total cellular content of deoxynucleoside triphosphates (dNTPs) in bacterial cell extracts by using DPA assay. When bacterial cell extracts adjusted to contain equal protein concentration were subjected to the assay, the level of dNTPs detected in the anaerobic cell extract was ∼60% of the level of dNTPs in the aerobic counterpart, further suggesting that synthesis of chromosomal DNA is likely hampered during anaerobic growth in P. aeruginosa.
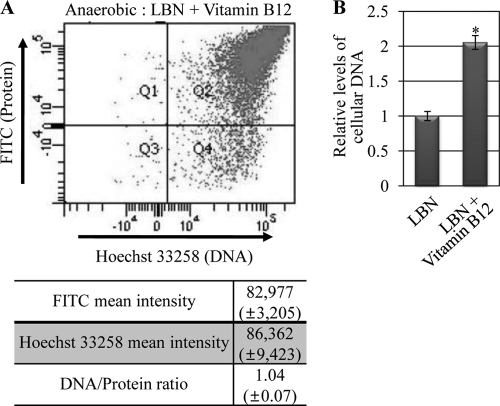
Fig 1.
Relative DNA content per cellular protein was decreased in elongated PAO1 cells. (A) Double-fluorescent dot plot analysis of PAO1. Bacterial cells (∼10,000 cells) grown aerobically or anaerobically in LBN for 16 h were stained with Hoechst 33258 and FITC for a quantitative presentation of cellular DNA and protein contents, respectively. In the dot plot, x and y axes represent the intensities derived from Hoechst 33258 (blue, for DNA) and FITC (green, for proteins), respectively. The plot is divided into four quadrants (Q1 to Q4). Quadrant Q2 contains PAO1 cells labeled with both dyes. (B) Average fluorescence intensities of PAO1 cells grown under either condition. The DNA/protein ratio was calculated by dividing the mean Hoechst 33258 intensity by the mean FITC intensity. The values shown are the means ± SDs from three independent experiments. *, P < 0.01 versus DNA/protein ratio of aerobically grown cells. (C) DPA assay of PAO1 cells grown in LBN under aerobic or anaerobic conditions. Bacterial cell extracts containing equal protein contents were used for the assay, and the DNA contents of anaerobically grown cells were normalized with those of aerobically grown PAO1 cells. Three independent experiments were performed, and values of means ± SDs are displayed in each bar. *, P < 0.01 versus DNA contents in aerobically grown PAO1 cells.
Transcript levels of genes encoding RNR were highly induced in anaerobically growing PAO1.
RNR is an enzyme that catalyzes the formation of deoxyribonucleotides from ribonucleotides, thereby providing the building blocks for DNA synthesis (24). In the P. aeruginosa genome, there are distinct gene clusters that encode three different classes of RNRs (Fig. 2A). Class I requires molecular oxygen to generate radicals for catalytic activity, while the activity of class III is known to be activated under anaerobic growth conditions (45a, 50). Class II RNR uses vitamin B12 as a cofactor to initiate catalytic activity and is active under both aerobic and anaerobic conditions (45a, 50). In bacteria, genes encoding RNRs are transcriptionally activated, when DNA synthesis is interrupted (12, 15), and therefore, measuring the transcript levels of RNR-coding genes provides a reliable method for monitoring the state of DNA synthesis. Our qRT-PCR analysis indicates that the mRNA levels of five selected genes, nrdA, nrdB, nrdJa, nrdJb, and nrdD, were invariably increased in PAO1 cells grown by anaerobic versus aerobic respiration, with nrdA and nrdJa being upregulated to the highest level at greater than 14- and 12-fold, respectively (Fig. 2B). Transcript levels of nrdB and nrdJb were ∼5.6- and ∼2.7-fold increased, respectively. These results, together with those shown in Fig. 1, suggest that DNA synthesis may not occur optimally in PAO1 during anaerobic growth and that such interrupted DNA synthesis likely accounts for the anaerobiosis-induced cell elongation.
Fig 2.
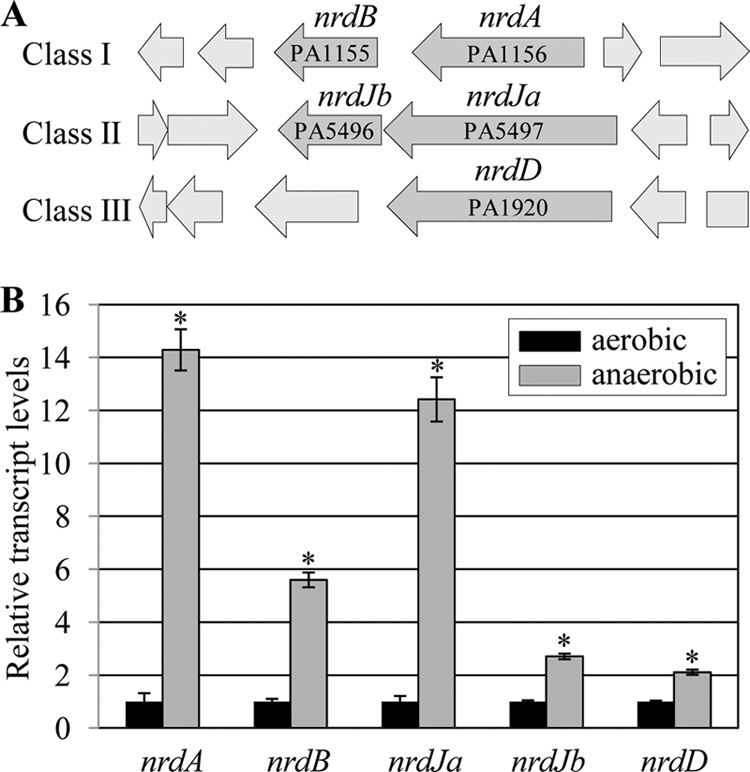
Quantitative RT-PCR analysis of genes encoding three different classes of RNR. (A) Open reading frame maps of RNR-coding regions in PAO1 genome. Three different genetic loci encoding a component(s) of each class of RNR are shown with corresponding PA numbers. (B) qRT-PCR was conducted on cDNA synthesized from 2 μg total RNA extracted from PAO1 cells grown either aerobically or anaerobically. Transcript levels of the five genes indicated at the bottom of each set of bars were normalized with levels of the rpoD transcript. Three independent experiments were performed, and values of means ± SDs are displayed in each bar. *, P < 0.05 versus transcript levels in PAO1 cells grown aerobically.
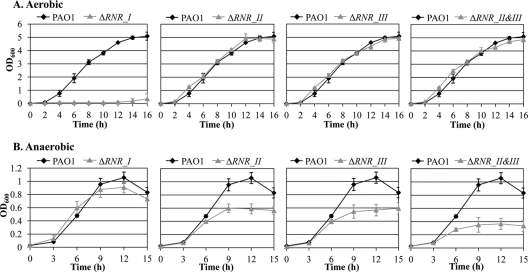
The deletion mutants of each RNR class have distinctive growth phenotypes under aerobic and anaerobic conditions.
Our results (Fig. 2B) demonstrated that transcription of genes for class I and II but not class III RNRs was highly induced upon anaerobiosis. This suggests that the expression of the former two genes is subject to more sensitive regulation and that each class of RNR may play a differential role depending on the oxygen tension in the growth environment. To examine the effects of deficiencies of each class of RNR on bacterial growth, we constructed a series of RNR mutant strains. No discernible growth was observed in a mutant PAO1 strain defective in RNR class I after 16 h of aerobic growth at 37°C (Fig. 3A). This particular mutant was recovered and maintained by anaerobic growth. In contrast, single mutants of class II or III and a class II/III double mutant exhibited completely normal growth by aerobic respiration (Fig. 3A), suggesting that DNA biogenesis under aerobic conditions is solely dependent on RNR class I, while the other two classes are dispensable. Under anaerobic conditions, however, bacterial growth of the class I mutant was only mildly affected (Fig. 3B, leftmost set of growth curves), further suggesting that the class I RNR plays a more dominant role under aerobic growth conditions. The extent of growth impairment resulting from mutations in class II or class III RNR was greater than that associated with the disruption of class I genes (Fig. 3B). Importantly, anaerobic growth of the class II/III double mutant was most severely affected (Fig. 3B). Together, these results demonstrate that (i) class I RNR is necessary and sufficient for DNA replication during aerobic growth and (ii) class II and class III RNRs play more significant roles in supporting anaerobic growth of P. aeruginosa.
Fig 3.
Growth curves of RNR-defective PAO1 mutants. Growth of various RNR mutant strains, indicated at the top of each graph, was compared with that of PAO1. Strains were grown in LBN either aerobically for 16 h (A) or anaerobically for 15 h (B). Anaerobic growth and aliquot samplings to measure OD600 were performed inside the anaerobic chamber. Each growth curve experiment was repeated for three times, and means ± SDs are displayed in each graph.
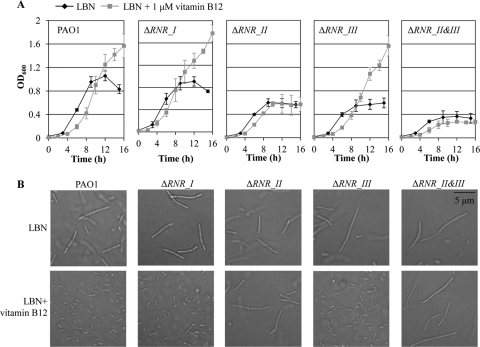
When grown with extraneous vitamin B12, cell shape returned to normal, and this morphological change resulted in robust anaerobic growth.
RNRs should have complete radical centers to function, and the activity of class II RNR is strictly dependent on the presence of its coenzyme, vitamin B12 (23). To better understand the degree to which the class II RNR could be activated upon anaerobic growth, we measured the level of vitamin B12 produced under such conditions. As shown in Fig. 4, the production of vitamin B12 was significantly suppressed during growth by anaerobic respiration. In contrast, the level of vitamin B12 secreted into the culture medium continued to increase with time during aerobic culture, demonstrating that the machinery to produce vitamin B12 was highly impaired upon anaerobiosis. Next, we evaluated the effects of added vitamin B12 on the anaerobic growth of P. aeruginosa strains. When grown with 1 μM vitamin B12, improved growth of PAO1 was clearly observed (Fig. 5A, first set). Similar growth enhancement was also observed in the mutants in which class I or class III RNR was inactivated (Fig. 5A, second and fourth sets). In the presence of extra vitamin B12, anaerobic growth reached an optical density at 600 nm (OD600) as high as 1.4 to 1.6 after a 16-h culture period, a value more than 2-fold higher than that after growth in LBN. Such an elevation in growth, however, was not detected when genes encoding class II RNR were disrupted (Fig. 5, third and fifth sets), further confirming the specific requirement of vitamin B12 for the activation of class II RNR.
Fig 4.
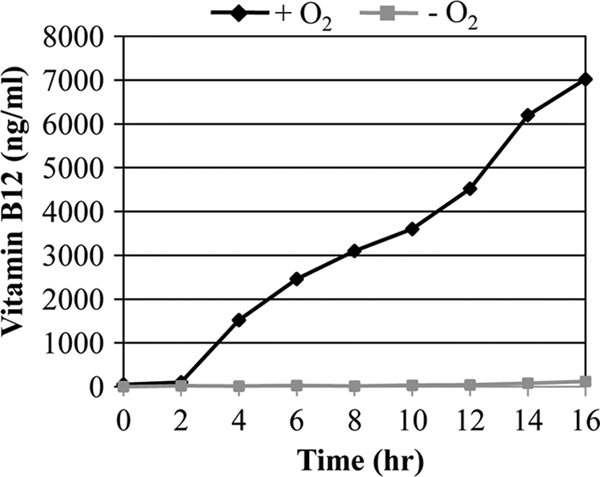
Vitamin B12 production profiles of PAO1 during aerobic and anaerobic growth. Culture supernatants removed every 2 h during aerobic and anaerobic growth were filter sterilized, and vitamin B12 contents were assessed as described in Materials and Methods.
Fig 5.
Effects of vitamin B12 on the anaerobic growth and cellular morphology of PAO1 and RNR-defective mutants. (A) Anaerobic growth of various RNR mutant strains was compared with that of PAO1. Strains were grown in LBN supplemented with 1 μM vitamin B12 or in LBN inside the anaerobic chamber. Each growth curve experiment was repeated for three times, and means ± SDs are displayed in each graph. (B) DIC images of P. aeruginosa strains grown in LBN and LBN plus 1 μM vitamin B12. All images were acquired at the same magnification.
Interestingly, enhanced anaerobic growth is accompanied by changes in cell shape. Upon anaerobic growth with the addition of vitamin B12, cell morphology returned to the normal rod shape in cells that possess uninterrupted class II RNR genes (Fig. 5B). Two strains that harbor mutations in class II RNR (ΔRNR_II and ΔRNR_II&III) remained elongated under the same growth conditions. These results suggest that restimulation of class II RNR by the addition of vitamin B12 helped bacteria undergo optimal cell division, which in turn resulted in a significant increase in the anaerobic growth of P. aeruginosa.
Next, we sought to examine whether the value of the DNA/protein ratio would reflect the cell shape change observed in our microscopic analysis (Fig. 5B). To investigate this, we repeated the FACS analysis using PAO1 cells grown anaerobically in LBN supplemented with vitamin B12. The average fluorescence intensities of Hoechst 33258 and FITC were 86,362 and 82,977, respectively, yielding a DNA/protein ratio of ∼1.04, a value comparable to that derived from aerobically grown PAO1 cells (Fig. 6A). The DPA assay also demonstrated that the relative DNA content per protein was increased ∼2-fold in PAO1 cells grown with added vitamin B12 (Fig. 6B). These results provide further evidence that the ability to synthesize DNA was restored by the sole addition of vitamin B12 in PAO1.
Fig 6.
Ability to replicate DNA was restored upon growth with added vitamin B12. (A) Double-fluorescent dot plot analysis was performed using PAO1 (∼10,000 cells) grown anaerobically in LBN supplemented with 1 μM vitamin B12. Experimental conditions were identical to those described for Fig. 1A. (B) DPA assay of PAO1 cells grown in LBN under anaerobic conditions in the absence or presence of added vitamin B12. Experimental conditions were identical to those described for Fig. 1C. *, P < 0.01 versus DNA contents in PAO1 cells grown in LBN.
Microarray analysis revealed significant changes in gene expression profiles in PAO1 cells grown with added vitamin B12.
Our results demonstrated that PAO1 cells, when grown with vitamin B12, underwent significant changes in growth-associated phenotypes, such as an enhanced growth rate and cell shape changes. To better understand the global changes in gene expression stimulated by the addition of vitamin B12, we performed microarray analysis. Figure 7 shows a list of genes that were highly upregulated or downregulated in PAO1 upon anaerobic growth in the presence versus absence of vitamin B12. Among the most upregulated genes, a substantial number are involved in fimbrial biogenesis (Fig. 7, top portion). In addition, the expression of genes encoding diverse metabolic enzymes (i.e., maiA, hmgA, hisF2, and hisH2) and surface molecules (i.e., wbpG and oprB) was also highly activated, when grown in medium supplemented with vitamin B12, and therefore, their rod-shaped morphology was steadily maintained. The transcription of a cluster of genes from PA0614 to PA0647 was invariably repressed upon growth with added vitamin B12. Most genes assembled in this cluster encode probable bacteriophage proteins and hypothetical proteins associated with bacteriophage (48). A subset of genes in this particular genetic locus was induced in their expression during anaerobic growth compared to the level of expression during aerobic growth (14). Importantly, anaerobic growth with vitamin B12 resulted in significant decreases in the expression of genes encoding class II and III RNRs (Fig. 7, black vertical line to the right), providing additional evidence that the ability to replicate DNA was restored in the presence of extraneous vitamin B12. It is also worthy of notice that the transcriptional levels of a group of stress-response genes (i.e., dnaK, grpE, recN, hslU, groES, and groEL; Fig. 7, red vertical line) were reduced, suggesting that a great deal of stress incurred during anaerobiosis is relieved by the addition of vitamin B12. Intriguingly, a significant decrease in the expression of genes encoding enzymes involved in anaerobic respiration was also detected in the microarray analysis (Fig. 7, blue vertical line).
To verify our microarray results, we selected a total of 11 genes, including 4 upregulated and 7 downregulated genes, and performed qRT-PCR analysis on those genes. Consistent with our microarray results, relative expression of all selected genes exhibited identical patterns in response to the growth with extraneously added vitamin B12 (Fig. 7, bar graphs).
Biofilm formation was reduced during anaerobic growth with extraneous vitamin B12.
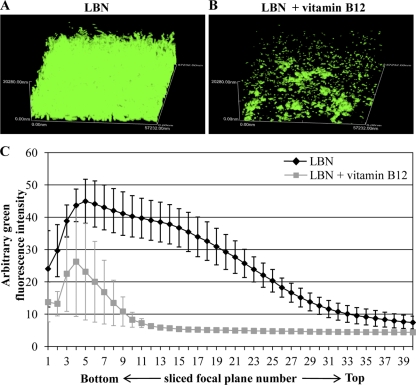
The biofilm formation of P. aeruginosa was significantly enhanced during anaerobic respiration (29, 60). Our previous research has demonstrated that a positive correlation exists between anaerobiosis-induced cell elongation and biofilm formation (57). To further demonstrate that enhanced biofilm formation is an event associated with cell elongation, we examined the effect of vitamin B12 on the anaerobic biofilm formation of P. aeruginosa. Figure 8 shows three-dimensional biofilm images (57.232 by 57.232 by 20.280 μm) of PAO1 cells harboring a plasmid that produces GFP. A robust biofilm with a considerable depth was formed by PAO1 cells, when grown anaerobically in LBN (Fig. 8A). On the basis of our biofilm image analysis, higher green fluorescence intensities were persistently detected with increasing biofilm height (Fig. 8C, black line). In contrast, biofilm formation was significantly reduced in PAO1 cells grown with extraneous vitamin B12 (Fig. 8B). In this biofilm, the green fluorescence intensity was detected only in the first 11 sliced images (Fig. 8C). These results clearly suggest that suppression of anaerobiosis-induced cell elongation by the addition of vitamin B12 negatively influenced the biofilm formation in PAO1.
Fig 8.
Effect of vitamin B12 on the anaerobic biofilm formation of PAO1. Confocal laser scanning microscopic analysis of anaerobic PAO1 biofilms. GFP-tagged PAO1 was grown statistically to form biofilms in LBN (A) or in LBN supplemented with vitamin B12 (B) for 24 h. Three-dimensional images are generated from a stack of 40 sliced images taken at 0.507-μm intervals for a total of 20.28 μm. Before image acquisition, 1-day-old anaerobic biofilms were washed with PBS for 2 times. (C) The average green fluorescence intensities in each of 40 sliced focal planes are compared between two biofilms. The values shown are the means ± SDs from four independent biofilm experiments.
Anaerobic growth rescue by vitamin B12 did not occur in a nitrite reductase-deficient mutant.
Our previous results demonstrated that a ΔnirS mutant lacking in the activity of nitrite reductase was not elongated during anaerobic growth in LBN (57). Given that (i) nitric oxide (NO) is the product of nitrite reductase and (ii) RNRs are highly susceptible to NO-mediated intoxication (18, 39), this result suggested that endogenously produced NO was ascribed to the inactivation of RNRs, resulting in the anaerobiosis-triggered cell elongation. We therefore sought to examine whether vitamin B12 can still rescue the limited anaerobic growth of the ΔnirS mutant, where NO-mediated intoxication would not occur. As shown in Fig. 9A, mutant cells maintained their rod shape regardless of the presence of vitamin B12, while PAO1 cells were repeatedly able to return to their normal shape. The anaerobic growth of the ΔnirS mutant was not elevated to any degree by the presence of added vitamin B12 (Fig. 9B, dashed lines), suggesting that vitamin B12 can effectively rescue only anaerobic growth arrested by NO-mediated RNR inactivation.
Fig 9.
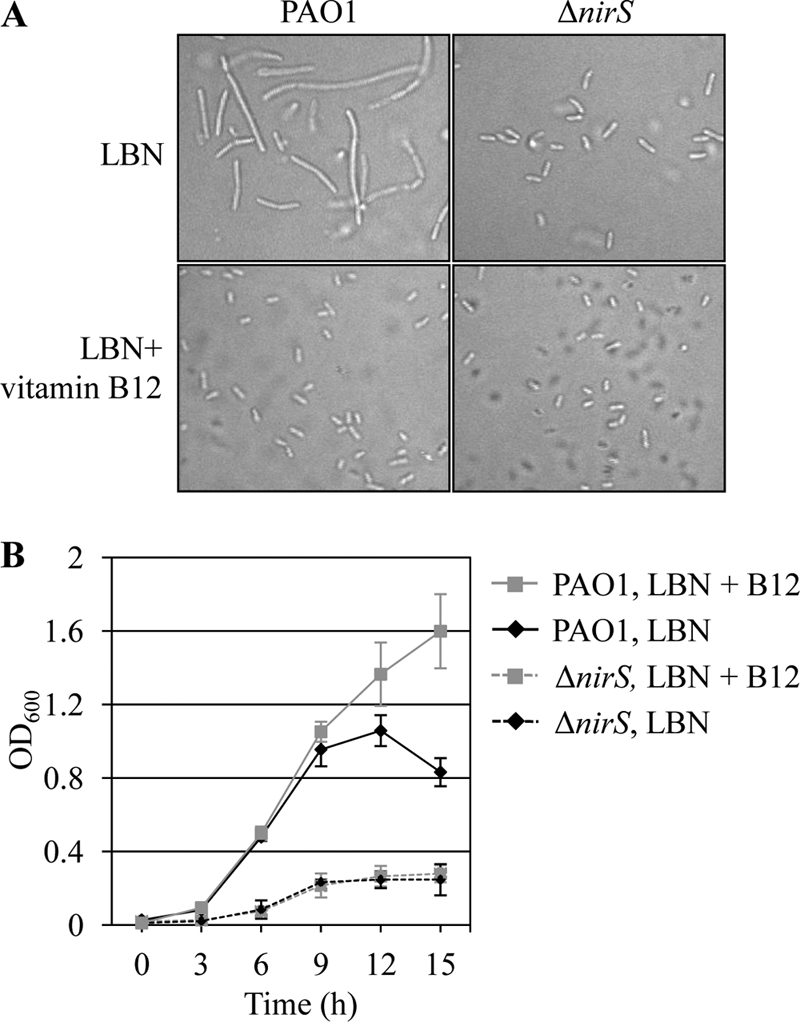
Vitamin B12-mediated anaerobic growth rescue was not observed in a ΔnirS mutant devoid of nitrite reductase. (A) DIC images of PAO1 and the ΔnirS mutant grown in LBN and LBN plus 1 μM vitamin B12. Experimental conditions were identical to those described for Fig. 5B. (B) Anaerobic growth of the ΔnirS mutant was compared with that of PAO1. Strains were grown in LBN or in LBN supplemented with 1 μM vitamin B12 inside the anaerobic chamber. Aliquots of cultures were withdrawn at designated times, and OD600 values (means ± SDs, n = 3) were plotted for growth curves.
DISCUSSION
P. aeruginosa, an obligate respirer, is capable of luxuriant growth by anaerobic respiration. Its genome harbors a series of enzymes involved in dissimilatory nitrate (NO3−) reduction, and several studies have demonstrated that fairly dense cell masses can readily be attained by anaerobic NO3− respiration (2, 13, 45, 51, 61, 62). In this study, we revealed for the first time that the anaerobic growth of P. aeruginosa is indeed accompanied by apparent defects in DNA replication. Binary fission of bacterial cells begins with chromosome replication, which should be completed before cytokinesis can take place for optimal cell division (35). Therefore, treatment that results in the inhibition of DNA replication can give rise to abnormal cell elongation in bacterial cells (5). Our initial hypothesis that DNA replication might be impaired in anaerobically growing P. aeruginosa was validated by the FACS analysis that compared the DNA/protein ratios of PAO1 cells grown under either condition. The DNA/protein ratio of PAO1 cells cultured by anaerobic NO3− respiration was less than half of the value obtained from aerobically grown PAO1. On the basis of findings by Odsbu and colleagues, the DNA/protein ratio was decreased by only 10% following treatment with 5 mM hydroxyurea (HU), which inhibits the activity of ribonucleotide reductase in E. coli (27). Thus, such a sharp decrease in the DNA/protein ratio indicates that DNA replication was substantially inhibited in anaerobically growing PAO1.
RNR plays an essential role in DNA biogenesis, and it is of particular interest that P. aeruginosa possesses three different classes of RNR that basically perform the same function (50). The presence of such redundancy accounts for the bacterial adaptability to survive under diverse environmental conditions (45a). The results presented in Fig. 3 clearly elucidated that (i) each class plays a distinct role under conditions of various oxygen concentrations and (ii) aerobic growth of PAO1 was strictly dependent on the presence of functional class I RNR. Fortunately, we managed to overcome the difficulty of growing a class I mutant aerobically by performing allelic exchange under strict anaerobic conditions. To our knowledge, the class I RNR mutant has not been previously constructed in any bacterial species, and this study has elucidated its growth-related phenotypes for the first time.
Vitamin B12, an essential cofactor for the class II RNR, is a tetrapyrrole-based aromatic macrocycle, and ∼30 enzymes are involved in its complicated biosynthesis process (36, 38). Due to the presence of genes encoding oxygen-dependent enzymes, such as cobG (PA2906) and cobN (PA2944), P. aeruginosa is considered to possess a vitamin B12 synthesis pathway that is dependent on the presence of molecular oxygen (19, 36). Consistent with this knowledge, we found that PAO1 produced very low levels of vitamin B12 during anaerobic growth (Fig. 4), likely rendering the class II RNR incompetent. Our growth curve experiments, results of which are shown in Fig. 3, appeared to suggest that class II and III RNRs contributed equally to the anaerobic growth of P. aeruginosa. Vitamin B12 add-back experiments, however, clearly demonstrated the dominant role of class II RNR in supporting the anaerobic growth of P. aeruginosa. When grown with the addition of 1 μM vitamin B12, a dramatic increase in anaerobic growth was apparently observed in strains with intact class II RNR, including wild-type PAO1. In addition, cell shape returned completely back to normal in the same set of strains that exhibited robust anaerobic growth. These results suggest that (i) inactivation of class II RNR due to the lack of sufficient production of vitamin B12 is responsible for the anaerobiosis-induced cell elongation and (ii) anaerobic growth is affected by such cell shape changes. These findings, therefore, also propose that LB supplemented with NO3−, which has been widely used as the “gold standard” to grow P. aeruginosa under anaerobic conditions, does not actually provide the best optimal conditions for such growth.
When cell elongation was suppressed in the presence of vitamin B12, biofilm formation was accordingly decreased to a significant extent, further supporting our conclusion that enhanced biofilm formation is a consequence of cell elongation incurred during anaerobic growth (57). Interestingly, these findings are similar to those of Gotoh and colleagues (17). When aerobically growing P. aeruginosa was treated with HU, a specific inhibitor of class I RNR, a high degree of cell elongation had occurred. Likewise, such cell shape changes also resulted in robust biofilm formation under aerobic conditions (17). Together with our results, these findings suggest that cell elongation invariably occurs by DNA replication inhibition, no matter whether it is due to anaerobiosis or to treatment with HU. Moreover, such changes in surface properties caused bacterial cells to form robust biofilms.
In our microscopic analyses, cell elongation became observable after ∼8 h of anaerobic growth (data not shown), supporting the idea that cell elongation may occur in response to an exposure to a molecule that accumulates over time during anaerobic respiration. Cell elongation and the growth-related phenotypes of the ΔnirS mutant provided a clue as to the involvement of NO in such a process. NO, a by-product of anaerobic respiration, has been reported to accumulate persistently during anaerobic NO3− respiration of P. aeruginosa (21, 61). Our results shown in Fig. 9 demonstrated that cells of the ΔnirS mutant, devoid of its ability to reduce nitrite (NO2−) to NO, were not elongated. In previous studies, we also found that cell elongation was suppressed in the presence of carboxy-PTIO (2-[4-carboxypheny]-4,4,5,5-tetramethylimidazoline-1-oxy-3-oxide), a stoichiometric NO scavenger (57). The ΔnirS mutant did not respond to vitamin B12 supplementation. Since this result implicates that vitamin B12 can boost the anaerobic growth rate only under conditions where endogenous NO is produced continuously, NO-mediated inactivation of RNR (especially class II RNR) accounts for the anaerobiosis-induced cell elongation of P. aeruginosa.
Mounting evidence has suggested that CF patients suffer from defective vitamin B12 absorption (7, 11, 25). Although vitamin B12 absorption can be corrected by administration of pancreatic supplements (7), such malabsorption provides a basis for the frequent occurrence of anemia among CF patients (54). It is not clear how much vitamin B12 is present in the CF patient mucous airway, which was reported to possess regions with reduced oxygen tension (56, 60). Our results demonstrate that the addition of vitamin B12 does not exert any noticeable effect on bacterial replication or protein synthesis profiles during aerobic growth (data not shown). Added vitamin B12, however, can significantly modulate the growth-related phenotypes and biofilm formation of P. aeruginosa under anaerobic conditions. In particular, biofilm formation during anaerobiosis was noticeably suppressed, suggesting that an adequate delivery of vitamin B12 may be useful in reducing the bacterial capability to form biofilm during anaerobic respiration, despite its positive effect on anaerobic growth. It will be important to further investigate the following questions. (i) Can vitamin B12 treatment also decrease the robustness of a preestablished anaerobic P. aeruginosa biofilm? (ii) Is there any difference in the relative antibiotic susceptibility between bacterial cells with contrasting cell shapes? (iii) Can we identify an optimal vitamin B12 concentration that can inhibit the biofilm formation of P. aeruginosa with only a marginal effect on its anaerobic growth? Answers to these questions will provide additional information with regard to the potential therapeutic applications of vitamin B12 for the treatment of an anaerobic biofilm infection of P. aeruginosa.
This study demonstrates an interesting phenotype that a major pathogen of patients with CF, P. aeruginosa, exhibits during anaerobic in vitro growth. Although the anaerobic nature of the airway mucus of patients with CF is well appreciated (32, 53, 56, 58, 60), it has to be stated that our results included the following limitations. (i) Our biofilm was formed on an abiotic surface for a short duration. This in vitro biofilm may not be biologically relevant to reflect P. aeruginosa biofilms in the chronically infected airway of CF patients. Moreover, in order to stimulate anaerobic growth of PAO1, a relatively large amount of NO3− was used. Although NO3− is presumed to be persistently provided, the largest amount of NO3− reported in the airway of a patient with CF was 700 μM (43). It remains yet to be addressed whether P. aeruginosa proliferating as a biofilm inside the airway mucus of a patient with CF encounters a similar level of anaerobiosis-induced stress. (ii) Multiple bacterial species are involved in the airway infection of patients with CF, rendering microbial lifestyle in the airway of patients with CF highly complicated (37, 53). Our results clearly showed that a prototype P. aeruginosa strain, PAO1, was unable to produce vitamin B12 during anaerobic respiration but responded dramatically to the exogenously supplied vitamin B12. It will be important to investigate whether other bacterial species can synthesize vitamin B12, which would help P. aeruginosa relieve anaerobic growth-associated stress.
In conclusion, we explored the molecular basis behind the anaerobiosis-induced cell elongation, and most importantly, we identified a molecule that can reverse such an abnormal morphological change, which can eventually influence the biofilm formation (Fig. 10). To establish effective treatment strategies for chronic P. aeruginosa infection of the airway of patients with CF, a molecular-level understanding of the anaerobiosis-induced modulation of bacterial virulence features is necessary. We anticipate that our current results will stimulate further investigations, with the ultimate goal of eradicating this clinically important opportunist from anaerobic mucous layers.
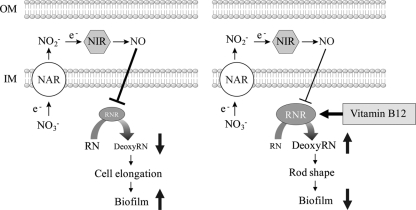
Fig 10.
Summary of anaerobiosis-induced cell elongation and role of vitamin B12 in the suppression of cell elongation. Nitrate reductase (NAR) reduces NO3−, an alternative electron acceptor to NO2−, which is further reduced to NO by periplasmic nitrite reductase (NIR). Endogenously accumulated NO intoxicates RNR, which in turn results in decreased synthesis of deoxyribonucleotides (downward-pointing arrow). Vitamin B12 restores RNR activity (especially class II type), and normal production of deoxyribonucleotides is achieved (upward-pointing arrow). Abbreviations: RN, ribonucleotide; DeoxyRN, deoxyribonucleotides; RNR, ribonucleotide reductase.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the South Korean Government (MEST), no. 2009-0087951 and no. 2011-0016210 (to S.S.Y.). This work was also supported by a grant from the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, A110096 (to S.S.Y.).
We thank Wasimul Bari for careful review of the manuscript and helpful discussion.
Footnotes
Published ahead of print 27 February 2012
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1. Arai H, Igarashi Y, Kodama T. 1994. Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 58:1286–1291 [DOI] [PubMed] [Google Scholar]
- 2. Arai H, Kodama T, Igarashi Y. 1998. The role of the nirQOP genes in energy conservation during anaerobic growth of Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 62:1995–1999 [DOI] [PubMed] [Google Scholar]
- 3. Blakley RL. 1966. Cobamides and ribonucleotide reduction. J. Biol. Chem. 241:176–179 [PubMed] [Google Scholar]
- 4. Boucher RC. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23:146–158 [DOI] [PubMed] [Google Scholar]
- 5. Burton P, Holland IB. 1983. Two pathways of division inhibition in UV-irradiated E. coli. Mol. Gen. Genet. 190:309–314 [DOI] [PubMed] [Google Scholar]
- 6. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 7. Deren JJ, Arora B, Toskes PP, Hansell J, Sibinga MS. 1973. Malabsorption of crystalline vitamin B 12 in cystic fibrosis. N. Engl. J. Med. 288:949–950 [DOI] [PubMed] [Google Scholar]
- 8. Donachie WD, Begg KJ. 1989. Cell length, nucleoid separation, and cell division of rod-shaped and spherical cells of Escherichia coli. J. Bacteriol. 171:4633–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donaldson SH, Boucher RC. 2003. Update on pathogenesis of cystic fibrosis lung disease. Curr. Opin. Pulm. Med. 9:486–491 [DOI] [PubMed] [Google Scholar]
- 10. Drapeau GR, Gariepy F, Boule M. 1984. Regulation and SOS induction of division inhibition in Escherichia coli K12. Mol. Gen. Genet. 193:453–458 [DOI] [PubMed] [Google Scholar]
- 11. Durie PR, Pencharz PB. 1989. A rational approach to the nutritional care of patients with cystic fibrosis. J. R. Soc. Med. 82(Suppl. 16):11–20 [PMC free article] [PubMed] [Google Scholar]
- 12. Elledge SJ, Davis RW. 1989. DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol. 9:4932–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Filiatrault MJ, Picardo KF, Ngai H, Passador L, Iglewski BH. 2006. Identification of Pseudomonas aeruginosa genes involved in virulence and anaerobic growth. Infect. Immun. 74:4237–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Filiatrault MJ, et al. 2005. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect. Immun. 73:3764–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filpula D, Fuchs JA. 1979. Increased synthesis of ribonucleotide reductase after deoxyribonucleic acid inhibition in various species of bacteria. J. Bacteriol. 139:694–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Formanek W, et al. 2002. Elevated nitrite in breath condensates of children with respiratory disease. Eur. Respir. J. 19:487–491 [DOI] [PubMed] [Google Scholar]
- 17. Gotoh H, et al. 2008. Pseudomonas aeruginosa, under DNA replication inhibition, tends to form biofilms via Arr. Res. Microbiol. 159:294–302 [DOI] [PubMed] [Google Scholar]
- 18. Haskin CJ, Ravi N, Lynch JB, Munck E, Que L., Jr 1995. Reaction of NO with the reduced R2 protein of ribonucleotide reductase from Escherichia coli. Biochemistry 34:11090–11098 [DOI] [PubMed] [Google Scholar]
- 19. Heldt D, et al. 2005. Aerobic synthesis of vitamin B12: ring contraction and cobalt chelation. Biochem. Soc. Trans. 33:815–819 [DOI] [PubMed] [Google Scholar]
- 20. Herrick J, Sclavi B. 2007. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 63:22–34 [DOI] [PubMed] [Google Scholar]
- 21. Hoffman LR, et al. 2010. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathog. 6:e1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horak F, Jr, et al. 2007. Longitudinal monitoring of pediatric cystic fibrosis lung disease using nitrite in exhaled breath condensate. Pediatr. Pulmonol. 42:1198–1206 [DOI] [PubMed] [Google Scholar]
- 23. Jordan A, et al. 1997. B12-dependent ribonucleotide reductases from deeply rooted eubacteria are structurally related to the aerobic enzyme from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 94:13487–13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jordan A, et al. 1999. Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181:3974–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindemans J, Neijens HJ, Kerrebijn KF, Abels J. 1984. Vitamin B12 absorption in cystic fibrosis. Acta Paediatr. Scand. 73:537–540 [DOI] [PubMed] [Google Scholar]
- 26. Linnane SJ, et al. 1998. Total sputum nitrate plus nitrite is raised during acute pulmonary infection in cystic fibrosis. Am. J. Respir. Crit. Care Med. 158:207–212 [DOI] [PubMed] [Google Scholar]
- 27. Odsbu I, Morigen Skarstad K. 2009. A reduction in ribonucleotide reductase activity slows down the chromosome replication fork but does not change its localization. PLoS One 4:e7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ojoo JC, Mulrennan SA, Kastelik JA, Morice AH, Redington AE. 2005. Exhaled breath condensate pH and exhaled nitric oxide in allergic asthma and in cystic fibrosis. Thorax 60:22–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'May CY, Reid DW, Kirov SM. 2006. Anaerobic culture conditions favor biofilm-like phenotypes in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. FEMS Immunol. Med. Microbiol. 48:373–380 [DOI] [PubMed] [Google Scholar]
- 30. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 31. Ozer I, Ozcetin M, Karaer H, Kurt SG, Sahin S. 2011. Retrospective approach to methylenetetrahydrofolate reductase mutations in children. Pediatr. Neurol. 45:34–38 [DOI] [PubMed] [Google Scholar]
- 32. Palmer KL, Brown SA, Whiteley M. 2007. Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 189:4449–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parsek MR, Tolker-Nielsen T. 2008. Pattern formation in Pseudomonas aeruginosa biofilms. Curr. Opin. Microbiol. 11:560–566 [DOI] [PubMed] [Google Scholar]
- 34. Philippe N, Alcaraz JP, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255 [DOI] [PubMed] [Google Scholar]
- 35. Prozorov AA. 2005. The bacterial cell cycle: DNA replication, nucleoid segregation, and cell division. Mikrobiologiia 74:437–451 (In Russian.) [PubMed] [Google Scholar]
- 36. Raux E, Schubert HL, Warren MJ. 2000. Biosynthesis of cobalamin (vitamin B12): a bacterial conundrum. Cell. Mol. Life Sci. 57:1880–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rogers GB, et al. 2004. Characterization of bacterial community diversity in cystic fibrosis lung infections by use of 16S ribosomal DNA terminal restriction fragment length polymorphism profiling. J. Clin. Microbiol. 42:5176–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roth JR, Lawrence JG, Rubenfield M, Kieffer-Higgins S, Church GM. 1993. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 175:3303–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy B, Lepoivre M, Henry Y, Fontecave M. 1995. Inhibition of ribonucleotide reductase by nitric oxide derived from thionitrites: reversible modifications of both subunits. Biochemistry 34:5411–5418 [DOI] [PubMed] [Google Scholar]
- 40. Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10:644–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sauer K, et al. 2004. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186:7312–7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schapiro JM, Libby SJ, Fang FC. 2003. Inhibition of bacterial DNA replication by zinc mobilization during nitrosative stress. Proc. Natl. Acad. Sci. U. S. A. 100:8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schobert M, Jahn D. 2010. Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:549–556 [DOI] [PubMed] [Google Scholar]
- 44. Schreiber K, et al. 2007. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 189:4310–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sias SR, Stouthamer AH, Ingraham JL. 1980. The assimilatory and dissimilatory nitrate reductases of Pseudomonas aeruginosa are encoded by different genes. J. Gen. Microbiol. 118:229–234 [DOI] [PubMed] [Google Scholar]
- 45a. Sjöberg BM, Torrents E. 2011. Shift in ribonucleotide reductase gene expression in Pseudomonas aerginosa during infection. Infect. Immun. 79:2663–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith A. 1997. Pathogenesis of bacterial bronchitis in cystic fibrosis. Pediatr. Infect. Dis. J. 16:91–95 [DOI] [PubMed] [Google Scholar]
- 47. Sriramulu DD, Lunsdorf H, Lam JS, Romling U. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667–676 [DOI] [PubMed] [Google Scholar]
- 48. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 49. Torheim NK, Boye E, Lobner-Olesen A, Stokke T, Skarstad K. 2000. The Escherichia coli SeqA protein destabilizes mutant DnaA204 protein. Mol. Microbiol. 37:629–638 [DOI] [PubMed] [Google Scholar]
- 50. Torrents E, Poplawski A, Sjoberg BM. 2005. Two proteins mediate class II ribonucleotide reductase activity in Pseudomonas aeruginosa: expression and transcriptional analysis of the aerobic enzymes. J. Biol. Chem. 280:16571–16578 [DOI] [PubMed] [Google Scholar]
- 51. Toyofuku M, et al. 2007. Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 189:4969–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Trunk K, et al. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ. Microbiol. 12:1719–1733 [DOI] [PubMed] [Google Scholar]
- 53. Tunney MM, et al. 2008. Detection of anaerobic bacteria in high numbers in sputum from patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 177:995–1001 [DOI] [PubMed] [Google Scholar]
- 54. von Drygalski A, Biller J. 2008. Anemia in cystic fibrosis: incidence, mechanisms, and association with pulmonary function and vitamin deficiency. Nutr. Clin. Pract. 23:557–563 [DOI] [PubMed] [Google Scholar]
- 55. Winteler HV, Haas D. 1996. The homologous regulators ANR of Pseudomonas aeruginosa and FNR of Escherichia coli have overlapping but distinct specificities for anaerobically inducible promoters. Microbiology 142(Pt 3):685–693 [DOI] [PubMed] [Google Scholar]
- 56. Worlitzsch D, et al. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoon MY, Lee KM, Park Y, Yoon SS. 2011. Contribution of cell elongation to the biofilm formation of Pseudomonas aeruginosa during anaerobic respiration. PLoS One 6:e16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yoon SS, et al. 2006. Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J. Clin. Invest. 116:436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yoon SS, Hassett DJ. 2004. Chronic Pseudomonas aeruginosa infection in cystic fibrosis airway disease: metabolic changes that unravel novel drug targets. Expert Rev. Anti Infect. Ther. 2:611–623 [DOI] [PubMed] [Google Scholar]
- 60. Yoon SS, et al. 2002. Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3:593–603 [DOI] [PubMed] [Google Scholar]
- 61. Yoon SS, et al. 2007. Two-pronged survival strategy for the major cystic fibrosis pathogen, Pseudomonas aeruginosa, lacking the capacity to degrade nitric oxide during anaerobic respiration. EMBO J. 26:3662–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zimmermann A, Reimmann C, Galimand M, Haas D. 1991. Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5:1483–1490 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.