Abstract
Outer membrane proteins (OMPs) serve as the permeability channels for nutrients, toxins, and antibiotics. In Escherichia coli, OmpA has been shown to be involved in bacterial virulence, and OmpC is related to multidrug resistance. However, it is unclear whether OmpC also has a role in the virulence of E. coli. The aims of this study were to characterize the role of OmpC in antimicrobial resistance and bacterial virulence in E. coli. The ompC deletion mutant showed significantly decreased susceptibility to carbapenems and cefepime. To investigate the survival of E. coli exposed to the innate immune system, a human blood bactericidal assay showed that the ompC mutant increased survival in blood and serum but not in complement-inactivated serum. These effects were also demonstrated in the natural selection of OmpC mutants. Also, C1q interacted with E. coli through a complex of antibodies bound to OmpC as a major target. Bacterial survival was increased in the wild-type strain in a dose-dependent manner by adding free recombinant OmpC protein or anti-C1q antibody to human serum. These results demonstrated that the interaction of OmpC-specific antibody and C1q was the key step in initiating the antibody-dependent classical pathway for the clearance of OmpC-expressing E. coli. Anti-OmpC antibody was detected in human sera, indicating that OmpC is an immunogen. These data indicate that the loss of OmpC in E. coli is resistant to not only antibiotics, but also the serum bactericidal effect, which is mediated from the C1q and anti-OmpC antibody-dependent classical pathway.
INTRODUCTION
Carbapenems are often used to treat serious infections caused by multidrug-resistant strains of Enterobacteriaceae. The spread of carbapenem resistance in Enterobacteriaceae may become a serious problem. In Escherichia coli, there are three major outer membrane proteins (OMPs), OmpA, OmpC, and OmpF, which function as passive diffusion channels for small molecules, like nutrients, toxic salts, and antibiotics. The expression of OmpC and OmpF is regulated by osmolarity (14, 42), and loss of OmpC or OmpF is related to antibiotic resistance, particularly when combined with production of extended-spectrum β-lactamases and AmpC (11, 24, 34). Previously, we have demonstrated that resistance to carbapenems in E. coli is related to the loss of OmpC expression (23). The loss of OmpC expression may be due to selection under antibiotic pressure.
OmpA in E. coli is well known as a virulence determinant for increased survival in macrophages, serum resistance, and invasion of brain microvascular endothelial cells (5, 18, 28, 36–38, 40). The deletion of ompC in E. coli isolated from Crohn's disease was shown to decrease adherence and the ability to invade intestinal cells (29, 30). OmpC is also characterized as a lactoferrin binding protein (13, 31). Recently, it has been shown that the loss of OmpK36 in Klebsiella pneumoniae leads to increased resistance to antibiotics and susceptibility to neutrophil phagocytosis (9). The aims of this study were to characterize the role of OmpC in antimicrobial resistance and bacterial virulence in E. coli. We show that the loss of OmpC not only increases resistance to carbapenems and 4th-generation cephalosporins, but also increases survival in human serum by escaping the OmpC-specific antibody-dependent classical complement activation pathway.
MATERIALS AND METHODS
Ethics statement.
The animals for antibody generation were raised and cared for according to the guidelines set up by the National Science Council, Taiwan. The protocol adhered to the rules of the Animal Protection Act of Taiwan and was approved by the Institutional Animal Care and Use Committee (IACUC) of National Cheng Kung University (IACUC approval no. 100048). All surgery was performed under chloral hydrate anesthesia, and all efforts were made to minimize suffering.
The clinical isolates of bacteria in this study were collected from patients as part of routing diagnostic screening by the Department of Pathology, National Cheng Kung University Hospital. The blood samples were obtained from healthy volunteers. This study was approved by the Institutional Biosafety Committee and the Institutional Ethics Committee of the College of Medicine, National Cheng Kung University and National Cheng Kung University Hospital, Tainan, Taiwan. Informed consent was obtained from healthy volunteers according to the relevant guidelines of the Ethics Committee (no. ER-98-143). Based on the retrospective strain collection, participants did not need to give consent according to the evaluation of the Ethics Committee (no. ER-99-330).
Bacterial strains.
E. coli 2837-2/05, producing the CMY-2 AmpC enzyme and TEM-1 narrow-spectrum β-lactamase, was isolated from an intra-abdominal drainage culture (23). This strain expressed only OmpA and OmpC, without OmpF, and was used for mutant construction and protein expression. E. coli JM110 and BL21 were used for the construction and expression of His6-tagged OmpC protein. E. coli was grown in Luria-Bertani (LB) broth with agitation at 37°C. When necessary, the antibiotics kanamycin (50 μg/ml) and chloramphenicol (20 μg/ml) were used for selection.
Construction of an ompC isogenic mutant and a complemented strain.
The ompC deletion mutant was constructed by allelic exchange mutagenesis (22). The oligonucleotide primers used for construction are listed in Table 1. The promoter region (800 bp) of ompC (accession no. EU372012) was amplified with primers com_ompC-2 (HindIII) and dpompC (SspI+StuI) and cloned into pUC18 to generate plasmid pMW618. The 800-bp fragment, including the 170-bp ompC coding region and the 630-bp downstream region of ompC, was amplified by primers dsompC (XbaI) and dsompC (SspI+StuI) and cloned into plasmid pMW618 to generate plasmid pMW619. For replacement of the ompC coding region, a chloramphenicol cassette (Cm) (807 bp) was cloned into the plasmid pMW619 digested with StuI, which generated plasmid pMW620. Finally, the 2.4-kb fragment, including the ompC promoter, Cm, and ompC downstream region, was amplified with primers com_ompC-2 (XbaI) and dsompC (XbaI) and subcloned into the temperature-sensitive plasmid pKO3 digested with XbaI to generate the ompC replacement plasmid pMW621. The plasmid pMW621 was introduced into E. coli 2837-2/05 by electroporation and counterselected with 5% sucrose. Finally, the ompC gene was replaced with Cm, and the strain was named SW307 and confirmed by Southern blotting. PCR also confirmed that the ompC gene of SW307 was removed from the chromosome (data not shown).
Table 1.
Primers used in the study
| Primer | Sequence (5′–3′)a |
|---|---|
| com_ompC-2 (HindIII) | CCCAAGCTTCAACAGTAAAAAGAAGCGGG |
| com_ompC-2 (XbaI) | GCTCTAGACAACAGTAAAAAGAAGCGGG |
| dpompC (SspI+StuI) | ACCTAATATTAGGCCTTTTTTATGCCACTGCATACTG |
| dsompC (XbaI) | GCTCTAGAATTGCTGATGCGACTGATTG |
| dsompC (SspI+StuI) | CCTAATATTAGGCCTGTGGCTACGACGACGAAGAT |
| com_ompC-1 (StuI) | AAGGCCTGCGGGTTGTGGTTTTTGATC |
| com_ompC-2 (StuI) | AAGGCCTCAACAGTAAAAAGAAGCGGG |
| rfOmpC-1 (BamHI) | CGGGATCCGATGAAAGTTAAAGTACTGTC |
| rfOmpC-2 (XhoI) | CCCGCTCGAGTTAGAACTGGTAAACCAGAC |
Introduced restriction enzyme sites are underlined.
The complementing plasmid pMW617 was constructed with wild-type ompC, including its promoter and the coding region, amplified with primers com_ompC-1 (StuI) and com_ompC-2 (StuI), in plasmid pACAC177 (New England BioLabs, Beverly, MA) and transformed into SW307 to generate a complemented strain, SW308. The expression of OmpC was demonstrated by Western blotting in 2837-2/05 and SW308, whereas SW307 failed to express the OmpC protein (data not shown).
Outer membrane protein analysis.
OMPs were prepared as described previously and separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) with 4 M urea (23). The gels were stained with 0.1% Coomassie blue.
Antimicrobial susceptibility testing.
The susceptibilities of various antimicrobial agents were determined by using Etest strips (AB Biodisk, Solna, Sweden) with interpretation according to the Clinical and Laboratory Standards Institute guidelines (10).
Expression and purification of the recombinant OmpC protein and generation of polyclonal antibody.
The full-length ompC gene (1,095 bp) was generated from the genomic DNA of wild-type 2837-2/05 with primers rfOmpC-1 (BamHI) and rfOmpC-2 (XhoI) (the sequences are shown in Table 1) and cloned into plasmid pET30b (Invitrogen, Cergy-Pontoise, France). The expression of the His6-tagged OmpC protein in E. coli BL21 was under induction with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16°C for 16 h. The purification of His6-tagged OmpC protein was done following the instructions of GE Amersham for Ni2+ affinity chromatography, with buffer containing 8 M urea to avoid protein aggregation. The recombinant OmpC protein (rOmpC) was dialyzed with phosphate-buffered saline (PBS) containing 6 M urea and concentrated by using an ultrafiltration cell (Amicon Corp., Lexington, MA) with a 10-kDa membrane. BALB/c mice were used for generation of OmpC polyclonal antibody by standard protocols. The protein rOmpC was directly recovered from SDS-10% PAGE gels and injected into mice subcutaneously, followed by 2 boosters every 7 days. Immune sera were collected 7 days after the second boost, and the titer of anti-OmpC was measured by enzyme-linked immunosorbent assay (ELISA) to rOmpC.
Bactericidal assay.
Blood from healthy volunteers was collected fresh with heparin as an anticoagulant, and normal human serum was stored at −80°C. Heat-inactivated serum was made by treatment at 56°C for 30 min to inactivate the complement activity. The human serum, including heat-inactivated serum, was diluted to 60% with sterile PBS. A bactericidal assay was performed with whole blood, serum, and heat-inactivated serum. Briefly, mid- to late-logarithmic-phase E. coli (1.2 × 106 CFU) was incubated with whole blood, 60% serum, or 60% heat-inactivated serum at 37°C for 1 h. The survival rate was determined by a plate count and calculated as the ratio of surviving CFU after treatment to the initial number of CFU.
For the competition assay, the 60% serum was preincubated with different concentrations of rOmpC or goat anti-human C1q polyclonal antibody (Calbiochem, La Jolla, CA) at 37°C for 30 min, followed by the bactericidal assay. The rOmpC was added to final concentrations of 6.1, 30.5, 152.7, and 458 μg/ml. The goat anti-human C1q polyclonal antibody was diluted with PBS 2-, 5-, or 50-fold and added at 1/15 of the total volume.
Natural selection of E. coli with ompC mutated.
E. coli 2837-2/05 was cultured in LB broth overnight and subcultured 1:50 into fresh LB broth. E. coli (mid- to late logarithmic phase; 107 CFU) was plated onto a Mueller-Hinton agar plate containing imipenem (3.2 μg/ml) and cultured at 37°C overnight. The colonies were maintained in Mueller-Hinton agar plates containing imipenem (3.2 μg/ml). The selected colonies were further checked by antimicrobial susceptibility testing, OMP profiles, ompC genotype (23), and bactericidal assay.
Inhibition of the classical pathway by human IgG cleavage.
The IdeS protein of Streptococcus pyogenes was chosen as a potential inhibitor of the classical pathway, as it cleaves human IgG specifically (1, 16, 17). After treatment with IdeS, human IgG is digested into one F(ab′)2 fragment and two Fc fragments of the heavy chain, and initiation of the classical pathway is blocked. Briefly, the serum was pretreated with IdeS (2 μg/20 μl 60% serum) at 37°C for 2 h to partially cleave the IgG, and the treated serum was used for the bactericidal assay.
Inhibition of the alternative pathway.
The alternative pathway has been shown to be inhibited by the absorption of properdin with bentonite (15, 35). Bentonite (10 mg) was washed with sterile PBS three times and incubated with serum (1 ml) at 37°C for 10 min. The serum treated with bentonite was separated by centrifugation and filter sterilized.
Ligand overlay assay.
The ligand overlay assay was used to assess the interaction between the molecules in human serum and E. coli. Based on the method of Alberti et al. (2), with modifications, the binding of C1q to any E. coli cell protein was detected by incubation with serum as a source of C1q and then detection with anti-C1q antibody. Briefly, the overnight culture of E. coli was washed with PBS twice and resuspended with PBS to the same volume. One milliliter of E. coli suspension was centrifuged at 6,000 rpm for 5 min and then concentrated in 100 μl of PBS and stored at −20°C. The frozen suspension was thawed at 4°C as the E. coli lysate, and the protein concentration of the lysate was determined with a Bio-Rad (Hercules, CA) protein assay kit. Each sample (3.5 μg/10 μl total protein) was boiled for 10 min in 10 μl 4× protein sample buffer (0.25 M Tris-HCl [pH 6.8], 8% SDS, 0.02% bromophenol blue, 30% glycerol, and 200 mM dithiothreitol [DTT]), separated by SDS-10% PAGE plus 4 M urea to enhance the separation of OMPs (23), and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking with 1% bovine serum albumin at 4°C overnight, the membrane was incubated with human serum (100-fold dilution with PBS) at 37°C for 10 min, goat anti-human C1q polyclonal antibody (1,000-fold dilution with PBS) at 37°C for 1 h, and horseradish peroxidase (HRP)-conjugated donkey anti-goat antibody (10,000-fold dilution with PBS; Santa Cruz, CA) as a secondary antibody at room temperature for 1 h. Finally, the membrane was visualized with Super Signal chemiluminescent substrate (Thermo Fisher Scientific Inc., IL) and imaged with a LAS3000 imaging system (Fuji Photo Film, Tokyo, Japan).
To test the antibody dependence of C1q binding to E. coli lysate, the ligand overlay was slightly modified by using IgG-depleted human serum. For IgG-depleted serum treatment, human serum diluted 100-fold was passed through HiTrap Protein A HP (GE Healthcare, Uppsala, Sweden) twice. The treated serum from the HiTrap Protein A HP was checked with a 12% SDS-PAGE gel to confirm the loss of IgG patterns.
Binding of C1q to E. coli.
The binding of C1q protein was analyzed by the method of Wooster et al. with modifications (40). Briefly, E. coli (1 × 107 CFU) was incubated with 5 μg purified human C1q protein (1.0 mg/ml; Complement Technology, Inc., TX) for 0.5 or 1 h at 37°C. After gentle incubation, the sample was washed three times with PBS by pelleting at 12,000 rpm for 5 min. Then, the pellet was resuspended in 15 μl PBS and boiled for 10 min in 15 μl 4× protein sample buffer before analysis. The 10-μl sample mixture was separated by 12% SDS-PAGE, and the C1q signal was detected by Western blot analysis.
Serological analysis of OmpC antibody.
The titers of human anti-OmpC IgG and IgM antibodies were measured by an ELISA using rOmpC-coated plates and detected with goat anti-human IgG and goat anti-human IgM, respectively, as secondary antibodies (Chemicon International, Temecula, CA). The specificity of human anti-OmpC antibody was also analyzed by Western blot analysis, with incubation of diluted human serum on membranes with bound E. coli lysate, and detected with goat anti-human IgG.
Statistical analysis.
Student's t test was used to determinant the difference in bactericidal killing of E. coli strains. Each experiment was repeated independently at least 3 times. A P value of less than 0.05 was considered statistically significant.
RESULTS
Bacterial growth rate and OMP profiles.
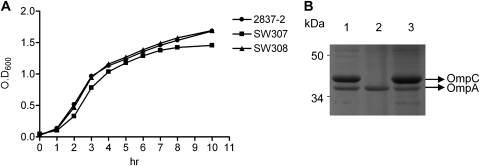
The growth rates (Fig. 1A) of the wild-type (2837-2/05), ompC mutant (SW307), and complemented (SW308) strains showed similar patterns (Fig. 1A). The OMP profile of SW307 showed the expression of OmpA, but not OmpC, whereas the wild-type strain 2837-2/05 and strain SW308 expressed OmpA and OmpC (Fig. 1B). All strains lacked OmpF expression.
Fig 1.
Bacterial growth curves and OMP profiles. (A) 2837-2/05, SW307, and SW308 were cultured in LB broth with agitation at 37°C, and bacterial growth rates were determined by the optical density at 600 nm (OD600). (B) OMP profiles of 2837-2/05, SW307, and SW308 with SDS-PAGE analysis. E. coli OMPs (10 μg/lane) were separated using SDS-10% PAGE plus 4 M urea. Lane 1, 2837-2/05; lane 2, SW307; lane 3, SW308.
Effect of OmpC on antimicrobial susceptibility.
Nine antimicrobial agents were used to determine the effect of OmpC on antimicrobial susceptibility. The results showed that 2837-2/05 was a multidrug-resistant E. coli strain susceptible only to imipenem, meropenem, and cefipime (Table 2). However, the ompC mutant, SW307, showed ≥32 μg/ml MICs of imipenem, meropenem, ertapenem, and cefipime. SW308 showed MIC results similar to those of 2837-2/05. No effects on the susceptibilities to ceftazidime, cefotaxime, aztreonam, gentamicin, and ciprofloxacin were found among 2837-2/05, mutant (SW307), and complementary (SW308) strains.
Table 2.
Antimicrobial susceptibility tests of strains 2837-2/05, SW307, and SW308
| Strain | MIC (μg/ml)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| GEM | CAZ | CTX | FEP | ATM | CIP | IPM | MEM | ETP | |
| 2837-2/05 | 256 | >256 | 256 | 3 | 128 | >256 | 0.38 | 0.25 | 1.5 |
| SW307 | 256 | >256 | >256 | 32 | 256 | >256 | >32 | 32 | >32 |
| SW308 | 256 | >256 | 128 | 2 | 128 | >256 | 0.38 | 0.19 | 0.5 |
GEM, gentamicin; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; CIP, ciprofloxacin; IPM, imipenem; MEM, meropenem; ETP, ertapenem.
Role of OmpC in human bactericidal activity.
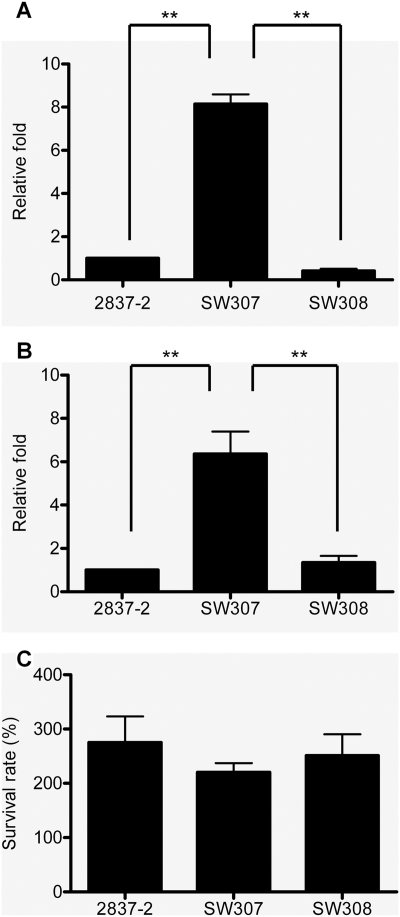
To demonstrate the role of OmpC in human bactericidal activity, E. coli was incubated with human whole blood, 60% serum, or complement-inactivated serum at 37°C for 1 h, followed by plate counts. In Fig. 2, the survival rates of 2837-2/05 and SW307 were significantly different in whole blood and serum. When E. coli was incubated with whole blood or serum, SW307 was 5- to 9-fold more resistant than 2837-2/05 (Fig. 2A and B), whereas no significant difference was found between 2837-2/05 and SW307 after incubation in heat-inactivated serum (Fig. 2C). Strain SW308 also showed the same scenario as the wild-type strain. The results of the bactericidal assay were similar when E. coli was incubated in whole blood or serum for 30 min (data not shown). No difference in ability to adhere to human polymorphonuclear leukocytes (PMNs) was observed between the wild type and SW307 (data not shown). These results suggested that OmpC is involved in complement-mediated serum killing of E. coli. Another clinical isolate, E. coli 2837-1/05, lacking expression of OmpC and resistant to carbapenems (23), also resisted bactericidal activity (data not shown). In addition, an ompC mutant was generated from a clinical isolate derived from blood and also showed OmpC-dependent bactericidal activity (data not shown). This indicated that the lack of OmpC leading to antibiotic and serum resistance was not strain dependent.
Fig 2.
Bactericidal activity of human blood and serum on E. coli. E. coli was incubated with human whole blood (A), 60% serum (B), or complement-inactivated serum (heated to 56°C) (C) at 37°C for 1 h. The viable bacteria were determined by plate count after serial dilution. (A and B) Relative fold survival is shown by normalization of the survival rates of SW307 and SW308 to that of 2837-2/05 for each independent experiment. The results are aggregates of more than three independent experiments, and the error bars represent the standard errors of the mean. **, P < 0.05.
Effects of naturally selective OmpC mutants on antimicrobial susceptibility and bactericidal activity.
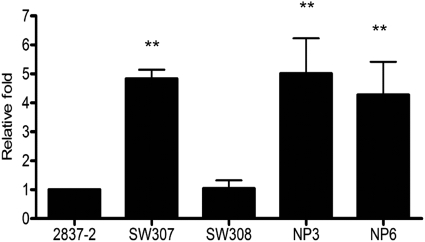
To further support the biological relevance of the loss of OmpC to antibiotic resistance and serum survival, an in vitro experiment was used to select E. coli isolates with mutated OmpC from imipenem-containing medium. Strains NP3 and NP6 were selected, and both strains showed reduced susceptibility to carbapenems and cefipime, like SW307 (Table 3). The sequence results also showed a truncation in the ompC coding region. In addition, strains NP3 and NP6 had bactericidal activities similar to that of SW307 (Fig. 3). These results demonstrated that ompC mutants can be selected under antibiotic pressure and showed both antibiotic and serum resistance.
Table 3.
Antimicrobial susceptibility tests and ompC genotypes of strains 2837-2/05, SW307, NP3, and NP6
| Strain | Disk zone size (mm)a |
ompC genotype | |||
|---|---|---|---|---|---|
| IPM | MEM | ETP | FEP | ||
| 2837-2/05 | 22 | 25 | 20 | 23 | Full length (1,095 bp) |
| SW307 | 6 | 6 | 6 | 15 | Deletion mutant |
| NP3 | 7 | 6 | 6 | 14 | Truncated form (G to A at position 371) |
| NP6 | 6 | 6 | 6 | 14 | Truncated form (frameshift at position 516) |
FEP, cefepime; IPM, imipenem; MEM, meropenem; ETP, ertapenem.
Fig 3.
Bactericidal activity of human serum on ompC-selected mutants. E. coli was incubated with 60% serum at 37°C for 1 h. The viable bacteria were determined by plate count after serial dilution. Relative fold survival is shown by normalization of the survival rates of SW307, SW308, NP3, and NP6 to that of 2837-2/05 for each independent experiment. The results are aggregates of more than three independent experiments, and the error bars represent the standard errors of the mean. **, P < 0.05, comparing 2837-2/05 and SW308.
OmpC-mediated antibody-dependent classical pathway.
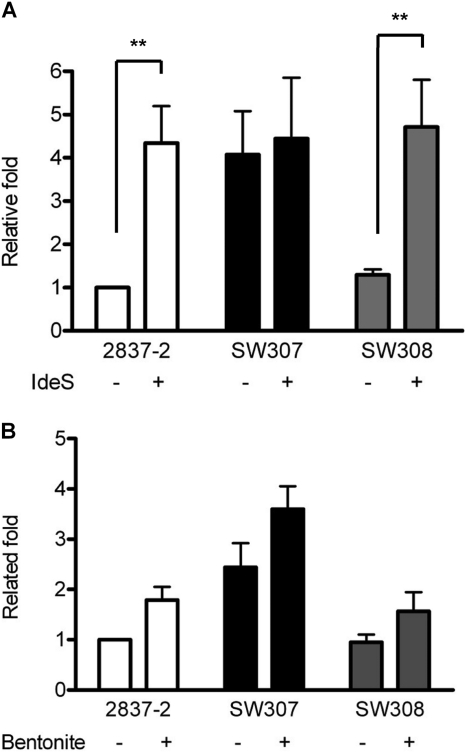
To investigate which complement pathway was involved in the OmpC-mediated bactericidal effect, the classical pathway was analyzed with the IgG cleavage protein, IdeS, while the role of the alternative pathway was further identified with the inhibitor bentonite. When serum was pretreated with IdeS, the survival of E. coli 2837-2/05 and SW308 was 4-fold increased, which was similar to the level of SW307. Only SW307 showed similar survival with or without IgG cleavage (Fig. 4A). This effect was also shown in 2837-1/05 (data not shown). These data suggested that OmpC is involved in the antibody-dependent bactericidal activity.
Fig 4.
Bactericidal activity of IgG-depleted (A) and bentonite-treated (B) human serum on E. coli. (A) Human serum was pretreated with IdeS (2 μg/20 μl 60% serum) at 37°C for 2 h to cleave IgG. The normal or treated 60% human serum was incubated with E. coli at 37°C for 1 h. (B) Bentonite was used to pretreat human serum at 37°C for 30 min to remove properdin and page the activation of the alternative pathway. The normal or treated 60% human serum was incubated with E. coli at 37°C for 1 h. The viable bacteria were determined by plate count after serial dilution, and the results are shown as relative fold survival. The results are from more than three independent experiments, and the error bars represent the standard errors of the mean. **, P < 0.05.
To identify whether the alternative pathway was also involved in OmpC-mediated serum bactericidal ability, bentonite was used to treat human serum. With the inhibition by bentonite of human serum, the viability of strains 2837-2/05, SW307, and SW308 increased 1.5-fold (Fig. 4B). Although the survival of E. coli was increased by the inhibition of the alternative pathway, there was no significant difference regardless of the strain's OmpC expression. These data suggested that the antibody-dependent classical pathway is the major mechanism in the OmpC-mediated bactericidal effect but that other molecular targets of E. coli maybe involved in the alternative pathway.
C1q binds to E. coli OmpC in an antibody-dependent manner.
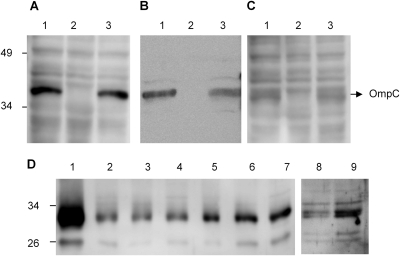
The result of IdeS pretreatment demonstrated the importance of the antibody-dependent classical pathway. To determine whether OmpC was involved in the activation of the classical pathway, a ligand overlay assay was used to determine the binding of C1q and anti-OmpC antibody in human serum to E. coli cell lysate. The results showed that there was a visible C1q signal at about 40 kDa in cell lysates from 2837-2/05 and SW308, but not from SW307 (Fig. 5A, lanes 1 and 3). The same size band in E. coli cell lysate was also detected by the anti-OmpC antibody, but not from the SW307 mutant strain (Fig. 5B, lanes 1 and 3). These results suggested that OmpC was the major antigen interacting with antibody and then binding C1q from human serum.
Fig 5.
Binding of C1q in human serum and C1q purified protein to E. coli cells. The ligand overlay assay was used to detect the binding of C1q in human serum on membranes with E. coli total lysate. Panels A, C, and D were developed with anti-C1q, and panel B was developed with anti-OmpC. (A) Normal serum diluted 100-fold and incubated with membranes containing E. coli proteins to detect the binding pattern of E. coli cell lysates. (B) The OmpC protein was detected by Western blotting with mouse anti-OmpC antibody on E. coli cell lysates. (C) Serum diluted 100-fold and interacted with HiTrap Protein A HP to deplete IgG was used for the ligand overlay assay. (A to C) Lane 1, 2837-2/05; lane 2, SW307; lane 3, SW308. (D) Purified C1q protein (5 μg) was directly incubated with bacteria at 37°C for 0.5 h (lanes 2 to 4 and 8) and 1 h (lanes 5 to 7 and 9). The binding ability of C1q protein was determined by Western blotting of the cell lysate. K. pneumoniae was used as the positive control for C1q binding ability. Lane 1, 100 ng C1q purified protein; lanes 2 and 5, 2837-2/05; lanes 3 and 6, SW307; lanes 4 and 7, SW308; lanes 8 and 9, K. pneumoniae.
To determine whether the interaction of C1q and OmpC was mediated by anti-OmpC antibody, a ligand overlay assay was performed with IgG depletion. The results showed a decreased interaction of C1q and OmpC in the ligand overlay assay after protein A pretreatment compared to normal serum (Fig. 5A and C).
In addition to the antibody-dependent classical pathway, the binding level of purified human C1q to E. coli directly was also investigated to assess the antibody-independent classical pathway. The results showed that the accumulation of C1q on E. coli increased from 0.5 to 1 h. However, the C1q binding level was no different among 2837-2/05, SW307, and SW308 (Fig. 5D), suggesting that OmpC is not a major binding site for C1q direct binding on E. coli. Moreover, the data also suggested that bactericidal activity mediated by IgG binding to OmpC could increase classical pathway activation, while direct binding of C1q to other molecules of bacteria may mediate a lower level of activation.
Role of the human anti-OmpC-specific antibody-mediated classical pathway in bactericidal activity.
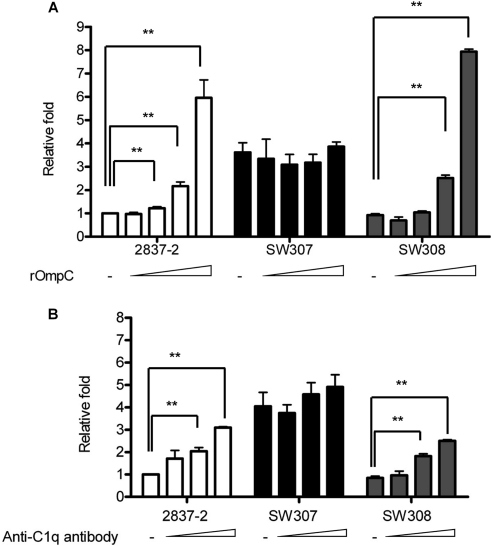
To determine whether human anti-OmpC antibody activated the classical pathway to kill OmpC-expressing E. coli, a competition assay with free rOmpC was used to neutralize the human anti-OmpC antibody in human serum. As shown in Fig. 6A, the survival of 2837-2/05 and SW308 after 1 h of incubation with pretreated serum was significantly increased in a dose-dependent manner. When these strains were incubated in serum pretreated with a low dose of rOmpC (6 μg/ml), survival was nearly the same as in serum without pretreatment. There was no effect on the survival of SW307 under any dose of rOmpC.
Fig 6.
Bactericidal ability of human serum with the competition of rOmpC or anti-C1q antibody. Human serum (60%) was preincubated at 37°C for 0.5 h with rOmpC in final concentrations of 6.1, 30.5, 152.7, and 458 μg/ml (A) or with goat-anti human C1q polyclonal antibody diluted with PBS 2-, 5-, or 50-fold (B) used as 1/15 of the total volume to neutralize anti-OmpC antibody or C1q in serum. The treated 60% human serum was further incubated with E. coli at 37°C for 1 h. The viable bacteria were determined by plate count after serial dilution, and the results are shown as relative fold survival. The results are from more than three independent experiments, and the error bars represent the standard errors of the mean. **, P < 0.05.
To test this hypothesis another way, the effect of neutralization of C1q by anti-C1q antibody was used to determine the role of C1q in activating the classical pathway to clear OmpC-expressing E. coli. After 1 h of incubation of E. coli in serum pretreated with anti-C1q antibody, survival was increased significantly in a dose-dependent manner in 2837-2 and SW308 (Fig. 6B), whereas no significant difference was detected in SW307. Based on the results in Fig. 6, it was demonstrated that the clearance of OmpC-expressing E. coli was mediated by the anti-OmpC antibody and the C1q-dependent classical pathway and that the ompC mutant survived by escaping this mechanism.
OmpC is a major immunogen.
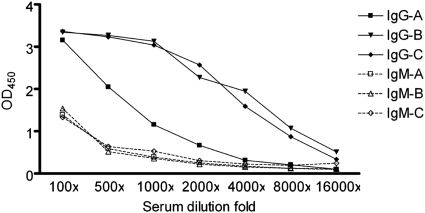
To determine whether human anti-OmpC antibody was present in serum, human antibody titers of IgG and IgM were determined by the ELISA method (Fig. 7). Human anti-OmpC antibody was detected, and the titer of IgG was higher than that of IgM among all tested sera. The titer of anti-OmpC IgG varied among different individuals, whereas anti-OmpC IgM was present in similar low titers among all tested sera. A Western blot also showed that IgG in human serum can target the OmpC in E. coli 2837-2/05 and SW308 (data not shown). This result suggested that OmpC in E. coli was a major immunogen generating the bactericidal antibody for the clearance of OmpC-positive E. coli.
Fig 7.
Identification of E. coli OmpC as an immunogen. Sera from 3 healthy volunteers (A, B, and C) were diluted 100-, 500-, 1,000-, 2,000-, 4,000-, 8,000-, and 16,000-fold, and the titers of anti-OmpC IgG and IgM were determined by ELISA as the OD450. The solid symbols are titers of IgG; the open symbols are titers of IgM. Squares, donor A; triangles, donor B; circles, donor C.
DISCUSSION
Outer membrane proteins in Gram-negative bacteria are known as the channels for nutrients crossing the bacterial outer membrane and have decreased expression in antibiotic resistance (6, 7, 11, 24, 34). In this study, we show that the loss of OmpC reduced susceptibility to carbapenems and 4th-generation cephalosporins in E. coli. Moreover, we demonstrated that OmpC of E. coli is a major immunogen inducing OmpC-specific antibody in human blood. The induced OmpC-specific antibody interacted with C1q to initiate the antibody-dependent classical pathway for the clearance of E. coli. To our knowledge, this is the first evidence that OmpC of E. coli is the target for the activation of the classical pathway, while loss of OmpC expression can allow E. coli to resist the bactericidal activity of human serum and also increase antibiotic resistance.
Several lines of evidence have shown that the outer membrane protein OmpA of E. coli can protect bacteria from bactericidal effects (28, 37, 38, 40). The target of OmpA is a C4-binding protein, avoiding complement activation and escaping immune clearance (but not in the mutant). However, the effect of OmpC we show here is different. The ompC mutant had a higher survival rate in whole blood and serum by escaping the anti-OmpC antibody and the C1q-dependent classical pathway. The recent study also demonstrated that OmpC can be recognized by signaling lymphocyte activation molecule (SLAM), being the microbial sensor to regulate the phagosome in macrophages for bacterial killing (8). It suggests that the loss of OmpC can promote survival of E. coli, including escaping the complement pathway and macrophage killing. Since the structures of OmpC and OmpF are similar, a cross-reaction of anti-OmpF antibody to OmpC may occur. However, based on Fig. 6 and strain character (without OmpF), this effect should be minor. As shown in Fig. 6A, the high concentration of rOmpC (458 μg/ml) not only neutralized anti-OmpC antibody in serum, but also bound to some OmpC target proteins, such as lactoferrin (13, 31), complement molecules, or unknown factors. This leads to a higher survival rate in the wild-type strain. In contrast, mutant (SW307) does not have OmpC to interact with the target proteins. Therefore, no dose-dependent effect was found.
An antibody-independent classical pathway through C1q has been demonstrated only by a 37-kDa porin of Salmonella enterica serovar Minnesota (20, 21), OmpK36 of K. pneumoniae (2–4), and a 40-kDa porin of Aeromonas salmonicida (26). However, our results showed that direct C1q binding to E. coli was similar among all strains tested (Fig. 5D), while OmpC was recognized by the anti-OmpC antibody and C1q to activate the classical pathway (Fig. 5 and 6). These data suggest that the activation of the antibody-dependent classical pathway via anti-OmpC IgG to OmpC in E. coli is more efficient due to direct binding of C1q. Moreover, we cannot exclude the possibility that other mechanisms are also involved in this event, such as K antigen (12), since there was a similar change in survival among all strains in the bentonite inhibition assay (Fig. 4B).
As E. coli bacteria are exposed to humans, they can induce many kinds of anti-E. coli antibody. A high level of antibody against OmpC has been demonstrated in 37 to 55% of Crohn's disease patients and their families (25, 27, 30). In Salmonella enterica serovar Typhi, OmpC is implicated in triggering strong, long-term IgG production in the antibody-mediated memory bactericidal response (32, 33). In addition, OmpK36 in K. pneumoniae is the immunogen for DNA vaccine development (19). Both porins share high similarity to E. coli OmpC protein. Our serological analysis was consistent with Salmonella and K. pneumoniae in that OmpC is the immunogen inducing an effective immune response for bactericidal ability in humans.
In Neisseria meningitidis serogroup B, a critical density of bactericidal antibodies and C1q needs to be reached in order to induce complement-mediated killing (39). Like the OmpC-mediated bactericidal model shown in this study, this may be a useful strategy to defend against low-virulence E. coli in healthy people. Unlike OmpA, the variation in OmpC may be much higher than we thought. Our previous data showed a higher mutation rate of the ompC gene (truncated forms or insertion) in multidrug-resistant E. coli (23, 41). We also found that the OmpC mutant can be detected from natural selection, and these mutants not only had similar carbapenems and cefepime resistance, but also had similar serum bactericidal activities (Fig. 3 and Table 3). This suggests that E. coli can adapt to different environments, such as antibiotic pressure or infectious tropism, by losing the expression of OmpC. The emergency occurs when multidrug resistance in E. coli is combined with the loss of OmpC expression. This may lead to E. coli not only surviving in antibiotic, but also escaping the immune response. However, it is still unknown how environmental pressures select for E. coli to survive by losing the expression of OmpC.
In conclusion, this study demonstrated that the loss of OmpC in E. coli can promote not only antimicrobial resistance, but also serum resistance. It suggests that E. coli OmpC has dual functions in pathogenesis when it is lost. When infecting recurrently, E. coli may benefit from a “side effect” with loss of OmpC expression to resist the immune response. Thus, treatment may be more challenging for patients under antibiotic pressure and without effective serum killing.
ACKNOWLEDGMENTS
We thank Robert Jonas for helpful comments on the manuscript.
This work was supported in part by the National Science Council, Taiwan (grant NSC99-3112-B-006-015); the Multidisciplinary Center of Excellence for Clinical Trial and Research, Department of Health, Executive Yuan, Taiwan (DOH100-TD-B-111-002); and National Cheng-Kung University Hospital, Taiwan (DOH101-TD-B-111-0022).
Footnotes
Published ahead of print 21 February 2012
REFERENCES
- 1. Agniswamy J, Lei B, Musser JM, Sun PD. 2004. Insight of host immune evasion mediated by two variants of group A Streptococcus Mac protein. J. Biol. Chem. 279:52789–52796 [DOI] [PubMed] [Google Scholar]
- 2. Alberti S, et al. 1993. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect. Immun. 61:852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alberti S, et al. 1996. Interaction between complement subcomponent C1q and the Klebsiella pneumoniae porin OmpK36. Infect. Immun. 64:4719–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alberti S, et al. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional model, and complement binding. Infect. Immun. 63:903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Badger JL, Kim KS. 1998. Environmental growth conditions influence the ability of Escherichia coli K1 to invade brain microvascular endothelial cells and confer serum resistance. Infect. Immun. 66:5692–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bannwarth M, Schulz GE. 2003. The expression of outer membrane proteins for crystallization. Biochim. Biophys. Acta 1610:37–45 [DOI] [PubMed] [Google Scholar]
- 7. Basle A, Rummel G, Storici P, Rosenbusch JP, Schirmer T. 2006. Crystal structure of osmoporin OmpC from E. coli at 2.0 A. J. Mol. Biol. 362:933–942 [DOI] [PubMed] [Google Scholar]
- 8. Berger SB, et al. 2010. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat. Immunol. 11:920–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen JH, et al. 2010. Contribution of outer membrane protein K36 to antimicrobial resistance and virulence in Klebsiella pneumoniae. J. Antimicrob. Chemother. 65:986–990 [DOI] [PubMed] [Google Scholar]
- 10. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 11. De E, et al. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189–198 [DOI] [PubMed] [Google Scholar]
- 12. Devine DA, Roberts AP. 1994. K1, K5 and O antigens of Escherichia coli in relation to serum killing via the classical and alternative complement pathways. J. Med. Microbiol. 41:139–144 [DOI] [PubMed] [Google Scholar]
- 13. Erdei J, Forsgren A, Naidu AS. 1994. Lactoferrin binds to porins OmpF and OmpC in Escherichia coli. Infect. Immun. 62:1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forst S, Delgado J, Ramakrishnan G, Inouye M. 1988. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J. Bacteriol. 170:5080–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joens LA, Nuessen ME. 1986. Bactericidal effect of normal swine sera on Treponema hyodysenteriae. Infect. Immun. 51:282–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johansson BP, Shannon O, Bjorck L. 2008. IdeS: a bacterial proteolytic enzyme with therapeutic potential. PLoS One 3:e1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kahn F, et al. 2008. Antibodies against a surface protein of Streptococcus pyogenes promote a pathological inflammatory response. PLoS Pathog. 4:e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim KS. 2002. Strategy of Escherichia coli for crossing the blood-brain barrier. J. Infect. Dis. 186(Suppl. 2):S220–S224 [DOI] [PubMed] [Google Scholar]
- 19. Kurupati P, Teh BK, Kumarasinghe G, Poh CL. 2006. Identification of vaccine candidate antigens of an ESBL producing Klebsiella pneumoniae clinical strain by immunoproteome analysis. Proteomics 6:836–844 [DOI] [PubMed] [Google Scholar]
- 20. Latsch M, Mollerfeld J, Ringsdorf H, Loos M. 1990. Studies on the interaction of C1q, a subcomponent of the first component of complement, with porins from Salmonella minnesota incorporated into artificial membranes. FEBS Lett. 276:201–204 [DOI] [PubMed] [Google Scholar]
- 21. Latsch M, Stemmer F, Loos M. 1992. Purification and characterization of LPS-free porins isolated from Salmonella minnesota. FEMS Microbiol. Lett. 69:275–281 [DOI] [PubMed] [Google Scholar]
- 22. Link AJ, Phillips D, Church GM. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu YF, Yan JJ, Ko WC, Tsai SH, Wu JJ. 2008. Characterization of carbapenem-non-susceptible Escherichia coli isolates from a university hospital in Taiwan. J. Antimicrob. Chemother. 61:1020–1023 [DOI] [PubMed] [Google Scholar]
- 24. Martinez-Martinez L, et al. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal beta-lactamase and showing altered porin profiles. Antimicrob. Agents Chemother. 44:2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mei L, et al. 2006. Familial expression of anti-Escherichia coli outer membrane porin C in relatives of patients with Crohn's disease. Gastroenterology 130:1078–1085 [DOI] [PubMed] [Google Scholar]
- 26. Merino S, Vilches S, Canals R, Ramirez S, Tomas JM. 2005. A C1q-binding 40 kDa porin from Aeromonas salmonicida: cloning, sequencing, role in serum susceptibility and fish immunoprotection. Microb. Pathog. 38:227–237 [DOI] [PubMed] [Google Scholar]
- 27. Mow WS, et al. 2004. Association of antibody responses to microbial antigens and complications of small bowel Crohn's disease. Gastroenterology 126:414–424 [DOI] [PubMed] [Google Scholar]
- 28. Prasadarao NV, Blom AM, Villoutreix BO, Linsangan LC. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169:6352–6360 [DOI] [PubMed] [Google Scholar]
- 29. Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. 2005. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 187:2286–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rolhion N, Carvalho FA, Darfeuille-Michaud A. 2007. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Mol. Microbiol. 63:1684–1700 [DOI] [PubMed] [Google Scholar]
- 31. Sallmann FR, et al. 1999. Porins OmpC and PhoE of Escherichia coli as specific cell-surface targets of human lactoferrin. Binding characteristics and biological effects. J. Biol. Chem. 274:16107–16114 [DOI] [PubMed] [Google Scholar]
- 32. Secundino I, et al. 2006. Salmonella porins induce a sustained, lifelong specific bactericidal antibody memory response. Immunology 117:59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh SP, Singh SR, Williams YU, Jones L, Abdullah T. 1995. Antigenic determinants of the OmpC porin from Salmonella typhimurium. Infect. Immun. 63:4600–4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stapleton PD, Shannon KP, French GL. 1999. Carbapenem resistance in Escherichia coli associated with plasmid-determined CMY-4 beta-lactamase production and loss of an outer membrane protein. Antimicrob. Agents Chemother. 43:1206–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Steele NP, Munson RS, Jr, Granoff DM, Cummins JE, Levine RP. 1984. Antibody-dependent alternative pathway killing of Haemophilus influenzae type b. Infect. Immun. 44:452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sukumaran SK, Shimada H, Prasadarao NV. 2003. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect. Immun. 71:5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang Y. 2002. The function of OmpA in Escherichia coli. Biochem. Biophys. Res. Commun. 292:396–401 [DOI] [PubMed] [Google Scholar]
- 38. Weiser JN, Gotschlich EC. 1991. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weynants VE, et al. 2007. Additive and synergistic bactericidal activity of antibodies directed against minor outer membrane proteins of Neisseria meningitidis. Infect. Immun. 75:5434–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wooster DG, Maruvada R, Blom AM, Prasadarao NV. 2006. Logarithmic phase Escherichia coli K1 efficiently avoids serum killing by promoting C4bp-mediated C3b and C4b degradation. Immunology 117:482–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yan JJ, Wu JJ, Lee CC, Ko WC, Yang FC. 2010. Prevalence and characteristics of ertapenem-nonsusceptible Escherichia coli in a Taiwanese university hospital, 1999 to 2007. Eur. J. Clin. Microbiol. Infect. Dis. 29:1417–1425 [DOI] [PubMed] [Google Scholar]
- 42. Yoshida T, Qin L, Egger LA, Inouye M. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114–17123 [DOI] [PubMed] [Google Scholar]