Abstract
The centrosome, a cytoplasmic organelle formed by cylinder-shaped centrioles surrounded by a microtubule-organizing matrix, is a hallmark of animal cells. The centrosome is conserved and essential for the development of all animal species described so far. Here, we show that, unlike the rest of animals, planarians and possibly other flatworms as well completely lack centrosomes. We found that in planarians, centrioles are only assembled in terminally differentiating ciliated cells through a so-called acentriolar pathway to trigger the assembly of cilia. This unique characteristic allowed us to identify a large set of conserved proteins required for centriole assembly in animals, as well as the centrosome signature proteins missing from the planarian genome. Our study uncovers the molecular architecture and evolution of the animal centrosome and emphasizes the plasticity of animal cell biology and development.
The centrosome controls essential cellular processes such as cell division, migration and polarity by anchoring microtubule nucleating factors and cell cycle regulators. In animals, it also plays a crucial role during development and organogenesis through its ability to control nucleus positioning and cell division orientation, and by nucleating primary cilia. The centrosome is found in all animals, and is absolutely essential to the development of all animal species studied so far. However, centrosomes are dispensable for cell division in some instances, in particular during mouse early embryogenesis and the later stages of Drosophila development, raising the question of how central the centrosome really is to fundamental cell biology and development (1, 2).
Animal centrosome reproduction relies on duplication of its core components, the centrioles, once per cell cycle. In addition to centriole duplication in proliferating cells, a second pathway, called acentriolar pathway, allows assembly of large numbers of centrioles in multiciliated cells undergoing terminal differentiation (3). In vertebrates, multiciliated cells drive mucus clearance, cerebrospinal fluid circulation and egg transportation along the oviduct. However, the molecular pathway underlying centriole assembly in multiciliated cells is poorly characterized. Flatworms like the freshwater planarian Schmidtea mediterranea use multiciliated cells for locomotion, suggesting they can serve as model systems for studying centriole assembly in multiciliated cells. Furthermore, the role played by centrioles in the regeneration of planarians remains unexplored thus far.
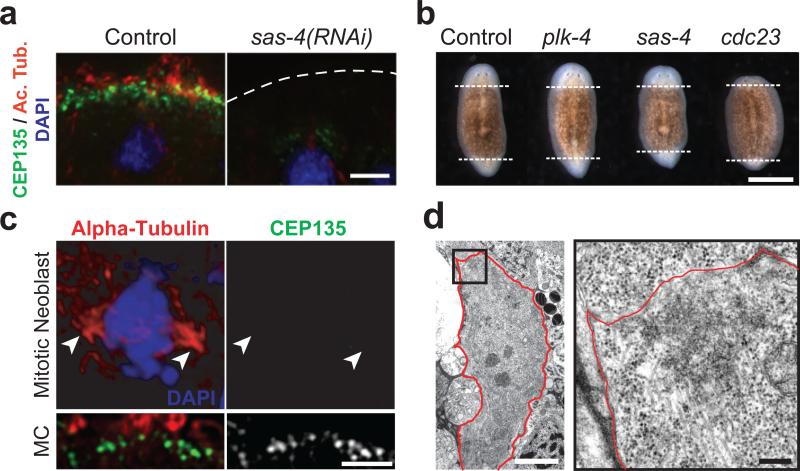
To characterize the phenotypes associated with the inhibition of centriole assembly in planarians, we targeted by RNAi the genes encoding planarian homologs of the conserved centriolar components SAS-4/CPAP and Plk4/SAK in asexual planarians (2, 4, 5). In contrast to untreated animals that glide smoothly on their ciliated epithelium, 100% of the plk4(RNAi) or sas-4(RNAi) animals exhibited inchworming locomotion (n = 100, 5 independent experiments; Supplemental Movies S1 and S2), a phenotype known to result from impaired ciliary function (6, 7). Using an antibody raised against SMED-CEP135, the planarian homolog of a conserved component of centrioles (5), we found that the ventral surface of plk4(RNAi) or sas-4(RNAi) animals was almost entirely devoid of centrioles and cilia (Fig. 1a, S1, S2). Thus, depleting the planarian homologs of proteins essential for centriole duplication within the centrosome abolished centriole assembly in planarian multiciliated cells.
Figure 1.
Centrioles in planarians are present in multiciliated cells but not proliferating cells. (a) Immunofluorescence staining of ventral epidermis in control or sas-4(RNAi) animals; centrioles (green, anti-SMED-CEP135), cilia (red, anti-acetylated tubulin), nuclei (blue, DAPI). (b) Regenerating trunk fragments. Dashed lines: boundary between newly formed and preexisting tissues. (c) Immunofluorescence staining of control mitotic neoblast; centrioles (green, anti-SMED-CEP135), mitotic spindle (red, anti-alpha-tubulin), chromosomes (blue, DAPI). Arrowheads: spindle poles. Lower panel: centrioles in a multiciliated cell (MC) from the same section. (d) TEM view of control mitotic neoblast (3,200x). Red line: plasma membrane. Right panel: high magnification view (15,000x) of boxed area. Bar is 5 μm in a, c and d-left panel, 1mm in b, and 0.5 μm in d-right panel.
Surprisingly however, sas-4(RNAi) and plk4(RNAi) animals showed no observable regeneration defect (Fig. 1b). The remarkable ability of planarians to regenerate whole animals from almost any part of their body is known to require division of the neoblasts, a population of totipotent stem cells and the only cells to undergo mitosis in asexual planarians (8). Blocking cell division by depleting the planarian homolog of CDC23, a component of the anaphase-promoting complex, resulted in regression of the regeneration blastemata as previously described (9). In contrast, animals depleted of centriole components regenerated missing tissues to a similar extent as control animals (Fig. 1b, S3). These results suggested that centrioles are dispensable for cell division and tissue formation in planarians. Remarkably, whereas centrioles were detected in the different ciliated tissues (Fig. 1a, S2, S4), they were absent from all other cell types. In particular, we did not observe centrioles in control neoblasts, neither by immunofluorescence nor by transmission electron microscopy (Fig. 1c, 1d, S4-S6), as also suggested by earlier ultrastructural studies (10, 11). We obtained the same result when analyzing S. mediterranea embryos by immunofluorescence, indicating that dividing embryonic cells also lack centrioles (Fig. S4). Thus, centrioles are missing from the only two cell types that can proliferate in planarians, and appear to be missing from all non-ciliated terminally differentiated cells that we have observed as well. This result might at first seem reminiscent of the case in Drosophila in which later stages of development can take place in mutants deficient for centriole duplication (2, 4). However, early Drosophila development is absolutely dependent on the presence of centrioles (12, 13). In contrast, our results show that planarians do not require centrioles at any stage of development. In animal cells, centrioles are essential for the duplication and stability of the centrosome (2, 4, 14). Our results thus indicate that planarians do not assemble centrosomes, but only assemble centrioles de novo during the differentiation of ciliated cells.
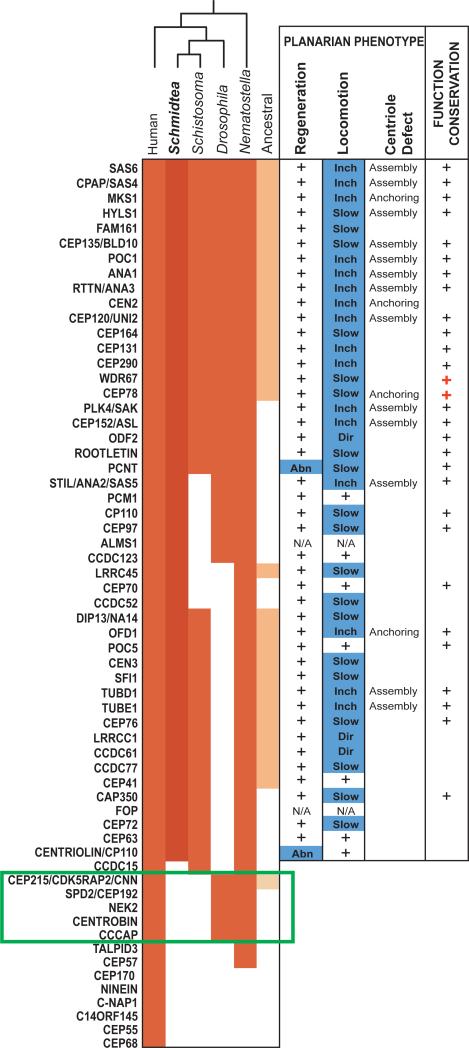
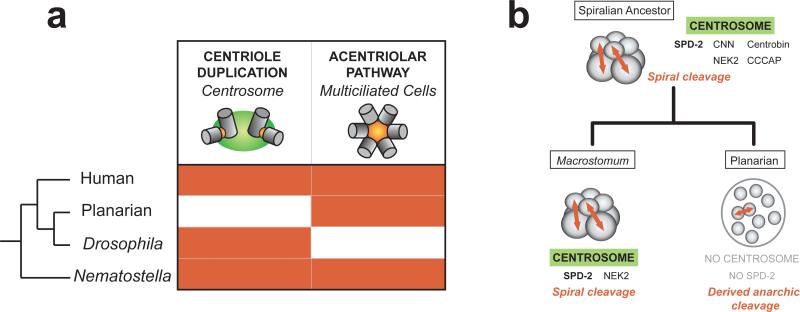
A first prediction based on this model is that proteins specifically required for centrosome assembly or function should have been eliminated from the planarian genome during evolution. To test this, we used a list of core centrosome components (Table S1) derived from the human centrosome proteome (15) and searched for potential homologs in the planarian genome. We found planarian homologs for a majority (47/55) of the centrosome components that were unequivocally present in the last common ancestor of humans and planarians (Fig. 2). Remarkably, five protein families were conserved in Drosophila but missing not only in Schmidtea but also in the parasitic flatworm Schistosoma mansoni. This subset included three protein families required for assembly or reproduction of the centrosome: SPD-2/Cep192, CNN/CDK5RAP2 and Nek2 (5, 16, 17). Thus, a key set of centrosome components essential to centrosome function is missing from the planarian genome. Because planarians evolved from ancestors that contain these proteins, we infer that they were eliminated from the planarian genome together with the centrosome itself during evolution, while an acentriolar pathway was retained to assemble centrioles during differentiation of ciliated cells (Fig. 3a).
Figure 2.
Planarian homologs of human centrosome components are required for centriole assembly in multiciliated cells. Left: conservation of centrosome components in animals (red). Ancestral eukaryotic genes are in orange. Green box: centrosome signature genes lost in the planarian Schmidtea mediterranea and the parasite flatworm Schistosoma mansoni but present in Drosophila. Middle: regeneration and locomotion phenotypes in planarians (+: no defect; Abn: abnormal; Inch: inchworming; Dir: abnormal direction of locomotion). Right: +: genes implicated in centriole duplication or ciliogenesis in other systems (red +: this study; S.O.M.).
Figure 3.
(a) Conservation of the two main pathways for centriole assembly in animals. Centrosome duplication and the acentriolar pathway are highlighted in red when present. (b) Model of the correlation between loss of the centrosome and loss of the spiral cleavage pattern.
A second prediction is that components of the human centrosome for which homologs are still present in planarians should be required for ciliogenesis through the acentriolar pathway. We targeted the homologs of centrosome components conserved in the planarian genome by RNAi and indeed we observed a locomotion defect in a majority of the knockdowns (38/45 genes tested) (Fig. 2). We identified a large set of conserved proteins required for ciliogenesis or optimal ciliary function in multicilated cells, revealing novel functions for the homologs of centrin 2 (Smed-cen2) and the uncharacterized protein Cep78 in centriole anchoring and ciliogenesis (Fig. 2, S7-S11). Only two genes coding for pericentrin and centriolin homologs were found to affect regeneration via a novel, cell proliferation-independent mechanism (Fig. 2, S12, S13), likely reflecting the fact that these proteins have centrosome-independent functions (18, 19). Thus, most centrosome components conserved in planarians are required for centriole assembly or ciliogenesis in multiciliated cells, which demonstrates that centrosome duplication in proliferating cells, and centriole assembly through the acentriolar pathway in multiciliated cells, rely on essentially the same mechanisms (Fig. S14). A key difference between the two pathways appears to be the involvement of SPD-2/Cep192 in centrosome duplication (5) but not in the acentriolar pathway, as supported by the loss of this protein family from the planarian genome.
The loss of the centrosome in planarians and possibly also in schistosomes is a remarkable event in the evolutionary history of animals. To determine when in evolution this event took place, we searched for homologs of the centrosome signature proteins in the genomes of species closely related to the planarian, in particular the basal flatworm Macrostomum lignano. Importantly, we found Macrostomum homologs for SPD-2/Cep192 and Nek2 (Fig. S15), which supports that the centrosome was still present during early stages of flatworm evolution, and was subsequently lost in the lineage leading to planarians and parasitic flatworms. In agreement with this hypothesis, centrioles are found at the poles of mitotic cells in Macrostomum embryos (20), in contrast to what we observed in planarians.
It is remarkable that the loss of such a conserved organelle as the centrosome occurred within non-parasitic flatworms, as cellular and developmental processes appear largely conserved between these species (21). A significant difference can be found in the mode of embryonic cleavage however. Macrostomum retained the ancestral spiral cleavage, also found in annelids and mollusks, which relies on a stereotypical pattern of cell division orientation (Fig. 3b). In contrast, planarian and Schistosoma embryos undergo divergent modes of embryonic cleavage, which apparently do not involve oriented cell divisions (22, 23). We hypothesize that centrosome loss occurred concomitantly with the loss of the spiral cleavage and oriented cell divisions in the ancestor of planarians and schistosomes (Fig 3b). Our results suggest that selective pressure to maintain the centrosome in animals comes from the need to coordinate specific developmental processes rather than a fundamental cellular requirement for the organelle. Our work furthermore uncovers an important aspect of cell division of the neoblasts, the totipotent stem cells that underlie planarian amazing regeneration ability.
Supplementary Material
Acknowledgements
We thank J. Rink for sharing reagents and helping with planarian embryo analysis; R. Zalpuri and K. McDonald for help with TEM sample preparation; N. Tang for help with cryostat sectioning; K. Thorn and the Nikon Imaging Center for help with confocal imaging; Y. Chen and M. Braunfeld for help with TEM acquisition; L. Holt, D. David, M. Bettencourt-Dias and M. Bornens for critical reading of the manuscript, the Marshall lab and the Sánchez lab for insightful comments. This work was supported by the W.M. Keck Foundation (W.F.M.), NIH grants GM077004 (W.F.M.) and GM57260 (A.S.A.). A.S.A. is a Howard Hughes Medical Institute investigator.
References
- 1.Calarco-Gillam PD, et al. Cell. 1983;35:621. doi: 10.1016/0092-8674(83)90094-6. [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt-Dias M, et al. Curr Biol. 2005;15:2199. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 3.Dawe HR, et al. J Cell Sci. 2007;120:7. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 4.Basto R, et al. Cell. 2006;125:1375. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 5.Azimzadeh J, Marshall WF. Current biology : CB. 2010;20:R816. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rink JC, et al. Science. 2009;326:1406. doi: 10.1126/science.1178712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rompolas P, et al. Molecular Biology of the Cell. 2010;21:3669. doi: 10.1091/mbc.E10-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner DE, et al. Science. 2011;332:811. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddien PW, et al. Dev Cell. 2005;8:635. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morita M, Best JB. J Exp Zool. 1974;187:345. doi: 10.1002/jez.1401870305. [DOI] [PubMed] [Google Scholar]
- 11.Sakai-Wada A, Mukaidani C. Chromosoma. 1985;93:43. [Google Scholar]
- 12.Stevens NR, et al. Curr Biol. 2007;17:1498. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues-Martins A, et al. Cell Cycle. 2008;7:11. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- 14.Bobinnec Y, et al. J Cell Biol. 1998;143:1575. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JS, et al. Nature. 2003;426:570. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 16.Kraemer N, et al. Cell Mol Life Sci. 2011;68:1719. doi: 10.1007/s00018-011-0635-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayor T, et al. Journal of Cell Science. 2002;115:3275. doi: 10.1242/jcs.115.16.3275. [DOI] [PubMed] [Google Scholar]
- 18.Gromley A, et al. Cell. 2005;123:75. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 19.Delaval B, Doxsey SJ. J Cell Biol. 2010;188:181. doi: 10.1083/jcb.200908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyler S. Hydrobiologia. 1981;84:231. [Google Scholar]
- 21.Egger B, et al. Dev Genes Evol. 2007;217:89. doi: 10.1007/s00427-006-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardona A, et al. Dev Genes Evol. 2006;216:667. doi: 10.1007/s00427-006-0094-3. [DOI] [PubMed] [Google Scholar]
- 23.Jurberg AD, et al. Dev Genes Evol. 2009;219:219. doi: 10.1007/s00427-009-0285-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.