Recent work by Kvon et al. in this issue of Genes & Development identifies highly occupied target (HOT) regions as active transcriptional enhancers. This Perspective discusses how HOT DNAs may direct cell-specific patterns of gene expression during development.
Keywords: HOT region, transcription factor, enhancer/cis-regulatory element, Drosophila embryo development
Abstract
Enhancers mediate localized patterns of gene expression during development. A common feature of “traditional” enhancers is the presence of clustered binding motifs for sequence-specific transcription factors (TFs). In this issue of Genes & Development, Kvon and colleagues (pp. 908–913) present new evidence that HOT (highly occupied transcription) DNAs direct specific patterns of gene expression, despite being depleted for TF-binding motifs.
Role of enhancers in development
Understanding how a fertilized egg produces a complex animal remains one of the great challenges in biology. Integral to this process is the precise and dynamic regulation of transcription. This is established by the integration of complex signaling and transcription networks converging on enhancer DNAs. Our current understanding is that such enhancers are composed of clusters of binding sites for sequence-specific transcription factors (TFs) that mediate combinatorial control of gene expression. Developmental enhancers are typically 200 base pairs (bp) − 1 kb in length and contain multiple binding sites for several TFs, both activators and repressors (Arnosti and Kulkarni 2005; Levine 2010).
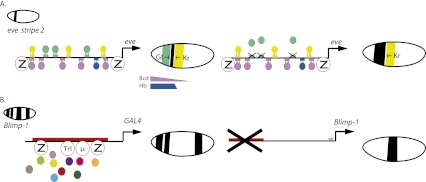
The Drosophila eve stripe 2 enhancer is one of the best-characterized enhancers in development. eve encodes a homeodomain protein expressed in a series of seven stripes that control segmentation of the Drosophila embryo (Goto et al. 1989; Harding et al. 1989). Like many patterning genes, it is regulated by multiple enhancers, each responsible for a subset (e.g., specific stripes) of the complete expression pattern. The minimal, 480-bp Bcd stripe 2 enhancer is located ∼1 kb upstream of the transcription start site (Small et al. 1992). It contains 12 TF-binding sites: six for the Bicoid (Bcd) and Hunchback (Hb) activators and six for the Kruppel (Kr) and Giant (Gt) repressors (Stanojevic et al. 1991). Bcd and Hb have the capacity to activate the enhancer in the anterior half of the embryo. The anterior and posterior stripe borders are established by the Gt and Kr repressors, respectively (Small et al. 1991, 1992). Thus, integrated input of repressors and activators controls the stripe 2 expression pattern. Optimal activation also depends on a global maternal activator, Zelda (Struffi et al. 2011) (summarized in Fig. 1A).
Figure 1.
Comparison of a traditional enhancer and HOT enhancer. (A) Traditional enhancer. eve stripe 2; the endogenous expression pattern is shown in the inset. The eve stripe 2 enhancer is ∼480 bp and has defined transcription factor-binding sites (shown as rectangles) activators are Bcd (purple) and Hb (blue), and repressors are Kr (yellow) and Gt (green). Each transcription factor binds its corresponding recognition sequence. The integration of the Bcd and Hb activators and Gt and Kr repressors establishes stripe 2 expression in a specific location. Loss of any of the binding sites affects this pattern. For example, loss of Gt (green) repressor sites causes an anterior expansion of the normal pattern. eve 2 contains noncanonical binding sites for Zelda (Struffi et al. 2011), consistent with its proposed role as a global activator (Liang et al. 2008). (B) HOT enhancer. Blimp-1; the endogenous expression pattern is shown in the inset. The HOT DNA region found ∼9–12 kb upstream of the Blimp-1 transcription start site contains binding sites for Zelda and Trithorax-like (Trl) (GAGA). Additional, unassigned motifs are also found in this region and are depicted by the binding of μ. More than 10 other TFs (multicolored circles) are associated with this region, but the corresponding binding motifs are generally absent. This HOT region can drive GAL4 expression in three of the four Blimp-1 stripes. A bacterial artificial chromosome (BAC) construct containing ∼45 kb of genomic DNA encompassing the endogenous Blimp-1 locus gives rise to the endogenous expression pattern (not shown). However, removal of the HOT DNA results in the loss of three of the four Blimp-1 stripes. A putative traditional enhancer might drive expression of the central Blimp-1 stripe (indicated in gray).
Whole-genome studies offer the promise of systematically identifying all of the enhancers engaged in specific developmental processes. A major goal of such studies is to “crack” cis-regulatory codes, whereby enhancer activities are predicted from the primary sequence of genomic DNAs. Most such efforts focus on the identification of clusters of TF-binding motifs (e.g., Markstein et al. 2002). In some cases, reduced nucleosome occupancy has been used to improve the accuracy of in silico enhancer predictions (Khoueiry et al. 2010). Such approaches have identified a number of novel enhancers engaged in a variety of processes. However, no explicit code or codes have been identified, and many TF genomic DNA clusters have no apparent enhancer function.
HOT (highly occupied transcription) DNAs are depleted for TF-binding motifs
A number of recent studies identified the distribution of a variety of TFs throughout several genomes. These investigations have identified a novel class of genomic DNAs in Caenorhabditis elegans (Gerstein et al. 2010), Drosophila (Moorman et al. 2006; Roy et al. 2010; Negre et al. 2011), and humans (unpublished ENCODE result cited in Negre et al. 2011) called HOT regions or HOT DNAs. Using the binding profiles of 41 different TFs, nearly 2000 HOT DNAs were identified in the Drosophila genome, each binding an average of 10 different TFs (Roy et al. 2010). In C. elegans, 22 different TFs identified 304 HOT DNAs containing 15 or more TFs (Gerstein et al. 2010). A large number of DNAs containing 10–14 TFs were also identified. Surprisingly, HOT DNAs do not appear to be enriched for the DNA motifs recognized by these TFs (Moorman et al. 2006; MacArthur et al. 2009; Gerstein et al. 2010; Roy et al. 2010). Moreover, Bcd proteins lacking the homeodomain nonetheless bind to certain HOT DNAs in Drosophila, suggesting that protein–protein interactions can be sufficient for their recruitment (Moorman et al. 2006).
Drosophila HOT DNAs share certain sequence features, including GAGA elements (see below) and the TAGteam motif, which binds Zelda (Liang et al. 2008; Satija and Bradley 2012). Like other regions containing these regulatory elements, HOT DNAs exhibit increased nucleosome turnover and histone H3.3, indicative of “open” chromatin (Jin et al. 2005). Genes proximal to HOT DNAs exhibit increased transcriptional activity during early development, a common feature of genes containing Zelda sites (Moorman et al. 2006; Satija and Bradley 2012).
Known and predicted functions
Prior to the study by Kvon et al. (2012), the only functional information about HOT DNAs came from C. elegans, where they were found to mediate ubiquitous expression in larvae (Liu et al. 2009; Gerstein et al. 2010). Furthermore, the genes associated with HOT DNAs in C. elegans are highly expressed and often encode essential functions. As seen in Drosophila, TF-binding motifs are not always required for the association of TFs with HOT DNAs (Gerstein et al. 2010). There are a few overrepresented sequence motifs, but the identities of the corresponding TFs are not known. As in Drosophila, C. elegans HOT DNAs are often associated with open chromatin and increased expression of linked genes (Gerstein et al. 2010).
Several additional functions were proposed for HOT DNAs. It was suggested that they might serve as sinks or buffers for sequestering excess TFs (MacArthur et al. 2009). Motifs similar to BEAF-32 and Trithorax-like (Trl) have been identified in HOT DNAs, suggesting that they might function as insulators (chromosomal boundary domains) (Roy et al. 2010). HOT DNAs were thought to be associated with DNA origins of replication. However, more recent studies suggest that such origins tend to be associated with open chromatin, rather than HOT DNAs (MacAlpine et al. 2010; Satija and Bradley 2012).
HOT DNAs function as tissue-specific enhancers
Kvon et al. (2012) found that HOT DNAs overlap with only 18% of known transcriptional enhancers in the Drosophila genome, which coincides with earlier findings (Negre et al. 2011). Nonetheless, 102 of 108 HOT DNAs were found to direct cell-specific patterns of gene expression during embryogenesis. The expression patterns are similar to those seen for neighboring genes and recapitulated the expression profiles of developmentally regulated genes. For example, the HOT DNA associated with the Blimp-1 gene directs three of the four expression stripes of the endogenous locus (Fig. 1B).
Spatially restricted expression patterns were seen in all major tissues, including the mesoderm, dorsal ectoderm, and neurogenic ectoderm. In contrast to findings in C. elegans, <10% of Drosophila HOT DNAs were found to direct ubiquitous expression patterns (Gerstein et al. 2010). However, genes located near HOT DNAs tend to be more strongly expressed than unlinked genes, as seen in C. elegans (Gerstein et al. 2010; Roy et al. 2010; Satija and Bradley 2012). Furthermore, HOT DNAs in both species contain GAGA elements. Thus, Drosophila and C. elegans HOT DNAs share some common features.
Kovn et al. (2012) note that some of the TFs associated with HOT DNAs have a neutral effect on enhancer activity. For example, not all of the HOT DNAs containing Twist mediate expression in the mesoderm, where it is a key activator of gene expression (Baylies and Bate 1996). How and why are neutral TFs maintained in HOT DNAs during evolution? Natural selection preserves the proper number, arrangement, and affinities of essential recognition sequences, and eliminates sites that might interfere with enhancer activity. However, TFs that exert minor effects on enhancer activities might be tolerated. Such neutral binding sites might be a rich source for de novo creation of enhancers from nonfunctional sequences (Birney et al. 2007). Whole-genome data sets, such as chromatin immunoprecipitation (ChIP) coupled with deep sequencing (ChIP-seq) and RNA sequencing (RNA-seq), from different species within a phylogeny will enable distinction of functional conservation, neutral divergence, and species-specific gene regulation. It is of interest to note that TF-binding sites within a HOT DNA influence enhancer activity, whereas there are no binding site motifs for neutral TFs.
What's next?
Drosophila HOT enhancers are enriched for Zelda and GAGA sites, which have been implicated in maintaining open chromatin (Nakayama et al. 2007; Harrison et al. 2011). These sites represent part of the genomic “signature” that permits prediction of additional HOT DNAs (Kvon et al. 2012). It will be important to determine their role in the formation or function of HOT enhancers. Both GAGA- and Zelda-binding sites are thought to be permissive and facilitate the binding or function of specific activator proteins (Nakayama et al. 2007; Harrison et al. 2011; Nien et al. 2011). It would be informative to determine whether HOT DNAs are lost in mutant embryos lacking Zelda or Trl, which binds GAGA.
Zelda is thought to regulate hundreds of genes during the maternal/zygotic transition (MZT), ∼2 h after fertilization (Liang et al. 2008). HOT enhancers might therefore represent an important class of early zygotic enhancers. However, HOT enhancers also direct gene expression during later stages of development, suggesting that they are not exclusively engaged in the MZT. Zelda sites in traditional enhancers that are active after the MZT have been proposed to coordinate the activation of gene batteries and to increase the expressivity of gene networks (Nien et al. 2011). It is conceivable that HOT DNAs also mediate such functions. While Zelda is present in both early embryonic enhancers and HOT DNAs, enrichment of GAGA sites is a more specific feature of HOT DNAs (in both C. elegans and Drosophila). Kvon et al. (2012) suggest that GAGA may be able to recruit other TFs, since Trl forms complexes with itself and Tramtrack (Bardwell and Treisman 1994).
HOT enhancers have a number of unusual properties; most notably, they bind many TFs via mass action despite the general depletion of specific TF-binding motifs. Future studies will identify TFs responsible for the localized expression patterns mediated by HOT enhancers. The lack of obvious binding motifs and the possible neutral binding of TFs to these regions will make this task challenging.
As we learn more about HOT enhancers, it will be interesting to see whether their mode of TF occupancy is qualitatively distinct from traditional enhancers. Alternatively, they could work like traditional enhancers, but the key TF sequence motifs are not yet known. Do HOT enhancers possess architectures that are more or less constrained than conventional enhancers? HOT enhancers might provide regulatory robustness in gene expression when conventional enhancers are compromised. Their removal from otherwise normal bacterial artificial chromosome (BAC) transgenes might help reveal such functions.
It is interesting to note that in yeast, binding of 10 or more TFs to a single cis-regulatory DNA is very rare (<1% of all regulatory elements), with the vast majority of promoter regions occupied by just a few TFs (Lee et al. 2002; Harbison et al. 2004). This observation raises the possibility that HOT DNAs are an adaptation of metazoan genomes, but what exactly are their roles? In time, we will find answers to these questions.
Conclusion
Until now, a common feature of all known enhancers was the presence of clustered binding motifs, mediating combinatorial patterns of gene expression. However, Kvon et al. (2012) have shown that HOT DNAs can function as enhancers and direct specific patterns of gene expression even though they are depleted for TF-binding motifs. It remains to be seen whether HOT enhancers possess distinctive properties as compared with conventional enhancers. This study provides a vivid example of the mysteries of the regulatory genome.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.192583.112.
References
- Arnosti DN, Kulkarni MM 2005. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898 [DOI] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R 1994. The POZ domain: A conserved protein–protein interaction motif. Genes Dev 8: 1664–1677 [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M 1996. twist: A myogenic switch in Drosophila. Science 272: 14811484. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Macdonald P, Maniatis T 1989. Early and late periodic patterns of even skipped expression are controlled by distinct regulatory elements that respond to different spatial cues. Cell 57: 413–422 [DOI] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K, Hoey T, Warrior R, Levine M 1989. Autoregulatory and gap gene response elements of the even-skipped promoter of Drosophila. EMBO J 8: 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 7: e1002266 doi: 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cai Y, Li B, Conaway RC, Workman JL, Conaway JW, Kusch T 2005. In and out: Histone variant exchange in chromatin. Trends Biochem Sci 30: 680–687 [DOI] [PubMed] [Google Scholar]
- Khoueiry P, Rothbacher U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P 2010. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol 20: 792–802 [DOI] [PubMed] [Google Scholar]
- Kvon EZ, Stampfel G, Yáñez-Cuna JO, Dickson BJ, Stark A 2012. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev (this issue) doi: 10.1101/gad.188052.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298: 799–804 [DOI] [PubMed] [Google Scholar]
- Levine M 2010. Transcriptional enhancers in animal development and evolution. Curr Biol 20: R754–R763 doi: 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Long F, Peng H, Aerni SJ, Jiang M, Sanchez-Blanco A, Murray JI, Preston E, Mericle B, Batzoglou S, et al. 2009. Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell 139: 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80 doi: 10.1186/gb-2009-10-7-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci 99: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. 2006. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci 103: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S 2007. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev 21: 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C 2011. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7: e1002339 doi: 10.1371/journal.pgen.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Bradley RK 2012. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res doi: 10.1101/gr.130682.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M 1991. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes Dev 5: 827–839 [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J 11: 4047–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254: 1385–1387 [DOI] [PubMed] [Google Scholar]
- Struffi P, Corado M, Kaplan L, Yu D, Rushlow C, Small S 2011. Combinatorial activation and concentration-dependent repression of the Drosophila even skipped stripe 3+7 enhancer. Development 138: 4291–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]