Abstract
Motile cilia create asymmetric fluid flow in the evolutionarily conserved ciliated organ of asymmetry (COA) and play a fundamental role in establishing the left-right (LR) axis in vertebrate embryos. The transcriptional control of the large group of genes that encode proteins that contribute to ciliary structure and function remains poorly understood. In this study we find that the winged helix transcription factor Rfx2 is expressed in motile cilia in mouse and zebrafish embryos. Morpholino knockdown of Rfx2 function in the whole embryo or specifically in cells of the zebrafish COA (Kupffer’s Vesicle, KV) leads to reduced KV cilia length and perturbations in LR asymmetry. LR patterning defects include randomization of the early asymmetric Nodal signaling pathway genes southpaw, lefty1 and lefty2 and subsequent reversals in the organ primordia of the heart and gut. Rfx2 is also required for ciliogenesis in zebrafish pronephric duct. We further show that by restoring Left-Right dynein (LRD) expression and motility specifically in a subset of ciliated cells of the mouse COA (posterior notochord, PNC), we can restore fluid flow, asymmetric expression of Pitx2 and partially rescue situs defects.
Introduction
Cilia are microtubule-based organelles found on the surface almost all cell types in vertebrates, where they carry out diverse sensory functions including the detection of light odor and fluid flow, or are motile and are responsible for moving cells over substrates or moving fluids over epithelial surfaces. Some cilia are both motile and sensory (Andrade et al., 2005; Shah et al., 2009). Cilia also play important roles in signal transduction and cellular homeostasis, regulating among other things intracellular calcium levels, and Wnt and Hedgehog signaling pathways (Gerdes et al., 2009). Defective cilia underlie a wide range of human disorders ranging from those localized to single tissues or organs, such as polycystic kidney disease and congenital heart defects, to others that are broadly pleiotropic, like BBS or laterality defects such as situs inversus (Bisgrove and Yost, 2006; Goetz and Anderson, 2010; Ishikawa and Marshall, 2011; Tobin and Beales, 2009).
The basic scaffold, the ciliary axoneme, is composed of nine microtubule doublets surrounded by the ciliary membrane. An additional central pair of inner microtubules is present in some types of cilia and absent in others. As protein synthesis does not occur in cilia, ciliary proteins are transported into and out of the cilium by kinesin and dynein motors and associated proteins in a process termed intraflagellar transport (IFT) (Pedersen et al., 2008; Rosenbaum and Witman, 2002). Specificity of ciliary function is governed in part by the selection of which motor, signaling and/or channel proteins are delivered into the cilium (Evans et al., 2006; Mukhopadhyay et al., 2007). Several bioinformatic, genomic and proteomic studies have identified a large number of proteins, perhaps as many as 1200, that are intimately involved in ciliary structure and function (Gherman et al., 2006).
Cilia play a vital role during early development in patterning the vertebrate embryo. Left-right asymmetry is established at the posterior notochord (PNC) in mouse (Supp et al., 1997), the notochordal plate in rabbit (Okada et al., 2005), gastrocoel roof plate in Xenopus (Schweickert et al., 2007) and Kupffer’s vesicle in zebrafish and Medaka (Essner et al., 2005; Kramer-Zucker et al., 2005; Okada et al., 2005). This “Ciliated Organ of Asymmetry” (COA) (Essner, Amack et al. 2005) comprises a small (50–250 cells) population of ciliated cells. Motile cilia in the COA are a specialized type of monocilia that usually have a 9+2 microtubule architecture and dynein-driven motility (Caspary et al., 2007; Feistel and Blum, 2006). Rotational beating of motile cilia generates directional leftward flow of extracellular fluid in the COA, which results in the asymmetric activation of the NODAL signaling cascade genes which are critical in breaking the early bilateral symmetry of the embryo (Nonaka et al., 1998a; Supp et al., 1999). The mechanism by which fluid flow across the COA triggers asymmetric gene expression remains unclear. Two models based on work in mice have been proposed: The ‘morphogen flow’ model proposes that signaling proteins such as FGF, SHH or NODAL, perhaps concentrated on small extracellular vesicles, are swept to the left side of the node where they are concentrated and trigger downstream signaling events (Nonaka et al., 1998a; Okada et al., 1999; Okada et al., 2005; Tanaka et al., 2005). A second ‘two cilia’ model postulates that two populations of cilia exist in the mouse COA and that fluid flow generated by a central group of LRD-expressing cilia is detected by mechanical bending of a second non-motile group of cilia on the periphery of the COA, which initiates a Ca2+ mediated signaling event (McGrath et al., 2003) leading to subsequent asymmetric expression of genes including Cerl2, Nodal and Pitx2 (Shiratori and Hamada, 2006). It appears that the teleost Kupffer’s Vesicle (KV) contains only motile cilia (Kramer-Zucker et al., 2005). Recent work from Kamura et al. suggests that the motile KV cilia in medaka fish have a dual function: to generate fluid flow and interpret it through Pkd1l1-Pkd2 sensor complexes associated with the cilia (Kamura et al., 2011).
Cell- and tissue-specific transcriptional control of distinct ciliary functions is only beginning to be understood (Thomas et al., 2010). The RFX family of transcription factors is defined by a wing-helix structure DNA binding domain that recognizes the X-Box promoter motif (Dorn et al., 1987; Emery et al., 1996; Sherman et al., 1987). Members of the RFX family have been implicated in transcriptional control of ciliogenesis in Drosophila, C. elegans, zebrafish and mouse (Bonnafe et al., 2004; Dubruille et al., 2002; Liu et al., 2007; Ma and Jiang, 2007; Swoboda et al., 2000). In Drosophila and C. elegans a single X-box transcription factor (dRFX and daf-19 respectively) regulates ciliogenesis in sensory neurons via transcriptional regulation of intraflagellar transport (IFT) proteins (Blacque et al., 2005; Dubruille et al., 2002; Efimenko et al., 2005; Li et al., 2004; Swoboda et al., 2000). In contrast, there are seven vertebrate RFX family members (RFX1–7) which can be categorized into three phylogenetic groups: RFX1–3; RFX4, 6; and RFX5, 7 (Aftab et al., 2008). The members are differentially expressed and have diverse functions, with RFX2, RFX3 and RFX4 linked to ciliogenesis. Unlike what is observed in invertebrates, loss of any single RFX transcription factors does not necessarily lead to loss of cilia, suggesting some redundancy in RFX function. RFX4 is expressed in neural tube and limb, and modulates hedgehog signaling by a regional effect on ciliogenesis (Ashique et al., 2009). RFX3 is more broadly expressed, including in ciliated cells of the embryonic COA, brain, lung, endocrine pancreas and testes (Baas et al., 2006; Bonnafe et al., 2004; El Zein et al., 2009). RFX3 has been shown to regulate the expression of a number of ciliary motility genes including the axonemal dynein heavy chains Dnahc5, 9 and 11. Mutation of RFX3 in mice results in congenital hydrocephalus, diabetes and defects in LR development correlated with stunting of motile cilia (Bonnafe et al., 2004). These observations led us to investigate whether the related transcription factor RFX2 played a significant role in the regulation ciliogenesis in the COA and the establishment of LR asymmetry in mice and zebrafish.
Here we show that Rfx2 is expressed at the COA in zebrafish and mouse. RFX2 is essential for development of left-right asymmetry: knockdown of Rfx2 in zebrafish results in shortened Kupffer’s vesicle cilia and subsequent randomization of heart and gut looping. Strikingly, Rfx2-expressing cells in the mouse embryo define a subpopulation of COA ciliated cells. Re-activation of LRD within this subpopulation is capable of generating adequate nodal flow for normal LR development, serving as the first demonstration that LRD function and cilia motility exclusively in COA cells is sufficient for mouse LR development. In summary, Rfx2 transcriptional regulation specifies motile cilia in the COA that are necessary and sufficient for the development of LR asymmetry.
Materials and Methods
Mice
Mice carrying the Lrd-GFPΔneo allele were described previously (McGrath et al 03) and carried on a mixed 129SVJ; C57BL/6J background. Lrd-GFP mice were produced by crossing Lrd-GFPΔneo mice with Actin-Cre mice as described previously; the Lrd-GFP mice are maintained as Lrd-GFP homozygous animals.
Rfx2-Cre mice were created by the Yale animal genomics service. The Rfx2-containing 280 kb BAC (Clone RP23-258K4) was obtained from BACPAC Resource Center (Children’s Hospital Oakland Research Institute). The Cre recombinase coding sequence was inserted at the start codon of Rfx2 by homologous recombination. PCR and Southern blotting confirmed correct insertion of the Cre recombinase into the Rfx2 gene. The resulting Rfx2-Cre BAC was introduced into (C57BL/6J X SJL/J) F2 embryos by pronuclear injection. Two Cre positive founder lines were created, bred to homozygosity and analyzed for Cre expression by crossing with reporter mice ROSA26 and R26-EYFP.
Zebrafish
Zebrafish, Danio rerio, were maintained at 28.5°C on a 14 hour/10 hour light/dark cycle. Embryos obtained from natural matings of Oregon AB strain zebrafish (Zebrafish International Resource Center, Oregon), were cultured and staged by developmental time and morphological criteria (Westerfield, 1995).
Putative homologues of mouse Rfx2 are present on zebrafish chromosome 8 in a region syntenic with mouse chromosome 17 that contains Rfx2. RT-PCR was used to generate cDNAs from late gastrula, or 24 hours post-fertilization (hpf) mRNA using the Superscript First Strand Synthesis System (Invitrogen). PCR products were ligated into pCRII TOPO (Invitrogen) and sequenced. Zebrafish rfx2 (GenBank BX664738) has two alternatively spliced protein isoforms of 704 and 734 amino acids differing in the inclusion or absence of the second translated exon. The predicted amino acid sequence of this exon is absent from the short (698 amino acids) and long (723 amino acids) protein isoforms of the mouse homologue. Zebrafish Rfx2 isoforms share 72% amino acid identity with those of mouse.
Morpholino knock-down of Rfx2 function and rescue with rfx2 RNA
Morpholino antisense oligonucleotides (Gene Tools, Oregon) were used to knock-down function of both Rfx2 splice isoforms in zebrafish embryos. Rfx2 translation-blocking morpholino (rfx2 AUGMO) is complementary to a region spanning the translation start site, 5’-CCTTCTGACCGCTGCATGGTTTCTC-3’, and rfx2 splice-blocking morpholino (rfx2 SBMO) 5’-TTGTGCTACTGAGTCTGCTCACTTG-3’overlaps the splice donor site of the first coding exon. A standard negative control morpholino, 5’-CCTCTTACCTCAGTTACAATTTATA-3’, was also used. All morpholinos were tagged with fluorescein to facilitate tracking their distribution in injected embryos.
Morpholinos were injected into the yolk under the blastoderm of 1- to 2-cell stage, mid-blastula or late blastula stages in a volume of 0.5 or 1.0 nl at the following doses: rfx2 AUGMO, 1 ng; rfx2 SBMO, 8 ng; control MO, 3 ng. To test the efficacy of the splice-blocking morpholino, RT-PCR was carried out on total RNA prepared from embryos injected with 8 ng of rfx2 SBMO (Supplemental Fig. 1). Sequencing of these RT-PCR products indicates that this morpholino blocks splicing at the splice donor site of the first coding exon (exon 2) and causes the retention of intronic sequences which encode 8 amino acids then a termination codon, predicted to truncate this chimeric protein at 43 amino acids (data not shown) which should lead to a substantial decrease in functional gene product.
To produce a rescuing RNA we amplified full length wild-type rfx2 using a 5’ primer (5’-GGTGGCGCCATGCAACGATCAGAA G-3’) that substituted 6 of the 25 bases targeted by the rfx2 AUG morpholino at the translation initiation site and cloned the product into pCS2+. Capped, in vitro synthesized RNA was produced from this ‘morpholino resistant’ cDNA using the mMessage Machine SP6 transcription kit (Ambion). Wild-type control and rfx2 AUGMO embryos were injected with 50 pg of RNA and scored at 48 hpf for orientation of the heart and gut.
Whole-mount RNA in situ localization
Riboprobes were synthesized from linearized DNA templates using SP6 or T7 polymerases and digoxigenin labeling mixes (Roche). In situ hybridizations of zebrafish embryos were carried out as previously described (Bisgrove et al., 1999). Anti-sense probes used include: rfx2 (this report); lefty1 (lft1), lefty2 (lft2) (Bisgrove et al., 1999); southpaw (spaw) (Long et al., 2003); cardiac myosin light chain 2 (cmlc2 or my17) (Yelon et al., 1999); foxa3 (fkd2) (Odenthal and Nusslein-Volhard, 1998); myod1 (Weinberg et al., 1996); Embryos were cleared in 70% glycerol/PBST and photographed with a Leica MZ12 dissecting microscope. Images were captured with a Nikon Coolpix 5000 digital camera and processed using Adobe Photoshop.
In-situ hybridizations of mouse embryos were carried out according as described previously (Lowe et al 96). Pitx2 probe was transcribed in vitro from a partial Pitx2 cDNA clone between primers 1Pitx2 (ACATGTAACTCGAGCCTGGC) and 2Pitx2 (ACAGGATGGGTCGTACATAGC) covering 3’ coding and 3’UTR sequence. The template for Nodal probe (Nodal cDNA missing the first 20 nt of the coding region) was obtained from M. Kuehn. Rfx2 probe spans the coding region of the cDNA between primers F (CGCAGCATCTTACTTTGAGACTC) and R (AAGACGTGGGACACATCTAGGTAC).
Immunohistochemistry
Mouse embryos were harvested from timed pregnancies. For immunohistochemistry (IHC) mouse and zebrafish embryos were fixed in 4% paraformaldehyde/PBS or overnight at 4°C. Mouse COA, zebrafish KV and pronephric duct cilia were labeled by immunohistochemistry with anti-acetylated tubulin antibody (1:300, Sigma), anti-phosphokinase C ζ (1:100, Santa Cruz Biotechnology) and fluorescent secondary antibodies (1:300, Molecular Probes) as previously described (Bisgrove et al., 2005). Zebrafish embryos were imaged on an Olympus Fluoview scanning laser confocal microscope, and images were processed using Image J software. Mouse embryos were imaged on a Zeiss Axiovert microscope equipped with Apotome optical interference imaging to obtain optical sections.
Live embryo imaging
Eight somite-stage zebrafish embryos were mounted with the tail bud oriented up in 1.5% low-melt agarose and the KV injected with a 1/50 dilution of Fluoresbrite polychromatic red 0.5 micron microshperes (Polysciences Inc.) The motions of these fluorescent beads were imaged at 60× on a Leica DMRA epifluorescence microscope equipped with a Coolsnap HQ video camera recording at 5 frames/sec. Five beads from each KV analyzed were tracked for approximately six seconds each using Metamorph software (Molecular Devices Corp.). Average bead velocities in 6 un-injected control KVs and 7 rfx2 AUGMO KVs were calculated and analyzed statistically with a Student’s two-tailed t-test.
E8.0 mouse embryos were dissected in DMEM with 5% FBS, incubated in 100% rat serum for 30 min in a tissue culture incubator at 37°C. For imaging of nodal flow, the embryo was positioned node up in the homemade imaging chamber containing 0.5 µm fluorescent beads (Molecular Probes) diluted 1:500 in 50% DMEM, 50% rat serum, covered with a #1 cover slip and visualized with a Zeiss Axiovert microscope and a 20X objective. Sequences of images were acquired with an AxioCam HRc at a rate of 15 frames per second with binning 3×3. The whole embryo was removed from the imaging chamber and PCR genotyped.
For visualization of GFP labeled cells in mouse embryos, Rfx2-Cre homozygous males or females were crossed with R26-EYFP mice. Embryos were harvested in DMEM, 5% FBS and incubated in 100% rat serum at 37°C. The imaging was performed as described above for nodal flow, except that the embryo was maintained during imaging in 100% rat serum on an environment-controlled stage at 37°C in 5% CO2. Z-stacks of images were acquired every 15–30 min over course of 6–8 hours with the AxioCam MRm. Obtained Images were processed using Axiovision 4.8 software from Carl Zeiss.
Results
Rfx2 expression patterns in mouse and zebrafish embryos
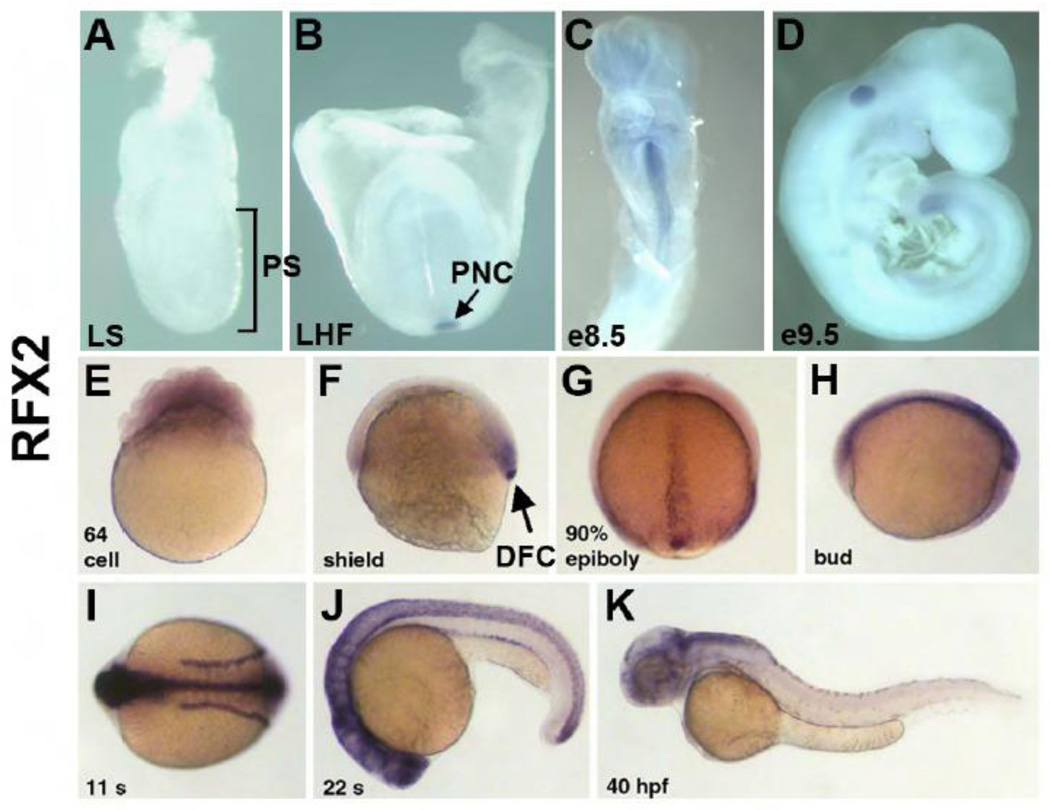
To begin to characterize the role of RFX2 in development, we analyzed the expression pattern of Rfx2 in mouse embryos from e7.5 through e9.0 and in zebrafish embryos from early cleavages stages through 2 days post-fertilization. Expression analyses of Rfx2 were performed by whole-mount in situ hybridization (WMISH) on mouse and zebrafish embryos using digoxigenin-labeled antisense probes. Sense stand controls were negative for expression (data not shown). mRNA for mouse Rfx2 was first detected in the posterior notochord (PNC) of the late headfold-stage embryo (pre-somite, Theiler stage 10) (Fig.1A and B), and then persisted at the midline at the turning stage (10 somites, Theiler stage 13) (Fig. 1C). Detection of Rfx2 mRNA with antisense ribobrobe followed by immunolabeling with anti-Arl13B antibodies (Supplemental Fig. 2A) shows that Rfx2mRNA localizes to cells at the posterior margin of the COA. This is also observed by two-color WMISH using Rfx2 and nodal antisense riboprobes (Supplemental Fig. 2B). After turning, Rfx2 disappeared except at the most posterior region of the tail (Fig. 1D). For comparison we also examined Rfx3 expression, a gene previously described to play a role in ciliogenesis in the mouse embryos (Bonnafe et al., 2004). Rfx3 expression occurred earlier than Rfx2 in the primitive streak, was more diffuse (Supplemental Fig. 3A), and persisted in the PNC of the late headfold embryo (Supplemental Fig. 3B). In the early somite embryo, Rfx3 was restricted to the head region (Supplemental Fig. 3C). After turning, low-level Rfx3 expression persisted throughout the embryo (Supplemental Fig. 3D). The earlier onset and more diffuse expression of Rfx3 in mouse indicate that it might regulate ciliogenesis in a broader range of cells than Rfx2.
Fig. 1.
Mouse and zebrafish homologues of rfx2 are expressed in ciliated organs of asymmetry and other ciliated tissues. In situ localization of mouse Rfx2 transcripts indicated no expression in late streak (LS) or earlier stage embryos (A). Expression was first detected in the posterior notochord (PNC) at late headfold stage (B), persists in the midline through e8.5 (C) and was downregulated in all but the posterior tail by e9.5 (D). In situ localization of zebrafish rfx2 indicated that transcripts are present maternally (E). At shield stage zygotic expression was limited to mesendoderm at the margin and dorsal forerunner cells (DFC) (F). During late epibiloy (G) and bud stages (H) rfx2 transcripts are restricted to the embryonic midline. During somite stages, expression was restricted to the neural tube (NT) and pronephric ducts (PND) (I,J) and becomes down-regulated everywhere except regions of the dorsal brain by 40 hr post-fertilization (K).
In zebrafish, rfx2 expression patterns in several tissues were consistent with mouse Rfx2 expression. Zebrafish rfx2 was expressed maternally and was detectable at early cleavage stages in the embryonic blastomeres (Fig. 1E). Zygotic rfx2 expression began during gastrulation at shield stage when transcripts were localized to mesendoderm in the germ band and in the dorsal forerunner cells (DFCs, Fig. 1F). During subsequent gastrulation stages rfx2 localized to the dorsal midline in cells that give rise to the spinal floorplate (Fig. 1G). At bud stage, prior to somitogenesis, DFCs aggregate to become Kupffer’s Vesicle (KV), the zebrafish ciliated organ of asymmetry (Cooper and D'Amico, 1996; Essner et al., 2005). At this stage rfx2 was expressed in KV and along the midline in the neural tube (Fig. 1H). Rfx2 expression in KV was extinguished by about the 3 somite stage, prior to inflation of KV and KV ciliogenesis, but continued to be expressed throughout the neural tube as well as in the pronephric ducts during somitogenesis (Fig. 1I, J). Expression gradually disappeared, and by 40 hours post-fertilization (hpf) remained only in limited regions of the brain (Fig. 1K).
In summary, Rfx2 in mouse and zebrafish was expressed in a related set of tissues including the midline and importantly the PNC and KV, the ciliated organs of asymmetry (COA) in the two species. This conservation of expression patterns is consistent with a conserved function for RFX2 in regulating expression of ciliary genes.
Rfx2 is required for early LR patterning in zebrafish
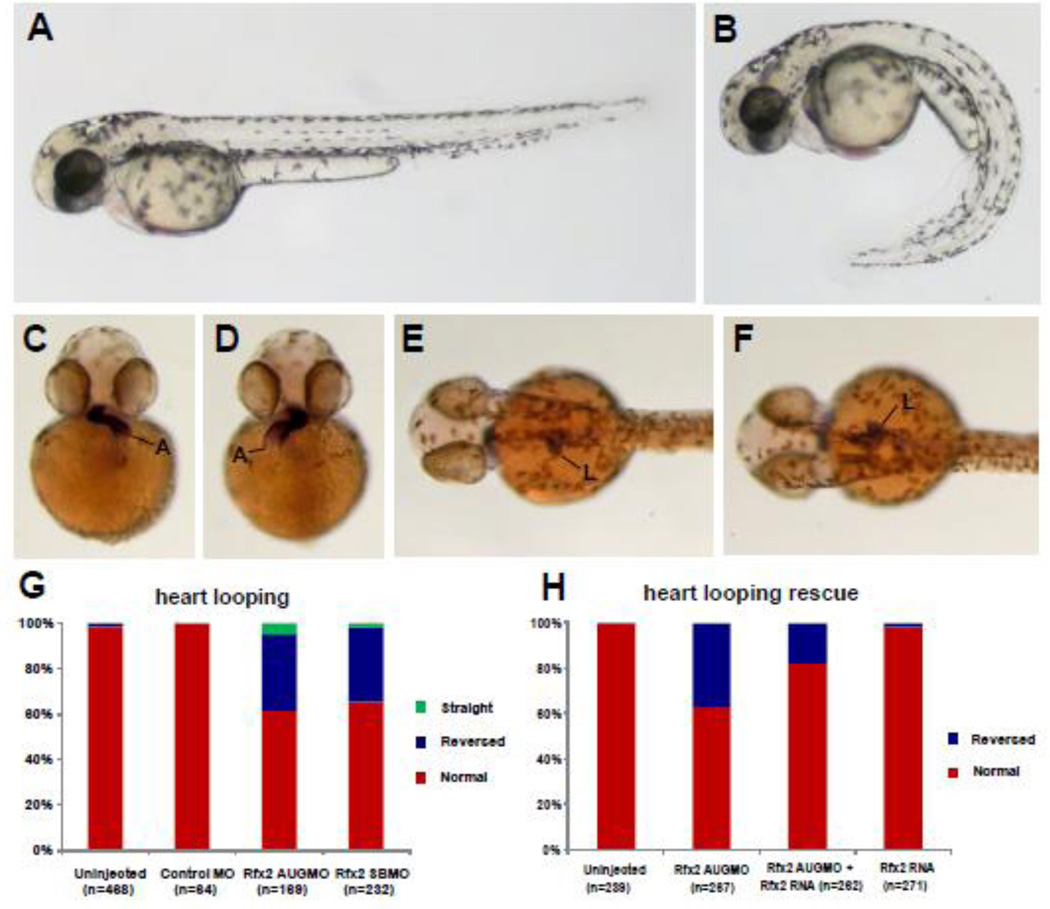
To analyze the role of Rfx2 in zebrafish development we knocked down gene function by injecting rfx2 morpholinos into 1–2 cell embryos. Both translation-blocking (rfx2 AUGMO) and splice-blocking (rfx2 SBMO) morpholinos gave similar phenotypes. The most obvious external morphological phenotype of rfx2 morphants was a strong ventral curvature of the trunk and tail (Fig. 2A, B). In normal embryos at 42 hpf the atrium of the heart loops to the left while the ventricle loops to the right, and the liver and intestinal bulb lie on the left side of the midline (Fig. 2C, E). In rfx2 morphants the orientation of heart and gut looping were frequently reversed (Fig. 2D, F). Organ reversal rates in un-injected embryos or embryos injected with Control Morpholino (Gene Tools, Inc) were less than 3%, while approximately 35% of rfx2 morphants had reversed orientation of the heart, with similar reversal rates in gut looping direction (Fig. 2G; Supplemental Fig. 4A). The frequency of heart reversal in rfx2 AUGMO embryos was rescued to approximately 18% by injection of in vitro synthesized rfx2 mRNA (Fig. 2H), with a similar rescue of gut looping (Supplemental. Fig. 4B). Injection of rfx2 RNA into wild-type embryos had no effect on organ laterality. In individual embryos, the orientation of heart and gut looping were almost always concordant.
Fig. 2.
Knockdown of rfx2 gene function results in altered body shape, and LR patterning defects in heart and gut. Rfx2 morphants (B) exhibited a strongly ventrally curved trunk and tail compared to un-injected wild-type embryos (A). The direction of heart and gut looping were often reversed in rfx2 morphants (C–H). In a wild-type embryo, the atrium of the heart loops toward the left side of the embryo (C) and the liver and intestinal bulb are located on the left side of the embryo (E) while in rfx2 morphants the position of these organs was often reversed (D, F). B, D, F – rfx2 AUGMO. Histogram depicting the percentage of heart and gut reversals in wild-type and rfx2 morphant embryos (G). The incidence of gut reversals in rfx2 AUGMO embryos was partially rescued by injection of rfx2 RNA. (H) Histogram depicting the percentage of heart reversals in wild-type and rfx2 AUGMO embryos injected with rfx2 RNA.
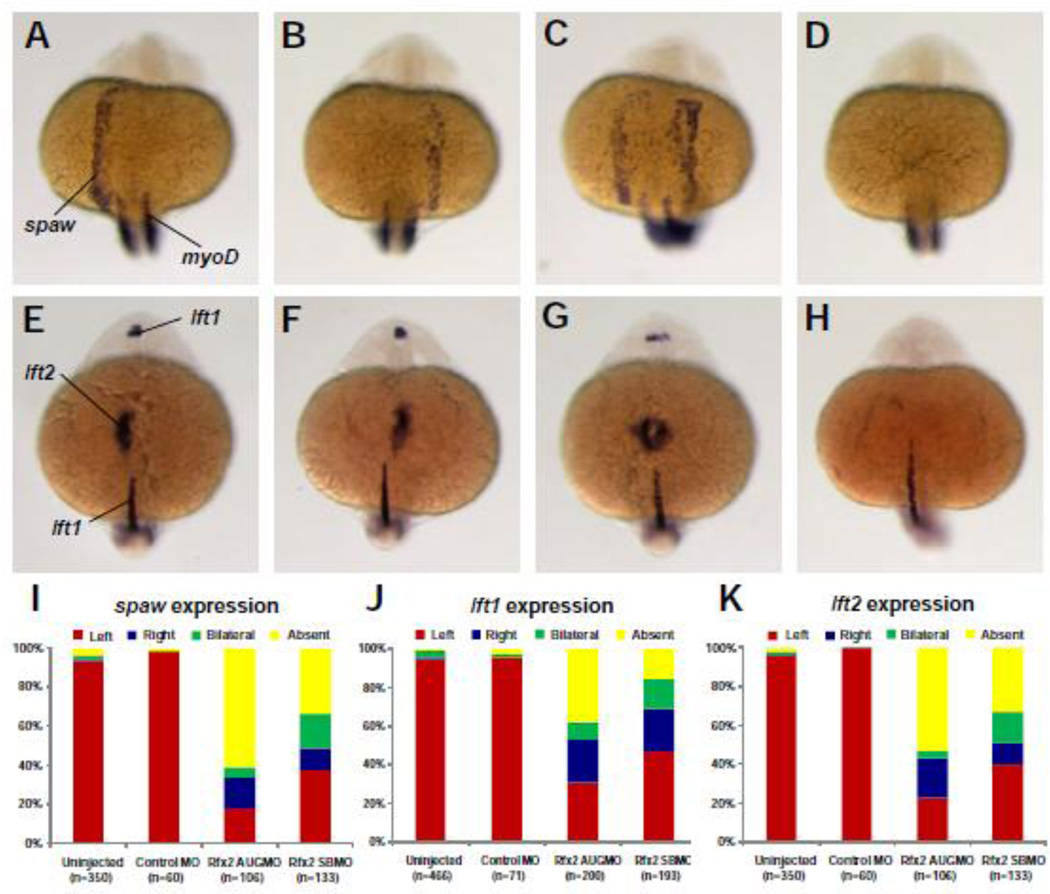
We assessed whether Rfx2 knockdown altered asymmetric gene expression, corresponding with alterations in organ asymmetries. Nodal signaling pathway genes southpaw (spaw), lefty1 (lft1), and lefty2 (lft2) are normally expressed only on the left side of the embryo (Fig. 3A–H) (Bisgrove et al., 1999; Bisgrove et al., 2000; Long et al., 2003). At the 19–21 somite stage spaw was normally expressed in the tailbud and asymmetrically in the left lateral plate mesoderm (LPM) (Fig. 3A). In contrast, spaw expression in the LPM of rfx2 morphants was randomized, with individual embryos displaying reversed, bilateral or absent spaw expression patterns (Fig. 3B–D). In embryos injected with either a translation-blocking (rfx2 AUGMO) or splice-blocking (rfx2 SBMO), 31 and 45% of morphant embryos respectively had normal left-sided spaw expression, while the remainder had right-sided, bilateral or no spaw expression in the LPM (Fig. 3I). In un-injected embryos and embryos injected with control morpholino, spaw was expressed in the left lateral plate mesoderm (LPM) in greater than 95% of embryos. The bilateral domain of spaw in the tailbud (Long et al., 2003) was unaffected in morphants.
Fig. 3.
Rfx2 functions upstream of early asymmetric LPM genes. In 19–21 somite-stage rfx2 morphants spaw was expressed in the lateral plate mesoderm on the left side (A, typical of wt embryos) as well as on the right side (B), bilaterally (C), or was not expressed (D). At the 22–24 somite-stage, the two lefty genes are expressed asymmetrically, with lft1 predominantly in the left diencephalon and lft2 expressed in the left heart field. Individual rfx2 morphants displayed this normal left-sided expression (E), or aberrant right-sided expression in both the diencephalon and heart field (F), bilateral expression in both (G), or an absence of expression (H). Midline expression of lft1 was normal in morphants. myoD expression in somites was used to stage embryos. Embryos shown are rfx2 AUGMO; rfx2 SBMO are similar. Histograms depicting the percentages of spaw (I), lft1 (J) and lft2 (K) expression patterns in wild-type and rfx2 morphant embryos.
Expression of the zebrafish lefty-related genes was also randomized in rfx2 morphants (Fig. 3E–H). Greater than 93% of un-injected embryos and embryos injected with control morpholino had normal, left-sided expression of lft1 in the left diencephalon, and lft2 in the left LPM (Fig. 3J, K). In rfx2 AUG and splice blocking morphants respectively, left-sided expression of lft1 was observed in 18–38% of embryos and left-sided expression of lft2 was observed in 23 and 40% of embryos. The midline, a source of left-right patterning information (Bisgrove et al., 2000), remained intact in these morphants as evident by the normal expression of lft1 (Fig. 3E–H) in the notochord. Expression of other midline marker genes including no tail in the notochord and sonic hedgehog in the floorplate was also normal in rfx2 morphants (data not shown). In summary, altered expression patterns of spaw, lft1 and lft2 were similar using two distinct rfx2 morpholinos, and were consistent with the number of reversals in heart and gut looping, indicating that rfx2 functions upstream of the Nodal/Lefty signaling pathway and, in turn, of organ laterality.
Rfx2 is important in Kupffer’s vesicle cilia
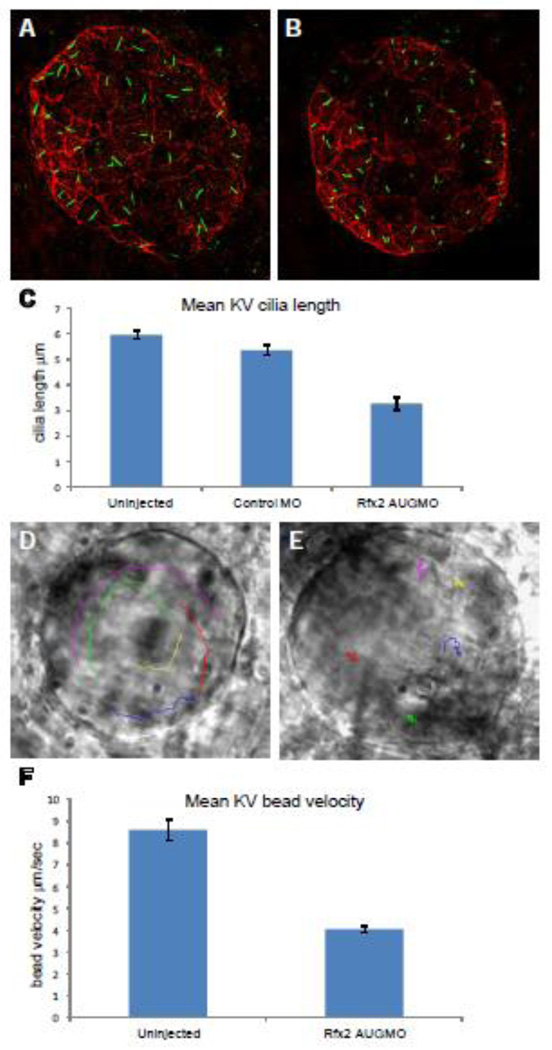
Kupffer’s vesicle in zebrafish functions as a ciliated organ of asymmetry (COA) to set up asymmetric Nodal signaling pathway in the left LPM, in an analogous manner to that proposed for the mouse PNC in the establishment of the LR axis (Essner et al., 2005). To determine whether rfx2 morphants had defects in KV formation or KV ciliogenesis, we used anti-acetylated tubulin immunohistochemistry to label cilia and anti-phosphokinase C ζ to label KV cell membranes in un-injected embryos, and embryos injected at the 1–2 cell stage with Control MO or rfx2 AUGMO. At the 8-somite stage KV is a hollow sphere lined with long cilia (Fig. 4A, B). Since the number of cilia varies with KV size and stage, we limited our analysis to 8–9-somite-stage embryos with similar size KV. Analysis of KV cilia at the 8–9 somite-stage in un-injected embryos, Control MO and rfx2 AUGMO morphant embryos showed that that un-injected and Control MO embryos had similar mean cilia number/KV (42.2 and 43.2, respectively) and lengths of KV cilia (5.97 µm and 5.34 µm, n=9 embryos each). In rfx2 AUG morphants, mean cilia number/KV was normal (41.6, n=9 embryos) but mean cilia length was significantly reduced (3.28 µm; p < 0.0001) (Fig. 4A, B, C), indicating that Rfx2 function is required for KV ciliogenesis.
Fig. 4.
Rfx2 knockdown disrupts ciliogenesis in KV and KV fluid flow. Cilia within Kupffer’s vesicle of 8–9 somite-stage embryos injected with control morpholino averaged 5.34 um in length (A) while those in embryos injected with rfx2 AUGMO were significantly shorter, averaging 3.28 um in length (B,). (C) Histogram of mean cilia length of KV cilia in un-injected, control MO and rfx2 AUGMO embryos (9 embryos each, analysis by one-way ANOVA, error bars are standard error) Fluorescent beads injected into KV of 8 somite-stage control embryos move in a counterclockwise direction (D; Supplemental Video 1A). In rfx2 AUGMO embryos there is no net directional flow and injected beads follow a random trajectory, often reversing direction (E; Supplemental Video 1B). The embryos in panels D, E are oriented with the embryonic notochord in the lower right-hand corner of the image. (F) Histogram of mean bead velocities in un-injected control embryos and rfx2 AUGMO embryos (6 control embryos, 7 rfx2AUGMO; 5 beads tracked in each, analysis by Student’s two tailed t-test, error bars are standard error).
To analyze the effect of shortened KV cilia on directional fluid flow we tracked the movement of fluorescent beads injected into the KV lumen (Fig. 4D, E; Supplemental Video 1A, B). In wild-type control embryos injected beads exhibited a counterclockwise flow with a mean velocity of 8.6 um/sec (6 embryos, 5 beads each) whereas in rfx2 AUGMO embryos beads exhibited a random motion and significantly reduced (p<0.0001) mean velocity of 4.1 um/sec (7 embryos, 5 beads each) (Fig. 4F).
rfx2 morpholinos targeted to COA cells perturb LR patterning and heart and gut orientation
To determine whether the LR defects in rfx2 morphants are due to perturbations in gene function specifically in ciliated KV cells, or whether disruptions of LR are due to functions in other tissues that result in body curvature, we used a technique that generates chimeric embryos by knocking down gene function exclusively in dorsal forerunner cells (DFCs) but not in other embryonic cell lineages (Amack and Yost, 2004). The DFC lineage contributes to KV (Cooper and D'Amico, 1996), and morpholinos injected into the yolk during mid-blastula stages (256–1024 cells) can diffuse throughout the yolk and are taken up by DFCs but excluded from the rest of the embryo (Amack and Yost, 2004). Using fluorescence microscopy we identified embryos in which fluorescein-tagged rfx2 morpholinos were specifically taken up by DFCs but not in other embryonic tissues and selected these chimeric embryos for analysis.
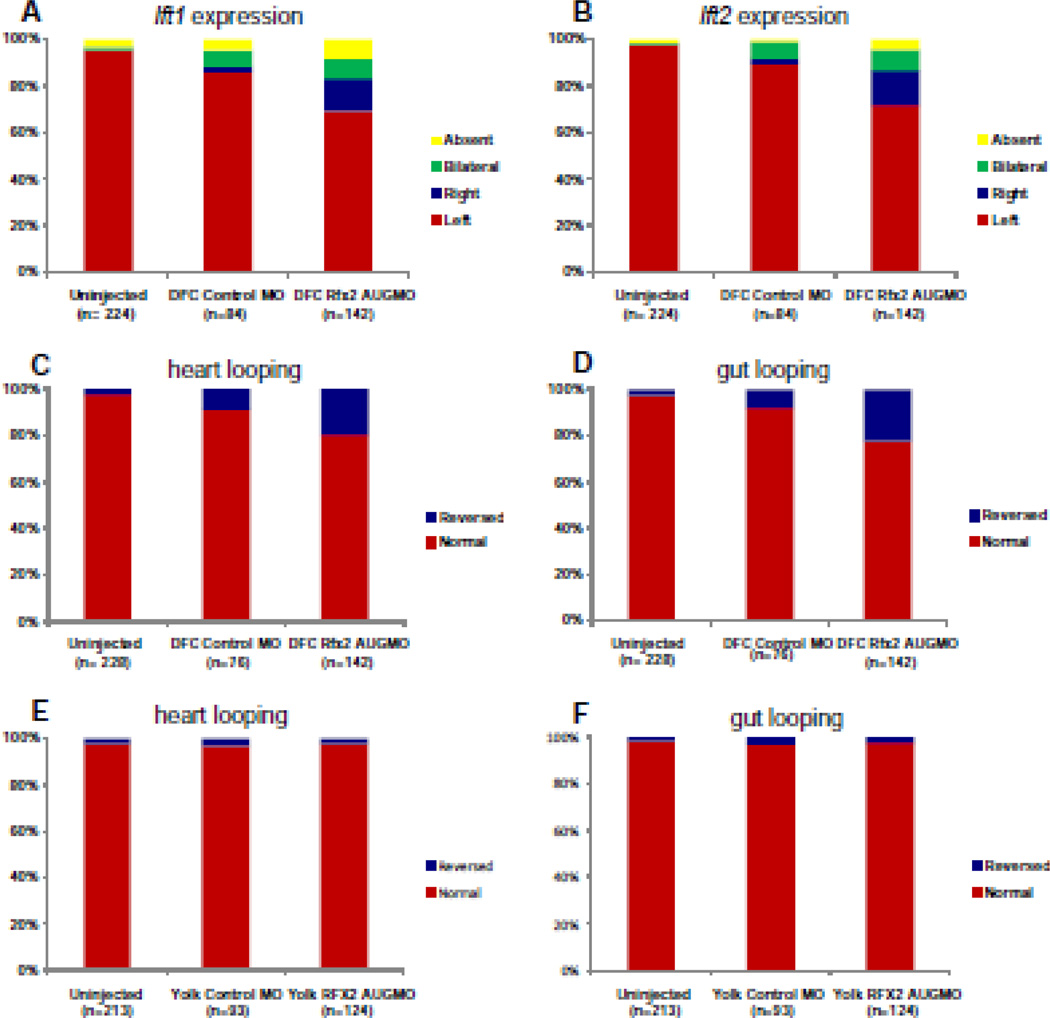
Mid-blastula stage injections of rfx2 AUGMO targeted to the DFC/KV (DFCrfx2 AUGMO) randomized asymmetric gene expression patterns (Fig. 5). Left-sided expression of lft1 in DFC-morphants was 69% versus 86% in embryos injected with control MO (DFCcontrolMO) (Fig. 5A) while left-sided expression of lft2 in the LPM of DFC-morphants was 72% versus 88% in embryos injected with control MO (Fig. 5B). DFC rfx2AUGMO also showed an increase in heart and gut reversals (20–22%) compared to embryos injected with control morpholino (9%) or to un-injected controls (3%) (Fig. 5C, D). Gross morphological phenotypes produced from 1–2 cell stage injections, including curled tails, were not seen in DFC-morphants produced from DFC-targeted injections (not shown). As an important control to confirm that rfx2 morpholinos must enter DFCs to perturb LR development, we injected morpholinos into the yolk at late blastula stages (dome stage to 30% epiboly), when connections between the yolk and DFCs are closed (YolkMO) (Cooper and D'Amico, 1996). In these embryos, yolk-injected rfx2 morpholino diffused throughout the yolk but did not accumulate in DFCs and did not cause significant reversals (3%) in heart and gut looping (Fig. 5E, F), indicating that Rfx2 function was required specifically in DFCs/KV, and not in yolk, for correct establishment of the LR axis.
Fig. 5.
DFC targeted rfx2 morpholinos perturb LR asymmetric gene expression and cardiac orientation. Histograms depicting the percentages of embryos with left, right, bilateral or no expression of lft1 (A) and lft2 (B) in un-injected embryos, or DFC-targeted control morpholino (DFCcontrolMO) or rfx2 AUGMO (DFCRfx2 AUGMO). Histograms depicting the percentages of embryos with normal or reversed orientation of cardiac (C) and gut looping (D) in un-injected embryos, DFCcontrolMO or DFCRfx2 AUGMO. As an important control in DFC-targeting experiments, morpholinos were injected into the yolk at a later stage during which uptake by DFCs no longer occurs either with control MO (yolkcontrolMO) or rfx2 AUGMO (yolkRfx2 AUGMO), and scored for orientation of heart (E) and gut looping (F).
The RFX2 promoter drives COA specific Cre expression
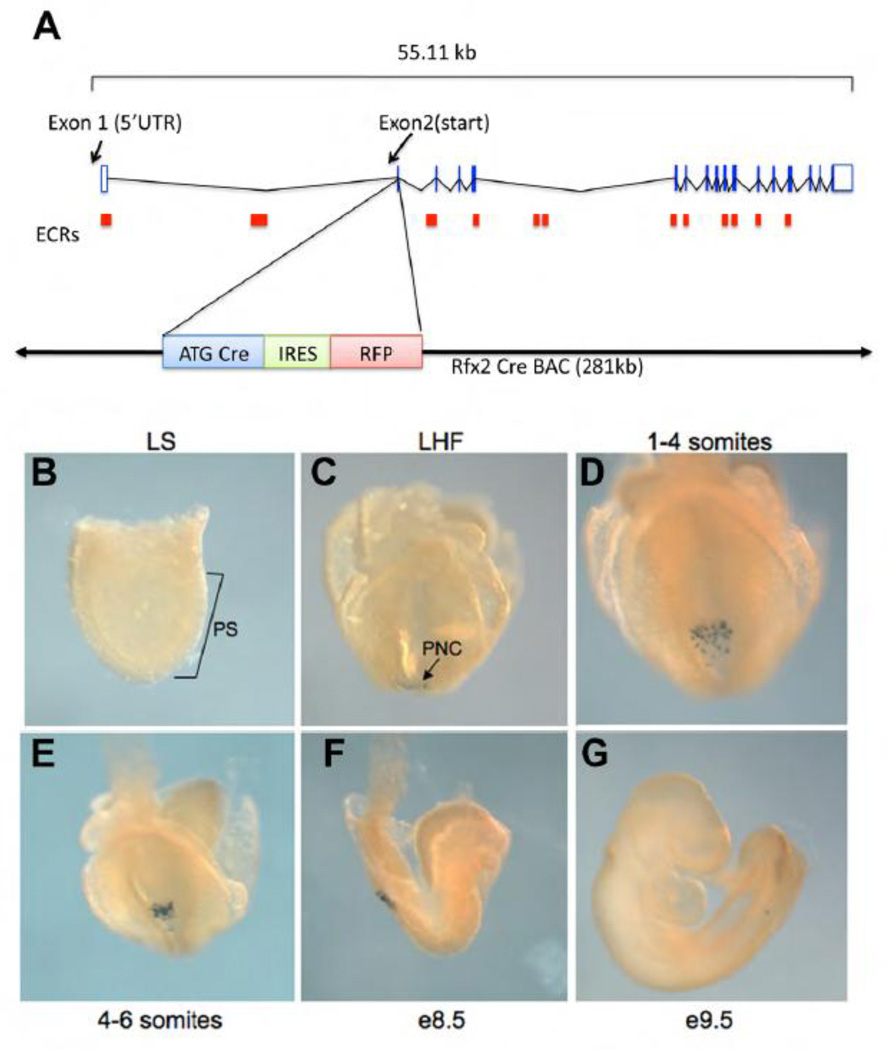
COA-specific Cre expressing mouse lines are an invaluable tool to dissect gene function in mouse COA, which includes posterior notochord (PNC) and organizer tissue. Three COA-specific Cre lines have previously been described: Foxa2-Cre, which drives Cre expression in the notochord and the PNC crown cells beginning at e7.5 (Kumar et al., 2007; Uetzmann et al., 2008), FoxJ1CreER, which expresses Cre in the ciliated cells of the COA beginning at 0–2 somites (Wang and Ware, 2009) and Nodal-Cre, which drives Cre expression in both crown and pit cells (Kumar et al., 2007). Full Cre expression in these lines is not established until the 0–2-somite stage, limiting their value in the analysis of earlier events including the first steps in LR patterning. Since we found that Rfx2 is expressed early and exclusively in the COA in mouse embryos (Fig. 1), and its role in LR development is dependent on function in analogous KV ciliated cells in zebrafish (Fig. 4, 5), we explored the possibility that a promoter derived from Rfx2 would serve as an excellent candidate to drive COA-specific Cre expression. Evaluation of the genomic region around the Rfx2 start site revealed conserved domains immediately upstream of the untranslated 1st exon, and within the 22.5 Kb 1st intron, preceding the Rfx2 translational start contained in exon 2 (Supplemental Fig. 5). To recapitulate the complete expression domain of Rfx2, a BAC construct was made which introduced Cre recombinase in-frame immediately following the Rfx2 translational start (Fig. 6A). The entire BAC construct was 280Kb in size, and encompassed the full genomic extent of Rfx2. One of two independent Rfx2-Cre transgenic mouse lines demonstrated the expected pattern of Cre expression. Homozygous Rfx2-Cre mice were found at expected Mendelian ratios, and were phenotypically normal and fertile, indicating that the BAC construct did not interrupt essential genes, and did not result in deleterious overexpression of genes within the BAC. Cre expression was evaluated by crosses between Rfx2-Cre mice and R26R conditional reporter mice, and by crosses between Rfx2-Cre mice and R26-EYFP fluorescent reporter mice. Expression was consistent over >3 generations. Rfx2-Cre did not demonstrate any Cre-induced expression of LacZ until the late headfold stage, when LacZ expression was observed in the COA (Fig. 6B, C). From late headfold to 6 somites, LacZ-expression was observed in the pit cells of the PNC (Fig. 6D, E). The expression was consistently spotty in the PNC and limited to a subset of PNC cells. Rarely, a few LacZ-expressing cells were found anterior to the PNC in the notochord. Since Rfx2 mRNA was detected at the midline at e8.5 (Fig 1C), it was not possible to determine whether the LacZ-expressing cells arose within the notochord, or migrated anteriorly from the PNC. A few LacZ-expressing cells were observed in the tail bud after embryonic turning (Fig. 6G).
Fig. 6.
Rfx2-Cre transgene recapitulates endogenous rfx2 expression. (A) Design of Rfx2-Cre transgene. The BAC encompasses the entire Rfx2 coding region. Cre recombinase is inserted in-frame behind the RFX2 start site located in exon 2 of the Rfx2 gene. (B–G) LacZ expression in RFX2Cre::ROSA26 embryos. (B) late primitive streak stage. (C) LacZ positive cells first become apparent in the posterior region of the node at late headfold stage. (D, E) Early somitogenesis, (F). During further development LacZ positive cells migrate to the caudal region of the embryo. (G) E9.5 LacZ positive cells are observed in the tail portion of the notochord.
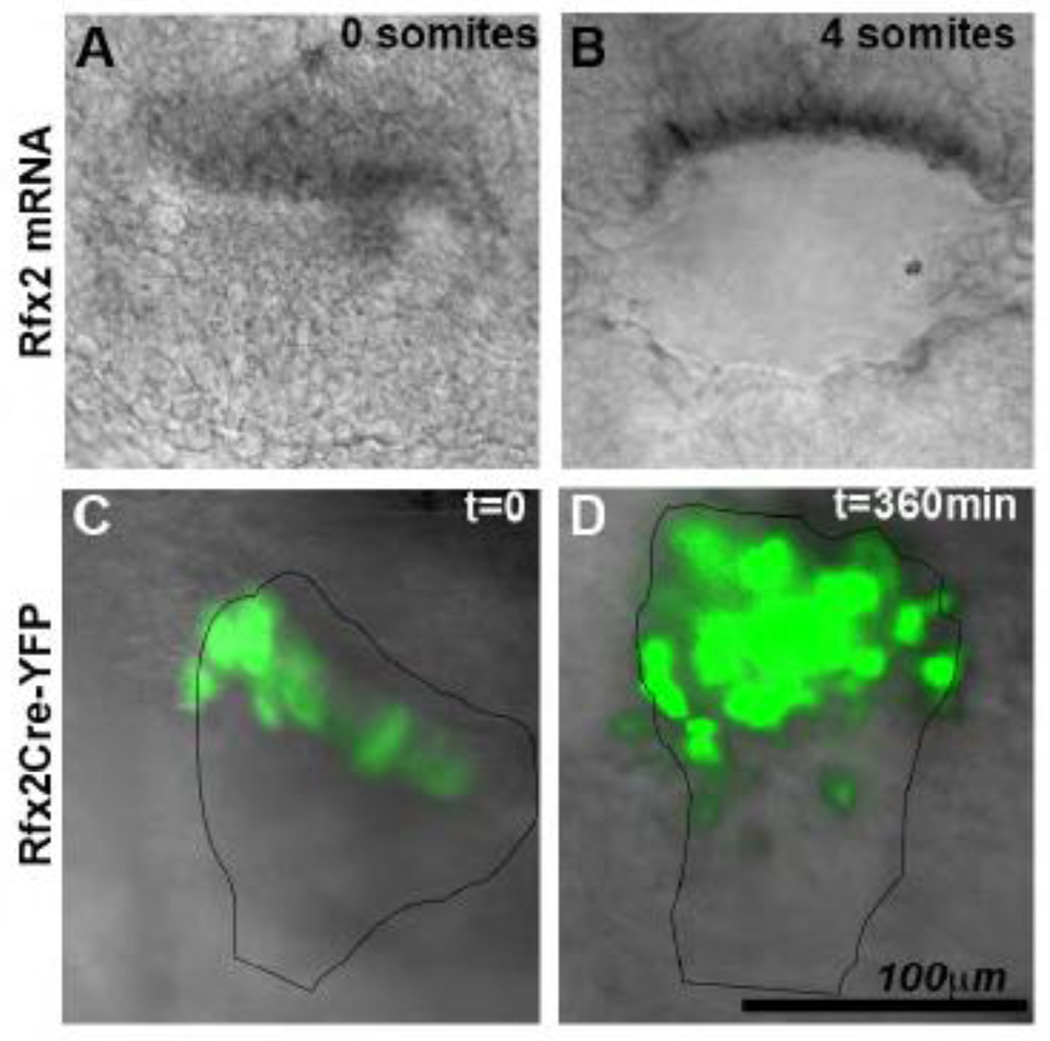
To further evaluate the fate of Rfx2-Cre expressing cells, we performed time-lapse live imaging of Rfx2-Cre-R26YFP embryos, which express YFP under the control of the Rfx2 promoter. Embryos from Rfx2-Cre-R26YFP matings were cultured in rat serum and imaged; embryos at equivalent stages were fixed and processed for WMISH with Rfx2 riboprobes. Rfx2 mRNA remains in the cells at the posterior margin of the PNC, and no RNA signal is observed in the PNC pit cells (Fig. 7A, B, and Supplemental Fig. 2). YFP-expressing cells are first observed at the posterior margin of the PNC at the late pre-somite through 2-somite stage (Fig. 7C). They subsequently appear in the PNC pit (Fig. 7D and Supplemental Fig. 6). In the absence of Rfx2 mRNA expression in the PNC pit cells, this observation suggests that the YFP-positive pit cells arise from cells from the posterior PNC margin. In summary, RFX2-Cre results in Cre expression in a subpopulation of posterior PNC cells in the COA. These cells migrate into the PNC pit during the 1–5 somite stages, which are crucial stages for cilia-driven fluid flow and development of LR asymmetry.
Fig. 7.
Live-imaging of COA in RFX2Cre :: R26YFP. (A–B) Rfx2 mRNA by WMISH in the COA of a fixed 0-somite (A) and 4-somite (B) embryo. (C–D) Live imaging of Rfx2Cre::R26YFP embryo for 360 min from 0 somites (C) to 4 somites (D)
COA specific rescue of Left-right dynein rescues the laterality phenotype of Lrd mutant mice
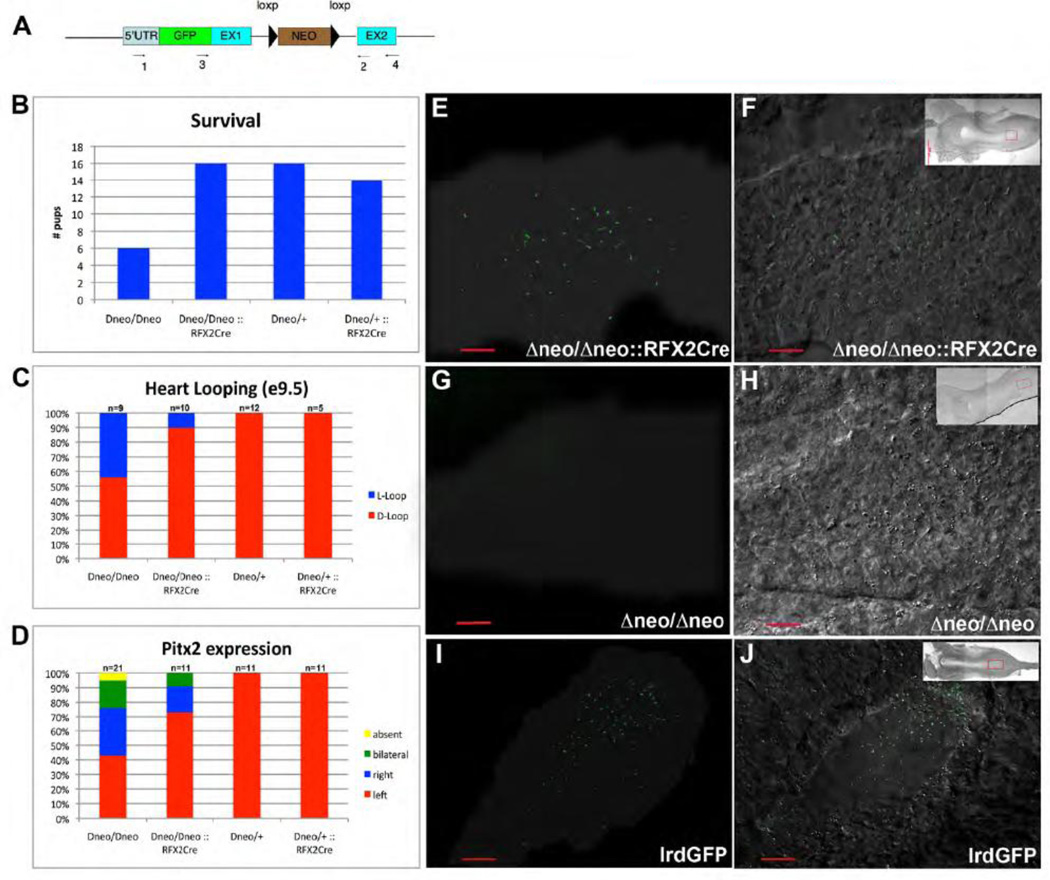
COA cells have a specialized motile cilium, which moves in a clockwise fashion to generate leftward flow of extraembryonic fluid, initially referred to as ‘nodal flow’ (Nonaka et al., 1998b). COA ciliary motility is powered by the outer arm dynein motors DNAHC5 and Left-Right Dynein (LRD, or DNAHC11). Mutants of either LRD or DNAHC5 have paralyzed, non-motile cilia, resulting in disrupted or absent nodal flow and abnormal development of left-right asymmetry (Ibanez-Tallon et al., 2002; Supp et al., 1997). Lrd mRNA is expressed in the COA beginning at e7.75, in the foregut pocket at e8.5 and the notochord and floor plate from e10.5 to e12.5 (Supp et al., 1999). We have previously generated mice that allow conditional activation of Lrd, Lrd-GFPΔneo (Fig.8A; Supplemental Fig. 7; (McGrath et al., 2003)). Briefly, EGFP was inserted in-frame immediately downstream of the Lrd translation start site. The neomycin resistance gene (NEO), flanked by loxP sites, was inserted into the 1st intron. Aberrant splicing from the NEO gene in the first intron of Lrd-GFPΔneo disrupts normal Lrd mRNA production. Thus, in homozygous lrd-GFPΔneo/Δneo embryos, no full-length Lrd transcript is detected, litter size is reduced, COA cilia are paralyzed, and approximately 50% of surviving offspring have situs inversus. Importantly, germ-line removal of NEO by actin-Cre-mediated recombination reconstitutes normal Lrd function, and results in Lrd-GFP mice expressing GFP-tagged, functional LRD (McGrath et al., 2003).
Fig. 8.
RFX2-Cre mediated rescue of LrdΔneo/Δneo. (A) Lrd-Δneo construct used to generate LrdΔneo/Δneo knock-in mice (B) RFX2-Cre rescues of embryonic lethality observed in LrdΔneo/Δneo mice. (C) RFX2-Cre rescue of situs inversus in LrdΔneo/Δneo liveborn mice. (D) RFX2-Cre mediated rescue of abnormal expression of Pitx2. (E–J) Node cilia shown via GFP-LRD fluorescence (left) overlaid onto DIC image of COA (right); shadow in (E,G,I) outlines COA in dark-field image. (E–F) LrdΔneo/Δneo :: RFX2-Cre (G–H) LrdΔneo/Δneo (I–J) Lrd-GFP.
To evaluate whether LRD function in COA cells is essential and sufficient for normal left-right development, Lrd-GFP Δneo/Δneo mice were crossed with Rfx2-Cre mice to rescue LRD specifically in RFX2-expressing PNC pit cells. LrdΔneo/+:: Rfx2-Cre stud males were evaluated for Cre expression by mating with ROSA26 females and evaluating lacZ expression in e8.0 embryos (analysis similar to Fig. 6). Males with offspring demonstrating robust COA-specific Cre expression were mated to lrdGFP Δneo/Δneo females. COA cell specific rescue of LRD resulted in rescue of the embryonic lethality observed in Lrd Δneo/Δneo mice (Fig. 8B), and no live-born Lrd Δneo/Δneo:: Rfx2-Cre mice had SI. While 45% of e9.5 Lrd Δneo/Δneo embryos had heart loop reversals, only 10% Lrd Δneo/Δneo:: Rfx2-Cre embryos had reversed heart loops (Fig. 8C). Left-right patterning was evaluated by analysis of Pitx2 mRNA expression at e8.5. In LrdΔneo/Δneo embryos, normal left-sided Pitx2 expression was observed in only 40%, while 70% of Lrd Δneo/Δneo:: Rfx2-Cre embryos had normal left-sided Pitx2 expression (Fig. 8D).
Since native LRD was replaced by GFP-LRD in Lrd Δneo/Δneo, and the non-recombined NEO prevents full-length mRNA accumulation from this construct, and thereby prevents expression of GFP-LRD, the efficiency of Cre-mediated recombination can be assessed by fluorescence imaging of the COA in Lrd Δneo/Δneo::Rfx2-Cre embryos and comparing the number of GFP-expressing cilia (i.e. ciliated cells in which recombination removed NEO and allowed expression of GFP-LRD) with those observed in embryos not expressing Cre (as a negative control) and in embryos expressing GFP-LRD (as a positive control). COA size and shape were indistinguishable between LrdΔneo/Δneo:: Rfx2Cre and Lrd-GFP embryos. GFP-tagged cilia were observed in both, however the number of GFP-LRD expressing cilia was lower in Lrd Δneo/Δneo:: Rfx2Cre embryos (60, 63; n=2) than Lrd-GFP embryos (128, 159; n=2; p=0.04) (Fig. 8E, F, I, J). Rescued embryos had GFP-LRD containing cilia throughout the COA without a left-right bias; however, at earlier stages, more GFP-positive cilia were found in the posterior half of the PNC. No GFP tagged cilia were observed in LrdΔneo/Δneo::Cre-minus littermate embryos (Fig. 8G,H). Together, these data indicate that COA specific rescue of LRD by Rfx2-Cre leads to significant rescue of the abnormal development of LR asymmetry observed in Lrd Δneo/Δneo mice, and suggests that Lrd mRNA expressed in other tissues including the foregut pocket was not required for LR development or embryonic survival.
Fluid flow was assessed by imaging movement of fluorescent beads across the COA in live Lrd-GFP and Lrd Δneo/Δneo::Rfx2-Cre embryos (Supplemental Fig. 8). Flow velocity was similar in both Lrd-GFP and lrd Δneo/Δneo::Rfx2-Cre embryos; however, flow in the rescued embryos was more prominent at the posterior PNC, while it was more uniform in the Lrd-GFP embryos. No flow was detected in the LrdΔneo/Δneo embryos. In summary, LRD-mediated ciliary motility in COA cells derived from RFX2-positive cells is sufficient to generate effective fluid flow across the COA and substantially rescue LR development in LrdΔneo/Δneo embryos. Similar to previous observation in Xenopus (Vick et al., 2009), LR development was substantially normal in the face of a reduced number of motile (GFP-LRD- expressing) COA cilia.
Discussion
The results presented here demonstrate that the X-box transcription factor RFX2 is expressed in the Ciliated Organ of Asymmetry (COA), specifically in the anterior tip of the primitive streak in mouse and the dorsal forerunner cells and Kupffer’s Vesicle (KV) cells in zebrafish. RFX2 is required for the development of left-right asymmetry, and motile cilia on RFX2-expressing cells are sufficient to establish adequate fluid flow across the COA to direct normal LR development in mouse. Together these results suggest that RFX2 expression defines an evolutionarily conserved population of ciliated cells in the COA.
The role of RFX2 in LR development
The RFX class of transcription factors exerts broad control over ciliogenesis in organisms including C. elegans and Drosophila. In vertebrates, the RFX family has 7 members, of which Rfx1–4 are known to be expressed in the developing mouse embryo and rfx1b, rfx2, rfx4 and rfx6 are expressed in the zebrafish embryo. Zebrafish and mouse homologues of Rfx2 share similar splice isoforms and 72% amino acid identity indicating that many aspects of protein function may be conserved. Knock-down of Rfx2 function in zebrafish results in embryos with ventrally curved tails, shorter KV cilia and defects in LR patterning. This cluster of phenotypes is consistent with previous reports of zebrafish mutants and morphants that affect ciliary formation and function (Becker-Heck et al., 2011; Bisgrove et al., 2005; Kramer-Zucker et al., 2005; Yu et al., 2008). Perturbations in LR patterning are manifest as randomization of the expression patterns of the asymmetric Nodal signaling pathway genes spaw, lft1 and lft2 which correlate with reversals in the direction of looping of the heart and gut primordia later in development.
By targeting morpholinos specifically to the DFC/KV we demonstrate that Rfx2 function is cell-autonomously required in DFC/KV cells for LR patterning. Rfx2 DFC-targeted morphants do not exhibit curved bodies, demonstrating that laterality defects in these morphants are independent of body curvature and are a consequence of reducing gene function only in DFCs/KV. While it is difficult to control the dose of morpholinos targeted to DFCs, and thus the penetrance of effect is often lower than whole-embryo targeting, this technique has provided a valuable approach to examining DFC/KV-specific gene function (Amack and Yost, 2004; Essner et al., 2005). The weaker penetrance of the laterality phenotype relative to that observed from morpholino injections at the 1–2 cell stage likely reflects inefficiencies of DFC-targeted injections, such as variability in the amount of morpholino that enters DFCs from the yolk, and variable number of DFCs that obtain morpholino in a given embryo. The important point is that the laterality effects are significantly above controls, demonstrating that Rfx2 is required in DFC/KV cells for LR patterning. Of course, as with any other gene to which this technology is applied, these observations do not exclude the possibility that Rfx2 has functions in other tissues in addition to its function for LR determination in DFC/KV. DFCs have no known developmental function (Warga and Stainier, 2002) other than their role in giving rise to ciliated KV which functions in the establishment of LR patterning (Essner et al., 2005). Reduced KV cilia length and LR patterning defects have been reported for knockdowns of several proteins expressed in KV cells, including Ift88/Polaris, Ift57, Fgf8, Fgf24, FoxJ1, DeltaD and are often correlated with impaired KV fluid flow (Bisgrove et al., 2005; Kramer-Zucker et al., 2005; Lopes et al., 2010; Neugebauer et al., 2009; Yu et al., 2008).
Similar to what we have observed in zebrafish rfx2 morphants, mice mutant for Rfx3 have markedly stunted cilia in the COA and embryos have LR patterning defects (Bonnafe et al., 2004). There is significant, but not complete overlap between the Rfx2 and Rfx3 expression pattern in the COA, suggesting that RFX2 and RFX3 may have cooperative function in LR development. RFX3 has previously been shown to have a role in LR development in mice via transcriptional control of IFT proteins including the axonemal dynein genes Dync2li1, Dnahc11, Dnahc9 and Dnahc5 (Bonnafe et al., 2004; El Zein et al., 2009). Interestingly, although both DNAHC11 and DNAHC5 are essential to LR development via an essential role in cilia motility in the COA, the LR phenotype in Rfx3−/− embryos is only partially penetrant, suggesting at least some functional redundancy. As RFX transcription factors can function as homo- or heterodimers, co-expression of both Rfx2 and Rfx3 in the COA could provide very fine control over ciliogenesis and function via temporo-spatial regulation of RFX 2 and RFX3 levels.
LRD drives LR asymmetry exclusively via fluid flow
One gene regulated by RFX transcription factors is the microtubule motor protein left-right dynein (LRD, DNAHC11) which has a conserved x-box located in the first intron. Lrd is expressed in the COA of multiple vertebrates including mice and zebrafish (Essner et al., 2002), is essential for cilia motility in the COA and for LR patterning of the brain, heart and gut in zebrafish (Essner et al., 2005), and has been implicated in human primary ciliary dyskinesia (Bartoloni et al., 2002). Mice with Lrd mutations also have paralyzed COA cilia and randomization of cardiac and visceral situs (Supp et al., 1999; Supp et al., 1997). In addition to prominent expression in the COA, it is also expressed at low levels in 1–8 cell embryos, and later in development is expressed in the foregut pocket and in the limb buds (Supp et al., 1999). Early LRD expression has been proposed to have a role in segregation of differentially imprinted chromatids, and it is suggested that this early function lies upstream of LRD-generated fluid flow (Armakolas and Klar, 2007; Armakolas et al., 2010). Our finding that LRD function restricted entirely to RFX2-expressing COA cells is sufficient to rescue LR development indicates that early LRD function in tissues outside of the COA is dispensable for the establishment of LR asymmetry, and that the primary role of LRD in LR development is a requirement for axonemal-dynein-driven ciliary motility to generate fluid flow across the COA. The incomplete rescue of organ asymmetries and Pitx2 expression in lrdΔneo/Δneo ::Rfx2-Cre mice could be due to minor contributions of non-RFX2-positive cells to fluid flow, or to incomplete rescue of LrdΔneo due to delay in Cre-mediated excision of the floxed LrdΔneo allele. The latter is quite likely, since LRD expression and subsequent COA fluid flow must be completed within a short developmental window extending from 0–4 somites. Whether LRD expression in the foregut pocket or in other domains functions to reinforce COA flow-initiated asymmetries remains to be investigated.
Origin and transcriptional regulation of ciliated COA cells
A functional COA has been identified in zebrafish, frog, rabbit and mouse. These structures all have motile cilia that produce directional fluid flow during a brief developmental window preceding the appearance of molecular or anatomic LR asymmetries. The zebrafish COA, Kupffer’s vesicle, derives from a migratory population of cells called dorsal forerunner cells (Cooper and D'Amico, 1996; Essner et al., 2005). The origin of cells that contribute to the COA in other vertebrates remains less defined. Based on observations of cell morphology and cilia function, the mammalian COA is not a homogeneous cell population, and the origin of COA cells, along with the mechanism by which COA cell subpopulations are specified remains unclear. The mouse COA, posterior notochord (PNC), consists of ~250 small columnar pit cells surrounded by a smaller group of flat crown cells (Sulik et al., 1994; Yamanaka et al., 2007). In mouse and rabbit, the PNC is a dynamic population of cells, marked by expression of Noto, Brachyury and FoxA2, surrounded by a small domain of Nodal and Cerl2 expression, and distinguished from the classical ‘node’ by the absence of organizer gene expression (Blum et al., 2007). Remarkably, although the cells in the PNC are mitotically inactive, the number of densely-packed ciliated PNC cells increases from 50 to 250 during the late headfold to early somite stages when the LR axis is specified. Our data indicates that at least some PNC cells in mouse arise either via movement from the anterior end of the primitive streak into the PNC, or are deposited in the PNC as the primitive streak regresses. In either case, cells destined for the PNC are bottle-shaped cells identified by expressing Rfx2. Our Rfx2-Cre-driven rescue of LRD-dependent cilia motility in this subset of PNC cells indicates that these cilia are sufficient to create fluid flow for the determination of LR asymmetry. We suggest that the Rfx2-expressing cells at the anterior end of the primitive streak in mouse correspond to the rfx2-expressing dorsal forerunner cells in zebrafish, providing a functional definition of COA cells.
The transcriptional control of ciliogenesis in the COA is complex. The homeobox protein Noto plays an important role in the formation of the COA in the mouse; mutants have defects in the PNC and laterality defects (Abdelkhalek et al., 2004), cilia are reduced in number and are shorter, and expression levels of the transcription factors FoxJ1 and Rfx3 are reduced or abolished (Beckers et al., 2007). RFX3 in turn has been shown to have an effect on cilia in the mouse COA: Rfx3−/− mice have laterality defects, and cilia in the COA are present but stunted (Bonnafe et al., 2004). Foxj1 also exerts control over COA ciliogenesis in zebrafish and in the specification of motile versus sensory cilia in the Xenopus COA by influencing expression of a wide range of genes required for ciliogenesis and ciliary motility including Rfx2 (Stubbs et al., 2008; Yu et al., 2008) (our unpublished observations). Rfx2 in the zebrafish pronephric duct (PND) exhibits a dynamic expression pattern: early during PND development, it is expressed in all ciliated cells, subsequently it becomes restricted to the multi-ciliated cells in response to the Notch signaling pathway (Liu et al., 2007; Ma and Jiang, 2007). We show here that knockdown of Rfx2 function results in the intermittent loss of cilia along the length of the PND, suggesting that these missing cilia may correspond to those of the rfx2-expressing multi-ciliated cells. Notch and Fgf signaling pathways have also recently been demonstrated to control KV cilia length through regulation of foxj1a and rfx2 expression (Lopes et al., 2010; Neugebauer et al., 2009). Thus, our and other studies suggest a complex gene regulatory network operating in COA and other ciliated structures with inputs from Notch and FGF signaling pathways modulating the expression of transcription factors including Foxj1 and Rfx2, which in turn regulate the expression of genes directly involved in cilia structure and function.
In summary, we demonstrate that the X-box transcription factor RFX2 is required for ciliogenesis and LR development. We define an evolutionarily-conserved group of cells that expresses Rfx2, populates the Ciliated Organ of Asymmetry, and is capable of generating fluid flow to establish LR asymmetry.
Highlights.
-
>
RFX2 expression is highly conserved in zebrafish and mice embryos
-
>
RFX2 is necessary for ciliogenesis in Kupffer’s Vesicle
-
>
RFX2 is required in KV for Left-Right patterning of brain, heart and gut
-
>
RFX2 is required for asymmetric fluid flow in the Ciliated Organ of Asymmetry in both zebrafish and mouse
-
>
mouse RFX2 transgene drives expression exclusively in subset of ciliated cells
-
>
reactivated cilia motility in subset of cilia is sufficient for LR development
Supplementary Material
Acknowledgements
We thank M. Boskovski and B. Basu for critical reading of the manuscript. Support for the production and analysis of transgenic mice was provided by J. McGrath and the Yale University Mouse Genomics Center. The technical assistance of B. S. Snarr, A. Emrazian Anderson, B. L. McMahon and Y-C. Su in the characterization of zebrafish rfx2 is greatly appreciated. J. Neugebauer provided advice and technical assistance with the analysis of KV cilia length and fluid flow. Assistance with the mouse work was provided by J. Drozd and M. Nash. MB and SM are supported by NIHRO1 HL093280-01A1, and work in HJY lab was supported by NIH R01 HL66292 and P01 HD048886.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelkhalek HB, Beckers A, Schuster-Gossler K, Pavlova MN, Burkhardt H, Lickert H, Rossant J, Reinhardt R, Schalkwyk LC, Muller I, Herrmann BG, Ceolin M, Rivera-Pomar R, Gossler A. The mouse homeobox gene Not is required for caudal notochord development and affected by the truncate mutation. Genes Dev. 2004;18:1725–1736. doi: 10.1101/gad.303504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aftab S, Semenec L, Chu JS, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–690. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Andrade YN, Fernandes J, Vazquez E, Fernandez-Fernandez JM, Arniges M, Sanchez TM, Villalon M, Valverde MA. TRPV4 channel is involved in the coupling of fluid viscosity changes to epithelial ciliary activity. J Cell Biol. 2005;168:869–874. doi: 10.1083/jcb.200409070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armakolas A, Klar AJ. Left-right dynein motor implicated in selective chromatid segregation in mouse cells. Science. 2007;315:100–101. doi: 10.1126/science.1129429. [DOI] [PubMed] [Google Scholar]
- Armakolas A, Koutsilieris M, Klar AJ. Discovery of the mitotic selective chromatid segregation phenomenon and its implications for vertebrate development. Curr Opin Cell Biol. 2010;22:81–87. doi: 10.1016/j.ceb.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashique AM, Choe Y, Karlen M, May SR, Phamluong K, Solloway MJ, Ericson J, Peterson AS. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Sci Signal. 2009 doi: 10.1126/scisignal.2000602. [DOI] [PubMed] [Google Scholar]
- Baas D, Meiniel A, Benadiba C, Bonnafe E, Meiniel O, Reith W, Durand B. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur J Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- Bartoloni L, Blouin JL, Pan Y, Gehrig C, Maiti AK, Scamuffa N, Rossier C, Jorissen M, Armengot M, Meeks M, Mitchison HM, Chung EM, Delozier-Blanchet CD, Craigen WJ, Antonarakis SE. Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc Natl Acad Sci U S A. 2002;99:10282–10286. doi: 10.1073/pnas.152337699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, Fliegauf M, Horvath J, Reinhardt R, Nielsen KG, Marthin JK, Baktai G, Anderson KV, Geisler R, Niswander L, Omran H, Burdine RD. The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat Genet. 2011;43:79–84. doi: 10.1038/ng.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers A, Alten L, Viebahn C, Andre P, Gossler A. The mouse homeobox gene Noto regulates node morphogenesis, notochordal ciliogenesis, and left right patterning. Proc Natl Acad Sci U S A. 2007;104:15765–15770. doi: 10.1073/pnas.0704344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Regulation of midline development by antagonism of lefty and nodal signaling. Development. 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Essner JJ, Yost HJ. Multiple pathways in the midline regulate concordant brain, heart and gut left-right asymmetry. Development. 2000;127:3567–3579. doi: 10.1242/dev.127.16.3567. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Snarr BS, Emrazian A, Yost HJ. Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer's vesicle are required for specification of the zebrafish leftright axis. Dev Biol. 2005;287:274–288. doi: 10.1016/j.ydbio.2005.08.047. [DOI] [PubMed] [Google Scholar]
- Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, McKay SJ, Huang P, Swoboda P, Jones SJ, Marra MA, Baillie DL, Moerman DG, Shaham S, Leroux MR. Functional genomics of the cilium, a sensory organelle. Curr Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Blum M, Andre P, Muders K, Schweickert A, Fischer A, Bitzer E, Bogusch S, Beyer T, van Straaten HW, Viebahn C. Ciliation and gene expression distinguish between node and posterior notochord in the mammalian embryo. Differentiation. 2007;75:133–146. doi: 10.1111/j.1432-0436.2006.00124.x. [DOI] [PubMed] [Google Scholar]
- Bonnafe E, Touka M, AitLounis A, Baas D, Barras E, Ucla C, Moreau A, Flamant F, Dubruille R, Couble P, Collignon J, Durand B, Reith W. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol Cell Biol. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Cooper MS, D'Amico LA. A cluster of noninvoluting endocytic cells at the margin of the zebrafish blastoderm marks the site of embryonic shield formation. Dev Biol. 1996;180:184–198. doi: 10.1006/dbio.1996.0294. [DOI] [PubMed] [Google Scholar]
- Dorn A, Durand B, Marfing C, Le Meur M, Benoist C, Mathis D. Conserved major histocompatibility complex class II boxes--X and Y--are transcriptional control elements and specifically bind nuclear proteins. Proc Natl Acad Sci U S A. 1987;84:6249–6253. doi: 10.1073/pnas.84.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubruille R, Laurencon A, Vandaele C, Shishido E, Coulon-Bublex M, Swoboda P, Couble P, Kernan M, Durand B. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- El Zein L, Ait-Lounis A, Morle L, Thomas J, Chhin B, Spassky N, Reith W, Durand B. RFX3 governs growth and beating efficiency of motile cilia in mouse and controls the expression of genes involved in human ciliopathies. J Cell Sci. 2009;122:3180–3189. doi: 10.1242/jcs.048348. [DOI] [PubMed] [Google Scholar]
- Emery P, Durand B, Mach B, Reith W. RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res. 1996;24:803–807. doi: 10.1093/nar/24.5.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Vogan KJ, Wagner MK, Tabin CJ, Yost HJ, Brueckner M. Conserved function for embryonic nodal cilia. Nature. 2002;418:37–38. doi: 10.1038/418037a. [DOI] [PubMed] [Google Scholar]
- Evans JE, Snow JJ, Gunnarson AL, Ou G, Stahlberg H, McDonald KL, Scholey JM. Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans. J Cell Biol. 2006;172:663–669. doi: 10.1083/jcb.200509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feistel K, Blum M. Three types of cilia including a novel 9+4 axoneme on the notochordal plate of the rabbit embryo. Dev Dyn. 2006;235:3348–3358. doi: 10.1002/dvdy.20986. [DOI] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137:32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Gorokhova S, Heintz N. Loss of function of axonemal dynein Mdnah5 causes primary ciliary dyskinesia and hydrocephalus. Hum Mol Genet. 2002;11:715–721. doi: 10.1093/hmg/11.6.715. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Kamura K, Kobayashi D, Uehara Y, Koshida S, Iijima N, Kudo A, Yokoyama T, Takeda H. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development. 2011;138:1121–1129. doi: 10.1242/dev.058271. [DOI] [PubMed] [Google Scholar]
- Kramer-Zucker AG, Olale F, Haycraft CJ, Yoder BK, Schier AF, Drummond IA. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- Kumar A, Yamaguchi T, Sharma P, Kuehn MR. Transgenic mouse lines expressing Cre recombinase specifically in posterior notochord and notochord. Genesis. 2007;45:729–736. doi: 10.1002/dvg.20346. [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pathak N, Kramer-Zucker A, Drummond IA. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Lopes SS, Lourenco R, Pacheco L, Moreno N, Kreiling J, Saude L. Notch signalling regulates left-right asymmetry through ciliary length control. Development. 2010;137:3625–3632. doi: 10.1242/dev.054452. [DOI] [PubMed] [Google Scholar]
- Ma M, Jiang YJ. Jagged2a-notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS Genet. 2007;3:e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. EMBO J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998a;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998a;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [published erratum appears in Cell 1999 Oct 1;99(1):117]. [DOI] [PubMed] [Google Scholar]
- Odenthal J, Nusslein-Volhard C. fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245–258. doi: 10.1007/s004270050179. [DOI] [PubMed] [Google Scholar]
- Okada Y, Nonaka S, Tanaka Y, Saijoh Y, Hamada H, Hirokawa N. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol Cell. 1999;4:459–468. doi: 10.1016/s1097-2765(00)80197-5. [DOI] [PubMed] [Google Scholar]
- Okada Y, Takeda S, Tanaka Y, Belmonte JC, Hirokawa N. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 2005;121:633–644. doi: 10.1016/j.cell.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Veland IR, Schroder JM, Christensen ST. Assembly of primary cilia. Dev Dyn. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Weber T, Beyer T, Vick P, Bogusch S, Feistel K, Blum M. Cilia-driven leftward flow determines laterality in Xenopus. Curr Biol. 2007;17:60–66. doi: 10.1016/j.cub.2006.10.067. [DOI] [PubMed] [Google Scholar]
- Shah AS, Ben-Shahar Y, Moninger TO, Kline JN, Welsh MJ. Motile cilia of human airway epithelia are chemosensory. Science. 2009;325:1131–1134. doi: 10.1126/science.1173869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PA, Basta PV, Ting JP. Upstream DNA sequences required for tissuespecific expression of the HLA-DR alpha gene. Proc Natl Acad Sci U S A. 1987;84:4254–4258. doi: 10.1073/pnas.84.12.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulik K, Dehart DB, Iangaki T, Carson JL, Vrablic T, Gesteland K, Schoenwolf GC. Morphogenesis of the murine node and notochordal plate. Dev Dyn. 1994;201:260–278. doi: 10.1002/aja.1002010309. [DOI] [PubMed] [Google Scholar]
- Supp DM, Brueckner M, Kuehn MR, Witte DP, Lowe LA, McGrath J, Corrales J, Potter SS. Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development. 1999;126:5495–5504. doi: 10.1242/dev.126.23.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P, Adler HT, Thomas JH. The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell. 2000;5:411–421. doi: 10.1016/s1097-2765(00)80436-0. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435:172–177. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- Thomas J, Morle L, Soulavie F, Laurencon A, Sagnol S, Durand B. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell. 2010;102:499–513. doi: 10.1042/BC20100035. [DOI] [PubMed] [Google Scholar]
- Tobin JL, Beales PL. The nonmotile ciliopathies. Genet Med. 2009;11:386–402. doi: 10.1097/GIM.0b013e3181a02882. [DOI] [PubMed] [Google Scholar]
- Uetzmann L, Burtscher I, Lickert H. A mouse line expressing Foxa2-driven Cre recombinase in node, notochord, floorplate, and endoderm. Genesis. 2008;46:515–522. doi: 10.1002/dvg.20410. [DOI] [PubMed] [Google Scholar]
- Vick P, Schweickert A, Weber T, Eberhardt M, Mencl S, Shcherbakov D, Beyer T, Blum M. Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog Xenopus laevis. Dev Biol. 2009;331:281–291. doi: 10.1016/j.ydbio.2009.05.547. [DOI] [PubMed] [Google Scholar]
- Wang S, Ware SM. Use of FOXJ1CreER2T mice for inducible deletion of embryonic node gene expression. Genesis. 2009;47:132–136. doi: 10.1002/dvg.20467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga RM, Stainier DY. The guts of endoderm formation. Results Probl Cell Differ. 2002;40:28–47. doi: 10.1007/978-3-540-46041-1_3. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Yamanaka Y, Tamplin OJ, Beckers A, Gossler A, Rossant J. Live imaging and genetic analysis of mouse notochord formation reveals regional morphogenetic mechanisms. Dev Cell. 2007;13:884–896. doi: 10.1016/j.devcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- Yelon D, Horne SA, Stainier DY. Restricted Expression of Cardiac Myosin Genes Reveals Regulated Aspects of Heart Tube Assembly in Zebrafish. Dev Biol. 1999;214:23–37. doi: 10.1006/dbio.1999.9406. [DOI] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, Roy S. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet. 2008;40:1445–1453. doi: 10.1038/ng.263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.