Abstract
Bcr is a serine/threonine kinase that is a critical regulator of vascular smooth muscle cell inflammation and proliferation. We have previously demonstrated that Bcr acts in part via phosphorylation and inhibition of PPARγ. We have identified the RNA helicase UAP56 as another substrate of Bcr. In this report we demonstrate that knockdown of UAP56 blocks Bcr induced DNA synthesis in vascular smooth muscle cells (VSMC). We also found that over expression of Bcr increased the expression of cyclin E and decreased the expression of p27. Knockdown of UAP56 reversed the effect of Bcr on cyclin E and p27 expression. Furthermore, we found that Bcr binds to UAP56 and demonstrate that binding of UAP56 to Bcr is critical for Bcr induced DNA synthesis in VSMC. Our data identify UAP56 as an important binding partner of Bcr and a novel target for inhibiting vascular smooth muscle cell proliferation.
Keywords: DNA synthesis, DExD/H box protein, RNA helicase
INTRODUCTION
Pathologic vascular smooth muscle cell (VSMC) proliferation occurs in many disease states including hypertension, atherosclerosis and restenosis after injury [1, 2]. This proliferation is mediated by growth factors such as platelet-derived growth factor (PDGF) and vasoconstrictive hormones such as Angiotensin II (Ang II), which induces protein synthesis and DNA synthesis and enhances PDGF induced DNA synthesis [1, 3, 4]. We have recently reported that breakpoint cluster region (Bcr), a serine/threonine kinase is an important mediator of Ang II and PDGF mediated responses in VSMC [5]. We found that knockdown of Bcr inhibited Ang II mediated NF-κB activation in VSMC. Specifically, we found that over expression of Bcr inhibits PPARγ transcriptional activation via phosphorylation of PPARγ, resulting in enhancement of NF-κB transcriptional activation. In addition to PPARγ, we found evidence of UAP56 as another substrate for Bcr.
UAP56 is an RNA helicase that was first identified in an analysis of genes centromeric to HLA-B in the human major histocompatibility complex and was named BAT1 (HLA B associated transcript 1) [6]. BAT1 was rediscovered as an essential RNA splicing factor recruited to mRNA precursors (pre-mRNA) by the splicing factor U2AF65 and was renamed UAP56 (56-kD U2AF associated protein) [7]. UAP56 is a member of the DExD/H box family of proteins (named after the amino acid sequence) and is an RNA dependent ATPase, hydrolyzing ATP into ADP [8, 9]. UAP56 is part of the TREX (transcription/export) complex [10] which is recruited to activated genes during transcription and travels the length of the gene with RNA polymerase during transcription elongation [10]. UAP56 plays a major role in several steps of RNA biology including spliceosome assembly, mRNA export and protein synthesis [11-14], and knockdown of UAP56 leads to down regulation of genes involved in the cell cycle, mitosis, cell division and DNA repair [15]. In the present study, we found that UAP56 is a key cell cycle regulator and is a novel interacting partner of Bcr in VSMC DNA synthesis.
MATERIALS AND METHODS
Cell culture
Rat VSMC were isolated as previously described [5] and were maintained in DMEM. HeLa cells were grown in DMEM containing 10% fetal bovine serum.
Plasmids and transfection
UAP56 wild-type plasmid was purchased from Origene. The single mutation of UAP56 was created with the QuikChange site-directed mutagenesis kit (Stratagene) as previously described [13]. For transient expression experiments, cells were transfected with Lipofectamine 2000 (Invitrogen) as previously described [13]. For siRNA experiments, VSMC were transfected with control or UAP56 siRNA oligonucleotides (Dharmacon) using Lipofectamine RNAiMAX reagent (Invitrogen).
Immunoprecipitation and Western blot
After treatment with reagents (indicated in the legends), the cells were washed twice with PBS and harvested in 0.5 mL of lysis buffer as previously described [5]. For immunoprecipitation, cell lysates were incubated with mouse anti-UAP56 antibody (1-2 μg) overnight at 40C, and then protein A/G beads were added and further incubated for 2 hours. The beads were then washed and boiled in 2X SDS sample buffer and western blotting was performed with primary antibodies as indicated in the legends followed by incubation with horseradish peroxidase-conjugated secondary antibody (Amersham Life Science).
[3H] Thymidine Incorporation assay
Cells were plated in 12 well plates and transfected with wild type (WT) Bcr or WT UAP56 plasmids using Lipofectamine 2000 according to manufacturer’s instructions. Cells were pulsed with [3H] thymidine during the last one-hour of incubation and DNA synthesis was measured as described before [5]. Briefly, cells were washed twice with cold PBS, then 500 μL of 10% ice-cold trichloroacetic acid was added to each well, and precipitates were collected on a micro-fiber filter using a manifold. Filters were washed twice with ice-cold 5% trichloroacetic acid, followed by 95% ethanol, allowed to air-dry, and then suspended in scintillation fluid. Acid precipitable counts were quantitated using a scintillation counter. Each experiment was performed at least 3 times, and triplicate wells were used in each experiment.
Mammalian Two-hybrid assay
HeLa cells were transfected in Opti-MEM (Invitrogen) with Lipofectamine mixture containing the pG5-luc vector and various pBIND and pACT plasmids (Promega) for 4 h. The pBIND vector contains the yeast GAL4–DNA-binding domain upstream of a multiple cloning region, and the pACT vector contains the herpes simplex virus VP16 activation domain upstream of a multiple cloning region. Bcr and various UAP56 fragments/mutants were cloned into the pBIND and pACT vector, respectively. Because pBIND also contains the Renilla luciferase gene, the expression and transfection efficiencies were normalized with the Renilla luciferase activity. Cells were collected 48 hours after transfection and the luciferase activity was assayed with the Dual-Luciferase kit (Promega) using a luminometer (TD-20/20; Turner Designs).
Statistics
Numerical data are expressed as mean ± SD. Statistical analysis was performed with the StatView 5.0 package (ABACUS Concepts, Berkeley, CA). Differences were analyzed with a one-way or a two way repeated–measure analysis of variance as appropriate, followed by Scheffé’s correction for multiple comparisons. A probability value < 0.05 was considered significant.
RESULTS
UAP56 is a substrate for Bcr
In an in vitro kinase assay using rat VSMC, in which Bcr was immunoprecipitated with Bcr antibody, we previously demonstrated that Bcr phosphorylates PPARγ [5]. In these studies, we also identified a protein around 60 kDa which we believed to represent another Bcr substrate that coimmunoprecipitated with Bcr in VSMC (Figure S1). We repeated these experiments (without P32) , cut out the band corresponding to the 60 kDa protein, and performed mass spectrometry. The highest hit was the protein UAP56 (BAT1) (Figure S2).
Knockdown of UAP56 blocks Bcr induced DNA synthesis
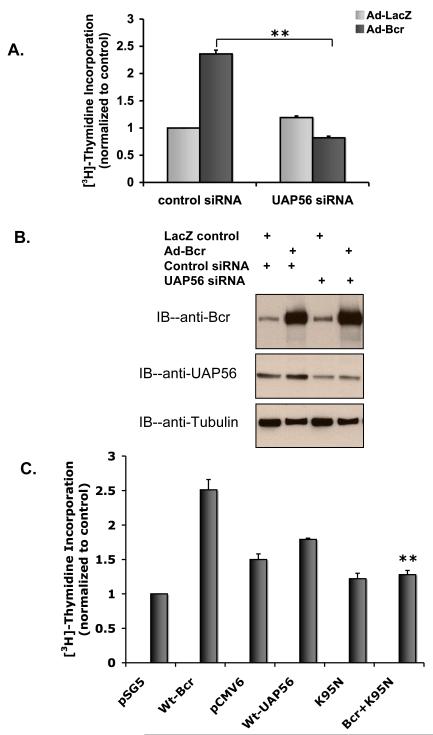
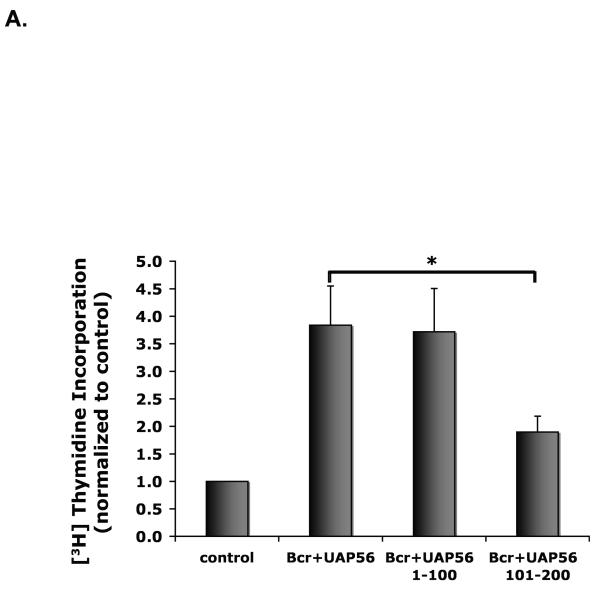
We previously demonstrated that knockdown of Bcr inhibits DNA synthesis [5]. As UAP56 was identified as a substrate of Bcr, we assessed the importance of UAP56 in Bcr induced DNA synthesis. Using UAP56 siRNA and WT Bcr adenovirus (Ad-Bcr), we demonstrated that knock down of UAP56 blocks Bcr induced DNA synthesis in rat VSMC (Figure 1A and 1B). Similarly, utilizing WT Bcr and a dominant negative UAP56 mutant (K95N, [16]), we demonstrated that knockdown of UAP56 blocks Bcr induced DNA synthesis in HeLa cells (Figure 1C).
Figure 1. Knockdown of UAP56 blocks Bcr induced DNA synthesis.
A. VSMC were transfected with UAP56 siRNA or control siRNA. 48 hours later the cells were treated with 50 MOI of Ad-Bcr or Ad-LacZ for 24 hours. During the last hour of incubation VSMC were pulse labeled with [3H] thymidine and incorporation of [3H] thymidine was measured. (**p<0.01). B. Western blots demonstrating Bcr and UAP56 expression in cells treated as in A. C. Following transfection with the indicated plasmids for 48 hours, HeLa cells were pulse labeled with [3H] thymidine for 1 hour. Cells were then harvested and [3H] thymidine incorporation was measured. (**p<0.01 compared with Wt-Bcr). pSG5 and pCMV6 are control plasmids for WT-Bcr and WT-UAP56 respectively. Results are means±SD. For all figures, the data are representative of triplicates using 2 or more different preparations of SMCs or HeLa cells.
UAP56 blocks cell cycle activation
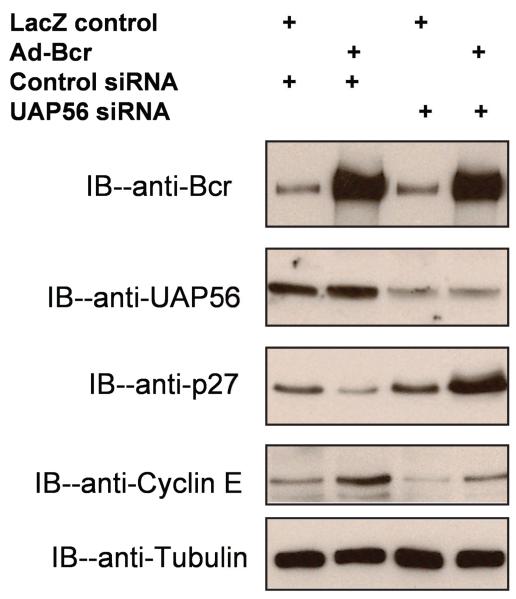
DExD/H box proteins are thought to affect cell growth via cell cycle regulation [17, 18]. For example, the RNA helicase p68 plays an important role in PDGF induced cell proliferation by up-regulating cyclin D1 expression [18]. We therefore assessed the effect of Bcr and UAP56 on the expression of several key cell cycle genes. Using WT Bcr adenovirus, we found that over expression of Bcr decreased the expression of the cyclin dependent kinase inhibitor p27 (Figure 2), a negative regulator of the cell cycle G1/S transition [19, 20]. This effect of WT Bcr was reversed by knockdown of UAP56 expression with UAP56 siRNA (Figure 2). While the RNA helicase p68 acts in part via cyclin D1 expression, we previously saw no effect of Bcr expression on cyclin D expression (unpublished data). We did find however that over expression of Bcr increased the expression of cyclin E, a positive regulator of cell cycle G1/S transition [21, 22], an effect that was reversed by knockdown of UAP56 expression with UAP56 siRNA (Figure 2).
Figure 2. Knockdown of UAP56 regulates expression of cyclin E and p27.
VSMC were transfected with UAP56 siRNA or control siRNA. 48 hours later the cells were treated with 50 MOI of Ad-Bcr or Ad-LacZ for 24 hours. Cells were harvested and Western blot was done with antibodies as listed.
Bcr binds to UAP56
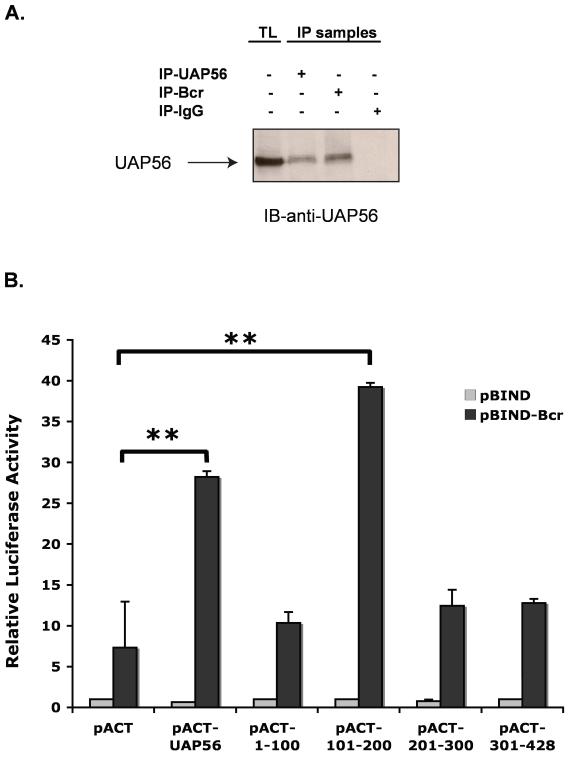
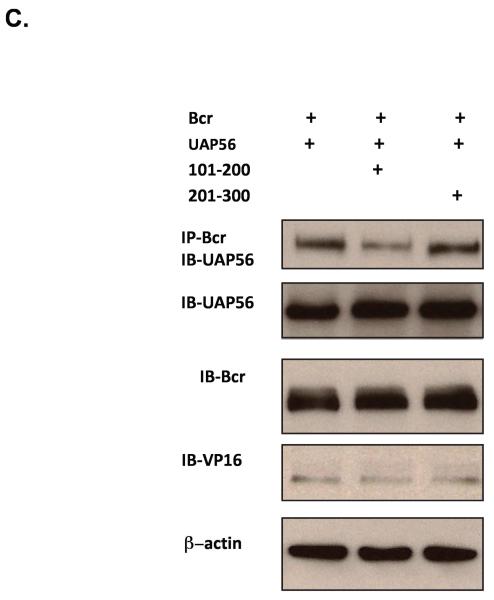
As our findings suggest an interaction between Bcr and UAP56 we next examined whether Bcr binds to UAP56. Immunoprecipitation/immunoblot studies demonstrated that Bcr does bind to UAP56 (Figure 3A). We also demonstrated that Bcr binds to UAP56 using a mammalian-two hybrid assay (Figure 3B). In addition to using full length UAP56, mammalian-two hybrid assay using UAP56 fragments (amino acids 1-100, 101-200, 201-300 and 301-428) demonstrated that fragment 2 (amino acids 101-200) of UAP56 binds to Bcr (Figure 3B). In addition, immunoprecipitation/immunoblot studies demonstrated that over expression of UAP56 fragment 2 blocked the binding of UAP56 and Bcr in HeLa cells (Figure 3C), further evidence of the importance of fragment 2 in Bcr/UAP56 binding.
Figure 3. Bcr binds to UAP56.
(A) After over expressing Bcr and UAP56 in HeLa cells, total cell lysates (TL) were prepared and UAP56 and Bcr were immunoprecipitated using respective antibodies and Western blotting was performed with UAP56 antibody. (B) (Mammalian two-hybrid assay). HeLa cells were transfected with pBIND or pBIND-Bcr and pACT UAP56 full length or UAP56 fragments. Cells were harvested and luciferase assay performed. Full length UAP56 binds to Bcr as does UAP56 fragment 2 (amino acids 101-200). (**p<0.01). (C) HeLa cells were transfected with pBIND-Bcr and pACT UAP56 full length or UAP56 fragments as indicated (the pACT vector contains the herpes simplex virus VP16 activation domain). Bcr was immunoprecipitated with Bcr antibody and immunblot done with UAP56 antibody. Over expression of UAP56 fragment 2 [amino acids (aa) 101-200] blocked Bcr/UAP56 binding. Over expression of fragment 3 (aa 201-300) had no effect.
UAP56/Bcr interaction is critical for Bcr induced DNA synthesis
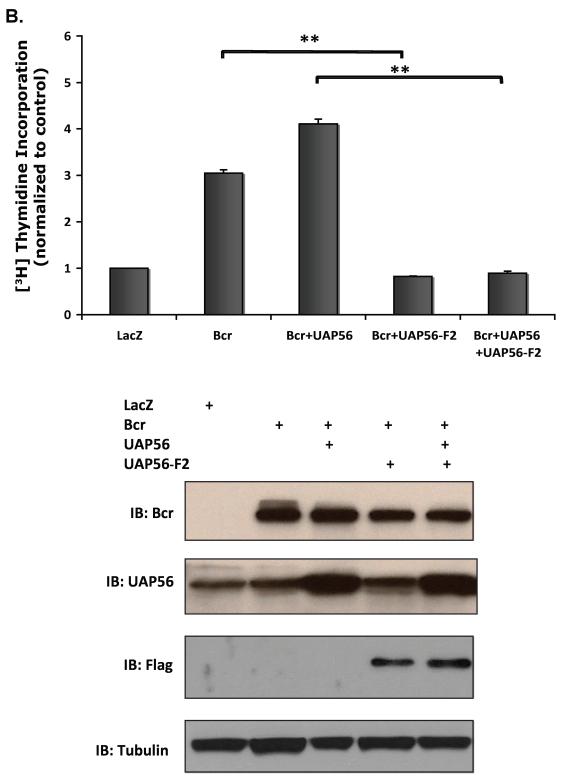
Having demonstrated that UAP56 binds to Bcr, we next examined whether the interaction between UAP56 and Bcr plays a role in Bcr induced DNA synthesis. Over expressing Bcr WT, UAP56 WT, UAP56 fragment 1 and UAP56 fragment 2 in HeLa cells, we found that over expression of UAP56 fragment 2 but not fragment 1, inhibited Bcr induced DNA synthesis (Figure 4A). Similarly, using Bcr WT, UAP56 WT, and UAP56 fragment 2 adenoviruses, we demonstrated that over expression of UAP56 fragment 2 inhibits Bcr induced DNA synthesis in VSMC (Figure 4B and 4C). These data demonstrate that UAP56/Bcr interaction is important for Bcr induced DNA synthesis.
Figure 4. Over expression of UAP56 fragment 2 inhibits Bcr induced DNA synthesis.
(A) HeLa cells were transfected with control, Bcr, and UAP56 full length or UAP56 fragment plasmids as indicated. Cells were harvested and [3H] thymidine incorporation was measured (*p<0.05). Over expression of fragment 1 (aa 1-100) had no effect. (B) VSMC were treated with Ad-Bcr, Ad-WT UAP56 , or Ad-Fragment 2 UAP56 as indicated. Cells were harvested and [3H] thymidine incorporation was measured. (**p<0.01).
DISCUSSION
The major findings of this study are that UAP56 binds to Bcr and interaction between UAP56 and Bcr is critical for Bcr induced DNA synthesis. We previously demonstrated that Bcr is a major regulator of SMC proliferation and inflammation. We demonstrated that this effect was in part via inhibition of PPARγ transcriptional activation by Bcr. Our data now show that Bcr also acts via PPARγ independent signaling. While our initial studies of the interaction between Bcr and UAP56 were in HeLa cells, using Ad-Bcr and Ad-WT UAP56 we demonstrated the importance of this interaction in VSMC as well. These findings demonstrate an important role of UAP56 in VSMC proliferation and identify UAP56/Bcr interaction as a potential target for treatment of vascular proliferative disease.
UAP56 is known to play an important role in RNA splicing, mRNA export, and protein synthesis. Our study now demonstrates that UAP56 plays an important role in DNA synthesis as well, and further defines the role of UAP56 in cellular proliferation. Yamazaki et al. recently reported that knockdown of UAP56 in HeLa cells was associated with down regulation of genes affecting the cell cycle, mitosis, mRNA transport, DNA replication, DNA repair and cell division, demonstrating the important role of UAP56 in cell growth [15]. Depletion of UAP56 causes mitotic delay and sister chromatid cohesion defects [15]. These findings suggest that UAP56 is a major control point for cell growth. UAP56 has a close homolog (URH49) which has 90% homology with UAP56 [12, 15]. Like UAP56, URH49 is also a DExD/H protein and an RNA helicase. The two helicases have different expression profiles in different tissues [12] but whether the two proteins have completely overlapping roles has been unknown. Using UAP56 and URH49 siRNA, Yamazaki et al demonstrated that similar to UAP56, URH49 is important in mitotic progression, but unlike UAP56, depletion of URH49 causes chromosome arm resolution defects and failure of cytokinesis. These findings demonstrate that UAP56 is an important regulator of mitosis, distinct from URH49. Our data extend these findings demonstrating the role of UAP56 in VSMC DNA synthesis.
Our results also further define the role of DExD/H box proteins and RNA helicases in cellular proliferation. DExD /H box proteins have been thought to control cell growth through regulation of the cell cycle but the mechanism has been unknown [17]. Yang et al. demonstrated that the RNA helicase p68 regulates PDGF induced cell proliferation by upregulating cyclin D1 and c-myc expression [18]. We have now demonstrated that UAP56 is another DExD/H box protein that is an important regulator of the cell cycle and cell proliferation. DExD /H box proteins are important in abnormal proliferation in tumors [23] and may well play an important role in pathological SMC proliferation.
In conclusion, the data presented demonstrate that UAP56 is a novel partner of Bcr in regulating VSMC DNA synthesis. This effect may occur in part due to UAP56-dependent modulation of cell cycle progression. Bcr/UAP56 interaction may be a target for inhibiting pathological VSMC proliferation.
Supplementary Material
UAP56 is an important regulator of DNA synthesis in vascular smooth muscle cells.
UAP56 binds to Bcr.
Interaction between Bcr and UAP56 is critical for Bcr induced DNA synthesis.
Acknowledgement
The authors thank Dr. Alan Friedman for his assistance with mass spectrometry.
Sources of Funding This study was supported by a grant from the National Institutes of Health to Dr. Alexis (HL80938).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Fujita N, Furukawa Y, Itabashi N, Okada K, Saito T, Ishibashi S. Differences in E2F subunit expression in quiescent and proliferating vascular smooth muscle cells. Am J Physiol Heart Circ Physiol. 2002;283:H204–12. doi: 10.1152/ajpheart.00545.2001. [DOI] [PubMed] [Google Scholar]
- [2].Jackson CL, Schwartz SM. Pharmacology of smooth muscle cell replication. Hypertension. 1992;20:713–36. doi: 10.1161/01.hyp.20.6.713. [DOI] [PubMed] [Google Scholar]
- [3].Bunkenburg B, van Amelsvoort T, Rogg H, Wood JM. Receptor-mediated effects of angiotensin II on growth of vascular smooth muscle cells from spontaneously hypertensive rats. Hypertension. 1992;20:746–54. doi: 10.1161/01.hyp.20.6.746. [DOI] [PubMed] [Google Scholar]
- [4].Naftilan AJ, Pratt RE, Dzau VJ. Induction of platelet-derived growth factor A-chain and c-myc gene expressions by angiotensin II in cultured rat vascular smooth muscle cells. J Clin Invest. 1989;83:1419–24. doi: 10.1172/JCI114032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Alexis JD, Wang N, Che W, Lerner-Marmarosh N, Sahni A, Korshunov VA, Zou Y, Ding B, Yan C, Berk BC, Abe J. Bcr kinase activation by angiotensin II inhibits peroxisome-proliferator-activated receptor gamma transcriptional activity in vascular smooth muscle cells. Circ Res. 2009;104:69–78. doi: 10.1161/CIRCRESAHA.108.188409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Spies T, Blanck G, Bresnahan M, Sands J, Strominger JL. A new cluster of genes within the human major histocompatibility complex. Science. 1989;243:214–7. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- [7].Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–72. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- [8].Allcock RJ, Williams JH, Price P. The central MHC gene, BAT1, may encode a protein that down-regulates cytokine production. Genes Cells. 2001;6:487–94. doi: 10.1046/j.1365-2443.2001.00435.x. [DOI] [PubMed] [Google Scholar]
- [9].Shen J, Zhang L, Zhao R. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem. 2007;282:22544–50. doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- [10].Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–8. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- [11].Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–21. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- [12].Kapadia F, Pryor A, Chang TH, Johnson LF. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene. 2006;384:37–44. doi: 10.1016/j.gene.2006.07.010. [DOI] [PubMed] [Google Scholar]
- [13].Sahni A, Wang N, Alexis JD. UAP56 is an important regulator of protein synthesis and growth in cardiomyocytes. Biochem Biophys Res Commun. 2010;393:106–10. doi: 10.1016/j.bbrc.2010.01.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008;22:1796–803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yamazaki T, Fujiwara N, Yukinaga H, Ebisuya M, Shiki T, Kurihara T, Kioka N, Kambe T, Nagao M, Nishida E, Masuda S. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol Biol Cell. 21:2953–65. doi: 10.1091/mbc.E09-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kota KP, Wagner SR, Huerta E, Underwood JM, Nickerson JA. Binding of ATP to UAP56 is necessary for mRNA export. J Cell Sci. 2008;121:1526–37. doi: 10.1242/jcs.021055. [DOI] [PubMed] [Google Scholar]
- [17].Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–15. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang L, Lin C, Zhao S, Wang H, Liu ZR. Phosphorylation of p68 RNA helicase plays a role in platelet-derived growth factor-induced cell proliferation by up-regulating cyclin D1 and c-Myc expression. J Biol Chem. 2007;282:16811–9. doi: 10.1074/jbc.M610488200. [DOI] [PubMed] [Google Scholar]
- [19].Attwooll C, Lazzerini E. Denchi, Helin K. The E2F family: specific functions and overlapping interests. EMBO J. 2004;23:4709–16. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–6. doi: 10.1038/nrm2510. [DOI] [PubMed] [Google Scholar]
- [22].Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153–66. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- [23].Yang L, Lin C, Liu ZR. Phosphorylations of DEAD box p68 RNA helicase are associated with cancer development and cell proliferation. Mol Cancer Res. 2005;3:355–63. doi: 10.1158/1541-7786.MCR-05-0022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.