Excessive caloric intake is associated with increased risk for cancer, while the nonobese state may be protective through mechanisms that reduce oxidative stress. In this review, Dang discusses the links between metabolism and cancer, which range from the low incidence of cancer in large mammals with low specific metabolic rates to altered cancer cell metabolism resulting from mutated enzymes or cancer genes.
Keywords: caloric restriction, cancer, glycolysis, metabolism, obesity, oncogenes, tumor suppressors
Abstract
Metabolism generates oxygen radicals, which contribute to oncogenic mutations. Activated oncogenes and loss of tumor suppressors in turn alter metabolism and induce aerobic glycolysis. Aerobic glycolysis or the Warburg effect links the high rate of glucose fermentation to cancer. Together with glutamine, glucose via glycolysis provides the carbon skeletons, NADPH, and ATP to build new cancer cells, which persist in hypoxia that in turn rewires metabolic pathways for cell growth and survival. Excessive caloric intake is associated with an increased risk for cancers, while caloric restriction is protective, perhaps through clearance of mitochondria or mitophagy, thereby reducing oxidative stress. Hence, the links between metabolism and cancer are multifaceted, spanning from the low incidence of cancer in large mammals with low specific metabolic rates to altered cancer cell metabolism resulting from mutated enzymes or cancer genes.
Ninety years ago, Otto Warburg published a body of work linking metabolism and cancer through enhanced aerobic glycolysis (also known as the Warburg effect) that distinguishes cancer from normal tissues (Warburg 1956; Hsu and Sabatini 2008; Vander Heiden et al. 2009a; Koppenol et al. 2011). The conversion of glucose to lactate, which can occur in hypoxic normal cells, persists in cancer tissues despite the presence of oxygen that would normally inhibit glycolysis through a process termed the Pasteur effect. We now know that sustained aerobic glycolysis (diminished Pasteur effect) in certain cancer cells is linked to activation of oncogenes or loss of tumor suppressors (Vander Heiden et al. 2009a; Levine and Puzio-Kuter 2010; Cairns et al. 2011; Koppenol et al. 2011). However, the Warburg effect in itself does not explain the persistence of mitochondrial respiration in many cancers or the role of aerobic glycolysis in cell mass accumulation and cell proliferation. Furthermore, glucose, which comprises carbon, hydrogen, and oxygen, could not provide all of the building blocks for a growing cell, which is composed of other elements such as nitrogen, phosphorus, and sulfur. In this regard, other nutrients are, a priori, required to build new cells. How growth signaling leads to nutrient uptake and building of a cell is discussed below.
As neoplastic cells accumulate in three-dimensional multicellular masses, local low nutrient and oxygen levels trigger the growth of new blood vessels into the neoplasm. The imperfect neovasculature in the tumor bed is poorly formed and inefficient and hence poses nutrient and hypoxic stress (Carmeliet et al. 1998; Bertout et al. 2008; Semenza 2010). In this regard, cancer cells and stromal cells can symbiotically recycle and maximize the use of nutrients (Sonveaux et al. 2008). Hypoxic adaptation by cancer cells is essential for survival and progression of a tumor. The role of hypoxia in cancer cell metabolism is discussed in the context of tumorigenesis (Semenza 2010).
In addition to cell-autonomous changes that drive a cancer cell to proliferate and contribute to tumorigenesis, it has also been observed that alterations in whole-organism metabolism such as obesity are associated with heightened risks for a variety of cancers (Khandekar et al. 2011). Although obesity triggers adult-onset diabetes and elevates glucose and insulin resistance, how obesity increases cancer risk is not simply a matter of increased circulating glucose. It stands to reason that the converse—nutrient deprivation—might be true; caloric restriction would be expected to result in protection from cancer risks. Despite the fact that the converse is true, our understanding of how caloric restriction limits tumorigenesis is still rudimentary (Hursting et al. 2010). Our current understanding of how organismal metabolism may be linked to tumorigenesis and major themes linking metabolism to cancer are discussed below in hope of provoking a new dialogue regarding the various connections between metabolism and cancer.
Negative entropy and building blocks for growing cells
As Erwin Schrodinger noted in What is Life? (Schrodinger 1992), life is a physical system that maintains structure and avoids decay by feeding on negative entropy through metabolism, a term derived from a Greek word describing the exchange of materials. However, a proliferating cell must capture enough energy and mass to replicate, in addition to the energy required to dampen entropy. In this regard, by studying batch cultures of L cells carefully fed and controlled, Kilburn et al. (1969) documented that the amount of additional energy (assuming that glucose is the main substrate) to produce a new cell is 50% above the baseline required to maintain cellular homeostasis. Hence, it is surmised that the amount of ATP in a proliferating cell is not dramatically different from a resting cell, but the proliferating cell must accumulate biomass, replicate DNA, and divide. In this context, glucose and glutamine are regarded as two major substrates for proliferating cells, providing both ATP and carbon skeletons for macromolecular synthesis (Locasale and Cantley 2011).
Building cells with glucose and glutamine
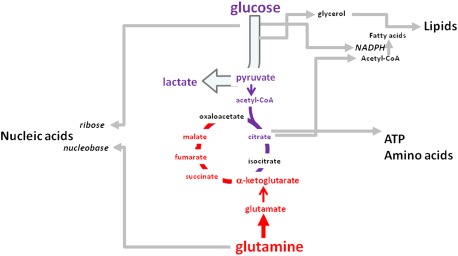
Glucose is transported into cells by facilitative transporters and then trapped intracellularly by glucose phosphorylation (Berg et al. 2002). The hexose phosphate is further phosphorylated and split into three-carbon molecules that are converted to glycerol for lipid synthesis or sequentially transformed to pyruvate. Pyruvate is converted to acetyl-CoA in the tricarboxylic acid (TCA) cycle, is transaminated to alanine, or becomes lactate, particularly under hypoxic conditions. Formation of citrate from acetyl-CoA and oxaloacetate permits a new round of TCA cycling, generating high-energy electrons, CO2, and carbon skeletons that could be used for biosynthesis or anaplerosis. Citrate itself could be extruded into the cytosol and then converted to acetyl-CoA by ATP citrate lyase (ACLY) for fatty acid synthesis and generation of biomembranes. Glucose, through the pentose phosphate pathway (PPP), generates ribose for nucleic acid synthesis and NADPH for reductive biosynthesis (Fig. 1).
Figure 1.
Glucose and glutamine feed cell growth and proliferation. Glucose and glutamine are depicted to contribute to glycolysis (conversion of glucose to pyruvate) and the TCA cycle, which is shown as a hybrid cycle comprising glucose and glutamine carbons. Carbon skeletons from glycolysis and the TCA cycle contribute to macromolecular synthesis for the growing cell.
Glutamine, which circulates with the highest concentration among amino acids, serves as a major bioenergetic substrate and nitrogen donor for proliferating cells (DeBerardinis and Cheng 2010). Glucose and glutamine are required for hexosamine biosynthesis (Wellen et al. 2010). Glutamine enters into the TCA via its conversion to glutamate and then to α-ketoglutarate (aKG), a key TCA cycle intermediate that is also a cofactor for dioxygenases (Chowdhury et al. 2011; Xu et al. 2011). Once in the TCA cycle, glutamine carbon skeletons contribute to a hybrid TCA cycle comprising carbons from glucose mixed with those of glutamine (Fig. 2). Under hypoxia, the hypoxia-inducible factor HIF-1 activates pyruvate dehydrogenase kinase (PDK1) that inhibits pyruvate dehydrogenase and the conversion of pyruvate to acetyl-CoA, thereby shunting pyruvate to lactate (Kim et al. 2006). In resting cells, this constitutes the canonical anaerobic glycolysis pathway that is well established in the didactic literature.
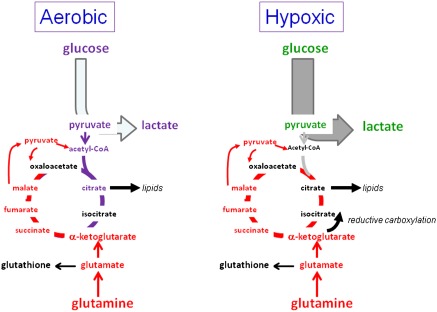
Figure 2.
Hypoxic rewiring of metabolism. While aerobic proliferating cells use glucose and glutamine for biomass production through the TCA cycle, hypoxic cells shunt glucose to lactate and rewire glutamine metabolism. Glutamine can be used to drive the TCA cycle independently of glucose or contribute to lipid synthesis via IDH-mediated reductive carboxylation of ketoglutarate generated from glutamine.
In proliferating cells, hypoxia, which diverts glucose to lactate, does not attenuate glutamine catabolism through the TCA cycle. In fact, glutamine could contribute to citrate and lipid metabolism through the reversal of the TCA cycle or reductive carboxylation of aKG by isocitrate dehydrogenase (IDH) to form citrate or through forward cycling of glutamine carbons (Fig. 2; Wise et al. 2011; Metallo et al. 2012; Mullen et al. 2012). Reductive carboxylation was first documented as a means for normal brown fat cells to synthesize lipids and was subsequently implicated as a way for hypoxic cancer cells to synthesize lipid from glutamine to grow (Yoo et al. 2008). Under glucose limitation, the TCA cycle could also be reprogrammed and driven solely by glutamine, generating citrate that consists of only glutamine carbons (Le et al. 2012). As such, hypoxic proliferating cells (perhaps as in the case of endothelial cells) reprogram the TCA cycle to maximize the use of glutamine for lipid synthesis.
It is notable that certain cells could also take up free fatty acids from media to support their macromolecular needs, whether for fatty acid oxidation (FAO) or direct insertion into the growing cells' membranes (Samudio et al. 2010; Zaugg et al. 2011). Quiescent primary human T cells and resting human B cells use FAO, but upon growth stimulation, these cells switch to glycolysis and glutaminolysis (Wang et al. 2011; Le et al. 2012). In this regard, inhibition of FAO in primary human acute myelogenous leukemia (AML) cells decreased quiescent leukemic progenitor cells (Samudio et al. 2010). Since ongoing fatty acid synthesis produces malonyl-CoA that inhibits mitochondrial import of fatty acids by CPT1, it remains unclear whether proliferating cells undergoing fatty acid synthesis could simultaneously use FAO. It is possible, as suggested by studies of human AML cells and of lymphocytes, that FAO may be used by cancer-initiating or resting cancer stem cells.
Oxygen radicals: signals, toxins, and stress
Part and parcel of cellular metabolism is the production of toxic by-products, which must be titrated for cell survival and maintenance of genome integrity (Ray et al. 2012). The major by-products, known collectively as the reactive oxygen species (ROS), comprise H2O2, superoxide O2−, and hydroxyl radical OH− (Finkel 2011). These ROS, which are produced from the mitochondria or NOX (NADPH oxidases), damage membranes and can be mutagenic and are hence titrated by glutathione and peroxiredoxins. Superoxide dismutases are essential for redox homeostasis through the conversion of superoxide to hydrogen peroxide, which is neutralized by catalase to water and oxygen. Oxidative stress resulting from altered cancer metabolism is expected to change the ability of cancer cells to handle ROS. Increased ROS was documented to modify a critical sulfhydryl group of pyruvate kinase M2 (PKM2), rendering it inactive and resulting in the shunting of glucose away from glycolysis toward the PPP (Anastasiou et al. 2011). The PPP generates NADPH, which reduces glutathione into an active antioxidant that protects the cell. In this manner, the shunting of glucose away from glycolysis toward the PPP is an essential element of redox homeostasis.
In addition to oxidation of PKM2, increased ROS can stabilize HIF-1. HIF-1, in turn, activates target genes such as PDK1, which diverts pyruvate away from mitochondrial oxidation, and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4), which degrades 2,6-fructose bisphosphate (2,6-FBP) (Keith et al. 2012; Semenza 2012). 2,6-FBP is a powerful allosteric activator of phosphofructose kinase 1 (PFK1), which converts fructose-1-phosphate to fructose-1,6-bisphosphate (1,6-FBP) at a rate-limiting step in glycolysis (Yalcin et al. 2009). Hence, increased PFKFB4, as observed in prostate cancer cell lines, would diminish PFK1 activity and divert glucose into the PPP shunt, elevating NAPDH to titrate ROS (Ros et al. 2012). It is notable, however, that hypoxia also elevates PFKFB3, which drives glycolysis and can oppose PFKFB4; as such, the balance between PFKFB3 and PFKFB4 activities is critical for shunting glucose into glycolysis versus the PPP.
It is also notable that ROS plays a role in intracellular signaling through alterations of the oxidative status of regulatory protein sulfhydryl moieties (Finkel 2011). In this regard, the antioxidant capability of cancer cells may profoundly influence their responses to metabolic stresses, with resistance to therapy linked to increased antioxidant capacity. Hence, a systematic way to measure cellular antioxidant capacity would be instructive and essential for any attempt to target cancer metabolism for therapy.
Nutrient sensing, signaling, and cell growth
The unicellular baker's yeast Saccharomyces cerevisiae is programmed to sense nutrients and activate signal transduction pathways that initiate biomass accumulation. Under glucose-limited growth conditions, yeast cells display oscillations in oxygen consumption alternating with reductive glycolytic phases. DNA replication is normally restricted to the oscillating reductive phase such that yeast cell cycle mutants that uncouple DNA replication from the reductive metabolic phase exhibited heightened spontaneous mutations. These observations suggest that coupling of circadian, metabolic, and cell division cycles is essential for genome integrity. (Chen et al. 2007). However, it should be noted that the coupling of these cycles is highly dependent on the experimental conditions because these cycles could be uncoupled under other nutrient-limiting conditions (Silverman et al. 2010; Slavov et al. 2011).
Cell growth or biomass accumulation occurs largely through the genesis of ribosomes, which are essential factories for building blocks of the growing cell and account for over half of the cellular dry mass. Mutations that cause constitutive expression of ribosome biogenesis genes result in mutant yeasts that are addicted to nutrients—glucose and glutamine, whose sensing by yeast are transmitted through Ras and mTORC1, respectively (Figs. 3, 4; Lippman and Broach 2009). With nutrient deprivation, yeast cells withdraw from the cell cycle (Klosinska et al. 2011). In contrast, mammals must feed to survive, unless they are capable of undergoing hibernation or a state of suspended animation with low metabolic rates. In this regard, certain mammals could store up enough energy as fat and slow metabolism sufficiently to survive long winter months (Dark 2005). Hydrogen sulfide, produced from cysteine via cystathione γ-lyase and cystathione β-synthase, has been implicated in reprogramming cellular metabolism by inhibiting cytochrome C oxidase, thereby lowering mitochondrial function for hibernation (Collman et al. 2009). Aside from hibernation, which is limited to certain species, other mammals can adapt to starvation or caloric restriction.
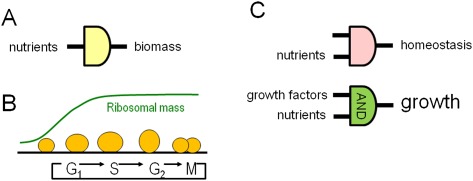
Figure 3.
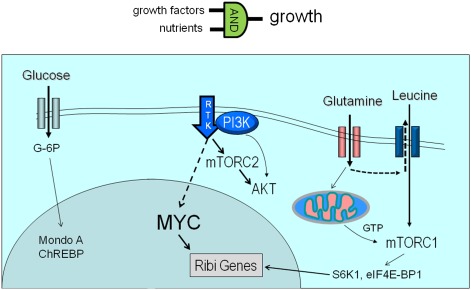
Nutrient signaling for biomass accumulation. (A) Yeasts could transmit nutrient sensing to biomass accumulation without specific growth factors. (B) A large fraction of cellular mass comprises ribosomes that accumulate in the G1- to S-phase period of the cell cycle. (C) Mammalian cells at rest use nutrients to maintain structure and homeostasis of membrane potentials. Upon stimulation with growth factors, signals from nutrients and growth factor receptors are integrated (via an AND logic gate) to stimulate cell growth or biomass accumulation.
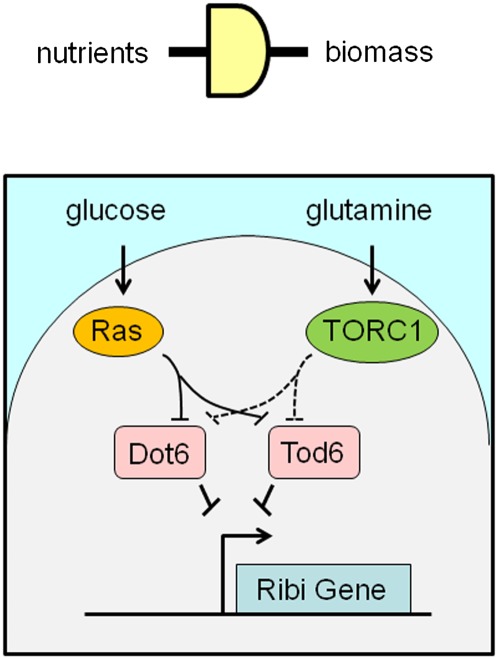
Figure 4.
Nutrient sensing and yeast cell growth. Glucose and glutamine are depicted to signal via Ras and TORC1, respectively, to inhibit repressors (Dot6 and Tod6) of ribosomal biogenesis (Ribi) genes.
The yin-yang nature of AMPK and mTOR pathways
With nutrient deprivation, mammals could mobilize glycogen from the liver and fat stores from adipose tissues to produce glucose for the brain and red cells. Upon starvation, they could mobilize amino acids from muscles, particularly alanine and glutamine (Berg et al. 2002). Glutamine released into the circulation is used by the kidney for gluconeogenesis by conversion to glutamate and then to aKG, which ends up as oxaloacetate and phosphoenol pyruvate for glucose synthesis (Owen et al. 2002). The ammonia released from glutamine is excreted into alkalinized urine. At the cellular level, low glucose or glutamine levels decrease ATP levels, and an increase in the AMP to ATP ratio is sensed by AMPK, which phosphorylates substrates to enhance energy production while diminishing processes that consume energy (Mihaylova and Shaw 2011). AMPK phosphorylates and inhibits acetyl-CoA carboxylase, which consumes ATP and produces malonyl-CoA for fatty acid synthesis. Additionally, AMPK-mediated phosphorylation of ULK-1 triggers autophagy, which recycles cellular components for energy production (Rabinowitz and White 2010; Singh and Cuervo 2011). Diminished malonyl-CoA levels relieve allosteric inhibition of CPT-1, which permits the translocation of fatty acids into the mitochondrion for oxidation to produce ATP. Furthermore, AMPK phosphorylates TSC2, which inhibits mTOR, the master stimulator of cell growth downstream from PI3K and AKT. Hence, under conditions of starvation, AMPK plays a critical role for cell survival by stimulating energy production and limiting the use of energy by active biosynthetic pathways usually operating in proliferating cells.
When energy supply is ample, particularly during development, mammalian cells bathed in nutrients must also be stimulated by growth factors to accumulate biomass and proliferate, which contrasts with yeast, which only needs to sense nutrients to trigger cell growth (Figs. 3, 5). As such, growth factors such as IGF-1, EGF, or PDGF participate in the stimulation of mammalian cellular biomass accumulation. Downstream from the growth factor-bound receptor tyrosine kinases is the activation of PI3K, which transmits the growth signal to AKT and mTOR (mTORC2) (Fig. 5; Zoncu et al. 2010). mTORC1 is activated by the availability of nutrients, particularly glutamine, which is taken up and then exported extracellularly in a fashion that is coupled with the import of leucine, a key amino acid that is necessary for the mobilization of mTORC1 to lysosomal membranes by G proteins for mTORC1 activation. The activated mTORC1 kinase phosphorylates a number of substrates, including S6K1 and eIF4E-BP1, to stimulate translation, ribosome biogenesis, and growth of the cell. mTORC1 phosphorylates ULK1 to inhibit autophagy when cells are replete with nutrients. Activated mTORC2, on the other hand, activates AKT, which phosphorylates a number of substrates, including hexokinase 2 (HK2), to stimulate glycolysis and activates FOXO3a to inhibit apoptosis and increase mitochondrial biogenesis to support a growing cell (Plas and Thompson 2005; Huang and Tindall 2007; Ferber et al. 2011). Glucose, when converted to glucose-6-phosphate, stimulates MondoA and ChREBP through nuclear translocation (Peterson and Ayer 2012). Under low anaplerotic flux, MondoA represses glucose uptake, whereas when glutamine elevates anaplerosis, MondoA represses TXNIP to stimulate glucose uptake (Fig. 5; Kaadige et al. 2009).
Figure 5.
Mammalian cell growth requires growth factors and nutrients. Glucose is shown to signal to MondoA, which down-modulates glucose metabolism. Glutamine contributes to mTORC1 activation through import of leucine and production of GTP via the TCA cycle. GTP is required for mTORC1 activation by its association with lysosomal membranes. mTORC1 activation of S6K1 stimulates ribosome biogenesis (Ribi) genes. Growth factor signaling through receptor tyrosine kinase (RTK) activates PI3K and mTORC2, resulting in AKT activation that stimulates glucose metabolism. Signal transduction via MEK to MYC initiates a transcriptional program that stimulates Ribi genes, coupled with increased glucose and glutamine metabolic gene expression.
Nutrients also modify the epigenome through metabolic intermediates such as acetyl-CoA, S-adenosylmethionine, NAD+, and aKG (Katada et al. 2012), thereby modifying gene expression. Furthermore, many metabolic enzymes are documented to be acetylated, and in some cases, their activities are modified (Zhao et al. 2010; Guan and Xiong 2011). Hence, metabolic intermediates contribute the complex tapestry of a network that links nutrients to metabolite intermediates, transcription, and regulation of enzyme activities in cell growth, proliferation, and homeostasis.
Growth factor-stimulated transcriptional responses
In addition to the PI3K–AKT–mTORC2 and amino acid–mTORC1 pathways, there is also an orderly growth factor-stimulated transcriptional program with activation of immediate early response genes, such as MYC, JUN, and FOS, and delayed genes that are stimulated by the early response transcription factors (Lau and Nathans 1987). It stands to reason, then, that transcriptional response to growth factor stimulation must trigger the expression of genes that are involved in metabolism and biomass accumulation (Fig. 5). Early studies of serum stimulation of fibroblasts documented Myc as an early response gene and lactate dehydrogenase A (LDHA) as a delayed response gene, but the link between MYC as a transcriptional activator and direct stimulation of LDHA as a Myc target gene was documented a number of years later using model cell lines, providing a direct link between a proto-oncogene and regulation of a gene involved in bioenergetics (Tavtigian et al. 1994; Shim et al. 1997). Recently, use of primary T cells permitted the molecular dissection of the roles of Myc versus HIF-1 in T-cell mitogenesis stimulated by CD3 and CD28 antibodies (Wang et al. 2011). This study documents that Myc is essential for the activation of genes involved in glycolysis and glutaminolysis for cell growth and proliferation such that conditional deletion of c-Myc in T cells results in cells incapable of mounting a growth program. HIF-1, which also stimulates glycolysis but not glutaminolysis, is not necessary for the early T-cell growth response program. Myc-dependent genes involved in polyamine biosynthesis are also highly stimulated in normal T cells. These findings corroborate early studies that link MYC to the regulation of metabolic genes, including ornithine decarboxylase, which is involved in polyamine synthesis and is the first reported metabolic gene directly regulated by Myc, particularly in cancer cells (Bello-Fernandez and Cleveland 1992).
Through the work of many laboratories, MYC emerges as a central regulator of cell growth and proliferation downstream from receptor signaling pathways and a key human oncogene that when deregulated could drive a constitutive transcriptional program for nutrient uptake and biomass accumulation (Dang 2010). Indeed, Myc target genes comprise those involved in glucose transport and glycolysis as well as genes involved in glutaminolysis and fatty acid synthesis (Morrish et al. 2009). Moreover, Myc stimulates genes that are involved in mitochondrial biogenesis and function. In this regard, the Myc-induced transcriptional metabolic program parallels those that are used to maintain the integrity of nonproliferating cells via other transcription factors, such as NRF1 (mitochondrial biogenesis), MondoA/ChREBP (carbohydrate metabolism), or SREBP (cholesterol and fatty acid synthesis). The switch from nonproliferative to proliferative states could be surmised as a switch from homeostatic E-box transcription factors to Myc, which is envisioned to co-opt the regulation of metabolic genes for a proliferating cell.
Because Myc stimulates genes involved in the acquisition of nutrients and the intermediary metabolism, it is hence not surprising that Myc also directly stimulates genes involved in ribosome biogenesis for biomass accumulation. The ability of Myc to stimulate ribosome biogenesis genes distinguishes it as a unique E-box transcription factor capable of coupling the expression of metabolic genes with genes involved in cell mass accumulation (van Riggelen et al. 2010; Ji et al. 2011). Moreover, Myc uniquely activates genes driven by RNA polymerases I and III, which are required for the expression of ribosomal RNAs (Gomez-Roman et al. 2006). Intriguingly, Myc stimulates and p53 opposes the expression of importin 7 (IPO7), which regulates the import of specific ribosomal proteins for ribosomal assembly, suggesting that cellular stresses regulate ribosome biogenesis through p53 (Golomb et al. 2012). In fact, Mdm2 senses nucleolar imbalance in ribosome biogenesis via binding of excess RPL11 and RPL5 with Mdm2, resulting in elevated p53 (Deisenroth and Zhang 2011). A mutation that eliminates Mdm2 binding to ribosomal proteins suppresses p53 tumor suppressor response and accelerates Myc-induced lymphomagenesis, suggesting that overexpression of Myc in cancers induces stress partly via imbalance in ribosomal biogenesis (Macias et al. 2010). Diminished RPL24 expression in mice, on the other hand, decreases Myc-induced lymphomagenesis, indicating that Myc's induction of ribosomal biogenesis is essential for tumorigenesis (Barna et al. 2008).
Myc further stimulates genes involved in nucleotide metabolism and specifically interacts with the E2F family of transcription factors to drive proliferating cells into S phase for DNA replication (Leone et al. 2001; Zeller et al. 2006; Rempel et al. 2009). As a pleiotropic transcription factor, Myc also directly stimulates cell cycle regulatory genes and those directly involved in DNA replication, such as CDK4, CDK6, and MCM genes (Zeller et al. 2006). To complete its job as a growth regulatory factor, Myc also regulates genes involved in G2 phase and mitosis, permitting the duplication of cells. The ability of Myc to stimulate genes involved in motility and repress genes encoding cell adhesion molecules probably reflects the need for mitotic cells to detach and divide (Dang et al. 2006).
Metabolism contributes to cancer
Why don't elephants get cancer?
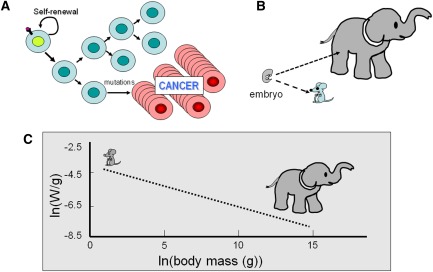
Somatic mutations resulting in oncogene activation and tumor suppressor inactivation are in part due to ROS produced as by-products of metabolism. The incidence of cancer as it relates to animal body size provides a potential link between metabolism and cancer in animals. The prevailing view of mutagenesis stipulates that mutations acquired with cell division could result in oncogenesis and cancer (Fig. 6A). As such, the number of cell divisions an animal sustains to reach adulthood should parallel the number of mutations acquired. Given that embryos start their developmental journey with similar sizes (Fig. 6B), an elephant or a whale would have to undergo many more cell divisions to reach adulthood than a mouse. It stands to reason, then, that the occurrence of cancer in large animals should be much higher in elephants and whales. Known as Peto's paradox, it has been observed that whales have been rarely found to have cancers (Nagy et al. 2007; Caulin and Maley 2011). Likewise, the veterinary literature notes that elephants also rarely have cancers, but that feral mice, with several orders of magnitude smaller body sizes, are estimated to have a lifetime frequency of cancer of 40%.
Figure 6.
(A) Diagram depicting clonal expansion of cancer cells after a hypothetical mutational event. (B) This cartoon illustrates the significantly different number of cell divisions needed to produce an adult elephant versus a mouse from similar-sized embryos. (C) Empirical measurements of specific metabolic rates (energy in watts per gram of tissue) reveal a power law relation with body mass (grams) as illustrated by a linear log–log relation (dashed line). Cartoons of the mouse and elephant are placed over the approximate body mass. Note the significant difference in specific metabolic rates (several orders of magnitude) between the mouse and elephant (see Savage et al. 2007 for details).
The amount of food consumed is inversely proportional to animal body size, which correlates also inversely with specific basal metabolic rates (metabolic rate per unit body mass) (Fig. 6C; Savage et al. 2007). Hence, on a unit mass basis, elephant tissues have much lower metabolic rates than those of mice. Studies of large numbers of animals have revealed a power law relationship between body mass (grams) and specific metabolic rates (watts per grams) (Fig. 6C). Because mammals maintain similar body temperatures, increased body surface area to body mass ratios in smaller animals require higher energy to maintain body temperature; the relationship of body surface area to body mass is inadequate, however, to account for the power law relationship between body mass and metabolic rates. A basis for the power law relation between body mass and specific metabolic rates has been derived theoretically by assuming that the cardiovascular tree branches from the heart sequentially as the body increases in size from mice to elephants. Hence, the ends of the vascular branches are separated farther and farther apart as body size increases, resulting in poorly perfused tissues in larger animals (Herman et al. 2011). As such, the tissue metabolic rates would be lower, most likely due to larger areas of hypoxia distal to blood vessels in large animal. With a small body size, well perfused by nutrients, the higher metabolic rate in mice could be linked to a higher incidence of cancer by means of higher oxidative stress and mutational rates.
Intriguingly, the power law relationship between body mass and specific basal metabolic rates also holds true for the correlation with sleep time (Lo et al. 2004; Siegel 2005). Mice sleep ∼12 h per day, while elephants sleep ∼4 h. Sleep is believed to provide a repair phase particularly in the brain, whose sizes tracks with animal body masses. Thus, metabolically more active animals require longer sleep or repair time. Intriguingly, disturbance of sleep, particularly in shift workers and night nurses, has been linked to higher incidences of cancers, with night shift nurses having a clear statistically higher incidence of breast cancer (Schernhammer et al. 2006; Hansen and Stevens 2011).
Circadian rhythm, metabolism, obesity, and cancer
Animal feeding and metabolism is intimately tied to the rotation of the earth through central and peripheral clocks that regulate metabolic genes, in keeping with the circadian feed and sleep cycle. In addition to the suprachiasmatic optic nuclei in the brain, which senses light and regulates rhythm centrally, individual cells have a transcription factor network, including CLOCK, Bmal, and Per proteins, that generates cyclic expression of genes that are dominated by those involved in metabolism (Sahar and Sassone-Corsi 2009; Bass and Takahashi 2010). Hepatic expression of metabolic genes is rhythmically phased with feeding cycles by circadian transcriptional factors. It stands to reason that the feed and sleep cycle would be regulated in a fashion to maximize energy utilization and storage for survival and repair of daily damages from ongoing oxidative phosphorylation and oxidative stresses. The circadian network of transcription factors is hence critical for daily life of an animal.
Intriguingly, disruption of the feeding cycle as it relates to the day–night cycle can contribute to obesity in mice. Mice entrained to light and dark cycles consume ∼80% of food at night when they are active. Alterations in food availability have been documented to have a significant impact on the circadian clock and body weight. When a high-fat diet is only available during the light or sleep cycle, mice gain more weight than those with the same diet available during the dark, active wake cycle. This observation indicates that circadian regulation of organismal and cellular metabolism is related to the availability of nutrients (Sahar and Sassone-Corsi 2009). It is speculated that the disruption of food intake and sleep by artificial light could be key factors contributing to the epidemic of childhood obesity. The old adages “early to bed and early to rise makes a man healthy, wealthy, and wise” (Benjamin Franklin) and “eat breakfast like a king, lunch like a prince, and dinner like a pauper” (Adelle Davis) may both be sage advice with a scientific foundation. Obese animals and human beings are more susceptible to tumorigenesis, linking circadian disruption to obesity to cancer.
Obesity results in fat tissues that produce adipokines, which in turn causes insulin insensitivity in peripheral tissues. Insulin insensitivity leads to elevated blood glucose levels, which stimulate the production of insulin and IGF-1 from pancreatic β cells (Khandekar et al. 2011). The heightened circulating levels of insulin and IGF-1 are thought to provide a tonic growth stimulation of cells, rendering them susceptible to oncogenic mutations. Studies in animal models provide evidence to support this view; however, key mechanisms contributing to increased mutagenesis remain unclear. Intervention at the insulin and IGF-1 level appears to curb tumorigenesis in animal models, suggesting that growth signaling downstream from insulin/IGF contributes to enhanced tumorigenesis in obese animals. Clearly, the simple tonic stimulation of cells must be accompanied by somatic mutations, which bypass cell cycle checkpoints or apoptotic signals, to trigger tumor formation. Detailed understanding of these mechanisms, including inflammation, will require additional studies (Khandekar et al. 2011).
Caloric restriction and cancer risk
The simple perspective that excess calories contribute to obesity, which in turn heightens tumorigenesis, would lead to the oversimplified conclusion that the converse must also be true. Indeed, caloric restriction in a number of animal models and in epidemiologic studies suggests that limited calories prolong life span (Hursting et al. 2010; Longo and Fontana 2010). Furthermore, caloric restriction in animal studies inhibits tumorigenesis, possibly through reduced IGF-1 levels. Notably, tumors that have activating PI3K mutations are resistant to caloric restriction, suggesting that diminished calories impact growth factor–receptor tyrosine kinase signaling through reduced IGF-1 levels such that mutations that activate the PI3K pathway render cancer cells resistant to caloric restriction (Kalaany and Sabatini 2009). The basis for diminished tumorigenesis in light of low calories is complex and may also be related to autophagy and mitophagy (the cellular process of lysosomally processing and recycling mitochondrial constituents) triggered through AMPK activation in a lowered energy state (Mihaylova and Shaw 2011). Activation of AMPK diminishes mTOR activity, leading to decreased cell growth. In fact, pharmacological inhibition of mTOR results in prolonged life span as well as diminished tumorigenesis. The effects on tumorigenesis, however, are complex because mTOR inhibition also affects inflammation and immune cells.
Inhibition of autophagy enhances senescence, which could be related to the inability of cells to clear defective mitochondria, thereby increasing oxidative stress and aging (Rabinowitz and White 2010; Rubinsztein et al. 2011). Caloric restriction, on the other hand, would stimulate mitophagy, clearing cells of defective mitochondria (Youle and Narendra 2011). Enhanced mitochondrial efficiency, through removal of poorly functioning mitochondria, decreases oxidative stress and mutagenesis that appear to underpin the way by which caloric restriction decreases tumorigenesis. Intriguingly, severe caloric restriction can also lower basal-specific metabolic rates, which is associated with reduced cancer frequency, as discussed above (Colman et al. 2009). Thus, the combination of lowered basal metabolic rates and more efficient mitochondrial function could decrease mutagenic oxidative stress with caloric restriction.
Oncogenes, tumor suppressors, metabolic enzymes, and tumorigenesis
Metabolic genes as cancer genes
Although the Warburg effect describes altered cancer metabolism, alterations of metabolic genes that could provide a direct genetic link to altered metabolism were not known until the identification of mutant TCA cycle enzymes that are associated with familial cancer syndromes (King et al. 2006). Specifically, mutations in fumarate hydratase were found in families afflicted with leimyomatosis and kidney cancers, and mutations in succinate dehydrogenase were found in patients with pheochromocytoma and paragangliomas. These mutations cause a disruption of the TCA cycle with the accumulation of fumarate or succinate, both of which can inhibit dioxygenases or prolyl hydrolases that mediate the degradation of HIF proteins (King et al. 2006). Elevation of HIF proteins as a consequence is likely to be pro-oncogenic, but it is also notable that these carboxylic acids can also affect dioxygenases that are involved in epigenetic modulation. As such, the contributions of TCA cycle intermediates to tumorigenesis are likely to be multifaceted. More recently, mutations in IDH stemming from cancer genome sequencing efforts uncovered remarkable connections between a mutant metabolic enzyme and tumorigenesis (Parsons et al. 2008; Yan et al. 2009). The mutant IDH enzyme possesses a neomorphic activity that converts aKG to 2-hydroxyglutarate (2-HG) as compared with the wild IDH activity, which converts isocitrate to aKG (Dang et al. 2009; Gross et al. 2010). 2-HG has been documented to inhibit dioxygenases that are involved in histone and DNA demethylation (Xu et al. 2011). In fact, studies of IDH mutations in AML linked them to a subset of AML that clusters together as a subgroup with a distinct epigenome (Figueroa et al. 2010). Likewise, glioblastomas grouped together according to methylation status correlate with IDH status (Noushmehr et al. 2010). In this regard, the associations provide a compelling case for an oncogenic mutant metabolic enzyme that drives tumorigenesis epigenetically.
Synthetic lethality screens aimed at metabolic enzymes uncovered an unsuspected oncogenic role for PHGDH (phosphoglycerate dehydrogenase), which catalyzes the first step in serine synthesis (Locasale et al. 2011; Possemato et al. 2011). PHGDH is involved in channeling glycolytic intermediates into a one-carbon metabolism involved in nucleotide biosynthesis. In fact, PHGDH is amplified in estrogen receptor-negative breast cancers, suggesting that it is an oncogenic enzyme when overexpressed. Loss of PHGDH decreases the level of a key TCA intermediate, aKG, but not serine, suggesting that PHGDH contributes to the TCA cycle anaplerotic flux (Possemato et al. 2011). Glycine decarboxylase (GLDC) is another enzyme that was recently implicated as an oncogenic enzyme, which is involved in glycine/serine metabolism and the one-carbon metabolic pathway (Zhang et al. 2012). Overexpression of GLDC is found in human lung cancer and experimentally promotes tumorigenesis. These two examples underscore the direct genetic evidence that altered metabolism contributes to tumorigenesis.
Mitochondrial DNA (mtDNA) mutations as tumorigenic drivers
In addition to oncogenic mutations in genes encoding enzymes, mutations in mtDNA could also contribute to tumorigenesis. A remarkable study of mtDNA mutations in normal tissues suggested that mtDNA heteroplasmy (a mixture of mutant and wild-type mtDNA in a population of cells) occurs during development without necessarily triggering cancer development (Polyak et al. 1998; He et al. 2010). However, when compared with normal tissues, cancer tissue have increased missense mtDNA mutations, suggesting a selective advantage in acquiring these mutations. In this regard, experimental evidence through cybrid (fusing heterologous nuclei and cytoplasm from different cells) experiments supports a role for mtDNA mutations in enhanced tumorigenesis and metastasis (Petros et al. 2005). These findings further underscore a role for mutations that affect metabolism directly in oncogenesis.
Oncogenes and tumor suppressors regulate metabolism
While mutations in metabolic enzymes hardwire metabolism to tumorigenesis, mutations that activate oncogenes or inactivate tumor suppressors appear to “softwire” cancer genes to metabolism, because metabolic enzymes are directly regulated by these cancer genes. Indeed, Myc was first linked to regulation of glycolysis in aerobic cells through the direct activations of LDHA and virtually all glycolytic genes (Shim et al. 1997; Dang et al. 2006). Myc was subsequently shown to activate genes involved in mitochondrial biogenesis and function as well as those involved in glutamine metabolism (Li et al. 2005; Wise et al. 2008; Gao et al. 2009). Mutated Ras also enhances glycolysis, partly through increasing the activity of Myc and HIF (Sears et al. 1999; Semenza 2010). HIF-1 could be elevated under aerobic conditions downstream from activated PI3K, which stimulates the synthesis of HIF-1. Loss of the tumor suppressor VHL in kidney cancer also stabilizes HIF-1, permitting it to activate glycolytic genes, which are normally activated by HIF-1 under hypoxic conditions. Intriguingly, HIF-1 could inhibit physiologic Myc function and provide a means to attenuate normal cell growth when oxygen is limited. HIF-2, however, could increase Myc function, which may be relevant in the context of normal cells that could proliferate under hypoxia, such as endothelial cells, which express high levels of HIF-2 rather than HIF-1 in hypoxia (Gordan et al. 2007, 2008). These interactions between Myc and HIFs could explain the existence of subsets of kidney cancers, and the occurrence of HIF-1α mutations in these cancers (Shen et al. 2011). When Myc is overexpressed in cancer cells, however, HIF-1 could not stoichiometrically inhibit the function of Myc (Kim et al. 2007). High levels of Myc not only increase HIF-1 levels, but also allow Myc (and N-Myc) to collaborate with HIF-1 (Qing et al. 2010).
Mutations of PI3K, PTEN, and p53 are prevalent in human cancers. Mutation of PI3K activates its function through the downstream activation of AKT and stabilization of HIF-1. PI3K is opposed by the tumor suppressor PTEN, which is frequently lost in human prostate cancer. Hence, activation of PI3K and loss of PTEN affects cellular metabolism because AKT and HIF-1 both profoundly increase glycolysis (Elstrom et al. 2004). In contrast to Myc, neither AKT nor HIF-1 enhances mitochondrial biogenesis and respiration. Myc is unique in that it drives parallel pathways that all contribute to the overall increased metabolic function of the cancer cell. In this regard, it is notable that resistance to PI3K pathway inhibition in human mammary cells and in a murine model of breast cancer is associated with MYC gene amplification, which bypasses signaling downstream from PI3K (Ilic et al. 2011; Liu et al. 2011). Intriguingly, the Myc target gene eIF4E, which is involved in protein synthesis, is also amplified in PI3K inhibition-resistant human mammary cells (Ilic et al. 2011), suggesting that the roles of Myc and eIF4E in biomass accumulation could underpin their lack of dependence on the PI3K pathway.
p53 is another prominent tumor suppressor that is eliminated in many human cancers. In addition to its role in cell cycle control, p53 also directly activates genes such as TIGAR, a PFKFB family member that inhibits glycolysis, shunting glucose into the PPP. p53 also activates genes such as SCO2 that enhance more efficient mitochondrial respiration (Bensaad et al. 2006; Matoba et al. 2006; Vousden and Ryan 2009; Wang et al. 2012). Hence, loss of p53 tends to favor glycolysis. p53 was also documented to activate the expression of the liver form of glutaminase (Gls2), in contrast to Myc, which increases the expression of the kidney form of glutaminase (Gls or Gls1) (Hu et al. 2010; Suzuki et al. 2010).
Although the links between oncogenes, tumor suppressors, and metabolism are being established in experimental cell models, oncogenic alterations of metabolism in vivo appear to depend on the specific oncogene and the tissue type. The recent study by Yuneva et al. (2012) illustrates that oncogenic drive and organ site profoundly influence the cellular usage of glucose or glutamine. Myc-driven murine liver cancer depends on high levels of glycolysis and glutaminolysis (Hu et al. 2011), while Met oncogene-driven liver cancer has markedly diminished glutaminolysis and displays an ability to synthesize glutamine (Yuneva et al. 2012). Myc-driven lung cancer cells also have glutamine synthetase activity as well as high glycolytic and glutaminolytic rates. It is notable that Myc-induced liver cancer is associated with an aggressive tumor phenotype and histology, while Met-induced liver cancer is relatively indolent and is associated with a more differentiated phenotype. As such, how oncogenes drive tumorigenesis and the resulting state of cellular differentiation can profoundly affect the metabolic profile of cancer cells.
Metabolic rewiring and the tumor microenvironment
Genetic alterations in the nuclear and mitochondrial genomes of cancer cells are linked to altered cancer metabolism. However, these cell-autonomous changes are modulated by the environment of the cancer cell, characterized by poor blood perfusion, hypoxia, and nutrient limitations. Hypoxia induces HIF-1 or HIF-2, which in turn activates a transcriptional program that alters the metabolic profile of cancer cells (Bertout et al. 2008; Semenza 2010). In particular, HIF-1 induces glycolysis and inhibits mitochondrial biogenesis, thereby superimposing its influence on the cell-autonomous metabolic changes caused by activation of oncogenes or loss of tumor suppressors. In this regard, one could imagine that there would be aerobic cells that undergo oxidative phosphorylation surrounding a blood vessel within a tumor bed (Semenza 2012). Cells distal to the blood vessel, however, would be robbed of an oxygen supply by cells located immediately around the blood vessel (Schroeder et al. 2005). These distal hypoxic cells would have a different metabolic profile than those located around the blood vessel. Indeed, one study supports the view that hypoxic cells distal to the blood vessel convert glucose to lactate, which could then be imported into aerobic cells and converted to pyruvate for oxidation in the mitochondrion (Sonveaux et al. 2008).
This concept of a symbiotic relationship between cells in the tumor microenvironment has been extended to suggest that the Warburg effect occurs in stromal cells, rather than in cancer cells that feed off of stromal cell-generated lactate (Martinez-Outschoorn et al. 2011). While this view is stimulating and provocative, it does not account for many observations that support cell-autonomous changes in cancer cell metabolism as discussed above. An area that needs further study is the occurrence of fibrotic material in the tumor bed and the role of immune cells in the metabolic milieu of the tumor microenvironment (Shiao et al. 2011). Hence, additional studies are necessary to delineate the contributions of the stroma and immune cells to tumor tissue metabolism. Additional insights will likely change our current oversimplified view of tumor metabolism.
Therapeutic opportunities
Given our current understanding of the contributions of glucose and glutamine to tumor metabolism, is there an opportunity to generate a new class of anti-tumor drugs that target altered metabolism in cancer cells? Are there differences between normal cell metabolism and cancer cell metabolism that provide clinically relevant therapeutic windows? These questions have been addressed by a number of recent excellent reviews (Vander Heiden 2011; Jones and Schulze 2012), and here we focus on several key issues.
It appears that normal T cells use metabolic programs very similar to those used by cancer cells to stimulate cell growth and proliferation. It is notable, however, that in the case of Myc oncogene-stimulated tumorigenesis, deregulated Myc renders Myc-transformed cells addicted to glucose and glutamine such that nutrient deprivation triggers Myc-transformed cell death. In contrast, MYC expression is attenuated in nutrient-deprived normal cells. As proof of concept that Myc-transformed cells' addiction to nutrients could be targeted, inhibitors of LDHA and glutaminase have been shown to have preclinical anti-tumor effects in vivo (Le et al. 2010, 2012; Wang et al. 2010).
The metabolic similarities between normal T cells and cancer cells suggest that anti-cancer metabolic inhibitors could modulate immune cells. It is hence not surprising that cyclosporine, which inhibits TOR, is an effective immunosuppressant. Mycophenolic acid, an inhibitor of IMPDH and pyrimidine biosynthesis, is yet another clinically used immunosuppressant. Both agents also display anti-tumor effects in animal studies. Thus, the question is whether there is a therapeutic window in the absence of mutations in specific metabolic enzymes such as IDH1 or IDH2. An animal model of MYC-induced hepatocellular carcinoma has elucidated one such candidate: In this model, liver tumor tissues have elevated Gls1 (kidney form) expression, while expression of Gls2 (liver form) is depressed in tumors (Hu et al. 2011). BPTES is a potent specific inhibitor of Gls1 but not Gls2, providing rational therapy in liver cancer (Wang et al. 2010; Delabarre et al. 2011; Cassago et al. 2012; Le et al. 2012). In this regard, an isozyme switch could also be targeted. Another example is the switch of pyruvate from PKM1 to PKM2 in tumor tissues; specific inhibitors or, counterintuitively, activators of PKM2 could be tumor-selective (Vander Heiden et al. 2009b; Jiang et al. 2010).
Mutant IDH1 or IDH2 enzyme poses a more tractable problem, as inhibitors specific for the mutant neoenzyme would conceptually provide a significant therapeutic opportunity because the mutant enzyme possesses a new enzymatic activity that could be specifically targeted. In other cases of increased expression—as in the case of LDHA, PHGDH, and GLDC—it is possible that there is a sufficient therapeutic window to target these enzymes. Fatty acid synthase (FASN), which catalyzes the synthesis of palmitate, was noted to be elevated in many human cancers and has been a target of interest for cancer therapy (Kuhajda et al. 1994; Zhou et al. 2003). Some cancers have amplicons that involve FASN and hence could provide a therapeutic window. The major metabolic enzyme targets have become key interests for many pharmaceutical companies. In addition, HIF is also another target of great interest (Semenza 2010). In fact, a number of HIF targets, such as carbonic anhydrase IX (CAIX) and the monocarboxylate transporter MCT4 (as well as the non-HIF target MCT1), are also of major interest as therapeutic targets (Brahimi-Horn et al. 2011; Morris et al. 2011). Hence, in the next 5–10 years, it is anticipated that we will see a number of metabolic inhibitors making it to the clinic.
Aside from targeted therapies based on tumor metabolic profiles, empirical observations of the effect of metformin on reducing cancer incidences led to intriguing leads for cancer metabolic therapy. Metformin inhibits mitochondrial complex I activity and hence is an example of mitochondrial metabolic inhibitor (Bost et al. 2012). Given the epidemiological evidence of reduction in cancer incidence for patients who took metformin for diabetes as compared with those treated with insulin, there are now a number of clinical trials aimed toward testing whether metformin could have an anti-tumor effect. The anti-malarial drug chloroquine is also being repurposed to block autophagy in cancer prevention and therapeutic clinical trials (Amaravadi et al. 2011). Targeting metabolism hence is a new strategy to develop a new class of anti-cancer drugs.
Conclusions
Metabolism is part and parcel of life, with plants and photosynthetic microorganisms capturing energy from sunlight to feed all other earth life forms. Development and growth of a mammal is inherently tied to the availability of nutrients such that mechanisms have evolved for animals to survive severe starvation. Intriguingly, energy deprivation prolongs life span, while excess calories are associated with obesity, human cancer, and shortened life span. At the cellular level, normal proliferating cells activate metabolic pathways and couple them with cell mass accumulation and DNA synthesis for cell reproduction. Normal cells sense nutrient cues and evolve mechanisms to diminish macromolecular synthesis and ATP consumption while enhancing ATP production pathways when deprived of nutrients. Autophagy evolved to sustain starved cells through self-eating to recycle cell components for energy production. The by-products of metabolism, specifically ROS, can damage cells and promote oncogenic DNA mutations; thus, metabolism can trigger tumorigenesis. Mutations of oncogenes and tumor suppressors, in turn, drive cell growth and proliferation coupled with import of adequate bioenergetic substrates. In this regard, mutant metabolic enzymes can drive tumorigenesis, and conversely, cancer genes regulate metabolism such that cellular machineries driving cell growth and proliferation are tightly coupled with the cell's ability to assimilate nutrients and energy.
The therapeutic windows for targeting cancer cell metabolism reside in differences between normal and mutant oncogenic enzymes and addiction of cancer cells to nutrients to support deregulated cell growth programs enforced by cancer genes. Hence, the complex regulatory networks involving cancer genes and metabolic pathways need to be defined for specific cancer types so that targeting of cancer cell metabolism could be strategically guided by somatic genetic changes in cancers. Given the explosion of interest and information on cancer metabolism, it is hoped that new therapies will emerge from the basic sciences of metabolism in the next decade.
Acknowledgments
I thank Brian Altman for comments. My original work is supported by an AACR Stand-Up-to-Cancer translational grant, Leukemia and Lymphoma Society, and NCI. I am also supported by the Abramson Family Cancer Research Institute at the University of Pennsylvania. Many original articles were omitted due to space limitations; for this, I apologize.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.189365.112.
References
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E 2011. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 17: 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. 2011. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science 334: 1278–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Takahashi JS 2010. Circadian integration of metabolism and energetics. Science 330: 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello-Fernandez C, Cleveland JL 1992. c-myc transactivates the ornithine decarboxylase gene. Curr Top Microbiol Immunol 182: 445–452 [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH 2006. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126: 107–120 [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L 2002. Biochemistry. W.H. Freeman and Company, New York [Google Scholar]
- Bertout JA, Patel SA, Simon MC 2008. The impact of O2 availability on human cancer. Nat Rev Cancer 8: 967–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost F, Sahra IB, Le Marchand-Brustel Y, Tanti JF 2012. Metformin and cancer therapy. Curr Opin Oncol 24: 103–108 [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn MC, Bellot G, Pouyssegur J 2011. Hypoxia and energetic tumour metabolism. Curr Opin Genet Dev 21: 67–72 [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW 2011. Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. 1998. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394: 485–490 [DOI] [PubMed] [Google Scholar]
- Cassago A, Ferreira AP, Ferreira IM, Fornezari C, Gomes ER, Greene KS, Pereira HM, Garratt RC, Dias SM, Ambrosio AL 2012. Mitochondrial localization and structure-based phosphate activation mechanism of Glutaminase C with implications for cancer metabolism. Proc Natl Acad Sci 109: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulin AF, Maley CC 2011. Peto's paradox: Evolution's prescription for cancer prevention. Trends Ecol Evol 26: 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL 2007. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science 316: 1916–1919 [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, Leung IK, Li XS, Woon EC, Yang M, et al. 2011. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep 12: 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collman JP, Ghosh S, Dey A, Decreau RA 2009. Using a functional enzyme model to understand the chemistry behind hydrogen sulfide induced hibernation. Proc Natl Acad Sci 106: 22090–22095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, et al. 2009. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science 325: 201–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2010. Rethinking the Warburg effect with Myc micromanaging glutamine metabolism. Cancer Res 70: 859–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F 2006. The c-Myc target gene network. Semin Cancer Biol 16: 253–264 [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. 2009. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462: 739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dark J 2005. Annual lipid cycles in hibernators: Integration of physiology and behavior. Annu Rev Nutr 25: 469–497 [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Cheng T 2010. Q's next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 29: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Zhang Y 2011. The ribosomal protein–Mdm2–p53 pathway and energy metabolism: Bridging the gap between feast and famine. Genes Cancer 2: 392–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delabarre B, Gross S, Fang C, Gao Y, Jha A, Jiang F, Song JJ, Wei W, Hurov JB 2011. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry 50: 10764–10770 [DOI] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, et al. 2004. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res 64: 3892–3899 [DOI] [PubMed] [Google Scholar]
- Ferber EC, Peck B, Delpuech O, Bell GP, East P, Schulze A 2011. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ doi: 10.1038/cdd.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T 2011. Signal transduction by reactive oxygen species. J Cell Biol 194: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, et al. 2009. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 458: 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L, Bublik DR, Wilder S, Nevo R, Kiss V, Grabusic K, Volarevic S, Oren M 2012. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol Cell 45: 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, Ramsbottom BA, Innes F, Kantidakis T, Kerr ER, et al. 2006. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem Soc Symp 2006: 141–154 [DOI] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC 2007. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 11: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordan JD, Lal P, Dondeti VR, Letrero R, Parekh KN, Oquendo CE, Greenberg RA, Flaherty KT, Rathmell WK, Keith B, et al. 2008. HIF-α effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell 14: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, et al. 2010. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med 207: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Xiong Y 2011. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci 36: 108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Stevens RG 2011. Case-control study of shift-work and breast cancer risk in Danish nurses: Impact of shift systems. Eur J Cancer doi: 10.1016/j.ejca.2011.07.005 [DOI] [PubMed] [Google Scholar]
- He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA Jr, Kinzler KW, Vogelstein B, Papadopoulos N 2010. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature 464: 610–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman AB, Savage VM, West GB 2011. A quantitative theory of solid tumor growth, metabolic rate and vascularization. PLoS ONE 6: e22973 doi: 10.1371/journal.pone.0022973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM 2008. Cancer cell metabolism: Warburg and beyond. Cell 134: 703–707 [DOI] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z 2010. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci 107: 7455–7460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, Kurhanewicz J, Vigneron DB, Goga A 2011. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 14: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Tindall DJ 2007. Dynamic FoxO transcription factors. J Cell Sci 120: 2479–2487 [DOI] [PubMed] [Google Scholar]
- Hursting SD, Smith SM, Lashinger LM, Harvey AE, Perkins SN 2010. Calories and carcinogenesis: Lessons learned from 30 years of calorie restriction research. Carcinogenesis 31: 83–89 [DOI] [PubMed] [Google Scholar]
- Ilic N, Utermark T, Widlund HR, Roberts TM 2011. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci 108: E699–E708 doi: 10.1073/pnas.1108237108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, Doan HM, Fan J, Cheadle C, Fallahi M, et al. 2011. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE 6: e26057 doi: 10.1371/journal.pone.0026057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, et al. 2010. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett 20: 3387–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NP, Schulze A 2012. Targeting cancer metabolism—aiming at a tumour's sweet-spot. Drug Discov Today 17: 232–241 [DOI] [PubMed] [Google Scholar]
- Kaadige MR, Looper RE, Kamalanaadhan S, Ayer DE 2009. Glutamine-dependent anapleurosis dictates glucose uptake and cell growth by regulating MondoA transcriptional activity. Proc Natl Acad Sci 106: 14878–14883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Sabatini DM 2009. Tumours with PI3K activation are resistant to dietary restriction. Nature 458: 725–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Imhof A, Sassone-Corsi P 2012. Connecting threads: Epigenetics and metabolism. Cell 148: 24–28 [DOI] [PubMed] [Google Scholar]
- Keith B, Johnson RS, Simon MC 2012. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer 12: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandekar MJ, Cohen P, Spiegelman BM 2011. Molecular mechanisms of cancer development in obesity. Nat Rev Cancer 11: 886–895 [DOI] [PubMed] [Google Scholar]
- Kilburn DG, Lilly MD, Webb FC 1969. The energetics of mammalian cell growth. J Cell Sci 4: 645–654 [DOI] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV 2006. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- Kim JW, Gao P, Liu YC, Semenza GL, Dang CV 2007. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol 27: 7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E 2006. Succinate dehydrogenase and fumarate hydratase: Linking mitochondrial dysfunction and cancer. Oncogene 25: 4675–4682 [DOI] [PubMed] [Google Scholar]
- Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR 2011. Yeast cells can access distinct quiescent states. Genes Dev 25: 336–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV 2011. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 11: 325–337 [DOI] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR 1994. Fatty acid synthesis: A potential selective target for antineoplastic therapy. Proc Natl Acad Sci 91: 6379–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Nathans D 1987. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: Coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci 84: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV 2010. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci 107: 2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, Tsukamoto T, Rojas CJ, Slusher BS, Zhang H, et al. 2012. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab 15: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone G, Sears R, Huang E, Rempel R, Nuckolls F, Park CH, Giangrande P, Wu L, Saavedra HI, Field SJ, et al. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol Cell 8: 105–113 [DOI] [PubMed] [Google Scholar]
- Levine AJ, Puzio-Kuter AM 2010. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330: 1340–1344 [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O'Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV 2005. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman SI, Broach JR 2009. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc Natl Acad Sci 106: 19928–19933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Cheng H, Santiago S, Raeder M, Zhang F, Isabella A, Yang J, Semaan DJ, Chen C, Fox EA, et al. 2011. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med 17: 1116–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CC, Chou T, Penzel T, Scammell TE, Strecker RE, Stanley HE, Ivanov P 2004. Common scale-invariant patterns of sleep-wake transitions across mammalian species. Proc Natl Acad Sci 101: 17545–17548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Cantley LC 2011. Metabolic flux and the regulation of mammalian cell growth. Cell Metab 14: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. 2011. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Fontana L 2010. Calorie restriction and cancer prevention: Metabolic and molecular mechanisms. Trends Pharmacol Sci 31: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y 2010. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell 18: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Outschoorn UE, Lin Z, Trimmer C, Flomenberg N, Wang C, Pavlides S, Pestell RG, Howell A, Sotgia F, Lisanti MP 2011. Cancer cells metabolically ‘fertilize’ the tumor microenvironment with hydrogen peroxide, driving the Warburg effect: Implications for PET imaging of human tumors. Cell Cycle 10: 2504–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM 2006. p53 regulates mitochondrial respiration. Science 312: 1650–1653 [DOI] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, et al. 2012. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ 2011. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13: 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JC, Chiche J, Grellier C, Lopez M, Bornaghi LF, Maresca A, Supuran CT, Pouyssegur J, Poulsen SA 2011. Targeting hypoxic tumor cell viability with carbohydrate-based carbonic anhydrase IX and XII inhibitors. J Med Chem 54: 6905–6918 [DOI] [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM 2009. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene 28: 2485–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ 2012. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JD, Victor EM, Cropper JH 2007. Why don't all whales have cancer? A novel hypothesis resolving Peto's paradox. Integr Comp Biol 47: 317–328 [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, Hanson RW 2002. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277: 30409–30412 [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. 2008. An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CW, Ayer DE 2012. An extended Myc network contributes to glucose homeostasis in cancer and diabetes. Front Biosci 17: 2206–2223 [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, et al. 2005. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci 102: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DR, Thompson CB 2005. Akt-dependent transformation: There is more to growth than just surviving. Oncogene 24: 7435–7442 [DOI] [PubMed] [Google Scholar]
- Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B 1998. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet 20: 291–293 [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. 2011. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476: 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G, Skuli N, Mayes PA, Pawel B, Martinez D, Maris JM, Simon MC 2010. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1α. Cancer Res 70: 10351–10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, White E 2010. Autophagy and metabolism. Science 330: 1344–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PD, Huang BW, Tsuji Y 2012. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell Signal 24: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempel RE, Mori S, Gasparetto M, Glozak MA, Andrechek ER, Adler SB, Laakso NM, Lagoo AS, Storms R, Smith C, et al. 2009. A role for E2F activities in determining the fate of Myc-induced lymphomagenesis. PLoS Genet 5: e1000640 doi: 10.1371/journal.pgen.1000640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros S, Santos CR, Moco S, Baenke F, Kelly G, Howell M, Zamboni N, Schulze A 2012. Functional metabolic screen identifies 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 (PFKFB4) as an important regulator of prostate cancer cell survival. Cancer Discovery doi: 10.1158/2159-8290.CD-11-0234 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Marino G, Kroemer G 2011. Autophagy and aging. Cell 146: 682–695 [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P 2009. Metabolism and cancer: The circadian clock connection. Nat Rev Cancer 9: 886–896 [DOI] [PubMed] [Google Scholar]
- Samudio I, Harmancey R, Fiegl M, Kantarjian H, Konopleva M, Korchin B, Kaluarachchi K, Bornmann W, Duvvuri S, Taegtmeyer H, et al. 2010. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J Clin Invest 120: 142–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH, West GB 2007. Scaling of number, size, and metabolic rate of cells with body size in mammals. Proc Natl Acad Sci 104: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schernhammer ES, Kroenke CH, Laden F, Hankinson SE 2006. Night work and risk of breast cancer. Epidemiology 17: 108–111 [DOI] [PubMed] [Google Scholar]
- Schrodinger E. 1992. What is life?: With ‘mind and matter’ and ‘autobiographical sketches.’ Cambridge University Press, Cambridge, UK. [Google Scholar]
- Schroeder T, Yuan H, Viglianti BL, Peltz C, Asopa S, Vujaskovic Z, Dewhirst MW 2005. Spatial heterogeneity and oxygen dependence of glucose consumption in R3230Ac and fibrosarcomas of the Fischer 344 Rat. Cancer Res 65: 5163–5171 [DOI] [PubMed] [Google Scholar]
- Sears R, Leone G, DeGregori J, Nevins JR 1999. Ras enhances Myc protein stability. Mol Cell 3: 169–179 [DOI] [PubMed] [Google Scholar]
- Semenza GL 2010. HIF-1: Upstream and downstream of cancer metabolism. Curr Opin Genet Dev 20: 51–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL 2012. Hypoxia-inducible factors in physiology and medicine. Cell 148: 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Beroukhim R, Schumacher SE, Zhou J, Chang M, Signoretti S, Kaelin WG Jr 2011. Genetic and functional studies Implicate HIF1α as a 14q kidney cancer suppressor gene. Cancer Discov 1: 222–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiao SL, Ganesan AP, Rugo HS, Coussens LM 2011. Immune microenvironments in solid tumors: New targets for therapy. Genes Dev 25: 2559–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV 1997. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc Natl Acad Sci 94: 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JM 2005. Clues to the functions of mammalian sleep. Nature 437: 1264–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman SJ, Petti AA, Slavov N, Parsons L, Briehof R, Thiberge SY, Zenklusen D, Gandhi SJ, Larson DR, Singer RH, et al. 2010. Metabolic cycling in single yeast cells from unsynchronized steady-state populations limited on glucose or phosphate. Proc Natl Acad Sci 107: 6946–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo AM 2011. Autophagy in the cellular energetic balance. Cell Metab 13: 495–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov N, Macinskas J, Caudy A, Botstein D 2011. Metabolic cycling without cell division cycling in respiring yeast. Proc Natl Acad Sci 108: 19090–19095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. 2008. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 118: 3930–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. 2010. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci 107: 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian SV, Zabludoff SD, Wold BJ 1994. Cloning of mid-G1 serum response genes and identification of a subset regulated by conditional myc expression. Mol Biol Cell 5: 375–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG 2011. Targeting cancer metabolism: A therapeutic window opens. Nat Rev Drug Discov 10: 671–684 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB 2009a. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 324: 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Christofk HR, Schuman E, Subtelny AO, Sharfi H, Harlow EE, Xian J, Cantley LC 2009b. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem Pharmacol 79: 1118–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Ryan KM 2009. p53 and metabolism. Nat Rev Cancer 9: 691–700 [DOI] [PubMed] [Google Scholar]
- Wang JB, Erickson JW, Fuji R, Ramachandran S, Gao P, Dinavahi R, Wilson KF, Ambrosio AL, Dias SM, Dang CV, et al. 2010. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell 18: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35: 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PY, Zhuang J, Hwang PM 2012. p53: Exercise capacity and metabolism. Curr Opin Oncol 24: 76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O 1956. On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB 2010. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev 24: 2784–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, et al. 2008. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci 105: 18782–18787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, Dematteo RG, Simon MC, Thompson CB 2011. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci 108: 19611–19616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin A, Telang S, Clem B, Chesney J 2009. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp Mol Pathol 86: 174–179 [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. 2009. IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, Kelleher JK 2008. Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 283: 20621–20627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP 2011. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Mates JM, Alonso FJ, Wang C, Seo Y, et al. 2012. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 15: 157–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, Huang P, Sawyer SK, Fuerth B, Faubert B, et al. 2011. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev 25: 1041–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci 103: 17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, et al. 2012. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148: 259–272 [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Simpson PJ, McFadden JM, Townsend CA, Medghalchi SM, Vadlamudi A, Pinn ML, Ronnett GV, Kuhajda FP 2003. Fatty acid synthase inhibition triggers apoptosis during S phase in human cancer cells. Cancer Res 63: 7330–7337 [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM 2010. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12: 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]