Abstract
Generalized epilepsy with febrile seizures plus (GEFS+), a clinical subset of febrile seizures (FS), is characterized by frequent episodes beyond 6 years of age (FS+) and various types of subsequent epilepsy. Mutations in β1 and αI-subunit genes of voltage-gated Na+ channels have been associated with GEFS+1 and 2, respectively. Here, we report a mutation resulting in an amino acid exchange (R187W) in the gene encoding the α-subunit of neuronal voltage-gated Na+ channel type II (Nav1.2) in a patient with FS associated with afebrile seizures. The mutation R187W occurring on Arg187, a highly conserved residue among voltage-gated Na+ channels, was not found in 224 alleles of unaffected individuals. Whole-cell patch clamp recordings on human embryonic kidney (HEK) cells expressing a rat wild-type (rNav1.2) and the corresponding mutant channels showed that the mutant channel inactivated more slowly than wild-type whereas the Na+ channel conductance was not affected. Prolonged residence in the open state of the R187W mutant channel may augment Na+ influx and thereby underlie the neuronal hyperexcitability that induces seizure activity. Even though a small pedigree could not show clear cosegregation with the disease phenotype, these findings strongly suggest the involvement of Nav1.2 in a human disease and propose the R187W mutation as the genetic defect responsible for febrile seizures associated with afebrile seizures.
Keywords: febrile seizures, epilepsy, channelopathy
Febrile seizures (FS) affect up to 6% of infants and young children in the age range from 6 mo to 6 yr (1–4). Extensive genetic studies have demonstrated that four loci are responsible for FS: FEB1 at chromosome 8q13-q21; FEB2, 19p; FEB3, 2q23–24; and FEB4, 5q14-q15 (5–9). A small population of individuals with FS has additional generalized epilepsy (1) or afebrile seizures. Genes for a β-subunit (β1) and an αI-subunit (Nav1.1: SCN1A) (10) of the neuronal voltage-gated Na+ channel have been identified to be responsible for generalized epilepsy with febrile seizures plus (GEFS+) type 1 and 2, respectively (11, 12). However, a large number of patients with GEFS+ still show no mutation for those genes. These, therefore, suggest that other genes might also be involved in GEFS+ and FS associated with afebrile seizures. The chromosomal locus 2q24, in which GEFS+ has been mapped, harbors not only Nav1.1 but also other α-subunits including Nav1.2 (SCN2A) (10, 13–15). Given that Nav1.2 is also expressed in high levels in the central nervous system with a tissue-specific profile (16), Nav1.2 is an intriguing candidate. In the present study, we report a mutation of Nav1.2 found in a patient with FS and afebrile seizures. A channel harboring the mutation shows abnormal electrophysiological properties that may underlie the neuronal hyperexcitability that triggers seizure activity.
Materials and Methods
Patients and Pedigrees.
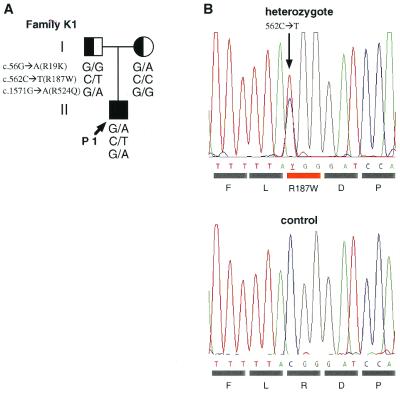
This study recruited nineteen unrelated Japanese families with members clinically diagnosed with GEFS+ or febrile seizures associated with afebrile seizures. Each participating subject or a responsible adult signed an informed consent form approved by the Ethics Review Committee of Fukuoka University or similar committees of the participating institutions. The proband of family K1 is a 6-yr-old boy with normal development (Fig. 1A). He had the first febrile seizure (FS) at 8 months of age and suffered 17 episodes of FS thereafter at both high and low grade fever. The FS were generalized tonic or tonic-clonic convulsions with duration of 1–5 min per episode. Since 4 yr of age, he also has experienced brief afebrile atonic seizures 5 times. The parents reported that head drops or forceful falls occurred suddenly without fever. The duration of seizures was less than 10 s. MRI showed no abnormality, but a recent electroencephalogram (EEG) showed single spikes over the right frontal region. His father and paternal uncle had febrile seizures twice in their childhood. His mother and maternal aunt also had febrile seizures three and two times, respectively.
Figure 1.
Missense mutations identified in a Japanese family with febrile seizures associated with afebrile seizures. (A) Pedigree of Japanese family K1 with FS and afebrile seizures (proband) and FS only (parents). c.562C → T (R187W) mutations appeared in proband P1 and his father. Circles = females, squares = males, arrow = proband, filled square = FS associated with afebrile seizures, half-filled square and circle = FS. (B) Electropherogram of the mutation in the gene for Nav1.2 identified in family K1. The nucleotide sequence of the relevant region of exon 4 of the proband is shown. Arrow indicates nucleotide 562, where a heterozygous C-to-T transition resulted in an amino acid substitution, R187W.
Mutational Analysis.
Genomic DNA was extracted from EDTA-Na2-treated blood samples of affected and unaffected individuals by using a commercial product (SepaGene, Sanko Jyunyaku, Tokyo, Japan). The intron-exon boundaries of Nav1.2 were elucidated by comparison between the sequences of a bacterial artificial chromosome (BAC) clone covering the coding region (Research Genetics, Huntsville, AL) and mRNA information (GenBank accession no. M94055, ref. 17). Primers were designed to amplify all 26 coding exons and the flanking intronic splice sites. The sequences of primers are available on request. Genomic PCR was performed using Pyrobest DNA polymerase (TaKaRa, Tokyo, Japan) according to the manufacturer's protocol. PCR products were directly sequenced with a dideoxy terminator kit and analyzed with an automated sequencer (model ABI3700, Perkin-Elmer). The sequences obtained from patients' DNA were compared with the revised Nav1.2 cDNA sequence (see Results).
Expression of Na+ Channels in a Cell Line.
The cDNA encoding rat Na+ channel αII subunit (rNav1.2, ref. 18) was inserted into the NotI site of a pCI-neo plasmid vector (Promega). Three mutations of rNav1.2 cDNA corresponding to R19K, R187W, and R524Q were introduced to the wild-type (WT) construct with a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All insert sequences as well as mutated sites were confirmed by dideoxynucleotide sequencing. Plasmid DNAs for transfection were isolated with a Plasmid Maxi kit (Qiagen, Hilden, Germany). For patch-clamp experiments, plasmids bearing WT, R19K, R187W, or R524Q full-length α-subunit cDNA as well as equimolar pEGFPN1 vector (CLONTECH) as a marker were transiently transfected into human embryonic kidney (HEK) 293 cells by Lipofectamine 2000 (Life Technologies, Rockville, MD) as recommended by the manufacturer. For the experiments of coexpression with Nav1.2 and β1 subunit (19), a molar ratio for Nav1.2:β1 was 1:1.
Patch-Clamp Experiments.
Cells were plated on poly-l-lysine-coated glass coverslips (Biocoat Cellware Poly-l-lysine 12-mm Coverslip, Becton Dickinson Labware) and maintained in culture medium (DMEM supplemented with 10% FBS) before patch-clamp experiments in an environment of humidified 5% CO2. Transient expression of functional Na+ channels was detected 24–48 h after plating.
Electrical recording.
Membrane currents were recorded by using the whole-cell configuration of the patch-clamp technique at room temperature (22°C). Pipette electrodes were made from borosilicate glass capillary tubes (0.8–1.0 mm inner diameter, Kimble Glass, Vineland, NJ) by using a multistep horizontal puller (P-97, Sutter Instruments, Novato, CA). Pipette resistance was 1.0–1.5 MΩ when filled with internal pipette solution. An Ag-AgCl pellet/3 M KCl-agar bridge was used for the reference electrode. Currents were recorded by using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA). Signals were filtered with a low-pass Bessel filter with a cut-off frequency at 10 kHz and stored on a hard disk in Macintosh G3/266 (Apple Computer, Cupertino, CA) by using Pulse + PulseFit 8.11 (HEKA Electronics, Lambrecht/Pfalz, Germany). Series resistance was compensated to 70–80%. Capacitative and leakage currents were digitally subtracted by using the P + P/4 procedure. All data were compensated for the liquid junction potential (−1.2 mV on average). Peak currents over 2.0 nA were considered as the response of expressed Na+ channels. The average Na current density was calculated to be 1252.7 pA/pF for WT channels and 1642.4 pA/pF for the R187W mutants. The peak Na+ current arising from endogenous Na+ channels in untransfected HEK cells in the present study was ≤100 pA (average Na current density ≤10 pA/pF).
Conductance-voltage relationship.
Currents were evoked by 10-ms depolarizations to various test potentials from a holding potential of −120 mV. The conductance (gNa) was calculated according to the equation, gNa = INa/(Vg − Vr), where INa is the peak amplitude of Na+ current, Vg is the test potential, and Vr is the reversal potential for Na+. The curves are drawn according to the equation, gNa/maxgNa = 1/{1 + exp[Vg0.5 − Vg]/kg]}, where maxgNa is the maximum value for gNa, Vg0.5 is the potential at which gNa is 0.5maxgNa, and kg is the slope factor.
Steady-state inactivation.
The membrane potential was held at various values for 2 s, and then Na+ current was evoked by a step depolarization to −10 mV. The curves were drawn according to the equation, I/Imax = 1/{1 + exp[(Vh − V1/2)/k]}, where Vh is the holding potential, V1/2 is the potential at which sodium current is one-half maximum, and k is the slope factor.
Solutions.
The internal pipette solution contained (in mM): CsF 135, NaCl 10, and Hepes-acid 5. The pH was adjusted to 7.0 with CsOH, and the osmolarity was 276 mOsm. The external solution contained (in mM): NaCl 135, CaCl2 2.0, MgCl2 1.0, and Hepes-acid 10. The solution was adjusted to pH 7.4 with NaOH, and the osmolarity was 281 mOsm and glucose 5.0.
Results
We recruited nineteen unrelated Japanese families in which affected individuals were diagnosed as GEFS+ or FS associated with afebrile seizures, and screened the proband of each family for genetic abnormalities of a β-subunit (β1) and two α-subunits (Nav1.1 and Nav1.2) of the neuronal voltage-gated Na+ channel. Initial screening of β1 gene did not show any mutation in the families (data not shown). We then analyzed all 26 coding exons and the flanking intronic splice sites of Nav1.1 and Nav1.2 in these families. We detected two disease-causing missense mutations and benign variants in the coding region for Nav1.1 (36). In the course of mutational analysis for Nav1.2, we found differences at two nucleotide positions between the sequence registered in GenBank (accession no. M94055, ref. 17) and the sequence we obtained from all available materials, including cDNA clones, bacterial artificial chromosome clones, patients, and healthy control individuals. One of them is position c.1571 (The first letter A of the start codon ATG was numbered as position c.1, which corresponds to the originally registered sequence position number 106). Although the nucleotide at c.1571 was registered as T (GenBank accession no. M94055), the bacterial artificial chromosome clone (Research Genetics) and 312 alleles from 156 volunteers of various ethnic origins (100 Japanese, 21 Caucasians, 18 Spanish, 9 Indians, and 8 Mexicans) bore G at the corresponding nucleotide position. Thus, G was considered as the WT sequence for the c.1571 position. This nucleotide substitution changed the amino acid leucine (L) to arginine (R) at the amino acid sequence position 524. For the position c.3974, the nucleotide was registered as C (GenBank accession no. M94055). However, the nucleotide was T in all our materials as well. We therefore consider T as a WT sequence for the position c.3974. This substitution changed the amino acid alanine (A) to valine (V) at the amino acid sequence position 1325.
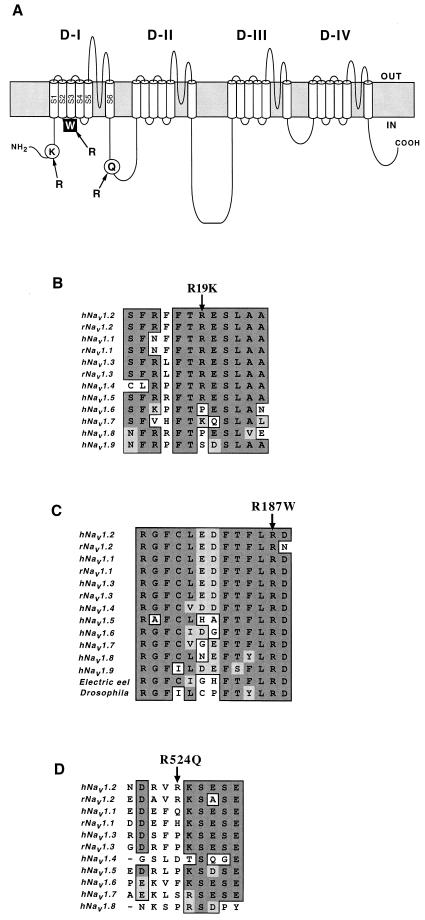
In the Nav1.2 genes of patients with FS associated with afebrile seizures, we found three single nucleotide substitutions, c.56G →A, c.562C →T, and c.1571G →A, resulting in an amino acid exchange, R19K, R187W, and R524Q, respectively. All probands that have nucleotide substitution(s) in Nav1.2 are summarized in Table 1. A proband of family K1 (P1) happened to have all these three missense mutations heterozygously. Analyses of fathers' and mothers' alleles revealed that R187W and R524Q mutations in P1 were paternally inherited, whereas R19K was maternally inherited (Fig. 1A). The proband P1 did not show any mutations in the coding regions of β1 and Nav1.1 (Table 1). The R19K mutation was found heterozygously in probands of five families, including the proband P1 of family K1 (Fig. 1A). R19K was also found in 11 of 224 alleles in unaffected individuals. Arginine-19, located in a cytoplasmic portion of the Nav1.2 protein (Fig. 2A), is a moderately conserved residue in the voltage-gated Na+ channel α-subunit family (Fig. 2B). These results suggest that R19K is most probably a benign variant; however, the high frequency of the R19K allele in patients (5/19 vs. 9/112, P = 0.0424) leaves open the possibility that the R19K is a modifying factor for epilepsy. Arginine-187, located in the cytoplasmic loop between S2 and S3 in domain I, is highly conserved among the voltage-gated Na+ channel α-subunit family members and through evolution (Fig. 2C). This R187W mutation did not appear in 224 alleles of unaffected Japanese individuals. R524Q mutation was rare and was found in only one of 324 alleles in unaffected individuals. Arginine-524, which is located in the loop linking domain I and II (Fig. 2A), is not conserved among the voltage-gated Na+ channel α-subunit family members (Fig. 2D). Furthermore, a proband P2 of another family, who had R524Q mutation of Nav1.2, also harbored a mutation in Nav1.1 for the conserved transmembrane helix, which is most likely responsible for the disease phenotype (Table 1). Together, these results suggest that R524Q of Nav1.2 is another benign variant.
Table 1.
Missense mutations of voltage-gated Na+ channel subunits identified in Japanese families with febrile seizures associated with afebrile seizures
| Patient | Missense
mutation
|
||||
|---|---|---|---|---|---|
|
Nav1.2
|
Nav1.1 | β1 | |||
| Exon 1 | Exon 4 | Exon 10 | |||
| P1 | Arg19Lys | Arg187Trp | Arg524Gln | ND† | ND |
| c.56G→A | c.562C→T* | c.1571G→A | |||
| P2 | Arg524Gln c.1571G→A | A missense mutation in a transmembrane helix‡* | ND | ||
| P3, P4, P5, P6 | Arg19Lys | ND | ND | ||
| c.56G→A | |||||
| Control | 9/112 | 0/112 | 1/162 | 0/112 | |
Bold characters indicate disease mutations.
ND, No mutation was detected.
Will be reported elsewhere (36).
Figure 2.
Diagram of Nav1.2 and amino acid sequence alignments of Na+ channel α-subunit family members. (A) Locations of missense mutations identified in this study on Nav1.2. Filled square indicates the mutation proposed to be responsible for the disease phenotype. The amino acid exchanges indicated by open circles are ones assumed to be benign variants. (B–D) Partial amino acid sequences of Nav1.2 and other α-subunit family members (GenBank accession nos.: M94055, M22253, Y00766, M81758, M77235, AB027567, X82835, AF117907, AF188679, D37977, L19979, M22252, and M32078). Dark shaded background indicates identical amino acids, light shaded background indicates conserved amino acids, and white background depicts nonconserved amino acids. Arrows above the sequences indicate the positions of the missense mutations, R19K (B), R187W (C), and R524Q (D). Arg187 is highly conserved among the α-subunits of voltage-gated Na+ channels (C).
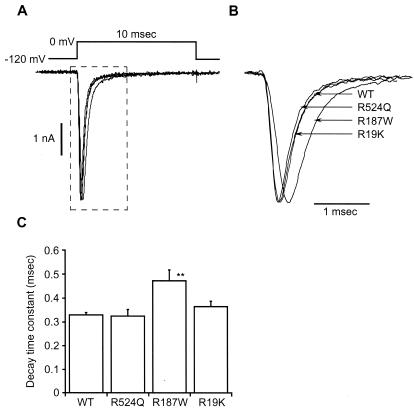
To examine whether these mutations affect Na+ channel function, we investigated the electrophysiological properties of rat Nav1.2 channels, with or without corresponding mutations, expressed in HEK 293 cells by using whole-cell patch clamp analysis. Analysis of the time course of Na+ current activation shows that the time to reach the peak current in R187W mutant channels was delayed compared with WT, R19K, and R524Q channels (Fig. 3 A and B), suggesting slowing of inactivation. Analysis of the time course of Na+ current inactivation (Fig. 3C) showed that the time constant of current decay during step depolarization in the R187W mutant was significantly slower than in WT, R19K, and R524Q channels.
Figure 3.
Differences in the kinetics between WT and mutant sodium channels. (A) Sodium currents of WT and mutant Na+ channels. Currents were induced by a step depolarization to 0 mV from a holding potential of −120 mV. (B) Enlargement of the area within dashed line in A. Peak amplitudes were 5.2 nA for WT, 3.9 nA for R524Q, 4.4 nA for R187W, and 4.1 nA for R19K. The peak currents were normalized. (C) Time constant of the decay during a step depolarization. Current decay was fitted by a single exponential function. The decay time constant was significantly slower in R187W channels than in WT, R19K, or R524Q channels. Each point is given as mean ± SEM (n = 7). Statistical test was done by ANOVA. **, the statistical difference at 1% level.
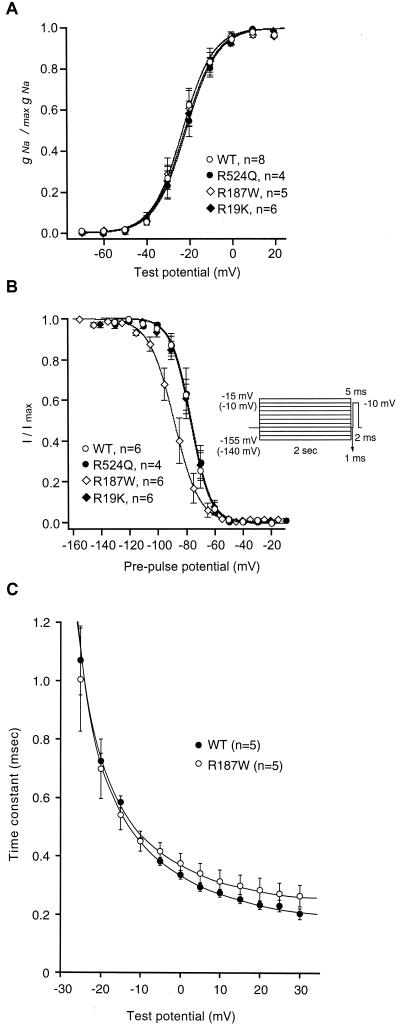
To clarify the mechanism of slowing of inactivation, we further examined peak conductance-voltage relationships, recovery from inactivation, and steady-state inactivation. None of the three mutations affected the potential difference required for an e-fold change in conductance compared with WT channels (Fig. 4A). There were no significant differences in the time course of recovery from inactivation among these Na+ channel mutants (data not shown). In contrast, the steady-state voltage dependence of inactivation was markedly affected by the R187W mutation (Fig. 4B). The steady-state inactivation curve was shifted in the hyperpolarizing direction by 11.7 mV compared with WT (Fig. 4B), and the slope factor was significantly reduced with respect to WT and the other two channel mutants. Fig. 4C shows the voltage dependence of the time constant of macroscopic current decay (τh/V). The maximal rates of inactivation for the R187W mutants were larger than those for WT channels at depolarized membrane potentials. Coexpression of the β1 subunit attenuated the difference between WT and mutant channels in all of the experimental protocols described (data not shown). A reduced slope factor is compatible with the notion that inactivated R187W mutant channels are less sensitive to voltage presumably because the coupling between the inactivation sensor and the transmembrane potential has been perturbed. The shift of the steady-state inactivation with larger slope factor could change the relative population of Na+ channels available for activation at physiological membrane potentials. Based solely on the modified kinetics and steady-state properties of Na+ channel inactivation, neurons expressing the dysfunctional channel would be expected to display anomalous inactivation. This dysfunction in and of itself, may allow the persistent repetitive firing during a depolarization to develop into a seizure (20–22). The alterations caused by R187W mutation may, therefore, explain the hyperexcitability leading to seizures at the neuronal level.
Figure 4.
Effects of missense mutations on conductance-voltage relationships and steady-state inactivation. (A) Peak conductance-voltage relationships for WT (open circle), R524Q (filled circle), R187W (open diamond), and R19K (filled diamond) Na+ channels expressed in HEK cells. No significant differences were detected between WT and mutant channels. Number of experiments was four, and all data were pooled. Curves are fitted to the data by using the equations given in Materials and Methods. (B) Steady-state voltage dependence of inactivation for WT (open circle), R524Q (filled circle), R187W (open diamond), and R19K (filled diamond) Na+ channels. Two different protocols were used. The prepulses were applied from −155 to −15 mV for WT, R524Q, and R19K mutant channels, and from −140 to −10 mV for R187W mutant channel with 10-mV increments. The half-inactivation potential and slope factors were estimated to be −76.9 mV and 6.7 for WT, −76.4 mV and 6.8 for R524Q, −88.6 mV and 8.7 for R187W, and −77.2 mV and 6.9 for R19K. All data were pooled and fitted as in A. (C) Voltage dependence of the inactivation time constant for WT and the R187W mutant channels. Note the clear difference in maximal rate of inactivation at the depolarized membrane potentials between WT and the R187W mutant.
Discussion
Recent extensive studies have uncovered that abnormal kinetics of mutated voltage-gated Na+ channels lead to various paroxysmal disorders, i.e., Na+ channelopathies, in humans as well as in animals (23, 24). Impairment of Na+ channel inactivation appears to be a recurrent defect in diseases characterized by hyperexcitability (25, 26). The III-IV cytoplasmic linker is currently considered to function as the inactivation gate and to contribute to the fast inactivation component of Na+ channels (27–29). In agreement, missense mutations in this inactivation gate domain and in its acceptors, which impair inactivation and affect voltage dependence, were found in permanent myotonia (30). Mutations in the III-IV cytoplasmic loop of the muscle and cardiac Na+ channel α-subunit have been found in potassium-aggravated myotonia and long QT syndrome 3 (26), respectively. Steady-state fast inactivation was shifted to hyperpolarizing potentials in some mutated channels associated with paramyotonia congenita (31–34). Remarkably, a missense mutation of β1-subunit of Na+ channel decreases the rate of inactivation for the α-subunit and induces the febrile seizures associated with afebrile seizure phenotypes (11). Similar effects on the kinetics and steady-state properties of inactivation were observed in the mutated Nav1.2 channels harboring R187W (Fig. 3 and 4). Unfortunately, the number of members for the family K1 that harbors the R187W mutation was not sufficient to establish unambiguously the cosegregation of the mutation with the disease phenotype. Appearances of the FS phenotypes in both parents also interfere with the assignment, even if the phenotype arises merely from the high frequency of FS phenotype (3–6%) in a general population. Even with these conditions, the finding of the electrophysiological abnormalities of channels with the R187W mutation similar to those observed in multiple other Na+ channelopathies strongly suggests that the R187W mutation is the molecular basis of febrile seizures associated with afebrile seizures in the patient studied in this report. We analyzed genes for three sodium channel subunits, β1, Nav1.1, and Nav1.2, in the mothers' genomic DNA, and only the missense mutation R19K in Nav1.2 gene was detected (Fig. 1A). Identification of R19K mutation in a number of control DNAs, as well as the electrophysiological study, suggested that R19K is a benign variant. Because the febrile seizure is the most commonly provoked seizure afflicting infants and children, it is likely that numerous factors contribute to generate heterogeneous seizure phenotypes. Actually, several loci are known to be responsible for FS (5–9). The febrile seizures of mother and maternal aunt may therefore most probably be accounted for by defects in other genes. However, the high frequency of occurrence of R19K among patients is statistically significant (P = 0.0424). The father with R187W mutation showed only febrile seizure; in contrast, the proband with R187W as well as R19K mutations showed febrile seizure associated with afebrile seizures. The possibility that the R19K mutation modifies or aggravates the disease phenotype therefore cannot be completely excluded, and the association study of the FS phenotype with R19K as well as R187W and even with R524Q mutations should be further pursued.
Paternal inheritance indicates that both the R187W and R524Q missense mutations are located on a single allele. The proximity relationship between R187W and R524Q in the three-dimensional structure of the sodium channel is unknown. Topological models assign the location of both residues at the cytoplasmic portion of the channel; however, they may be distant from each other. To the extent that the kinetics of R524Q mutant channels is not distinguishable from WT channels, the kinetics of the double mutant channel are predicted to be similar to those of the R187W mutant channel. The kinetics of the double mutant channel have not been hitherto characterized; however, the mutant may not merely exhibit the aggregate effect of the two point mutations.
Nav1.1 and Nav1.2 show different distributions within neurons and also in several regions in rat brain (35). Nav1.1 is mainly localized in the soma of neurons whereas Nav1.2 is predominantly expressed in axons. Genetic abnormalities of these α-subunits may hence lead to differences in seizure phenotypes. In fact, the father of P1 in family K1, who shared the same R187W mutation with proband P1, had only FS (Fig. 1A), whereas the GEFS+2 phenotype resulting from Nav1.1 mutations associates with high penetrance (6). Further analyses of the genotype-phenotype relationship in febrile seizures associated with afebrile seizures are therefore necessary.
In summary, we have identified a missense mutation in Nav1.2 in a patient with febrile seizures associated with afebrile seizures, and the electrophysiological abnormalities of channels harboring the mutation lead us to conclude its involvement in the disease phenotype. To our knowledge, there have been no previous reports of a mutation of Nav1.2 proposed to be responsible for human disease; further identification of mutations in other families with epilepsy should corroborate our findings and deepen our understanding of the molecular pathology of epilepsy.
Acknowledgments
We thank all members of the families in this study for their helpful cooperation. We are grateful to Dr. R. Kado, [Brain Science Institute (BSI), The Institute of Physical and Chemical Research (RIKEN), Japan] and Dr. T. Narahashi (Northwestern University) for helpful discussions. This study was conducted as part of a comprehensive project organized by The Epilepsy Genetic Study Group, Japan (Chairperson, S.K.) and supported in part by grants for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, The Epilepsy Research Foundation (M.I., A.M., S.K., and S.H.), Uehara Memorial Foundation (S.K. and S.H.), Heiwa Nakajima Foundation, International Research Fund of Kyushu University School of Medicine Alumni (S.H.), The Clinical Research Foundation (G.F., A.M. and S.H.), The Foundation for the Advancement of Clinical Medicine, Japan, and The Central Research Institute of Fukuoka University (A.M. and S.H.). Research at the University of California is supported by National Institutes of Health Grant GM-49711 (M.M.).
Abbreviations
- FS
febrile seizures
- FS+
febrile seizures plus
- GEFS+
generalized epilepsy with febrile seizures plus
- WT
wild-type
- HEK
human embryonic kidney
Footnotes
References
- 1.Scheffer I E, Berkovic S F. Brain. 1997;120:479–490. doi: 10.1093/brain/120.3.479. [DOI] [PubMed] [Google Scholar]
- 2.Singh R, Scheffer I E, Crossland K, Berkovic S F. Ann Neurol. 1999;45:75–81. doi: 10.1002/1531-8249(199901)45:1<75::aid-art13>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.O'Donohoe N V. In: Epileptic Syndromes in Infancy, Childhood and Adolescence. Roger J, Dravet C, Bureau M, Dreifuss F E, Wolf P, Perret A, editors. London: John Libby Eurotext; 1992. pp. 45–52. [Google Scholar]
- 4.Bird T D. Epilepsia. 1987;28,Suppl.:S71–S81. doi: 10.1111/j.1528-1157.1987.tb05761.x. [DOI] [PubMed] [Google Scholar]
- 5.Wallace R H, Berkovic S F, Howell R A, Sutherland G R, Mulley J C. J Med Genet. 1996;33:308–312. doi: 10.1136/jmg.33.4.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson E W, Dubovsky J, Rich S S, O'Donovan C A, Orr H T, Anderson V E, Gil-Nagel A, Ahmann P, Dokken C G, Schneider D T, Weber J L. Hum Mol Genet. 1998;7:63–67. doi: 10.1093/hmg/7.1.63. [DOI] [PubMed] [Google Scholar]
- 7.Kugler S L, Stenroos E S, Mandelbaum D E, Lehner T, McKoy V V, Prossick T, Sasvari J, Swannick K, Katz J, Johnson W G. Am J Med Genet. 1998;79:354–361. doi: 10.1002/(sici)1096-8628(19981012)79:5<354::aid-ajmg5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 8.Peiffer A, Thompson J, Charlier C, Otterud B, Varvil T, Pappas C, Bernitz C, Gruenthal K, Kuhn R, Leppert M. Ann Neurol. 1999;46:671–678. doi: 10.1002/1531-8249(199910)46:4<671::aid-ana20>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakayama J, Hamano K, Iwasaki N, Nakahara S, Horigome Y, Saitoh H, Aoki T, Maki T, Kikuchi M, Migita T, et al. Hum Mol Genet. 2000;9:87–91. doi: 10.1093/hmg/9.1.87. [DOI] [PubMed] [Google Scholar]
- 10.Goldin A L, Barchi R L, Caldwell J L, Hofmann F, Howe J R, Hunter J C, Kallen R G, Mandel G, Meisler M H, Netter Y B. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 11.Wallace R H, Wang W W, Singh R, Scheffer I E, George A L, Jr, Phillips H A, Saar K, Reis A, Johnson E W, Sutherland G R, et al. Nat Genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- 12.Escayg A, MacDonald B T, Meisler M H, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, et al. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- 13.Baulac S, Gourfinkel-An I, Picard F, Rosenberg-Bourgin M, Prud'homme J-F, Baulac M, Brice A, LeGuern E. Am J Hum Genet. 1999;65:1078–1085. doi: 10.1086/302593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moulard B, Guipponi M, Chaigne D, Mouthon D, Buresi C, Malafosse A. Am J Hum Genet. 1999;65:1396–1400. doi: 10.1086/302621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes-Cendes I, Scheffer I E, Berkovic S F, Rousseau M, Andermann E, Rouleau G A. Am J Hum Genet. 2000;66:698–701. doi: 10.1086/302768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plummer N W, Meisler M H. Genomics. 1999;57:323–331. doi: 10.1006/geno.1998.5735. [DOI] [PubMed] [Google Scholar]
- 17.Ahmed C M I, Ware D H, Lee S C, Patten C D, Ferrer-Montiel A V, Schinder A F, McPherson J P, Wagner-McPherson K, Wasmuth J J, Evans G A, Montal M. Proc Natl Acad Sci USA. 1992;89:8220–8224. doi: 10.1073/pnas.89.17.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuhmer W, Conti F, Suzuki H, Wang X D, Noda M, Yahagi N, Kubo H, Numa S. Nature (London) 1989;339:597–603. doi: 10.1038/339597a0. [DOI] [PubMed] [Google Scholar]
- 19.Patton D E, Isom L L, Catterall W A, Goldin A L. J Biol Chem. 1994;269:17649–17655. [PubMed] [Google Scholar]
- 20.Narahashi T. J Pharmacol Exp Ther. 2000;294:1–26. [PubMed] [Google Scholar]
- 21.Song J H, Narahashi T. J Pharmacol Exp Ther. 1995;275:1402–1411. [PubMed] [Google Scholar]
- 22.Narahashi T. Ann N Y Acad Sci. 1986;479:133–151. doi: 10.1111/j.1749-6632.1986.tb15566.x. [DOI] [PubMed] [Google Scholar]
- 23.Lehmann-Horn F, Jurkat-Rott K. Physiol Rev. 1999;79:1317–1372. doi: 10.1152/physrev.1999.79.4.1317. [DOI] [PubMed] [Google Scholar]
- 24.Hirose S, Okada M, Kaneko S, Mitsudome A. Epilepsy Res. 2000;41:191–204. doi: 10.1016/s0920-1211(00)00141-8. [DOI] [PubMed] [Google Scholar]
- 25.Catterall W A. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 26.Bennet P B, Yazawa K, Makita N, George A L., Jr Nature (London) 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 27.West J W, Patton D E, Scheuer T, Wang Y, Goldin A L, Catterall W A. Proc Natl Acad Sci USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patton D E, West J W, Scheuer T, Wang Y, Goldin A L, Catterall W A. Proc Natl Acad Sci USA. 1992;89:10905–10909. doi: 10.1073/pnas.89.22.10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McPhee J C, Ragsdale D S, Scheuer T, Catterall W A. J Biol Chem. 1998;273:1121–1129. doi: 10.1074/jbc.273.2.1121. [DOI] [PubMed] [Google Scholar]
- 30.Lerche H, Heine R, Pika U, George A L, Jr, Mitrovic N, Browatzki M, Weiss T, Rivert-Bastide M, Franke C, Lomonako M, et al. J Physiol. 1993;470:13–22. doi: 10.1113/jphysiol.1993.sp019843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang N, Ji S, Zhou M, Ptacek L J, Barchi R L, Horn R, George A L., Jr Proc Natl Acad Sci USA. 1994;91:12785–12789. doi: 10.1073/pnas.91.26.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richmond J E, Featherstone D E, Ruben P C. J Physiol. 1997;499. 3:589–600. doi: 10.1113/jphysiol.1997.sp021952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendahhou S, Cummins T R, Kwiecinski H, Waxman S G, Ptácek L J. J Physiol. 1999;518:337–344. doi: 10.1111/j.1469-7793.1999.0337p.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Featherstone D E, Fujimoto E, Ruben P C. J Physiol. 1998;506:627–638. doi: 10.1111/j.1469-7793.1998.627bv.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong B, Rhodes K J, Bekele-Arcuri Z, Trimmer J S. J Comp Neurol. 1999;412:342–352. [PubMed] [Google Scholar]
- 36.Sugawara, T., Mazaki-Miyazaki, E., Ito, M., Nagafuji, H., Fukuma, G., Mitsudome, A., Wada, K., Kaneko, S., Hirose, S. & Yamakawa, K. (2001) Neurology, in press. [DOI] [PubMed]