While plants have thousands of nearly identical rRNA genes, not all of them are constantly transcribed. At certain developmental stages, when the cellular demand is low, rRNA gene dosage is controlled via down-regulation of a subset of the genes. Pikaard and colleagues now identify the histone H3K9 and H3K27 methyltransferases that promote rRNA gene silencing and influence which genes are silenced. Interestingly, the identities of these chromatin modifiers vary with genomic context.
Keywords: DNA methylation, chromatin modification, epigenetic, gene expression, histone methylation, nucleolar dominance, transcription
Abstract
Eukaryotes have hundreds of nearly identical 45S ribosomal RNA (rRNA) genes, each encoding the 18S, 5.8S, and 25S catalytic rRNAs. Because cellular demands for ribosomes and protein synthesis vary during development, the number of active rRNA genes is subject to dosage control. In genetic hybrids, one manifestation of dosage control is nucleolar dominance, an epigenetic phenomenon in which the rRNA genes of one progenitor are repressed. For instance, in Arabidopsis suecica, the allotetraploid hybrid of Arabidopsis thaliana and Arabidopsis arenosa, the A. thaliana-derived rRNA genes are selectively silenced. An analogous phenomenon occurs in nonhybrid A. thaliana, in which specific classes of rRNA gene variants are inactivated. An RNA-mediated knockdown screen identified SUVR4 {SUPPRESSOR OF VARIEGATION 3-9 [SU(VAR)3-9]-RELATED 4} as a histone H3 Lys 9 (H3K9) methyltransferase required for nucleolar dominance in A. suecica. H3K9 methyltransferases are also required for variant-specific silencing in A. thaliana, but SUVH5 [SU(VAR)3-9 HOMOLOG 5] and SUVH6, rather than SUVR4, are the key activities in this genomic context. Mutations disrupting the H3K27 methyltransferases ATXR5 or ATXR6 affect which rRNA gene variants are expressed or silenced, and in atxr5 atxr6 double mutants, dominance relationships among variants are reversed relative to wild type. Interestingly, these changes in gene expression are accompanied by changes in the relative abundance of the rRNA gene variants at the DNA level, including overreplication of the normally silenced class and decreased abundance of the normally dominant class. Collectively, our results indicate that histone methylation can affect both the doses of different variants and their differential silencing through the choice mechanisms that achieve dosage control.
In eukaryotes, 45S ribosomal RNA (rRNA) genes are tandemly repeated at chromosomal loci known as nucleolus organizer regions (NORs) (Long and Dawid 1980; Haaf et al. 1991; Raska et al. 2006; Saez-Vasquez and Echeverria 2007). Each rRNA gene has the potential to be transcribed by RNA polymerase I (Pol I) to produce a 45S pre-rRNA transcript that is then processed into the 18S, 5.8S, and 25S rRNAs present at the catalytic core of ribosomes, the cellular machines that carry out protein synthesis (Granneman and Baserga 2005; Moss et al. 2007; Kressler et al. 2010). Of the hundreds (sometimes thousands) of rRNA genes present in eukaryotic genomes, only a subset is transcribed by Pol I in most cells, with changes in DNA methylation and post-translational histone modifications affecting rRNA gene “on” or “off” states in both mammals and plants (Grummt and Pikaard 2003; Lawrence and Pikaard 2004; Grummt 2007; McStay and Grummt 2008; McKeown and Shaw 2009)
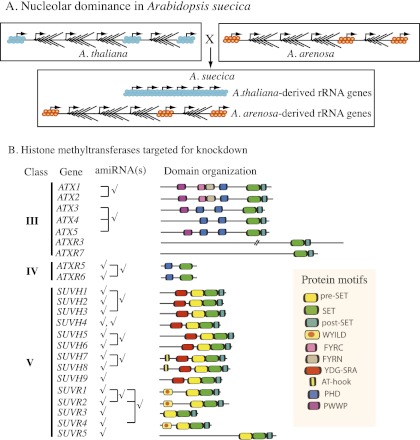
Nucleolar dominance is a phenomenon that occurs in interspecific hybrids and describes the epigenetic silencing of the rRNA genes contributed by one of the progenitors (McStay 2006; Tucker et al. 2010). First described in plants, nucleolar dominance also occurs in hybrids of insects, invertebrates, amphibians, and mammals (Pikaard 2000). In Arabidopsis suecica, the allotetraploid hybrid of Arabidopsis thaliana and Arabidopsis arenosa, the A. thaliana-derived rRNA genes are selectively silenced during early vegetative development such that only A. arenosa-derived rRNA genes are expressed in leaves of mature plants (Fig. 1A; Chen et al. 1998; Pontes et al. 2007). Initial studies with chemical inhibitors showed that rRNA gene silencing in nucleolar dominance involves concerted changes in DNA methylation and histone modification (Chen and Pikaard 1997; Lawrence et al. 2004). Subsequent RNAi-mediated knockdown screens have identified histone deacetylase 6 (HDA6), methylcytosine-binding domain protein 6 (MBD6), MBD10, the de novo DNA methyltransferase DRM2, and proteins of the siRNA-directed DNA methylation pathway as activities required for nucleolar dominance in A. suecica (Earley et al. 2006a; Preuss et al. 2008).
Figure 1.
Nucleolar dominance and HKMTs in Arabidopsis. (A) Cartoon depicting the selective epigenetic silencing of A. thaliana-derived rRNA genes in A. suecica, the allotetraploid hybrid of A. thaliana and A. arenosa. (B) Putative HKMTs in Arabidopsis, and amiRNA constructs designed to target the corresponding HKMT mRNAs (check marks). Brackets indicate instances where a single amiRNA targeted multiple mRNAs. Domain organizations of the HKMT proteins are depicted in the diagrams to the right.
A nucleolar dominance-like phenomenon also occurs in nonhybrid A. thaliana, in which distinct rRNA gene variants are selectively inactivated during early vegetative development, following a time course similar to the silencing of A. thaliana-derived rRNA genes in A. suecica (Earley et al. 2010; Abou-Ellail et al. 2011). Variant-specific rRNA gene silencing in A. thaliana involves HDA6 (Earley et al. 2010), as in A. suecica, as well as nucleolin (AtNUC-L1), a protein implicated in multiple aspects of rRNA gene expression and rRNA processing (Ginisty et al. 1999; Mongelard and Bouvet 2007; Pontvianne et al. 2007, 2010a). The involvement of HDA6 in rRNA gene silencing in both A. suecica and A. thaliana suggests that rRNA gene silencing mechanisms in hybrids and nonhybrids might be fundamentally similar.
Histone lysine methyltransferases (HKMTs) play important roles in eukaryotic gene regulation and chromatin organization. Depending on the lysine that is modified, HKMT activities can repress or activate transcription (Lachner and Jenuwein 2002; Bell et al. 2011). For example, dimethylation of histone H3 on Lys 9 (H3K9me2) is a mark typical of inactive heterochromatin. Accordingly, silent A. thaliana-derived rRNA genes in hybrid A. suecica are associated with nucleosomes enriched for H3K9me2. Conversely, the dominant A. arenosa-derived rRNA genes in A. suecica are preferentially associated with H3 that is trimethylated on Lys 4 (H3K4me3), a hallmark of active genes (Lawrence et al. 2004; Earley et al. 2006a). In hda6 mutants of A. thaliana, in which selective silencing of rRNA gene variants fails to occur during development, reduced association of rRNA genes with H3K9me2 and increased H3K4me3 association is observed (Earley et al. 2010). Therefore, H3K9me2 enrichment correlates with gene silencing in both hybrid and nonhybrid Arabidopsis, but the identities of the H3K9 methyltransferases involved are unknown.
The catalytic activity of lysine methyltransferases resides in their SET domains, named for the founding members of the family: the Drosophila proteins Suppressor of Variegation 3-9 [Su(var)3-9], Enhancer of Zeste, and Trithorax (Rea et al. 2000). In Arabidopsis, 49 genes encode proteins with a SET domain, 30 of which are homologous to enzymes known to methylate the N-terminal tails of histones (Pontvianne et al. 2010b). Among these are a class of 14 enzymes {class V; SUVH1–9 [SU(VAR)3-9 HOMOLOG 1–9] and SUVR1–5 [SU(VAR)3-9-RELATED 1–5]} whose known or predicted substrates are H3K9 (Fig. 1B; Baumbusch et al. 2001; Pontvianne et al. 2010b). SUVH proteins are distinguishable from SUVR proteins by the presence of a RING finger-associated (SRA) domain in addition to a SET domain. The SRA domain binds methylated DNA, thereby linking DNA methylation to H3K9 methylation in order to reinforce the silent epigenetic state (Fig. 1B; Johnson et al. 2007; Law and Jacobsen 2010; Rajakumara et al. 2011). Mutations in the SRA domain can disrupt gene silencing, indicating that the domain is essential for SUVH function (Johnson et al. 2007, 2008; Rajakumara et al. 2011).
SUVH4/KRYPTONITE (KYP) is thought to be the major H3K9 methyltransferase enzyme in Arabidopsis (Jackson et al. 2002; Johnson et al. 2002; Malagnac et al. 2002; Jasencakova et al. 2003). However, SUVH5 and SUVH6 act redundantly with SUVH4 to silence transcription at some loci (Ebbs et al. 2005; Ebbs and Bender 2006). SUVR4 catalyzes di- and trimethylation of H3K9 in vitro and is implicated in transposon silencing (Thorstensen et al. 2006; Veiseth et al. 2011). SUVR5 plays a role in flowering time regulation (Krichevsky et al. 2007).
Histone H3 can be monomethylated on Lys 27 (H3K27me1) by ATXR5 and ATXR6 in Arabidopsis, and this modification plays an important role in gene silencing and genome stability (Jacob et al. 2009). In atxr5 atxr6 (atxr5/6) double mutants, repetitive elements that are normally silenced are derepressed without affecting H3K9 methylation or DNA methylation, demonstrating that H3K27me1 is a critical modification for silencing these elements. Moreover, mutations disrupting ATXR5 and ATXR6 activity cause the overreplication of heterochromatic regions enriched for silenced repetitive elements, implicating the H3K27me1 chromatin mark in the negative control of origin of replication activity (Jacob et al. 2010).
In this study, we sought to identify specific histone methyltransferases that regulate the number of actively transcribed rRNA genes in the allopolyploid hybrid A. suecica and in diploid, nonhybrid A. thaliana. We demonstrate that rRNA gene silencing is mediated by distinct H3K9 methyltransferases in these two genomic contexts. Moreover, we show that the H3K27me1 methyltransferases ATXR5 and AXTR6 influence variant-specific rRNA gene expression patterns in diploid A. thaliana, both by affecting the relative copy number (dose) of variants and by separable effects on the dosage control system.
Results
SUVR4 is required for nucleolar dominance in A. suecica
We conducted a systematic screen of Arabidopsis H3K9 methyltransferases involved in nucleolar dominance in A. suecica by expressing transgenes encoding artificial microRNAs (amiRNAs) (Schwab et al. 2006) that target the mRNAs of class V (SUVH and SUVR) enzymes. Some amiRNAs were designed to knock down mRNAs of as many as three closely related genes, and others targeted individual mRNAs (Fig. 1B).
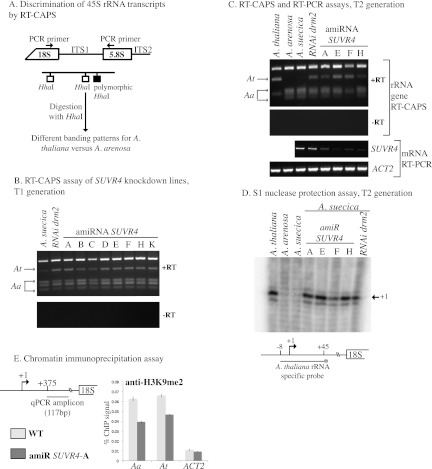
In A. suecica, pre-rRNA transcripts from the A. arenosa- and A. thaliana-derived rRNA genes can be discriminated from one another using an RT-CAPS (reverse transcription-cleaved amplified polymorphic sequence) assay (Lewis and Pikaard 2001). This PCR-based assay exploits a single-nucleotide polymorphism that generates an additional Hha I restriction endonuclease site within the internal transcribed spacer 1 (ITS1) region of A. arenosa rRNA genes, relative to A. thaliana rRNA genes, such that Hha I digestion allows A. thaliana and A. arenosa genes or their transcripts to be discriminated from one another (Fig. 2A).
Figure 2.
SUVR4 is implicated in nucleolar dominance. (A). Diagram outlining the CAPS assay that distinguishes the A. arenosa- or A. thaliana-derived rRNA genes or their transcripts in A. suecica. (B) RT-CAPS analysis of rRNA gene variant expression in SUVR4 knockdown lines (T1 generation), nontransgenic A. suecica (negative control), or a DRM2 RNAi knockdown line (positive control). Control reactions in which reverse transcriptase was omitted (−RT) are also shown. (C) Persistence of disrupted nucleolar dominance in T2 siblings of a SUVR4 knockdown line. Also shown are SUV4 mRNA levels in the knockdown lines relative to A. suecica and DRM2-RNAi controls. RT–PCR of actin (ACT2) served as a control, showing that equivalent amounts of RNA were assayed. (D) S1 nuclease protection assay following hybridization of total RNA with an end-labeled A. thaliana-specific ssDNA probe. Following S1 nuclease digestion, products were subjected to denaturing polyacrylamide gel electrophoresis and autoradiography. (E) ChIP analysis of H3K9me2 levels in the region downstream from the transcription start site in wild-type A. suecica and SUVR4 amiRNA plants. Quantitative PCR used TaqMan probes. ACTIN2 served as a H3K9me2-depleted control.
RT-CAPS assays were performed using RNA isolated from six or more independent transgenic individuals for each of the 22 different amiRNAs that were expressed in transgenic A. suecica plants. Nucleolar dominance was unaffected by the majority of amiRNA constructs. However, an amiRNA that concurrently targeted mRNAs of SUVR1, SUVR2, and SUVR4, the most closely related genes within the SUVR subfamily, disrupted silencing of A. thaliana rRNA genes in multiple transformants (Supplemental Fig. S1A,B), similar to a previously characterized RNAi line (Preuss et al. 2008) that knocks down the de novo DNA methyltransferase DRM2.
We next engineered and expressed amiRNAs targeting SUVR1, SUVR2, or SUVR4 individually and tested eight primary transformants (T1 plants) for each construct by both RT-CAPS and quantitative PCR to see whether silencing of A. thaliana-derived rRNA genes was disrupted. Significant expression of A. thaliana-derived pre-rRNA was detected in plants expressing the amiRNA targeting SUVR4 compared with nontransformed A. suecica (Fig. 2B; Supplemental Fig. S3). In these lines, SUVR4 mRNA levels are reduced relative to the nontransformed plants (Fig. 2C), but nontargeted mRNAs, including SUVR1, SUVR2, SUVH4, SUVH5, and SUVH6 mRNAs, are unaffected, demonstrating the specificity of the SUVR4 amiRNA (Supplemental Fig. S1C). Likewise, amiRNAs targeting SUVH4, SUVH5, and/or SUVH6 mRNAs knocked down their targets several-fold (Supplemental Fig. S2A,B). However, no disruption of nucleolar dominance occurred in SUVH4, SUVH5, or SUVH6 knockdown lines (Supplemental Fig. S2C–E).
The disruption of nucleolar dominance observed in SUVR4 amiRNA T1 plants (Fig. 2B) persisted in the T2 generation, as shown using RT-CAPS and S1 nuclease protection using a probe specific for A. thaliana rRNA gene transcripts that are accurately initiated at the gene promoter (Fig. 2C,D). Approximately half of the plants expressing the amiRNA that targets only SUVR1 showed some loss of nucleolar dominance (Supplemental Fig. S1B), suggesting that SUVR1 may also participate in rRNA gene silencing. However, the lower frequency of an effect compared with SUVR4 knockdown lines suggests that SUVR1 may be less critical than SUVR4 for nucleolar dominance.
To determine whether SUVR4 knockdown affects H3K9 methylation at rRNA genes, chromatin immunoprecipitation (ChIP) was performed using antibodies specific for dimethylated H3K9 (H3K9me2). A significant decrease in H3K9me2 associated with both A. thaliana- and A. arenosa-derived rRNA genes was detected in SUVR4 amiRNA plants (line A) compared with wild-type A. suecica plants (Fig. 2E). This change in H3K9me2 levels at both progenitors' rRNA genes is consistent with prior studies that indicated that dosage control of the dominant set of rRNA genes (A. arenosa) involves the same chromatin modifications responsible for the nearly complete silencing of the underdominant set of genes (Lawrence et al. 2004). For instance, the failure to silence underdominant rRNA genes upon treatment of plants with 5-aza-deoxycytosine, a cytosine methylation inhibitor, or trichostatin A, a HDA inhibitor, is accompanied by the up-regulation of the dominant rRNA genes (Lawrence et al. 2004).
SUVH5 and SUVH6, but not SUVR4, regulate rRNA gene expression in A. thaliana
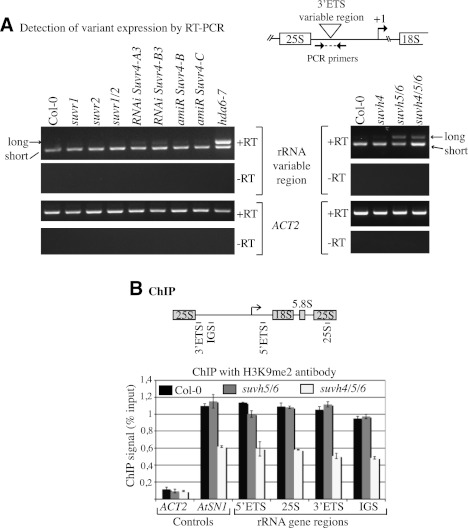
We next asked whether H3K9 methyltransferases involved in uniparental rRNA gene silencing in allotetraploid hybrids mediate the silencing of specific rRNA gene variants in nonhybrid diploids. Previous work showed that a class of “long” rRNA gene variants, representing ∼50% of the total rRNA gene pool in A. thaliana ecotype Col-0, is transcriptionally inactivated in mature leaves (Earley et al. 2010; Pontvianne et al. 2010a; Abou-Ellail et al. 2011). The presence or absence of an 81-base-pair (bp) insertion at the 3′ end of the pre-rRNA transcripts distinguishes long variants from short variants such that expression of both variant types can be monitored by RT–PCR using primers that flank the insertion/deletion site (Fig. 3A). In leaves of mature wild-type plants (Col-0), only short variants are abundantly expressed. However, in hda6 mutants, repression of long variant transcription is disrupted such that long and short variants are both expressed (Fig. 3A; Earley et al. 2010). Only short variant transcripts are detected in suvr1- or suvr2-null mutants or suvr1 suvr2 (suvr1/2) double mutants. Likewise, knocking down SUVR4 using amiRNAs or RNAi-inducing dsRNAs did not disrupt long variant repression (Fig. 3A, left panel), in contrast to the disruption of nucleolar dominance that occurs upon SUVR4 knockdown in A. suecica hybrids.
Figure 3.
SUVH5 and SUVH6 contribute to long variant silencing in A. thaliana. (A) RT–PCR detection of long and short rRNA gene variant transcripts in wild type or suvr or hda6 mutants (left panel) or suvh mutants (right panel). (B) H3K9me2 association with rRNA genes, measured by ChIP. The diagram depicts a 45S rRNA gene repeat and shows the regions that were queried by PCR following ChIP: 25S rRNA, 3′ ETS, 5′ ETS, or IGS. Actin (ACT2) and AtSN1 retroelements served as controls for active, H3K9me2-depleted or silent, H3K9me2-enriched loci, respectively.
We next examined mutants ablating members of the SUVH subfamily of histone methyltransferases. No change in rRNA long variant expression was detected in suvh4 single mutants, relative to wild-type plants, but long variant silencing is disrupted in suvh5 suvh6 double mutants (suvh5/6) or suvh4 suvh5 suvh6 triple mutants (suvh4/5/6) (Fig. 3A, right panel). Interestingly, short variant expression is elevated in suvh4/5/6 triple mutants, relative to the suvh5/6 double mutants or wild-type Col-0. These data suggest that SUVH5 and/or SUVH6 are important in the choice mechanism(s) responsible for selective silencing of long rRNA gene variants, with SUVH4 playing a role in regulating the total amount of gene expression from specific subsets of rRNA genes.
ChIP was employed to examine H3K9me2 at rRNA genes in wild-type Col-0, suvh5/6, and suvh4/5/6 mutant plants (Fig. 3B). These ChIP data report on the chromatin states of both the long and short variants combined. Surprisingly, H3K9me2 was only found to be reduced within transcribed (5′ ETS [external transcribed sequence], 25S, and 3′ ETS) and intergenic spacer (IGS) regions in suvh4/5/6 triple mutants, whereas no change from wild type was observed in the suvh5/6 double mutant (Fig. 3B). These data indicate that SUVH4 mediates H3K9 dimethylation of rRNA genes, consistent with a role in modulating their expression levels, despite not being essential in the choice mechanisms that discriminate among gene variants.
Loss of CG methylation at rRNA genes in suvh5/6 mutants
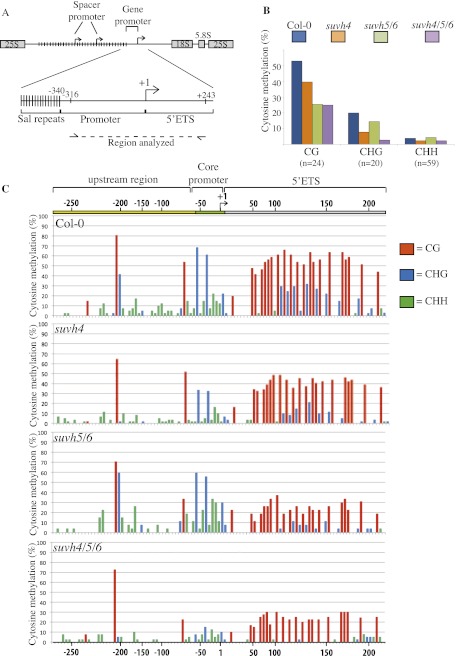
We reported previously that long variant expression correlates with reduced CG and CHG methylation flanking the rRNA gene transcription initiation site (Earley et al. 2010; Pontvianne et al. 2010a). Using bisulfite treatment followed by DNA sequencing (Frommer et al. 1992), we analyzed the cytosine methylation status of rRNA gene promoter regions (positions −316 to +243 relative to the transcription start site, defined as +1) in wild-type plants and suvh mutants (Fig. 4A). Importantly, the sequences of all rRNA genes are identical in this region, such that variants cannot be discriminated within their promoter regions. Promoter region CG methylation is reduced ∼20% in suvh4 mutants and ∼50% in suvh5/6 double mutants or suvh4/5/6 triple mutants relative to wild-type Col-0 (Fig. 4B). Changes in CG methylation are mainly restricted to a 150-bp region beginning 50 bp downstream from +1 within the transcribed region (Fig. 4C). No significant change in overall CHH methylation was detected in suvh5/6 compared with Col-0, but CHG methylation was reduced in the double mutant, mostly within the transcribed region where CG methylation is also reduced (Fig. 4B,C). More dramatic reductions in CHG and CHH methylation were observed in suvh4 and suvh4/5/6 mutants (Fig. 4B,C) and are therefore attributable to suvh4. Because silencing of long rRNA gene variants is disrupted in suvh5/6 but not suvh4 mutants, we conclude that SUVH5 and SUVH6 are important for CG and CHG methylation in the transcribed region downstream from +1 and that decreased cytosine methylation in this region correlates with long variant expression.
Figure 4.
SUVH function affects rRNA gene cytosine methylation. (A) Diagram of the rRNA gene intergenic region highlighting the promoter region analyzed by bisulfite sequencing. (B) Histograms documenting the mean cytosine methylation levels at CG, CHG, or CHG motifs in wild type (Col-0) and suvh4, suvh5/6, and suvh4/5/6 mutants based on sequencing of at least 30 independent clones following bisulfite treatment of genomic DNA. The number of CG, CHG, or CHH motifs per clone is indicated (n). (C) Positions of methylcytosines in the promoter region, displayed according to sequence context: CG (red), CHG (blue), and CHH (green). The height of each bar denotes the frequency at which that cytosine is methylated.
A role for H3K27 monomethyltransferases ATXR5 and ATXR6 in rRNA gene regulation
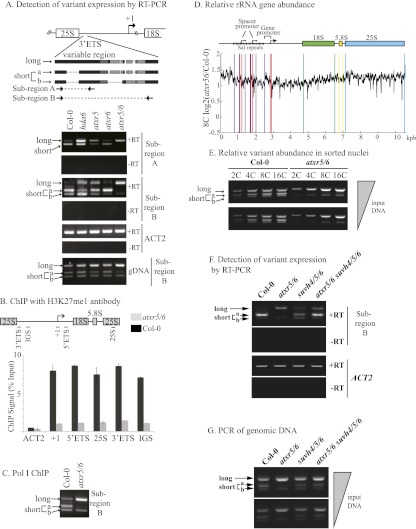
To determine whether histone H3K27me1 plays a role in rRNA gene variant silencing, we analyzed long and short variant transcript accumulation by RT–PCR in atxr5 or atxr6 single mutants and atxr5/6 double mutants, comparing them with wild type (Col-0) and hda6 mutants (Fig. 5A). Unlike hda6 mutants, in which long and short rRNA gene variants are both highly expressed, atxr6 single mutants resemble wild-type plants in that the short variants remain dominant (Fig. 5A, top panels, RT–PCR analysis of subregion A). However, in atxr5 single mutants, long variant transcripts become more abundant than short variant transcripts. Remarkably, in atxr5/6 mutants, only long variants are expressed at high levels. In these double mutants, the once-dominant short variants are expressed in only trace amounts.
Figure 5.
The H3K27 monomethylases ATXR5 and ATXR6 affect which rRNA gene variants are expressed or silenced. (A) The diagram depicts the polymorphic region in which long and short (a and b) rRNA gene variants differ due to indels. The panels below show RT–PCR analyses of rRNA gene variant expression in wild type (Col-0) or hda6, atxr5, atxr6, or atxr5/6 mutants using PCR primers spanning two different polymorphic intervals (subregions A or B). The ACT2 panel shows that equal amounts of RNA were subjected to RT–PCR for each genotype. Reactions in which reverse transcriptase was omitted (−RT) control for DNA contamination. The bottom panel shows PCR of genomic DNA (gDNA) using the primers that amplify subregion B. (B) H3K27me1 ChIP using chromatin isolated from 2-wk-old Col-0 or atxr5/6 plants. Following ChIP, the indicated regions of the rRNA genes (or the ACT2 control) were amplified and measured using quantitative PCR. (C) ChIP using an antibody recognizing the N-terminal domain of the RNA Pol I second-largest subunit, NRPA2. Following ChIP, polymorphic subregion B was amplified by PCR. (D) Relative rRNA gene copy number in atxr5/6 double mutants relative to wild type (Col-0). The graph plots the log base 2 value for the ratio of normalized read counts for genomic DNA of 8C nuclei subjected to Illumina deep sequencing (Jacob et al. 2010). (E) Relative rRNA gene variant abundance in FACS-sorted nuclei of different ploidy (2C to 16C). Wild type (Col-0) and atxr5/6 double mutants are compared. DNA was isolated from an equal number of nuclei for each ploidy level. PCR amplification of genomic DNA was conducted using primers specific for polymorphic subregion B. Reactions shown in the top panel were conducted using twice as much genomic DNA as in the bottom panel. Twofold increases in PCR products with each twofold increase in ploidy (and therefore DNA content) indicate that the amplification conditions are within the linear range. (F) RT–PCR analyses of rRNA gene variant expression patterns in wild type (Col-0), atxr5/6 double mutants, suvh4/5/6 triple mutants, or atxr5/6 suvh4/5/6 quintuple mutants. The PCR primers amplify subregion B. ACT2 and −RT controls are included. (G) Relative rRNA gene variant abundance in atxr5/6 double mutants, suvh4/5/6 triple mutants, or atxr5/6 suvh4/5/6 quintuple mutants. Reactions shown in the bottom panel used one-half the genomic DNA used in the top panel, revealing that results are semiquantitative.
Short variants can be further subdivided into two subclasses, short-a and short-b, based on small insertions/deletions (indels) that occur downstream from the duplicated repeat that distinguishes long from short variants (see the diagram in Fig. 5A). PCR primers that amplify this larger region reveal additional complexity in the patterns of rRNA gene variant expression. In wild-type plants and atxr6 single mutants, short-a transcripts are more abundant than short-b transcripts (Fig. 5A, RT–PCR analysis of subregion B). In atxr5 mutants, long variants are now expressed, but the short-a and short-b transcripts are still detected in the same proportion as in wild-type plants. In hda6 mutants, short-b transcripts become more abundant than short-a transcripts. The most dramatic change in rRNA gene variant expression occurs in atxr5/6 double mutants in which short-a transcripts are undetectable and short-b variants are detected only in trace amounts (Fig. 5A). Although the atxr6 mutation alone has little effect on rRNA gene variant expression patterns, its synergistic effect when combined with the atxr5 mutation suggests that ATXR5 and ATXR6 are partially redundant in their effects on the rRNA gene variant choice mechanism(s).
We used ChIP to determine whether histones bearing the H3K27me1 mark were depleted at rRNA genes in atxr5/6 mutants (Fig. 5B; Supplemental Fig. S4). Indeed, consistent with ATXR5 and ATXR6 being the key enzymes for H3K27 monomethylation in Arabidopsis, H3K27me1 levels are very low throughout all regions of the rRNA genes in atxr5/6 double mutants (Jacob et al. 2009).
We also performed ChIP using an antibody against the NRPA2 subunit of RNA Pol I to determine whether production of long, short-a, or short-b transcripts is consistent with transcription by Pol I within the polymorphic region that distinguishes these rRNA gene variant types (Fig. 5C). Comparison of wild type and atxr5/6 double mutants shows that Pol I associates with the different variants within the polymorphic region in proportion to their relative expression levels (Fig. 5, cf. A and C, subregion B PCR results).
Remarkably, the absence of short-a transcripts in atxr5/6 double mutants correlates with a substantial decrease in the abundance of the rRNA gene variants that encode these transcripts. This change at the DNA level is apparent upon PCR amplification of region B using genomic DNA as the template (Fig. 5A [bottom panel], E). Moreover, PCR analyses of genomic DNA indicate that the abundance of long variants, relative to short variants, is increased in atxr5 and atxr5/6 mutants.
Jacob et al. (2010) showed that heterochromatic regions are overreplicated in atxr5/6 mutants, providing a possible explanation for the changes in rRNA gene variant abundance that we observed upon PCR analysis of genomic DNA (see Fig. 5A, bottom panel). The overreplication of heterochromatin in atxr5/6 mutants is most apparent in endopolyploid cells, which are common in Arabidopsis leaves. We therefore asked whether rRNA gene copy number, relative to other genomic sequences, is affected in atxr5/6 mutants based on Illumina sequencing read counts (Jacob et al. 2010). Isolation of 8C nuclei (octaploid) from wild type and atxr5/6 using fluorescence-activated cell sorting (FACS) followed by genome-wide deep sequencing revealed an approximately twofold increase in overall rRNA gene abundance in atxr5/6 double mutants relative to wild-type Col-0 plants (Fig. 5D).
PCR amplification of rRNA gene variants was conducted using DNA isolated from equal numbers of FACS-sorted 2C (diploid), 4C, 8C, or 16C nuclei at different input dilutions (Fig. 5E). In general, the relative band intensities for the different variants are similar at all ploidy levels for both wild type and atxr5/6 double mutants such that the increased abundance of long variants relative to short variants, especially short-a variants, is apparent even in 2C nuclei of atxr5/6 mutants. Importantly, the twofold increases in PCR products with each twofold increase in ploidy (and therefore nuclear DNA content) indicates that the PCR conditions are within the linear range and therefore are semiquantitative. Collectively, the data of Figure 5, A–E, suggest that in atxr5/6 mutants, rRNA gene long variants increase in copy number at the expense of short variants. The change in abundance of the variants correlates with the apparent switch in the dominance relationship such that long variants are now expressed.
The changes in rRNA gene variant expression in suvh4 suvh5 suvh6 triple mutants and atxr5 atxr6 double mutants prompted an examination of a suvh4 suvh5 suvh6 atxr5 atxr6 quintuple mutant (Fig. 5F,G). Interestingly, the effects of the mutations are additive such that all rRNA gene variants are expressed (Fig. 5F) without any apparent loss of the short-a variants at the DNA level (Fig. 5G).
Subnuclear distribution of NORs is altered in atxr5/6
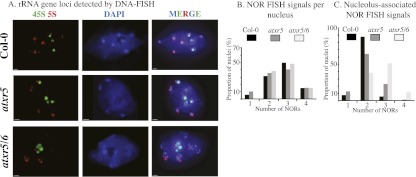
In A. thaliana leaf nuclei at interphase, the inactive rRNA genes within the NORs are highly condensed and yield punctate signals at the external periphery of the nucleolus upon fluorescent in situ hybridization (FISH) using rRNA gene probes (Fig. 6A, green signals). These condensed portions of the NORs coincide with chromocenters that are composed of a variety of repetitive elements and that stain intensively with the fluorescent DNA-binding dye DAPI (4′, 6-diamidino-2-phenylindole) (Fig. 6A). Repetitive arrays of 5S rRNA genes are included among these repeats (Fig. 6A, red FISH signals). There are two NORs in each haploid A. thaliana genome, one located on chromosome 2 (NOR2) and the other located on chromosome 4 (NOR4), each composed of ∼375 tandemly repeated rRNA genes and each spanning ∼4 Mb of chromosomal DNA (Copenhaver and Pikaard 1996a,b). NORs tend to coalesce during interphase such that it is relatively rare to observe four NOR-FISH signals in diploid nuclei. Instead, two or three NOR-FISH signals are most commonly observed (Fig. 6B). Pol I transcription of rRNA genes takes place in the nucleolus, which is enriched in RNA but contains relatively little DNA, such that the nucleolus appears as a dark hole in DAPI-stained nuclei (Fig. 6A). Active rRNA genes within a NOR are highly decondensed within the nucleolus such that their FISH signals are below the limits of our detection but emanate from NORs associated with the nucleolus (Raska et al. 2006; McStay and Grummt 2008).
Figure 6.
rRNA subnuclear organization is altered in atxr5/6. (A) FISH localization of 45S loci (NORs; green signals) and 5S (red) rRNA gene loci in leaf interphase nuclei, comparing wild type (Col-0), atxr5 mutants, and atxr5/6 double mutants. Nuclei were counterstained with DAPI (blue). Bar, 5 μm. (B,C) Histograms representing the number of NOR-FISH signals observed per nucleus (B) or the proportion of nucleolus-associated NOR-FISH signals (C).
Because short rRNA gene variants are predominantly expressed in wild type, long rRNA gene variants are expressed in atxr5/6 mutants, and both are expressed in atxr5, we compared the NOR-FISH patterns in nuclei of the three genotypes. The number of NOR-FISH signals is the same in Col-0, atxr5, and atxr5/6 double mutants, with two to three signals commonly observed in each (Fig. 6B). However, the number of NOR-FISH signals that associate with the nucleolus increases in atxr5 mutants and further increases in atxr5/6 double mutants (Fig. 6C). Whereas the vast majority of wild-type nuclei have two nucleolus-associated NOR-FISH signals, and only ∼5% have three, the number of nucleolus-associated NOR-FISH signals increases to 24% in atxr5 and 62% in atxr5/6 double mutants. These observations indicate that changes in variant expression in the mutants correlate with changes in rRNA gene localization within the nucleus, as was previously demonstrated for hda6 mutants, too (Earley et al. 2010).
Discussion
Studies in yeast, mammals, and plants suggest that rRNA gene dosage control is achieved through a combination of chromosome-level adjustments that dictate the number of active (or silenced) genes, as well as gene-level fine-scale adjustments that regulate promoter activity or RNA polymerase elongation rates. It is clear that epigenetic mechanisms, including histone modifications and cytosine methylation, are associated with the repression of excess rRNA genes at the chromosomal level (Grummt and Pikaard 2003; Lawrence and Pikaard 2004; Grummt 2007; McStay and Grummt 2008; McKeown and Shaw 2009). However, the mechanisms by which specific rRNA genes are chosen for activation or repression remain unclear.
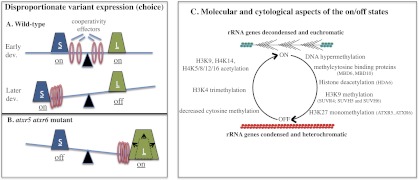
Our study provides new insights into both the chromosome-scale and gene-scale mechanisms involved in rRNA gene choice and dosage control, with a focus on the roles of histone H3 methylation in these processes. On the chromosomal scale, we found that the H3K27 monomethylases ATXR5 and ATXR6 influence which sets of rRNA gene variants are expressed or silenced. Importantly, the discovery by Jacob et al. (2010) that heterochromatic regions that are normally enriched in repressive H3K27me1 and H3K9me2 marks are overreplicated in atxr5 and atxr6 mutants provides a plausible explanation for our observations. The inactive fraction of rRNA genes that is external to the nucleolus is assembled into heterochromatin and condensed, along with other heterochromatic sequences, within the chromocenters that stain intensively with DAPI (see Fig. 6). Our studies suggest that the inactive rRNA gene pool is greatly enriched for the long rRNA gene variants, which account for half of the total number of rRNA genes, yet are expressed at very low levels compared with short variants (a and b subclasses combined) during vegetative growth (see Fig. 5). In contrast, the active gene pool is biased toward the short-a variants, which are preferentially expressed despite the fact that their abundance is similar to the abundance of short-b variants. In atxr5 mutants, PCR amplification of genomic DNA indicates that the dose of long variants increases in the genome, and in the atxr5 atxr6 double mutants, this increase is even more apparent and is accompanied by a decrease in the normally dominant short-a variants. One hypothesis to explain these observations is that the normally silenced class of rRNA genes (the long variants) is overreplicated relative to the normally active class (the short-a variants) in atxr5 and atxr6 mutants as a consequence of the loss of the H3K27me1 chromatin mark. In contrast, the normally active set of rRNA genes (short-a, in particular) might not overreplicate. Based on previous studies (Copenhaver and Pikaard 1996a,b), we predict that the different variants are clustered and not interspersed randomly at NORs. Therefore, intra-NOR recombination events involving endoreduplicated long variants might then lead to stable increases in their relative abundance while deleting short variants, explaining the changes in the relative abundance of the long, short-a, and short-b variants in atxr5 atxr6 double mutants (see Fig. 5). The change in rRNA gene variant abundance presumably tips the balance toward long variant expression during dosage control (Fig. 7A,B). We envision that the tipping process is a consequence of cooperative effects among active genes experienced at a subchromosomal level, rather than intrinsic differences in promoter strength or activator-binding affinities among the different variants, as variants are essentially identical in sequence except for the short indels that define the variants at the 3′ ends of their transcribed regions.
Figure 7.
rRNA gene dose and dosage control. (A) The model depicts the roles of rRNA gene dose and unknown cooperative effects in the disproportionate expression of specific gene variants. The number of long (L) and short (S) variants are similar in A. thaliana Col-0 and are thus depicted as weights of equal mass in wild-type cells. Molecular or cellular factors that assist in the cooperative expression of groups of rRNA genes are depicted as ring weights that can slide in either direction. Early in development, long and short variants are each expressed. Short variants become progressively more dominant as development proceeds (Earley et al. 2010). (B) In atxr5 atxr6 double mutants, long variants increase in copy number relative to short variants, altering the balance in favor of long variant expression. (C) Summary of chromatin modifications and cytological changes associated with the “on” or “off” states of rRNA genes in Arabidopsis.
The scenario above suggests that ATXR5 and ATXR6 might only affect rRNA gene expression indirectly as a consequence of suppressing heterochromatin overreplication and rRNA gene dose. However, our finding that quintuple suvh4 suvh5 suvh6 atxr5 atxr6 mutants express all rRNA gene variants without any apparent changes in variant abundance (dose) suggests that H3K27 monomethylation, like H3K9 dimethylation, also affects rRNA gene silencing and dosage control, independent of any effects on endoreduplication. The fact that the suvh4 suvh5 suvh6 triple mutant suppresses the loss of the short-a variants and the relative increase in long variants in atxr5 atxr6 mutants is also intriguing and worthy of further study in order to understand the mechanisms responsible.
Our finding that SUVR4 is required for silencing of A. thaliana-derived rRNA genes subjected to nucleolar dominance in A. suecica is consistent with a study showing that SUVR4 localizes within the nucleolus, suggesting a direct role in rRNA gene regulation (Thorstensen et al. 2006). Moreover, our previous study implicating RNA-directed DNA methylation (RdDM) by the de novo DNA methyltransferase DRM2 in nucleolar dominance (Preuss et al. 2008), coupled with a recent study implicating SUVR4 in RdDM of transposable elements (Veiseth et al. 2011), is consistent with these activities working in collaboration to silence repetitive elements that include rRNA genes.
Interestingly, two different H3K9 methyltransferases, SUVH5 and SUVH6, are required for rRNA gene dosage control during vegetative development in nonhybrid A. thaliana (Fig. 3). Studies of duplicated phosphoanthranilate isomerase (PAI) genes first revealed the cross-talk between SUVH5- and SUVH6-mediated histone modifications and cytosine methylation (Ebbs and Bender 2006). Our finding that cytosine methylation is decreased at rRNA genes in suvh5/6 double mutants is generally consistent with these studies. However, thus far, we have been unable to detect changes in the H3K9 methylation at rRNA genes in mature leaves of suvh5/6 double mutants (see Fig. 3B). One possibility is that SUVH5 and SUVH6 may exert an effect on rRNA gene H3K9 methylation early in vegetative development, at a time when the choice mechanism for long variant-specific silencing is operational. Cross-talk with the cytosine methylation machinery might then establish cytosine methylation patterns that persist among clonally related cells as a result of maintenance methylation, even if the initial histone modifications involved in establishing these cytosine methylation patterns no longer persist. Alternatively, it is plausible that a function distinct from H3K9 methylation might be required for SUVH5 and SUVH6 to affect rRNA gene silencing. For instance, SUVH2 and SUVH9 lack catalytically functional SET domains, yet are required for silencing at some loci. Interestingly, disruption of the methylcytosine-binding SRA domains of SUVH2 and SUVH9 releases silencing and correlates with loss of DNA methylation at affected loci (Johnson et al. 2008). Therefore, recruitment of other chromatin modifiers, rather than histone methylation, could be an important aspect of SUVH5 and SUVH6 function, too.
Our comparative studies of differential rRNA gene silencing in allotetraploid A. suecica and nonhybrid A. thaliana suggest that the epigenetic mechanisms involved in the two genomic contexts are similar, but not identical. Although both require HDA6 as well as involvement of H3K9 methyltransferases, the specific H3K9 methylases involved are different. Likewise, cytosine methylation is implicated in rRNA gene silencing in both A. suecica and A. thaliana, but CHH methylation upstream of the transcription start site appears to correlate with silencing in A. suecica, and CG methylation in the region just downstream from the transcription start site correlates with silencing in A. thaliana. Whether these differences are superficial or reflective of different mechanisms or pathways is not yet clear.
Materials and methods
Plant materials
Genotypes used in the study were A. thaliana ecotype Col-0 and A. suecica laboratory strain LC1. Homozygous mutants in the A. thaliana Col-0 genetic background were hda6-7 (also known as rts1-1) (Aufsatz et al. 2002); suvh4 (kyp-6), suvh5/6, and suvh4/5/6 (Ebbs and Bender 2006); suvr1, suvr2, suvr1/2, RNAi SUVR4-A3, and RNAi SUVR4-B3 (Veiseth et al. 2011); and atxr5, atxr6, and atxr5/6 (Jacob et al. 2009).
Generation of amiRNA lines
amiRNAs were designed using the Web MicroRNA Designer tool (WMD3, http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) as described in Ossowski et al. (2008). Resulting amiRNA constructs were cloned into the expression vector pEarleyGate100 (Earley et al. 2006b). Agrobacterium-mediated transformation of A. suecica or A. thaliana was performed as previously described (Lawrence and Pikaard 2003).
RNA assays
Total RNA was isolated from 2-wk-old rosettes of A. thaliana or 4-wk-old rosettes of A. suecica using the RNAeasy Plant minikit (Qiagen). The RNA was treated with Turbo DNase (Ambion) for 45 min. Semiquantitative RT–PCR and quantitative RT–PCR (qRT–PCR) were performed using random-primed cDNA generated from 1.5 μg of total RNA using SuperScript III reverse transcriptase (Invitrogen). TaqMan probes for qRT-PCR were used to specifically and simultaneously detect A. thaliana- or A. arenosa-derived rRNA genes of A. suecica using an Applied Biosystems model 7500 thermocycler. PCR primers are listed in the Supplemental Material. S1 nuclease protection using an end-labeled oligonucleotide probe matching the A. thaliana gene promoter start site was performed as previously described (Lawrence et al. 2004).
Cytosine methylation analyses
Genomic DNA was extracted using an Illustra DNA “phytopure” extraction kit (GE Healthcare). After digestion with BamHI, 2 μg of DNA was bisulfite-treated using an Epitect Bisulfite kit (Qiagen). The region of interest was amplified using primers described in Pontvianne et al. (2010a); resulting PCR products were cloned into pGEM-T-Easy (Promega) and sequenced using an ABI3730 machine. Sequenced clones were analyzed using CyMATE (Hetzl et al. 2007) and graphed using a custom Perl script and Microsoft Excel.
ChIP
ChIP was performed as described previously (Wierzbicki et al. 2008) using anti-H3K9me2 (Abcam), anti-Pol I, or anti-H3K27me1 antibodies as described (Jacob et al. 2009; Earley et al. 2010). DNA was amplified using Platinum Taq Polymerase (Invitrogen), SYBR Green I (Invitrogen), Internal Reference dye (Sigma), and an Applied Biosystems model 7500 thermocycler. Results were analyzed using the comparative CT method (Livak and Schmittgen 2001) relative to input.
For the ChIP coupled with deep sequencing (ChIP-seq) analyses shown in Supplemental Figure S4, genomic sequencing reads were mapped to a previously described rDNA sequence (Cokus et al. 2008) using Bowtie (Langmead et al. 2009) by allowing up to two mismatches and retaining reads that mapped to multiple locations.
Relative rRNA gene copy number quantitation
For the copy number analysis in Figure 5, Illumina sequencing read counts at each rRNA gene position were normalized by the total number of mapping reads in the Arabidopsis genome to correct for differences in sequencing depths, as described in Jacob et al. (2010).
DNA-FISH
DNA-FISH analyses were performed using nuclei from leaves of 4-wk-old plants, as previously described (Pontvianne et al. 2007). Briefly, 1 g of leaves from 4-wk-old plants was fixed for 20 min in 4% formaldehyde in Tris buffer (10 mM Tris-HCL at pH 7.5, 10 mM EDTA, 100 mM NaCl), then chopped as finely as possible with a razor blade in 0.5 mL of LB01 buffer (15 mM Tris-HCl at pH 7.5, 2 mM NaEDTA, 0.5 mM spermine, 80 mM KCl, 20 mM NaCl, 0.1% Triton X-100). The lysate was filtered through a 40-μm cell strainer (BD Falcon), and 12 μL of sorting buffer (100 mM Tris-HCl at pH 7.5, 50 mM KCl, 2 mM MgCl2, 0.05% Tween-20, 5% sucrose) was added per 3 μL of cell/nuclei suspension; this mixture was then spread on a round coverslip. After fixation in 2% formaldehyde in PBS (phosphate-buffered saline), slides were incubated for 30 min at 60°C prior to RNase A digestion (60 min at 37°C with 100 μg/mL RNase A). The sample on the coverslip was then washed three times in 2× SSC and then dehydrated in an ethanol series (70%, 90%, and 100% for 2 min each) prior to hybridization in formamide buffer (50% formamide, 2× SSC, 50 mM sodium phosphate at pH 7.0, 10% dextran sulphate) with 5S or 45S rRNA gene probes labeled with biotin-16-UTP (Roche) or digoxigenin-11-UTP (Roche). After overnight hybridization, slides were washed successively as follows: in 2× SSC for 5 min at 42°C, in 0.1× SSC for 5 min at 42°C, in 2× SSC for 3 min at 42°C, and in 2× SSC/0.1% Tween-20 at room temperature. Avidin conjugated with Texas Red (1:500; Vector Laboratories) followed by goat anti-avidin conjugated with biotin (1:100; Vector Laboratories) and avidin-Texas Red (1:500) were used for the detection of the biotin-labeled probe; mouse anti-digoxigenin (1:125; Roche) followed by rabbit anti-mouse fluorescein isothiocyanate (FITC) (1:500; Sigma) and Alexa 488-conjugated goat anti-rabbit (Molecular Probes) were used for the detection of the digoxigenin-labeled probe. Nuclei were then counterstained with DAPI. An Applied Precision DeltaVision personal DV microscope and SoftWorx and Imaris imaging software were used to analyze the data.
Acknowledgments
We thank Tage Thorstensen and Reidunn Aalen for providing suvr1, suvr2, suvr1/2, and SUVR4 RNAi lines, as well as anti-SUVR4 antisera. We thank Judith Bender for suvh5/6 and suvh4/5/6 seeds. We thank Ross Cocklin for ChIP experiments not shown, Chantal Leblanc for help with DNA-FISH, and Yannick Jacob for fruitful discussions. We thank Jim Powers and the Indiana University Bloomington (IUB) Light Microscopy Imaging Center and Christiane Hassel of the IUB Flow Cytometry Core Facility for their expert help. This work was supported by National Institutes of Health grants GM60380 (to C.S.P) and GM075060 (to S.D.M.). T.B. was supported by a Ruth L. Kirschstein National Research Service Award. S.E.J. is an Investigator of the Howard Hughes Medical Institute. C.S.P. is an Investigator of the Howard Hughes Medical Institute and Gordon and Betty Moore Foundation. T.B. performed the analyses of bisulfite sequencing data and the PCR analyses in Figure 5, E–G, and Supplemental Figure S1C. C.C. performed bisulfite sequencing analyses of Figure 4, B and C. W.F. performed ChIP for Figure 5B, isolated RNA for Figure 5F, and helped with FACS sorting of nuclei for Figure 5E. H.S. and S.E.J. provided Illumina sequence data in Figure 5D and Supplemental Figure S4. H.S. generated the atxr5/6 suvh4/5/6 quintuple mutant. F.P. performed all other experiments. F.P., T.B., and C.S.P. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.182865.111.
References
- Abou-Ellail M, Cooke R, Saez-Vasquez J 2011. Variations in a team: Major and minor variants of Arabidopsis thaliana rDNA genes. Nucleus 2: 204–299 [DOI] [PubMed] [Google Scholar]
- Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ 2002. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 21: 6832–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB 2001. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res 29: 4319–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thoma NH, Schubeler D 2011. Determinants and dynamics of genome accessibility. Nat Rev Genet 12: 554–564 [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS 1997. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev 11: 2124–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Comai L, Pikaard CS 1998. Gene dosage and stochastic effects determine the severity and direction of uniparental ribosomal RNA gene silencing (nucleolar dominance) in Arabidopsis allopolyploids. Proc Natl Acad Sci 95: 14891–14896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS 1996a. RFLP and physical mapping with an rDNA-specific endonuclease reveals that nucleolus organizer regions of Arabidopsis thaliana adjoin the telomeres on chromosomes 2 and 4. Plant J 9: 259–272 [DOI] [PubMed] [Google Scholar]
- Copenhaver GP, Pikaard CS 1996b. Two-dimensional RFLP analyses reveal megabase-sized clusters of rRNA gene variants in Arabidopsis thaliana, suggesting local spreading of variants as the mode for gene homogenization during concerted evolution. Plant 9: 273–282 [DOI] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS 2006a. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev 20: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS 2006b. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS 2010. Mechanisms of HDA6-mediated rRNA gene silencing: Suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24: 1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bender J 2006. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18: 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs ML, Bartee L, Bender J 2005. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol Cell Biol 25: 10507–10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginisty H, Sicard H, Roger B, Bouvet P 1999. Structure and functions of nucleolin. J Cell Sci 112: 761–772 [DOI] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ 2005. Crosstalk in gene expression: Coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol 17: 281–286 [DOI] [PubMed] [Google Scholar]
- Grummt I 2007. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum Mol Genet 16: R21–R27 doi: 10.1093/hmg/ddm020 [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS 2003. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4: 641–649 [DOI] [PubMed] [Google Scholar]
- Haaf T, Hayman DL, Schmid M 1991. Quantitative determination of rDNA transcription units in vertebrate cells. Exp Cell Res 193: 78–86 [DOI] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O 2007. CyMATE: A new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J 51: 526–536 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jacob Y, Feng S, LeBlanc CA, Bernatavichute YV, Stroud H, Cokus S, Johnson LM, Pellegrini M, Jacobsen SE, Michaels SD 2009. ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat Struct Mol Biol 16: 763–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Y, Stroud H, Leblanc C, Feng S, Zhuo L, Caro E, Hassel C, Gutierrez C, Michaels SD, Jacobsen SE 2010. Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466: 987–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Soppe WJ, Meister A, Gernand D, Turner BM, Schubert I 2003. Histone modifications in Arabidopsis—high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33: 471–480 [DOI] [PubMed] [Google Scholar]
- Johnson L, Cao X, Jacobsen S 2002. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12: 1360–1367 [DOI] [PubMed] [Google Scholar]
- Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE 2007. The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LM, Law JA, Khattar A, Henderson IR, Jacobsen SE 2008. SRA-domain proteins required for DRM2-mediated de novo DNA methylation. PLoS Genet 4: e1000280 doi: 10.1371/journal.pgen.1000280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J 2010. Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Krichevsky A, Gutgarts H, Kozlovsky SV, Tzfira T, Sutton A, Sternglanz R, Mandel G, Citovsky V 2007. C2H2 zinc finger-SET histone methyltransferase is a plant-specific chromatin modifier. Dev Biol 303: 259–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, Jenuwein T 2002. The many faces of histone lysine methylation. Curr Opin Cell Biol 14: 286–298 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence RJ, Pikaard CS 2003. Transgene-induced RNA interference: A strategy for overcoming gene redundancy in polyploids to generate loss-of-function mutations. Plant J 36: 114–121 [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Pikaard CS 2004. Chromatin turn ons and turn offs of ribosomal RNA genes. Cell Cycle 3: 880–883 [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS 2004. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell 13: 599–609 [DOI] [PubMed] [Google Scholar]
- Lewis MS, Pikaard CS 2001. Restricted chromosomal silencing in nucleolar dominance. Proc Natl Acad Sci 98: 14536–14540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 −Δ Δ C(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Long EO, Dawid IB 1980. Repeated genes in eukaryotes. Annu Rev Biochem 49: 727–764 [DOI] [PubMed] [Google Scholar]
- Malagnac F, Bartee L, Bender J 2002. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21: 6842–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown PC, Shaw PJ 2009. Chromatin: Linking structure and function in the nucleolus. Chromosoma 118: 11–23 [DOI] [PubMed] [Google Scholar]
- McStay B 2006. Nucleolar dominance: A model for rRNA gene silencing. Genes Dev 20: 1207–1214 [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I 2008. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol 24: 131–157 [DOI] [PubMed] [Google Scholar]
- Mongelard F, Bouvet P 2007. Nucleolin: A multiFACeTed protein. Trends Cell Biol 17: 80–86 [DOI] [PubMed] [Google Scholar]
- Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V 2007. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell Mol Life Sci 64: 29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D 2008. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Pikaard CS 2000. The epigenetics of nucleolar dominance. Trends Genet 16: 495–500 [DOI] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Silva M, Preuss S, Costa-Nunes P, Earley K, Neves N, Viegas W, Pikaard CS 2007. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS ONE 2: e1157 doi: 10.1371/journal.pone.0001157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J 2007. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell 18: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al. 2010a. Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 6: e1001225 doi: 10.1371/journal.pgen.1001225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Pikaard CS 2010b. Arabidopsis histone lysine methyltransferases. Adv Bot Res 53: 1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, et al. 2008. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell 32: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Law JA, Simanshu DK, Voigt P, Johnson LM, Reinberg D, Patel DJ, Jacobsen SE 2011. A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev 25: 137–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D 2006. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18: 325–334 [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Saez-Vasquez J, Echeverria M 2007. Polymerase I transcription. InRegulation of transcription in plants. Annual plant reviews vol. 29 (ed. KD Grasser), pp. 162–183. Wiley-Blackwell, Oxford, UK [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorstensen T, Fischer A, Sandvik SV, Johnsen SS, Grini PE, Reuter G, Aalen RB 2006. The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res 34: 5461–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S, Vitins A, Pikaard CS 2010. Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol 22: 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiseth SV, Rahman MA, Yap KL, Fischer A, Egge-Jacobsen W, Reuter G, Zhou MM, Aalen RB, Thorstensen T 2011. The SUVR4 histone lysine methyltransferase binds ubiquitin and converts H3K9me1 to H3K9me3 on transposon chromatin in Arabidopsis. PLoS Genet 7: e1001325 doi: 10.1371/journal.pgen.1001325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS 2008. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648 [DOI] [PMC free article] [PubMed] [Google Scholar]