Highly occupied target (HOT) regions are bound by numerous, often functionally unrelated, transcription factors and are an intriguing discovery of the modENCODE project. In investigating their elusive function, Stark and colleagues find that 94% of the assayed HOT regions function as active transcriptional enhancers that strongly up-regulate neighboring genes. The work also identifies a cis-regulatory signature that enables identification of these regions purely by sequence.
Keywords: HOT region, transcription factor, enhancer/cis-regulatory element, Drosophila embryo development
Abstract
HOT (highly occupied target) regions bound by many transcription factors are considered to be one of the most intriguing findings of the recent modENCODE reports, yet their functions have remained unclear. We tested 108 Drosophila melanogaster HOT regions in transgenic embryos with site-specifically integrated transcriptional reporters. In contrast to prior expectations, we found 102 (94%) to be active enhancers during embryogenesis and to display diverse spatial and temporal patterns, reminiscent of expression patterns for important developmental genes. Remarkably, HOT regions strongly activate nearby genes and are required for endogenous gene expression, as we show using bacterial artificial chromosome (BAC) transgenesis. HOT enhancers have a distinct cis-regulatory signature with enriched sequence motifs for the global activators Vielfaltig, also known as Zelda, and Trithorax-like, also known as GAGA. This signature allows the prediction of HOT versus control regions from the DNA sequence alone.
Recent studies have revealed genomic regions that are bound by surprisingly many and often functionally unrelated transcription factors (TFs) in humans, the fruit fly Drosophila melanogaster (Moorman et al. 2006; MacArthur et al. 2009; Gerstein et al. 2010; The modENCODE Consortium 2010; Nègre et al. 2011), and the nematode Caenorhabditis elegans (Gerstein et al. 2010). These so-called HOT (highly occupied target) regions or “hot spots” are depleted for TF motifs compared with regions occupied by single TFs, which suggests that factors are recruited nonspecifically or via protein–protein interactions (Gerstein et al. 2010; The modENCODE Consortium 2010). In D. melanogaster, HOT regions represent 5% of all identified TF-bound regions (1962 out of 38,562) (The modENCODE Consortium 2010). The presence of these regions in humans, flies, and worms suggests they might reflect a general property of regulatory genomes. Remarkably, HOT regions correlate with decreased nucleosome density and increased nucleosome turnover and are primarily associated with open chromatin (Gerstein et al. 2010; The modENCODE Consortium 2010; Nègre et al. 2011). However, the function of HOT regions has remained unclear (Blaxter 2010; Furlong 2011), and proposed roles include a putative function in DNA replication (The modENCODE Consortium 2010), an interplay with boundary elements (The modENCODE Consortium 2010), and the regulation of ubiquitously expressed genes (Gerstein et al. 2010). In D. melanogaster, only ∼19% of known transcriptional enhancers overlap with HOT regions, and most known enhancers are bound by few TFs, such that they would not classify as hot spots (Table 1; Nègre et al. 2011). In addition, it is unknown whether these regions share other features beyond TF binding, such as characteristic sequence signatures.
Table 1.
Only 19% of known enhancers correspond to HOT regions
Here, we show that Drosophila HOT regions function as transcriptional enhancers with diverse activity patterns. While a large number of bound TFs is the defining feature of HOT regions, many TFs seem to be bound neutrally without any apparent contribution to enhancer activity. HOT enhancers are characterized by a distinct and predictive cis-regulatory signature, which includes motifs for Vielfaltig/Zelda (ZLD), a recently reported activator of the early Drosophila genome (Liang et al. 2008; Harrison et al. 2011; Nien et al. 2011), and Trithorax-like/GAGA (GAGA), a TF known to form homomeric and heteromeric complexes (Bardwell and Treisman 1994) and to be required for the generation and maintenance of nucleosome-free regions (Croston et al. 1991; Nakayama et al. 2007).
Results and Discussion
HOT regions function as early embryonic enhancers with diverse patterns
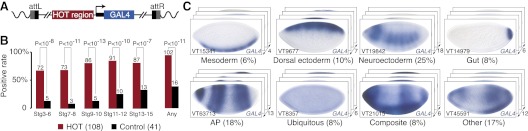
We tested a representative set of 108 D. melanogaster HOT regions (see the Materials and Methods; Supplemental Tables 1, 2) in transgenic embryos with site-specifically integrated transcriptional reporters (Fig. 1A). Strikingly, 94% (102) of these HOT regions drove reporter expression in a specific pattern during embryogenesis. In contrast, only 39% of control regions (16 of 41) functioned as enhancers, including 11 of 21 regions chosen to contain TF-binding sites and five of 20 regions chosen to contain no known binding sites (see the Materials and Methods; Supplemental Fig. S1; Supplemental Table 1). This enrichment of HOT over control regions is highest at stages 3–10 (corresponding to 1–5 h after fertilization) (Fig. 1B), at which most of the chromatin immunoprecipitation (ChIP) experiments leading to the definition of the HOT regions had been performed (MacArthur et al. 2009; The modENCODE Consortium 2010).
Figure 1.
HOT regions act as enhancers with diverse activity patterns. (A) Transcriptional reporter to test enhancer activity of candidate regions (Pfeiffer et al. 2008). (B) A large majority of the 108 HOT regions (red) function as active transcriptional enhancers during Bownes stages 3–15 (1–13 h after fertilization), while much fewer of the 41 control regions are active (black). Shown are the positive rates (bar heights), the number of positive regions (numbers above bars), and the hypergeometric P-values. (C) HOT enhancers display diverse spatial patterns in the blastoderm embryo. Shown are seven representatives of manually grouped patterns that reoccurred at least three times and “other” patterns, the number of embryos in each group (each bottom right corner), and the corresponding percentage (see Supplemental Fig. S1 for all patterns).
Contrary to expectations that HOT enhancers might constitute a particular class of enhancers (for example, with ubiquitous activity), they display highly diverse spatial activity patterns in all major presumptive tissues of the blastoderm embryo (Fig. 1C; Supplemental Fig. S1). In particular, we found enhancers that are active in the early mesoderm (four HOT enhancers), dorsal ectoderm (seven), neurogenic ectoderm (18), and gut (six). Thirteen HOT enhancers display characteristic anterior–posterior (AP) patterns, six have composite patterns, and 12 have other diverse patterns. Interestingly, only six of the 72 HOT enhancers that function at the blastoderm stage (8%) are ubiquitously active (Fig. 1C), and even fewer (3%) are ubiquitously active during the entire embryogenesis (Supplemental Fig. S1). This shows that HOT regions can function as transcriptional enhancers that recapitulate well-studied expression patterns of developmentally regulated genes.
HOT enhancers up-regulate nearby genes in vivo
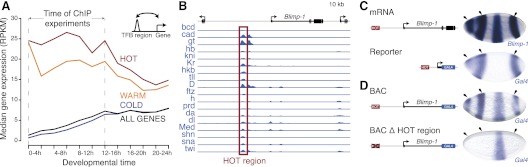
A developmental time series of gene expression throughout Drosophila embryogenesis (Graveley et al. 2011) further supports that HOT regions function as transcriptional enhancers in vivo: When we assigned each HOT and control region to the neighboring gene with the closest transcription start site (TSS), we found that genes assigned to HOT regions are more highly expressed than other genes (Fig. 2A). The strong up-regulation of these genes is specific for the first 12 h of embryo development, when the ChIP experiments had been performed (The modENCODE Consortium 2010). These data suggest that HOT enhancers also function in their genomic context to regulate neighboring genes. Indeed, 12 out of 60 intronic HOT enhancers recapitulate the entire expression pattern of their host gene or characteristic parts thereof, 19 HOT enhancers match to one of the two immediately flanking genes (first-degree neighbors [not considering the host gene for intronic enhancers]), and four match to a second-degree neighbor (Supplemental Fig. S1). For example, one of the HOT enhancers is located ∼10 kb upstream of the Blimp-1 gene (Fig. 2B) and is sufficient to recapitulate three of the four distinct stripes of Blimp-1 expression in the early embryo (Fig. 2C). To test whether this HOT enhancer regulates Blimp-1 expression in vivo, we constructed a bacterial artificial chromosome (BAC) that contained ∼45 kb of genomic DNA surrounding the Blimp-1 locus, replaced the Blimp-1 coding sequence with a GAL4 transcriptional reporter gene, and created transgenic flies that carried this BAC at a defined genomic position (see the Materials and Methods). This BAC transgene allows the direct readout of Blimp-1 transcription from the BAC locus via in situ hybridization independent of endogenous Blimp-1 expression, which it indeed faithfully reproduced (Fig. 2D). In contrast, a variant BAC in which we replaced the HOT enhancer with neutral DNA resulted in the full loss of the three Blimp-1 stripes above (Fig. 2D). Taken together, our data indicate that this HOT enhancer is both sufficient and required for three of the four stripes of Blimp-1 expression in vivo (Fig. 2C).
Figure 2.
HOT enhancers regulate nearby genes. (A) Genes next to HOT regions are up-regulated during early Drosophila development. Shown are median RNA expression levels (reads per kilobase per million reads [RPKM]) (Graveley et al. 2011) for all genes (black line) and for genes assigned to regions bound by one to three factors (COLD; blue), four to 10 factors (WARM; orange), and >10 factors (HOT; red). (B) The HOT enhancer ∼10 kb upstream of Blimp-1 recapitulates three of the four stripes of the Blimp-1 expression pattern (University of California at Santa Cruz Genome Browser screenshot shows chr3L: 5,600,000–5,652,000, including published ChIP-on-chip and ChIP-seq [ChIP coupled with deep sequencing] profiles) (MacArthur et al. 2009; The modENCODE Consortium 2010). (C) The top right embryo shows the in situ hybridization against the Blimp-1 transcript from BDGP (Tomancak et al. 2002), and the bottom right embryo highlights the HOT enhancers' activity by in situ hybridization to the GAL4-reporter. (D) HOT enhancer is required for correct Blimp-1 expression. A BAC construct with an ∼45-kb region surrounding the endogenous Blimp-1 locus in which the coding sequence was replaced by GAL4 and integrated in the fly genome. The top embryo shows in situ hybridization to the wild-type GAL4-reporter, which fully reproduces the endogenous Blimp-1 expression pattern. The bottom embryo shows the same BAC after we deleted the HOT region, leading to a lack of the first, second, and third Blimp-1 stripes.
Many bound TFs appear neutral with respect to HOT enhancer activity
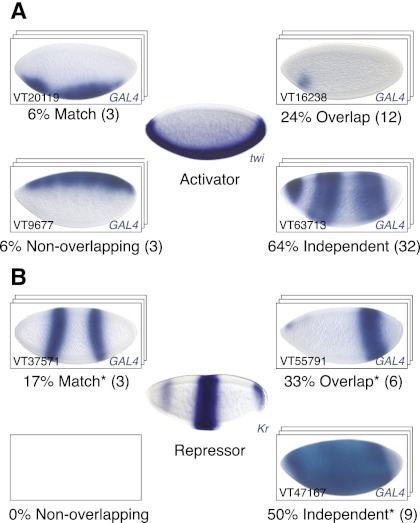
While TFs generally contribute activating and repressing cues to enhancers, it is known that many genomic TF-binding sites do not function as transcriptional enhancers, indicating that neutral or nonfunctional TF binding is possible (Li et al. 2008). Our study provides the potential to reveal the contribution of bound TFs to enhancer activity by comparing the spatial activity pattern of each HOT enhancer with the expression patterns of the bound TFs (data from Tomancak et al. 2002). Surprisingly, the majority of TF binding to active HOT enhancers appeared to be functionally neutral, as judged from largely uncorrelated enhancer activity and TF expression patterns (Supplemental Fig. S2): For example, Twist (Twi), a master regulator of mesoderm development, is expressed in the early presumptive mesoderm of the fly embryo, where it activates genes involved in mesoderm specification (Baylies and Bate 1996). To our surprise, only 15 out of 50 (30%) Twi-bound HOT enhancers matched or overlapped the Twi expression pattern, while 32 (64%) appeared to be independent and three (6%) were entirely nonoverlapping (Fig. 3A; Supplemental Fig. S2). Similar numbers of nonoverlapping patterns were observed for other transcriptional activators for which expression patterns were available (Supplemental Fig. S2).
Figure 3.
Many transcriptional activators appear neutral with respect to HOT enhancer activity. Shown are the expression patterns of the transcriptional activator Twist (A) and the repressor Kruppel (B), surrounded by blastoderm stage embryos for representative HOT enhancers bound by Twist (A, middle) or Kruppel (B, middle) (see the text for details; see Supplemental Fig. S2 for other factors). (*) For repressors such as Kr, “Match” means a fully complementary pattern and “Overlap” means a partially complementary pattern.
Interestingly, “functional footprints” were more evident for repressors: For example, nine out of 18 (50%) HOT enhancers bound by the gap repressor Kruppel (Kr) were inactive in a domain that matched or overlapped the characteristic Kr expression pattern. Another nine (50%) did not show signs of Kr-mediated repression and appeared independent, and none was specifically active in the Kr expression domain (Fig. 3B). Similar results were also obtained for other repressors, such as Knirps (kni) or Snail (sna) (see Supplemental Fig. S2).
As TF binding detected by ChIP is restricted to cells that express the TF, the extent of nonoverlapping patterns suggests that neutral binding of TFs to enhancers is abundant and that TF binding can be neutral not only at nonfunctional genomic sites, but also at transcriptional enhancers. It further suggests that enhancers can be accessible and bound by TFs in cells in which they are not actively enhancing transcription.
HOT regions have a distinct sequence signature
Compared with regions bound by single TFs, HOT regions have been found to be depleted in the bound TFs' motifs (Gerstein et al. 2010; The modENCODE Consortium 2010; Nègre et al. 2011), raising the question of whether HOT regions have characteristic sequence features. We systematically compared the motif content (known and predicted motifs from Stark et al. 2007) between HOT regions (>10 TFs), regions bound by fewer TFs (COLD regions; one to three TFs), and known early embryonic enhancers (EEES) (a subset of the manually curated CAD database from Bonn et al. 2012; see also Table 1).
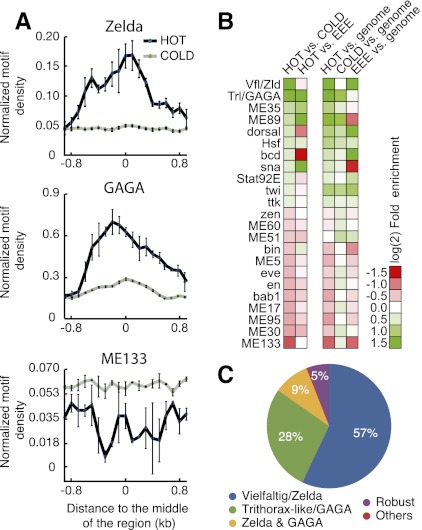
We found 48 motifs to be significantly differentially distributed between HOT and COLD regions (P < 10−2) (Fig. 4A,B), some of which had been reported before (The modENCODE Consortium 2010). HOT regions were, for example, enriched for motifs of the global transcriptional activators ZLD (CAGSTAR; 2.8-fold; P < 10−90) and GAGA (RSWGAGMRHRR; 2.4-fold; P < 10−303). In contrast, COLD regions were enriched for the computationally identified motifs ME133 (AAAAGCT; 2.0-fold; P < 10−16) and ME51 (RCAAWTTR; 1.3-fold; P < 10−9), which did not match to any known TF. Interestingly, ZLD motifs were about equally abundant in HOT regions and EEEs (1.1-fold enriched in EEEs; P = 0.34) (Supplemental Table 3). In contrast, GAGA and the computationally predicted motif ME89 (CACRCAC) were strongly enriched in HOT regions compared with EEEs (3.6- and 3.9-fold, respectively; P < 10−17). EEEs were enriched in motifs for Bicoid, Schnurri, Hunchback, Dorsal, or Caudal (all >1.4-fold; P < 10−2), which likely reflects a database bias toward well-studied early embryonic patterning systems.
Figure 4.
HOT regions are characterized by a unique cis-regulatory signature that is predictive. (A) Distribution of ZLD, GAGA, and ME133 motifs (motif count per 200-nucleotide bin) around the HOT and COLD regions aligned by their center. Lines represent means, and error bars represent standard deviations from three nonoverlapping subsets of the data (30% each). Motifs matching to ZLD and GAGA are strongly enriched in HOT compared with COLD regions, while the ME133 motif is depleted from HOT regions. (B) Heat map showing the most differentially distributed motifs (multiple testing-corrected P-value < 0.01) between HOT and COLD regions (first column), HOT regions and EEEs (second column), and their enrichments in HOT and COLD regions and EEEs compared with the genome average values (third through fifth columns). (C) HOT regions are dependent on ZLD and GAGA motifs. Shown are well-predicted HOT regions (score ≥75) that drop substantially (≥20) after in silico motif mutations or are robust (violet).
We next asked whether the motif content of HOT regions is sufficiently distinct to allow their successful classification versus other regions solely based on DNA sequences. With an established machine learning approach (a support vector machine [SVM]), we classified HOT versus COLD regions and versus EEEs using leave one out cross-validation (LOOCV) based entirely on the regions' motif content (i.e., using the number of motif instances as features) (see the Materials and Methods). This classification worked surprisingly well, correctly predicting 72% of HOT versus COLD regions (area under the receiver operating characteristic [ROC] curve [AUC], 0.77) (see Supplemental Fig. S3A) and 67% of HOT regions versus EEEs (AUC, 0.72) (Supplemental Fig. S3B). Importantly, when we shuffled the regions' assignments to the HOT and COLD or EEE classes, the predictions dropped to ∼50%, as expected for binary classification (50% and 55%, respectively; AUC, 0.53 and 0.54) (Supplemental Fig. S3A,B). This indicates that the prediction success stemmed from characteristic sequence differences of HOT regions and not from our computational approach per se. Notably, discriminating between HOT regions and all known Drosophila enhancers from the REDfly database (Gallo et al. 2010), which includes enhancers that function at other developmental stages, yielded a higher accuracy of 72% (AUC, 0.78, vs. 51% [AUC, 0.55] after shuffling). The successful classification of HOT regions based on their motif content shows that they share characteristic sequence features that distinguish them from regions bound by fewer TFs and from other enhancers and suggests that the information about hot spots is encoded in the DNA sequence.
Motifs for the global activators ZLD and GAGA are characteristic of enhancers and HOT regions in the early Drosophila embryo
To better understand cis-regulatory requirements for individual HOT enhancers, we scored the dependence of each individual region's successful classification on each of the Drosophila TF motifs (for details, see the Materials and Methods).
Strikingly, for 239 (57%) out of 419 well-predicted HOT regions, successful classification versus COLD regions depended on motifs for the TF ZLD, 116 (28%) depended on GAGA motifs, and 39 (9%) depended on the presence of both (Fig. 4C). In fact, motifs for these two TFs alone were sufficient to discriminate HOT from COLD regions (accuracy, 69%; AUC, 0.69). Interestingly, however, ZLD motifs were not important for the successful classification of HOT regions versus EEEs, which only depended on motifs for GAGA. Both findings are in agreement with the differential motif distribution (Fig. 4A,B) and suggest that ZLD is more generally important for enhancers in early Drosophila embryos, while GAGA might be more specifically important for HOT regions that are bound by many different TFs. Both TFs are maternally deposited into Drosophila embryos and are ubiquitously present at early stages. The transcriptional activator ZLD was recently shown to be an essential key activator of the early Drosophila zygotic genome (ten Bosch et al. 2006; Liang et al. 2008; Harrison et al. 2011; Nien et al. 2011) and a facilitator of overlapping TF-binding patterns (Satija and Bradley 2012), while GAGA is known as an enhancer of position effect variegation (PEV) (Farkas et al. 1994), an anti-repressor (Croston et al. 1991), and a factor required for creating and maintaining nuclease-hypersensitive regions (Lu et al. 1993).
Taken together, our data show that Drosophila HOT regions function as cell type-specific transcriptional enhancers to up-regulate nearby genes during early embryo development. In contrast to prior expectations, HOT enhancers display diverse spatial and temporal activity patterns, which are reminiscent of expression patterns of important developmental genes. We further found that the activity of many HOT enhancers appears to be unrelated to the expression of the bound transcriptional activators, suggesting that neutral TF binding to HOT regions is frequent. Interestingly, for Twi, Kr, and five additional TFs, we found that HOT enhancers with functional footprints of the TFs are significantly enriched in the TFs' motifs compared with HOT enhancers to which the TFs seem to bind neutrally (e.g., 2.2-fold for Twi [P < 10−3]) (Supplemental Fig. S2; Supplemental Table S5). This supports previous suggestions that the recruitment of TFs to HOT regions might be independent of the TFs' motifs and mediated by protein–protein interactions or nonspecific DNA binding (Moorman et al. 2006; The modENCODE Consortium 2010). This seems to be particularly true for (HOT) regions to which the TFs bind neutrally without impact on the regions' transcriptional enhancer activity.
By uncovering a distinct cis-regulatory signature that is characteristic and predictive of HOT regions, our computational analysis establishes a link between HOT regions, EEEs, and maternal TFs that are ubiquitously present in the early Drosophila embryo. Specifically, our results suggest that ZLD might be more generally important for the establishment of regulatory elements in the early embryo, while GAGA appears to be a distinguishing feature of HOT regions. This is supported by an analysis of genome-wide data on ZLD and GAGA binding in early Drosophila embryos (data from Harrison et al. 2011 and Nègre et al. 2011, respectively): While 71.4% of HOT regions and 75.0% of EEEs are bound by ZLD (compared with 42.2% and 13.0% of control WARM and COLD regions), GAGA binds to 53.4% of HOT regions but only 20.0% of EEEs (compared with 28.3% and 7.8% for WARM and COLD regions). Even when considering only regions that are functioning as transcriptional enhancers in the early embryo (all EEEs from CAD and this study combined), GAGA binds to significantly more HOT enhancers than to enhancers that are not HOT (38.8% vs. 15.8%; 2.5-fold; P < 0.01). An instructive role for ZLD in defining chromatin that is open and accessible to other factors (Harrison et al. 2011) is further supported by its unusual property to bind to the majority (64%) of all occurrences of its sequence motif in the Drosophila genome (Harrison et al. 2011). ZLD might thus be a prerequisite for both HOT regions (Nien et al. 2011; Satija and Bradley 2012) and EEEs more generally. Similarly, a role for GAGA in nucleating or promoting the formation of TF complexes is consistent with its ability to self-oligomerize via its BTB/POZ domain (Espinás et al. 1999) and also form heteromeric complexes with the TF Tramtrack (Bardwell and Treisman 1994) and potentially other BTB/POZ domain-containing TFs (e.g., Abrupt, Bric-a-brac, Broad complex, and others). GAGA, with its ability to recruit other TFs by protein–protein interactions, might contribute to HOT regions independent of the specific cellular or developmental context. Interestingly, C. elegans HOT regions (Gerstein et al. 2010) are also strongly enriched in the GAGA motifs (Supplemental Table 4), and the motif is the most important sequence feature when classifying C. elegans HOT versus control regions (Supplemental Fig. S4). GAGA-like factors or their putative homologs or functional analogs across species might be a conserved feature of metazoan HOT regions.
Materials and methods
Cloning, BAC recombineering, and transgenesis
BAC recombineering was performed as described in Venken et al. (2006). All BACs were integrated into attP40 landing site on chromosome 2 (Markstein et al. 2008). The transgenic flies to test HOT and control regions are a subset of a large resource that is currently being built by the Dickson laboratory VT project (C Masser, SS Bidaye, A Stark, and BJ Dickson, unpubl.). Briefly, candidate HOT and control regions were cloned in pBPGUw reporter vector and integrated into attP2 landing site on chromosome 3 (see Supplemental Table 1; Pfeiffer et al. 2008). The tested HOT regions comprise a representative set across the full range of complexity scores defined by modENCODE (Supplemental Table 1; Supplemental Fig. 1), which is unbiased with respect to the expression of neighboring genes (see Supplemental Table 2).
Whole-mount in situ hybridization and imaging
Colometric in situ hybridization was performed using standard methods (Lécuyer et al. 2008). Probes against GAL4 were generated using primers described earlier (Pfeiffer et al. 2008). Embryos were imaged on a Zeiss Axiophot microscope using Nomarski optics.
Definition of HOT, WARM, and COLD regions
We used D. melanogaster HOT regions as defined previously (complexity score strictly >8) (The modENCODE Consortium 2010). As controls, we defined WARM regions as genomic regions with a complexity score ≤8 and strictly >3, and COLD regions as genomic regions with a complexity score ≤3. HOT regions are, on average, bound by 11.9 TFs, WARM regions are bound by 5.7 TFs, and COLD regions are bound by 1.6 TFs
Comparison with known enhancers
We obtained the genomic coordinates of known transcriptional enhancers in Drosophila from REDfly (Gallo et al. 2010) and CAD2, which is restricted to embryonic enhancers (Bonn et al. 2012). We restricted both data sets to enhancers of lengths ≤2 kb and removed redundancy by merging overlapping enhancers. We then intersected the enhancers' genomic coordinates with the coordinates of HOT, WARM, and COLD regions and required that matches overlapped by at least 50% of the shorter region's length.
Peak to gene assignment and gene expression analysis
Each region was assigned to the gene with the closest TSS. We calculated a median reads per kilobase per million reads (RPKM) value (Mortazavi et al. 2008) of all genes uniquely assigned to one of the classes HOT, WARM, and COLD using modENCODE RNA sequencing data (Graveley et al. 2011).
Motif analysis and predictions
We scanned HOT and control regions for occurrences of known and predicted TF motifs from Stark et al. (2007) with a position weight matrix (PWM) cutoff P ≤ 2.44 × 10−4 (1/4096) (for details, see the Supplemental Material). The predictions were performed with SVM light (Joachims 1999) using a linear kernel and default parameters and a manual implementation of the LOOCV (for details, see the Supplemental Material). Features were the motifs, and as attributes, the number of motif instances within each region. The AUC was computed by the R package ROCR (Sing et al. 2005).
Supplemental Material
Supplemental Material is also available at at http://www.starklab.org/data/kvon-stampfel_genesdev_2012.
Acknowledgments
We thank members of the Dickson laboratory VT project for cloning the candidate regions and generating transgenic flies, and Katharina Schernhuber, Katarina Bartalska, Michaela Pagani, and IMP/IMBA services for technical support. A.S. is supported by a European Research Council (ERC) Starting Grant from the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement number 242922. Generation of transgenic lines was supported in part by a European Research Council (ERC) Advanced Investigator Grant to B.J.D. Basic research at the IMP is supported by Boehringer Ingelheim GmbH.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.188052.112.
References
- Bardwell VJ, Treisman R 1994. The POZ domain: A conserved protein-protein interaction motif. Genes Dev 8: 1664–1677 [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bate M 1996. twist: A myogenic switch in Drosophila. Science 272: 1481–1484 [DOI] [PubMed] [Google Scholar]
- Blaxter M 2010. Revealing the dark matter of the genome. Science 330: 1758–1759 [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczyński B, Riddell A, Furlong EEM 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Croston GE, Kerrigan LA, Lira LM, Marshak DR, Kadonaga JT 1991. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science 251: 643–649 [DOI] [PubMed] [Google Scholar]
- Espinás ML, Jiménez-García E, Vaquero A, Canudas S, Bernués J, Azorín F 1999. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biol Chem 274: 16461–16469 [DOI] [PubMed] [Google Scholar]
- Farkas G, Gausz J, Galloni M, Reuter G, Gyurkovics H, Karch F 1994. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature 371: 806–808 [DOI] [PubMed] [Google Scholar]
- Furlong EEM 2011. Molecular biology: A fly in the face of genomics. Nature 471: 458–459 [DOI] [PubMed] [Google Scholar]
- Gallo SM, Gerrard DT, Miner D, Simich M, Soye Des B, Bergman CM, Halfon MS 2010. REDfly v3.0: Toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res 39: D118–D123 doi: 10.1093/nar/gkq999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, Baren MJV, Boley N, Booth BW, et al. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li X-Y, Kaplan T, Botchan MR, Eisen MB 2011. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet 7: e1002266 doi: 10.1371/journal.pgen.1002266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims T 1999. Making large-scale SVM learning particle. In Advances in kernel methods: Support vector learning (B Schölkopf et al.), pp. 169–184 MIT Press, Cambridge, MA [Google Scholar]
- Lécuyer E, Parthasarathy N, Krause HM 2008. Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. Methods Mol Biol 420: 289–302 [DOI] [PubMed] [Google Scholar]
- Li X-Y, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Hendriks CLL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27 doi: 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Wallrath LL, Granok H, Elgin SC 1993. (CT)n (GA)n repeats and heat shock elements have distinct roles in chromatin structure and transcriptional activation of the Drosophila hsp26 gene. Mol Cell Biol 13: 2802–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li X-Y, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keränen SVE, et al. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80 doi: 10.1186/gb-2009-10-7-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N 2008. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet 40: 476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Consortium 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu X-J, White KP, Bussemaker HJ, et al. 2006. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci 103: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S 2007. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev 21: 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C 2011. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet 7: e1002339 doi: 10.1371/journal.pgen.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo T-TB, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci 105: 9715–9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Bradley RK 2012. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res 22: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T 2005. ROCR: Visualizing classifier performance in R. Bioinformatics 21: 3940–3941 [DOI] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, et al. 2007. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Bosch JR, Benavides JA, Cline TW 2006. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133: 1967–1977 [DOI] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3: research0088.1–research0088.14 doi: 10.1186/gb-2002-3-12-research0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJT, He Y, Hoskins RA, Bellen HJ 2006. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314: 1747–1751 [DOI] [PubMed] [Google Scholar]