Mammalian development begins with the formation of a totipotent embryo; however, the molecular mechanisms underlying the acquisition of totipotency are largely unknown. In this study by Peters and colleagues, the authors investigate the role of mammalian Polycomb proteins in the initiation of mammalian development. By using conditional gene disruption approaches, the authors demonstrate that embryos deficient for maternal and zygotic gene function of Ring1 and Rnf2, two core components of the PRC1 complex, arrest their development at the two-cell stage. In addition, the authors show that Ring1 and Rnf2 define cytoplasmic and chromosomal maternal contributions during oogenesis and that this is essential for proper initiation of embryonic development. Thus, this study provides novel insights into the role of Polycomb proteins in the female germline and early mammalian development.
Keywords: Polycomb-repressive complex 1, maternal effect, intergenerational inheritance, epigenetic memory, nuclear transfer, intra-S-phase checkpoint
Abstract
In mammals, totipotent embryos are formed by fusion of highly differentiated gametes. Acquisition of totipotency concurs with chromatin remodeling of parental genomes, changes in the maternal transcriptome and proteome, and zygotic genome activation (ZGA). The inefficiency of reprogramming somatic nuclei in reproductive cloning suggests that intergenerational inheritance of germline chromatin contributes to developmental proficiency after natural conception. Here we show that Ring1 and Rnf2, components of Polycomb-repressive complex 1 (PRC1), serve redundant transcriptional functions during oogenesis that are essential for proper ZGA, replication and cell cycle progression in early embryos, and development beyond the two-cell stage. Exchange of chromosomes between control and Ring1/Rnf2-deficient metaphase II oocytes reveal cytoplasmic and chromosome-based contributions by PRC1 to embryonic development. Our results strongly support a model in which Polycomb acts in the female germline to establish developmental competence for the following generation by silencing differentiation-inducing genes and defining appropriate chromatin states.
In mammals, fusion of two dimorphic gametes generates a totipotent embryo that has the ability to form all different cell types of the embryonic and extraembryonic lineages. Initially, both parental genomes are transcriptionally silent, and early embryonic events are controlled by “maternal” transcripts and proteins, stored during oogenesis, and provided to the embryo (Tadros and Lipshitz 2009). However, the role of potentially inherited germline chromatin states is largely unknown.
Classical work on genomic imprinting shows that DNA methylation established in oocytes confers intergenerational epigenetic inheritance (Gill et al. 2012). For certain repetitive sequences and many genes, however, DNA methylation is reprogrammed in early embryos (Lane et al. 2003; Blewitt et al. 2006; Smallwood et al. 2011). Nuclear transfer experiments revealed the capacity of the cytoplasm of metaphase II (M-II) oocytes and mitotic one-cell embryos to reprogram chromatin states of somatic nuclei, thereby promoting embryonic development (Egli et al. 2007; Inoue et al. 2008). Nonetheless, reprogramming of germ cell nuclei is more effective than that of somatic cell nuclei (Hochedlinger and Jaenisch 2003), suggesting that germ cell chromatin is more compatible with the reprogramming abilities of oocytes. This may be due to the fact that mammalian germline chromatin is prepatterned for early embryonic development, as in zebrafish and Caenorhabditis elegans (Arico et al. 2011; Lindeman et al. 2011).
Several recent studies on mammalian systems have suggested the existence of intergenerational (between) or transgenerational (across multiple) epigenetic inheritance of acquired traits (Anway et al. 2005; Anderson et al. 2006; Carone et al. 2010; Ng et al. 2010). Other studies indicate that proper chromatin regulation in the germline is required for gene regulation or other chromatin-based processes in the next generation (Blewitt et al. 2006; Chong et al. 2007; Puschendorf et al. 2008). The later studies suggest the existence of so-called “intrinsic” (nonacquired) intergenerational epigenetic programs that support early embryonic development in mammals (Gill et al. 2012). Here we study whether Ring1/Rnf2 and Polycomb-repressive complex 1 (PRC1) constitute such an intrinsic program that is essential for mediating transmission of epigenetic information between generations.
Polycomb group (PcG) proteins are evolutionarily conserved transcriptional repressors that were originally identified in Drosophila as factors required for the maintenance but not establishment of transcriptional silencing of homeotic genes during embryonic development (Jürgens 1985). More recently, PcG proteins have been implicated in more dynamic modes of gene silencing during development, dosage compensation, and genomic imprinting and in tumorigenesis (Sparmann and van Lohuizen 2006; Schuettengruber and Cavalli 2009).
Mammalian PcG proteins function in at least two major classes of complexes—termed PRC1 and PRC2—that catalyze monoubiquitination of H2A (H2A119ub1) and trimethylation of H3K27 (H3K27me3) (Simon and Kingston 2009). It has been shown that methylation by the PRC2 components E(Z)/EZH2 and recognition of H3K27me3 by ESC/EED are required for propagation of the repressed state (Hansen et al. 2008; Margueron et al. 2009), providing, in principle, a mechanism for epigenetic inheritance. H3K27me3 is further thought to contribute to chromatin targeting of canonical PRC1 complexes containing different Cbx and Pcgf proteins (Gao et al. 2012; Morey et al. 2012; Tavares et al. 2012). Finally, PRC1 complexes may repress transcription by compacting chromatin and/or blocking RNA polymerase elongation, the latter possibly through H2A119ub (Simon and Kingston 2009).
In mouse embryonic stem cells (ESCs), PcG proteins and their associated histone modifications occupy genes encoding transcription and signaling factors required later during development (Boyer et al. 2006; Mikkelsen et al. 2007; Endoh et al. 2008; Ku et al. 2008; Mohn et al. 2008). Similar genes are marked by H3K27me3 in pluripotent inner cell mass (ICM) cells from blastocyst stage embryos (Dahl et al. 2010). However, it is unknown whether PcG-mediated gene silencing in pluripotent ICM cells and ESCs is newly established during preimplantation embryonic development or originates from PcG-based repression in the germline. Compatible with the second option, mature oocytes and spermatozoa contain H3K27me3. In sperm, H3K27me3 marks genes that serve developmental functions, reminiscent of Polycomb-binding profiles in somatic cells types (Hammoud et al. 2009; Brykczynska et al. 2010).
In mice, zygotic deficiency of the core PRC1 component Rnf2 (Ring1b) results in embryonic lethality during gastrulation (Valk-Lingbeek et al. 2004). In contrast, the Rnf2 paralog Ring1 (Ring1a) is not essential (del Mar Lorente et al. 2000). Recently, we showed that various PRC1 components are expressed in oocytes and maternally provided to the embryo. As shown by immunofluorescence analyses, Rnf2 is required for propagation and establishment of global patterns of repressive chromatin on maternal and paternal genomes, respectively, in early embryos (Puschendorf et al. 2008). However, maternal deficiency for Rnf2 does not aggravate the developmental defects observed in embryos zygotically deficient for Rnf2, suggesting no major role for maternally provided PRC1 for early embryonic development (Terranova et al. 2008). Then again, Ring1, although lowly expressed, may compensate for the loss of Rnf2 function during oogenesis and early embryonic development, as observed in ESCs (Endoh et al. 2008).
Here we address the maternal function of PRC1 in early embryogenesis by deleting Rnf2 and Ring1 in growing oocytes. We show that Ring1 does indeed compensate for Rnf2 deficiency during oogenesis. Genetic ablation of both paralogs results in loss of chromatin-bound PRC1 in oocytes, induction of massive transcriptional misregulation during oocyte growth, and a developmental arrest at the two-cell stage of embryogenesis. Importantly, by performing nuclear transfer experiments, we dissect the components underlying this strong maternal effect. Our data indicate that PRC1 functions during oogenesis to specify maternal contributions in the cytoplasm as well as on maternal chromosomes, both of which contribute to the developmental competence of preimplantation embryos.
Results
Maternal Ring1/Rnf2-deficient embryos do not develop beyond the two-cell stage
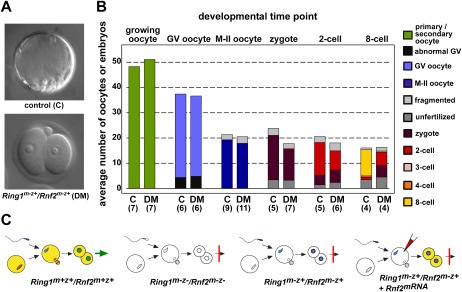
To investigate the function of maternal PRC1 in early embryogenesis, we deleted Ring1 and Rnf2 in developing oocytes. To generate Ring1/Rnf2 double homozygous mutant (dm) oocytes, we intercrossed animals that were constitutively deficient for Ring1 with mice carrying floxed alleles of Rnf2 (Rnf2F/F) and a transgenic allele of Cre recombinase, which is specifically expressed in growing oocytes under the control of the Zona pellucida 3 promoter (Zp3-cre) (Supplemental Fig. S1A,B). We used a Prm1-cre transgene, expressed in late haploid spermatids, to generate Ring1/Rnf2 dm sperm (Supplemental Fig. S1A,B). We subsequently fertilized Ring1/Rnf2 dm oocytes with Ring1/Rnf2 dm sperm to obtain embryos deficient for both maternal (m−) and zygotic (z−) expression of both paralogs (Ring1m−z−/Rnf2m−z−). We observed that development of Ring1m−z−/Rnf2m−z embryos was abrogated at the two-cell stage. Similarly, Ring1/Rnf2 dm oocytes fertilized with wild-type sperm (Ring1m−z+/Rnf2m−z+ embryos) also arrested at the two-cell stage, suggesting a maternal origin of the developmental phenotype (Fig. 1A,B).
Figure 1.
Loss of maternal Ring1 and Rnf2 impairs early embryonic development beyond the two-cell stage. (A) Differential interference contrast images of Ring1m−z+/Rnf2m+z+ (further used as control; C) and Ring1m−z+/Rnf2m−z+ (DM) embryos at embryonic day 3.5. (B) Average number of Ring1−/− (C) and Ring1/Rnf2 dm (DM) oocytes and Ring1−/− (C) and Ring1m−z+/Rnf2m−z+ (DM) embryos isolated per female mouse at the indicated developmental stages. The number of female mice analyzed is shown in brackets. (C) Cartoons illustrating developmental potential of embryos with different maternal and/or zygotic deficiencies for Ring1 and Rnf2. (m−) Maternal deficiency; (z−) zygotic deficiency; (z+) zygotic proficiency from either paternal or both parental origins. Coloring of (pro)nuclei indicates wild-type alleles of Ring1 and Rnf2 ([red] maternal; [blue] paternal; [green] maternal and paternal). The right panel shows microinjection of Rnf2 mRNA. Yellow and white cytoplasms indicate the presence or absence of Ring1/Rnf2 proteins, respectively.
PRC1 function during oogenesis is required for early embryogenesis
The early arrest of Ring1m−z+/Rnf2m−z+ embryos could reflect a function of PRC1 during oogenesis, or alternatively, maternally provided Ring1 and Rnf2 transcripts and proteins may be required during early embryogenesis. To address these possibilities, we first analyzed transcript and protein expression of PRC1 components in growing oocytes and early embryos (Supplemental Fig. S1C–F). We found that in contrast to early embryos, where only Rnf2 mRNA is detected, growing oocytes express both Ring1 and Rnf2 (Supplemental Fig. S1C). Furthermore, in oocytes, expression of either paralog is sufficient for nuclear localization of PRC1 core components (e.g., Cbx2 and Bmi1) and Rybp, a Rnf2-interacting protein (Gao et al. 2012; Hisada et al. 2012; Tavares et al. 2012), and for supporting early embryogenesis (Supplemental Fig. S1D). Interestingly, in Rnf2 single-mutant oocytes, we observed increased Ring1 protein levels, while transcript levels were unaltered, arguing for a post-transcriptional compensation mechanism operating during oocyte growth, as in Rnf2 mutant ESCs (Supplemental Fig. S1D; Endoh et al. 2008). These results suggest that Ring1-supported PRC1 function during oocyte growth enables Rnf2m−z+ embryos to develop beyond the two-cell stage.
We subsequently tested whether reconstitution of PRC1 function in Ring1m−z+/Rnf2m−z+ embryos could alleviate their two-cell arrest. After microinjection of Rnf2 mRNA into early zygotes, we observed nuclear localization of myc-tagged Rnf2 in control and Ring1m−z+/Rnf2m−z+ late zygotes (Fig. 2A) and two-cell embryos (data not shown) as well as reappearance of Cbx2 and Bmi1 (Fig. 2A; Supplemental Fig. S2A–D). All three PRC1 members showed wild-type-like chromatin localization patterns as described before (Puschendorf et al. 2008), arguing for reconstitution of a de novo chromatin-bound PRC1 complex. Nonetheless, irrespective of the amount of Rnf2 mRNA injected, we never observed a developmental rescue of Ring1m−z+/Rnf2m−z+ embryos (Fig. 2B; Supplemental Fig. S2E). Thus, although we cannot exclude the possibility that chromatin localization of reconstituted PRC1 occurred too late during pronuclear formation in mutant zygotes, the data suggest that Ring1/Rnf2 function is likely required during oocyte growth to ensure proper early embryonic development (Fig. 1C).
Figure 2.
PRC1 function during oogenesis is required for embryonic development. (A) Microinjection of myc-tagged Rnf2 mRNA into Ring1m−z+/Rnf2m−z+ zygotes leads to de novo PRC1 complex formation. Immunofluorescence analyses of control and Ring1m−z+/Rnf2m−z+ zygotes microinjected with water or with myc-tagged Rnf2 mRNA. Embryos were stained with anti-myc antibody to detect injected myc-tagged Rnf2, and with anti-Cbx2 and anti-Bmi1 antibodies to visualize reconstitution of chromatin-bound PRC1. For some embryos, focal planes of two parental pronuclei were merged into one image. Bar, 20 μm. (B) Microinjection of 25 ng of myc-tagged Rnf2 mRNA into early zygotes does not alleviate the developmental arrest at the two-cell stage. Diagram shows developmental potential of control and Ring1m−z+/Rnf2m−z+ embryos at embryonic day 3.5 that had been microinjected at the early zygote stage with water or Rnf2 mRNA.
Maternal Ring1/Rnf2 deficiency delays meiotic maturation and embryonic development
To dissect the cause of the embryonic arrest, we studied cell cycle progression and transcription in mutant oocytes and embryos. In contrast to embryogenesis, Ring1/Rnf2 double deficiency in growing oocytes did not majorly impair oogenesis (Fig. 1B; Supplemental Fig. S3A). Ring1−/−/Rnf2F/F/Zp3-cre and control females generated similar numbers of phenotypically normal germinal vesicle (GV) oocytes (Fig. 1B). Meiotic maturation was affected, however, with delays in GV breakdown and in alignment of chromosomes during the first and second meiotic divisions, possibly due to impaired spindle formation (Supplemental Fig. S3B,C). Nonetheless, Ring1/Rnf2 dm oocytes complete meiosis, as we isolated equivalent numbers of one-cell embryos from Ring1−/−/Rnf2F/F/Zp3-cre and control littermates (Fig. 1B).
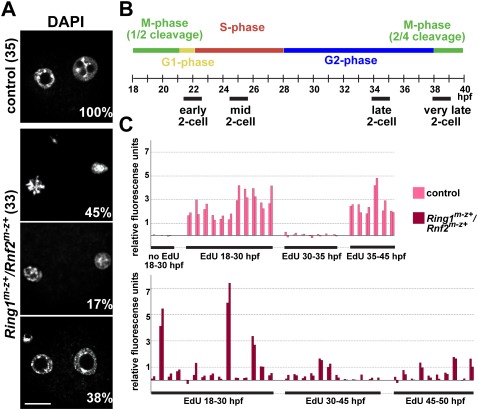
Upon fertilization, the formation of maternal and paternal pronuclei was delayed in Ring1m−z+/Rnf2m−z+ zygotes compared with control embryos (Fig. 3A; Supplemental Fig. S3D). Correspondingly, the first cleavage division was delayed (Supplemental Fig. S3E), and slightly fewer mutant embryos entered the two-cell stage in vitro and in vivo (Fig. 1B). Finally, development of Ring1m−z+/Rnf2m−z+ embryos ceased before the second cleavage division, as we failed to detect signs of chromatin condensation, spindle formation, genome-wide acquisition of phosphorylation at histone H3 Ser 10 (a marker of late G2/M-phase chromatin), or nuclear localization of the M-phase marker CyclinB1 (Supplemental Fig. S3F,G; Ohashi et al. 2001).
Figure 3.
Aberrant cell cycle progression in Ring1m−z+/Rnf2m−z+ embryos. (A) Ring1m−z+/Rnf2m−z+ zygotes stained with DAPI show a delay in pronuclear formation at 5 h post-insemination. For some embryos, focal planes of two parental pronuclei were merged into one image. Bars, 20 μm. (B) Timing of cell cycle phases of two-cell embryos and definitions for early, mid, late, and very late two-cell stages. (C) Quantification of DNA synthesis in control and Ring1m−z+/Rnf2m−z+ two-cell and four-cell embryos cultured in the presence of EdU for the indicated intervals. EdU was quantified by measuring total fluorescent signal in both nuclei of two-cell embryos or in two randomly chosen nuclei of a four-cell embryo (indicated by two adjacent bars).
Defective replication and S-phase checkpoint activation in Ring1m−z+/Rnf2m−z+ embryos
To further delineate the time point of cell cycle arrest, we studied DNA replication in Ring1m−z+/Rnf2m−z+ embryos. Detailed time-course analysis of BrdU incorporation (a deoxyribonucleotide analog) revealed that Ring1m−z+/Rnf2m−z+ zygotes entered S phase with a delay and in a less synchronous fashion compared with control embryos. Notably, the majority of Ring1m−z+/Rnf2m−z+ embryos also entered the second S phase but did not exit it, as BrdU incorporation continued even up to the time when control embryos were engaged in the third round of replication (Supplemental Fig. S4A). In line with this, quantification of EdU incorporation (another deoxyribonucleotide analog) showed that DNA synthesis was markedly impaired in most Ring1m−z+/Rnf2m−z+ two-cell embryos compared with control embryos (Fig. 3B,C).
To address whether impaired replication would be due to replication fork stalling, we stained two-cell embryos with anti-Ser 139-phospho H2AX (γH2AX) antibody, a marker of DNA damage known to accumulate in response to replication stress (Smith et al. 2010). In control two-cell embryos, γH2AX labeling changed dynamically during cell cycle progression (Supplemental Fig. S4B). While G1/early S-phase embryos exhibited only a few γH2AX foci, the number increased drastically by mid/late S phase, declined again by G2 phase, and disappeared completely before M phase. Mitotic chromatin was heavily labeled with γH2AX, as reported before (Ziegler-Birling et al. 2009). In Ring1m−z+/Rnf2m−z+ embryos, we observed similar γH2AX patterns at G1/early S and mid-S phases (Supplemental Fig. S4C). However, during the time at which 85% (n = 53) of control embryos showed a G2-phase-like γH2AX pattern, almost 80% (n = 27) of Ring1m−z+/Rnf2m−z+ embryos still showed an S-phase-like γH2AX pattern. These results support the notion that most Ring1m−z+/Rnf2m−z+ embryos do not finish S phase.
To test whether sustained γH2AX in Ring1m−z+/Rnf2m−z+ embryos would activate S-phase checkpoint kinases, we stained embryos for the phosphorylated forms of Chk1 and Chk2 and of proteins phosphorylated by Atr/Atm kinases (Supplemental Fig. S4D–F). As controls, we used γ-irradiated and hydroxyurea (HU)-treated control embryos that displayed a strong and intermediate activation, respectively, of all checkpoint proteins examined and a corresponding increase in γH2AX levels. Interestingly, in mid-two-cell (S-phase) control embryos, we did not detect checkpoint kinase activation, although γH2AX was abundant. In contrast, the kinases were activated in 16 out of 19 Ring1m−z+/Rnf2m−z+ late two-cell stage embryos. Thus, these findings suggest that the prolonged S-phase in Ring1m−z+/Rnf2m−z+ embryos is in part due to reduced DNA synthesis, triggering an intra-S-phase checkpoint response that may underlie the two-cell arrest observed in these embryos.
Zygotic genome activation (ZGA) is severely impaired in Ring1m−z+/Rnf2m−z+ two-cell embryos
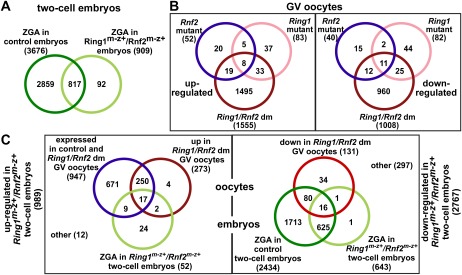
In mice, major ZGA occurs in two-cell embryos and is essential for progression beyond the two-cell stage. To examine whether ZGA is affected, we carried out genome-wide expression profiling of control (Ring1m−z+/Rnf2m+z+) and Ring1m−z+/Rnf2m−z+ late two-cell embryos cultured with or without the transcriptional elongation inhibitor α-amanitin (Fig. 4A). We found 3676 probe sets that were α-amanitin-sensitive in control embryos, representing de novo activated genes. In contrast, only 909 probe sets were α-amanitin-sensitive in Ring1m−z+/Rnf2m−z+ embryos, with 92 being inappropriately de novo activated. Together, ZGA is severely impaired in Ring1m−z+/Rnf2m−z+ embryos. We speculate that this impairment contributes to the developmental arrest.
Figure 4.
Aberrant gene expression in Ring1/Rnf2 dm GV oocytes and impaired ZGA in Ring1m−z+/Rnf2m−z+ two-cell embryos. (A) Analyses of ZGA by transcriptome profiling of untreated, α-amanitin-treated control, and Ring1m−z+/Rnf2m−z+ late two-cell embryos. Venn diagram shows overlap among α-amanitin-sensitive probe sets (≥1.5-fold in untreated vs. α-amanitin-treated; P-value < 0.05) in control (dark-green) and Ring1m−z+/Rnf2m−z+ (light green) embryos. (B) Analyses of gene expression in wild-type, Ring1 mutant, Rnf2 mutant, and Ring1/Rnf2 dm GV oocytes. Venn diagrams show overlap among probe sets either up-regulated or down-regulated in Rnf2 mutant, Ring1 mutant, and Ring1/Rnf2 dm GV oocytes compared with wild type (≥1.5-fold; P-value < 0.05). (C) Origin of transcripts misregulated in Ring1m−z+/Rnf2m−z+ late two-cell embryos. (Left diagram) For probe sets up-regulated in Ring1m−z+/Rnf2m−z+ versus control two-cell embryos, the majority was expressed in control and Ring1/Rnf2 dm GV oocytes (blue), with some up-regulated in Ring1/Rnf2 dm GV oocytes (dark red). Only a minority is de novo transcribed (α-amanitin-sensitive) in Ring1m−z+/Rnf2m−z+ two-cell embryos (light green). (Right diagram) For probe sets down-regulated in Ring1m−z+/Rnf2m−z+ versus control two-cell embryos, the majority is not de novo transcribed (α-amanitin-sensitive) to a level observed in control embryos (light and dark green). Only a few probe sets were down-regulated in Ring1/Rnf2 dm GV oocytes (light red). For all comparisons, a ≥1.5-fold difference with a P-value of <0.05 was used.
Genome-wide transcriptional misregulation in Ring1/Rnf2 dm oocytes
To elucidate the mechanism underlying aberrant replication and ZGA in Ring1m−z+/Rnf2m−z+ embryos, we subsequently investigated the effect of Ring1/Rnf2 deficiency on transcription during oogenesis. We observed that transcription is properly shut down in Ring1/Rnf2 dm GV oocytes, as assessed by immunofluorescence staining for RNA polymerase II (RNAPII) (Abe et al. 2010) as well as microinjected BrUTP, a ribonucleotide analog incorporated into nascent RNA (Supplemental Fig. S5A,B). To examine gene-specific expression defects during oocyte growth, we determined genome-wide mRNA levels in GV oocytes, which naturally store the majority of transcripts produced during the growing phase for subsequent meiotic maturation and early embryogenesis. We observed that 2563 probe sets were misexpressed in Ring1/Rnf2 dm oocytes, 60% of which were up-regulated. In contrast, only 165 and 92 probe sets were misregulated in Ring1 and Rnf2 single mutants, respectively, suggesting, at least for some genes, Ring1- or Rnf2-specific regulatory functions (Fig. 4B; Supplemental Fig. S6A,B). Single-gene analyses confirmed the mRNA profiling results (Fig. 6A, below; Supplemental Figs. S6C, S7A). These findings demonstrate that Ring1 and Rnf2 serve similar, mostly redundant, gene regulatory functions during oogenesis.
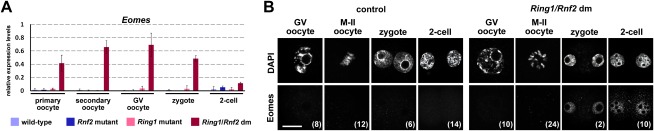
Figure 6.
Transcripts of the developmental regulator Eomes, derepressed in Ring1/Rnf2 dm oocytes, are only translated after fertilization. (A) Quantitative real-time PCR analysis of Eomes transcripts in growing oocytes (from primary and secondary follicles), GV oocytes, zygotes, and two-cell embryos that are wild type or single or double deficient for maternal Ring1 and/or Rnf2. Transcript levels were normalized to LnmB1 control. Error bars indicate standard deviation based on two to three biological replicates. (B) Immunofluorescence staining for Eomes in control and Ring1/Rnf2 dm GV and M-II oocytes, zygotes, and two-cell embryos. Numbers of oocytes or embryos analyzed at each stage are indicated in brackets. Bar, 20 μm.
Aberrant maternal transcripts are transmitted to the embryo
To address whether transcripts misregulated in Ring1/Rnf2 dm GV oocytes may contribute to the two-cell embryonic arrest, we compared transcriptomes of Ring1/Rnf2 dm GV oocytes and Ring1m−z+/Rnf2m−z+ two-cell embryos. Among the 989 probe sets up-regulated in Ring1m−z+/Rnf2m−z+ embryos, just 24 were only de novo transcribed in the embryo (being α-amanitin-sensitive). In contrast, 953 had been expressed in Ring1/Rnf2 dm GV oocytes, with 273 even being up-regulated in Ring1/Rnf2 dm versus control oocytes (Fig. 4C). Thus, the great majority of transcripts up-regulated in double-mutant two-cell embryos are indeed inherited from the Ring1/Rnf2 dm oocyte.
In contrast, among 2767 probe sets down-regulated in Ring1m−z+/Rnf2m−z+ embryos, only 34 showed reduced transcript levels in Ring1/Rnf2 dm oocytes. Instead, 641 probe sets were zygotically activated in Ring1m−z+/Rnf2m−z+ and control embryos, while the remaining 1793 were part of the ZGA program in control embryos only. These data argue that reduced transcript levels in Ring1m−z+/Rnf2m−z+ embryos mainly result from a failure to activate gene expression in the course of ZGA (Fig. 4C). In sum, Ring1/Rnf2 deficiency in oocytes effectively alters the transcriptome of Ring1m−z+/Rnf2m−z+ two-cell embryos by providing extra maternal transcripts while impairing ZGA.
Genes up-regulated in Ring1/Rnf2 dm GV oocytes are likely Polycomb targets
Gene ontology (GO) analyses indicated that genes up-regulated in Ring1/Rnf2 dm oocytes are overrepresented for developmental gene functions, similar to genes bound by PRC1 and PRC2 proteins in ESCs and differentiated somatic cells (Fig. 5A; Supplemental Table S1; Boyer et al. 2006). Additionally, transcriptome analyses showed that a significant number of genes are commonly misregulated in Ring1/Rnf2 dm GV oocytes and in mouse ESCs deficient for the PcG components Eed and Rnf2 (P-value < 2 ×10−16) (Supplemental Table S2; Leeb et al. 2010). Furthermore, significantly more of the up-regulated genes than down-regulated genes were bound by Rnf2 in ESCs (P-value = 4.9 × 10−8 versus P-value = 0.00192) (Supplemental Table S3; Endoh et al. 2008).
Figure 5.
Transcriptional misregulation of canonical PRC1 target genes in Ring1/Rnf2 dm oocytes. (A) GO analysis of genes up-regulated (red) or down-regulated (green) in Ring1/Rnf2 dm GV oocytes. Fold overrepresentation indicates the observed percentage of up-regulated genes in a particular GO category over the percentage expected on the basis of all GO-annotated genes on the array. P-values indicate significance of enrichment. (B) Box plot showing log2 fold change of expression in Ring1/Rnf2 dm over control GV oocytes for all probe sets (black) and for probe sets classified as promoter H3K27me3-positive (dark gray) or H3K27me3-negative (light gray) in chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) studies in human sperm (1:1 orthologs) (Hammoud et al. 2009), mouse ESCs, and MEFs (Mikkelsen et al. 2007). (C) Percent of probe sets detected (ON) or not detected (OFF) during normal oogenesis (Pan et al. 2005) and early embryonic development (Zeng et al. 2004; Zeng and Schultz 2005) among not changed, up-regulated, or down-regulated probe sets in Ring1/Rnf2 dm GV oocytes compared with control (≥1.5-fold; P-value < 0.05).
Finally, genes marked by H3K27me3 in human sperm, mouse ESCs, or mouse embryonic fibroblasts (MEFs) (Mikkelsen et al. 2007; Hammoud et al. 2009) are more frequently up-regulated in Ring1/Rnf2 dm oocytes than genes lacking H3K27me3 on their promoters in these cells (Fig. 5B). Together, these data support the notion that many genes up-regulated in Ring1/Rnf2 dm GV oocytes are direct targets of PRC1 during oogenesis, elucidating for the first time a function for PRC1 in the mammalian germline.
Constitutive and dynamic gene repression by PRC1 during oogenesis and embryogenesis
To investigate during which developmental periods Ring1 and Rnf2 regulate transcription, we compared the expression status of probe sets misregulated in Ring1/Rnf2 dm fully grown GV oocytes to the temporal expression pattern of genes at subsequent stages of normal oogenesis (Supplemental Fig. S1A; Zeng et al. 2004; Pan et al. 2005; Zeng and Schultz 2005). We classified loci as either not expressed or expressed early, late, or stably during normal oogenesis. We observed that 44% of probe sets up-regulated in Ring1/Rnf2 dm GV oocytes are not expressed during normal oogenesis and that 28% are only active at early stages of oogenesis (Supplemental Fig. S6D). GO term studies revealed a clear overrepresentation of developmental gene functions among up-regulated transcripts that are normally never expressed during oogenesis (Supplemental Fig. S6E; Supplemental Table S4). Quantitative PCR (qPCR) analyses confirmed that transcript levels of three lineage markers normally expressed during embryogenesis (Eomes, Gata4, and Pax6) are up-regulated in Ring1/Rnf2 dm oocytes from the primary follicle stage onward, concurrent with Zp3-cre expression driving deletion of Rnf2 (Fig. 6A; Supplemental Fig. S7A). Functions in the cytoplasm, organelles, and apoptosis were overrepresented for up-regulated genes that normally become repressed during oocyte growth (Supplemental Fig. S6E; Supplemental Table S4), indicating special physiological functions for PRC1-mediated gene repression during oogenesis as well. In all, the majority of up-regulated transcripts are normally repressed by PRC1 throughout oogenesis or become repressed during oocyte growth.
Furthermore, we correlated expression states of probe sets during normal oogenesis and preimplantation embryogenesis (Fig. 5C; Supplemental Fig. S1A, S6D; Zeng et al. 2004; Zeng and Schultz 2005). These analyses indicate that Ring1/Rnf2 repress different gene sets either constitutively or dynamically during oogenesis and likely early embryogenesis. Hox genes are examples of constitutively repressed loci, while genes like Sox2, Klf4, Eomes, Gata4, and Cebpa, marking lineage specification during preimplantation development, are repressed by PRC1 in oocytes (Supplemental Figs. S6C, S7A).
Translational repression of Ring1/Rnf2 controlled maternal transcripts
We subsequently investigated the fate of transcripts up-regulated in Ring1/Rnf2 dm GV oocytes. We analyzed protein expression of key developmental regulators Eomes, Gata4, and Pax6 in oocytes, zygotes, and two-cell embryos (Fig. 6B; Supplemental Fig. S7B). Consistent with their role in blastocyst and embryonic differentiation, we did not detect protein expression for any gene in control oocytes and early embryos. Despite elevated transcript levels in Ring1/Rnf2 dm GV oocytes (Fig. 6A; Supplemental Fig. S7A), we also failed to detect these proteins in Ring1/Rnf2 dm GV and M-II oocytes. In Ring1m−z+/Rnf2m−z+ zygotes and two-cell embryos, however, we detected nuclear localization of Eomes, Gata4, and Pax6 proteins, indicating translation of these aberrant maternal messengers only upon fertilization. These data indicate that the impairment of Ring1/Rnf2-mediated transcriptional repression during oogenesis is at least in part functionally suppressed via translational repression of aberrant maternal transcripts, a widely conserved gene regulatory mechanism functioning during oogenesis and maternal-to-embryonic transition in a variety of species (Stitzel and Seydoux 2007; Chen et al. 2011). The delay in translation of aberrant transcripts may explain the timing of the arrest during early embryogenesis and not during oogenesis.
Both cytoplasmic and chromatin contributions defined by Ring1 and Rnf2 in oocytes are needed for proper embryonic development
Finally, we asked whether the developmental arrest of Ring1m−z+/Rnf2m−z+ embryos is mediated by abnormal transcripts and proteins present in the cytoplasm of Ring1/Rnf2 dm oocytes, by inheritance of maternal chromosomes present in an aberrantly programmed chromatin state, or by a combination of these two.
To test this, we exchanged maternal pronuclei (matPN) between control and Ring1m−z+/Rnf2m−z+ early zygotes, thereby constructing diploid hybrid embryos (Fig. 7A). Control experiments showed that embryos obtained by exchanging matPNs between either control zygotes or Ring1m−z+/Rnf2m−z+ zygotes largely recapitulated the developmental phenotypes observed in naturally generated embryos (Figs. 1B, 7B; Supplemental Fig. S8A), underscoring the technical feasibility of the transfers (as demonstrated previously in Egli et al. 2007). Importantly, 87% of hybrid embryos composed of double-mutant cytoplasm and a control matPN (cyto-DM/matPN-C) failed to develop into morulae or blastocysts (P-value = 3.315 × 10−12). Furthermore, development of 69% of hybrid embryos reconstructed with control cytoplasm and a double-mutant matPN (cyto-C/matPN-DM) was impaired (P-value = 7.085 × 10−8) (Fig. 7B). In both conditions, we ruled out that the reductions in developmental potential were due to the cytoplasm fraction transferred along with a control or mutant matPN (Supplemental Fig. S7B,C). These data reveal the importance of PRC1 in defining proper cytoplasmic and possibly nuclear contributions for embryonic development.
Figure 7.
Ring1/Rnf2 expression during oogenesis defines maternal cytoplasmic and chromatin contributions required for embryonic development. (A) Cartoon illustrating generation of hybrid embryos by exchanging matPN between early zygotes with different genotypes. Wild-type paternal pronuclei (patPN) are not exchanged. (B) Diagram showing developmental potential (scored at embryonic day 4.5) of reconstructed zygotes. Control groups include (1) control (Ring1m−z+) zygote containing a matPN from another control zygote (cyto-C/matPN-C) and (2) Ring1m−z+/Rnf2m−z+ zygote containing a matPN from another Ring1m−z+/Rnf2m−z+ zygote (cyto-DM/matPN-DM). Experimental groups include (1) matPN of control zygote transferred into Ring1m−z+/Rnf2m−z+ zygote (cyto-DM/matPN-C) and (2) matPN of Ring1m−z+/Rnf2m−z+ zygote transferred into control zygote (cyto-C/matPN-DM). Numbers of reconstructed embryos analyzed are shown in brackets. (C) Cartoon illustrating generation of hybrid embryos by exchanging M-II chromosomes between M-II oocytes with different genotypes. M-II oocytes were activated to produce parthenogenetic embryos. (D) Diagram showing developmental potential (scored at embryonic day 4.5) of parthenogenetically activated reconstructed M-II oocytes. Control group includes (enucleated) control (Ring1 mutant) M-II oocyte containing chromosomes from another control M-II oocyte (cyto-C/M-II-C). Experimental groups include (1) chromosomes of control M-II oocyte transferred into (enucleated) Ring1/Rnf2 dm M-II oocyte (cyto-DM/M-II-C) and (2) chromosomes of Ring1/Rnf2 dm M-II oocyte transferred into (enucleated) control M-II oocyte (cyto-C/M-II-DM). Numbers of reconstructed embryos analyzed are shown in brackets. Note that parthenogenic activation causes a developmental arrest more frequently at the zygotic than at the two-cell stage compared with embryonic activation by sperm fusion. (E,F) Immunofluorescence staining for H2AK119ub1 in wild-type GV and MII oocytes and in Ring1/Rnf2 dm GV oocytes. Numbers of oocytes analyzed are indicated in brackets.
However, since pronuclear formation is likely controlled by maternal cytoplasmic factors, our experiments do not exclude the possibility that the poor developmental outcome of cyto-C/matPN-DM embryos is due to aberrant cytoplasm present in mutant early zygotes, affecting pronuclear formation and impairing the developmental potential of matPN-DMs before their transfer into the cytoplasm of control embryos. To circumvent this possibility, we exchanged M-II chromosomes between control and Ring1/Rnf2 dm M-II oocytes, followed by parthenogenetic activation of reconstructed oocytes (Fig. 7C; Supplemental Fig. S8).
We observed that 87% of unmanipulated and 78% of reconstructed (cyto-C/M-II-C) control embryos developed to the morula and blastocyst stages following parthenogenetic activation (Fig. 7D; Supplemental Fig. S8A). Comparable with the pronuclei exchange experiments, 98% of hybrid oocytes composed of double-mutant cytoplasm and control M-II chromosomes (cyto-DM/M-II-C) failed to develop into morulae and blastocysts (P-value < 2.2 × 10−16), confirming the importance of proper cytoplasm for early embryogenesis.
Furthermore, 50% of hybrid embryos reconstructed with control cytoplasm and double-mutant M-II chromosomes (cyto-C/M-II-DM) failed to develop into morulae and blastocysts. Intriguingly, morphological analyses of cyto-C/M-II-DM- versus cyto-C/M-II-C-arrested zygotes showed that equal percentages of mutant and control reconstructed embryos failed to be activated. However, three times more cyto-C/M-II-DM than control embryos arrested at the one-cell stage after having formed pronuclei (P-value = 0.0002) (data not shown), revealing a strong detrimental effect of double-mutant chromosomes on early development, even prior to ZGA. These data may reflect structural alterations in chromatin organization of Ring1/Rnf2 dm M-II oocytes.
Together, our data indicate that Ring1/Rnf2-mediated gene regulation during oogenesis is essential to provide oocytes with proper maternal cytoplasmic factors that support preimplantation development. Furthermore, the chromosomal transfer data argue that Ring1 and Rnf2 program maternal chromatin to sustain preimplantation development. Importantly, the low percentage of cyto-C/matPN-DM and cyto-C/M-II-DM embryos progressing to the blastocyst stage suggests that an aberrant chromatin state inherited from Ring1/Rnf2 dm oocytes can only be partially reprogrammed in early embryos by wild-type maternal cytoplasmic factors.
Discussion
In this study, we demonstrate that two PRC1 core components, Ring1 and Rnf2, serve redundant gene regulatory functions during mouse oogenesis that are essential for early embryogenesis. Loss of Ring1/Rnf2 function in oocytes causes a multitude of developmental defects. Importantly, reconstitution of chromatin-bound PRC1 in maternal mutant zygotes does not alleviate the developmental arrest at the two-cell stage. Instead, we show that maternal cytoplasmic and chromosomal contributions defined by PRC1 during oogenesis confer intergenerational control of early embryonic development.
In Ring1m−z+/Rnf2m−z+ two-cell embryos, we observed a prolonged S phase with reduced DNA synthesis rates and activation of checkpoint kinases. Although PRC1 has been linked to the DNA damage response (Liu et al. 2009), the two-cell arrest is unlikely to be due to checkpoint activation alone, since this was shown to only delay cell cycle progression in two-cell embryos but not arrest development, even if repair of DNA damage is incomplete (Shimura et al. 2002; Yukawa et al. 2007). Instead, impairment of ZGA is known to cause a two-cell arrest (Aoki et al. 1997), which is indeed majorly affected in late Ring1m−z+/Rnf2m−z+ two-cell embryos. We speculate that both attenuation of cell cycle and defective ZGA cause the developmental failure of Ring1m−z+/Rnf2m−z+ two-cell embryos.
We found that PRC1-mediated repression is needed to limit transcriptional activity in growing oocytes. Many of the genes up-regulated in Ring1/Rnf2 dm oocytes are regulated by PcG proteins in a variety of somatic cells, including ESCs. For example, homeotic genes and lineage specification factors such as Eomes, Gata4, Gata6, and Krt8 are repressed by PRC1 in oocytes and ESCs. The transcriptional up-regulation of such genes in Ring1/Rnf2 dm oocytes that normally control embryogenesis may reflect the developmentally primed state of oocytes (similar to ESCs) and underscores the importance of Polycomb function during oogenesis in suppressing aberrant gene activation, presumably driven by a variety of inducing signals. Furthermore, several genes controlled by PRC1 during oogenesis are marked by H3K27me3 in human spermatozoa and mouse round spermatids and spermatozoa (Hammoud et al. 2009; Brykczynska et al. 2010; S Erkek, M Hisano, M Stadler, and AHFM Peters, unpubl.). Thus, our data show for the first time that PRC1 controls repression of classes of genes in the female germline similar to those in soma.
To determine whether changes in the pool of maternal transcripts in the cytoplasm of fully grown GV oocytes, resulting from transcriptional misregulation in Ring1/Rnf2-deficient growing oocytes, would underlie the observed embryonic defects, we performed nuclear transfer experiments. We observed that cytoplasm of Ring1/Rnf2 maternally deficient oocytes or zygotes indeed triggered a robust early developmental arrest in reconstructed embryos. Protein analyses of three differentiation-inducing factors transcriptionally up-regulated in Ring1/Rnf2 dm oocytes demonstrated that such ectopic transcripts were only translated after fertilization. These data argue that translational repression mechanisms are not perturbed in Ring1/Rnf2 dm growing oocytes and that aberrant messages are recruited for translation during meiotic maturation or after fertilization, concomitant with the timing of the appearance of phenotypic defects. Similarly, Ring1/Rnf2 deficiency in oocytes may cause, directly or indirectly, down-regulation of maternal factors required for embryogenesis. In all, although repressing transcription in the nucleus, PcG proteins in oocytes are essential factors to define the proper dormant maternal cytoplasmic program that is executed during subsequent meiotic maturation and early embryonic development. Likewise, PcG proteins are required in ESCs for defining the proper cytoplasm that enables reprogramming of nuclei of differentiated cells upon cellular fusion (Pereira et al. 2010). Thus, similar cytoplasmic feedback mechanisms may operate at other stages of development to reinforce chromatin-based repressive mechanisms in suppressing changes in cell identity otherwise induced by stochastic variations in transcriptional regulatory circuitries.
Our chromosome transfer experiments also strongly suggest that intergenerational transmission of chromatin states specified in oocytes by PRC1 is required for full developmental competence of early embryos. In support of this concept, comparison of wild-type expression states between oocytes and early embryos shows that 37% of genes up-regulated in Ring1/Rnf2-deficient GV oocytes are normally repressed throughout oogenesis and early embryogenesis. Furthermore, another 12% of up-regulated genes become repressed during the course of normal oocyte growth and remain repressed during preimplantation development. Thus, these data strongly suggest that PRC1 drives transcriptional repression all through the reproductive life cycle of gametogenesis and early embryogenesis, until expression is induced later during development.
Our data challenge the classical paradigm, based on studies in Drosophila, which states that Polycomb functions in maintaining transcriptionally repressed states that were established by gene-specific transcriptional repressors during embryonic development. Instead, our chromosomal transfer data raise the intriguing possibility that for a number of genes, PcG function is already required in the mammalian germline to safeguard the repressed state during early embryogenesis. In other words, besides possible de novo establishment of a PRC1-controlled transcriptional repressive state by sequence-specific transcription factors and/or noncoding RNAs during embryogenesis (Margueron and Reinberg 2011), inheritance of germline prepatterned chromatin states is likely required for faithful gene repression in the early embryo. We therefore propose that Ring1/Rnf2 and PRC1 constitute an intrinsic intergenerational epigenetic program, which is essential for mediating transmission of epigenetic information between generations.
What is the molecular entity of such an intrinsic intergenerational epigenetic program? In somatic cells, “Polycomb-repressed” chromatin is generally characterized by the presence of PRC1 and PRC2 proteins, H2AK119ub1 H3K27me3, and, at times, long noncoding RNAs and the absence of elongating RNAPII and associated active chromatin marks (Stock et al. 2007; Ku et al. 2008; Leeb et al. 2010; Chu et al. 2011). In contrast to growing oocytes, PRC1 and PRC2 proteins are dissociated from chromatin in fully grown GV oocytes and during meiotic maturation in a genome-wide manner, possibly limiting a direct role in inheritance. Only upon fertilization do PRC1 components become reloaded onto chromatin (Puschendorf et al. 2008). The significance of H2AK119ub1 in epigenetic memory is also unclear, given that global down-regulation of this mark during mitosis by the deubiquitinase Ubp-M/USP16 is required for mitotic progression (Joo et al. 2007) and that the E3 ligase activity of Rnf2 is dispensable for higher-order chromatin compaction and silencing of Hox genes (Eskeland et al. 2010). In wild-type GV oocytes, H2AK119ub1 is present on chromatin, while the immunofluorescence signal is absent in Ring1/Rnf2 dm GV oocytes, indicating that Ring1 and Rnf2 are the main enzymes responsible for H2AK119ub1 in growing oocytes (Fig. 7E). Intriguingly, we failed to detect H2AK119ub1 on chromosomes of wild-type M-II oocytes (Fig. 7F), arguing that this modification may not be involved in intergenerational transmission of PRC1-mediated gene repression. In contrast, germline PRC2-mediated H3K27me3 may function in the intergenerational epigenetic memory of PRC1. The chromodomain of the maternally provided PRC1 component Cbx2 binds to H3K27me3 (Bernstein et al. 2006; Puschendorf et al. 2008). Consistently, PRC1 binding to euchromatin in zygotes is Ezh2-dependent and correlates with levels of H3K27me3 (Puschendorf et al. 2008).
It is currently unclear what changes in chromatin underlie the reduced developmental competence of reconstructed embryos carrying chromosomes from double-mutant oocytes/zygotes. Possibly, changes may be restricted to genes misregulated in mutant oocytes, having acquired active chromatin characteristics (modifications and proteins) or aberrant repressive chromatin (e.g., H3K9 and DNA methylation) at up-regulated or down-regulated genes, respectively. Other PRC1 targets, however, with unaltered expression in mutant oocytes, may still be marked by H3K27me3, thereby potentially reducing prospective deleterious effects on embryonic development. In agreement, we found that chromatin-localized levels of PRC2 components (Ezh2, Eed, and Suz12) were unchanged in Ring1/Rnf2 dm growing oocytes. Likewise, H3K27me3 was present in growing and GV Ring1/Rnf2 dm oocytes (data not shown).
It is well appreciated that the cytoplasm of oocytes has a remarkable capacity to reprogram chromatin. We found that only 18% of cyto-C/matPN-DM and 30% of cyto-C/M-II-DM reconstructed embryos (vs. 70%–80% of control embryos) were able to develop to the blastocyst stage, arguing that aberrant chromatin inherited from Ring1/Rnf2-deficient oocytes can only be in part reprogrammed by wild-type maternal cytoplasmic factors. Qualitative differences in the kind of cytoplasm used and the duration of the reprogramming period will affect the reprogramming efficiency, possibly explaining why more cyto-C/M-II-DM than cyto-C/matPN-DM reconstructed embryos developed into morulae and blastocysts. For example, it is thought that upon M-phase entry and nuclear membrane breakdown, nuclear factors are released in the cytoplasm that would promote reprogramming (Egli et al. 2007; Inoue et al. 2008; Egli and Eggan 2010). However, the extent to which epigenetic reprogramming and thus modulation of intrinsic intergenerational epigenetic programs occur during normal early embryonic development remains to be determined.
Materials and methods
Mice
Mice maternally and/or zygotically deficient for Rnf2 were generated using Zp3-cre and Prm1-cre transgenes to mediate deletion in growing oocytes or maturing spermatids, respectively. We further used a constitutive mutant allele of Ring1 (for details, see the Supplemental Material).
Collection, in vitro fertilization (IVF), and culture of mouse oocytes and embryos
Oocytes/embryos were harvested from 5- to 12-wk-old females in M2 (Sigma) or FHM medium (Chemicon) at the indicated time points after hCG injection: GV oocyte, 46 h after PMSG, 2.5 μM milrinone in medium; M-II oocyte, 14 or 18 h; late zygotes, 26 h; early two-cell, 36 h; mid-two-cell, 42 h; late two-cell, 48 h; and blastocyst stage, 94 h. For meiotic maturation experiments, GV oocytes were transferred into M16 medium (Sigma) without milrinone and harvested at indicated time points. For IVF, sperm capacitation was carried out in human tubular fluid (HTF) containing 9 mg/mL BSA for 2 h. IVF of M-II oocytes was performed in capacitation medium for 2 h, and embryos were cultured in FHM or KSOM + AA (Chemicon) in a humidified atmosphere of 5% CO2 in air until required. Meiotically incompetent growing oocytes (primary [diameter 50–60 μm] or secondary [diameter >60 μm]) were collected from 12- to 14-d-old mice. Ovaries were dissected in Ca2+- and Mg2+-free CZBT medium (CMF-CZBT) with 1 mg/mL collagenase (Worthington Biochemical Corp.) and 0.2 mg/mL DNase I (Sigma) and dissociated by repeated pipetting. For inhibition of de novo transcription, 24 μg/mL α-amanitin was added at 6 h post-fertilization (hpf) (after IVF) to culture medium. For positive controls of checkpoint activation, control embryos were subjected to 10 Gy of γ-radiation or placed into culture medium containing 0.02 M HU (Sigma, H8627) at 24 hpf.
Immunofluorescence
Immunofluorescence stainings of GV and M-II oocytes and embryos were carried out as described before (Puschendorf et al. 2008).
Ovaries from 12- to 14-d-old mice were frozen in Tissue-Tek O.C.T. compound (Sakura Finetek) on dry ice. Ten-micrometer-thick cryosections were cut with Microm HM355S. Cryosections were fixed on slides with 2% PFA in PBS (pH 7.4) for 10 min on ice, permeabilized in 0.1% Triton-X100 in 0.1% sodium citrate for 15 min, and blocked for 30 min at room temperature in 0.1% Tween-20 in PBS containing 2% BSA and 5% normal goat serum. Incubation with primary and secondary antibodies as well as mounting were the same as for embryos.
Microscopy and image analysis
Immunofluorescence stainings were analyzed using a laser scanning confocal microscope LSM510 META (Zeiss) and LSM510 software. Either a Z-series of 1.3-μm slices or one confocal slice through the maximal radius of each (pro)nucleus was scanned. Images were analyzed using Imaris (Bitplane) software and exported as TIFF files. For data presentation, in case the planes of maximal radius of maternal and paternal pronuclei were in different focal planes, separate images of both pronuclei were merged into a single image using Photoshop.
Differential interference contrast images were recorded with a 2.45 Zeiss Z1 microscope.
Quantification of Rnf2 signal was done using ImageJ software by summing fluorescent intensities of all Z-slices into one plane and quantifying total fluorescent signal. Nuclear fluorescent signal was corrected for background levels (cytoplasmic signal).
DNA replication analysis by BrdU or EdU incorporation
Time intervals for culture in the presence of BrdU (500 μM; Sigma) and EdU (100 nM; Invitrogen Click-iT Alexa Fluor 488) are indicated in Supplemental Figure S4A and Figure 3C, respectively. Embryos were fixed at the end of each indicated interval. For BrdU analysis, standard immunofluorescence protocol was used with the addition of a denaturing step (25 min. at room temperature, 4 M HCl in PBS/0.1% Triton X-100) and a neutralizing step (0.1 M Tris-HCl at pH 8.5) after permeabilization. EdU was detected according to the manufacturer's instructions (Invitrogen Click-iT Alexa Fluor 488). Quantification of BrdU signals was performed by eye (+ for strong, +/− for weak, and − for no BrdU incorporation), while EdU signals were quantified using ImageJ software.
Microinjection of Rnf2 mRNA into early zygotes
N-terminally myc-tagged Rnf2 (NM_011277) cloned into a pcDNA3.1-polyA vector (Yamagata et al. 2005) was in vitro transcribed using the mMessage mMachine T7 kit (Ambion, AM1344). Two picoliters to 4 pL of mRNA in nuclease-free water (Ambion, AM9937) (0, 0.1, 1, 10, and 50 ng/μL for quantification experiments; 0, 2, 25, and 50 ng/μL for developmental experiments) was microinjected into early zygotes using the Eppendorf FemtoJet injector system.
Microinjection of BrUTP into oocytes
GV oocytes were injected with 2–4 pL of 100 mM BrUTP (Sigma) in TE and cultured in M16 (Sigma) with milrinone for 2 h before fixation.
Pronuclear and M-II chromatin transfer experiments
matPN were exchanged between 24 and 28 h post-hCG zygotes, and M-II chromatin was exchanged in 13-h post-hCG M-II oocytes in M2 medium + 5 μg/mL Cytochalasin B using an inverted microscope with micromanipulators (Olympus-Narishige Micromanipulators MO-188, Nikon, and Burleigh PiezoDrill system). For pronuclear transfers, the polar bodies were removed, and the smaller matPN were aspirated and subsequently reinjected into the cytoplasm of receiver embryos from which the matPN had been previously removed. For M-II transfers, chromatin and spindle were aspired and injected into previously enucleated oocytes and allowed to recover for 15 min in FHM. For parthenogenetic activation, oocytes were placed into Ca++-free CZB medium containing 10 mM strontium chloride (CZB-Sr) and 5 μg/mL Cytochalasin B for 5–6 h. Embryos were cultured in FHM under mineral oil at 37°C under 5% CO2.
Quantitative real-time RT–PCR
Oocytes or embryos were pooled from several mice, and RNA was isolated from batches of five to 50 using the PicoPure RNA Isolation kit (KIT0202), adding 100 ng of Escherichia coli rRNA as carrier and a bacterial probe set as spike (GeneChipEukaryotic Poly-A RNA Control kit). Reverse transcription of RNA corresponding to 20–25 oocytes or embryos was done using random primers and SuperScript III Reverse Transcriptase (Invitrogen). cDNA corresponding to 0.4 oocytes or embryos was used for each qPCR reaction using SYBR Green PCR Master mix (Applied Biosystem) and ABI Prism 7000 Real-Time PCR machine. Measurements were performed on at least two biological replicates from independent isolations and were normalized against endogenous LnmB1 and to exogenous bacterial spike gene Thr (data not shown).
Expression profiling of late two-cell embryos and GV oocytes and data analysis
IVF and in vitro cultured (in the presence of α-amanitin [as described before] or not) late two-cell embryos from several mice were harvested at 35 hpf in batches of 40 embryos, three biological replicates per genotype/treatment condition. GV oocytes were pooled from several mice in batches of 50 oocytes, three biological replicates per genotype. RNA was isolated using the PicoPure RNA Isolation kit (KIT0202, Stratagene). RNA quality was assessed with the Agilent 2100 Bioanalyzer and RNA 6000 Pico Chip. RNA was converted into OmniPlex WTA cDNA libraries and amplified by WTA PCR using the TransPlex Whole Transcriptome Amplification kit (WTA1, Sigma) following the manufacturer's instructions with minor modifications. cDNA was purified using the GeneChip cDNA Sample Cleanup module (Affymetrix). The labeling, fragmentation, and hybridization of cDNA were performed according to Affymetrix's instructions (GeneChip Whole Transcription Sense Target Labeling technical manual, revision 2) with minor modifications. Samples were hybridized to Mouse Gene 1.0 arrays from Affymetrix. Expression data are available at NCBI Gene Expression Omnibus (GEO).
Microarray quality control and analysis were carried out in R 2.10.0 and Bioconductor 2.5. Briefly, array quality was assessed using the “ArrayQualityMetrics” package. GV oocyte raw data were normalized with RMA using the “affy” package, and differentially expressed genes were identified using the empirical Bayes method (F test) (LIMMA) and P-values adjusted for false discovery rate (FDR) with the Benjamini and Hochberg correction. Probe sets with a log2 average contrast signal of at least 3, a P-value of <0.05, and an absolute linear fold change of at least 1.5-fold were selected. Two-cell raw data were normalized by the vsnrma function of the “vsn” package. Probe sets with an average contrast signal of at least 4.5, a P-value of <0.05, and an absolute linear fold change of at least 1.5-fold were selected.
P-values for enriched GO terms were obtained using GO Stat (http://gostat.wehi.edu.au). All overrepresented and underrepresented GO terms with a P-value of <0.05 were considered. Clustering criteria 2 was used, meaning that GO categories that do not differ by two or more genes are shown together. The raw expression data of GV oocytes and two-cell embryos are available at GEO (GSE23033 and GSE28710).
Acknowledgments
We thank Arie Otte (Swammerdam Institute for Life Sciences, University of Amsterdam, The Netherlands) for providing antisera, and Kazuo Yamagata (CDB, RIKEN, Japan) for the pcDNA3.1-polyA vector. We are grateful to Laurent Gelman (microscopy and imaging), Stephane Thiry (functional genomics), Michael Stadler (bioinformatics), Fred Zilbermann, and the FMI animal facility for excellent assistance, and Serap Erkek for computational support. We thank Peter de Boer and members of the Peters laboratory for critical reading of the manuscript, and Dirk Schübeler and coworkers for fruitful discussions. Research in the Peters laboratory has been supported by the Novartis Research Foundation, the Swiss National Science Foundation (31003A_125386 and NRP 63–Stem Cells and Regenerative Medicine), SystemsX.ch (Cell Plasticity), the Japanese Swiss Science and Technology Cooperation Program, the European Network of Excellence “The Epigenome,” and the EMBO YIP program. E.P., R.K., J.S., N.B., and A.H.F.M.P. conceived and designed the experiments. E.P., R.K., V.B., J.S., E.C., Z.L., and M.T. performed the experiments. E.P., R.K., T.C.R., J.S., N.B., and A.H.F.M.P. analyzed the data. M.v.L. provided conditionally deficient Rnf2 mice and antibodies. M.V. provided Ring1-deficient mice and antibodies. E.P. and A.H.F.M.P. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.188094.112.
References
- Abe K, Inoue A, Suzuki MG, Aoki F 2010. Global gene silencing is caused by the dissociation of RNA polymerase II from DNA in mouse oocytes. J Reprod Dev 56: 502–507 [DOI] [PubMed] [Google Scholar]
- Anderson LM, Riffle L, Wilson R, Travlos GS, Lubomirski MS, Alvord WG 2006. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition 22: 327–331 [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308: 1466–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM 1997. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 181: 296–307 [DOI] [PubMed] [Google Scholar]
- Arico JK, Katz DJ, van der Vlag J, Kelly WG 2011. Epigenetic patterns maintained in early Caenorhabditis elegans embryos can be established by gene activity in the parental germ cells. PLoS Genet 7: e1001391 doi: 10.1371/journal.pgen.1001391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26: 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E 2006. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet 2: e49 doi: 10.1371/journal.pgen.0020049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17: 679–687 [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M 2011. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 25: 755–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Vickaryous N, Ashe A, Zamudio N, Youngson N, Hemley S, Stopka T, Skoultchi A, Matthews J, Scott HS, et al. 2007. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet 39: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JA, Reiner AH, Klungland A, Wakayama T, Collas P 2010. Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos. PLoS ONE 5: e9150 doi: 10.1371/journal.pone.0009150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127: 5093–5100 [DOI] [PubMed] [Google Scholar]
- Egli D, Eggan K 2010. Recipient cell nuclear factors are required for reprogramming by nuclear transfer. Development 137: 1953–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Rosains J, Birkhoff G, Eggan K 2007. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature 447: 679–685 [DOI] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. 2008. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development 135: 1513–1524 [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill ME, Erkek S, Peters AHFM 2012. Parental epigenetic control of embryogenesis: A balance between inheritance and reprogramming? Curr Opin Cell Biol doi: 10.1016/j.ceb.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K 2008. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 10: 1291–1300 [DOI] [PubMed] [Google Scholar]
- Hisada K, Sanchez C, Endo TA, Endoh M, Roman-Trufero M, Sharif J, Koseki H, Vidal M 2012. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol Cell Biol 32: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R 2003. Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N Engl J Med 349: 275–286 [DOI] [PubMed] [Google Scholar]
- Inoue A, Nakajima R, Nagata M, Aoki F 2008. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod 23: 1377–1384 [DOI] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H 2007. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449: 1068–1072 [DOI] [PubMed] [Google Scholar]
- Jürgens G 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153–155 [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242 doi: 10.1371/journal.pgen.1000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W 2003. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35: 88–93 [DOI] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A 2010. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 24: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Muller F, Alestrom P, et al. 2011. Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell 21: 993–1004 [DOI] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. 2009. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L 2012. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10: 47–62 [DOI] [PubMed] [Google Scholar]
- Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ 2010. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467: 963–966 [DOI] [PubMed] [Google Scholar]
- Ohashi A, Minami N, Imai H 2001. Nuclear accumulation of cyclin B1 in mouse two-cell embryos is controlled by the activation of Cdc2. Biol Reprod 65: 1195–1200 [DOI] [PubMed] [Google Scholar]
- Pan H, O'Brien MJ, Wigglesworth K, Eppig JJ, Schultz RM 2005. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol 286: 493–506 [DOI] [PubMed] [Google Scholar]
- Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, et al. 2010. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell 6: 547–556 [DOI] [PubMed] [Google Scholar]
- Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, Kolb C, Otte AP, Koseki H, Orkin SH, et al. 2008. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet 40: 411–420 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G 2009. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Shimura T, Inoue M, Taga M, Shiraishi K, Uematsu N, Takei N, Yuan ZM, Shinohara T, Niwa O 2002. p53-dependent S-phase damage checkpoint and pronuclear cross talk in mouse zygotes with X-irradiated sperm. Mol Cell Biol 22: 2220–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G 2011. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43: 811–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Tho LM, Xu N, Gillespie DA 2010. The ATM–Chk2 and ATR–Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 108: 73–112 [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M 2006. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G 2007. Regulation of the oocyte-to-zygote transition. Science 316: 407–408 [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD 2009. The maternal-to-zygotic transition: A play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15: 668–679 [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M 2004. Stem cells and cancer; the polycomb connection. Cell 118: 409–418 [DOI] [PubMed] [Google Scholar]
- Yamagata K, Yamazaki T, Yamashita M, Hara Y, Ogonuki N, Ogura A 2005. Noninvasive visualization of molecular events in the mammalian zygote. Genesis 43: 71–79 [DOI] [PubMed] [Google Scholar]
- Yukawa M, Oda S, Mitani H, Nagata M, Aoki F 2007. Deficiency in the response to DNA double-strand breaks in mouse early preimplantation embryos. Biochem Biophys Res Commun 358: 578–584 [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM 2005. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol 283: 40–57 [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz RM 2004. Transcript profiling during preimplantation mouse development. Dev Biol 272: 483–496 [DOI] [PubMed] [Google Scholar]
- Ziegler-Birling C, Helmrich A, Tora L, Torres-Padilla ME 2009. Distribution of p53 binding protein 1 (53BP1) and phosphorylated H2A.X during mouse preimplantation development in the absence of DNA damage. Int J Dev Biol 53: 1003–1011 [DOI] [PubMed] [Google Scholar]