Planarians are capable of regenerating body parts after injury; however, the first molecular events and the responses that lead to regeneration after an animal is injured are unclear. In this study from Reddien and colleagues, the authors discovered a molecular program involved in regeneration initiation, expanding the set of known wound-induced genes. The genes identified in this wound-induced program occurred in two waves in differentiated tissues. The first wave includes conserved immediate early genes, and many second-wave genes encode conserved patterning factors required for proper regeneration. This work provides novel insight into the activation of regeneration and how wounds mobilize stem cells for tissue repair.
Keywords: regeneration, planarians, neoblast, stem cell, wound response, immediate early
Abstract
Planarians are capable of regenerating any missing body part and present an attractive system for molecular investigation of regeneration initiation. The gene activation program that occurs at planarian wounds to coordinate regenerative responses remains unknown. We identified a large set of wound-induced genes during regeneration initiation in planarians. Two waves of wound-induced gene expression occurred in differentiated tissues. The first wave includes conserved immediate early genes. Many second-wave genes encode conserved patterning factors required for proper regeneration. Genes of both classes were generally induced by wounding, indicating that a common initial gene expression program is triggered regardless of missing tissue identity. Planarian regeneration uses a population of regenerative cells (neoblasts), including pluripotent stem cells. A class of wound-induced genes was activated directly within neoblasts, including the Runx transcription factor-encoding runt-1 gene. runt-1 was required for specifying different cell types during regeneration, promoting heterogeneity in neoblasts near wounds. Wound-induced gene expression in neoblasts, including that of runt-1, required SRF (serum response factor) and sos-1. Taken together, these data connect wound sensation to the activation of specific cell type regeneration programs in neoblasts. Most planarian wound-induced genes are conserved across metazoans, and identified genes and mechanisms should be important broadly for understanding wound signaling and regeneration initiation.
Responses to wounds must be rapid and elicit the proper reparative outcome. In highly regenerative animals, wound responses trigger the initiation of missing body part regrowth. However, the wound-induced gene programs that control regeneration initiation remain largely uncharacterized. Because common wound repair pathways can be activated at animal wounds (e.g., grainy head and extracellular signal-regulated kinase [ERK] are active at both Drosophila and mouse wounds) (Mace et al. 2005; Ting et al. 2005), genetic study of regenerative model systems has the potential to elucidate broadly used wound response mechanisms that elicit regeneration.
Planarians (freshwater flatworms) are well known for their capacity to regenerate any missing body part (Reddien and Sánchez Alvarado 2004). Given the ease with which gene expression and gene function can be assessed in planarians using newly developed molecular tools (Reddien et al. 2005a; Robb et al. 2008), planarians present a powerful system for studying wound response programs that mediate regeneration. Proliferating regenerative cells in adult Schmidtea mediterranea planarians are called neoblasts (Reddien and Sánchez Alvarado 2004) and include pluripotent stem cells (Wagner et al. 2011). Neoblasts respond to injuries with two distinct phases of cell division (Saló and Baguñà 1984; Wenemoser and Reddien 2010), migration to wounds (Dubois 1949; Wenemoser and Reddien 2010), and local differentiation (Eisenhoffer et al. 2008; Wenemoser and Reddien 2010). The first described neoblast response to injury (elevated mitotic numbers) occurs within 3–6 h, is body-wide, and is generically induced by wounding. This first response is followed by a local, regeneration-specific proliferative response (i.e., injuries that do not require significant new tissue formation do not robustly elicit this response). Molecular responses to wounds in planarians, however, have not previously been systematically explored.
In addition to closing wounds and triggering neoblast responses, injury sites must specify regeneration programs appropriate to the identity of the lost tissue, a process that remains mysterious. It is known that Wnt signaling is activated to specify tail rather than head regeneration at posterior-facing planarian wound sites (Gurley et al. 2008; Iglesias et al. 2008; Petersen and Reddien 2008, 2009; Adell et al. 2009). However, wnt1, which activates the pathway, is generically induced at all wound sites within 6 h following injury (Petersen and Reddien 2009). The selectivity of Wnt activity for posterior-facing wounds is accomplished by feedback activation of the Wnt inhibitor notum at anterior-facing wound sites in response to local tissue polarity (Petersen and Reddien 2011). These observations indicate that generic wound signaling might have an active role in specification of tissue regeneration programs.
Using differential expression analysis, we identified the transcriptional changes that occur in pre-existing, differentiated tissue and in neoblasts following wounding in planarians. We identified four categories of wound-induced genes, differing in timing and location of expression. We show that RNAi of wound-induced genes can lead to a variety of regeneration defects and identify a connection between wounding and gene expression changes in neoblasts for cell type specification in regeneration.
Results
Wound-induced genes in differentiated tissues
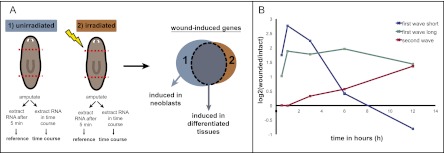
To identify genes associated with planarian regeneration initiation, we performed expression microarray analyses with RNA isolated from transversely amputated animals at different time points post-injury (30 min to 12 h) (Fig. 1A). We reasoned that many important factors signaling information about injuries would be induced in differentiated cells adjacent to wounds. To distinguish between wound-induced genes expressed in differentiated tissue versus the neoblasts, we also performed expression microarray analyses on lethally irradiated (neoblast-depleted) (Dubois 1949) animals (Fig. 1A). Three-hundred-seventy-four genes were significantly up-regulated at any time point in both irradiated and nonirradiated conditions and were selected as candidates to be induced in differentiated tissues. Three-hundred-eighty-eight genes were significantly down-regulated and were not investigated further. Two temporal gene expression waves were observed within the first 12 h following wounding. The first wave (215 genes) (Supplemental Table S1) initiated ∼30 min following wounding and reached maximum expression by 3 h (Figs. 1B, 2A). The second wave (159 genes) (Supplemental Table S1) initiated within 3–12 h following wounding, reaching maximum expression at 6 h or later (Figs. 1B, 2C).
Figure 1.
(A) Cartoon illustrating microarray design and analysis. Irradiation eliminates neoblasts and their descendants. (B) Two temporal waves of wound-induced gene expression occur in the differentiated tissue. The graph depicts three examples of wound-induced expression in differentiated tissues. (First wave short) Genes are expressed within 30 min, but expression ceases within 6–12 h following wounding (blue, Smed-egr-3). (First wave long) Genes are expressed within 30 min to 1 h, but expression is maintained beyond 12 h (green, Smed-fos-1). (Second wave) Genes are expressed within 3–12 h following wounding (red, TFPI-1).
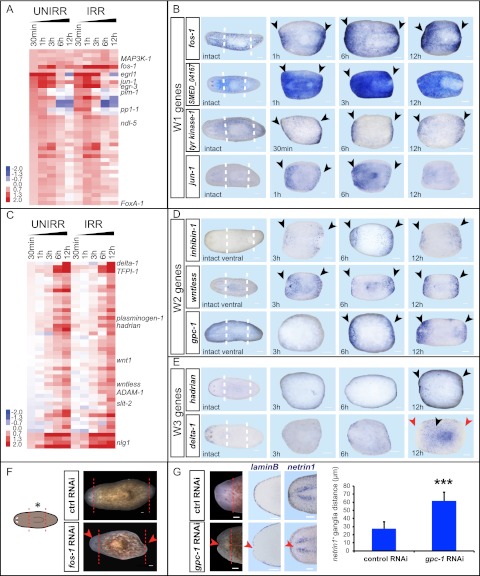
Figure 2.
Wound-induced genes activated in differentiated tissues are divided into three categories. (A) Heat map of selected genes that are up-regulated early (30 min to 1 h) in differentiated tissues; log2(time point/intact); (red) up, (blue) down; P < 0.05. (B) W1 genes. In situ hybridizations probing for genes in which wound-induced expression peaked within 3 h, typically initiating within 30 min to 1 h. (C) Heat map of selected genes that are up-regulated late (3–12 h) in differentiated tissues; log2(time point/intact); (red) up, (blue) down; P < 0.05. (D) W2 genes. In situ hybridizations probing for wound-induced genes between 3 and 12 h and with subepidermal expression at the wound site. (E) W3 genes. In situ probing for wound-induced genes with epidermal expression between 3 and 12 h. (F,G) RNAi of wound-induced genes can perturb regeneration. Animals were fed control, fos-1, or glypican-1 (gpc-1) RNAi food four times (day 0, day 4, day 8, and day 12) and were amputated 6 d later. (F) fos-1(RNAi) caused cyclopic animals with small blastemas and asymmetric posterior blastemas (10 of 30); control (0 of 30). (G) gpc-1(RNAi) animals had indented blastemas (33 of 37), lacked dorsal–ventral boundary marker expression (laminB, n = 8 of 8), and had midline defects (netrin1, n = 5 of 5) as shown in the graph to the right (distance between netrin1+ ganglia, average ± SD, P < 0.001, two-tailed Student's t-test). Anterior region of animals is shown. netrin1 (Cebria and Newmark 2005) labels a subset of the planarian nervous system. Live images, day 9; in situ hybridizations, day 8. (Red and white lines) Amputation plane; (black arrows) up-regulated expression; (red arrows) down-regulated expression and regeneration defects; (UNIRR) unirradiated; (IRR) 6000-rad irradiated. Intact animals and amputated trunk fragments are shown; anterior is to the left. Bars, 100 μm.
To validate the microarray data and determine the spatial expression domains of wound-induced genes, we assessed the expression of 108 of 374 identified wound-induced genes using in situ hybridizations on amputated fragments fixed at different times following injury. Ninety-four tested genes displayed detectable wound-induced expression (69 of 94, strong induction) (Supplemental Table S2). Genes that were activated within 30 min to 1 h and reached maximum expression by 3 h were named “W1” genes (wound-induced class 1). Some W1 genes displayed persisting (past ∼6 h) expression, and others showed transient (declining after ∼1–3 h) expression (Figs. 1B, 2A). For most tested W1 genes, expression was near wounds and in many cell types, including prominent expression in subepidermal cells and epidermis (Fig. 2B; Supplemental Fig. S1). Multiple W1 genes are homologous to “immediate early genes” in other organisms, including those encoding transcription factors such as jun-1, fos-1, egr-2, egr-3, egr-4, and egrl1 (Muller et al. 1984; Chavrier et al. 1988; Lamph et al. 1988) and others encoding signaling proteins such as pi3k-like protein, protein phosphatase 1-1, pim-1, pim-2, pim-3, and multiple GTPase-encoding genes. Immediate early genes are rapidly transcribed following exposure to various stimuli in a translation-independent manner, can mediate long-term expression changes (Greenberg et al. 1986; Almendral et al. 1988; Sonnenberg et al. 1989), and are involved in processes ranging from serum response to learning (Abraham et al. 1991; Iyer et al. 1999).
The 159 genes expressed during the second wave (expression peaking ∼6 h after injury) were divided into two categories, named W2 and W3, based on differences in spatial expression patterns by in situ hybridizations (Fig. 2D,E; Supplemental Fig. S1). W2 genes were mostly expressed subepidermally and at the wound site. W3 gene expression, in contrast, occurred in the epidermis and even far from wounds. Many W2 and W3 genes (Fig. 2C,D) are predicted to encode secreted factors, such as signaling proteins and matrix remodeling factors [e.g., W2: wntless, wnt1, inhibin-1, noggin-like1 (nlg1), follistatin, glypican-1 (gpc-1), ADAM-1, and ADAM-2; W3: delta-1, plasminogen-1, and tissue factor pathway inhibitor-1] (Ogawa et al. 2002; Adell et al. 2009; Molina et al. 2009; Petersen and Reddien 2009; Yan and Lin 2009). Some of the eight W3 genes, including delta-1 (encoding a Delta-like Notch ligand homolog) and a gene we named “ston” (encoding a novel protein), were densely expressed in epidermal cells throughout amputated body fragments between 6 and 12 h, but, notably, expression was excluded from the wound site (Fig. 2E; Supplemental Fig. S1). A W3 gene we named “hadrian,” which encodes a novel protein, was expressed around the entire periphery (at the dorsal–ventral midpoint) of injured fragments between 6 and 12 h following wounding, including at the wound site (Fig. 2E; Supplemental Fig. S1) (hadrian and ston are named after well-known walls). Homologs of tissue factor pathway inhibitor-1 and plasminogen-1 (other W3 genes) were expressed similarly to hadrian but were not expressed at the wound site (Supplemental Fig. S1).
W1, W2, and W3 gene expression, which is irradiation-insensitive (Fig. 2A,C), was found to occur in differentiated cells (Fig. 2B,D,E; Supplemental Figs. S1, S2). Little to no overlap was found between W1 and W3 or W2 and W3 genes (Supplemental Fig. S2A). However, W1 and W1, W2 and W2, and W1 and W2 genes were coexpressed in numerous subepidermal cells at wounds (Supplemental Fig. S2B–D). This suggests that a subepidermal population of wound-responsive cells might mediate early regeneration events. Moreover, these findings raise the possibility of a functional relationship between W1 and W2 genes.
Roles for W1 and W2 wound-induced genes in regeneration
Potential roles of W1 genes include wound closure, apoptosis, pathogen defense, activation of neoblast proliferation, or regulation of W2 gene expression. Many W1 genes failed to display an RNAi phenotype, potentially reflecting redundancy between the many activated genes. RNAi of the W1 gene fos-1, however, which encodes a member of the AP-1 family of transcription factors (Lee et al. 1987), led to impaired regeneration, including cyclopic blastemas and asymmetric tails (Fig. 2F).
We identified several W2 genes encoding members of Wnt and TGF-β signaling pathways, some of which have roles in body patterning during homeostasis and regeneration (Fig. 2C,D; Supplemental Table S1). Early wound-induced expression of wnt1 (6 h) has been previously described (Petersen and Reddien 2009); the W2 genes wnt1 and wntless are known to be required for the decision to regenerate a head or tail at transverse amputation planes (Adell et al. 2009; Petersen and Reddien 2009). Several W2 genes encode candidate TGF-β signaling regulators, such as gpc-1, inhibin-1, nlg1, and follistatin. Previous studies found roles for Bmp signaling (a subtype of TGF-β signaling) during regeneration and patterning (Molina et al. 2007, 2011; Orii and Watanabe 2007; Reddien et al. 2007; Gaviño and Reddien 2011). RNAi of gpc-1, which encodes a member of a class of extracellular proteins (glypicans) that can be involved in TGF-β, Hedgehog, or Wnt signaling (Yan and Lin 2009), led to indented blastema regeneration lacking midline expression for genes found at the dorsal–ventral boundary (Fig. 2G; Supplemental Fig. S3). This phenotype is similar to that observed previously following Bmp pathway inhibition (Molina et al. 2007, 2011; Orii and Watanabe 2007; Reddien et al. 2007; Gaviño and Reddien 2011). gpc-1(RNAi) animals also displayed midline abnormalities with the nervous system, similar to a phenotype caused by RNAi of the BMP pathway component tolloid (Reddien et al. 2007). These results regarding W2 genes, together with prior work on wnt1 and the Wnt inhibitor notum (Petersen and Reddien 2009, 2011), raise the possibility that generic wounding activates body position control genes, which subsequently regulate the regeneration outcome (i.e., the identity of tissue regenerated) depending on the tissue context of the wound.
W1 and W3 gene induction does not require de novo protein synthesis
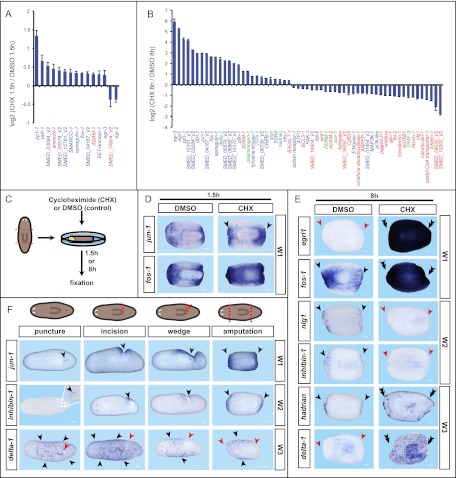
As described above, many W1 genes encode homologs of immediate early genes, which by definition are expressed independently of protein translation. We therefore tested whether new protein synthesis was required for expression of planarian wound-induced genes. We chose the NanoString nCounter platform (Geiss et al. 2008) to allow quantification of expression and assessment of a defined set of wound-induced genes under many conditions. We amputated animals in water containing the translation inhibitor cycloheximide (CHX) and quantified transcript levels of 72 selected wound-induced genes (33 W1, 31 W2, and eight W3) (Supplemental Table S3). We also included seven constitutively expressed “housekeeping” genes as controls and for normalization purposes in our probe set.
CHX treatment did not affect wound closure, and treated animals appeared normal (Supplemental Fig. S4A). At 1.5 h following wounding, induced expression of most W1 genes tested (n = 32 of 33) was not inhibited by CHX (Fig. 3A). Eleven of these W1 genes were overinduced following amputation in CHX. This phenomenon has been observed for immediate early genes in other species (Kelley-Loughnane et al. 2002), and can be explained if new protein synthesis is required for down-regulation of expression after induction (Lau and Nathans 1987). W1 genes with translation-independent expression can be considered classical immediate early genes. To control for CHX efficiency, we performed control experiments involving two rounds of amputation, allowing longer CHX incubation times (Supplemental Fig. S4B). Our findings were confirmed by in situ hybridization on animals amputated in CHX (Fig. 3C,D; Supplemental Fig. S4C), and we obtained similar results using anisomycin (Supplemental Fig. S4A,D), a second well-established translation inhibitor (Grollman 1967).
Figure 3.
W1 and W3 gene expression does not require de novo protein synthesis, and injury is sufficient to induce the transcriptional wound response. (A–E) Animals were amputated and kept in CHX (0.1 μg/μL) or DMSO (1:1000; control) until RNA was extracted at the indicated time points following wounding. RNA hybridizations and counts were performed using the NanoString platform. Expression changes, log2 ratios (P < 0.05); values, average ± SEM. (Blue) W1; (red) W2; (green) W3. (A) Effects of CHX treatment at 1.5 h following amputation. (B) W1 (n = 27 of 33) and W3 (n = 5 of 8) genes are unaffected or overinduced at 8 h following wounding in CHX, whereas most W2 (n = 25 of 31) genes fail to induce strongly (see also Supplemental Table S3). (C) Cartoon illustrating the experimental design. (D,E) In situ hybridizations of amputated trunk fragments; treatments, RNA probes, and time points are as indicated. (D) W1 genes are either unaffected or overinduced (n = 11) at 1.5 h following wounding in CHX. (E) In situ hybridizations confirm the effects of CHX on W gene expression. (F) In situ hybridizations probing for expression of W1, W2, and W3 genes following different degrees of injury at 3 h, 6 h, and 23 h, respectively. Probes are as indicated. Surgeries are illustrated on top; (red dotted line/circle) injury sites; (black arrows) up-regulated expression; (double black arrows) overinduced expression; (red arrows) lack of/decreased expression; anterior to the left. (Insets) Magnified version of the white rectangle area. Bars, 100 μm.
At 8 h following injury in CHX, 27 (20 of 33 W1) genes tested were hyperinduced (Fig. 3B,E). W3 genes (n = 5 of 8) were among the hyperinduced genes (Fig. 3B,E), which was unexpected because these genes are induced much later than W1 genes (between 6 and 12 h). The wound-induced expression of the majority of tested W2 genes (n = 25 of 31 genes) was strongly inhibited by CHX treatment (Fig. 3B,E), indicating that intermediate steps between wound detection and W2 induction exist that require de novo protein synthesis. Some W1 gene expression was sensitive to CHX treatment at this time point (seven of 33) (Fig. 3B), indicating that maintenance of their wound-induced gene expression requires new protein synthesis. In summary, wound induction of W1 and W3 genes is largely translation-independent, whereas wound induction of W2 genes is largely dependent on new protein synthesis.
A common program of gene expression is activated at diverse injuries, including those that do not elicit blastema formation
To determine whether simple wounding (not requiring blastema formation) or tissue loss (requiring blastema formation for repair) are key determinants of wound-induced gene expression, we generated a series of different injury types. W1 and W2 genes were induced at every tested wound type, ranging from a simple needle puncture to transverse amputation, albeit to a lesser degree at minor wounds (Fig. 3F; Supplemental Fig. S5A,B). Seventeen of 17 genes tested showed this generic wound induction. This is consistent with described findings for the wnt1 gene (Petersen and Reddien 2009). Therefore, W1 and W2 gene expression is part of the general response to wounding and is not specific to regeneration per se. This does not exclude the potential existence of factors expressed only at wounds where substantial regeneration is required for repair. Long-term W1 and W2 expression (at 23 h) was only maintained at wounds that required substantial regeneration, and expression was restricted to certain areas or certain wound types (nlg1, ventral [Supplemental Fig. S5C; Ogawa et al. 2002]; wnt1, during tail regeneration [Petersen and Reddien 2009]). W3 genes were also induced by both simple wounding and tissue loss, with expression maintained beyond 23 h for all injury types. Several W3 genes were expressed broadly, except directly proximal to the wound site, even at minor wounds (Supplemental Fig. S5D). We conclude that simple wounding is sufficient to induce the transcriptional changes associated with regeneration initiation. We therefore hypothesize that later events and/or the tissue context in which a common wound response program is induced serve to tailor the regeneration program to the identity of the missing tissue.
Wound-induced gene expression in neoblasts
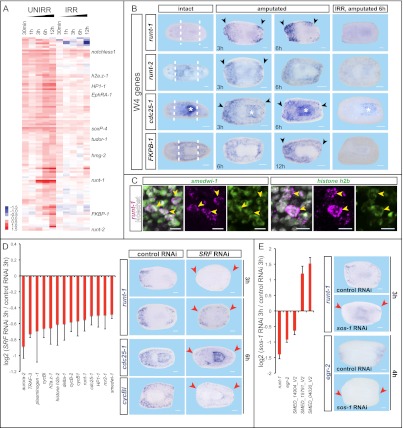
The microarray experiment described above allowed identification of 454 genes that were candidates to be induced in neoblasts or to require the presence of neoblasts for their induction. These genes were up-regulated in injured but otherwise untreated animals, but not in injured irradiated animals lacking neoblasts, and were classified as “W4” genes (Fig. 4A; Supplemental Table S4). W4 genes include those predicted to encode transcription factors, chromatin remodeling proteins, cell cycle factors, and histone methyltransferases. Thirty-three of 43 tested W4 genes were indeed expressed in irradiation-sensitive parenchymal cells near wounds following injury (Fig. 4B), consistent with expression occurring in neoblasts and/or their immediate descendants. Thirty of 43 W4 genes were normally expressed in neoblasts, with array data therefore indicating that wounding simply increases expression. Three of 43 genes tested—runt-1, runt-2, and cdc25-1—displayed minimal to no detectable expression prior to injury (Fig. 4B) (cdc25-1 also displays pharynx expression with an unknown role). runt-1 was recently found to be expressed near planarian wounds, included in a large set of genes activated in the first 3 d of head regeneration (Sandmann et al. 2011), but the cells in which it is expressed were unknown. Wound-induced expression of all W4 genes tested was sensitive to translation inhibition by CHX, except for runt-2 (Supplemental Fig. S6A). Taking runt-1 as a sample W4 gene, we observed, with quantitative PCR (qPCR), that runt-1 is wound-induced, whereas a canonical neoblast gene (smedwi-1) is not (Supplemental Fig. S6B). Furthermore, runt-1 expression was not robustly induced following feeding (another mitotic stimulus) (Baguñà 1974), indicating specificity of response to injury (Supplemental Fig. S6B). Double fluorescent in situ hybridizations with RNA probes for neoblast markers and multiple W4 genes confirmed expression in neoblasts (e.g., 319 of 319 runt-1+ cells were smedwi-1+) (Fig. 4C; Supplemental Fig. S6C). These data indicate clear heterogeneity in gene expression in neoblasts near wounds and provide some of the first genetic insights into the gene expression changes that occur in neoblasts during regeneration initiation.
Figure 4.
Identification of wound-induced genes in neoblasts and their immediate descendants following wounding. (A) Heat map of genes up-regulated (P < 0.05) in untreated amputated, but not irradiated amputated, animals [log2(time point/intact); (red) up, (blue) down]. (UNIRR) Unirradiated; (IRR) 6000-rad irradiated. (B) W4 genes. In situ hybridizations probing for wound-induced genes in neoblasts. (*) The pharynx. (Right column) Fragments were irradiated 5 d prior to amputation. (C) Fluorescent in situ hybridizations; W4 genes (runt-1, magenta) were coexpressed with neoblast markers (smedwi-1 and histone h2b) (n = 319 of 319); bars, 10 μm. (D,E) RNA hybridizations and counts were performed using the NanoString platform at 3 h following wounding. In situ hybridizations are shown at the right. (D) SRF RNAi is required for induction of multiple W4 genes. (Right) SRF RNAi impact on runt-1 expression was strong at 3 h, but weak expression was detected at 6 h; therefore, blind scoring was performed at 6 h, and 31 of 34 were scored correctly. (E) sos-1 RNAi affected wound-induced gene expression. Expression changes, log2 ratios (P < 0.01); values, average ± SEM. Probes and time points are indicated. (Black arrowheads) Up-regulated expression; (red arrowheads) absent/decreased expression; (white dotted lines) amputation planes. Intact and amputated trunk fragments are shown; anterior is to the left. Bars, 100 μm.
Smed-SRF (serum response factor) and Smed-sos-1 mediate wound-induced gene expression in neoblasts
What connects wounds to gene expression changes in the neoblasts? We performed RNAi of 15 genes that were candidates to mediate wound sensation, selected based on similarity to transcription factors and signaling proteins that can mediate responses to stimuli in other animals. We assessed potential requirements for these genes in wound-induced gene expression using the NanoString nCounter system. Eighteen W4 genes were selected as probes (Supplemental Table S5). Moreover, probes for 72 other wound-induced transcripts and seven housekeeping genes were included (Supplemental Table S3). RNAi of two genes, Smed-SRF and Smed-sos-1, resulted in specific defects in wound-induced gene expression.
Smed-SRF RNAi led to impaired induction of many W4 genes, such as cycB-1, cycB -2, cycBl, cdc25-1, and runt-1 (Fig. 4D). In situ hybridizations confirmed SRF RNAi effects for runt-1, cdc25-1, and cycBl induction (Fig. 4D); in contrast, W1 genes were typically not affected by SRF RNAi (Supplemental Fig. S6D). Neoblasts can be recognized by their >2N DNA content during replication and division by flow cytometry (known as X1 cells) (Hayashi et al. 2006). There were no significant differences in X1 numbers between control and SRF(RNAi) animals, indicating that the effect of SRF RNAi on W4 gene expression levels is not explained by decreased neoblast numbers (Supplemental Fig. S6E). The manner by which SRF activity impacts wound induction is unknown, but SRF encodes a homolog of serum response factor, a MADS-box transcription factor (Norman et al. 1988) that can regulate responses to stimuli (e.g., serum), consistent with the possibility that SRF transduces wound stimuli to transcriptional changes. SRF expression was not wound-induced (Supplemental Fig. S6F), as can occur in other species (Latasa et al. 2007). SRF(RNAi) animals regenerated smaller than normal blastemas (Supplemental Fig. S6F; Reddien et al. 2005a). By 48 h after injury, expression of W4 genes (e.g., runt-1) in SRF(RNAi) animals ultimately reached normal levels, indicating incomplete SRF inhibition or the action of additional W4-activating pathways (Supplemental Fig. S6G). This might provide a candidate explanation for the mild regeneration defects observed in SRF(RNAi) animals (Supplemental Fig. S6F).
Smed-sos-1 RNAi specifically affected wound induction of the W4 gene runt-1 and the W1 gene egr-2 in the Nanostring nCounter data; these specific defects were confirmed by in situ hybridizations (Fig. 4E). Sos is a guanine nucleotide exchange factor (GEF) that can activate MAP kinase signaling pathways. RNAi of Smed-sos-1 caused smaller blastemas and asymmetric tail formation during regeneration (Supplemental Fig. S6H). sos-1 is expressed in irradiation-sensitive cells following wounding, among others, and is therefore itself a W4 gene (Supplemental Fig. S6H). We conclude that SRF and sos-1 have important roles in the induction of wound-induced gene expression in neoblasts.
Roles for W4 genes in regeneration
Following wounding, neoblasts change cell division properties (Saló and Baguñà 1984; Wenemoser and Reddien 2010), migrate to wounds (Dubois 1949; Wenemoser and Reddien 2010), and begin differentiation programs that eventually lead to regeneration of new tissues (Eisenhoffer et al. 2008; Wenemoser and Reddien 2010; Lapan and Reddien 2011; Scimone et al. 2011). A wound-induced program in neoblasts presumably regulates aspects of these transitions. We studied two W4 genes predicted to encode Runt/Runx family transcription factors (Supplemental Fig. S7A)—the first transcription factors known to be specifically wound-induced in neoblasts rapidly (within 3–6 h) following injury—as case studies for understanding the linkage between injury and neoblast regeneration programs. Runt transcription factors are conserved throughout the Metazoa (Sullivan et al. 2008) and have been implicated in the developmental balance between cell proliferation and differentiation in many systems (Coffman 2003). runt-1 was found recently to be expressed at planarian wounds and required for eye regeneration (Sandmann et al. 2011). Here, we found runt-1 and runt-2 to be wound-induced in a subset of neoblasts, raising the possibility that runt-1 and/or runt-2 activation in neoblasts near wounds helps transition undifferentiated cells to differentiation.
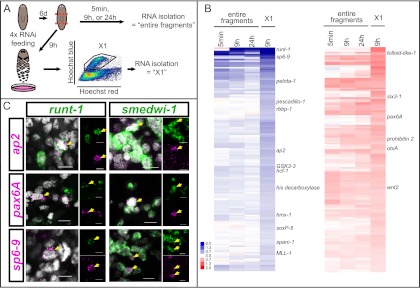
To identify the role(s) of wound-induced runt-1 in neoblasts, we performed expression microarray experiments using RNAi animals and X1 neoblasts from RNAi animals (Fig. 5A). Most X1 cells (∼90%) express smedwi-1 (Reddien et al. 2005b); because all runt-1 cells express smedwi-1 (see above), the X1 fraction from wounded animals will contain the runt-1+ cells. The array data demonstrated that inhibition of wound-induced runt-1 expression by RNAi was efficient (Fig. 5B). We found no significant changes in numbers of X1s in runt-1(RNAi) animals (Supplemental Fig. S7B). However, many genes displayed aberrant expression in X1s from wounded runt-1(RNAi) animals. Genes that failed to be up-regulated following injury in runt-1(RNAi) X1 cells encode homologs of transcription factors that have been implicated in neuronal development: sp6-9 (Lapan and Reddien 2011), hmx-1 (Wang et al. 2004), ap2 (Mitchell et al. 1991), and soxP-5 (Fig. 5B; Lai et al. 2008). Other transcription factors associated with neuronal development, such as six3-1 (Pineda and Salo 2002), otxA (Umesono et al. 1999; Lapan and Reddien 2011), and pax6A (Pineda et al. 2002), were significantly overexpressed in runt-1(RNAi) neoblasts (Fig. 5B). Additional gene types, such as those encoding cell cycle regulators, were also affected by runt-1 RNAi. Several of the identified transcription factors (ap2, pax6A, and sp6-9) were expressed within a subset of runt-1+ cells in wounded animals (Fig. 5C). Because all runt-1+ cells express neoblast markers (smedwi-1) (Fig. 4C), these additional transcription factors must also be induced in neoblasts but were not broadly expressed in neoblasts in intact animals (Supplemental Fig. S8A). Indeed, sp6-9 is known to be expressed in neoblasts during regeneration of eyes (Lapan and Reddien 2011). ap2 and pax6A were also found to be expressed in smedwi-1+ cells early (within 6–24 h) following wounding (Fig. 5C). These data demonstrate that substantial heterogeneity is induced in neoblasts by injuries.
Figure 5.
Wound-induced runt-1 expression is required for proper expression of developmental regulators in neoblasts during regeneration. (A) Cartoon depicts the expression microarray design. (B) Heat maps showing all genes that were differentially expressed (P < 0.05) in X1s from amputated runt-1(RNAi) animals 9 h post-wounding [(left column) all genes down in X1s; (right column) all genes up in X1s; (red) up, (blue) down; log2(time point/intact)]. Data from entire fragments are shown as comparison. (C) Fluorescent in situ hybridizations 6 h (for pax6A and sp6-9) or 24 h (for ap2) following wounding; runt-1 is coexpressed with multiple transcription factors (ap2, pax6A, and sp6-9) in neoblasts near wounds, and ap2, pax6A, and sp6-9 are coexpressed with the neoblast marker smedwi-1. Nuclear labeling with Hoechst (gray). Probes are indicated; (yellow arrows) colabeled cells; bars, 10 μm.
runt-1 is required for specification of neuron and eye precursors in the neoblast population after injury
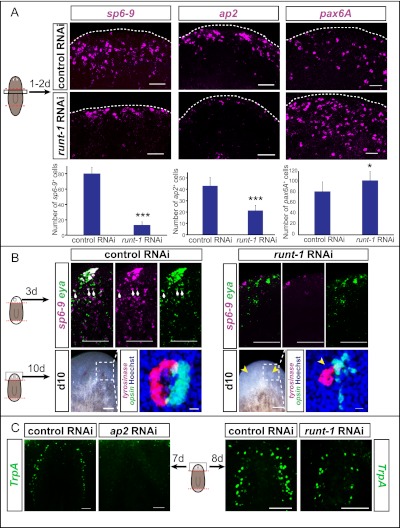
In situ hybridizations revealed that runt-1 RNAi resulted in decreased formation of sp6-9+ and ap2+ cells following wounding, but increased formation of pax6A+ cells (Fig. 6A), as predicted by the microarray data described above. Because X1 numbers are normal in runt-1(RNAi) animals (Supplemental Fig. S7B), we conclude that this effect is not simply explained by changes in numbers of neoblasts in runt-1(RNAi) animals. The rapid defects in wound-induced sp6-9, pax6A, and ap2 (detected by 9 h after wounding in the microarray) is consistent with a defect in initial wound-induced expression of these transcription factors in runt-1(RNAi) neoblasts, as opposed to a defect in proliferative expansion of such cells. Whether these transcription factor+ neoblasts expand through cell division, with runt-1 also involved in this process, is unknown.
Figure 6.
Wound-induced runt-1 expression in neoblasts is required for neuronal and photoreceptor cell type formation during regeneration. (A) In situ hybridizations on amputated trunk fragments (anterior top) from control and runt-1(RNAi) animals, probing for wound-induced transcription factors identified by microarray (see Fig. 5). Graphs show number of positive cells per animal. Time points for quantification: sp6-9 (image 24 h), 40 h (n = 3); ap2, 48 h (n = 3); pax6A, 24 h (n = 10). (*) P < 0.05, (***) P < 0.001. (B) runt-1 RNAi leads to impaired optic cup progenitor formation (sp6-9+/eya+ cells) on regeneration day 3 (n = 12) and, consequently, eye formation defects on day 10, as shown here by live images and labeling with tyrosinase (magenta) and opsin (green) (n = 10). (Blue) Hoechst. tyrosinase and opsin expression mark optic cup and photoreceptor neurons, respectively. Bars: 100 μm; for tyrosinase/opsin labeling, 10 μm. (White arrows) sp6-9+/eya+ cells; (yellow arrows) eye regeneration defects. (C) ap2 RNAi leads to a strong decrease in TrpA+ neuron (green) regeneration. TrpA+ neuron regeneration is also strongly impaired in runt-1(RNAi) animals (n = 4). Bars, 50 μm.
sp6-9 is required for formation of planarian pigmented optic cup progenitor cells and, consequently, optic cup regeneration (Lapan and Reddien 2011). runt-1(RNAi) animals had reduced numbers of sp6-9+/eya+ optic cup progenitors at day 3 following amputation (Fig. 6B), providing a cellular explanation for impaired eye regeneration in runt-1(RNAi) animals (Fig. 6B; Supplemental Fig. S8B). Despite being important for eye formation during regeneration, runt-1 was not visibly expressed in eye precursor cells or eyes themselves (Supplemental Fig. S8C), in contrast to all previously reported planarian eye regulatory genes (Saló et al. 2002; Mannini et al. 2004; Lapan and Reddien 2011). Moreover, as described above, induction of runt-1 expression is specific to wounding (Supplemental Fig. S6B). These observations suggest that it is the wound-induced expression of runt-1 in neoblasts that explains its role in optic cup regeneration. To further test this hypothesis, we performed long-term RNAi experiments in unamputated animals, comparing RNAi of runt-1, sp6-9, and six1/2 (six1/2 is required for planarian eye regeneration) (Mannini et al. 2004; Lapan and Reddien 2011). six1/2(RNAi) intact animals completely lost eyes within 40 d of gene inhibition, and sp6-9(RNAi) animals lost their optic cups within 70 d; in contrast, no eye defects were observed in runt-1(RNAi) animals during 93 d of observation (Supplemental Fig. S8D). At this time point, we were still able to identify eya+/dlx+/SMEDWI+ cells within the optic cup cells of runt-1(RNAi) animals, indicating that active turnover of these cells was normally occurring (Supplemental Fig. S8E). These data indicate that runt-1 has a regeneration-specific role for the planarian eye.
Because runt-1 RNAi impacts expression of many genes in neoblasts from wounded animals, we reasoned that multiple cell types would be aberrantly regenerated in runt-1(RNAi) animals. We therefore characterized the function of a second candidate runt-1 target: ap2. RNAi of the ap2 gene does not affect eye regeneration, but instead disrupts regeneration of a subtype of neurons that are TrpA+ in the planarian brain (Fig. 6C) (TrpA encodes an ion channel expressed centrally in the bilobed cephalic ganglia). Accordingly, runt-1 RNAi also impaired TrpA+ neuron formation during regeneration (Fig. 6C; Supplemental Fig. S8F). We conclude that wound-induced runt-1 expression in neoblasts regulates neuron- and eye-specific cell type formation during regeneration.
Discussion
All animals with long life suffer from injuries and require robust wound response and repair programs. Relatively simple anatomy, rapid regeneration, and ease of gene function studies combine to make planarians an attractive model for investigation of the molecular programs acting at wounds to control regeneration. Many genes were found to be activated in a transcriptome analysis during the first 3 d of planarian head regeneration, with two (egrl1 and runt-1) showing expression at wound sites early in regeneration (Sandmann et al. 2011). We focused on the first 12 h following wounding (including anterior- and posterior-facing wounds, and including comparison with wound-induced expression in irradiated animals lacking neoblasts) and investigated gene expression of up-regulated genes with in situ hybridizations. We identified three major classes of planarian wound-induced genes that are expressed in differentiated tissues (W1, W2, and W3 genes) and a class of genes induced in the proliferative cells that drive regeneration, called neoblasts (W4 genes) (Fig. 7). These findings expand the list of wound-induced genes with described in vivo expression pattern at planarian wounds from about five to more than a hundred (with several hundred more present in microarray data, and a recent head regeneration transcriptome analysis [Sandmann et al. 2011] providing an additional resource). Given that many of the genes we identified are evolutionarily conserved, study of planarian wound response factors could shed light on broadly used wound response and regeneration mechanisms.
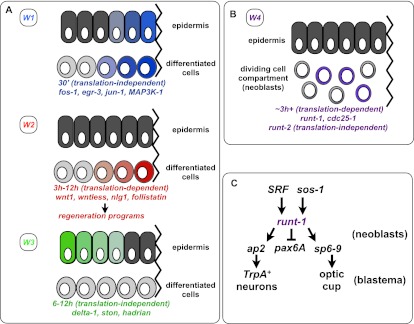
Figure 7.
Model of the four different classes of wound-induced genes in the differentiated tissue (W1, W2, and W3) (A) and in the neoblasts (W4) (B). Some W3 genes display expression restricted from the wound site (as shown), and others (e.g., hadrian) do not. (C) runt-1 is activated in neoblasts following wounding and is important for proper eye and TrpA+ neuron regeneration. SRF and sos-1 are required for normal wound-induced runt-1 expression (arrows). “Translation-independent” indicates that most genes in this class did not require new protein synthesis for activation, whereas the “translation-dependent” genes did. See the text for details.
Wound-induced immediate early gene expression (W1 and W3 genes) is observed at diverse wound types
W1 genes were expressed within 30 min following wounding in many tissues, and many encode homologs of established immediate early genes, such as transcription and signaling factors (e.g., jun-1, fos-1, egrl1, egr-2, egr-3, egr-4, and pp1-1) (Fig. 7A). Like immediate early genes, many W1 genes are activated in a translation-independent manner. W3 genes were expressed between 6 and 12 h in the epidermis following wounding (expression could occur far from wounds and, for some, be excluded from wounds) and encode candidate extracellular matrix remodeling proteins (plasminogen-1 and TFPI-1) as well as a Notch signaling pathway ligand (delta-1) (Fig. 7A). Like W1 genes, many W3 genes also displayed translation-independent gene activation and can therefore also be considered as immediate early genes by this criterion; this was unexpected because these genes were induced much later (between 6 and 12 h) following wounding.
Immediate early genes are expressed in many different biological contexts and are important mediators of rapid genomic responses to numerous stimuli (Abraham et al. 1991; Schreiber et al. 1991; Iyer et al. 1999; Cooper et al. 2005). Many factors/pathways can activate immediate early genes, suggesting that complex and potentially redundant factors can activate these genes in planarians. Immediate early gene induction at wounds in regenerative tissues may be widespread. For example, >100 immediate early genes are up-regulated in injured livers (Mohn et al. 1991; Su et al. 2002). Gene expression studies of wounded mice and Drosophila larvae similarly revealed “activation genes” mainly encoding transcription factors up-regulated early (i.e., within 30 min to 3 h) and transiently at wounds (Cooper et al. 2005; Stramer et al. 2008). Immune response genes can also be activated at animal wounds (Cooper et al. 2005; Stramer et al. 2008), and future identification of planarian infection-induced genes could allow comparison with planarian wound-induced genes.
The roles of early wound-induced gene expression are mostly unexplored, with a few exceptions (Galko and Krasnow 2004; Mace et al. 2005; Ting et al. 2005). Our results raise the possibility that conserved mechanisms involving a diverse cassette of translation-independent, rapidly induced genes are important for initiating aspects of the regenerative response to injuries, exemplified by the phenotype of fos-1(RNAi) animals. This phenotype is similar to the phenotype of DjerkA(RNAi) planarians (Tasaki et al. 2011). Given the potential of planarians for studying wound responses, future experiments based on these findings should shed light on the mechanisms of regeneration initiation.
A late wave of wound-induced gene expression—W2 genes—includes factors that are required for proper patterning during regeneration
W2 genes were induced between 3 and 12 h following wounding and were expressed subepidermally. W2 genes include homologs of patterning factors (e.g., wntless, wnt1, nlg1, inhibin-1, gpc-1, and follistatin) (Fig. 7A). inhibin-1 and follistatin, regulators of Activin signaling, are expressed following partial hepatectomy in mice and also have been implicated in tissue repair and scar formation following skin injury (Hübner et al. 1996; Wankell et al. 2001; Su et al. 2002). Wnt genes are expressed at wounds in other organisms as well, ranging from skin wounds and during hair follicle regeneration to cardiac injury in mice (Ito et al. 2007; Duan et al. 2012). These findings suggest that several common wound-induced genes might exist among animals.
Several W2 genes, although not required for regeneration per se, are required for the initiation of the proper type of planarian regeneration or for proper patterning during regeneration, such as wntless, wnt1 (Adell et al. 2009; Petersen and Reddien 2009), and gpc-1. Notably, W2 genes were generically induced by all tested wound types involving epidermal damage. This result is surprising because different wounds must execute different regeneration programs for tissue repair and might therefore be expected to have highly divergent expression of signaling/patterning factors. In contrast, our results suggest that early in the process of regeneration, wounds display very similar expression of signaling/patterning factors regardless of the nature of the missing tissue or whether there is significant missing tissue requiring blastema formation at all. These results are consistent with the hypothesis (Petersen and Reddien 2009) that wounding has a role in missing tissue identity determination in regeneration by activating signaling factors, which then have different effects depending on the local tissue environment at wounds. Further investigation of W2 genes will be an important direction for exploration of this idea.
runt-1 is a wound-induced Runt transcription factor in neoblasts required for cell type specification during regeneration
During planarian head regeneration, neoblasts proliferate predominantly at the base of the blastema, with the growing blastema consisting of nonmitotic neoblast progeny cells (Wenemoser and Reddien 2010). Two models could explain how neoblasts promote formation of specific missing cell types in blastemas: (1) In a “naive neoblast model,” neoblasts are naive to the fate of their progeny cells, with post-mitotic neoblast progeny acquiring identity exclusively by the position at which they find themselves in the blastema. (2) In a “specified neoblast model,” regeneration of different cells is accomplished by neoblasts that are specified to be different in their fate prior to production of post-mitotic progeny cells. Neoblasts are the only somatic dividing cells in adult planarians and have historically been discussed as a homogeneous population of cells. If true, this would favor the naive neoblast model. In contrast, our data with wound-induced expression of runt-1 in neoblasts, considered together with data demonstrating that planarian eye and protonephridia progenitors initiate in the neoblast (Lapan and Reddien 2011; Scimone et al. 2011), favor the specialized neoblast model (for at least some blastema cells) (Fig. 7B,C). An important future direction will be to determine whether the specified neoblast model applies to many or even all lineages or whether the naive neoblast model applies to the regeneration of other cell types. runt-1 is activated within 3 h of injury and even at injuries that do not require substantial regeneration, indicating that runt-1 is generically activated by wounds. runt-1 is required for the appearance of normal numbers of sp6-9+ and ap2+ neoblasts near wounds, promoting optic cup and TrpA+ neuron regeneration, respectively. Based on these observations, we propose a model in which generic wounding induces neoblasts to be competent for specialization by activation of the gene runt-1.
Runt proteins can be important for the decision-making process between cell cycle and differentiation (Coffman 2003) and are involved in many developmentally important processes in other species, including segmentation, hematopoiesis, and eye development in Drosophila (Lebestky et al. 2000; Coffman 2003); lateral hypodermal stem cell (seam cell) divisions in Caenorhabditis elegans (Kagoshima et al. 2007); and hematopoesis, osteogenesis, and neurogenesis in the dorsal root ganglia in mammals (Speck and Gilliland 2002; Coffman 2003). Activation of runt-1 and other W4 genes requires Smed-SRF. SRF is a MADS-box transcription factor (Norman et al. 1988) that, together with TCF (ternary complex factor) proteins, binds serum response elements of target genes (Gille et al. 1992). SRF is also required for induction of cell cycle components during liver regeneration (Latasa et al. 2007). Smed-sos-1 was also required for normal induction of runt-1 (and egr-2) following wounding; Sos is a GEF, indicating that receptor tyrosine kinase (RTK) signaling might also be involved in mediation of planarian wound signals. RTK family signaling has been implicated in fin and heart regeneration in zebrafish, midgut regeneration in Drosophila, and tail regeneration in Xenopus tadpoles (Whitehead et al. 2005; Lepilina et al. 2006; Lin and Slack 2008; Jiang et al. 2011). A candidate pathway of action thus connects wounding to regeneration of eyes and specific brain neurons mediated by runt-1 in planarians (Fig. 7C): Following wounding, SRF and Sos are required for wound-induced activation of runt-1 in a subset of neoblasts. RUNT-1 then acts as a transcription factor to promote formation of specialized neoblasts: sp6-9+ neoblasts for the eye, and ap2+ neoblasts for TrpA+ neurons. Understanding how undifferentiated cells, such as neoblasts, can transition to committed progenitors that can replace missing cell types in response to injuries is an important challenge. Our results indicate that Runt transcription factors can be wound-induced to promote such transitions.
Planarians represent an ideal system for investigating the genetic underpinnings of wound responses for regeneration initiation. Here we found that wounding generically induces a common program of regeneration factors (patterning/signaling genes in differentiated tissue and runt-1 in neoblasts) that can be involved in the regeneration of different types of tissue depending on the wound context. With the discovery and classification of hundreds of wound-induced genes, wound responses for regeneration programs can now be systematically dissected using planarians.
Materials and methods
Exposure to γ-irradiation
For lethal irradiation (elimination of all neoblasts), planarians were exposed to 6000 rad (6K, ∼72 min) using a cesium source (∼83 rad/min).
Gene cloning and names
For RNA probes, genes were cloned into pGEM and amplified using nested PCR with T7 promoter-containing primers or existing cDNA clones (Supplemental Table S6). For RNAi, genes were cloned into pPR244 as described (Reddien et al. 2005a). A list of gene names with BLAST data is provided in Supplemental Table S7, and nomenclature is described in the Supplemental Material.
Flow cytometry
Animals were amputated in cold CMFB, and cells were prepared as described (Scimone and Reddien 2010). For quantification of X1 cells, five animals were used per RNAi condition, and triplicate experiments were performed. Analyses and sorting were performed using a Moflo3 FACS sorter and FlowJo.
In situ hybridizations
Fluorescence and NBT/BCIP in situ hybridizations were performed as described (Pearson et al. 2009). For double/triple labeling, HRP inactivation was performed between labelings in 4% formaldehyde for 30 min.
Immunostaining
Animals were killed in 10% NAC in PBS and labeled with anti-SMEDWI-1 (1:2000) as described (Newmark and Sánchez Alvarado 2000). Anti-SMEDWI-1 antibody was generated in rabbits using the peptide previously described (Guo et al. 2006).
Imaging and analyses
For quantification of pax6A/sp6-9/ap2/TrpA-expressing cells, anterior blastemas were photographed (7 × 1-μm stacks) from the wound site to the pharynx (for TrpA only the brain region). Numbers were determined using AxioVision software (Zeiss) and/or manually. Images were acquired using an AxioImager with an Apotome (Zeiss) or an LSM 700 (Zeiss).
RNAi
RNAi feeding was performed as described (Reddien et al. 2005a), except bacteria were resuspended 1:30 with a mixture of calf liver and planaria water (7:3), and no agarose was added. pPR244 containing the C. elegans gene unc-22 was used for control RNAi (Reddien et al. 2005a). For nCounter experiments and runt-1 microarray experiments, RNA was extracted from animals that had been subjected to four RNAi feedings (10 animals per RNAi, in three biological replicates) for the respective gene. RNA was extracted 3, 9, and 24 h following amputation from RNAi-treated animals.
Microarray experiments
All microarray experiments used biological triplicates, with RNA extracted from 10 animals per condition, using TRIzol and DNase treatment. RNA was amplified using a MessageAmp II aRNA amplification kit (Ambion) and was labeled using an ASAP labeling kit (Perkin-Elmer). For the analysis using sorted cells, 600,000 cells were collected for each condition and replicate. Custom Agilent arrays of 44,000 probes, representing all known S. mediterranea ESTs and gene predictions (Scimone et al. 2011), were hybridized according to the manufacturer's instructions and scanned using an Agilent scanner. Agilent two-color arrays were within-array-normalized by loess, followed by between-array quantile normalization of average intensities across channels (Aquantile). Differential expression analysis was performed using a moderated t-test, as implemented in the limma package of Bioconductor, with P-value correction by false discovery rate. Log2 ratios of mean expression levels (treatment/control) and false discovery rate (FDR)-adjusted P-values were used for further analysis. For the differentiated tissue data set, up-regulation was defined by a P-value <0.05 and log2 ratios >0.3 in the untreated data set and >0.6 in the irradiated data set. W1 genes are defined as peaking in expression before or within 3 h following wounding; W2 genes peak in expression after 3 h following wounding; and W3 genes peak after 3 h following wounding but are expressed epidermally. For the W4 genes, up-regulation was defined by a P-value <0.05 and log2 ratios >0.4 in the untreated data and <0.4 in the irradiated data. For analysis of the Smed-runt-1 RNAi data, all significantly (P < 0.05) differentially expressed genes were selected. Cluster 3.0 and Java TreeView were used for clustering and microarray data visualization. Similarity to genes from other organisms was determined using BLAST with an E-value cutoff of E−10. Microarray data have NCBI accession numbers GSE36945 and GSE36869.
NanoString experiments
For NanoString nCounter experiments (Geiss et al. 2008), total RNA (10 animals per condition, three biological replicates) was isolated using TRIzol and DNase treatment as performed for the microarray experiments. S. mediterranea-specific code sets were designed by NanoString, and hybridization and analysis procedures were performed according to the manufacturer's protocol. Differential expression was assayed from NanoString counts using an overdispersed Poisson model with a Fisher-like exact test, as implemented in Bioconductor's edgeR package (Robinson et al. 2010). A gene was considered differentially regulated if the FDR-adjusted P-value was P < 0.01 for RNAi experiments; for all other experiments, P < 0.05.
CHX treatment
Animals were amputated in a dilution of CHX (Sigma) in planaria water (0.1 μg/μL; 1:1000 dilution of 100 mg/mL in DMSO) or a 1:1000 dilution of DMSO in planaria water as control and kept in this solution until fixation or RNA extraction.
Acknowledgments
We thank Reddien laboratory members for comments and discussion, Irving Wang for cartoons, and the Genome Technology Core. P.W.R. is an early career scientist of the Howard Hughes Medical Institute and an associate member of the Broad Institute of Harvard and MIT. We acknowledge support from the NIH (R01GM080639) and the Keck Foundation.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.187377.112.
References
- Abraham W, Dragunow M, Tate WP 1991. The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol 5: 297–314 [DOI] [PubMed] [Google Scholar]
- Adell T, Salo E, Boutros M, Bartscherer K 2009. Smed-Evi/Wntless is required for β-catenin-dependent and -independent processes during planarian regeneration. Development 136: 905–910 [DOI] [PubMed] [Google Scholar]
- Almendral JM, Sommer D, Macdonald-Bravo H, Burckhardt J, Perera J, Bravo R 1988. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol 8: 2140–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguñà J 1974. Dramatic mitotic response in planarians after feeding, and a hypothesis for the control mechanism. J Exp Zool 190: 117–122 [DOI] [PubMed] [Google Scholar]
- Cebria F, Newmark PA 2005. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132: 3691–3703 [DOI] [PubMed] [Google Scholar]
- Chavrier P, Zerial M, Lemaire P, Almendral J, Bravo R, Charnay P 1988. A gene encoding a protein with zinc fingers is activated during G0/G1 transition in cultured cells. EMBO J 7: 29–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA 2003. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int 27: 315–324 [DOI] [PubMed] [Google Scholar]
- Cooper L, Johnson C, Burslem F, Martin P 2005. Wound healing and inflammation genes revealed by array analysis of ‘macrophageless’ PU.1 null mice. Genome Biol 6: R5 doi: 10.1186/gb-2004-6-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan JZ, Gherghe C, Liu DX, Hamlett E, Srikantha L, Rodgers L, Regan JN, Rojas M, Willis M, Leask A, et al. 2012. Wnt1/β catenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J 31: 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois F 1949. Contribution á l 'ètude de la migration des cellules de règènèration chez les Planaires dulcicoles. Bull Biol Fr Belg 83: 213–283 [Google Scholar]
- Eisenhoffer GT, Kang H, Sánchez Alvarado A 2008. Molecular analysis of stem cells and their descendants during cell turnover and regeneration in the planarian Schmidtea mediterranea. Cell Stem Cell 3: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galko MJ, Krasnow MA 2004. Cellular and genetic analysis of wound healing in Drosophila larvae. PLoS Biol 2: 1114–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviño MA, Reddien PW 2011. A Bmp/Admp regulatory circuit controls maintenance and regeneration of dorsal–ventral polarity in planarians. Curr Biol 21: 294–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26: 317–325 [DOI] [PubMed] [Google Scholar]
- Gille H, Sharrocks AD, Shaw PE 1992. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature 358: 414–417 [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Hermanowski AL, Ziff EB 1986. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol 6: 1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grollman AP 1967. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J Biol Chem 242: 3226–3233 [PubMed] [Google Scholar]
- Guo T, Peters AH, Newmark PA 2006. A bruno-like gene is required for stem cell maintenance in planarians. Dev Cell 11: 159–169 [DOI] [PubMed] [Google Scholar]
- Gurley KA, Rink JC, Sánchez Alvarado A 2008. β-Catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319: 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A 2010. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev Biol 347: 24–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Asami M, Higuchi S, Shibata N, Agata K 2006. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ 48: 371–380 [DOI] [PubMed] [Google Scholar]
- Hübner G, Hu Q, Smola H, Werner S 1996. Strong induction of activin expression after injury suggests an important role of activin in wound repair. Dev Biol 173: 490–498 [DOI] [PubMed] [Google Scholar]
- Iglesias M, Gomez-Skarmeta JL, Salo E, Adell T 2008. Silencing of Smed-βcatenin1 generates radial-like hypercephalized planarians. Development 135: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Ito M, Yang ZX, Andl T, Cui C, Kim N, Millar SE, Cotsarelis G 2007. Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447: 316–320 [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Boguski MS, et al. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283: 83–87 [DOI] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA 2011. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H, Nimmo R, Saad N, Tanaka J, Miwa Y, Mitani S, Kohara Y, Woollard A 2007. The C. elegans CBFβ homologue BRO-1 interacts with the Runx factor, RNT-1, to promote stem cell proliferation and self-renewal. Development 134: 3905–3915 [DOI] [PubMed] [Google Scholar]
- Kelley-Loughnane N, Sabla GE, Ley-Ebert C, Aronow BJ, Bezerra JA 2002. Independent and overlapping transcriptional activation during liver development and regeneration in mice. Hepatology 35: 525–534 [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JRL, Macklis JD 2008. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron 57: 232–247 [DOI] [PubMed] [Google Scholar]
- Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM 1988. Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature 334: 629–631 [DOI] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW 2011. dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet 7: e1002226 doi: 10.1371/journal.pgen.1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa MU, Couton D, Charvet C, Lafanechere A, Guidotti JE, Li Z, Tuil D, Daegelen D, Mitchell C, Gilgenkrantz H 2007. Delayed liver regeneration in mice lacking liver serum response factor. Am J Physiol Gastrointest Liver Physiol 292: G996–G1001 doi: 10.1152/ajpgi.00493.2006 [DOI] [PubMed] [Google Scholar]
- Lau LF, Nathans D 1987. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: Coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci 84: 1182–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebestky T, Chang T, Hartenstein V, Banerjee U 2000. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288: 146–149 [DOI] [PubMed] [Google Scholar]
- Lee W, Haslinger A, Karin M, Tjian R 1987. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature 325: 368–372 [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD 2006. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127: 607–619 [DOI] [PubMed] [Google Scholar]
- Lin G, Slack JM 2008. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol 316: 323–335 [DOI] [PubMed] [Google Scholar]
- Mace KA, Pearson JC, McGinnis W 2005. An epidermal barrier wound repair pathway in Drosophila is mediated by grainy head. Science 308: 381–385 [DOI] [PubMed] [Google Scholar]
- Mannini L, Rossi L, Deri P, Gremigni V, Salvetti A, Salo E, Batistoni R 2004. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev Biol 269: 346–359 [DOI] [PubMed] [Google Scholar]
- Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R 1991. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev 5: 105–119 [DOI] [PubMed] [Google Scholar]
- Mohn KL, Laz TM, Hsu JC, Melby AE, Bravo R, Taub R 1991. The immediate-early growth-response in regenerating liver and insulin-stimulated H-35 cells: Comparison with serum-stimulated 3T3 cells and identification of 41 novel immediate-early genes. Mol Cell Biol 11: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MD, Salo E, Cebria F 2007. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev Biol 311: 79–94 [DOI] [PubMed] [Google Scholar]
- Molina MD, Salo E, Cebria F 2009. Expression pattern of the expanded noggin gene family in the planarian Schmidtea mediterranea. Gene Expr Patterns 9: 246–253 [DOI] [PubMed] [Google Scholar]
- Molina MD, Neto A, Maeso I, Gómez-Skarmeta JL, Saló E, Cebrià F 2011. Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr Biol 21: 300–305 [DOI] [PubMed] [Google Scholar]
- Muller R, Bravo R, Burckhardt J, Curran T 1984. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature 312: 716–720 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Sánchez Alvarado A 2000. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol 220: 142–153 [DOI] [PubMed] [Google Scholar]
- Norman C, Runswick M, Pollock R, Treisman R 1988. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 55: 989–1003 [DOI] [PubMed] [Google Scholar]
- Ogawa K, Ishihara S, Saito Y, Mineta K, Nakazawa M, Ikeo K, Gojobori T, Watanabe K, Agata K 2002. Induction of a noggin-like gene by ectopic DV interaction during planarian regeneration. Dev Biol 250: 59–70 [DOI] [PubMed] [Google Scholar]
- Orii H, Watanabe K 2007. Bone morphogenetic protein is required for dorso–ventral patterning in the planarian Dugesia japonica. Dev Growth Differ 49: 345–349 [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sánchez Alvarado A 2009. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn 238: 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2008. Smed-βcatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319: 327–330 [DOI] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2009. A wound-induced Wnt expression program controls planarian regeneration polarity. Proc Natl Acad Sci 106: 17061–17066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Reddien PW 2011. Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332: 852–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda D, Salo E 2002. Planarian Gtsix3, a member of the Six/so gene family, is expressed in brain branches but not in eye cells. Gene Expr Patterns 2: 169–173 [DOI] [PubMed] [Google Scholar]
- Pineda D, Rossi L, Batistoni R, Salvetti A, Marsal M, Gremigni V, Falleni A, Gonzalez-Linares J, Deri P, Salo E 2002. The genetic network of prototypic planarian eye regeneration is Pax6 independent. Development 129: 1423–1434 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sánchez Alvarado A 2004. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 20: 725–757 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sánchez Alvarado A 2005a. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 8: 635–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A 2005b. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 310: 1327–1330 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, Sánchez Alvarado A 2007. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134: 4043–4051 [DOI] [PubMed] [Google Scholar]
- Robb SM, Ross E, Sánchez Alvarado A 2008. SmedGD: The Schmidtea mediterranea genome database. Nucleic Acids Res 36: D599–D606 doi: 10.1093/nar/gkm684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saló E, Baguñà J 1984. Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J Embryol Exp Morphol 83: 63–80 [PubMed] [Google Scholar]
- Saló E, Pineda D, Marsal M, Gonzalez J, Gremigni V, Batistoni R 2002. Genetic network of the eye in Platyhelminthes: Expression and functional analysis of some players during planarian regeneration. Gene 287: 67–74 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Vogg MC, Owlarn S, Boutros M, Bartscherer K 2011. The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol 12: R76 doi: 10.1186/gb-2011-12-8-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SS, Tocco G, Shors TJ, Thompson RF 1991. Activation of immediate early genes after acute stress. Neuroreport 2: 17–20 [DOI] [PubMed] [Google Scholar]
- Scimone ML, Reddien PW 2010. The Mi-2-like Smed-CHD4 gene is required for stem cell differentiation in the planarian Schmidtea mediterranea. Development 137: 1231–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW 2011. A regulatory program for excretory system regeneration in planarians. Development 138: 4387–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg JL, Rauscher FJ III, Morgan JI, Curran T 1989. Regulation of proenkephalin by Fos and Jun. Science 246: 1622–1625 [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG 2002. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer 2: 502–513 [DOI] [PubMed] [Google Scholar]
- Stramer B, Winfield M, Shaw T, Millard TH, Woolner S, Martin P 2008. Gene induction following wounding of wild-type versus macrophage-deficient Drosophila embryos. EMBO Rep 9: 465–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG 2002. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci 99: 11181–11186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JC, Sher D, Eisenstein M, Shigesada K, Reitzel AM, Marlow H, Levanon D, Groner Y, Finnerty JR, Gat U 2008. The evolutionary origin of the Runx/CBFβ transcription factors—studies of the most basal metazoans. BMC Evol Biol 8: 228 doi: 10.1186/1471-2148-8-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki J, Shibata N, Nishimura O, Itomi K, Tabata Y, Son F, Suzuki N, Araki R, Abe M, Agata K, et al. 2011. ERK signaling controls blastema cell differentiation during planarian regeneration. Development 138: 2417–2427 [DOI] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao LL, Ellis S, Kaur P, Uchida Y, Holleran WM, et al. 2005. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308: 411–413 [DOI] [PubMed] [Google Scholar]
- Umesono Y, Watanabe K, Agata K 1999. Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev Genes Evol 209: 31–39 [DOI] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW 2011. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 332: 811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WD, Grimmer JF, Van De Water TR, Lufkin T 2004. Hmx2 and Hmx3 homeobox genes direct development of the murine inner ear and hypothalamus and can be functionally replaced by Drosophila Hmx. Dev Cell 7: 439–453 [DOI] [PubMed] [Google Scholar]
- Wankell M, Munz B, Hubner G, Hans W, Wolf E, Goppelt A, Werner S 2001. Impaired wound healing in transgenic mice overexpressing the activin antagonist follistatin in the epidermis. EMBO J 20: 5361–5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D, Reddien PW 2010. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 344: 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead GG, Makino S, Lien CL, Keating MT 2005. fgf20 is essential for initiating zebrafish fin regeneration. Science 310: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Yan D, Lin X 2009. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol 1: a002493 doi: 10.1101/cshperspect.a002493 [DOI] [PMC free article] [PubMed] [Google Scholar]